Significance
TRAF4 is a signaling molecule involved in cancer by direct interaction with the TGF-β receptor and in ROS production in platelets that initiates thrombosis. Previous studies indicated that disruptions of the TRAF4 interaction with platelet receptors and the TGF-β receptor are a potential target for therapeutic intervention by antithrombotic drugs and anticancer drugs, respectively. In this study, we report a crystal structure of TRAF4 in complex with a peptide from the GPIbβ receptor. The structure revealed a TRAF4-binding motif of Arg–Leu–X–Ala. We also found that the TGF-β receptor contains the TRAF4-binding motif and showed a direct interaction between TRAF4 and the receptor. The current structure will provide a template for furthering our understanding of the receptor-binding specificity of TRAF4, TRAF4-mediated signaling, and related diseases.
Keywords: platelet, TRAF4, TRAF domain, glycoprotein Ib, protein structure
Abstract
Tumor necrosis factor (TNF)-receptor associated factor 4 (TRAF4), an adaptor protein with E3-ligase activity, is involved in embryogenesis, cancer initiation and progression, and platelet receptor (GPIb-IX-V complex and GPVI)-mediated signaling for reactive oxygen species (ROS) production that initiates thrombosis at arterial shears. Disruption of platelet receptors and the TRAF4 interaction is a potential target for therapeutic intervention by antithrombotic drugs. Here, we report a crystal structure of TRAF4 (amino acid residues 290∼470) in complex with a peptide from the GPIbβ receptor (amino acid residues 177∼181). The GPIbβ peptide binds to a unique shallow surface composed of two hydrophobic pockets on TRAF4. Further studies revealed the TRAF4-binding motif Arg–Leu–X–Ala. The TRAF4-binding motif was present not only in platelet receptors but also in the TGF-β receptor. The current structure will provide a template for furthering our understanding of the receptor-binding specificity of TRAF4, TRAF4-mediated signaling, and related diseases.
The glycoprotein (GP) Ib-IX-V complex and GPVI are two major platelet receptors that mediate the initial adhesion of platelets to the injured vascular wall for initiating thrombus formation, and they are involved in thrombotic diseases such as stroke and myocardial infarction (1, 2). GPVI is known as a primary collagen receptor, while the GP Ib-IX-V complex binds to the von Willebrand factor and diverse ligands including coagulation factor XI (FXI), factor XII, thrombin, and P-selectin (3, 4). The existence of various ligands of the GP Ib-IX-V complex indicates that this receptor complex governs platelet function via multiple signaling pathways (5). The GP Ib-IX-V complex, consisting of GPIbα (the major ligand-binding subunit), GPIbβ, GPIX, and GPV, is the second most abundant receptor in platelets and is physically and functionally linked to another platelet receptor, GPVI (3).
Tumor necrosis factor (TNF)-receptor associated factor 4 (TRAF4) is one of the seven identified TRAF1∼TRAF7 factors in mammals and was initially identified as a regulator of embryogenesis in mice, especially in the central and peripheral nervous system (6). TRAF4 is involved in gross tracheal, neural tube, and skeletal formation (7–9). The involvement of TRAF4 in the initiation and progression of many types of cancer has also been reported, suggesting that TRAF4 is a tentative therapeutic target for cancer treatment (10, 11). Unlike other TRAF family members, TRAF4 contains a nuclear localization signal (NLS) and does not interact with canonical TRAF-binding receptors such as TNFR2, CD30, and CD40, indicating that TRAF4 is not involved in cellular signaling that is mediated by the typical TRAF family (12, 13).
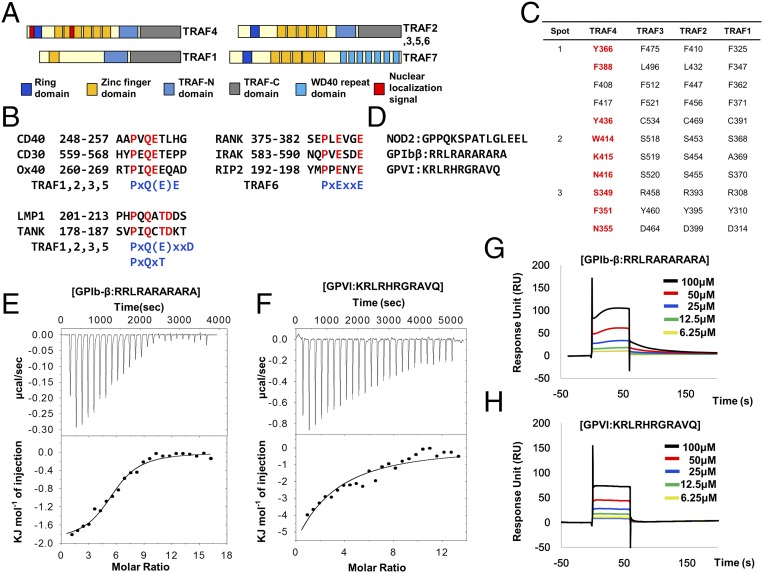
Because of the critical function of the TRAF family in various signaling events, including immunity, inflammation, and apoptosis, these have been the main focus of functional, structural, and biochemical analyses of these proteins (13). Intensive study has revealed that the TRAF family, except TRAF7, contains conserved TRAF domains at the C terminus that mediate their interaction with upstream receptors and downstream effectors (14) (Fig. 1A). Despite the structural similarity of the TRAF domain, each domain on different TRAF proteins is specific to interacting upstream receptors. For example, the TRAF domain of TRAF6 binds specifically to the TRAF6 consensus-binding motif, PxExxZ (Z: acidic or aromatic residues), and TRAF1, -2, -3, and -5 recognize other binding motifs, including Px(Q/E)E, Px(Q/E)xxD, and Px(Q/E)xT (15–20) (Fig. 1B). Although three different groups, including ours, have recently determined the structure of the TRAF4 TRAF domain (21–23), it is not known how TRAF4 can accommodate various receptors using a limited interaction interface. Interestingly, sequence comparison with other TRAF family members revealed that receptor-interacting hot spots are not conserved in TRAF4, indicating that TRAF4 might use a novel interaction mode for binding with different receptors (Fig. 1C). Based on studies that sought to find TRAF4 interacting receptors and TRAF4-mediated signaling events, three candidate receptors have been identified, including NOD2 and two platelet receptors (GPIbβ and GPVI) (24, 25). Although the putative TRAF4 interaction region on NOD2 contains a typical TRAF-binding motif-like motif (PxQxS) (25), the mostly positively charged region on these two platelet receptors, which have been mapped as TRAF4-binding regions, do not contain any known TRAF-binding motif (Fig. 1D).
Fig. 1.
