Significance
Spatial structure is postulated to have a powerful influence on establishing and sustaining the signaling and metabolic exchanges that define relationships among members of the gut microbiota and host. However, information about gut community spatial structure is limited. Simultaneous imaging of components of a 15-member model human gut bacterial community over a range of spatial scales in gnotobiotic mice revealed that the colon is better conceptualized as an incompletely mixed bioreactor, rather than having sharply stratified luminal and mucosal compartments. Identifying host and microbial factors that constrain the ability of community members to establish sizeable single or oligotaxon agglomerations should yield new insights about how “micro”-scale mixing defines community function.
Keywords: gut microbial ecology, community biogeography, bacterial–bacterial interactions, microbiome function, multiplex fluorescence imaging
Abstract
Knowledge of the spatial organization of the gut microbiota is important for understanding the physical and molecular interactions among its members. These interactions are thought to influence microbial succession, community stability, syntrophic relationships, and resiliency in the face of perturbations. The complexity and dynamism of the gut microbiota pose considerable challenges for quantitative analysis of its spatial organization. Here, we illustrate an approach for addressing this challenge, using (i) a model, defined 15-member consortium of phylogenetically diverse, sequenced human gut bacterial strains introduced into adult gnotobiotic mice fed a polysaccharide-rich diet, and (ii) in situ hybridization and spectral imaging analysis methods that allow simultaneous detection of multiple bacterial strains at multiple spatial scales. Differences in the binding affinities of strains for substrates such as mucus or food particles, combined with more rapid replication in a preferred microhabitat, could, in principle, lead to localized clonally expanded aggregates composed of one or a few taxa. However, our results reveal a colonic community that is mixed at micrometer scales, with distinct spatial distributions of some taxa relative to one another, notably at the border between the mucosa and the lumen. Our data suggest that lumen and mucosa in the proximal colon should be conceptualized not as stratified compartments but as components of an incompletely mixed bioreactor. Employing the experimental approaches described should allow direct tests of whether and how specified host and microbial factors influence the nature and functional contributions of “microscale” mixing to the dynamic operations of the microbiota in health and disease.
The functions expressed by members of a microbial community are impacted by their neighbors (1–4) and by physiological features of their environment (5–9). A substantial body of theory suggests that spatial structure has a powerful influence on the evolutionary stability of mutualistic relationships among members of a microbial community and between the community and its host (10–17). Depending on the details of the model or the experimental system, cooperative interactions can be either stabilized or destabilized by spatial structure (reviewed in refs. 12, 16, and 18). Recently, Coyte et al. used modeling to predict that the host would benefit from compartmentalizing microbial species in the gut to weaken interactions between species and promote community stability (16).
Microbes display diverse adherence properties and growth rates that could contribute to spatially structured communities. In the gut, microbes can adhere to the epithelium and mucins (19–21); these components of the ecosystem are arranged nonrandomly in ways that could lead to spatial structuring of adherent community members (22, 23). Similarly, partially digested food particles in the lumen could serve as sites of attachment (24–28). Differential replication of a microbe based on its localization in the mucus layer or the lumen (29) could itself generate a spatially structured microbial consortium or could amplify differences established by differential adherence.
The lumen of the gut is considered to be a compartment inhabited by a microbiota distinct from that of the mucus layer (23, 29–33). This view is largely based on studies that have compared mucosal samples to feces (30, 34–36). In contrast, studies that have directly compared mucosa-associated communities with adjacent luminal contents have generally shown only modest differences in the relative proportions of taxa (29, 33, 37–39), although exceptions occur. For example, Yasuda et al. (33) reported that Helicobacteraceae dominated the colonic mucosa in rhesus macaques but were a minor constituent in the luminal community.
The complexity and dynamism of the gut microbiota pose considerable challenges for quantitative analysis of its spatial organization. The most comprehensive analysis to date used gnotobiotic mice colonized with a human fecal community and fluorescence in situ hybridization (FISH) with probes having specificities that ranged from phylum level to genus level (5). Other studies have used FISH, antibody staining, or labeling of polysaccharides to investigate the distribution of particular taxa within the microbiota, or of the microbiota as a whole in the presence of host perturbations (38, 40–48). Using microbes genetically engineered to express distinct combinations of two fluorescent proteins, Whitaker et al. (49) were able to discriminate six engineered strains of Bacteroides in the gut of gnotobiotic mice.
Adherence to available substrates such as mucus, food particles, or other microbes, including to the polysaccharide-rich capsular structures that some community members produce, may localize an organism to a preferred microhabitat but may only modestly prolong its residence time in the gut. Interestingly, modeling studies performed in experimental bioreactors, notably recently developed gut-on-a-chip technology, have shown that peristaltic mixing is a key factor in maintaining high bacterial densities, counteracting the tendency of flow to cause rapid depletion of bacteria (50). The importance of “precise” spatial positioning of microbiota members relative to their metabolic partners to the healthy functioning of the microbiota is largely unknown. Moreover, published studies have yet to examine the biogeography of a diverse microbiota within the colon at a species level, and in a manner where most members of a complex community could be targeted simultaneously without their prior genetic engineering to produce reporter proteins. In the present report, we use a FISH approach that allows simultaneous identification of many bacterial species (51, 52) to study the spatial organization of a defined 15-member community of sequenced and phylogenetically diverse human gut-derived taxa installed in the guts of gnotobiotic mice. While this artificial community is simplified relative to natural gut communities, it was complex enough to allow for a variety of spatial distributions and metabolic interactions between its members (4, 53, 54). Moreover, our multiplexed imaging approach provided an opportunity to examine the degree of spatial organization of this defined community at multiple scales, ranging from hundreds of micrometers across the diameter of the gut, to a mesoscale of tens of micrometers, to just a few micrometers at the highest resolution.
Results
Strategy for Imaging the Distribution of Microbes in the Gnotobiotic Colon.
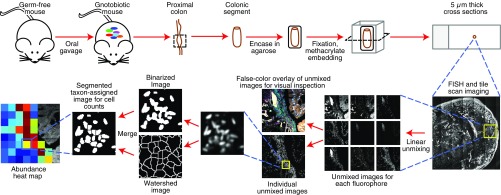
The experimental design and workflow of our strategy are shown in Fig. 1. Adult germ-free mice were colonized with a consortium of 15 human gut-derived bacterial species (Table S1), and gut segments were fixed and embedded in a glycol methacrylate (GMA) resin. The GMA remained in place not only during sectioning but also during subsequent in situ hybridization and imaging steps, preserving the 3D structure of the gut contents. The proportional representation of members of this community, whose genome sequences had been defined, was initially estimated by shotgun sequencing of DNA prepared from fecal samples (Table S2 for the results of Community Profiling by Sequencing; COPRO-Seq).
Fig. 1.
Experimental design and workflow. Germ-free mice were gavaged with a 15-member consortium of human gut bacterial strains and killed 14 d later. Segments of proximal colon were encased in agarose, fixed with formaldehyde, embedded in glycol methacrylate (GMA) resin, sectioned, and hybridized with fluorescent probes. The resin remained in place during hybridization and imaging steps, preserving 3D spatial structure. Gut cross-sections were imaged as a tile scan of multiple fields of view to image entire sections at high resolution. Each field of view was imaged by excitation with six laser lines sequentially and processed by linear unmixing to create separate images for each of nine fluorophores and for autofluorescence from host tissue and ingested food particles. Each image was then segmented into binary images to allow for an automated count of cells in each square of an 8 × 8 grid for each field of view. The results are displayed as a heat map of cell abundance. The unmixed fluorophore channels were also false-colored and overlaid to produce the final unmixed image.
Table S1.
