Significance
A subset of human papillomaviruses (HPVs) causes 5% of human cancers, including virtually all cancers of the cervix. In a mouse model of cervical cancer, estrogen is a necessary cofactor that contributes to disease by signaling through the underlying tumor microenvironment. In this study, we discovered that epithelial expression of the HPV oncoproteins reprograms the cervical tumor microenvironment and its response to estrogen. These changes involve the elicitation of paracrine-acting factors implicated in carcinogenesis, and the expression of a subset of these factors was also induced in cocultures of human cervical cancer cells and stromal fibroblasts. We hypothesize that HPV oncogenes cause cancer in part by creating a unique tumor microenvironment that synergizes with estrogen in the cervix.
Keywords: human papillomavirus, cervical cancer, estrogen, stroma, paracrine signaling
Abstract
High-risk human papillomaviruses (HPVs) infect epithelial cells and are causally associated with cervical cancer, but HPV infection is not sufficient for carcinogenesis. Previously, we reported that estrogen signaling in the stromal tumor microenvironment is associated with cervical cancer maintenance and progression. We have now determined how HPV oncogenes and estrogen treatment affect genome-wide host gene expression in laser-captured regions of the cervical epithelium and stroma of untreated or estrogen-treated nontransgenic and HPV-transgenic mice. HPV oncogene expression in the cervical epithelium elicited significant gene-expression changes in the proximal stromal compartment, and estrogen treatment uniquely affected gene expression in the cervical microenvironment of HPV-transgenic mice compared with nontransgenic mice. Several potential estrogen-induced paracrine-acting factors were identified in the expression profile of the cervical tumor microenvironment. The microenvironment of estrogen-treated HPV-transgenic mice was significantly enriched for chemokine/cytokine activity and inflammatory and immune functions associated with carcinogenesis. This inflammatory signature included several proangiogenic CXCR2 receptor ligands. A subset of the same CXCR2 ligands was likewise increased in cocultures of early-passage cells from human cervical samples, with levels highest in cocultures of cervical fibroblasts and cancer-derived epithelial cells. Our studies demonstrate that high-risk HPV oncogenes profoundly reprogram the tumor microenvironment independently of and synergistically with estrogen. These observations illuminate important means by which HPVs can cause cancer through alterations in the tumor microenvironment.
Viruses cause ∼15% of human cancers (1), and high-risk human papillomaviruses (HPVs) alone are responsible for nearly 5% of human malignancies (2). HPVs infect poorly differentiated, basal keratinocytes within stratified squamous epithelia where they can establish persistent infections. The high-risk mucosotropic HPVs cause malignancies in the stratified epithelia lining the cervix, vagina, and other organs of the lower female reproductive tract, as well as the anus, penis, and epithelia lining the oropharynx. We previously developed transgenic mice expressing the high-risk type HPV16 oncogenes E6 and E7 (3, 4). In these mice, the keratin 14 (K14) promoter directs expression of the HPV viral oncogenes in the basal layer of stratified squamous epithelia, which is the natural site of papillomavirus infection. We have used this in vivo model to show how HPV E6 and E7 oncogenes and cellular cofactors contribute to HPV-associated carcinogenesis in various anatomical sites (5–8), including the cervix (9, 10).
Despite the strong etiological link between HPV and cervical cancer, persistent HPV infection is not sufficient for human cervical cancer development (11, 12), and thus other cofactors likely contribute to carcinogenesis. One such cofactor is estrogen (17β-estradiol) (13, 14). Elevated levels of circulating estrogens have been detected in women with HPV+ lesions and cancers (15, 16), and multiparity and long-term oral contraceptive use are significant cervical cancer risk factors (17–20). Experiments in HPV transgenic mice have directly linked estrogen and cervical cancer. In K14E6/E7 mice, expression of HPV16 E6 and E7 in the cervical epithelium is essential but not sufficient to cause cervical cancer, and additional systemic delivery of exogenous 17β-estradiol and expression of its receptor ERα are required for the onset, maintenance, and progression of neoplastic disease (9, 10, 21, 22). Furthermore, treatment with estrogen-signaling inhibitors promotes regression of cervical cancer and precancerous lesions in these mice (23, 24). While caution is reasonable in extrapolating the efficacy of drugs in murine models to therapeutic treatment of human disease (25), human population-based data do correlate long-term antiestrogen use with lower risk of cervical neoplasia (26).
An important observation is that estrogen contributes to cervical carcinogenesis through the underlying stroma. Continued expression of ERα in the cervical stroma of HPV transgenic mice is required for the maintenance of neoplastic disease in the cervical epithelium (27). In human cervical cancers, expression of ERα is, in fact, retained in the stroma but is lost in epithelial cancer cells (28), and human cervical cancer-associated fibroblasts have been shown to mediate estrogen-dependent signaling (29). Moreover, stromal–epithelial interactions mediate the effects of estrogen on female reproductive tract morphogenesis, and stromal ERα facilitates the proliferative effects of estrogen on the adjacent epithelium (30–33). The tumor microenvironment directs the biology of many cancers, as bidirectional communication between epithelial cells and the tumor microenvironment affects tumor initiation, neoplastic progression, metastasis, and therapeutic response (34). These insights suggest that stromal estrogen signaling drives cervical carcinogenesis in concert with epithelial HPV oncogene expression. In the present report, we describe how we used our estrogen-dependent K14E6/E7 cervical cancer mouse model to investigate the independent and synergistic effects of epithelial HPV oncogene expression and estrogen signaling on genome-wide gene-expression levels in the cervical stroma. We discovered an extensive reprogramming of the cervical stroma in the context of mice expressing the HPV16 E6 and E7 oncogenes in the epithelia. Unique changes were also noted in the microenvironment of these mice when given exogenous estrogen to promote cervical carcinogenesis. Paracrine factors were identified whose expression was increased in the cervical stroma of K14E6/E7 mice and was enhanced in these mice when treated with estrogen. The expression of these same paracrine factors, which were previously implicated in carcinogenesis, was further enhanced when human cervical cancer-derived cell strains were cocultured with cervical fibroblasts, defining potential modulators of cervical carcinogenesis arising from the tumor microenvironment.
Results
Gene-Expression Profiling of Cervical Epithelium and Adjacent Stroma.
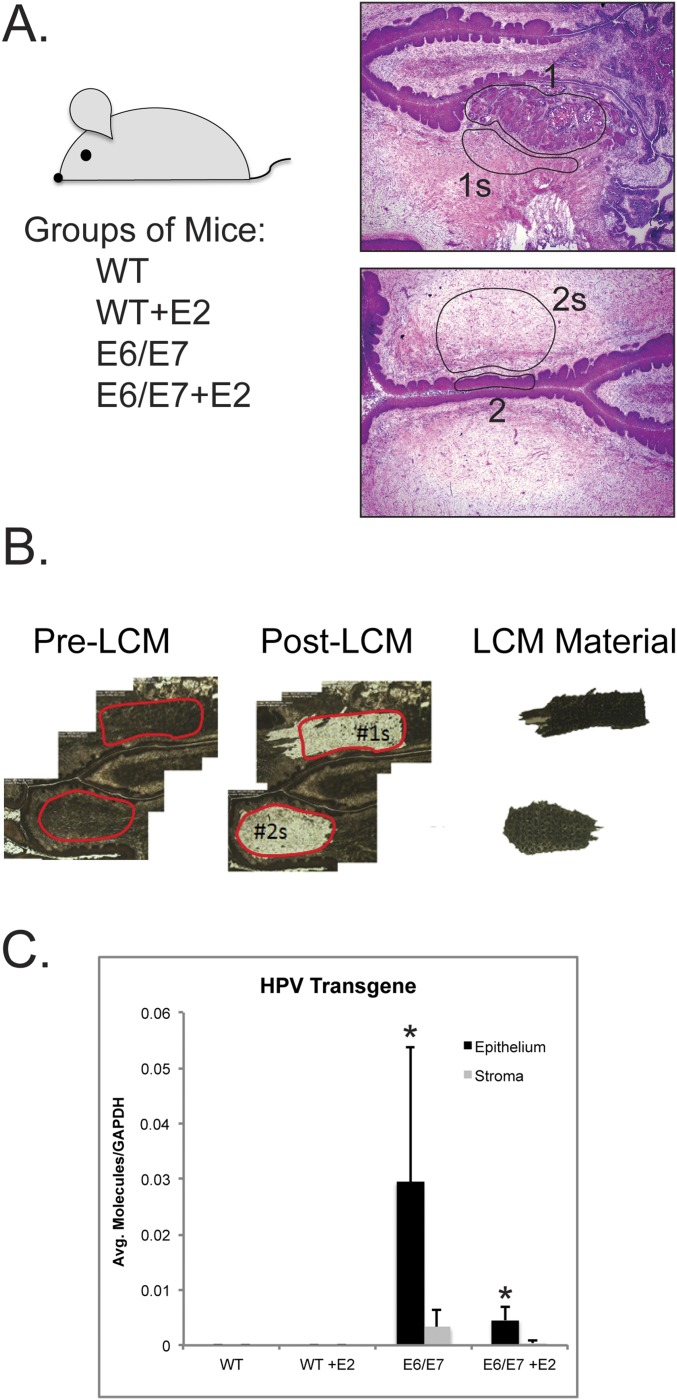
We used our mouse model of cervical carcinogenesis to identify HPV oncogene- and estrogen-driven gene changes in the stroma associated with cervical cancer development. Four groups of adult female mice representing different grades of disease were included: (i) nontransgenic mice (WT; no disease); (ii) nontransgenic mice treated with estrogen (hereafter abbreviated as “E2”) for 1 mo, a duration sufficient to induce benign hyperplasia (WT+E2; hyperplasia); (iii) K14E6/E7 bitransgenic mice [E6/E7; largely cervical intraepithelial neoplasia grade 1/2 (CIN1/2) precancerous lesions]; and (iv) K14E6/E7 bitransgenic mice treated with estrogen for 6 mo, a duration necessary to induce cervical cancers (E6/E7+E2; cancer) (Fig. S1A and Table S1). From the reproductive tracts of the above mice, 20 epithelial regions of histopathological interest (i.e., no disease, hyperplasia, precancerous lesions, or cancer) and matched regions of proximal stroma were laser-capture microdissected. Pre–laser-capture microdissection (LCM) and post-LCM images were used to assess the accuracy of tissue capture before RNA extraction (Fig. S1B). RT-PCR was used to measure levels of the K14E6/E7 transgene (Fig. S1C), expression of which should be restricted to the epithelial compartment due to the tissue specificity of K14 promoter activity (3, 4). The average number of transgene transcripts was significantly higher in E6/E7 (P = 0.0007) and E6/E7+E2 (P = 0.03) cervical epithelia than in the adjacent stroma, confirming accurate capture of epithelia and stroma. A significantly higher level of transgene-specific mRNA was measured in E6/E7 epithelia than in E6/E7+E2 epithelia (P = 0.006), likely because the squamous cell carcinomas in the E6/E7+E2 group are a more heterogeneous mixture of cells compared with the more homogenous stratified epithelium in the E6/E7 group. Total RNA was then analyzed using Affymetrix GeneChip Mouse Genome 430 2.0 microarrays comprehensively measuring nearly 40,000 murine transcripts. Significant gene-expression changes between pairwise group comparisons were defined as having a false-discovery rate (FDR) ≤0.05 and a fold change ≥2.