Interaction of TRAF4 with two platelet receptors. (A) Domain boundary of the TRAF family. (B) TRAF-binding receptors and previously identified TRAF-binding motifs. (C) Conserved amino acid residues on TRAF1, -2, and -3, which are involved in the interaction with various receptors. Three hot spots on TRAF1, -2, and -3 are used for binding with TRAF-binding motifs. The amino acid residues on TRAF4, which are not conserved, are colored in red. (D) Previously reported putative TRAF4-binding regions on NOD2 and two platelet receptors. (E and F) ITC experiments showing titration of GPIbβ (E) and GPVI (F) peptides into a TRAF4 solution. The raw calorimetric titration data are shown in Upper, and experimental fitting of the data to a single site interaction model is shown in Lower. (G and H) SPR characterization of GPIbβ (G) and GPVI (H) peptides binding to immobilized TRAF4. Both peptides (6.25–100 μM) were applied to a TRAF4-coupled sensor chip. Data represent the average of two independent experiments.
Generation of reactive oxygen species (ROS) is one of the hallmarks of GPIb-IX-V complex and GPVI platelet receptor-mediated signaling events that initiate thrombosis at arterial shears (2). A previous study indicates that ROS production by platelets at the site of thrombosis via the GPIb-IX-V complex and GPVI signaling is mediated by direct interaction between TRAF4 and the GPIb-IX-V complex and GPVI, and disruption of this interaction might be a potential target for therapeutic intervention by antithrombotic drugs that can prevent thrombosis without affecting hemostasis (24, 26).
Results and Discussion
Interaction of TRAF4 with Two Platelet Receptors.
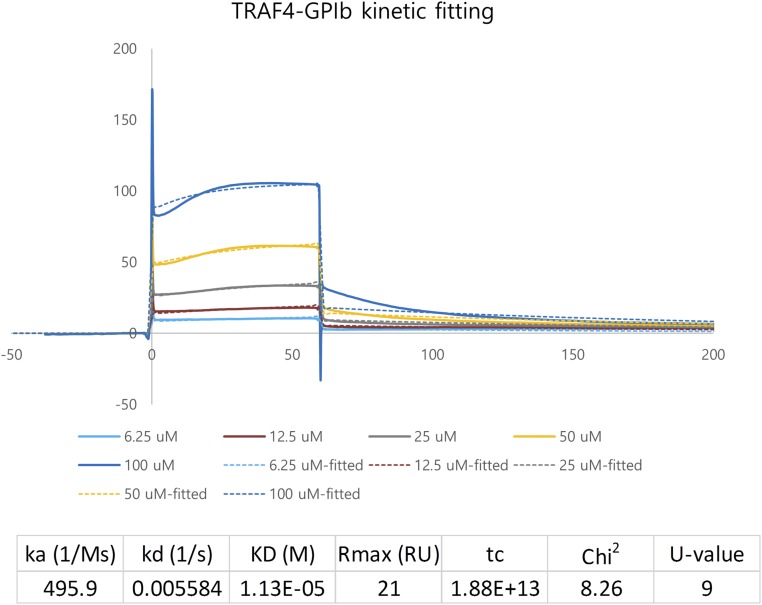
In an effort to shed light on the molecular mechanism of receptor recognition by TRAF4 and its potential application for therapeutic intervention, we performed a structural study of TRAF4 with several known receptors. Since the TRAF4 interactions with NOD2 and two platelet receptors have been mapped, we synthesized the receptor peptides based on previous studies (24, 25). We had previously quantitatively analyzed the interactions between TRAF4 and the following receptor peptides—NOD2 and two platelet receptors, NOD2 (GPPQKSPATLGLEEL), GPIbβ (RRLRARARARA), and GPIV (KRLRHRGRAVQ)—using surface plasmon resonance (SPR). As a preliminary interaction study in vitro, one fixed concentration of three peptides (50 μM) was tested for interaction with TRAF4. According to the initial screening, GPIbβ and GPIV seem to interact with TRAF4, while NOD2 did not. Based on these preliminary binding data, we thoroughly analyzed the interaction between TRAF4 and the platelet receptors (GPIbβ and GPIV) using isothermal titration calorimetry (ITC) and SPR. Each peptide (1 mM) was titrated into 20 μM of TRAF4 with 25 injections for incremental ITC experiments (Fig. 1 E and F). In both cases, the released heat was in agreement with ideal interaction values, indicating a single type of binding site without distinct cooperation in the interaction. The control titration, performed without binding peptides, was measured and subtracted from the experimental titration curve. The dissociation constants were 46 μM for GPIbβ and 35 μM for GPIV, indicating that both GPIbβ and GPIV peptides interact with TRAF4 with similar affinities (Fig. 1 E and F). For the SPR experiment, purified 6xHis-tagged TRAF4 was coupled with a Ni-NTA sensor chip using a His-tag affinity system. The binding of different concentrations of each peptide, ranging from 1 μM to 100 μM, with TRAF4 was analyzed. Both GPIbβ and GPIV showed clear concentration-dependent interactions (Fig. 1 G and H). Since the Ni-NTA sensor chip could not keep a stable baseline and peptides showed large bulk response, data were difficult to fit. In the case of GPIbβ peptide, the Kd value could be calculated from kinetic fitting, while the GPIV data could not be fitted either with the kinetic or equilibrium fitting method. A Kd value of 11.3 μM was obtained for the GPIbβ peptide interaction (Fig. S1). The binding affinity between TRAF4 and receptor peptides, with a Kd value around 10∼100 μM, was comparable to those between other TRAF family members and their respective receptor peptides previously studied (Table S1).
Fig. S1.
Kinetic fitting of SPR data from TRAF4 and GPIbβ receptor peptide interaction.
Table S1.
Receptor peptide-binding affinities of TRAF family
| TRAF family | Receptor/adaptor sequence | Kd | Method |
| TRAF4 | GPIbβ (RRLRARARAR) | Kd = 11.3 μM | SPR |
| Kd = 46.1 μM | ITC | ||
| GPVI (KRLRHRGRAVQ) | Kd = 35.6 μM | ITC | |
| TRAF1 | TANK (SVPIQCTDKT) | Kd = 16 μM | ITC |
| Kd = 27.9 μM | SPR | ||
| TRAF2 | CD30 (SDVMLSVEEEG) | Kd = 40 μM | ITC |
| Ox40 (PIQEE) | Kd = 50 μM | ITC | |
| TRAF3 | TANK (SVPIQCTDKTDKQEALFK) | Kd = 23.9 μM | ITC |
| TRAF6 | CD40 (KQEPQEIDF) | Kd = 84.0 μM | ITC |
| IRAK (RQGPEESDEF) | Kd = 54.3 μM | ITC |
Crystal Structure of the TRAF4/GPIbβ Complex.