Bacterial species, probes, and probe sets used in this study
| Species | Strain | Phylum | Class | Probe name | Probe oligonucleotide 5′–3′ | Fluorophore, probe set 1 | Fluorophore, probe set 2 | Fluorophore, probe set 3 |
| B. thetaiotaomicron | VPI 5482 | Bacteroidetes | Bacteroidia | Bthe577 | TAACTGTCCACCTACGCT | Alexa 488 | Rhodamine Red X | Alexa 488 |
| B. cellulosilyticus | WH2 | Bacteroidetes | Bacteroidia | Bcel85 | GGTCGCCATCAACCTATTGCT | Alexa 555 | Alexa 594 | Alexa 555 |
| B. vulgatus | ATCC 8482 | Bacteroidetes | Bacteroidia | Bvul264 | CCATCGAAGACTAGGTGGGCC | Alexa 660 | Alexa 660 | Alexa 488 |
| B. ovatus | ATCC 8483 | Bacteroidetes | Bacteroidia | Bova412 | TACGACCCATAGAGCCTTC | Alexa 555 | Alexa 488 | |
| B. caccae | ATCC 43185 | Bacteroidetes | Bacteroidia | Bcac218 | GCATCCCCATCTCATACCG | Alexa 594 | ||
| B. uniformis | ATCC 8492 | Bacteroidetes | Bacteroidia | Buni81 | GTCGCCATCAAACTTAGCAAGC | Pacific Orange | Alexa 488 | |
| P. distasonis | ATCC 8503 | Bacteroidetes | Bacteroidia | Pdis156 | GCGGTATTAGTCCGACTTTCGC | Rhodamine Red X | ||
| E. rectale | ATCC 33656 | Firmicutes | Clostridia | Erec1259 | GCTCGGCTTCACAGCTTTGCTT | Rhodamine Red X | Alexa 532 | |
| R. torques | ATCC 27756 | Firmicutes | Clostridia | Rtor88 | GCTCAGTCACAATCCTCTTCA | Alexa 647 | Alexa 647 | Alexa 647 |
| C. scindens | ATCC 35704 | Firmicutes | Clostridia | Csci87 | TCAGTCGCAAGGCTCCTCGT | Alexa 660 | ||
| C. spiroforme | DSM 1552 | Firmicutes | Erysipelotrichia | Cspi159 | GCGGTCTTAGCTGCCGTTT | Alexa 532 | ||
| F. prausnitzii | M21/2 | Firmicutes | Clostridia | Fpra197 | CTCAAAGCGGATTGCTCCTTT | Alexa 532 | ||
| R. obeum | ATCC 29174 | Firmicutes | Clostridia | Robe76 | AGACCAAATCTGCCGAAGCTTCA | Alexa 532 | ||
| D. longicatena | DSM 13814 | Firmicutes | Clostridia | Dlon180 | CCATGCGGTACCGTGGTCTT | Alexa 532 | ||
| C. aerofaciens | ATCC 25986 | Actinobacteria | Coriobacteriia | Caer280 | TCAACCCGGCTACCCGTT | Alexa 594 | Alexa 405 | Alexa 405 |
| Most bacteria | Eub338 | GCTGCCTCCCGTAGGAGT | Alexa 514 |
All probe oligonucleotide sequences were designed in this study except for the Eub338 probe for most bacteria (81). Probes were labeled at the 5′ end with the fluorophore indicated. Probe set 1 is shown in Figs. 3B and 6; probe set 2 is shown in Fig. 3D; probe set 3 is shown in Figs. 3 A and C and 5. In cases where a probed taxon was not detected in an image, the taxon name was omitted from the figure legend (e.g., E. rectale in Figs. 3B and 6).
Table S2.
Proportional representation of bacterial groups
| Bacterial groups | FISH, % | COPRO-Seq, % | Dense fields, % | Sparse fields, % |
| B. cellulosilyticus | 40.6 ± 2.1 | 40.0 ± 1.7 | 42.5 ± 3.7 | 45.8 ± 3.8 |
| B. thetaiotaomicron; B. ovatus; B. vulgatus; B. uniformis | 35.9 ± 5.7 | 43.8 ± 2.6 | 31.7 ± 4.5 | 35.6 ± 3.7 |
| B. caccae | 5.1 ± 2.7 | 6.4 ± 2.0 | 4.2 ± 3.6 | 2.5 ± 0.9 |
| P. distasonis | 0.1 ± 0.1 | 4.1 ± 1.0 | — | — |
| C. scindens | 7.0 ± 2.7 | 0.6 ± 0.2 | 7.7 ± 2.0 | 8.1 ± 3.7 |
| R. torques | 3.1 ± 1.0 | 1.1 ± 0.3 | 2.8 ± 0.7 | 2.1 ± 1.1 |
| E. rectale; C. spiroforme; F. prausnitzii; R. obeum; D. longicatena | 0.6 ± 0.7 | 3.0 ± 1.3 | — | — |
| C. aerofaciens | 7.6 ± 2.6 | 1.4 ± 0.7 | 11.0 ± 1.3 | 5.9 ± 2.1 |
Mean values ± SD are shown. FISH, bacterial cell counts from 372 fields of view across five sections from two mice. COPRO-Seq, taxon abundance based on analysis of fecal samples from 10 mice. For comparison, values are shown that were obtained from 11 fields of view containing primarily densely populated regions and 12 fields of view containing sparsely populated regions from the same section.
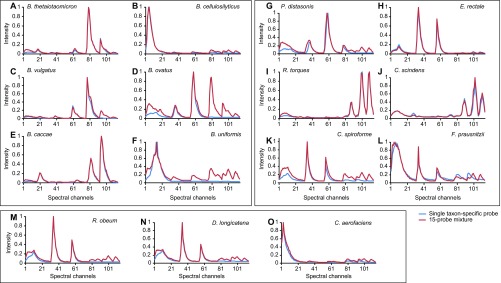
To construct a probe set that would provide information on the distribution of all 15 taxa simultaneously, we initially employed a combinatorial labeling and spectral imaging strategy where each taxon was labeled with a unique binary combination chosen from among six fluorophores (51, 52). However, we found that because of the high microbial density in the colon, adjacent bacterial cells frequently overlapped one another in the same pixel of the image, despite confocal optical sectioning. This overlap resulted in ambiguous combinations of binary signals. Therefore, we reverted to a strategy of labeling each microbe with a single fluorophore. To visualize the distribution of each taxon, we hybridized some sections with mixtures of oligonucleotides targeting six or seven taxa individually (e.g., probe sets 1 and 2 in Table S1). To evaluate the distribution of the entire community, we used a mixture (probe set 3) comprising three sets of oligonucleotides: (i) six probes, each labeled with a different fluorophore, targeting six species (Bacteroides cellulosilyticus, Bacteroides caccae, Parabacteroides distasonis, Ruminococcus torques, Clostridium scindens, and Collinsella aerofaciens); (ii) four probes, all labeled with Alexa 488, targeting four moderately abundant members of Bacteroides (Bacteroides thetaiotaomicron, Bacteroides vulgatus, Bacteroides ovatus, Bacteroides uniformis); and (iii) five probes, all labeled with Alexa 532, targeting five low abundance Firmicutes (Eubacterium rectale, Clostridium spiroforme, Faecalibacterium prausnitzii, Ruminococcus obeum, and Dorea longicatena) (Methods and Fig. S1 for details of probe design and for validation of their taxon specificities). In addition, we labeled all bacteria with the universal bacterial probe Eub338 conjugated to Alexa 514 and used DAPI to label host and bacterial DNA. In some experiments, wheat germ agglutinin (WGA), which binds to N-acetyl-d-glucosamine and sialic acid, or an antibody to mouse colonic mucin (MCM) was included to localize mucus. Finally, we made use of endogenous autofluorescence to image food particles and host tissue.
Fig. S1.
Spectral profiles demonstrating the specificity of FISH probes. A pure culture of each bacterial strain was hybridized with the probe targeting that strain and with a mixture of all 15 probes. Images of cells were acquired with excitation wavelengths of 633, 594, 561, 514, 488, and 405 nm sequentially. The resulting six image stacks were concatenated, and a single reference spectrum was measured from the concatenated stack. The fluorescence emission spectrum obtained from the single-probe hybridization (blue) is shown plotted with the emission spectrum from the probe mixture hybridization (red). Where spectra are identical, no cross-hybridization occurs. Additional peaks in the probe mixture spectrum indicate cross-hybridization; e.g., the spectrum in D indicates cross-hybridization of the B. thetaiotaomicron probe to B. ovatus. Despite cross-hybridization, strains are clearly differentiable based on the distinct shapes of the emission spectra. (A) B. thetaiotaomicron. (B) B. cellulosilyticus. (C) B. vulgatus. (D) B. ovatus. (E) B. caccae. (F) B. uniformis. (G) P. distasonis. (H) E. rectale. (I) R. torques. (J) C. scindens. (K) C. spiroforme. (L) F. prausnitzii. (M) R. obeum. (N) D. longicatena. (O) C. aerofaciens.
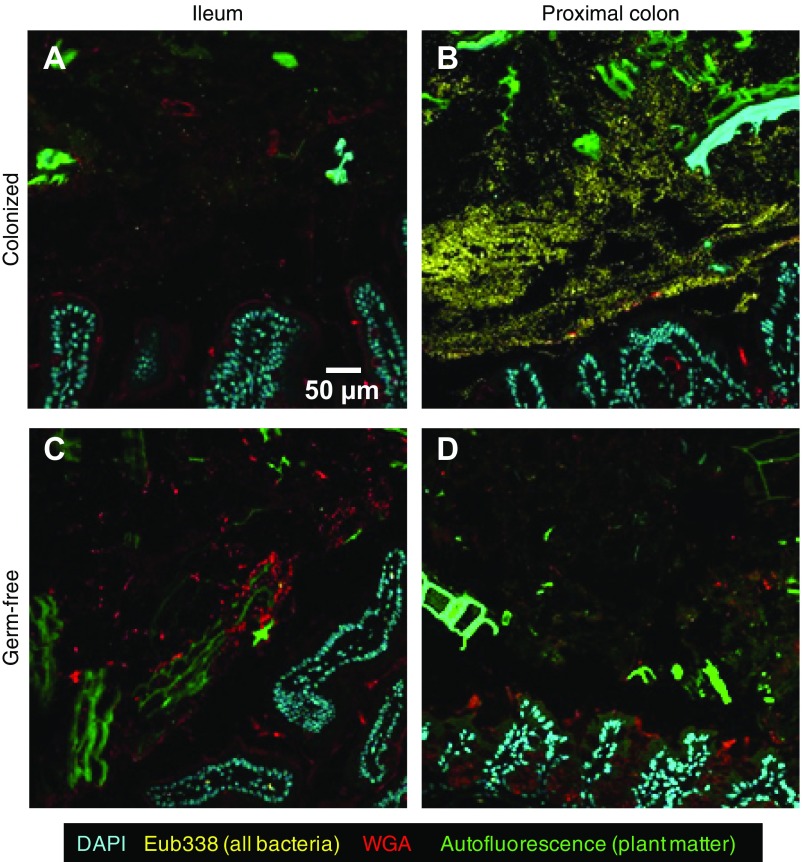
When gut samples are imaged at a resolution sufficient to distinguish individual microbial cells, the field of view is necessarily limited (∼100 × 100 μm) and the depth of field is shallow (∼1 μm), so that ∼10,000 cubic micrometers of material are visualized in a single image. The density of microbes in the small intestine has been reported to be 104 to 107 cells per mL of luminal contents, whereas in the colon and feces it is orders of magnitude greater (as much as 1011–1012 cells per g; refs. 55 and 56). This translates to 1,000 microbes per field of view in the colon and feces, but this number decreases 10,000- to 1 million-fold in the jejunum and ileum. The FISH probe that reacted with all 15 members of the community (Eub338) confirmed that the distal small intestine of these gnotobiotic mice was sparsely colonized (Fig. S2A), whereas the colon was densely colonized (Fig. S2B; compare with the absence of microbes in the ileum and colon of germ-free controls in Fig. S2 C and D). Because of its significantly higher microbial density, we chose to focus our multiscale multiplex imaging analysis on the proximal colon.
