Significance
Recent advances in cancer epigenetics have shown the involvement of epigenetic abnormalities in the initiation and progression of cancer, but their role in cancer-specific aberrant splicing is not clear. The identification of upstream epigenetic regulators of cancer-specific splicing will enable us to therapeutically target aberrant splicing and provide an approach to cancer therapy. Here we have demonstrated a mechanism of intragenic DNA methylation-mediated regulation of alternative splicing by Brother of Regulator of Imprinted Sites (BORIS), which can contribute to breast cancer tumorigenesis by favoring the Warburg effect. The reversal of the Warburg effect was achieved by the inhibition of DNA methylation or down-regulation of BORIS, which may serve as a useful approach to inhibit the growth of breast cancer cells.
Keywords: alternative splicing, BORIS, DNA methylation, cancer, Warburg effect
Abstract
Aberrant alternative splicing and epigenetic changes are both associated with various cancers, but epigenetic regulation of alternative splicing in cancer is largely unknown. Here we report that the intragenic DNA methylation-mediated binding of Brother of Regulator of Imprinted Sites (BORIS) at the alternative exon of Pyruvate Kinase (PKM) is associated with cancer-specific splicing that promotes the Warburg effect and breast cancer progression. Interestingly, the inhibition of DNA methylation, BORIS depletion, or CRISPR/Cas9-mediated deletion of the BORIS binding site leads to a splicing switch from cancer-specific PKM2 to normal PKM1 isoform. This results in the reversal of the Warburg effect and the inhibition of breast cancer cell growth, which may serve as a useful approach to inhibit the growth of breast cancer cells. Importantly, our results show that in addition to PKM splicing, BORIS also regulates the alternative splicing of several genes in a DNA methylation-dependent manner. Our findings highlight the role of intragenic DNA methylation and DNA binding protein BORIS in cancer-specific splicing and its role in tumorigenesis.
Alternative splicing of pre-mRNA is a process that can generate multiple protein-coding isoforms through the combinatorial use of splice sites, leading to the regulated expansion of the transcriptome. The aberrant splicing has been implicated in various hallmarks of cancer, including the abnormal metabolism (1). Similarly, global changes in epigenetic modifications also contribute significantly to the metabolic rewiring associated with tumorigenesis (2). Though aberrant splicing and epigenetic changes are individually well known to be associated with the reprogramming of metabolism in cancer, whether and how epigenetic alterations contribute to the cancer-specific splicing and thereby abnormal cancer metabolism is not clear.
The reprogramming of metabolism is an emerging hallmark of cancer (3), and increased glycolysis regardless of oxygen availability—termed as aerobic glycolysis, or the Warburg effect—is the primary metabolic change, which confers a proliferative advantage to cancer cells (4). The alternative spliced isoform Pyruvate Kinase M2 (PKM2) contributes to the Warburg effect by promoting aerobic glycolysis, whereas PKM1 isoform promotes oxidative phosphorylation (5). The PKM gene contains two mutually exclusive exons, exons 9 and 10, which are alternatively included in the final transcript to give rise to PKM1 and PKM2 isoforms, respectively (6) (Fig. 1A). PKM2 is preferentially expressed in highly proliferating cells, including embryonic stem cells (7) and cancer cells (8).
Fig. 1.
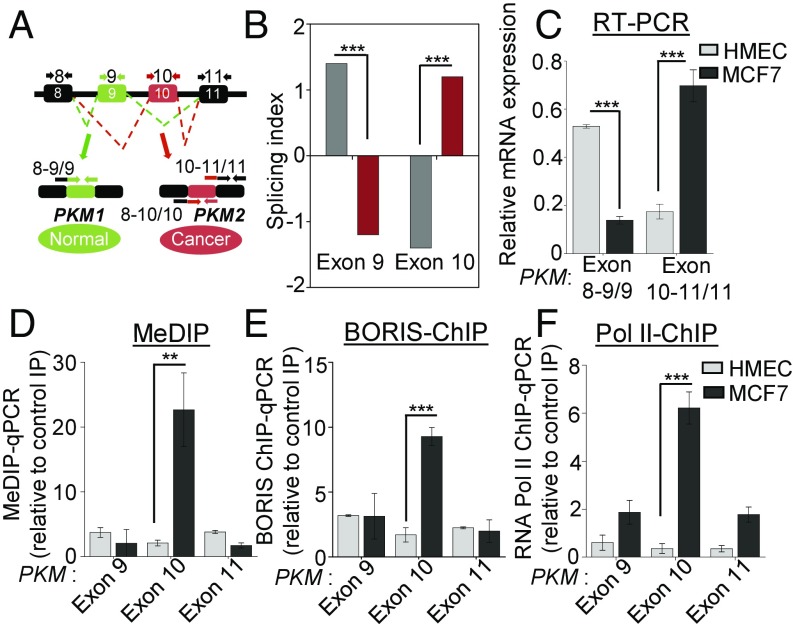
Elevated DNA methylation at exon 10 of PKM gene correlates with increased PKM2 isoform expression in breast cancer. (A) Schematic of PKM-spliced isoform found in normal and cancer cells. PKM gene has two isoforms, PKM1 and PKM2. PKM1 contains exon 9, whereas, PKM2 contains exon 10 in the processed mRNA. In addition to primer sets directed against the specific exon as a whole, exon junction-specific primer sets were used to specifically measure spliced products, as indicated above. The exon junction-spanning primers (SI Appendix, Table S1) exon 8–9/9 indicates the inclusion of exon 9, whereas exon 8–10/10 and exon 10–11/11 indicate the inclusion of exon 10. (B) Splicing index is the measure of exon inclusion and exclusion levels in alternative splicing analysis. It measures a quantitative change in the level of exon 9 and exon 10 in PKM (Affymetrix Transcript ID: TC15002610.hg.1) between normal and breast cancer tissues (GSE76250). Positive splicing index (≥+1.2) represents inclusion, and negative splicing index (≤−1.2) represents exclusion. (C) Splicing pattern of PKM gene in MCF7 breast cancer cell line and HMECs was measured by qRT-PCR using indicated junction-specific primers for PKM gene. Relative expression was normalized with RPS16 and constitutive exon (n = 3). (D) MeDIP in MCF7 cells and in HMECs genomic DNA using 5-methyl-cytosine antibody followed by qRT-PCR, relative to input and control IgG (n = 3). (E and F) ChIP in MCF7 cells and in HMECs using (E) BORIS and (F) RNA Pol II antibody, followed by qRT-PCR, relative to input and control IgG (n = 3). Error bar shows mean values ± SD. As calculated using two-tailed Student’s t test, **P < 0.01, ***P < 0.001.