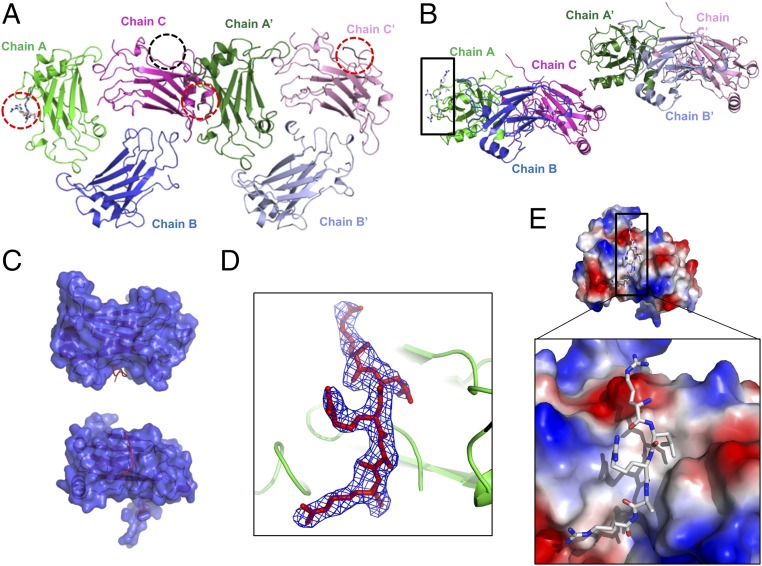
Because the in vitro interaction study confirmed the interaction between TRAF4 and designed platelet receptor peptides, which do not contain any known typical TRAF-binding motifs, we proceeded to investigate how TRAF4 recognizes these peptides by solving their complex structure. TRAF4 crystals were soaked with various concentrations of platelet receptor peptides at different time scales. The crystal soaked in 100 μM GPIbβ peptide for 24 h diffracted to 2.5 Å, and the density of the peptide was clearly detected after solving the structure by molecular replacement. Although the TRAF4 crystals were obtained at similar buffer conditions to the crystals that produced the native structure, the peptide-soaked crystals used for this complex structure study have slightly larger cell dimensions with a different space group, P21, and contain six molecules (two trimers): chain A, B, and C for one trimer and chain A′, B′, and C′ for another, in the asymmetric unit (ASU) (Fig. 2A). The structure was refined to an Rwork of 21.1% and Rfree of 26.4%. Crystallographic and refinement statistics are summarized in Table S2. Among the six chains, three (chain A, A′, and C′) contained GPIbβ peptide at the typical receptor interaction region of other TRAF family members (Fig. 2 A and B). The receptor-binding pocket was well-defined with a deep groove, and receptor peptides were bound in the pocket (Fig. 2C). Although the length and clearance of electron densities for each peptide was different, the GPIbβ peptide in chain A was the most clearly modeled (Fig. 2D). A small untraceable electron density that might be a peptide was also detected in chain C. Although we designed, synthesized, and used the amino acid sequence RRLRARARAR for the GPIbβ receptor peptide, only the RLRAR portion, which might be a core part of the interaction, was modeled because the remaining residues were not observed in the electron density (Fig. 2D). Although the overall surface electrostatic features of TRAF4 were composed of dispersed charged residues, the receptor peptide-binding region exhibits a distinct hydrophobic groove along with acidic surfaces at both ends that can interact with a basically charged residue, Arg, from the peptide (Fig. 1E).
Fig. 2.
Crystal structure of the TRAF4/GPIbβ complex. (A) Cartoon of the hexameric (two trimers in the ASU) TRAF4/GPIbβ complex. Chain A, chain B, and chain C make up one trimer, and chain A′, chain B′, and chain C′ make up another trimer. GPIbβ peptides were detected at three positions (red-dot circle). Unclear peptide density was detected at one position (black circle). (B) Side view of hexameric TRAF4/GPIbβ complex observed in the ASU of the crystal. The model of the clearest peptide was built at chain A position (black box). (C) Surface figure of monomeric TRAF4 (blue color) with bound GPIbβ peptide (Red line). Chain A was used as a representative monomeric molecule. (D) A simulated annealing Fo-Fc omit density map contoured at the 2.5 σ level around the GPIbβ peptide shown as a blue-colored mesh. (E) Charge surface representation of TRAF4 with the GPIbβ peptide in stick model. Upper and Lower showed the overall surface and magnified surface focused on the peptide binding pocket, respectively.
Table S2.
Crystallographic statistics
| Native data | TRAF4/GPIbβ |
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c, Å | 56.7, 88.6, 120.7 |
| α, β, γ, ° | 90.0, 93.6, 90.0 |
| Resolution, Å | 50.0–2.5 |
| Rsym, % | 7.7 (26.9)† |
| Mean I/σ, I | 21.7 (3.4)† |
| Completeness, % | 95.0 (95.9)† |
| Redundancy | 3.1 (3.1)† |
| Refinement | |
| Resolution, Å | 33.0–2.5 |
| Molecules/au* | 6 (proteins) and 3 (peptides) |
| No. reflections | 38,763 |
| Rwork/Rfree, % | 21.1/26.4 |
| No. atoms | |
| Protein | 7,805 |
| Water | 29 |
| Average B-factors, Å2 | |
| Chain A | 49.1 |
| Chain B | 60.4 |
| Chain C | 55.1 |
| Chain D | 52.6 |
| Chain E | 56.1 |
| Chain F | 55.0 |
| Chain G | 58.7 |
| Chain I | 60.7 |
| Chain K | 84.8 |
| Water | 52.7 |
| rmsd | |
| Bond lengths, Å | 0.010 |
| Bond angles, ° | 1.390 |
| Ramachandran plot, % | |
| Most favored regions | 96.1 |
| Additional allowed regions | 3.9 |
Asymmetric unit.
Statistics for the highest resolution shell are shown in parentheses.
Detailed TRAF4-GPIbβ Interaction Mode and Its Comparison with Other TRAF Family Members.