Fig. S2.
Bacterial colonization is dense in the proximal colon and sparse in the ileum in the gnotobiotic mouse model. (A) A small number of bacterial cells are visible in the ileum of a mouse colonized with the 15-strain consortium. (B) The proximal colon, by contrast, shows dense colonization in the lumen. (C and D) Imaging verified the absence of bacteria from both the ileum and colon of a germ-free mouse. Samples were embedded in methacrylate resin, sectioned, subjected to FISH with the Eub338 probe, and stained with DAPI and fluorophore-conjugated WGA.
Characterizing the Distribution of Bacteria in the Gut Is a Multiscale Problem.
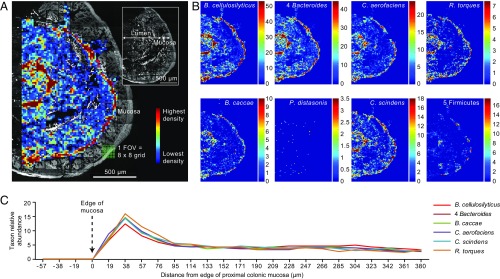
To quantitatively evaluate the overall distribution of bacteria in the proximal colon, we tile-scanned five nearly complete cross-sections and a total of 372 microscopic fields from this region in two mice and represented the data as heat maps of bacterial cell abundance (Fig. 1 for a summary of the approach). As demonstrated by the universal Eub338 probe, bacteria were not distributed homogeneously across cross-sections; rather, they were concentrated at the border between the lumen and mucosa, as well as in patches in the interior of the lumen (Fig. 2A). These regions of highest density contained ∼50 cells per 100 μm2. Substantially lower densities occurred in the remainder of the lumen, consisting primarily of regions in and surrounding large autofluorescent particles that we interpret as remnants of food (Fig. 2A).
Fig. 2.
Microbes are most abundant near host tissue and in patches in the lumen. Microbial density in each region of the cross-section is shown as a heat map (A) representing the number of cells hybridizing to the Eub338 probe and ranging from zero (dark blue) to 158 cells (red) per 19 × 19 μm grid square. The heat map is overlaid on a tile scan (Inset) of the section showing autofluorescence from host tissue and ingested food particles. (B) The density of individual taxa or groups of taxa shows a similar distribution. (C) A line-scan analysis of cell abundance along transects perpendicular to host tissue illustrates that taxa have similar distributions at this scale. Values shown are the percent of cells observed in each quantum of the transect, and are the mean of 245 transects from a total of five intestinal cross-sections from two mice. Transects consisted of 24 adjacent grid squares placed so that the fifth grid square contained the first bacterial cell labeled with the universal probe Eub338. Thus, the first four grid squares (76 µm) of each transect line would be empty unless occupied with a bacterial cell hybridizing to a specific probe but not to Eub338.
Heat maps of the individual bacterial taxa showed a broadly similar pattern of distribution at the scale of tens to hundreds of micrometers (Fig. 2B). This similarity was evaluated quantitatively by line-scan analysis of transects running from the edge of the mucosa into the lumen (Fig. 2C). In these scans, the transects extended far enough into the lumen to determine the width of the high density microbial zone near the mucosa but not so far as to intersect patches of high luminal concentration. Data were collected for 245 transects, and abundances were expressed as the fraction of bacterial cells present in discrete segments of the transect. When averaged across the 245 transects, the abundance pattern for each taxon was similar, peaking close to the mucosa and then falling off sharply to a low level at distances greater than 75 μm into the lumen.
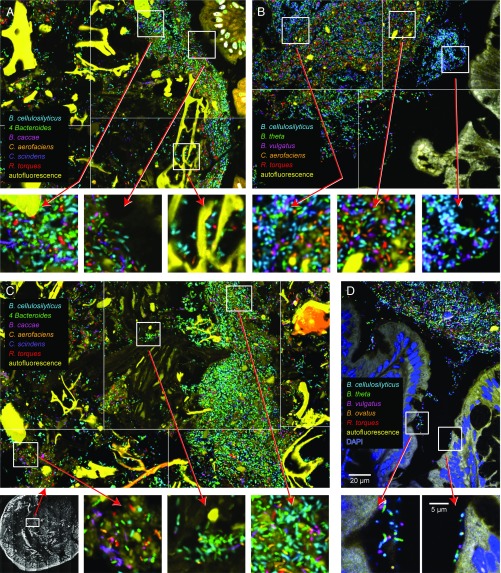
We focused our analysis on four “microhabitats”: (i) densely colonized regions adjacent to the colonic mucosa, (ii) dense patches of microbes within the lumen of the colon, (iii) sparsely colonized areas within the lumen, and (iv) sparsely colonized crypts of Lieberkühn. All of the abundant taxa were detectable in each of these microhabitats. In some images, large autofluorescent particles of food pressed close to the mucosa so that the microbe-rich region was only 10–20 µm thick (Fig. 3A). Other images show a wider, densely colonized region at the edge of the mucosa (Fig. 3B). The lumen contained densely colonized regions bordered by irregularly shaped autofluorescent food particles of variable size and brightness (Fig. 3C). A diverse microbial community was thinly arrayed on some of these particles and colonized the edges of cavities within them (Fig. 3 A and C). This latter observation is consistent with the notion that food particles can serve as platforms for attachment of bacterial taxa (including potential syntrophic partners). Crypts were colonized by a sparse, taxonomically mixed community (Fig. 3D), suggesting that they were colonized from the lumen and did not serve as microhabitats for any select groups within the 15-member model human gut microbiota.
Fig. 3.
Colonization patterns in distinct microhabitats in the colon. Tiled images show the distribution of microbes relative to host tissue and large autofluorescent food particles. Images shown are representative of the region proximal to the epithelium (A and B), the region distal to the epithelium (lumen; C), and crypts (D). White boxes show the positions of higher magnification views (Lower) where individual bacterial cells are visible; low-magnification image in C shows the image location in the lumen. Microbes were spatially mixed at micrometer scales in all microhabitats. Legend in A also applies to C. Scale bars in D apply throughout the figure.
The overall composition of the microbial community differed only modestly from microhabitat to microhabitat within a cross-section when measured at a mesoscale (i.e., on the order of 100 µm). To quantify these differences, we compared, from a single cross-section, 11 fields of view (each 152 × 152 µm) containing primarily the dense community and 12 fields containing almost exclusively the sparser, food particle-associated community. The six Bacteroides species collectively dominated in all microhabitats; for example, B. cellulosilyticus made up 43 ± 4% (mean ± SD) of the cells in the densely colonized fields and 46 ± 4% in the sparsely colonized fields (Table S2). C. aerofaciens was most abundant in the densely colonized areas, where it made up 11 ± 1% of bacteria cells versus 6 ± 2% within the sparsely colonized areas. Of the two members of Clostridia targeted with specific probes, C. scindens comprised 8 ± 2–4% of the community in both microhabitats, while R. torques made up 3 ± 1% in dense fields and 2 ± 1% in sparsely colonized regions of the lumen. Thus, modest differences in overall microbial community composition in mucus-rich and food-rich regions were detectable. Summing FISH results across all of the sections yielded relative abundances that in many cases were comparable to results obtained from COPRO-Seq analysis of fecal DNA (Table S2).
Communities observed at 10-µm scales consisted of a mixture of diverse species, with the relative proportions of species varying from region to region. The autofluorescent particles we observed ranged in size from distinctively shaped objects several hundreds of micrometers long (Fig. 2) down to small “blobs” only a few micrometers in diameter. While the larger objects were generally only thinly colonized, smaller particles were present in densely colonized patches, with the exception of some patches near the mucosa (Fig. 3).