The PKM2 isoform contributes to the Warburg effect by promoting the regeneration of nicotinamide adenine dinucleotide (NAD+) during the conversion of pyruvate to lactate to support high glycolytic flux in cancer cells (9). The glycolytic intermediates generated during aerobic glycolysis are then utilized for macromolecule synthesis to promote the proliferation of cancer cells (10). Thus, it is imperative to understand the regulation of splicing switch that facilitates the preferential expression of PKM2 over PKM1 in cancer cells.
Previous studies have described a role of the c-Myc inducible splicing repressors hnRNPA1/A2 and PTB (11–13), and SRSF3 (14), in PKM exon 9 exclusion and exon 10 inclusion, respectively. However, epigenetic regulation of PKM alternative splicing has not been reported. We have earlier shown a role of intragenic DNA methylation in the regulation of alternative pre-mRNA splicing due to the modulation of the binding of CTCF during lymphocyte development (15). The emerging evidence suggests that intragenic DNA methylation can regulate alternative pre-mRNA splicing via various mechanisms (16, 17). In addition, the deregulation of DNA methylation is an early event in breast tumorigenesis (18). However, the role of DNA methylation in abnormal metabolism due to alternative splicing has not been reported. Considering that breast cancer poses a significant challenge worldwide with the highest rate of incidence, mortality, and 5-y prevalence (19), here we systematically investigated the role of DNA methylation in the regulation of alternative pre-mRNA splicing of PKM in breast cancer.
Results
Intragenic DNA Methylation at PKM Exon 10 Correlates with BORIS and Pol II Enrichment at Exon 10 and PKM2 Expression in Breast Cancer.
The M2 isoform of the PKM gene is the primary isoform expressed in the mouse model of a breast tumor (5). We also observed PKM splicing switch between normal and tumor breast cancer profiles (Fig. 1B) and a higher PKM2/PKM1 ratio in breast cancer (SI Appendix, Fig. S1 A–C) leading to the increased PKM2 expression in breast cancer patients’ gene expression profiles (20) (SI Appendix, Fig. S1D and Table S2). Notably, increased PKM2 expression is also associated with poor overall and recurrence-free survival in breast cancer patients (SI Appendix, Fig. S1 E–G). To examine the role of DNA methylation in PKM splicing, we assayed the splicing pattern of the PKM gene in the two breast cancer cell lines MCF7 and MDA-MB-231 and normal human mammary epithelial cells (HMECs) by performing qRT-PCR using isoform-specific exon junction primers (Fig. 1A). We observed an increased inclusion of alternative exon 10 in both the breast cancer cell lines compared with HMECs; conversely, mutually exclusive exon 9 was primarily included in HMECs compared with the breast cancer cell lines (Fig. 1C and SI Appendix, Fig. S1H). This leads to higher PKM2 isoform expression compared with PKM1 isoform in the breast cancer cell lines relative to HMECs (SI Appendix, Fig. S1 I and J). We have earlier shown the role of intragenic DNA methylation in alternative splicing of CD45 exon 5 during lymphocyte development (15). To determine whether the DNA methylation is associated with the PKM alternative splicing, we carried out methylated DNA immunoprecipitation (MeDIP) using an antibody specific for 5-methyl cytosine. Interestingly, we observed higher DNA methylation at PKM exon 10 in MCF7 cells compared with HMECs (Fig. 1D), which correlates with the inclusion of exon 10 of the PKM gene. In addition, we have analyzed whole genome bisulfite sequencing data from MCF7 cells (21), which also suggests that exon 10 has higher methylation compared with the surrounding exons (SI Appendix, Fig. S1K). Furthermore, similar results were obtained in MDA-MB-231 breast cancer cells (SI Appendix, Fig. S1L), indicating that the correlation of methylation with the inclusion of exon 10 is not limited to a specific cell line. DNA methylation at the exonic region can modulate the recognition of the alternative exon by controlling the recruitment of various methyl-sensitive DNA binding proteins such as MeCP2 or CTCF (15, 16). Considering that PKM exon 10 has a CTCF binding site, we hypothesized that the DNA methylation at exon 10 might be playing an important role in the preferential expression of the PKM2 isoform by modulating the binding of a methyl-sensitive DNA binding protein. The binding of CTCF to DNA is inhibited by DNA methylation (22), whereas the CTCF paralog Brother of Regulator of Imprinted Sites (BORIS) preferentially binds to the methylated DNA (23). Moreover, BORIS is classified as a cancer/testis antigen, which is predominantly expressed in primary spermatocytes but gets reexpressed in cancer cells (24). Further, BORIS was detected at the protein level in MCF7 cells but not in the HMECs (SI Appendix, Fig. S1M). To examine whether BORIS binds to methylated PKM exon 10, we carried out BORIS chromatin immunoprecipitation (ChIP) and observed increased binding of BORIS to methylated PKM exon 10 in MCF7 and MDA-MB-231 cells compared with HMECs (Fig. 1E and SI Appendix, Fig. S1N).
The increased binding of BORIS to methylated exon 10 further correlates with increased RNA polymerase II (Pol II) enrichment at exon 10 in MCF7 cells compared with HMECs (Fig. 1F). According to the kinetic coupling model of regulation of alternative pre-mRNA splicing (25), an increase in Pol II enrichment downstream of the 3′ splice site of the alternative exon during transcription elongation may favor the inclusion of the alternative exon by promoting its spliceosomal recognition (26). Thus, the DNA methylation-mediated binding of BORIS and associated Pol II enrichment at exon 10 might allow the inclusion of exon 10 and thereby increased PKM2 expression in breast cancer. In addition, we did not find a significant change in PKM2 expression in CTCF-depleted cells (SI Appendix, Fig. S1 O and P), suggesting a specific role of BORIS enrichment at methylated exon 10 in the regulation of PKM alternative pre-mRNA splicing.