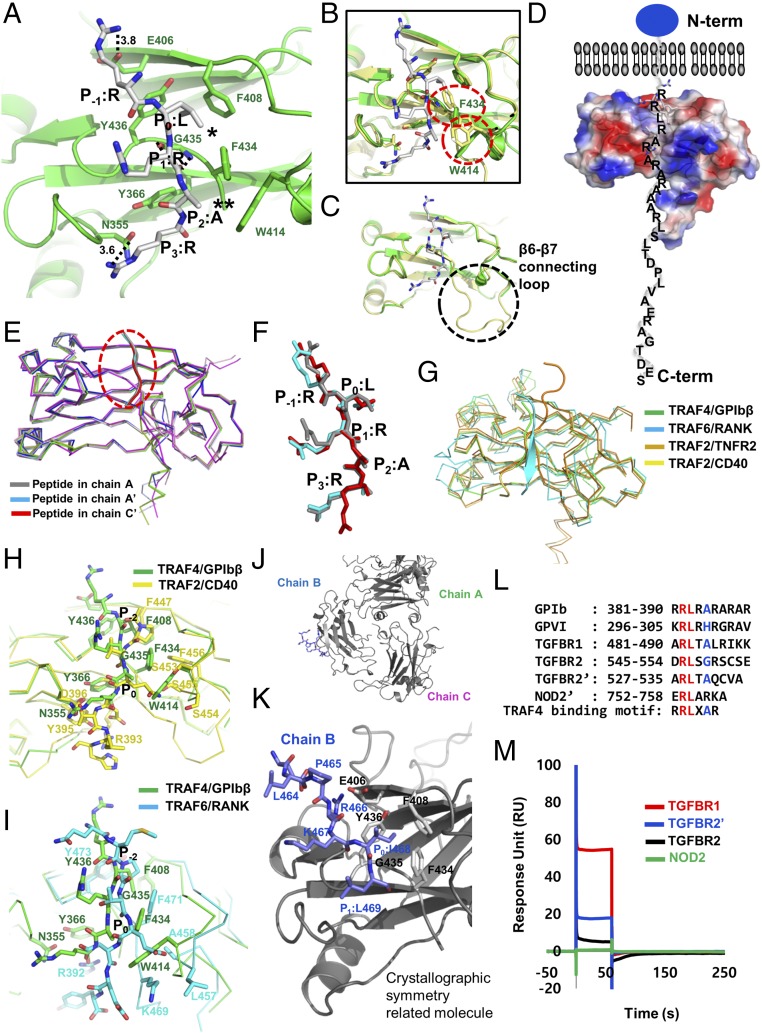
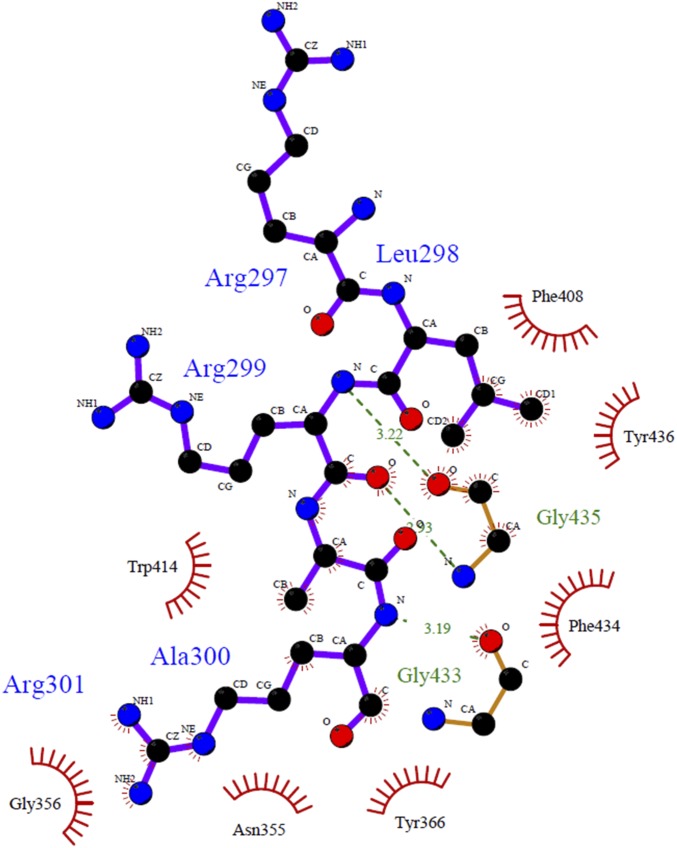
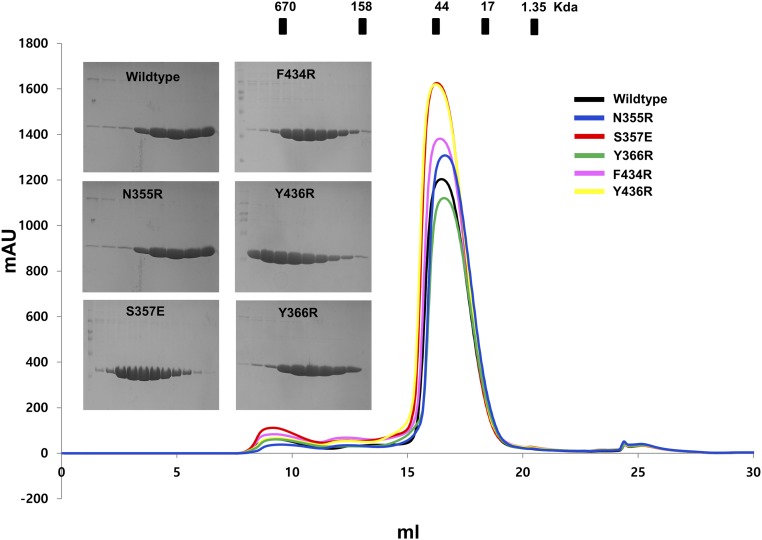
Previous structural studies of the TRAF family with various peptide complexes, including CD40, CD30, TNFR2, and TANK, denoted the most conserved amino acid in the TRAF-binding motif as P0, or the zero position of the TRAF-binding motif. Based on this labeling strategy, in the GPIbβ receptor peptide, the RLRAR motif was named as R (P−1), L (P0), R (P1), A (P2), and R (P3) (Fig. 3A). Our complex structure indicated that the side chains of R (P−1), L (P0), A (P2), and R (P3) on the GPIbβ receptor were involved in the TRAF4 interaction. R at the P−1 position and another R at the P3 position formed a hydrogen bond with the E406 and N355 side chains on TRAF4, respectively. The main stable hydrophobic interaction is formed by L at GPIbβ P0 with F408, F434, and Y436 from TRAF4. A second minor hydrophobic pocket formed by W414 and F434 with TRAF4 was also detected and participated in the interaction with A at GPIbβ P2 (Fig. 3A and Fig. S2). To confirm the interaction between TRAF4 and the GPIbβ receptor peptide in our structure, we generated five recombinant variants of TRAF4 (N355R, S357E, Y366R, F434R, and Y436R) that could disrupt the interaction, based on the current complex structure. All five TRAF4 variants were expressed and purified an equal amount compared with wild type (Fig. S3), indicating that mutagenesis did not affect the expression yield and correct folding of the target proteins. The GPIbβ receptor peptide interaction between TRAF4 and these mutants was studied via ITC and SPR, which revealed that three mutants—N355R, Y366R, and Y436R—disrupt the interaction, while S357E and F434R mutants reduced the interaction affinity, indicating that the GPIbβ receptor peptide interacts with the shallow binding pocket on TRAF4 formed by N355, S357, Y366, F434, and Y436 (Table S3). These five residues were completely conserved in TRAF4 across species (Fig. S4), indicating that those residues might be functionally important by being involved in the receptor interaction.
Fig. 3.