Quantitative Analysis of Spatial Organization at Micrometer Scales.
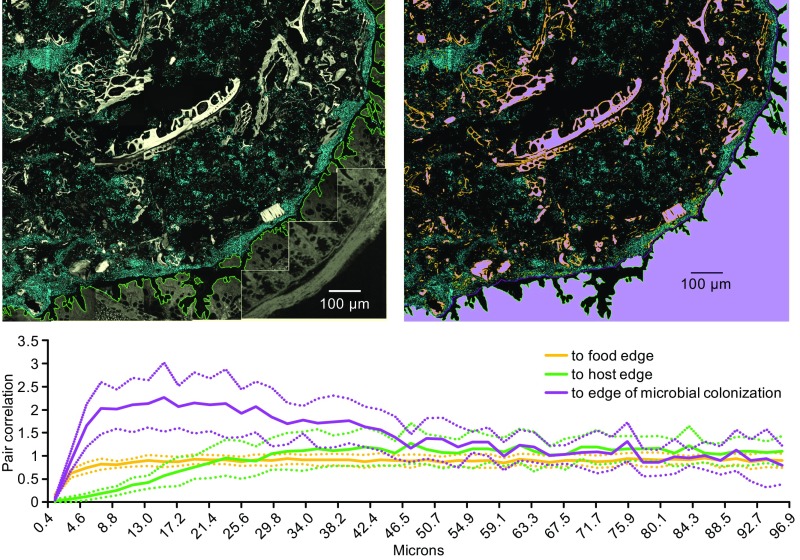
To assess whether the spatial arrangement of microbes at micrometer scales differed relative to the visible landmarks of the epithelial border and large food particles, we carried out an analysis of spatial arrangement using the method of linear dipoles as implemented in DAIME (57). Linear dipole analysis calculates a pair cross-correlation function for two categories of objects by estimating the probability, normalized to their density in the image, that the ends of a line segment of a given length will touch both of them.
Linear dipole analysis demonstrated a tendency of microbes to be in low abundance near crypts and invaginations in the mucosal epithelium (Fig. 4). This result is not surprising because these regions are generally occupied by a dense mucus layer resistant to bacterial penetration (58). Bordering this largely microbe-free zone there was dense colonization of microbes. For purposes of analysis, we marked this border of dense colonization with a hand-drawn line and calculated the spatial cross-correlation between the microbes and this line (Fig. 4). The results revealed a concentration of microbes within ∼30 µm of this line (Fig. 4). By contrast, and counterintuitively, microbes were underrepresented within 5–10 micrometers of large food particles (Fig. 4). It is possible that this underrepresentation was due in part to a failure to detect the edges of food particles accurately, as food particles were detected by virtue of their endogenous fluorescence rather than by specific staining. However, the images also give no visual evidence of any clustering of microbes close to these autofluorescent particles.
Fig. 4.
Spatial analysis of bacteria relative to visible landmarks. A tiled image (Upper Left) was segmented to identify bacterial cells and to outline the edge of large food particles, host tissue, and the edge of dense colonization (Upper Right; orange, green, and purple lines). Spatial correlation analysis was carried out using the method of linear dipoles (57), which calculates the pair cross-correlation function as the probability that two categories of object are located at a given distance from one another, normalized to their density in the image. This analysis revealed that bacteria tend to localize within 30 µm of the marked edge of colonization, but are underrepresented within 20 µm of the epithelium and within 5–10 µm of large food particles. Interiors of food particles and host tissue (Upper Right, magenta) were excluded from the analysis. Dotted lines indicate 95% confidence intervals generated by dividing the image into 10 radial sectors for analysis. Food particles and host tissue were identified by autofluorescence with 405-nm excitation, and their edges were outlined by eroding the image by seven pixels (∼1 µm) using FIJI. The edge of dense colonization was outlined by hand.
Micrometer-Scale Analysis Reveals Differences in the Organization of Specific Taxa with Respect to the Mucosa and One Another.
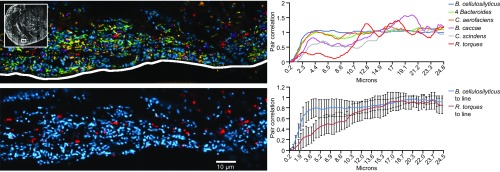
Turning from the distribution of bacteria overall to the distribution of individual taxa within the community, we detected an underrepresentation of R. torques in the dense community at the border between the mucosa and the lumen. Bacteroides were abundantly represented throughout this microbe-dense region, but R. torques exhibited marked scarcity in the region closest to the mucosa (Fig. 5). To quantify the distribution of taxa relative to the edge of the mucosa, we marked the limit of dense microbial colonization with a 1-μm-thick line (Fig. 5) and used linear dipole analysis to calculate the cross-correlation between each taxon and the line. The use of the line, rather than the mucosa itself, was necessary because of crypts and invaginations in the mucosa that resulted in variable distances between the epithelial border and microbial community, whereas the edge of microbial colonization itself was relatively smooth. The results confirmed the visual observation that R. torques is frequently, although not universally, underrepresented in the microbial community positioned in the 5- to 10-μm region closest to host cells (Fig. 5).
Fig. 5.
Distinctive community organization in the 10 µm closest to the mucosa. The microbial community near the mucosal epithelium is abundantly populated by B. cellulosilyticus, but R. torques is underrepresented in a narrow band close to the mucosa. The white boxed area in Inset shown in Upper Left denotes the field shown at higher magnification in Upper Left and Lower Left. (Upper Left) A representative image near the mucosa showing all taxa and a 1-µm-thick line representing the edge of microbial colonization. (Upper Right) Pair cross-correlation (PCC) analysis showing the probability of detecting a cell at each distance from the line, normalized to the density of cells in the image. Analysis was carried out using the method of linear dipoles as implemented in DAIME (57). (Lower Left) The B. cellulosilyticus and R. torques channels are shown separately for clarity and to demonstrate the rarity of R. torques in the 5- to 10-µm zone at the edge of the microbe-dense region. (Lower Right) Results of PCC analysis depicted as the mean of all 11 images from two sections in which a 100 µm length and 40 µm width of mucosal border was visible. Confidence intervals of 95% are shown for these two bacterial strains.
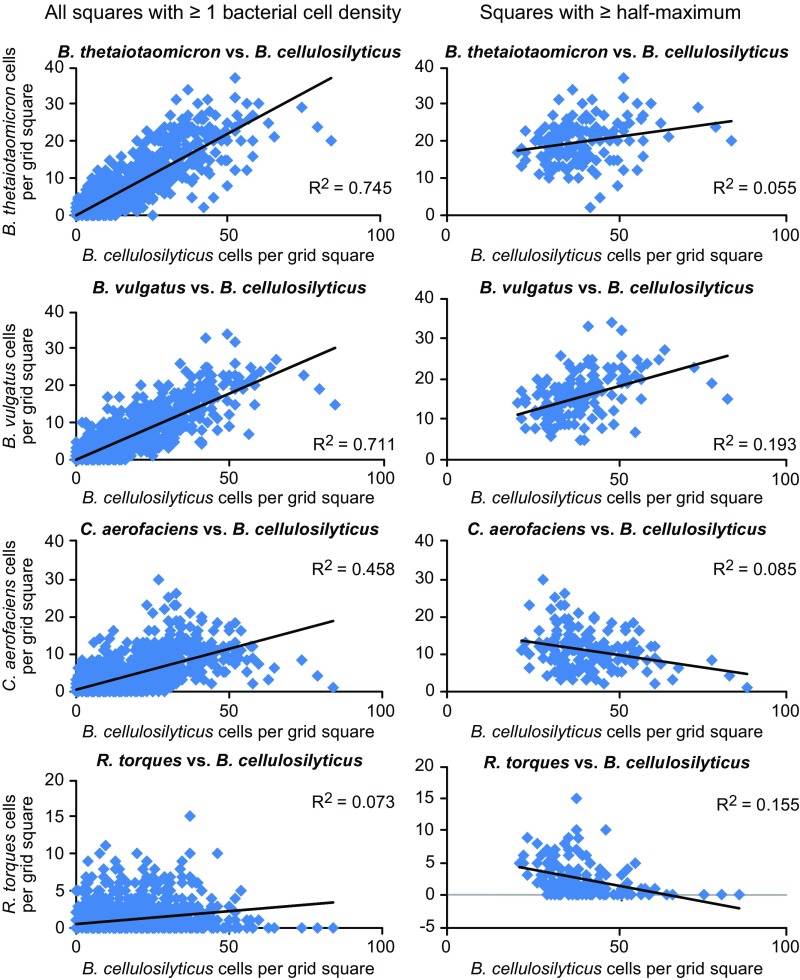
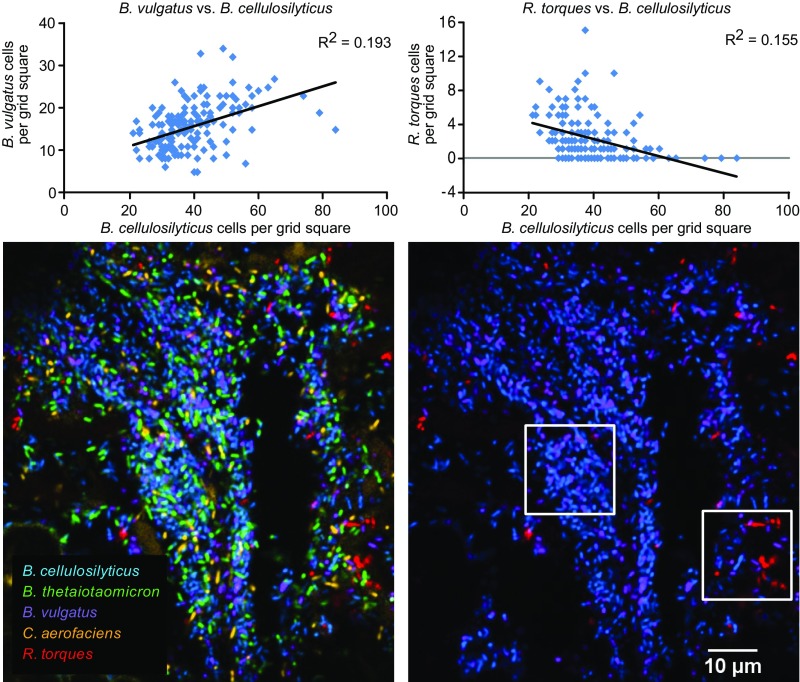
Differences in the distribution of individual taxa relative to one another were also evident at micrometer scales. Images were divided into grid squares of 19 × 19 µm, and bacterial cells within each grid square were tallied. Scatter plots comparing the abundance of pairs of taxa within these grid squares showed a linear regression with a positive slope (Fig. S3), likely reflecting the presence of a mixed microbial community in regions of both high and low microbial abundance. One possibility is that the primary driver of correlations in abundances among taxa is simply the overall abundance of bacterial cells in any given 19 × 19 µm grid square. However, analyzing only “densely populated” grid squares (defined as those containing at least half the maximum number of bacterial cells) revealed differences in taxon distribution (Fig. 6 and Figs. S3 and S4). For example, abundance of the most prominent taxon, B. cellulosilyticus, was positively correlated with that of B. vulgatus but negatively correlated with R. torques. Among the possible explanations for these observed distributional differences are cooperative (attractive) versus competitive (repulsive) interactions between taxa, or an indirect effect of binding or proliferation of taxa in distinct microenvironments.