Depletion of DNMT3B Results in the Reduced Expression of Cancer-Specific PKM2 Isoform.
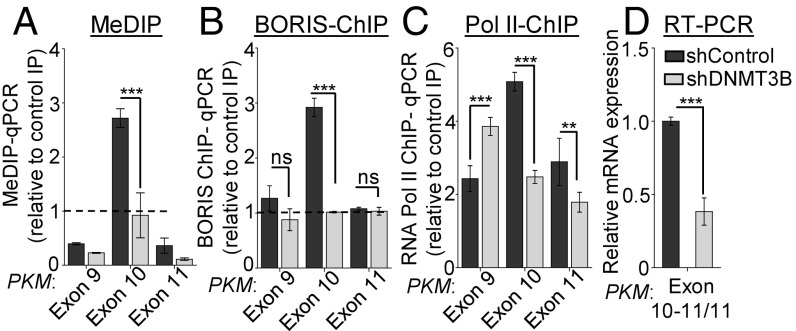
To investigate the role of DNA methylation in PKM alternative splicing, we used shRNA to deplete DNA methyltransferase (DNMT) 1, 3A, and 3B levels in the MCF7 cells. All three DNMTs are overexpressed in breast cancer compared with normal breast tissue in various breast cancer profiles analyzed (SI Appendix, Fig. S2 A–C and Table S3). We did not observe any change in DNA methylation at exon 10 and PKM alternative pre-mRNA splicing upon depletion of DNMT1 and 3A (SI Appendix, Fig. S2 D–K). Interestingly, shRNA-mediated depletion of de novo DNA methyltransferase DNMT3B (SI Appendix, Fig. S3 A and B) resulted in reduced DNA methylation at PKM exon 10 (Fig. 2A), which correlates with the reduced BORIS enrichment at the exon 10 region (Fig. 2B). Notably, reduced BORIS enrichment after DNMT3B depletion correlates with decreased Pol II occupancy (Fig. 2C), consequently leading to the reduced inclusion of exon 10 (Fig. 2D and SI Appendix, Fig. S3 C and D). Furthermore, the reduced inclusion of exon 10 in DNMT3B-depleted cells correlates with the mutually exclusive inclusion of exon 9 in these cells (SI Appendix, Fig. S3E). In addition, depletion of DNMT3B also leads to reduced PKM2 and increased PKM1 protein expression compared with the control MCF7 cells (SI Appendix, Fig. S3F). Interestingly, depletion of DNMT3B also results in increased Pol II occupancy at exon 9 (Fig. 2C). The change in DNA methylation at exon 10 in DNMT3B-depleted cells might also lead to changes in local chromatin structure at exon 9, which may lead to increased Pol II occupancy. In support, we observed increased nucleosome occupancy at exon 9 in DNMT3B-depleted cells (SI Appendix, Fig. S3G). Similarly, inhibition of DNA methylation by 5-Aza 2′-deoxycytidine (27) results in reduced DNA methylation at PKM exon 10 (SI Appendix, Fig. S3H) and mutual inclusion and exclusion of exons 9 and 10, respectively (SI Appendix, Fig. S3 I–K). Together our data suggest that higher DNA methylation at exon 10 leads to the recruitment of BORIS, which causes RNA Pol II enrichment and leads to the inclusion of exon 10, and is thus associated with increased expression of cancer-specific spliced isoform PKM2.
Fig. 2.
DNMT3B depletion leads to reduced exon 10 inclusion in PKM transcripts. (A) MeDIP in MCF7 cells transfected with shRNA against DNMT3B versus shControl cells, followed by qRT-PCR relative to input and control IgG (n = 3). (B and C) ChIP in MCF7 cells transfected with shRNA against DNMT3B versus shControl using (B) BORIS and (C) RNA Pol II antibody, followed by qRT-PCR relative to input and control IgG (n = 6). (D) qRT-PCR to measure the splicing pattern of PKM gene after down-regulation of DNMT3B. Indicated exon junction-specific primers were used and normalized with RPS16 and constitutive exon of PKM gene (n = 3). Error bar shows mean values ± SD. As calculated using two-tailed Student’s t test, **P < 0.01, ***P < 0.001; ns, nonsignificant.
BORIS Recruitment Is Critical for Pol II Enrichment and PKM2 Expression.
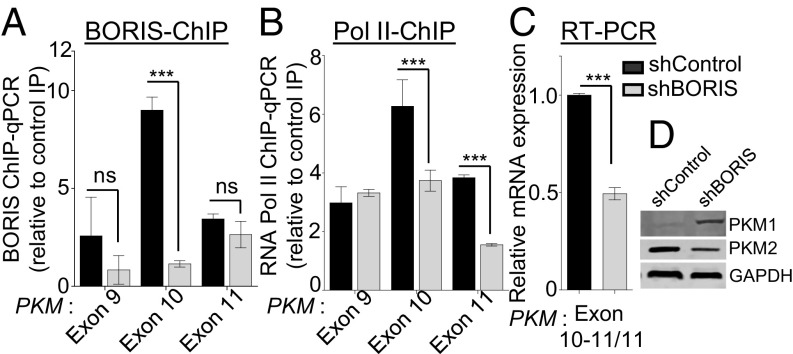
BORIS is considered an epigenetic oncogene that inhibits the tumor suppressor activity of CTCF (28) and gets overexpressed in many cancers, including breast cancer (29). Interestingly, BORIS ChIP-sequencing (ChIP-seq) data (GSM1817668 and GSM1817669) from MCF7 cells also showed enrichment of BORIS at the exon 10 region of the PKM gene (SI Appendix, Fig. S4A). To address the direct role of BORIS in PKM alternative pre-mRNA splicing, we carried out shRNA-mediated depletion of BORIS in MCF7 cells (SI Appendix, Fig. S4 B and C) and observed reduced BORIS binding (Fig. 3A), and decreased Pol II enrichment at exon 10 (Fig. 3B). The reduced Pol II enrichment, in turn, favors decreased inclusion of exon 10 (Fig. 3C and SI Appendix, Fig. S4D) with a concomitant increase in the inclusion of mutually exclusive exon 9 (Fig. 3D and SI Appendix, Fig. S4 E and F).