Fig. S1.
Experimental overview of gene-expression profiling of cervical stroma. (A, Left) Reproductive tracts were harvested from four groups of mice, and histopathology analysis was performed on H&E-stained murine endocervical tissue sections. (Right) Representative regions of interest (ROIs) for LCM. Matched sets of ROIs share the same numerical label, with epithelial ROIs indicated by numbers only (i.e., 1 and 2) and stromal ROIs labeled with a number and the letter “s” (i.e., 1s and 2s). (B) Representative pre-LCM and post-LCM images showing the accuracy of LCM of stromal tissue. Extracted stromal tissue is shown on the far right. (C) Results of RT-PCR analysis specific for the HPV transgene performed using RNA extracted from epithelial (black bars) and stromal (gray bars) tissue isolated from FVB or K14E6/E7 transgenic mice. Values were normalized to those for GAPDH and then averaged among groups. A Wilcoxon rank-sum test was used to compare epithelial versus stroma values within each group. *P < 0.03 compared with the stroma sample within each group. Error bars indicate SD.
Table S1.
Overview of samples included in analysis
| Genotype | Compartment | Treatment (duration) | No. of samples | Grade of worst disease* | Pathology | Location | GEO database accession number (GSE102232) |
| FVB | Epithelia | No estrogen | 5 | No disease | Normal | Endocervix | GSM2730977 |
| No disease | Normal | Outer cervix | GSM2730978 | ||||
| No disease | Normal | Endocervix | GSM2730979 | ||||
| No disease | Normal | Endocervix | GSM2730980 | ||||
| No disease | Normal | Outer cervix | GSM2730981 | ||||
| K14E6/E7 | Epithelia | No estrogen | 7 | CIN2 | CIN1/2 | Endocervix | GSM2730982 |
| CIN2 | CIN1/2 | Outer cervix | GSM2730983 | ||||
| No disease | Normal | Endocervix | GSM2730984 | ||||
| CIN1/2 | CIN1/2 | Endocervix | GSM2730985 | ||||
| CIN1/2 | CIN1/2 | Outer cervix | GSM2730986 | ||||
| CIN1/2 | CIN1/2 | Endocervix | GSM2730987 | ||||
| CIN1/2 | CIN1/2 | Endocervix | GSM2730988 | ||||
| FVB | Stroma | No estrogen | 5 | No disease | Normal | Endocervix | GSM2730989 |
| No disease | Normal | Outer cervix | GSM2730990 | ||||
| No disease | Normal | Endocervix | GSM2730991 | ||||
| No disease | Normal | Endocervix | GSM2730992 | ||||
| No disease | Normal | Outer cervix | GSM2730993 | ||||
| K14E6/E7 | Stroma | No estrogen | 7 | CIN2 | CIN1/2 | Endocervix | GSM2730994 |
| CIN2 | CIN1/2 | Outer cervix | GSM2730995 | ||||
| No disease | Normal | Endocervix | GSM2730996 | ||||
| CIN1/2 | CIN1/2 | Endocervix | GSM2730997 | ||||
| CIN1/2 | CIN1/2 | Outer cervix | GSM2730998 | ||||
| CIN1/2 | CIN1/2 | Outer cervix | GSM2730999 | ||||
| CIN1/2 | CIN1/2 | Endocervix | GSM2731000 | ||||
| K14E6/E7 | Stroma | Estrogen (1 mo) | 5 | Cancer | Proximal to cancer | Endocervix | GSM2731001 |
| Cancer | Proximal to cancer | Endocervix | GSM2731002 | ||||
| Cancer | Proximal to cancer | Endocervix | GSM2731003 | ||||
| Cancer | Proximal to cancer | Endocervix | GSM2731004 | ||||
| Cancer | Proximal to cancer | Endocervix | GSM2731005 | ||||
| FVB | Stroma | Estrogen (6 mo) | 3 | Hyperplasia | Normal | Endocervix | GSM2731006 |
| Hyperplasia | Normal | Endocervix | GSM2731007 | ||||
| Hyperplasia | Normal | Endocervix | GSM2731008 |
Grade of worst disease determined by disease present in epithelial compartment.
Epithelial Expression of HPV Oncogenes Dramatically Alters Host Gene Expression in Adjacent Cervical Stroma.
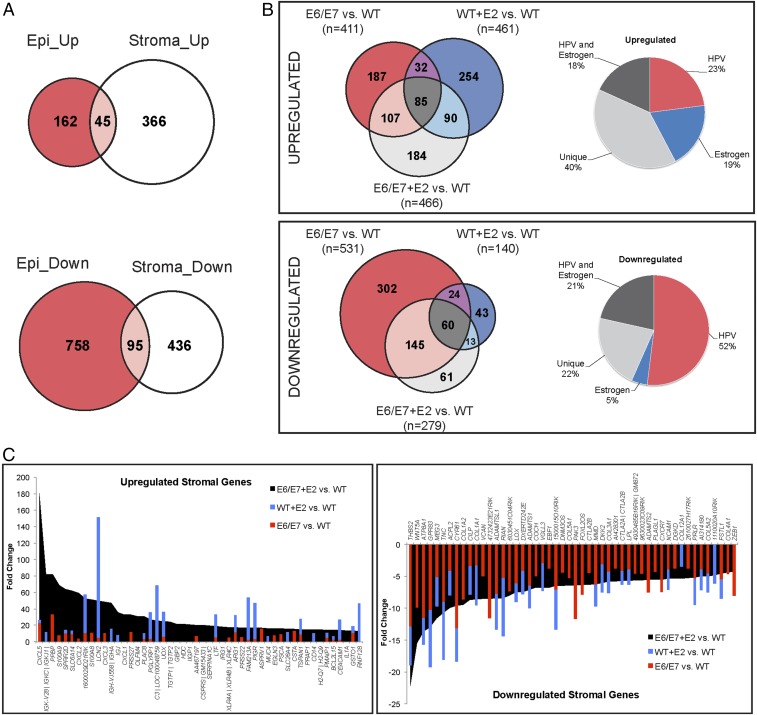
We first compared gene expression in the cervical epithelium and stroma of E6/E7 and WT mice. HPV E6 and E7 significantly altered gene expression in the epithelium: 207 unique annotated genes were up-regulated, and 853 genes were down-regulated, in the K14E6/E7 cervical epithelium (Fig. 1A and Dataset S1). Strikingly, the HPV oncogenes had substantial effects on gene expression in the surrounding stroma: 411 genes were up-regulated, and 532 genes were down-regulated, in the cervical stroma adjacent to K14E6/E7 epithelia (Fig. 1A and Dataset S2 A and B). Differentially expressed genes in the K14E6/E7 epithelium and its adjacent stroma were largely exclusive to each compartment, suggesting that the HPV oncogenes cause gene-expression changes in the stromal microenvironment distinct from those in the epithelium.