Fig. S3.
Correlation of bacterial abundance overall and in densely populated regions of images. Abundance of each bacterial strain was tabulated within 19 × 19 μm grid squares from a section hybridized with probe set 1, and the percent representation of the most abundant taxon, B. cellulosilyticus, was plotted against each of four less-abundant taxa. Left presents data for all 1,572 grid squares containing at least one bacterial cell; Right shows the 160 grid squares with at least 67 bacterial cells (equals half the maximum bacterial abundance of 134 cells per grid square). Taxon abundances are positively correlated when all data are considered (Left) but show variable relationships when considering only grid squares that contain a high density of bacterial cells (at least 50% of the maximum density).
Fig. 6.
Distinctive organization of microbes relative to one another. Abundance of each taxon was tabulated within 1,572 grid squares measuring 19 × 19 μm (cf. individual squares in the heat map in Fig. 2) from a section hybridized with the species-specific probe set 1. Scatter plots of individual taxa show that the abundance of B. cellulosilyticus and B. vulgatus are positively correlated (Upper Left) while the abundance of B. cellulosilyticus and R. torques are negatively correlated (Upper Right). Scatter plots include only those grid squares that contain a high density of bacterial cells (at least 50% of the maximum density). An image of such a densely populated region (Lower) shows that B. cellulosilyticus and B. vulgatus are abundant in the same region of the image, while the abundance of R. torques is highest where abundance of the Bacteroides is low. These spatial relationships are consistent across mice, as demonstrated by analysis of both mice with the comprehensive probe set 3 (Fig. S4).
Fig. S4.
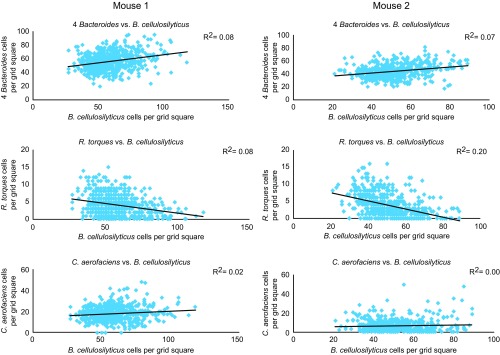
Distinctive organization of microbes relative to one another is consistent across mice and probe sets. The abundance of each taxon was tabulated within 19 × 19 μm grid squares from three sections from mouse 1 and two sections from mouse 2, all hybridized with probe set 3. The abundance of the most abundant taxon, B. cellulosilyticus, was plotted against the abundance of the set of B. thetaiotaomicron, B. ovatus, B. vulgatus, and B. uniformis all hybridized with the same fluorophore (“four Bacteroides”) and against R. torques and C. aerofaciens. Scatter plots include only grid squares that contained a high density of bacterial cells (at least 50% of the maximum density). Relative abundance relationships are modest but consistent across mice and consistent with those found using probe set 1 (Fig. 6).
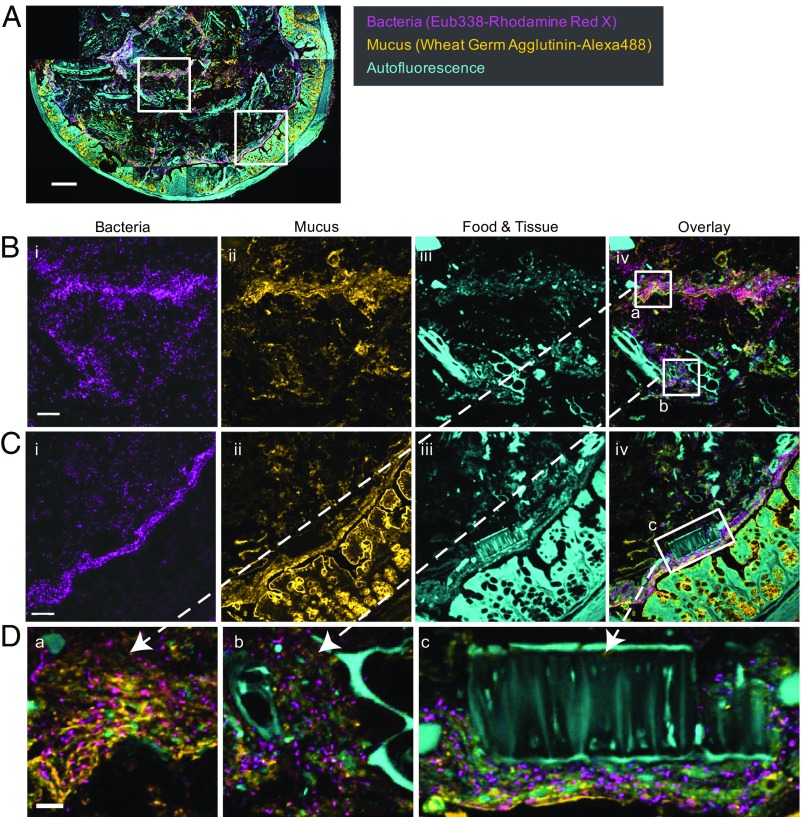
To further investigate factors underlying differences in taxon density and distribution, we imaged the distribution of bacteria, mucus, and food in additional cross-sections using the Eub338 bacterial probe in conjunction with WGA and a mouse colonic mucin antibody (anti-MCM) to localize mucus (Fig. 7 and Figs. S5 and S6). As before, bacteria were detected in high abundance in regions of the lumen as well as close to the mucosa (Fig. 7 B and C). WGA and anti-MCM showed similar staining patterns to one another (Fig. S5), and a qualitative inspection of the images revealed dense concentrations of mucus in regions corresponding to areas of high bacterial density, as well as in goblet cells (Fig. 7 B and C and Fig. S6). Large autofluorescent particles, by contrast, generally occupied regions in which bacteria were not abundant, although small autofluorescent particles were mixed with mucus and bacteria even in regions of highest bacterial density (Fig. 7D). These results are consistent with the notion that dense concentrations of mucus support high bacterial density, both in the loose mucus layer at the mucosal border and in the interior of the lumen.
Fig. 7.
Dense bacterial aggregations occupy regions rich in mucus. (A) Cross-section of colon highlighting location of panels shown at higher magnification below. (B) Bacterial density (i) is heterogeneous in the lumen. Staining with fluorophore-labeled WGA (ii) shows high density of mucus in areas of the lumen that contain abundant bacteria. Large autofluorescent food particles (iii) occupy areas of the lumen in which bacterial density is low. (iv) Overlay of i–iii. (C) Bacterial density (i) is also high in a narrow zone located at the edge of the mucosa. WGA staining (ii) shows a high density of mucus in this zone. Autofluorescent food particles (iii) are located within micrometers of the mucosa. (D) High-magnification views showing regions of the lumen with abundant mucus (a), large food particles (b), and a large food particle pressed close to the mucosa (c). The cross-section shown is adjacent to the one presented in Fig. 2. (Scale bars: A, 200 μm; B and C, 50 μm; D, 10 μm.)