Fig. 3.
BORIS is critical for DNA methylation-dependent exon 10 inclusion in PKM. (A and B) ChIP in MCF7 cells transfected with shRNA against BORIS versus shControl using (A) BORIS and (B) RNA Pol II antibody, followed by qRT-PCR relative to input and control IgG (n = 5). (C) qRT-PCR to measure the splicing pattern of PKM gene after down-regulation of BORIS. Indicated exon junction-specific primers were used and normalized with RPS16 and constitutive exon of PKM gene (n = 3). (D) Western blot for PKM1 and PKM2 protein level expression in shBORIS and shControl MCF7 cells. Error bar shows mean values ± SD. As calculated using two-tailed Student’s t test, ***P < 0.001; ns, nonsignificant.
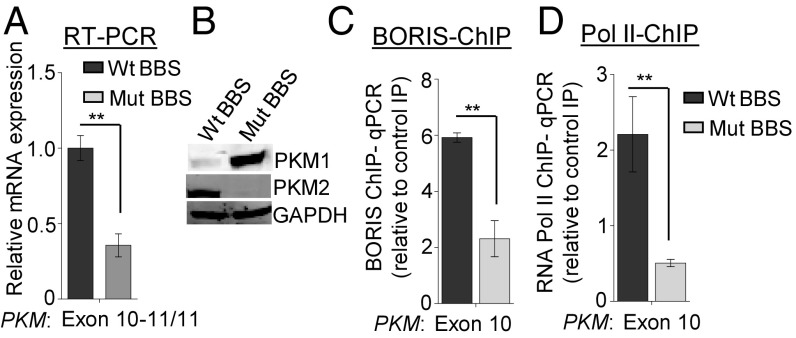
Having demonstrated the relationship between BORIS binding, Pol II enrichment, and PKM exon 10 inclusion, it was essential to exclude the possibility of additional factors responsible for Pol II enrichment and alternative splicing of PKM. PKM exon 10 contains two sequences that are similar to the consensus BORIS binding site (BBS) (30, 31) (SI Appendix, Fig. S5A), and the analysis of BORIS ChIP-seq data in MCF7 cells also suggested that exon 10 has two BBSs (SI Appendix, Fig. S5 A and B). However, BORIS ChIP-qPCR showed a higher enrichment of BORIS to the first BBS compared with the second (SI Appendix, Fig. S5C). Therefore, we decided to mutate the first BBS at the PKM exon 10 region. Thus, we have generated a cell line that has a mutation in the endogenous BBS at PKM exon 10 using CRISPR/Cas9 technology (SI Appendix, Fig. S5 D and E). The disruption of BBS did not affect BORIS expression in the cells with a mutated BBS (Mut BBS) compared with the cells with a wild-type BBS (Wt BBS) (SI Appendix, Fig. S5F). The mutation of BBS did not disrupt the splicing factor binding site as predicted by a human splicing finder tool (www.umd.be/HSF3/HSF.shtml) (SI Appendix, Table S4). To investigate the specific role of BORIS in the regulation of PKM alternative splicing, the splicing pattern of the PKM gene was analyzed in cells either with a Mut BBS or a Wt BBS. Interestingly, we observed reduced inclusion of exon 10 and a concomitant increase in the inclusion of exon 9 in Mut BBS compared with Wt BBS cells (Fig. 4 A and B and SI Appendix, Fig. S5 G and H). We also demonstrated a reduced BORIS enrichment upon mutation of the BBS (Fig. 4C and SI Appendix, Fig. S5I), which resulted in reduced Pol II occupancy (Fig. 4D and SI Appendix, Fig. S5J) and decreased exon 10 inclusion in the Mut BBS cells compared with Wt BBS cells (Fig. 4A). Collectively, these results provide strong evidence to support our hypothesis that BORIS is required for PKM alternative splicing.
Fig. 4.
CRISPR/Cas9-mediated BORIS binding site mutation in PKM exon 10 region and its effect on PKM splicing. (A) RPS16 normalized qRT-PCR data from Wt BBS and Mut BBS in PKM gene using exon 10-specific junction primers in MCF7 cells (n = 3). (B) Level of PKM1 and PKM2 protein in Wt BBS and Mut BBS MCF7 cells by Western blot analysis using GAPDH as a loading control. (C and D) ChIP in Wt BBS and Mut BBS MCF7 cells using (C) BORIS and (D) RNA Pol II antibody, followed by qRT-PCR relative to input and control IgG (n = 3). Error bar shows mean values ± SD. As calculated using two-tailed Student’s t test, **P < 0.01.
Clinical Relevance of Intragenic DNA Methylation-Dependent BORIS Binding on PKM2 Expression in Breast Cancer Patients.