Fig. 1.
Epithelial expression of HPV oncogenes alters host gene expression in the stroma and affects the transcriptional response to estrogen. (A) Venn diagrams showing comparative analyses of differentially expressed genes up-regulated (Upper) and down-regulated (Lower) in the K14E6/E7 cervical epithelium (red) and stroma (white) compared with nontransgenic (E6/E7 vs. WT). Genes shared between the epithelium and stroma are shown in pink. (B, Left) Three-way Venn diagram analysis of differentially expressed genes in the stroma of E6/E7 (red), WT+E2 (blue), and E6/E7+E2 (light gray) compared with WT mice. The total number of differentially expressed genes for each given comparison is indicated in parentheses. See Dataset S2 G and H for detailed lists of genes in this comparative analysis. (Right) Pie charts showing the percentage of total genes up-regulated (n = 466) (Upper) and down-regulated (n = 279) Lower) in E6/E7+E2 versus WT stroma that are also differentially expressed in E6/E7 (HPV, red segment), WT+E2 (Estrogen, blue segment), all three groups (HPV and Estrogen, black segment), or are unique to the E6/E7+E2 stroma (Unique, gray segment). (C) The 50 genes with the highest increased (Left) and decreased (Right) fold changes between E6/E7+E2 and WT cervical stroma are shown in black. Gene names are given on the x axis, and fold change is indicated on the y axis. For each gene, the fold changes measured for the WT+E2 versus WT comparison (blue bars) and E6/E7 versus WT comparison (red bars) are superimposed. Genes unique to the E6/E7+E2 cervical stroma lack superimposed blue or red bars.
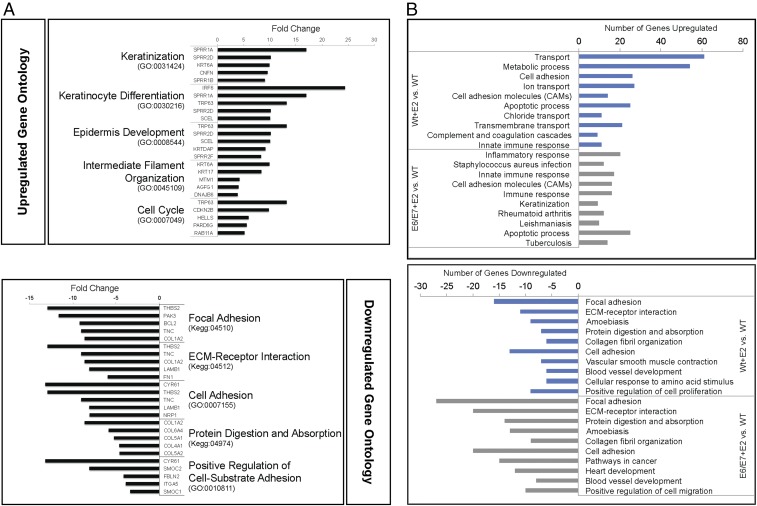
Gene ontology (GO) analysis (Fig. 2A and Table S2) showed that many genes up-regulated in the K14E6/E7 stroma are involved in epithelial processes such as keratinization, keratinocyte differentiation, and epidermis development as well as intermediate filament organization, cell-cycle, metabolic, and apoptotic processes. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with genes up-regulated in the transgenic stroma included the p53 signaling pathway, cell cycle, and DNA replication. Ontology associated with down-regulated genes in the K14E6/E7 stroma highlighted dysregulation of genes involved in stromal tissue architecture, such as cell adhesion, extracellular matrix (ECM) organization, and collagen fibril organization. KEGG pathway analysis reinforced this observation, indicating significant association with focal adhesion, ECM–receptor interaction, and gap junction. We conclude that HPV oncogene expression in the epithelial compartment changes the expression of genes in the microenvironment that are largely involved in both epithelial and stromal tissue dynamics.
Fig. 2.
GO analysis of differentially expressed genes. (A) GO of up-regulated (Upper) and down-regulated (Lower) genes differentially expressed in K14E6/E7 versus nontransgenic stroma. The top five most significant GO groups are shown (for P values, see Table S2) along with GO identifiers in parentheses. The five genes classified in each GO group with the highest fold-change values measured between E6/E7 and WT groups are shown in the bar graphs. (B) GO comparisons of genes differentially expressed in WT+E2 versus WT stroma (blue bars) and E6/E7+E2 versus WT stroma (gray bars). The top 10 most significant GO groups are shown (for more details, see Tables S3 and S4) for the up-regulated genes (Upper) and down-regulated genes (Lower). The number of genes in each GO group that are differentially expressed in each comparison is indicated in the bar graphs.
Table S2.
GO analysis of genes differentially expressed in K14E6/E7 (E6/E7) stroma versus nontransgenic (WT) stroma
| GO identifier | GO description | Count* | Percentage† | P value‡ |
| Up-regulated | ||||
| Biological processes | ||||
| GO:0031424 | Keratinization | 12 | 42.86 | 2.24 E -14 |
| GO:0030216 | Keratinocyte differentiation | 11 | 23.91 | 4.27 E -10 |
| GO:0008544 | Epidermis development | 8 | 24.24 | 3.49 E -07 |
| GO:0045109 | Intermediate filament organization | 6 | 42.86 | 6.81 E -07 |
| GO:0007049 | Cell cycle | 23 | 4.32 | 1.26 E -06 |
| GO:0008152 | Metabolic process | 43 | 2.58 | 2.63 E -06 |
| GO:0006915 | Apoptotic process | 20 | 3.97 | 3.40 E -05 |
| GO:0006810 | Transport | 38 | 2.35 | 1.12 E -04 |
| GO:0018149 | Peptide cross-linking | 5 | 26.32 | 1.13 E -04 |
| GO:0010466 | Negative regulation of peptidase activity | 9 | 8.57 | 1.19 E -04 |
| KEGG pathways | ||||
| KEGG:04115 | p53-signaling pathway | 9 | 13.43 | 3.51 E -06 |
| KEGG:04110 | Cell cycle | 9 | 7.32 | 3.31 E -04 |
| KEGG:03030 | DNA replication | 5 | 14.29 | 1.24 E -03 |
| KEGG:00480 | Glutathione metabolism | 5 | 10.00 | 5.36 E -03 |
| KEGG:05200 | Pathways in cancer | 10 | 3.12 | 4.00 E -02 |
| Down-regulated | ||||
| Biological processes | ||||
| GO:0007155 | Cell adhesion | 38 | 8.44 | 3.74 E -17 |
| GO:0010811 | Positive regulation of cell-substrate adhesion | 11 | 34.38 | 8.16 E -11 |
| GO:0030198 | ECM organization | 15 | 17.65 | 9.42 E -11 |
| GO:0007507 | Heart development | 17 | 10.83 | 6.54 E -09 |
| GO:0030199 | Collagen fibril organization | 9 | 32.14 | 1.23 E -08 |
| GO:0006355 | Regulation of transcription, DNA-dependent | 53 | 3.05 | 5.35 E -07 |
| GO:0030335 | Positive regulation of cell migration | 13 | 11.30 | 6.10 E -07 |
| GO:0070208 | Protein heterotrimerization | 5 | 71.43 | 1.20 E -06 |
| GO:0006351 | Transcription, DNA-dependent | 49 | 3.04 | 2.02 E -06 |
| GO:0045893 | Positive regulation of transcription, DNA-dependent | 24 | 4.71 | 6.04 E -06 |
| KEGG pathways | ||||
| KEGG:04510 | Focal adhesion | 33 | 16.58 | 5.54 E -25 |
| KEGG:04512 | ECM–receptor interaction | 23 | 26.74 | 1.65 E -22 |
| KEGG:04974 | Protein digestion and absorption | 14 | 17.95 | 4.59 E -11 |
| KEGG:05146 | Amoebiasis | 14 | 12.28 | 5.59 E -09 |
| KEGG:05200 | Pathways in cancer | 22 | 6.85 | 5.84 E -09 |
| KEGG:04270 | Vascular smooth muscle contraction | 13 | 11.30 | 5.89 E -08 |
| KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 9 | 12.16 | 8.12 E -06 |
| KEGG:04540 | Gap junction | 9 | 10.47 | 2.59 E -05 |
| KEGG:05215 | Prostate cancer | 9 | 10.11 | 2.77 E -05 |
| KEGG:05414 | Dilated cardiomyopathy | 9 | 10.23 | 2.80 E -05 |
Items listed are restricted to the top 10 most significant GO terms where applicable.
Number of annotated genes in input gene list included in GO term.