Detailed TRAF4-GPIbβ interaction mode and identification of the TRAF4-binding motif. (A) Close-up view of the GPIbβ peptide bound to the TRAF4 TRAF domain. GPIbβ peptide bound on chain A is represented. The TRAF4 residues involved in the GPIbβ peptide interaction are indicated. H-bonds are shown as black-dashed lines. Amino acid positions on the GPIbβ peptide labeled as R (P−1), L (P0), R (P1), A (P2), and R (P3) are shown. * and ** indicate major and minor hydrophobic pockets, respectively. (B and C) Structural comparison of receptor-bound TRAF4 (green color) and apo TRAF4 (yellow color) by superposition. Red circles indicate structurally moved amino acid residues upon binding with GPIbβ peptide (B). Black circle indicates the disordered loop (β6–β7 connecting loop: amino acid 421–432) upon binding with GPIbβ peptide (C). (D) Surface electrostatic representation of TRAF4/GPIbβ peptide complex. Blue circle indicates the extracellular domain of GPIbβ platelet receptor. The C-terminal tail of GPIbβ platelet receptor located in the intracellular part recognized by TRAF4 was modeled. N-term and C-term indicate N terminus and C terminus of TRAF4, respectively. (E and F) Structural comparison of TRAF4-bound receptor peptides (gray, peptide from chain A; cyan, peptide from chain A′; and red, peptide from chain C′) by superposition. Overall main chain conformation of the peptides is almost identical (E). The structural conservation of the side-chain conformation as well as the main-chain conformation at P−1, P0, and P2 positions (F). (G) Structural comparison of the TRAF4/GPIbβ receptor peptide complex with other TRAF/receptor peptide complexes. TRAF6/RANK complex, TRAF2/TNFR2 complex, and TRAF2/CD40 were superposed with TRAF4/GPIbβ complex. (H and I) Pair-wise comparison of the peptide-binding site of TRAF4 with TRAF2 (H) and TRAF6 (I). Amino acid residues that are involved in the interaction with peptides were labeled. P−2 and P0 indicated previously identified positions on the peptides of CD40 and RANK. P−2 and P0 positions of peptides are the same as P0 and P2 positions of GPIbβ peptide, respectively. (J) The interaction between five C terminus residues of chain B and the crystallographic symmetry-related molecule. (K) Close-up view of the binding interface formed between five C terminus residues of chain B (blue color) and the crystallographic symmetry-related molecule (gray color). The P−2, P−1, P0, and P1 positions on chain B and interacting residues on the crystallographic symmetry-related molecule are indicated. (L) Putative TRAF4-binding motif identified by sequence comparison via alignment and the structurally defined motif. Red and blue colors indicate completely and partially conserved residues, respectively. (M) SPR characterization of the peptides containing a putative TRAF4-binding motif binding to immobilized TRAF4; 100 μM peptides were applied to TRAF4-coupled sensor chip. Data represent the average of two independent experiments.
Fig. S2.
Ligplot+ diagram showing molecular interaction between TRAF4 and GPIbβ peptide in detail.
Fig. S3.
Sequence alignment of TRAF4 across species. Five resides that are involved in the interaction with the GPIbβ peptide are shown with red color. All five residues are completely conserved.
Table S3.
Quantitative binding analysis of TRAF4 variants with receptor peptides
| TRAF4 variants | Peptides | SPR | ITC | Interaction |
| Wild type | GPIbβ (RRLRARARAR) | Kd = 11.3 ± 10.8 | Kd = 46.1 ± 28.4 | Interact |
| GPVI (KRLRHRGRAVQ) | N.D. | Kd = 35.6 ± 14.2 | Interact | |
| F434R | GPIbβ (RRLRARARAR) | Kd = 27.2 ± 21.1 | Kd = 82.6 ± 24.4 | Reduced |
| GPVI (KRLRHRGRAVQ) | N.D. | Kd = 108.2 ± 52.1 | ||
| S357E | GPIbβ (RRLRARARAR) | Kd = 90.6 ± 64.8 | Kd = 95.8 ± 35.6 | Reduced |
| GPVI (KRLRHRGRAVQ) | N.D. | N.D. | ||
| Y366R | GPIbβ (RRLRARARAR) | N.R. | N.R. | Disrupt |
| GPVI (KRLRHRGRAVQ) | N.R. | N.R. | ||
| Y436R | GPIbβ (RRLRARARAR) | N.R. | N.R. | Disrupt |
| GPVI (KRLRHRGRAVQ) | N.R. | N.R. | ||
| E406R | GPIbβ (RRLRARARAR) | N.R. | N.R. | Disrupt |
| GPVI (KRLRHRGRAVQ) | N.R. | N.R. | ||
| N355R | GPIbβ (RRLRARARAR) | N.R. | N.R. | Disrupt |
| GPVI (KRLRHRGRAVQ) | N.R. | N.R. | ||
| Wild type | NOD2 (GPPQKSPATLGLEEL) | N.R. | N.R. | Not interact |
| NOD2′ (ERLARKA) | N.R. | N.R. | Not interact | |
| Wild type | TGFBR1 (ARLTALRIKK) | N.D. | Kd = 115.3 ± 48.2 | Interact |
| Wild type | TGFBR2 (DRLSGRSCSE) | N.D. | N.D. | Interact (low affinity) |
| TGFBR2′ (ARLTAQCVA) | N.D. | N.D. | Interact (low affinity) |
Kd is the dissociation constant in micromolar. Data are the mean values from two independent experiments. N.R. and N.D. indicate no response and not determined, respectively.
Fig. S4.
Purification of TRAF4 and its five variants. The profile of size-exclusion chromatography was shown. SDS-PAGEs of peak fractions were shown at the left side of peaks.
To analyze the structural changes of TRAF4 upon binding of the GPIbβ peptide, we superimposed a previously solved TRAF4 native structure [Protein Data Bank (PDB) ID code 4K8U] with the currently solved TRAF4/GPIbβ complex structure. The superposition study demonstrated that the overall structures of TRAF4 and the TRAF4/GPIbβ peptide complex are nearly identical, with a root-mean-square deviation (rmsd) of 0.8 Å (Fig. 3B). However, clear movement of the side chains of several residues on TRAF4, including W414 and F434, was detected at the interaction interface upon binding of GPIbβ (Fig. 3B). Two hydrophobic pockets, formed by F408, Y436, and F434 for the major pocket and by W414 and F434 for the minor pocket, are premature forms without receptor interaction. Once GPIbβ interacts, two hydrophobic pockets are formed completely by moving the side chains of W414 and F434 and accommodating L at the P0 position and A at the P2 position of the GPIbβ peptide via hydrophobic interactions (Fig. 3B). This indicates that GPIbβ binding induced structural modification of TRAF4 to accommodate the receptor. Another distinct feature of the structure of peptide-bound TRAF4 was a highly disordered β6–β7 connecting loop, which was not properly modeled in the TRAF4/peptide complex structure due to poor density but was well defined in the native TRAF4 structure (Fig. 3C). Increasing flexibility of the β6–β7 connecting loop upon binding with the receptor might influence signal transduction and should be investigated further. Since the TRAF4-binding region of GPIbβ is located to the right next to the transmembrane domain, TRAF4 might be located near the intracellular membrane during GPIbβ and GPIV-mediated platelet signaling (Fig. 3D). A recent study demonstrated that TRAF4 destabilizes tight junctions (TJs) of malignant mammary epithelial cells (MECs) and increases cell migration for cancer progression via binding to phosphoinositide (PIP) on the intracellular membrane, supporting our finding that TRAF4 is attached to its proper receptor and the intracellular membrane during signaling events (22).
To compare the structures of three peptides in the chain—A, A′, and C′—all six molecules in the ASU were superposed. The structure of all six molecules was nearly identical, with an rmsd of 0.84 ∼ 1.82 Å between the molecules. The backbones of A, A′, and C′ were identical in the TRAF4 structure, with an rmsd of less than 0.2 Å, indicating that the conformation of the main chain of the receptor peptide is structurally conserved (Fig. 3E). Although the conformation of the main chain was totally conserved, the side chain conformation was diverse. The side chain conformation of three residues at position P−1, P0, and P2 out of the five residues in the peptide were highly conserved, while the side-chain conformation of the other two residues at P1 and P3 were variable (Fig. 3F).