Fig. S5.
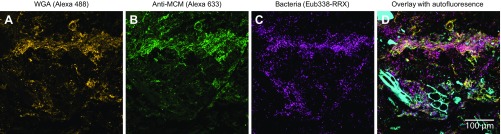
Visualizing mucus, bacteria, and partially digested food particles in a colonic section. (A and B) Visualization of mucus with wheat germ agglutinin labeled with Alexa 488 (A) and Alexa 633-labeled antibodies directed at mouse colonic mucin (B). (C) Bacteria, identified by hybridization to the Rhodamine Red X-conjugated Eub338 probe, are most abundant in regions with abundant mucus. (D) Overlay of mucus stains, bacteria, and partially digested food particles (cyan).
Fig. S6.
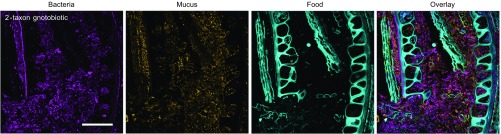
Dense bacterial aggregations occupy areas rich in mucus. Images from a two-taxon gnotobiotic mouse colonized with B. thetaiotaomicron and E. rectale are comparable to images from the gnotobiotic mouse colonized with 15 taxa (Fig. 7) and show heterogeneous bacterial density in the lumen (magenta), with a high density of mucus labeled with wheat germ agglutinin (orange) in areas that contain abundant bacteria. Large autofluorescent food particles (cyan) occupy areas of the lumen where bacterial density is low. Methacrylate sections were hybridized with the Eub338 oligonucleotide probe labeled with Alexa 647 as well as species probes labeled with Rhodamine Red X and Alexa 594. Mucus was visualized with wheat germ agglutinin labeled with Alexa 488. Food autofluorescence spectra were read directly from the images for use in linear unmixing. (Scale bar: 100 μm.)
Discussion
We investigated the spatial arrangement of members of a defined artificial 15-member human gut microbiota in the proximal colon of gnotobiotic mice at macroscale, mesoscale, and micrometer scale. At a macrolevel scale of hundreds of micrometers, bacterial cell density was heterogeneous throughout the colon but was highest in regions rich in mucus, both in a layer near the epithelium and in patches in the lumen. Compositional “patchiness” was observed at a mesoscale (tens of micrometers), while at micrometer scales, each microhabitat was generally occupied by a complex community of microbes intermingled with one another. This extensive mixing at micrometer scales, the absence of large microcolonies associated with the mucosa or food particles, and the broad similarity of luminal and mucosa-adjacent communities lead us to view the lumen and mucosa in the proximal colon not as stratified compartments but as components of a partially mixed bioreactor. By “bioreactor,” we mean an environment constructed to harbor microbes and harness their metabolism of available nutrients, where the degree of spatial homogenization of community members is a manifestation of a complex dynamic involving many factors including flux of mucus and epithelial cells into the lumen, the affinities of community members for these host constituents and food particles, cooperative/competitive (attractive/repulsive) interactions between microorganisms, and peristaltic flow of bulk material through the lumen.
One might have expected a priori that differences between taxa in their binding affinities for substrates such as mucus or food particles, combined with more rapid replication in a preferred microhabitat, would lead to localized clonal expansion. The resulting communities would have spatial structure in the form of microcolonies and a distinctive community composition within the mucus layer compared with luminal contents. Instead, the spatial distribution of both substrates and microbes suggests a dynamic model for the proximal colon in which mixing and dispersal by host factors tend to homogenize the community. Such mixing could also explain why large clusters with strongly distinctive taxonomic composition were not observed between the mucus layer and the lumen. The turnover time for mucus is ∼6 h for mucus in goblet cells and as little as a single hour for the inner mucus layer in the distal colon (59). The replication time of two common gut symbionts, B. thetaiotaomicron and Escherichia coli, has been estimated at 3 h in the mucus layer and 3–8 h in colonic contents (29). Thus, these and other gut bacterial taxa may carry out only a few rounds of cell division within the mucus layer before being shed along with mucus into the lumen. Adhesion to the epithelium could support persistence of microbes for a longer time, but epithelial cells are continuously discarded; with the exception of Paneth cells, the other three mouse gut epithelial lineages turn over every 3–5 d (60–64). Microbes in the lumen also have limited opportunity for replication before being expelled, as the contents of the gut traverse the mouse intestine in 4–6 h (65–67). We hypothesize that through rapid turnover and mixing of the mucus layer, the epithelium, and the gut contents as a whole, the mammalian host acts to diminish the ability of the microbiota to establish spatially segregated communities or sizeable single-taxon agglomerations both in the lumen and adjacent to the mucosa. Whether the physiology of the microbes themselves also fosters mixing is a question raised by the extensive micrometer-scale taxonomic intermingling in densely populated mucus-rich regions of our images. The bacteria comprising the artificial human gut microbiota characterized in this study are nonflagellated. Diffusion is unlikely to be a significant force within the viscous gel of the mucus layer (29), although there is little information about how foraging of mucus glycans by resident microbes affects mucus viscosity. Regardless of the means by which mixing is achieved, its effect is to diminish micrometer-scale spatial structure in the community.
The microbial assemblage we employed was simple enough so most of the abundant taxa could be simultaneously visualized with multilabel FISH and complex enough to permit a variety of possible taxon–taxon interactions and spatial distributions. Inevitably, our findings are dictated by the microbes that we chose to create this synthetic community. We do not know whether the consortium of 15 taxa studied display a spatial organization that is representative of the native mouse gut microbiota. It is possible that this low diversity consortium of human gut-derived bacterial strains is less likely to form spatially structured communities than the native mouse gut microbiota. Moreover, members of a more complex community of human gut microbes, representing lineages from Bacteria and other domains of life (e.g., methanogenic archaeons and eukaryotes) could demonstrate more pronounced differences in their replication rates and stronger spatial associations with one another or with food particles, host cells, mucus, or other features of the gut habitat.
The observed spatial arrangement differs dramatically from the highly ordered and more clustered arrangements visible in human dental plaque (68). These dissimilarities could reflect a number of factors including differences in flow rates in the two ecosystems and the absence of a surface in the gut to which microbes can stably adhere. In the mouth, continuous rapid flow of saliva ensures that adherence to a surface, either directly or indirectly via binding to other adherent microbes, is critical for remaining in the habitat. Further, chemical communication in salivary flow is most effective at distances on the order of micrometers (1) so precise positioning relative to metabolic partners is critical.
Deeper understanding of spatial relationships in the gut should be gained as methods are developed for quantifying the distribution of nutrients and metabolic products over different spatial scales. Gnotobiotic animal models harboring defined consortia of microbes and fed diets of known composition and physical (e.g., particulate) properties represent a starting point for these types of studies. For example, follow-up experiments in which gnotobiotic mice are fed purified dietary fibers of defined composition and size could provide an opportunity to identify taxa that exhibit reproducible patterns of cooccurrence or spatial association with the population of food particles represented in their colons. In addition, the relative effects of propulsive contractions, nonpropulsive mixing, and/or the rate of renewal of the mucus layer can be explored using gnotobiotic animals with mutations that affect their gut motility and/or the composition of their mucus (e.g., ref. 67). These genetic manipulations can also be applied to gnotobiotic zebrafish where the transparency of the organism can support in vivo imaging of microbial communities (69, 70). This latter feature avoids a potentially confounding variable; namely, that mesoscale and microscale spatial structure may be disrupted in unknown ways during the processing of gut tissues taken from euthanized animals. The resulting datasets should enable modeling both in silico and in ex vivo experimental bioreactors, including “gut-on-a-chip” systems (71). Together, these efforts should provide information about forces that promote or retard mixing at microscale levels. Put another way, studies of microscale mixing provide an opportunity to determine how this parameter is related to the niches (“jobs”) of community members (both symbionts and pathogens), the expressed functional properties of a microbiota (including its resiliency to perturbations), and whether changes in this spatial feature are a reflection of, or causally related to, various types of dysbioses.
Materials and Methods
Collection of Samples from Gnotobiotic Mice and Preparation for Imaging.
All experiments involving mice were performed using protocols approved by the Animal Studies Committee of the Washington University School of Medicine. Two 8-wk-old, male, germ-free C57BL/6J mice were gavaged with a 15-member bacterial consortium prepared and administered using procedures described in earlier reports (4, 54), and then maintained in a gnotobiotic isolator under a strict light cycle (lights on at 0600 hours and off at 1800 hours). Two additional mice were gavaged with a two-member bacterial community composed of B. thetaioatomicron VPI-5482 and E. rectale ATCC 33656, and two control mice were maintained as germ-free. All animals were fed a sterilized, low-fat, plant polysaccharide-rich chow (Product 7378000; B&K Universal) ad libitum. All mice were killed 14 d after gavage of their bacterial consortium.