Percentage of genes within GO term associated with input gene list.
Corrected hypergeometric P value.
Estrogen Affects Stromal Gene Expression Differently in K14E6/E7 Versus Nontransgenic Mice.
To examine how estrogen affects stromal gene expression, differentially expressed genes were determined by comparing WT+E2 and E6/E7+E2 mice with WT mice. All E6/E7+E2 mice developed squamous cell carcinomas, and these stroma samples therefore represented the tumor microenvironment.
Estrogen treatment up-regulated 461 genes in the WT+E2 stroma and 466 genes in the E6/E7+E2 cervical stroma (Dataset S2 C and E). E6/E7+E2 stroma and WT+E2 stroma shared some Gene Ontology (GO) associations, but a large proportion in the E6/E7+E2 stroma was distinct (Fig. 2B and Tables S3 and S4). For instance, inflammatory response, keratinization, antigen processing and presentation, neutrophil chemotaxis, cellular response to IFN-γ, and NOD-like receptor signaling were significantly associated with up-regulated genes in E6/E7+E2 stroma but not in WT+E2 stroma. Estrogen treatment down-regulated 140 genes in WT+E2 stroma and 279 genes in E6/E7+E2 stroma (Dataset S2 D and F). A large overlap in processes negatively affected by estrogen in the WT+E2 and E6/E7+E2 cervical stroma included GO groups such as focal adhesion, ECM–receptor interaction, collagen fibril organization, and cell adhesion. However, these processes were more significantly associated with down-regulated genes in E6/E7+E2 mice than in the WT+E2 mice (compare Tables S3 and S4). Other GO associations were unique to genes down-regulated in E6/E7+E2 stroma, including cell migration, negative regulation of epithelial cell proliferation, and the TGF-β–signaling pathway. Collectively, the gene-expression patterns indicated that estrogen treatment affected distinct biological processes in the cervical stroma of K14E6/E7 versus nontransgenic mice.
Table S3.
GO analysis of genes differentially expressed in estrogen-treated nontransgenic mice (WT+E2) stroma versus nontransgenic (WT) stroma
| GO identifier | GO description | Count* | Percentage† | P value‡ |
| Up-regulated | ||||
| Biological processes | ||||
| GO:0006810 | Transport | 61 | 3.77 | 1.90 E-13 |
| GO:0008152 | Metabolic process | 54 | 3.25 | 2.19 E-09 |
| GO:0007155 | Cell adhesion | 26 | 5.78 | 5.84 E-09 |
| GO:0006811 | Ion transport | 27 | 4.88 | 7.77 E-08 |
| GO:0006915 | Apoptotic process | 25 | 4.96 | 2.09 E-07 |
| GO:0006821 | Chloride transport | 11 | 13.41 | 4.26 E-07 |
| GO:0055085 | Transmembrane transport | 21 | 4.73 | 7.55 E-06 |
| GO:0045087 | Innate immune response | 11 | 7.75 | 9.94 E-05 |
| GO:0006508 | Proteolysis | 20 | 3.58 | 0.0008 |
| GO:0034220 | Ion transmembrane transport | 15 | 4.53 | 0.0008 |
| KEGG pathways | ||||
| KEGG:04514 | Cell adhesion molecules (CAMs) | 14 | 9.79 | 1.50 E-07 |
| KEGG:04610 | Complement and coagulation cascades | 9 | 11.84 | 1.44 E-05 |
| KEGG:04530 | Tight junction | 10 | 7.58 | 9.98 E-05 |
| KEGG:04972 | Pancreatic secretion | 9 | 8.91 | 0.0001 |
| KEGG:04670 | Leukocyte transendothelial migration | 9 | 7.69 | 0.0002 |
| KEGG:04978 | Mineral absorption | 6 | 13.33 | 0.0003 |
| KEGG:04970 | Salivary secretion | 7 | 9.46 | 0.0004 |
| KEGG:04142 | Lysosome | 8 | 6.56 | 0.0013 |
| KEGG:05150 | Staphylococcus aureus infection | 5 | 10.00 | 0.0040 |
| KEGG:00330 | Arginine and proline metabolism | 5 | 9.26 | 0.0051 |
| Down-regulated | ||||
| Biological processes | ||||
| GO:0030199 | Collagen fibril organization | 6 | 21.43 | 3.72 E-07 |
| GO:0007155 | Cell adhesion | 13 | 2.89 | 2.29 E-06 |
| GO:0001568 | Blood vessel development | 6 | 10.17 | 1.36 E-05 |
| GO:0071230 | Cellular response to amino acid stimulus | 6 | 5.66 | 3.37 E-04 |
| GO:0008284 | Positive regulation of cellular proliferation | 9 | 2.59 | 0.0005 |
| GO:0006813 | Potassium ion transport | 5 | 3.55 | 0.0078 |
| GO:0045893 | Positive regulation of transcription, DNA-dependent | 8 | 1.57 | 0.0115 |
| GO:0043066 | Negative regulation of apoptotic process | 6 | 1.76 | 0.0195 |
| GO:0055085 | Transmembrane transport | 6 | 1.35 | 0.0311 |
| GO:0006811 | Ion transport | 7 | 1.27 | 0.0424 |
| KEGG pathways | ||||
| KEGG:04510 | Focal adhesion | 16 | 8.04 | 1.24 E-15 |
| KEGG:04512 | ECM–receptor interaction | 11 | 12.79 | 4.41 E-13 |
| KEGG:05146 | Amoebiasis | 9 | 7.89 | 6.87 E-09 |
| KEGG:04974 | Protein digestion and absorption | 7 | 8.97 | 2.16 E-07 |
| KEGG:04270 | Vascular smooth muscle contraction | 7 | 6.09 | 2.58 E-06 |
| KEGG:05200 | Pathways in cancer | 7 | 2.18 | 1.77 E-03 |
| KEGG:04080 | Neuroactive ligand–receptor interaction | 5 | 1.81 | 2.00 E-02 |
Items listed are restricted to the top 10 most significant GO terms where applicable.
Number of annotated genes in input gene list included in GO term.
Percentage of genes within GO term associated with input gene list.
Corrected hypergeometric P value.
Table S4.
GO analysis of genes differentially expressed in estrogen-treated K14E6/E7 mice (E6/E7+E2) stroma versus nontransgenic (WT) stroma
| GO identifier | GO description | Count* | Percentage† | P value‡ |
| Up-regulated | ||||
| Biological processes | ||||
| GO:0006954 | Inflammatory response | 20 | 11.17 | 2.16 E -11 |
| GO:0045087 | Innate immune response | 17 | 11.97 | 3.30 E -10 |
| GO:0006955 | Immune response | 16 | 11.35 | 2.47 E -09 |
| GO:0031424 | Keratinization | 9 | 32.14 | 4.60 E -09 |
| GO:0006915 | Apoptotic process | 25 | 4.96 | 1.80 E -07 |
| GO:0008152 | Metabolic process | 47 | 2.82 | 1.24 E -06 |
| GO:0019882 | Antigen processing and presentation | 7 | 26.92 | 2.12 E -06 |
| GO:0030593 | Neutrophil chemotaxis | 7 | 25.93 | 2.20 E -06 |
| GO:0071346 | Cellular response to IFN-γ | 6 | 37.50 | 2.25 E -06 |
| GO:0042832 | Defense response to protozoan | 6 | 35.29 | 2.51 E -06 |
| KEGG pathways | ||||
| KEGG:05150 | S. aureus infection | 12 | 24.00 | 3.77 E -11 |
| KEGG:04514 | Cell adhesion molecules (CAMs) | 16 | 11.19 | 5.12 E -10 |
| KEGG:05323 | Rheumatoid arthritis | 12 | 14.81 | 5.37 E -09 |
| KEGG:05140 | Leishmaniasis | 10 | 15.63 | 8.61 E -08 |
| KEGG:05152 | Tuberculosis | 14 | 8.09 | 3.06 E -07 |
| KEGG:04670 | Leukocyte transendothelial migration | 11 | 9.40 | 1.82 E -06 |
| KEGG:04380 | Osteoclast differentiation | 11 | 9.48 | 1.94 E -06 |
| KEGG:04621 | NOD-like receptor-signaling pathway | 8 | 14.29 | 3.25 E -06 |
| KEGG:04620 | Toll-like receptor-signaling pathway | 10 | 10.00 | 3.45 E -06 |
| KEGG:05160 | Hepatitis C | 11 | 8.09 | 5.92 E -06 |
| Down-regulated | ||||
| Biological processes | ||||
| GO:0030199 | Collagen fibril organization | 9 | 32.14 | 1.03 E -10 |
| GO:0007155 | Cell adhesion | 20 | 4.44 | 8.43 E -09 |
| GO:0007507 | Heart development | 12 | 7.64 | 1.70 E -07 |
| GO:0001568 | Blood vessel development | 8 | 13.56 | 9.04 E -07 |
| GO:0030335 | Positive regulation of cell migration | 10 | 8.70 | 1.06 E -06 |
| GO:0043588 | Skin development | 7 | 16.67 | 1.62 E -06 |
| GO:0071230 | Cellular response to amino acid stimulus | 9 | 8.49 | 3.99 E -06 |
| GO:0016477 | Cell migration | 9 | 7.76 | 6.37 E -06 |
| GO:0001501 | Skeletal system development | 8 | 9.76 | 6.40 E -06 |
| GO:0050680 | Negative regulation of epithelial cell proliferation | 6 | 13.33 | 3.57 E -05 |
| KEGG pathways | ||||
| KEGG:04510 | Focal adhesion | 27 | 13.57 | 8.40 E -26 |
| KEGG:04512 | ECM–receptor interaction | 20 | 23.26 | 3.81 E -24 |
| KEGG:04974 | Protein digestion and absorption | 14 | 17.95 | 3.29 E -15 |
| KEGG:05146 | Amoebiasis | 13 | 11.40 | 1.41 E -11 |
| KEGG:05200 | Pathways in cancer | 15 | 4.67 | 6.44 E -08 |
| KEGG:04270 | Vascular smooth muscle contraction | 9 | 7.83 | 1.08 E -06 |
| KEGG:04060 | Cytokine–cytokine receptor interaction | 11 | 4.51 | 8.81 E -06 |
| KEGG:05222 | Small cell lung cancer | 7 | 8.24 | 1.76 E -05 |
| KEGG:04360 | Axon guidance | 8 | 6.15 | 2.50 E -05 |
| KEGG:04350 | TGF-β–signaling pathway | 6 | 7.14 | 1.89 E -04 |
Items are listed restricted to the top 10 most significant GO terms where applicable.