Our structure of TRAF4 with its receptor GPIbβ revealed a mode of binding, which is in agreement with the receptor specificity of TRAF4, in which nonconserved amino acid residues are critical for the interaction with various receptors. The lower part of the GPIbβ receptor peptide (P1, P2, and P3) binds to TRAF4 at a similar position to where receptor peptides bind to TRAF2, while the upper part is 38° away from the receptor peptide-binding site on TRAF2 (Fig. 3G). Compared with the receptor-binding site on TRAF6, the GPIbβ peptide-binding site on TRAF4 does not overlap with the RANK peptide on TRAF6, indicating that the mode of receptor association with TRAF4 is slightly different to that of TRAF6 (Fig. 3G). Pair-wise comparison showed the unique receptor-binding site of TRAF4 in detail. Major interactions between TRAF2 and its receptors are hydrogen-binding interactions of Q(E) at P0 position of peptides with a serine triad pocket formed by S453, S454, and S455 in the TARF2. In the case of TRAF4, however, serine triad is absent, and this region is replaced by a tryptophan residue (W414) (Fig. 3H). Serine triad is also not detected on TRAF6. This region is replaced by L457 and A458. Interestingly, replaced W414 on TRAF4 forms a small minor hydrophobic pocket with F434 and is involved in the accommodation of an alanine residue on the GPIbβ peptide (Fig. 3H). In TRAF6, the main chain amide nitrogen atoms of L457 and A458 from hydrogen bond with E at P0 position of peptides (Fig. 3I). L at the GPIbβ P0 position and P at the CD40 and RANK P−2 position were correlated (Fig. 3 H and I). They form a hydrophobic interaction with surrounding residues on TRAF4 and TRAF2/TRAF6. In this regard, the two hydrophobic pockets on the surface of TRAF4 may be a critical determining factor for its different mode of receptor binding compared with that of other TRAF family members.
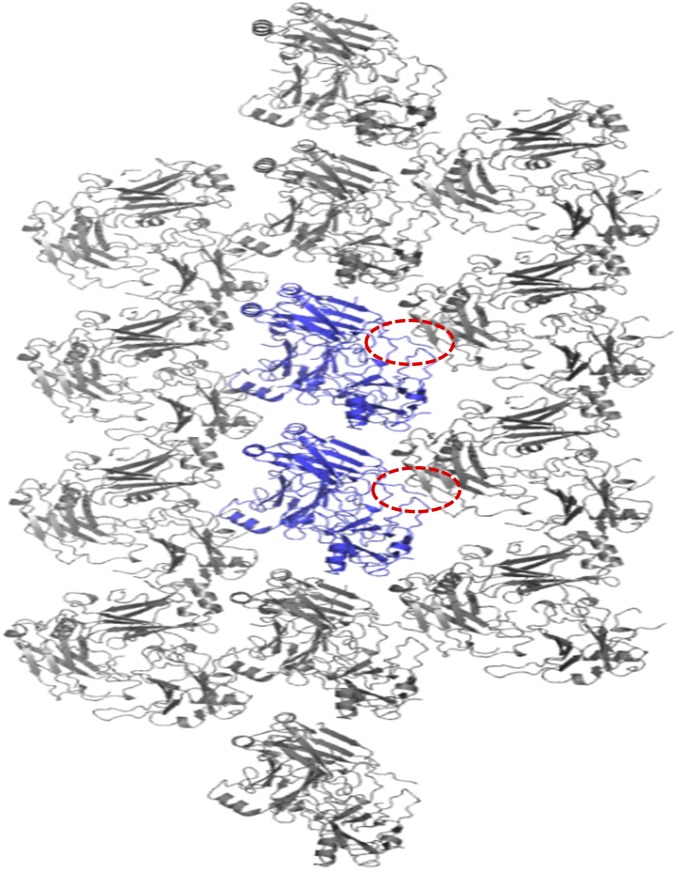
A peptide-like electron density was found at the receptor peptide-binding site on chain B and B′, caused by a crystallographic symmetry-related molecule. Five residues from the C terminus of chain B (465-PRKIL-469) were located at the typical peptide receptor-binding site of another chain B in the crystallographic symmetry-related molecule (Fig. 3 J and K and Fig. S5). Interestingly, the mode of interaction formed by crystallographic packing is similar to that of the interaction between TRAF4 and the GPIbβ receptor peptide. I468 from chain B was surrounded by a hydrophobic pocket formed by F408, F434, and Y436 of the crystallographic symmetry-related molecule, all of which are involved in the formation of the major hydrophobic pocket for the interaction of TRAF4 with the GPIbβ receptor peptide (Fig. 3K). Unlike the TRAF4–GPIbβ receptor peptide interaction that uses R at position P−1 to form a hydrogen bond with E406 on TRAF4, the interaction between TRAF4 and the crystallographic symmetry-related molecule uses R at position P−2 to form a hydrogen bond with E406 on TRAF4 (Fig. 3K). The charged residue at position P−2 and Ile at position P0 might be an alternative TRAF4-binding motif. Although R from the GPIbβ receptor peptide at position P−2 was not involved in the current complex structure, two peptides from the two platelet receptors contain R at position P−2 and had higher affinity than the peptide without R at position P−2, indicating that positively charged residues at position P−2 might be important for proper affinity and specificity.
Fig. S5.
Crystallographic packing of TRAF4/GPIbβ receptor peptide complex. Two trimer molecules formed in ASU are colored in blue. Other gray molecules are crystallographic symmetry-related molecules. Red-dot circles indicate the C termini of TRAF4 that are involved in the crystal packing.
Identification of the TRAF4-Binding Motif.