The distal fourth of the small intestine and the proximal third of the colon were snap-frozen in optimal cutting temperature compound and stored at −80 °C. These frozen segments were cut into 5- to 10-mm-long pieces, and molten 0.5% agarose was pipetted onto the two cut ends of each piece. Samples were then fixed in 2% paraformaldehyde in PBS for 12 h at 4 °C, then washed, resubmerged in molten 0.5% agarose, dehydrated in acetone for 1 h, infiltrated with Technovit 8100 glycol methacrylate resin with several changes over 12 h at 4 °C, and were then transferred to embedding solution where they were allowed to solidify for 12 h at 4 °C. Embedded samples were sectioned with a Sorvall JB-4 microtome (Dupont Instruments) to 5–10 µm thickness and subjected to fluorescent labeling experiments. We employed paraformaldehyde rather than Carnoy’s fixative because paraformaldehyde is commonly used for microbial FISH (72, 73). Moreover, our direct comparison of the two methods revealed similar preservation of mucus and other spatial landmarks in methacrylate-embedded colonic segments fixed with either Carnoy’s solution or paraformaldehyde (74).
Probe Design.
Candidate probes identified using the “probe design” function of the ARB software package (75) were further analyzed by calculating the overall free energy of hybridization (ΔG0overall; ref. 76) for each. A set of 16S rRNA-directed oligonucleotides was selected that were predicted to be taxon-specific at the same hybridization stringency (arbitrarily chosen to be 20% formamide). Probes were synthesized (Invitrogen) with a 5′ fluorophore and their specificity evaluated by hybridization to pure cultures of target and nontarget bacterial strains.
FISH Analysis of Colonic Sections.
FISH was carried out at 46 °C for 6 h in 0.9 M NaCl, 0.02 M Tris pH 7.5, 0.01% SDS, 20% HiDi formamide (Applied Biosystems), and 2 µM of each probe. Sections were then washed twice for 10 min each at 48 °C in wash buffer (0.215 M NaCl, 0.02 M Tris pH 7.5, 5 mM EDTA), incubated in DAPI in PBS at room temperature for 15 min in the dark, washed in PBS, dehydrated through a series of ethanol washes [3 min each in 50%, 80%, and 96% (vol/vol) ethanol], mounted in ProLong Gold antifade reagent (catalog no. P36934; Invitrogen) with a No. 1.5 coverslip, and placed in the dark at room temperature for at least 24 h before imaging. FISH on pure cultures was carried out in the same way, but without DAPI staining.
Lectin and Mucin Staining.
Labeling of methacrylate sections with WGA was carried out after the FISH hybridization and washing steps. Slides were incubated in 40 µg/mL WGA conjugated to Alexa Fluor 488 (catalog no. W11261; Invitrogen) in PBS at room temperature for 15 min in the dark. Slides were washed briefly in PBS, dipped in distilled H2O, dehydrated through a series of ethanol washes and mounted as above. Slides to be labeled with anti-mouse colonic mucin (anti-MCM, a gift of Dr. Ingrid B. Renes, Erasmus MC-Josephine Nefkens Institute, Rotterdam, The Netherlands) were treated with blocking buffer (2% goat serum; 1% BSA; 0.2% Triton X-100; 0.05% Tween 20) for 1 h at room temperature, incubated with anti-MCM diluted 1:50 in blocking buffer for 12 h at 4 °C, rinsed three times for 3 min each in PBS, incubated with blocking buffer at room temperature for 1 h, incubated with a 1:1,000 dilution of Alexa Fluor 633 goat anti-rabbit IgG (catalog no. A21070; Invitrogen) in blocking buffer for 1 h, rinsed three times for 3 min each in PBS, incubated with WGA, washed, and mounted as above.
Image Acquisition and Linear Unmixing.
Spectral image stacks were acquired using a Zeiss LSM 710 confocal microscope equipped with a QUASAR spectral detector and a Plan-Apochromat 63×, 1.4 N.A. objective lens or a Zeiss LSM 780 confocal microscope equipped with a GaAsP spectral detector and a Plan-Apochromat 40×, 1.4 N.A. objective. Excitation wavelengths of 633 nm, 594 nm, 561 nm, 514 nm, 488 nm, and 405 nm were employed sequentially. Linear unmixing was carried out using ZEN software (Carl Zeiss) or using a custom algorithm on the Mathematica platform (Wolfram Research) using the concatenated reference spectra shown in Fig. S1. Details of the unmixing procedure and figure preparation are given in SI Materials and Methods.
Spatial Arrangement Analysis.
Images of each taxon were segmented in FIJI using the IsoData or RenyiEntropy global thresholding algorithm (77) and were then imported into DAIME version 2.1 for analysis of spatial arrangement using linear dipoles.
SI Materials and Methods
Linear Unmixing.
Spectral images were unmixed using ZEN software (Carl Zeiss) or using a custom algorithm on the Mathematica platform (Wolfram Research) that permitted concatenation of spectral images sequentially acquired from the same field of view using six different excitation wavelengths. This concatenation effectively made use of information about the excitation spectrum as well as the emission spectrum of each fluorophore (78). Images from probe sets 1 and 2 were unmixed using ZEN, and images from probe set 3 were unmixed using Mathematica. Reference spectra for the Mathematica unmixing consisted of nine FISH probe-conferred spectra, two autofluorescence spectra from food particles, one autofluorescence spectrum from the host tissue, and a background spectrum from a position that was not occupied by any object. Image stacks from probe set 3 acquired with 594 nm excitation were also unmixed using ZEN with six reference spectra (for fluorophores Alexa 594, Alexa 647, and Alexa 660; an autofluorescence spectrum from a food particle; an autofluorescence spectrum from the host tissue; and a background spectrum from a position that was not occupied by any object) and the unmixed Alexa 660 image used in place of that unmixed on the Mathematica platform because of its better signal-to-noise ratio.
Acquisition of Reference Spectra.
For unmixing in ZEN of cells hybridized with probe sets 1 and 2, reference spectra were acquired from cells grown as monocultures and hybridized with taxon-specific probes. Regions of interest (ROI) were drawn on fluorescently labeled cells in spectral image stacks acquired with each excitation wavelength. All autofluorescence and background spectra included in the reference spectral libraries were obtained from ROIs selected in experimental images of intestinal sections. For unmixing in Mathematica of cells hybridized with probe set 3, reference spectra of FISH probe-targeted bacterial species were acquired using a pure culture of each taxon incubated with a mixture of all 15 target-specific FISH probes (Fig. S2). Images of the FISH probe-labeled bacterial strains were acquired using the same image acquisition conditions employed for intestinal sections, except that when cultured bacterial cells emitted significantly stronger fluorescent signals than their counterparts in intestinal sections, laser intensities were reduced for cultured cells without changing the relative intensities of the six lasers. Reference spectra were sampled from concatenated spectral image stacks of individual FISH probe-labeled bacterial cultures. An averaged spectrum from most or all cells in an image was recorded as a reference spectrum.
Image Binarization and Bacterial Cell Count.
Spectrally unmixed images were digitally segmented into binary images using the Bernsen local thresholding algorithm (79) in ImageJ (80) followed by the “find maxima” watershed algorithm. Particles outside the size range of 2–150 pixels were digitally removed from the segmented binary images. Segmented binary images were screened individually to ensure that the positions of the cells defined in the binary images reflected bacterial cells visible in presegmentation images. For automated cell counting in Mathematica, segmented images were divided into an 8 × 8 grid, and objects in each grid square were counted; the dimensions of each grid square were 19 μm × 19 μm.
Pseudocolored Images for Visual Inspection.
For multicolor FISH image presentation, the image from each unmixed fluorophore channel was assigned a pseudocolor, and then images of each FOV showing different bacterial groups and autofluorescence were merged and brightness and contrast adjusted for clarity. Images of autofluorescent food particles shown in Figs. 3 B and D, 4, and 7 and Figs. S2 and S4 are from single unmixed autofluorescence images; those in Fig. 3 A and C are an average intensity projection of three autofluorescence images corresponding to three different reference spectra read from food particles in images acquired with 405-nm excitation. A median filter, radius two pixels, was applied to the unmixed Alexa 405 image in Fig. 3 A and C for noise reduction.
Supplementary Material
Acknowledgments
We thank David O’Donnell and Maria Karlsson for help with gnotobiotic animal husbandry; Louie Kerr for microscopy support; and Christopher Rieken, Blair Rossetti, and Liping Xun for outstanding technical support. This work was supported in part by NIH Grants DE 022586, DK30292, DK70977, and DK78669.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711596114/-/DCSupplemental.