Number of annotated genes in input gene list included in GO term.
Percentage of genes within GO term associated with input gene list.
Corrected hypergeometric P value.
Interestingly, 40% (184 of 466) of the up-regulated genes and 22% (61 of 279) of the down-regulated genes in the E6/E7+E2 cervical stroma were unique, meaning their differential expression required both E6/E7 and estrogen (Fig. 1B and Dataset S2 G and H). Of the down-regulated genes, 52% (145 of 279) and 5% (13 of 279) were shared exclusively with E6/E7 and WT+E2 conditions, respectively, suggesting that HPV oncogenes drive much of the gene down-regulation in the E6/E7+E2 stroma. We conclude that the HPV oncogenes influence how estrogen affects gene expression in the microenvironment, resulting in a subset of genes responding differently to estrogen in K14E6/E7 versus nontransgenic mice.
To determine the relative magnitude of the effect of the HPV oncogenes or estrogen on gene dysregulation in the E6/E7+E2 cervical stroma, the 50 highest-fold gene-expression changes between the E6/E7+E2 and WT cervical stroma (Fig. 1C, black) were compared with their respective fold changes in E6/E7 (red) and WT+E2 (blue) stroma. These fold changes in the E6/E7+E2 cervical stroma reflected either their unique nature, indicated by genes shown in black that have no overlap with genes in red or blue (e.g., CXCL1, GBP2, HDC) or strong synergy compared with either E6/E7 or estrogen alone (e.g., CXCL5, SPRR2D, IGJ). Conversely, nearly all the top 50 genes most down-regulated in the E6/E7+E2 stroma showed overlap with both factors, particularly with E6/E7. We conclude that the HPV oncogenes and estrogen not only function individually but also cooperate to alter stromal gene expression.
Candidate Paracrine Factors in the Cervical Microenvironment.
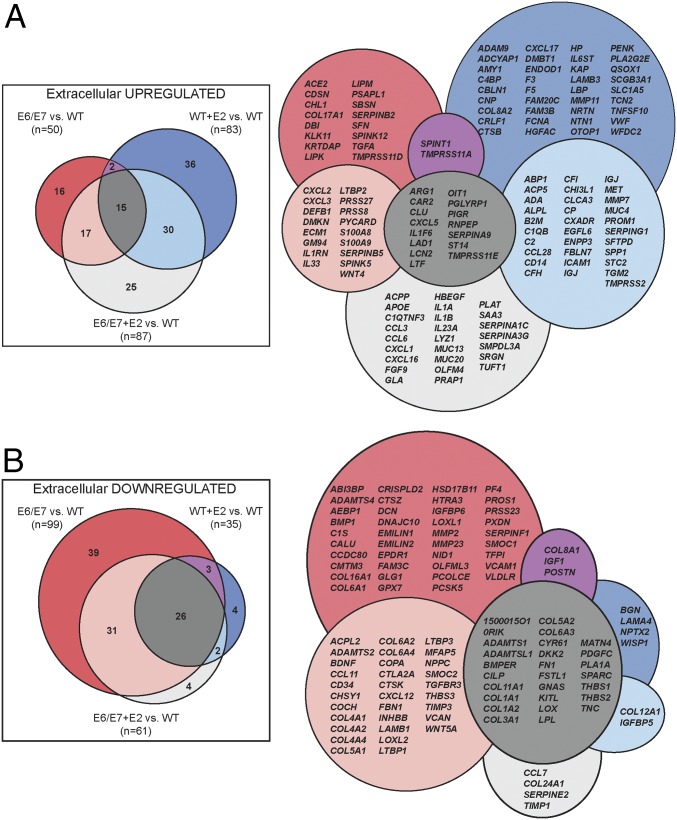
We hypothesized that estrogen induces paracrine factors in the K14E6/E7 stroma to promote cervical carcinogenesis in the adjacent HPV+ epithelia. Therefore, we compared differentially expressed stromal genes ontologically annotated as components of the extracellular space, extracellular region, ECM, and/or proteinaceous ECM (Fig. 3). In E6/E7+E2 cervical stroma, 87 extracellular genes were up-regulated, and 61 genes were down-regulated. Consistent with the unique effects of estrogen on gene expression in the K14E6/E7 stroma, more than 25% (25 of 87) of the up-regulated extracellular genes were unique to the E6/E7+E2 condition. With this list of “extracellular” genes, we thus identified a collection of candidate paracrine factors in the cervical microenvironment regulated by estrogen and/or epithelial HPV oncogene expression.
Fig. 3.
Identification of candidate paracrine factors in the cervical stroma. Overview of analysis of differentially expressed extracellular genes. Significantly differentially expressed genes determined previously for E6/E7, WT+E2, and E6/E7+E2 compared with WT were used as input for cellular component GO analysis. Results that were classified as extracellular (GO:0005578, GO:0005576, GO:0031012, and GO:0005615) were pooled to generate a consolidated list of extracellular genes. The lists of up-regulated (A) and down-regulated (B) extracellular genes were compared using Venn diagrams. The total numbers of significantly differentially expressed extracellular genes are shown in parentheses under each comparison label. The diagrams on the right contain gene symbols corresponding to the Venn diagrams.
Genes Involved in Inflammatory Response Are Increased by Epithelial E6/E7 Expression and Further Enhanced by Estrogen.
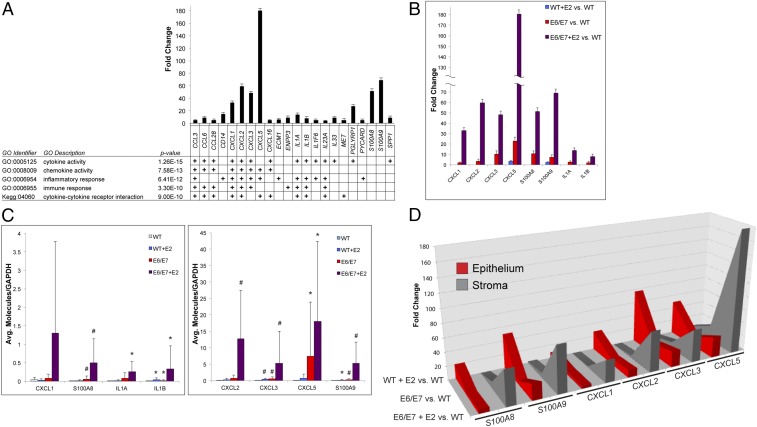
We narrowed our focus to the extracellular genes up-regulated in the E6/E7+E2 stroma, as they could function as positive-acting factors on the nearby epithelia. GO of these genes identified a strong inflammatory and immune response signature (Fig. 4A). The top five most significant GO classes included cytokine activity, chemokine activity, inflammatory response, immune response, and cytokine–cytokine receptor interaction. A core group of genes driving these signatures were proinflammatory cytokines, including CCL3, CCL6, CCL28, CXCL1, CXCL2, CXCL3, CXCL5, IL1A, and IL1B, as well as the S100A8 and S100A9 genes that have well-characterized roles in inflammation and carcinogenesis (Fig. 4A) (35). CXCL1, CXCL2, CXCL3, and CXCL5 were among the genes most up-regulated in the E6/E7+E2 stroma (Dataset S2E). These members of the ELR+ CXC family of chemokines are CXCR2 receptor ligands and comprise a signaling axis involved in several facets of tumorigenesis (36). Their identification as some of the most highly up-regulated genes in the E6/E7+E2 cervical stroma makes them prime candidates for paracrine factors in epithelial–stroma communication.