To further study the role of BORIS in alternative splicing, we examined its clinical relevance in breast cancer patients. We analyzed breast cancer profiles available in The Cancer Genome Atlas (TCGA) and Oncomine and found overexpression of BORIS in breast cancer subtypes compared with normal breast tissues (SI Appendix, Fig. S6 A–H and Tables S5 and S6). This correlates with the increased protein expression of BORIS observed in tumor tissues compared with adjacent normal tissues by Western blot analysis (SI Appendix, Fig. S6I). This is consistent with the notion that BORIS acts as an oncogene in cancer progression (28). Furthermore, BORIS overexpression is associated with a poor survival outcome in breast cancer patients (SI Appendix, Fig. S6 J–L). Next, we performed qRT-PCR in paired breast tumor and adjacent normal tissues surgically collected from breast cancer patients (SI Appendix, Table S6). We found that all breast tumor tissues have significantly increased PKM2 isoform expression compared with normal tissues, at both the RNA as well as the protein level (Fig. 5A and SI Appendix, Fig. S7 A and B). Also, MeDIP in tissue samples showed increased DNA methylation at exon 10 in the tumor tissue compared with normal tissues (Fig. 5B), whereas no significant difference was observed at the adjacent exons in breast tumor and normal tissues (SI Appendix, Fig. S7C). Subsequently, when we performed BORIS ChIP, we found significant enrichment of BORIS at exon 10 in breast tumor tissue (Fig. 5C and SI Appendix, Fig. S7D), which notably also correlates with higher DNA methylation at exon 10 (Fig. 5B). Finally, RNA Pol II ChIP demonstrated enriched Pol II at PKM exon 10 in breast cancer compared with adjacent tissue (Fig. 5D and SI Appendix, Fig. S7E). Taken together, these clinical observations strongly support the role of BORIS and DNA methylation in PKM alternative splicing and are consistent with our cell culture studies. An important question that emerges from the fact that BORIS and CTCF share the common binding site (23) is whether CTCF binds to unmethylated exon 10 in normal cells where it might promote the inclusion of exon 10 in normal cells. Interestingly, we see an enrichment of BORIS and Pol II at exon 10 in breast cancer cells compared with normal breast tissue (Fig. 5 C and D), but we did not observe significant recruitment of CTCF in breast cancer and normal breast tissue (SI Appendix, Fig. S7 F and G). Though CTCF and BORIS share the same binding site, however, all of the binding sites of CTCF and BORIS do not overlap, due to nucleosome composition (30, 31). Moreover, the binding of CTCF and BORIS to the chromatin may also depend on the underlying epigenetic modifications (30, 31), and the mechanism that precludes CTCF from binding exon 10 in normal cells may be the subject of future investigation. Thus, BORIS specifically promotes exon 10 inclusion in cancer cells in a DNA methylation-dependent manner.
Fig. 5.
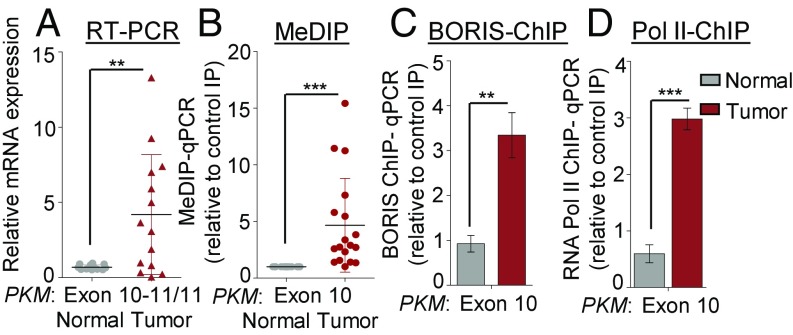
Clinical relevance of BORIS-mediated PKM splicing in breast cancer patients. (A) RPS16 normalized qRT-PCR data from normal and breast tumor tissue samples using the indicated exon junction-specific primers (n = 14). (B) MeDIP in normal and breast tumor tissue genomic DNA and qRT-PCR of PKM exon 10 region relative to input and control IgG (n = 20). (C and D) ChIP in normal and breast tumor tissues using (C) BORIS and (D) RNA Pol II antibody and qRT-PCR relative to input and control IgG (n = 3). Error bar shows mean values ± SD. As calculated using two-tailed Student’s t test, **P < 0.01, ***P < 0.001.
Loss of BORIS Suppresses the Warburg Effect and Growth of Breast Cancer Cells.
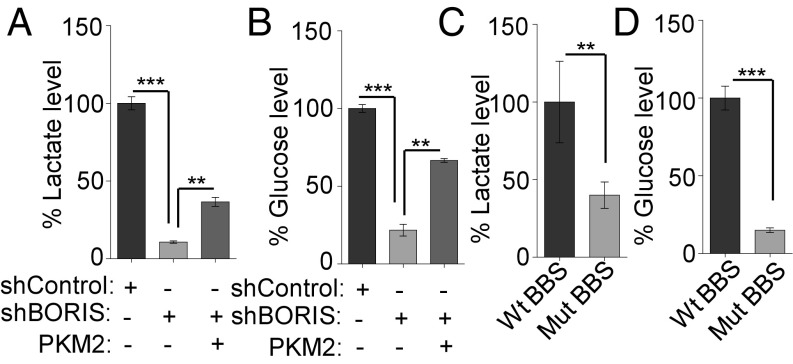
Based on previous reports, a switch from PKM1 to PKM2 isoform is known to contribute to the metabolic phenotype known as the Warburg effect (5). Having shown the role of DNA methylation-dependent binding of BORIS in PKM2 isoform expression, we made a hypothesis that BORIS depletion may cause a reversal of the Warburg effect, due to a switch in PKM splicing from PKM2 to PKM1. As the up-regulated glycolysis results in increased lactate production (8) (SI Appendix, Fig. S8A) and increased glucose consumption in cancer cells (8), we assessed the effect of BORIS on the Warburg effect using a lactate and glucose uptake assay. Interestingly, BORIS depletion resulted in reduced lactate production and reduced glucose consumption in breast cancer cells. However, overexpression of PKM2 significantly rescued the lactate production and glucose uptake in BORIS-depleted cells, suggesting that BORIS is involved in the regulation of the Warburg effect through influencing PKM splicing (Fig. 6 A and B and SI Appendix, Fig. S8B). Additionally, we also observed reduced lactate production and reduced glucose uptake in Mut BBS compared with the Wt BBS cells (Fig. 6 C and D), which suggested the specific role of BORIS in PKM splicing and thereby on the Warburg effect. As the observed effect of BORIS on the Warburg effect is partly due to a change in PKM splicing, we expected a similar effect on aerobic glycolysis by changes in DNA methylation. Accordingly, we observed reduced lactate production and decreased glucose consumption upon DNMT3B depletion as well (SI Appendix, Fig. S8 C and D). Similarly, inhibition of DNA methylation by 5-Aza-2′-deoxycytidine also led to reduced lactate production and decreased glucose uptake (SI Appendix, Fig. S8 E and F). Significantly, an increase in aerobic glycolysis in cancer cells is also associated with reduced oxidative phosphorylation (8). In this aspect, Gene Set Enrichment Analysis (GSEA) in breast cancer profiles showed that genes comprising an oxidative phosphorylation signature were highly enriched in BORIS-negative cells versus BORIS-positive cells (SI Appendix, Fig. S8G). In addition to BORIS, the expression of DNMT3B and PKM2 also showed a significant inverse correlation with the enrichment of genes belonging to oxidative phosphorylation in breast cancer patient profiles (SI Appendix, Fig. S8 H–J and Table S7).