References
- 1.Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 2.Fischbach MA, Sonnenburg JL. Eating for two: How metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio. 2013;4:e00459-13. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu M, et al. Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science. 2015;350:aac5992. doi: 10.1126/science.aac5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Earle KA, et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 2015;18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abreu NAA, Taga ME. Decoding molecular interactions in microbial communities. FEMS Microbiol Rev. 2016;40:648–663. doi: 10.1093/femsre/fuw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol. 2016;14:93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proctor DM, Relman DA. The landscape ecology and microbiota of the human nose, mouth, and throat. Cell Host Microbe. 2017;21:421–432. doi: 10.1016/j.chom.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33:206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 11.Mitri S, Xavier JB, Foster KR. Social evolution in multispecies biofilms. Proc Natl Acad Sci USA. 2011;108:10839–10846. doi: 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estrela S, Brown SP. Metabolic and demographic feedbacks shape the emergent spatial structure and function of microbial communities. PLOS Comput Biol. 2013;9:e1003398. doi: 10.1371/journal.pcbi.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein RR, et al. Ecological modeling from time-series inference: Insight into dynamics and stability of intestinal microbiota. PLOS Comput Biol. 2013;9:e1003388. doi: 10.1371/journal.pcbi.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovács AT. Impact of spatial distribution on the development of mutualism in microbes. Front Microbiol. 2014;5:649. doi: 10.3389/fmicb.2014.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambert G, Vyawahare S, Austin RH. Bacteria and game theory: The rise and fall of cooperation in spatially heterogeneous environments. Interface Focus. 2014;4:20140029. doi: 10.1098/rsfs.2014.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: Networks, competition, and stability. Science. 2015;350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 17.Peña J, Wu B, Traulsen A. Ordering structured populations in multiplayer cooperation games. J R Soc Interface. 2016;13:20150881. doi: 10.1098/rsif.2015.0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Momeni B, Waite AJ, Shou W. Spatial self-organization favors heterotypic cooperation over cheating. Elife. 2013;2:e00960. doi: 10.7554/eLife.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derrien M, et al. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes. 2010;1:254–268. doi: 10.4161/gmic.1.4.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naughton JA, et al. Divergent mechanisms of interaction of Helicobacter pylori and Campylobacter jejuni with mucus and mucins. Infect Immun. 2013;81:2838–2850. doi: 10.1128/IAI.00415-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Etzold S, Juge N. Structural insights into bacterial recognition of intestinal mucins. Curr Opin Struct Biol. 2014;28:23–31. doi: 10.1016/j.sbi.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Gunning AP, et al. Mining the “glycocode”–Exploring the spatial distribution of glycans in gastrointestinal mucin using force spectroscopy. FASEB J. 2013;27:2342–2354. doi: 10.1096/fj.12-221416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLoughlin K, Schluter J, Rakoff-Nahoum S, Smith AL, Foster KR. Host selection of microbiota via differential adhesion. Cell Host Microbe. 2016;19:550–559. doi: 10.1016/j.chom.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 24.Shipman JA, Berleman JE, Salyers AA. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J Bacteriol. 2000;182:5365–5372. doi: 10.1128/jb.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 26.Macfarlane S, Macfarlane GT. Composition and metabolic activities of bacterial biofilms colonizing food residues in the human gut. Appl Environ Microbiol. 2006;72:6204–6211. doi: 10.1128/AEM.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker AW, et al. The species composition of the human intestinal microbiota differs between particle-associated and liquid phase communities. Environ Microbiol. 2008;10:3275–3283. doi: 10.1111/j.1462-2920.2008.01717.x. [DOI] [PubMed] [Google Scholar]
- 28.Nagara Y, Takada T, Nagata Y, Kado S, Kushiro A. Microscale spatial analysis provides evidence for adhesive monopolization of dietary nutrients by specific intestinal bacteria. PLoS One. 2017;12:e0175497. doi: 10.1371/journal.pone.0175497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun. 2015;6:8292. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stearns JC, et al. Bacterial biogeography of the human digestive tract. Sci Rep. 2011;1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 2012;10:e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sommer F, Bäckhed F. The gut microbiota–Masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 33.Yasuda K, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe. 2015;17:385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoetendal EG, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albenberg L, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627–638. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pédron T, et al. A crypt-specific core microbiota resides in the mouse colon. MBio. 2012;3:e00116-12. doi: 10.1128/mBio.00116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malmuthuge N, Griebel PJ, Guan L. Taxonomic identification of commensal bacteria associated with the mucosa and digesta throughout the gastrointestinal tracts of preweaned calves. Appl Environ Microbiol. 2014;80:2021–2028. doi: 10.1128/AEM.03864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuppler M, Lötzsch K, Waidmann M, Autenrieth IB. An abundance of Escherichia coli is harbored by the mucosa-associated bacterial flora of interleukin-2-deficient mice. Infect Immun. 2004;72:1983–1990. doi: 10.1128/IAI.72.4.1983-1990.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan V, et al. Visualization of Helicobacter species within the murine cecal mucosa using specific fluorescence in situ hybridization. Helicobacter. 2005;10:114–124. doi: 10.1111/j.1523-5378.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 42.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmed S, et al. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol. 2007;73:7435–7442. doi: 10.1128/AEM.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cullender TC, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geva-Zatorsky N, et al. In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat Med. 2015;21:1091–1100. doi: 10.1038/nm.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okumura R, et al. Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature. 2016;532:117–121. doi: 10.1038/nature17406. [DOI] [PubMed] [Google Scholar]
- 49.Whitaker WR, Shepherd ES, Sonnenburg JL. Tunable expression tools enable single-cell strain distinction in the gut microbiome. Cell. 2017;169:538–546.e12. doi: 10.1016/j.cell.2017.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cremer J, et al. Effect of flow and peristaltic mixing on bacterial growth in a gut-like channel. Proc Natl Acad Sci USA. 2016;113:11414–11419. doi: 10.1073/pnas.1601306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valm AM, et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci USA. 2011;108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valm AM, Mark Welch JL, Borisy GG. CLASI-FISH: Principles of combinatorial labeling and spectral imaging. Syst Appl Microbiol. 2012;35:496–502. doi: 10.1016/j.syapm.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McNulty NP, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNulty NP, et al. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 2013;11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cummings JH, et al. The amount and composition of large bowel contents in man. Gastroenterology. 1990;98:A408. [Google Scholar]
- 56.Sarma-Rupavtarm RB, Ge Z, Schauer DB, Fox JG, Polz MF. Spatial distribution and stability of the eight microbial species of the altered schaedler flora in the mouse gastrointestinal tract. Appl Environ Microbiol. 2004;70:2791–2800. doi: 10.1128/AEM.70.5.2791-2800.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daims H, Lücker S, Wagner M. Daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol. 2006;8:200–213. doi: 10.1111/j.1462-2920.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- 58.Johansson MEV, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johansson MEV. Fast renewal of the distal colonic mucus layers by the surface goblet cells as measured by in vivo labeling of mucin glycoproteins. PLoS One. 2012;7:e41009. doi: 10.1371/journal.pone.0041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am J Anat. 1974;141:461–479. doi: 10.1002/aja.1001410403. [DOI] [PubMed] [Google Scholar]
- 61.Cheng H. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. II. Mucous cells. Am J Anat. 1974;141:481–501. doi: 10.1002/aja.1001410404. [DOI] [PubMed] [Google Scholar]
- 62.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. III. Entero-endocrine cells. Am J Anat. 1974;141:503–519. doi: 10.1002/aja.1001410405. [DOI] [PubMed] [Google Scholar]
- 63.Cheng H. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. IV. Paneth cells. Am J Anat. 1974;141:521–535. doi: 10.1002/aja.1001410406. [DOI] [PubMed] [Google Scholar]
- 64.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian theory of the origin of the four epithelial cell types. Am J Anat. 1974;141:537–561. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- 65.Degen LP, Phillips SF. Variability of gastrointestinal transit in healthy women and men. Gut. 1996;39:299–305. doi: 10.1136/gut.39.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Padmanabhan P, Grosse J, Asad ABMA, Radda GK, Golay X. Gastrointestinal transit measurements in mice with 99mTc-DTPA-labeled activated charcoal using NanoSPECT-CT. EJNMMI Res. 2013;3:60. doi: 10.1186/2191-219X-3-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dey N, et al. Regulators of gut motility revealed by a gnotobiotic model of diet-microbiome interactions related to travel. Cell. 2015;163:95–107. doi: 10.1016/j.cell.2015.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 2016;113:E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Melancon E, et al. Best practices for germ-free derivation and gnotobiotic zebrafish husbandry. Methods Cell Biol. 2017;138:61–100. doi: 10.1016/bs.mcb.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci USA. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moter A, Göbel UB. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J Microbiol Methods. 2000;41:85–112. doi: 10.1016/s0167-7012(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 73.Amann R, Fuchs BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Microbiol. 2008;6:339–348. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- 74.Hasegawa Y, Mark Welch JL, Rossetti BJ, Borisy GG. Preservation of three-dimensional spatial structure in the gut microbiome. bioRxiv. 2017 doi: 10.1101/175224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yilmaz LS, Noguera DR. Mechanistic approach to the problem of hybridization efficiency in fluorescent in situ hybridization. Appl Environ Microbiol. 2004;70:7126–7139. doi: 10.1128/AEM.70.12.7126-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valm AM, Oldenbourg R, Borisy GG. Multiplexed spectral imaging of 120 different fluorescent labels. PLoS One. 2016;11:e0158495. doi: 10.1371/journal.pone.0158495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bernsen J. Proceedings of the Eighth International Conference on Pattern Recognition. IEEE Computer Society; Los Alamitos, CA: 1986. Dynamic thresholding of grey-level images; pp. 1251–1255. [Google Scholar]
- 80.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Amann RI, et al. Combination of 16S rRNA-targeted oligonuleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]