Fig. 4.
Inflammation-associated gene expression is increased by epithelial E6/E7 expression and is exacerbated by chronic estrogen treatment. (A) GO was performed on the 87 extracellular genes up-regulated in the E6/E7+E2 stroma identified in Fig. 3A. The most significant GO groups associated with these genes are shown. Individual genes driving the significant GO associations are shown; a plus sign denotes that the gene is classified in the corresponding GO group. The fold change of each gene in the E6/E7+E2 stroma compared with WT stroma is shown in the bar graph. Error bars indicate SD. (B) Fold-change values are shown for a representative group of core genes (x axis) in the inflammatory signature identified in A. The fold change of each gene in the stroma of WT+E2 (blue bars), E6/E7 (red bars), and E6/E7+E2 (purple bars) compared with WT stroma is shown. Error bars indicate SD. (C) RT-PCR validation of selected inflammation-associated genes. RNA from laser-captured stroma was used to measure transcript levels of the indicated genes using RT-PCR. Statistical analysis was performed using a Wilcoxon rank-sum test. #P ≤ 0.01, *P ≤ 0.05 compared with WT. Error bars indicate SD. (D) Comparison of fold-change values for selected inflammation-associated genes in the epithelium (red) and stroma (gray) in WT+E2 (rearmost values), E6/E7 (middle values), and E6/E7+E2 (foremost values) compared with WT stroma.
Interestingly, many of the inflammation-associated genes were highly up-regulated in the E6/E7 stroma even in the absence of estrogen, and their expression was further increased by estrogen treatment. Estrogen treatment alone in nontransgenic mice had little, if any, effect on inflammation-associated gene expression in the stroma, although CXCL5 and S100A9 were increased slightly over WT stroma (Fig. 4B). Real-time qPCR specific to several of the inflammation-associated genes performed on independently laser-captured cervical stroma validated these observations (Fig. 4C).
We next compared the effects of HPV oncogenes and estrogen on inflammation-associated gene expression in the cervical epithelium and stroma (Fig. 4D). In WT mice, estrogen treatment had minimal effect on inflammation-associated gene expression in the stroma but significantly up-regulated expression in the epithelia. In E6/E7 mice, inflammation-associated gene expression increased in both the cervical epithelia and surrounding stroma. Expression of inflammation-associated genes in the epithelia of E6/E7+E2 mice increased modestly compared with E6/E7 cervical epithelia but increased dramatically in the stromal compartment. The level of inflammation-associated gene expression was much higher in the E6/E7+E2 stroma than in the adjacent E6/E7+E2 epithelia or in either WT+E2 or E6/E7 stroma. Thus, HPV oncogenes induced the expression of proinflammatory genes in both the epithelial and stromal compartments of the cervix. However, estrogen appeared to function synergistically with epithelial HPV oncogene expression to specifically enhance stromal proinflammatory gene expression.
Chemokine Expression Increases upon Coculture of Primary Human Cervical Cancer Cells with Cervical Fibroblasts.
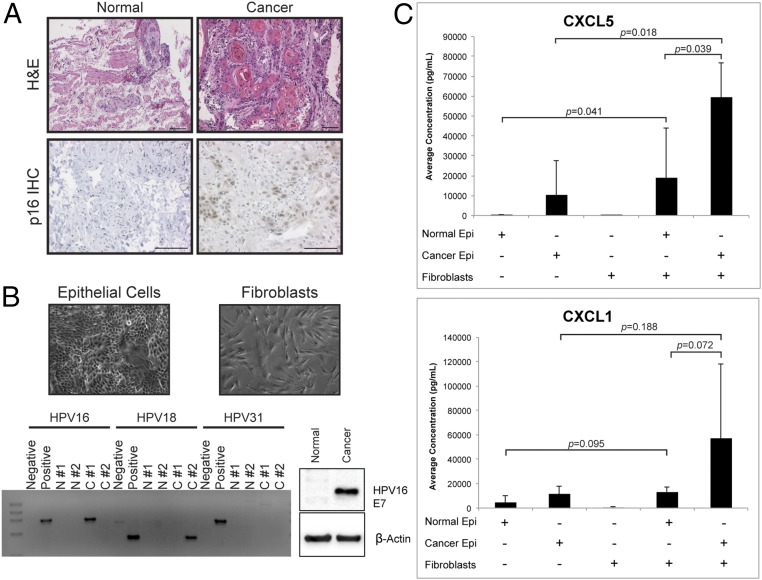
Since stromal chemokine expression in E6/E7+E2 mice increased significantly (Fig. 4), we evaluated whether it was likewise affected in human cervical cancer cells. Fresh normal human cervix and cervical cancers were evaluated for tissue histopathology and immunohistochemistry for p16, a well-validated marker of HPV infection (Fig. 5A). The cancers generally showed histopathological markers of HPV+ cervical carcinomas, such as keratin pearls and p16+ cells, whereas normal cervical samples did not. From these same tissue samples, primary epithelial and fibroblast cell cultures were established (Fig. 5B). HPV-specific PCR and immunoblots verified that normal samples were HPV− and that cancer samples were positive for high-risk HPV16 (cancer tissue no. 1) or HPV18 (cancer tissue no. 2) (Fig. 5B).
Fig. 5.
Chemokine expression increases upon coculture of primary human cervical cancer cells with cervical fibroblasts. (A) Tissue sections of representative primary human normal cervical and cervical cancer samples (normal tissue no. 1 and cancer tissue no. 1) were stained with H&E (Upper) or were used for immunohistochemistry for the human p16 protein (Lower). (Scale bars, 100 μM.) (B) Characterization of primary human cervical cell strains. (Upper) Representative brightfield images of epithelial (Left) and fibroblast (Right) cell strains isolated from primary human cervical samples. (Lower Left) HPV16-, HPV18-, and HPV31-specific PCR analysis of total DNA isolated from primary epithelial cell strains. Negative controls were DNA from untransfected normal cervical epithelial cells. Positive controls were DNA from normal cervical epithelial cells transfected with HPV16, HPV18, or HPV31 genomes. N#1, normal cervical epithelial cells, sample WICVX-1; N#2, normal cervical epithelial cells, sample WICVX-3; C#1, cervical cancer epithelial cells, sample WICVX-2; C#2, cervical cancer epithelial cells, sample no. 6204. (Lower Right) Immunoblot analysis of protein lysates harvested from normal (N#2) and cervical cancer epithelial (C#1) cells probed with an antibody specific to HPV16 E7 protein. β-Actin is shown as a loading control. (C) In vitro coculture experiments and analysis of secreted chemokines. The average concentration (pg/mL) of CXCL5 (Upper) and CXCL1 (Lower) in the CM of monocultured and cocultured cervical normal (normal tissue no. 2) and cancer (cancer tissue no. 2) epithelial cells and fibroblasts measured using a multiplexed bead-based screening assay. Shown are composite data from normal fibroblasts and cancer-associated fibroblasts, as no significant difference was observed between the different sources in either monoculture or coculture. Results represent three independent experiments. Epithelial-only and fibroblast-only concentration values were compared with coculture values using a multiple-experiment statistical test to preserve each experiment structure. Normal coculture and cancer coculture values were compared using a two-sided Wilcoxon rank-sum test. Error bars indicate SD.