Fig. 6.
BORIS is required for the maintenance of the Warburg effect in breast cancer. (A and B) Percentage of lactate production and glucose uptake level in MCF7 cells after BORIS depletion. Decreased lactate production and decreased glucose uptake were rescued after overexpression of PKM2 in BORIS-depleted MCF7 cells (n = 3). (C and D) Measurement of lactate and glucose uptake levels after BBS mutation in MCF7 cells (n = 3). Error bar shows mean values ± SD. As calculated using two-tailed Student’s t test, **P < 0.01, ***P < 0.001.
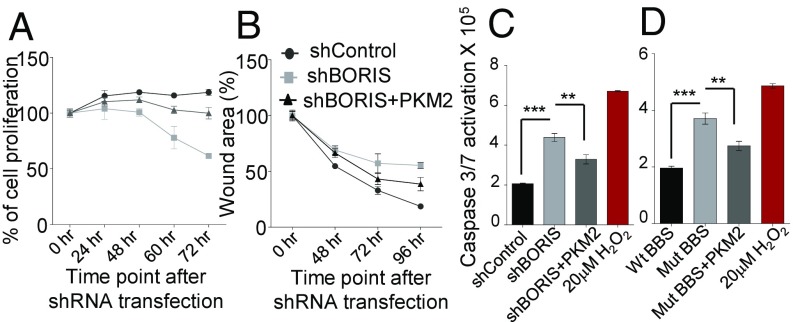
As we have demonstrated that BORIS-mediated expression of PKM2 promotes the Warburg effect, and the Warburg effect is known to be associated with the growth of cancer cells (10) (SI Appendix, Fig. S8 K and L), we therefore investigated the role of BORIS in tumorigenesis. The shRNA-mediated depletion of BORIS results in reduced cell viability and incomplete wound healing compared with the control cells, which was rescued by the overexpression of PKM2 (Fig. 7 A and B and SI Appendix, Fig. S8M). Moreover, a colony-forming assay showed decreased number of colonies in BORIS-depleted cells (SI Appendix, Fig. S8N). BORIS is known to maintain the tumor cell viability by inhibiting the apoptosis in breast cancer cells (32). Consistent with this, we found activation of caspase 3/7 in BORIS-depleted MCF7 cells (Fig. 7C and SI Appendix, Fig. S8 O and P). The overexpression of PKM2 in BORIS-depleted MCF7 cells partially rescues the inhibition of apoptosis (Fig. 7C). Moreover, mutated BBS cells also showed increased apoptosis, which was partially inhibited by overexpression of PKM2 (Fig. 7D), suggesting its role in the apoptosis of cancer cells. Considering that the Warburg effect correlates with suppression of apoptosis (33), BORIS-mediated inhibition of apoptosis might be in part due to its role in promoting the Warburg effect through the preferential expression of the PKM2 isoform.
Fig. 7.
BORIS down-regulation suppresses growth of breast cancer cells. (A) Percentage of relative cell proliferation in shBORIS versus shControl transfected MCF7 cells by MTT assay. The absorbance at 570 nm at 0 h was considered 100%. Overexpression of PKM2 partially rescued the cell viability in BORIS-depleted cells (n = 6). (B) MCF7 cells were transfected with shBORIS and shControl, and scratch was applied to cells. The graph is expressed as a percentage of wound area, which is calculated by measuring the gap distance between the edges of the scratch at the different time point compared with gap distance at 0 h. Overexpression of PKM2 partially rescued the gap distance in shBORIS transfected cells (n = 3). (C) Apoptotic death via caspase 3/7 activation in shBORIS versus shControl transfected cells at 96 h posttransfection (n = 5). Overexpression of PKM2 partially reduces the apoptotic cell death in shBORIS transfected cells. Hydrogen peroxide (20 μM) was used as a positive control for caspase activation and apoptosis. (D) Apoptotic death via caspase 3/7 activation in Wt BBS versus Mut BBS MCF7 cells. Overexpression of PKM2 partially rescues the apoptotic cell death in BORIS-depleted cells (n = 5). Error bar shows mean values ± SD. As calculated using two-tailed Student’s t test, **P < 0.01, ***P < 0.001.
Global Effect of BORIS on Alternative Splicing in Breast Cancer Cells.