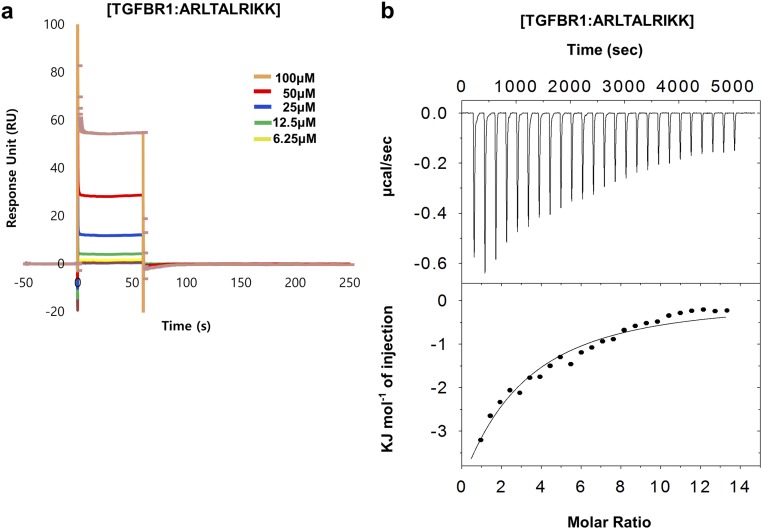
Because of the biological importance of the TRAF family in various signaling pathways, TRAF-binding consensus motifs from TRAF-binding receptors have been identified in structural studies. However, the TRAF4-binding motif has not previously been determined. Based on the current TRAF4–GPIbβ receptor complex structure and the structure-based sequence alignment, we have determined the TRAF4-binding consensus motifs. Because NOD2 and the TGF-β receptor have been identified as TRAF4-binding receptors in a previous study (24, 25), we analyzed their amino acid sequences and found GPIbβ receptor peptide-like sequences at the C-terminal regions (Fig. 3L). We designed peptides for NOD2, TGF-β receptor 1 (TGFBR1), and TGF-β receptor 2 (TGFBR2) containing putative TRAF4-binding motifs and tested their binding with TRAF4 by SPR. SPR measurements indicated that TGFBR1 (ARLTALRIKK) binds to TRAF4 with the highest response unit, while NOD2 (ERLARKA) did not bind to TRAF4 (Fig. 3M). Two TGFBR2 peptides (TGFBR2: DRLSGRSCSE and TGFBR2′: ARLTAQCVA) interact with TRAF4 with a low response unit (Fig. 3M). This result correlates with a previous cell-based interaction study, which showed that TRAF4 associated with activated TGF-β receptors has a higher affinity with TGFBR1 than TGFBR2 (10). Based on this preliminary interaction study, we quantitatively analyzed the interaction between TRAF4 and TGFBR1 (ARLTALRIKK) via SPR and ITC. For SPR, purified 6xHis-tagged TRAF4 was coupled with a Ni-NTA sensor chip using a His-tag affinity system. Different concentrations of each peptide, ranging from 6.25 μM to 100 μM, were analyzed for their binding with TRAF4 fixed to the sensor chip. Although the data were not well-fitted, the results suggested concentration-dependent interactions (Fig. S6A), which were confirmed by ITC. TGFBR1 peptide (ARLTALRIKK) was titrated into TRAF4, and the released heat was in agreement with the ideal interaction value, indicating the existence of a single type of binding site without distinct cooperativity in the interaction (Fig. S6B). The measured dissociation constant was 115.3 μM, which suggests a lower affinity than the interaction between TRAF4 and the GPIbβ receptor peptide (RRLRARARARA). Those results with a corresponding structural analysis indicate that the Arg–Leu motif at position P−1 and P0 is critical for the TRAF4 interaction, and the Ala residue at position P2 influences the affinity. Replacement of the Ala residue at position P2 with His (GPVI peptide) and Gly (TGFBR2 peptides) reduced the binding affinity with TRAF4, and replacement with Arg (NOD2 peptide) abolished the interaction. This led to the definition of a TRAF4-binding motif for P−1 to P2 of Arg–Leu–X–Ala, where X can be any amino acid and Ala can be replaced by small, uncharged residues (Fig. 3L).
Fig. S6.
Interaction of TRAF4 with TGFBR1. (A) SPR characterization of TGFBR1 binding to immobilized TRAF4. Peptides (6.25–100 μM) were applied to a TRAF4-coupled sensor chip. Data represent the average of three independent experiments. (B) ITC experiment showing titration of the TGFBR1 peptide into a TRAF4 solution. The raw calorimetric titration data are shown in Upper, and experimental fitting of the data to a single site interaction model is shown in Lower.
In summary, the current structural and biochemical study revealed a molecular mechanism by which TRAF4 participates in platelet-mediated thrombosis by direct interaction with two platelet receptors, the GPIb-IX-V complex and GPVI, and participates in cancer initiation and progression by interaction with the TGF-β receptor. This will provide a template for further understanding of the receptor-binding specificity of TRAF4, TRAF4-mediated signaling events, and related diseases and for drug development for treatment of thrombotic diseases and cancers.
Methods
Sequence Alignment.
The amino acid sequence of TRAFs was analyzed using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/).
Peptide Synthesis.
Eight peptides were synthesized, including GPIbβ (RRLRARARARA), GPVI (KRLRHRGRAVQ), GPIbβ-5 (RLRAR), TGFBR1 (ARLTALRIKK), TGFBR2 (DRLSGRSCSE), TGFBR2′ (ARLTAQCVA), NOD2 (GPPQKSPATLGLEEL), and NOD2′ (ERLARKA).
Protein Expression and Purification.
The expression and purification methods for TRAF4 have been described in detail elsewhere (21). The TRAF4 TRAF domain (amino acid 290–470) was expressed in Escherichia coli BL21 (DE3) under overnight induction at 20 °C. The protein contained a carboxyl terminal His-tag and was purified by nickel affinity and size-exclusion chromatography using a Superdex 200 gel filtration column 10/30 (GE healthcare). TRAF4 was concentrated to 9–10 mg/mL for structural studies. Site-directed mutagenesis was conducted using a QuikChange kit (Stratagene) according to the manufacturer’s protocols. Mutagenesis was confirmed by sequencing, and mutant proteins were expressed and purified using the same method as described above.
ITC.
A NanoITC (TA Instruments) was used for ITC experiments. The TRAF4 TRAF domain was dialyzed against PBS buffer, and the eight lyophilized peptides, including GPIbβ peptide (RRLRARARARA), were dissolved in the same buffer to minimize the heat of dilution values. Before titration, the protein sample and the peptides were centrifuged at 10,000 × g at 4 °C for 5 min to remove any precipitants. For incremental injection in ITC, 2 μL of a concentrated peptide solution (1 mM) was injected into a sample cell, containing 190 μL of the TRAF4 TRAF domain at a concentration of ∼20 μM. All titrations were carried out at 15 °C with 25 injections at 160-s intervals. The area under each titration peak was integrated, plotted against the number of injections, and fitted to a one-site independent binding model, by using the software provided by TA Instruments. Experimental data were subtracted from appropriate baselines acquired by injecting peptides into the buffer without the TRAF4 TRAF domain.
SPR.
Physical interactions between peptides and TRAF4 were analyzed by SPR. TRAF4 was immobilized onto an NTA sensor chip, using a 6xHis-tag from Biacore T200 (GE Healthcare). First, the sensor chip surface was coated by injecting 100 mM nickel. Then, TRAF4 was diluted in PBS to a concentration of 100 μg/mL and injected at a rate of 10 μL/min for 1 min for tandem immobilization on the NTA chip surface, resulting in around 3,000 RU (Response Unit) after stabilization. Concentrations of peptides ranging from 6.25 μM to 100 μM were prepared by dilution in PBS and injected in the flow channel at a flow rate of 30 μL/min for 1 min, followed by a dissociation time of 300 s and regeneration with a mixture of EDTA and guanidine HCl. Raw sensorgrams were double blanked by subtracting responses from the reference flow channel and blank injection.
Crystallization, Peptide Soaking, and Data Collection.
Crystallization of TRAF4 was conducted at 20 °C by the hanging drop vapor-diffusion method using a preidentified condition for the structural study of TRAF4 (21). The final crystals used for the X-ray diffraction study were grown on plates by equilibrating a mixture containing 1 μL of protein solution (9–10 mg/mL protein in 20 mM Tris⋅HCl at pH 8.0 and 150 mM NaCl) and 1 μL of reservoir solution containing 0.14 M magnesium formate dehydrate and 13% polyethylene glycol 3350 against 0.4 mL of reservoir solution. The crystals grew to maximum dimensions of 0.1 × 0.2 × 0.2 mm in 3 d. Crystals were soaked at 100 μM GPIbβ peptide-containing buffer for 24 h before freezing in liquid nitrogen. Diffraction data were collected at the SB-II (5C) beamline at the Pohang Accelerator Laboratory (PAL), Republic of Korea. Data processing and scaling were performed using HKL2000 (27).