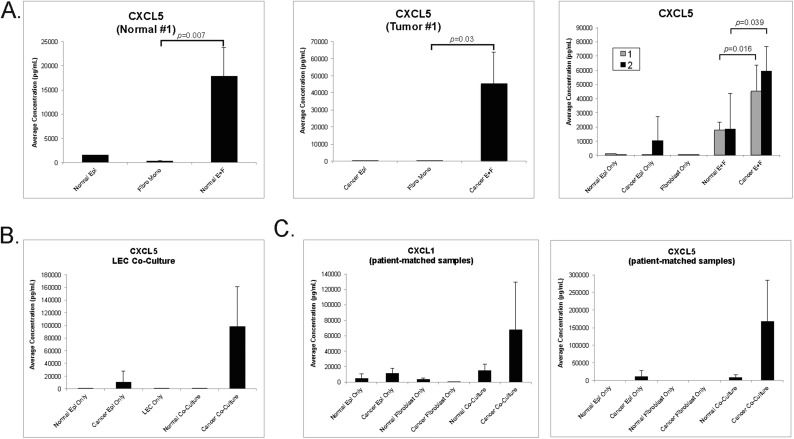
We hypothesized that interaction between HPV+ cervical epithelia and the surrounding microenvironment contributes to the inflammatory signature we observed in mice. To test this, 3D cultures were generated from primary epithelial cells only, primary cervical fibroblasts only, or cocultures of both cell types. After 72 h of culture, levels of two representative chemokines, CXCL1 and CXCL5, were measured in conditioned medium (CM). Results from one representative normal cervical sample and one cervical tumor (normal tissue no. 2 and HPV18+ cancer tissue no. 2 in Fig. 5B) are shown in Fig. 5C. The CXCL5 level in CM from cocultures was significantly higher than in CM from monocultured normal epithelial cells (P = 0.041) and cancer epithelial cells (P = 0.018). Likewise, CXCL5 levels were higher in cocultures containing both normal (P = 0.001) and tumor (P = 9.1 × 10−5) epithelial cells and fibroblasts than in cultures of fibroblasts alone. The CXCL5 concentration was also significantly higher in CM from cocultured cancer epithelial cells and fibroblasts than in CM from cocultured normal epithelial cells and fibroblasts (P = 0.039). An additional comparison of an early-passage normal and a cervical cancer sample (normal tissue no. 1 and cancer tissue no. 1 in Fig. 5B and Fig. S2A) showed similar results. CXCL5 secretion was also increased when cervical cancer epithelial cells were cocultured with human dermal lymphatic endothelial cells (HDLECs) (Fig. S2B), suggesting that, in addition to fibroblasts, other cell types may contribute to the inflammatory signature. The amount of CXCL1 produced from cocultured normal epithelial cells and fibroblasts (P = 0.005) and cancer epithelial cells and fibroblasts (P = 0.0007) was significantly higher than that produced from fibroblast monocultures. The level of CXCL1 produced from cocultured cells was also increased over that from epithelial cell monocultures, but this difference did not reach statistical significance (normal epithelial cell coculture, P = 0.095; cancer cell epithelial cell coculture, P = 0.188) (Fig. 5C). These results indicated that the expression of two representative inflammation-associated chemokines, CXCL5 and CXCL1, increases when early-passage human cervical epithelial cells and fibroblasts are cocultured. Interestingly, levels of CXCL1 and CXCL5 secreted from cocultured cells were much higher when early-passage cancer epithelial cells were cultured with their autologous fibroblasts (Fig. S2C), suggesting an inherent relationship between an individual tumor and its microenvironment that promotes high chemokine secretion. The analysis of primary human cervical cells showed that HPV+ human cervical tumors interact with their surrounding microenvironment to increase inflammation-associated gene expression, as is consistent with the observations in the cervix of K14E6/E7 mice.
Fig. S2.
Chemokine analysis in cocultured primary human cervical cells. (A) Average CXCL5 concentration (pg/mL) measured in the CM of monocultured and cocultured cervical normal (normal tissue no. 1) (Left) and cancer (cancer tissue no.1) (Center) epithelial cells and fibroblasts measured using a multiplexed bead-based screening assay. The sample size of monocultured epithelial cells was insufficient for statistical analysis (n = 1 sample each). A two-sided Wilcoxon rank-sum test was performed to compare the monocultured fibroblasts with cocultured cervical normal (normal E+F; n = 5 replicates each) and cancer epithelial cells (cancer E+F; n = 4 replicates each). (Right) A comparison of two normal and two cervical cancer epithelial samples (normal tissue no. 1 and cancer tissue no. 1 are shown in gray; normal tissue no. 2 and cancer tissue no. 2 are shown in black and are the same as those shown in Fig. 5C). A Wilcoxon rank-sum test was used to compare normal cocultured values with cancer cocultured values (at least n = 4 replicates for each). (B) Average CXCL5 concentration (pg/mL) measured in the CM of monocultured and cocultured cervical normal (normal tissue no. 2) and cancer (cancer tissue no. 2) epithelial cells and lymphatic endothelial cells (LEC) measured using a multiplexed bead-based screening assay. Sample sizes were insufficient for statistical analysis. (C) Analysis of patient-matched epithelial cells and fibroblasts. Normal tissue no. 2 and cancer tissue no. 2 epithelial cells and fibroblasts were monocultured or were cocultured with only patient-matched counterpart cells. CXCL1 and CXCL5 average concentrations were measured in the CM as described previously. Sample size was insufficient for statistical analysis. All error bars in A–C represent SDs for samples in which two or more samples were tested. Where error bars are absent, only one sample was tested. Where P values are given, the sample size in each group was at least four.
Discussion
The activities of the multifunctional HPV oncogenes E6 and E7 (37) are not sufficient for cervical carcinogenesis (11). Previous work identified estrogen as a cocarcinogen that appears to function through signaling in the stroma (9, 10, 21, 27–29). In this study, we show that epithelial expression of the HPV oncogenes dramatically affects gene expression in the cervical stroma of K14E6/E7 transgenic mice and influences the effects of estrogen on gene expression in the microenvironment (Figs. 1 and 2). Thus, HPV oncogenes and estrogen, individually and synergistically, affect gene expression in the murine cervical stroma. We reasoned that HPV-dependent, estrogen-induced paracrine factors in the stroma contribute to cervical carcinogenesis, and we identified differential expression of several candidates in the cervical stroma (Fig. 3). The stroma of estrogen-treated K14E6/E7 mice had a strong inflammatory and immune-related gene-expression signature, specifically for chemokine and cytokine activity (Fig. 4). Primary human cervical cancer epithelial cells and fibroblasts similarly showed increased chemokine expression, but only when cocultured (Fig. 5). Therefore, inflammation-associated chemokines in K14E6/E7 cervical stroma are candidate genes that may function in estrogen-driven epithelial–stromal crosstalk during carcinogenesis. While several studies have analyzed gene expression in cervical cancer epithelial cells (28, 38–40), few have performed such analyses on the cervical stroma (41–43). Our results define potentially important mechanisms through which HPV-infected epithelia and the microenvironment cooperate during cervical cancer development.
The primary objective of our study was to determine mechanisms of estrogen-dependent stroma-to-epithelium signaling. Surprisingly, we discovered additional epithelium-to-stroma signaling. Because we detected (i) low K14E6/E7 transgene transcript levels in the stroma (Fig. S1C), (ii) little overlap in differentially expressed genes between stroma and epithelium (Fig. 1A), and (iii) strong opposing trends in gene-expression patterns, such as for cytokines, between the epithelium and stroma (Fig. 4D), the profound changes in the stroma of K14E6/E7 mice cannot be due to epithelial contamination of laser-captured stroma but rather is due to epithelium-to-stroma signaling between the HPV+ epithelia and the surrounding microenvironment.
At least two nonexclusive hypotheses could explain this paracrine effect. First, the HPV oncogenes might alter the epithelial secretion repertoire in vivo as they do in keratinocytes cultured in organotypic rafts (44), leading to stromal changes. Second, changes in the number and/or content of extracellular vesicles secreted by HPV+ epithelial cells and delivered to stromal cells might alter stromal gene expression. Exosomes, for instance, deliver proteins, RNA, miRNA, and/or DNA to recipient cells to modulate gene expression (45, 46). Virus infections can alter vesicle profiles (47). For example, Epstein–Barr virus–positive cancer cells release exosomes containing viral proteins, miRNAs, and signal transduction molecules that can activate protumorigenic signaling pathways in recipient cells (48, 49). Exosomes from HPV+ keratinocytes contain several antiapoptotic proteins (50), unique miRNAs (51, 52), and even E6 and E7 transcripts (52), suggesting transferable oncogenic potential. Thus, HPV-infected epithelial cells could use exosomes to alter gene expression in the microenvironment to support tumorigenesis. It will be important to determine not only the extent to which HPV+ epithelial cells alter gene expression in a paracrine fashion but also the functions of individual HPV oncogenes in this process and whether these effects are reversible, as has been done previously in epithelial cells (53). Likewise, establishing whether exosomes are involved and which stromal cell types contribute to the differentially expressed gene patterns in the K14E6/E7 cervical stroma will provide further mechanistic insight.
The striking increase in markers for epithelium-associated processes in the K14E6/E7 stroma indicated that HPV16 E6 and E7 induce an epithelium-like stromal signature (Fig. 2A and Table S2). While surprising, epithelium-specific gene expression has been reported in cervical cancer stroma. Gius et al. (42) observed an increase in the transcription of DSG3-encoding desmoglein 3, involved in epithelial cell–cell contacts, in the stromal compartment of human cervical cancers. We did not detect an increase in DSG3, but DSG1 and DSG2 were increased significantly in the K14E6/E7 stroma compared with nontransgenic stroma. Explanations for this epithelial signature in the stroma include the possibility that the HPV oncogenes elicit mesenchymal-to-epithelial transition or epithelial–mesenchymal plasticity in the stroma (54, 55). Another possibility is that HPV oncogenes increase the abundance of normally infrequent cervical mesenchymal stem cells expressing cytokeratins (56). The HPV oncogenes may induce such changes in the stroma to promote tissue regeneration through reepithelialization (57), a process that can occur by reprogramming stromal cells to become epithelial cells (58). This could also reflect a stromal response to the metabolic demands of HPV+ epithelia, as suggested by signatures observed in a previous report (42). Future studies will investigate this epithelial signature and determine whether the HPV oncogenes induce cellular plasticity in the stroma to facilitate neoplastic progression.