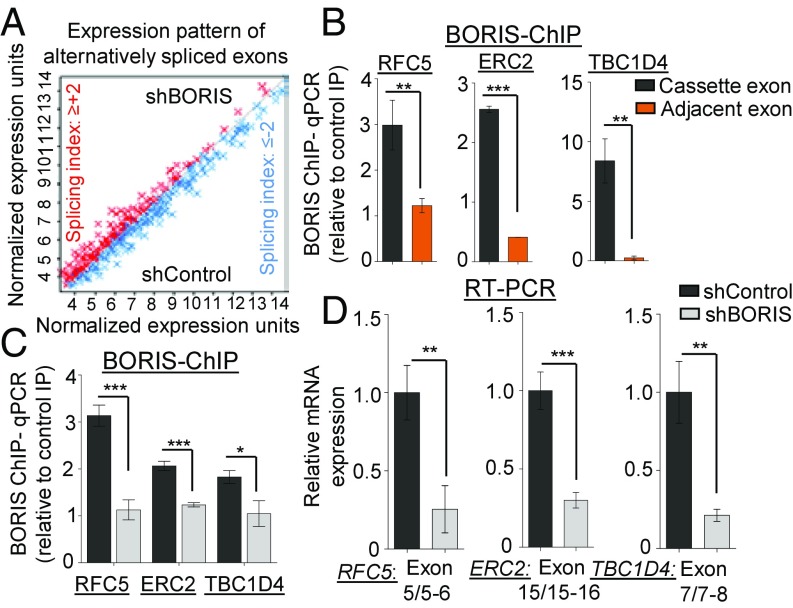
Having demonstrated that the DNA methylation-dependent binding of BORIS can regulate the alternative pre-mRNA splicing of PKM, we next examined the BORIS-mediated global changes in alternative pre-mRNA splicing using Human Transcriptome Array 2.0 (HTA 2.0) in BORIS-depleted MCF7 cells compared with the control MCF7 cells (SI Appendix, Fig. S9A). BORIS depletion results in the differential expression as well as exclusion of 361 and the inclusion of 221 alternative exons compared with the control MCF7 cells (Fig. 8A and SI Appendix, Fig. S9 B and C and Dataset S1). Collectively, these results support the notion that BORIS promotes exon recognition and inclusion in breast cancer. To validate the alternative splicing events identified from the HTA 2.0 array, we selected 10 candidate genes based on the splicing index score (SI Appendix, Fig. S9C). BORIS ChIP in MCF7 cells confirmed the enrichment of BORIS at the alternative exon of candidate genes RFC5, ERC2, and TBC1D4 (Fig. 8B). Additionally, analysis of BORIS ChIP-seq data from MCF7 cells stably expressing exogenous BORIS (31) also shows BORIS enrichment at the alternative exon of candidate genes compared with the adjacent exon (SI Appendix, Fig. S9D). This leads us to our next question of whether the BORIS-mediated inclusion of exons is methylation dependent or not. Interestingly, we observed increased DNA methylation at the cassette exons of RFC5, ERC2, and TBC1D4, which is reduced upon DNMT3B depletion (SI Appendix, Fig. S9E). Subsequently, we found reduced BORIS enrichment at the cassette exons after BORIS and DNMT3B depletion (Fig. 8C and SI Appendix, Fig. S9F), which further correlates with the exclusion of cassette exons in BORIS- and DNMT3B-depleted cells (Fig. 8D and SI Appendix, Fig. S9G). These results suggest that the DNA methylation and BORIS-mediated regulation of alternative splicing is a more common mechanism than just being limited to the PKM gene. In addition to this, gene ontology analysis of BORIS-mediated alternatively spliced events showed a higher enrichment of genes involved in cell death, cell cycle, DNA metabolism, and other cellular processes. This implies the possible role of BORIS in the alternative pre-mRNA splicing of genes involved in the major hallmarks of cancer (SI Appendix, Fig. S9H and Table S8).
Fig. 8.
Global identification of BORIS-mediated alternative splicing. (A) Global differential expression pattern of alternatively spliced exons in shBORIS versus shControl transfected MCF7 cells [n = 2; excluded exons = 361 (splicing index ≤ −2), included exons = 221 (splicing index ≥ +2); and P < 0.05]. The graph represents normalized expression levels (signal space transformation–robust multiarray average algorithm) of differentially expressed exon probes. Red dots represent the exon inclusion; blue dots represent exon exclusion. (B) Validation of BBS in alternative exons by BORIS ChIP in MCF7 cells (n = 3). (C) BORIS ChIP-qPCR in MCF7 cells transfected with shBORIS and shControl cells to detect BORIS binding at alternative exons of genes from shBORIS versus shControl HTA 2.0 profile (n = 3). (D) Validation of BORIS-regulated alternative splicing events by qRT-PCR in shControl versus shBORIS transfected MCF7 cells (n = 3). Error bar shows mean values ± SD. As calculated using two-tailed Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Here we describe that the differential intragenic DNA methylation in breast cancer cells is associated with cancer-specific splicing. Our results show increased DNA methylation specifically at PKM exon 10 in breast cancer tissues compared with normal tissues where it correlates with the inclusion of exon 10 and consecutively to higher PKM2 expression. Remarkably, depletion of DNMT3B (34) resulted in the loss of DNA methylation at exon 10, loss of BORIS binding, and Pol II enrichment followed by exon 10 exclusion. The specific effect of DNMT3B on PKM splicing suggests the locus-specific role of DNMT3B in regulating the DNA methylation (35). The DNMT3B is overexpressed in breast cancer (36), which may be associated with aberrant DNA methylation at PKM exon 10 and thereby PKM2 isoform expression in breast cancer cells.
Earlier, three different mechanisms each involving different proteins such as CTCF (15), MeCP2 (16), and HP1 (17) have been described, by which DNA methylation can be linked to alternative splicing, but these mechanisms may explain the alternative splicing of only a subset of all of the alternative splicing controlled by DNA methylation (37). Our results reveal the role of BORIS in the regulation of DNA methylation-mediated alternative splicing in cancer cells. We found that BORIS specifically binds to methylated exon 10 and causes RNA Pol II enrichment, and thereby inclusion of exon 10 in breast cancer cells. The mutation of the BBS at exon 10 endogenously by CRISPR/Cas9 system also revealed the specific role of BORIS on the alternative splicing of PKM. BORIS is not expressed by the normal somatic cells, but the reexpression of BORIS in cancer (24) along with aberrantly expressed DNMT3B may be associated with the cancer-specific splicing of PKM. In view of previous studies on splicing factor-mediated regulation of PKM alternative splicing (11–14), our study suggests a cross-talk between epigenetics and splicing factor-mediated splicing regulation that needs further investigation.
The preferential expression of PKM2 is associated with increased aerobic glycolysis and is associated with growth advantage to the cancer cells (5). Notably, the splicing switch from PKM2 to PKM1 isoform upon depletion of DNMT3B or BORIS as well as upon the mutation of BBS at PKM exon 10 is associated with the reversal of the Warburg effect, which further inhibits the growth of breast cancer cells. Though the observed reversal of the Warburg effect may not be completely attributed to the splicing switch induced by DNMT3B and BORIS, it may partially explain the reason for poor prognosis or growth of breast cancer associated with the overexpression of DNMT3B (38) and BORIS (32).
Importantly, our results show that in addition to PKM splicing, BORIS also regulates the alternative splicing of several genes in a DNA methylation-dependent manner. BORIS is also known to inhibit the apoptosis of breast cancer (32) and colorectal cancer cells (39). In accordance, we have also shown the inhibition of cell proliferation and apoptosis upon BORIS depletion, which may in part be due to its role in the alternative splicing of PKM and other genes.