Structure Determination and Analysis.
The complex structure was determined by the molecular replacement phasing method using Phaser (28). The previously solved TRAF4 structure (PDB ID code 4K8U) (21) was used as a search model. The structure was completed by iterative manual building and refinement in Coot (29) and PHENIX Refine (30), respectively. The quality of the model was checked using PROCHECK (31). Molecular structure images were generated using the PyMOL Molecular Graphics System (32).
Supplementary Material
Acknowledgments
This study was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Ministry of Education, Science and Technology (NRF-2017M3A9D8062960) and a grant from the Korea Healthcare Technology R&D project, Ministry of Health & Welfare, Republic of Korea (HI13C1449).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5YC1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708688114/-/DCSupplemental.
References
- 1.Jandrot-Perrus M, et al. Cloning, characterization, and functional studies of human and mouse glycoprotein VI: A platelet-specific collagen receptor from the immunoglobulin superfamily. Blood. 2000;96:1798–1807. [PubMed] [Google Scholar]
- 2.Mackman N. Triggers, targets and treatments for thrombosis. Nature. 2008;451:914–918. doi: 10.1038/nature06797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li R, Emsley J. The organizing principle of the platelet glycoprotein Ib-IX-V complex. J Thromb Haemost. 2013;11:605–614. doi: 10.1111/jth.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nieswandt B, Watson SP. Platelet-collagen interaction: Is GPVI the central receptor? Blood. 2003;102:449–461. doi: 10.1182/blood-2002-12-3882. [DOI] [PubMed] [Google Scholar]
- 5.Savage B, Almus-Jacobs F, Ruggeri ZM. Specific synergy of multiple substrate-receptor interactions in platelet thrombus formation under flow. Cell. 1998;94:657–666. doi: 10.1016/s0092-8674(00)81607-4. [DOI] [PubMed] [Google Scholar]
- 6.Masson R, et al. Tumor necrosis factor receptor associated factor 4 (TRAF4) expression pattern during mouse development. Mech Dev. 1998;71:187–191. doi: 10.1016/s0925-4773(97)00192-5. [DOI] [PubMed] [Google Scholar]
- 7.Kedinger V, Rio MC. TRAF4, the unique family member. Adv Exp Med Biol. 2007;597:60–71. doi: 10.1007/978-0-387-70630-6_5. [DOI] [PubMed] [Google Scholar]
- 8.Shiels H, et al. TRAF4 deficiency leads to tracheal malformation with resulting alterations in air flow to the lungs. Am J Pathol. 2000;157:679–688. doi: 10.1016/S0002-9440(10)64578-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Régnier CH, et al. Impaired neural tube closure, axial skeleton malformations, and tracheal ring disruption in TRAF4-deficient mice. Proc Natl Acad Sci USA. 2002;99:5585–5590. doi: 10.1073/pnas.052124799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, et al. TRAF4 promotes TGF-β receptor signaling and drives breast cancer metastasis. Mol Cell. 2013;51:559–572. doi: 10.1016/j.molcel.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Li W, et al. TRAF4 is a critical molecule for Akt activation in lung cancer. Cancer Res. 2013;73:6938–6950. doi: 10.1158/0008-5472.CAN-13-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krajewska M, et al. TRAF-4 expression in epithelial progenitor cells. Analysis in normal adult, fetal, and tumor tissues. Am J Pathol. 1998;152:1549–1561. [PMC free article] [PubMed] [Google Scholar]
- 13.Häcker H, Tseng PH, Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol. 2011;11:457–468. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- 14.Chung JY, Park YC, Ye H, Wu H. All TRAFs are not created equal: Common and distinct molecular mechanisms of TRAF-mediated signal transduction. J Cell Sci. 2002;115:679–688. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- 15.Park YC, Burkitt V, Villa AR, Tong L, Wu H. Structural basis for self-association and receptor recognition of human TRAF2. Nature. 1999;398:533–538. doi: 10.1038/19110. [DOI] [PubMed] [Google Scholar]
- 16.Zhang P, et al. Single amino acid substitutions confer the antiviral activity of the TRAF3 adaptor protein onto TRAF5. Sci Signal. 2012;5:ra81. doi: 10.1126/scisignal.2003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye H, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418:443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 18.Ni CZ, et al. Molecular basis for CD40 signaling mediated by TRAF3. Proc Natl Acad Sci USA. 2000;97:10395–10399. doi: 10.1073/pnas.97.19.10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu S, et al. LMP1 protein from the Epstein-Barr virus is a structural CD40 decoy in B lymphocytes for binding to TRAF3. J Biol Chem. 2005;280:33620–33626. doi: 10.1074/jbc.M502511200. [DOI] [PubMed] [Google Scholar]
- 20.Li C, et al. Downstream regulator TANK binds to the CD40 recognition site on TRAF3. Structure. 2002;10:403–411. doi: 10.1016/s0969-2126(02)00733-5. [DOI] [PubMed] [Google Scholar]
- 21.Yoon JH, Cho YJ, Park HH. Structure of the TRAF4 TRAF domain with a coiled-coil domain and its implications for the TRAF4 signalling pathway. Acta Crystallogr D Biol Crystallogr. 2014;70:2–10. doi: 10.1107/S139900471302333X. [DOI] [PubMed] [Google Scholar]
- 22.Rousseau A, et al. TRAF4 is a novel phosphoinositide-binding protein modulating tight junctions and favoring cell migration. PLoS Biol. 2013;11:e1001726. doi: 10.1371/journal.pbio.1001726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu F, Ru H, Ding W, Ouyang S, Liu ZJ. Structural biology study of human TNF receptor associated factor 4 TRAF domain. Protein Cell. 2013;4:687–694. doi: 10.1007/s13238-013-3068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arthur JF, et al. TNF receptor-associated factor 4 (TRAF4) is a novel binding partner of glycoprotein Ib and glycoprotein VI in human platelets. J Thromb Haemost. 2011;9:163–172. doi: 10.1111/j.1538-7836.2010.04091.x. [DOI] [PubMed] [Google Scholar]
- 25.Marinis JM, Homer CR, McDonald C, Abbott DW. A novel motif in the Crohn’s disease susceptibility protein, NOD2, allows TRAF4 to down-regulate innate immune responses. J Biol Chem. 2011;286:1938–1950. doi: 10.1074/jbc.M110.189308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruber A, Hanson SR. Potential new targets for antithrombotic therapy. Curr Pharm Des. 2003;9:2367–2374. doi: 10.2174/1381612033453938. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 32.DeLano WL, Lam JW. PyMOL: A communications tool for computational models. Abstr Pap Am Chem Soc. 2005;230:U1371–U1372. [Google Scholar]