Collectively, we have described a mechanism based on BORIS binding to explain the alternative splicing events regulated by DNA methylation in breast cancer (SI Appendix, Fig. S10). As the Warburg effect and PKM2 overexpression are the characteristics of most cancers (40), it is possible that the DNA methylation-dependent BORIS binding may regulate PKM2 expression in other cancers as well. Furthermore, this study provides the mechanism to switch the PKM splicing from cancer-specific to normal isoform by controlling the upstream DNA methylation or BORIS, which may provide a route for therapeutic management of breast cancer in future.
Materials and Methods
The study was approved by the Institute Ethics Committee of Indian Institute of Science Education and Research Bhopal. Informed consent was obtained from all the patients. For details on cell culture, patients, plasmids, shRNAs, qRT-PCR, ChIP, MeDIP, Western blot, CRISPR/Cas9, cell viability, wound healing, colony forming, lactate, glucose uptake, caspase 3/7, HTA 2.0 array, TCGA data analysis, MNase assay, and immunofluorescence, please see SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank N. Kalra for critical reading of this manuscript. This work is supported by Science and Engineering Research Board (SERB) Grant EMR/2014/000716 and Wellcome Trust/Department of Biotechnology (DBT) India Alliance Fellowship Grant IA/I/16/2/502719 (to S. Shukla). S. Singh and A.G. were supported by an Indian Institute of Science Education and Research Bhopal fellowship. N.A. and S.Y. were supported by a Council of Scientific and Industrial Research fellowship. S.P.N. was supported by SERB-National Postdoctoral Fellowship PDF/2015/000560.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The HTA2.0 array data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE81001).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708447114/-/DCSupplemental.
References
- 1.Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311–5318. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- 2.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 5.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 6.Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–13812. [PubMed] [Google Scholar]
- 7.Lee J, Kim HK, Han YM, Kim J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int J Biochem Cell Biol. 2008;40:1043–1054. doi: 10.1016/j.biocel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feron O. Pyruvate into lactate and back: From the Warburg effect to symbiotic energy fuel exchange in cancer cells. Radiother Oncol. 2009;92:329–333. doi: 10.1016/j.radonc.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 10.Lunt SY, Vander Heiden MG. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 11.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clower CV, et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci USA. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M, David CJ, Manley JL. Concentration-dependent control of pyruvate kinase M mutually exclusive splicing by hnRNP proteins. Nat Struct Mol Biol. 2012;19:346–354. doi: 10.1038/nsmb.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, et al. Exon-centric regulation of pyruvate kinase M alternative splicing via mutually exclusive exons. J Mol Cell Biol. 2012;4:79–87. doi: 10.1093/jmcb/mjr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yearim A, et al. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 2015;10:1122–1134. doi: 10.1016/j.celrep.2015.01.038. [DOI] [PubMed] [Google Scholar]
- 18.van Hoesel AQ, et al. Assessment of DNA methylation status in early stages of breast cancer development. Br J Cancer. 2013;108:2033–2038. doi: 10.1038/bjc.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferlay J, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 20.Desai S, et al. Tissue-specific isoform switch and DNA hypomethylation of the pyruvate kinase PKM gene in human cancers. Oncotarget. 2014;5:8202–8210. doi: 10.18632/oncotarget.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menafra R, et al. Genome-wide binding of MBD2 reveals strong preference for highly methylated loci. PLoS One. 2014;9:e99603. doi: 10.1371/journal.pone.0099603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen P, et al. CTCFL/BORIS is a methylation-independent DNA-binding protein that preferentially binds to the paternal H19 differentially methylated region. Cancer Res. 2008;68:5546–5551. doi: 10.1158/0008-5472.CAN-08-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin-Kleiner I. BORIS in human cancers: A review. Eur J Cancer. 2012;48:929–935. doi: 10.1016/j.ejca.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Naftelberg S, Schor IE, Ast G, Kornblihtt AR. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem. 2015;84:165–198. doi: 10.1146/annurev-biochem-060614-034242. [DOI] [PubMed] [Google Scholar]
- 26.Shukla S, Oberdoerffer S. Co-transcriptional regulation of alternative pre-mRNA splicing. Biochim Biophys Acta. 2012;1819:673–683. doi: 10.1016/j.bbagrm.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christman JK. 5-azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 28.Link PA, Zhang W, Odunsi K, Karpf AR. BORIS/CTCFL mRNA isoform expression and epigenetic regulation in epithelial ovarian cancer. Cancer Immun. 2013;13:6. [PMC free article] [PubMed] [Google Scholar]
- 29.D’Arcy V, et al. BORIS, a paralogue of the transcription factor, CTCF, is aberrantly expressed in breast tumours. Br J Cancer. 2008;98:571–579. doi: 10.1038/sj.bjc.6604181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sleutels F, et al. The male germ cell gene regulator CTCFL is functionally different from CTCF and binds CTCF-like consensus sites in a nucleosome composition-dependent manner. Epigenetics Chromatin. 2012;5:8. doi: 10.1186/1756-8935-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pugacheva EM, et al. Comparative analyses of CTCF and BORIS occupancies uncover two distinct classes of CTCF binding genomic regions. Genome Biol. 2015;16:161. doi: 10.1186/s13059-015-0736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dougherty CJ, et al. Selective apoptosis of breast cancer cells by siRNA targeting of BORIS. Biochem Biophys Res Commun. 2008;370:109–112. doi: 10.1016/j.bbrc.2008.03.040. [DOI] [PubMed] [Google Scholar]
- 33.Ruckenstuhl C, et al. The Warburg effect suppresses oxidative stress induced apoptosis in a yeast model for cancer. PLoS One. 2009;4:e4592. doi: 10.1371/journal.pone.0004592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 35.Linhart HG, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girault I, Tozlu S, Lidereau R, Bièche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415–4422. [PubMed] [Google Scholar]
- 37.Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31:274–280. doi: 10.1016/j.tig.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Roll JD, Rivenbark AG, Jones WD, Coleman WB. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol Cancer. 2008;7:15. doi: 10.1186/1476-4598-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. Brother of Regulator of Imprinted Sites (BORIS) suppresses apoptosis in colorectal cancer. Sci Rep. 2017;7:40786. doi: 10.1038/srep40786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang W, Lu Z. Regulation and function of pyruvate kinase M2 in cancer. Cancer Lett. 2013;339:153–158. doi: 10.1016/j.canlet.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.