The ontology of down-regulated genes in K14E6/E7 stroma was associated with ECM organization and stromal architecture (Fig. 2A and Table S2), driven by fibronectin (FN1), more than 15 collagen genes, tenascin C (TNC), thrombospondin 2 (THBS2), and cysteine-rich protein 61 (CYR61). These data are consistent with the reported association of precancerous lesions in K14HPV16 mice with stromal collagen fibril degradation and extensive ECM remodeling (59). HPV16 E7 inhibits the fibronectin promoter (60), and the HPV oncogenes reduce THBS1 gene expression in human keratinocytes (61). Interestingly, the potent angiogenesis inhibitors THBS1, THBS2, and THBS3 thrombospondins (62) were all down-regulated in K14E6/E7 stroma, suggesting a proangiogenic effect of HPV oncogenes on the microenvironment. However, the proangiogenic CYR61 gene was among the most down-regulated genes in the K14E6/E7 cervical stroma, in contrast to its previously reported increased expression in cultured fibroblasts isolated from the stroma of K14HPV16 transgenic mice and human cervical cancers (29, 41). We postulate that HPV oncogenes reorganize the underlying stroma to accommodate processes such as vasculature reorganization, immune cell recruitment, and epithelial cell invasion. Our results indicate that, in addition to altering the local microenvironment through enzymatic means (63), the HPV oncogenes may also regulate ECM dynamics through transcriptional regulation.
In addition, our results show that the HPV oncogenes condition the stroma to respond differently to estrogen (Figs. 1 B and C and 2). Many differentially expressed genes, particularly those up-regulated, were uniquely expressed in the estrogen-treated K14E6/E7 stroma (Fig. 1B). Moreover, the HPV oncogenes and estrogen synergistically increased the expression of several genes, including proinflammatory chemokines (Figs. 1C and 4), and may cooperate to dysregulate genes involved in collagen/ECM dynamics and focal adhesion in the cervical microenvironment of E6/E7+E2 mice (compare Tables S3 and S4). Therefore, while broadly affecting some pathways, estrogen distinctly affects biological processes in the cervical stroma of K14E6/E7 mice compared with nontransgenic mice (Fig. 2B). We expect that among these HPV-dependent, estrogen-induced stromal genes will be microenvironmental factors critical for cervical carcinogenesis.
Many of the up-regulated extracellular genes in the estrogen-treated K14E6/E7 cervical stroma encode proinflammatory cytokine/chemokines (Figs. 3 and 4), including CCL3, CCL6, CCL28, CXCL1, CXCL2, CXCL3, CXCL5, IL1A, IL1B, S100A8, and S100A9. We confirmed that CXCL1 and CXCL5 are similarly increased in CM of cocultured early-passage human cervical cancer cells and fibroblasts (Fig. 5), suggesting that cell-to-cell signaling elevates these chemokines in human cervical cancer. Cytokines have well-characterized roles in immune cell recruitment, tumor cell proliferation, angiogenesis, and modulation of the tumor microenvironment (36, 64), in large part through their effects on immune and endothelial cells. The cluster of CXCL1, CXCL2, CXCL3, and CXCL5 genes in the inflammation signature are members of the ELR+ C-X-C family of chemokines that bind the CXCR2 receptor. Despite the increase in expression of several CXC ligands in the E6/E7+E2 stroma, we did not observe significant variation in CXCR2 receptor gene expression among various conditions. The CXCL/CXCR2 signaling axis is considered proangiogenic and is implicated in a variety of human cancers (65–70). Strikingly similar to our report here, proinflammatory signatures have been observed in stromal fibroblasts adjacent to prostate cancer (71), melanoma (72), breast cancer (73), and HPV-associated skin and cervical cancers (29, 41).
Inflammation is implicated in several facets of HPV-associated carcinogenesis (74). In K14HPV16 skin, an influx of inflammatory cells was proposed to cause stromal ECM remodeling (75), and inflammation regulates the angiogenic switch (76). Our results contrast with an earlier report by Erez et al. (41), who identified a highly similar proinflammatory signature including CXCR2 ligands in fibroblasts adjacent to dysplastic skin, but not the cervix, of HPV transgenic mice. Conversely, moderate expression of these proinflammatory genes was reported in fibroblasts isolated from human cervical cancers (29). Our results identified inflammation-associated genes as some of the most highly differentially expressed genes in the E6/E7 and E6/E7+E2 cervical stroma and revealed the origins of this response. Our tissue culture-based experiments further support the hypothesis that communication between epithelial cells and multiple stromal cell types, including fibroblasts (Fig. 5) and endothelial cells (Fig. S2B), contributes to proinflammatory gene expression. Therefore, the discordant observations between reports may be due to evaluating a single cell type rather than the microenvironment in its entirety. Nonetheless, our results show that proinflammatory cytokines and chemokines are clear candidate extracellular genes that are induced synergistically by the HPV oncogenes and estrogen in the K14E6/E7 stroma. There is evidence that prostate epithelial cell-derived IL-1 can induce the expression of several of the same CXCR2 ligands in prostate stromal cells (71). Interestingly, the IL-1 gene was significantly increased in the K14E6/E7 cervical epithelia in our study, and this factor could therefore be part of a mechanism that induces proinflammatory gene expression in the cervical stroma. Antiestrogen drugs reduced the expression of several genes in the proinflammatory signature in human cervical cancer-associated fibroblasts (29), suggesting that estrogen signaling may also contribute to chemokine expression. It will be interesting to determine the contribution of proinflammatory chemokines to cervical cancer development in estrogen-treated K14E6/E7 mice and how their stromal expression correlates with cervical cancer regression and recurrence following antiestrogen therapy (23, 77).
Additional candidate paracrine factors unique to the estrogen-treated K14E6/E7 cervical stroma were identified (Fig. 3), including IL1A and IL1B, fibroblast growth factor 9 (FGF9), and heparin-binding epidermal growth factor-like growth factor (HBEGF). IL-1α, a proinflammatory cytokine, has been suggested to provide a selective growth advantage to HPV+ epithelia (78). Secreted FGF9 is highly induced in gastric cancer-associated fibroblasts and promotes invasion and antiapoptotic mechanisms in gastric cancer epithelial cells (79). Moreover, estrogen and FGF9 synergize to increase the number of breast cancer stem cells (80). HBEGF, an EGF receptor (EGFR) ligand, is detected in and produced by stromal fibroblasts adjacent to cervical cancers (29, 81). Such fibroblasts increase cervical cancer epithelial cell proliferation in an HBEGF-dependent manner (81). Antiestrogen drugs reduce HBEGF gene expression in cervical cancer-associated fibroblasts, suggesting that estrogen promotes its induction (29).
In conclusion, using our in vivo mouse model of HPV-associated cervical cancer, we have shown that the HPV oncogenes and estrogen dramatically alter host gene expression in the cervical stroma. With the extensive prior results supporting stromal-to-epithelium signaling (27, 28), this reveals that the epithelial–stromal crosstalk in cervical carcinogenesis is bidirectional. These results provide a foundation for future investigations in several important facets of the progression and maintenance of cervical cancers and potentially other HPV+ cancers. This includes further focus on key paracrine factors and molecular pathways implicated here; their effects on angiogenesis, immune cell recruitment, and so forth; and the sequence of these events. Since our principal aim was to analyze genome-wide gene expression in the tumor microenvironment of HPV+, estrogen-driven cervical cancer, the current study analyzed time points soon after cancers appeared, i.e., after 6-mo estrogen treatment of K14E6/E7 mice. Our recent preliminary evidence from 1-mo estrogen treatment of K14E6/E7 mice indicates that at least some of the changes in stromal gene expression observed here are already apparent well before the appearance of cancers. Thus, in the future it will be useful to analyze intermediate estrogen treatment times to more fully define the order of stromal and epithelial changes that accompany neoplastic progression. Such studies should further illuminate how HPV oncogenes and estrogen interact across tissue compartments to drive progression and maintenance of cervical cancer and further assist the identification of therapeutic interventions.
Materials and Methods
Animals, Treatment, and Tissue Processing.
K14E6/K14E7 (referred to as K14E6/E7 herein) bitransgenic mouse lines maintained on the FVB/N genetic background have been described previously (3, 82). Mice were housed and treated in the American Association of Laboratory Animal Care-approved Wisconsin Institute for Medical Research Vivarium of the University of Wisconsin School of Medicine and Public Health (Madison, WI) according to a protocol approved by the University of Wisconsin Institutional Animal Care and Use Committee. Nontransgenic FVB/N and K14E6/E7 transgenic female mice were either untreated or treated with exogenous estrogen (17β-estradiol). Frozen reproductive tracts were cryosectioned and used for histopathological analysis to assign lesion location and grade. Regions of interest (ROIs) in the epithelial or stromal compartments were sampled using LCM.
RNA Processing and Microarray-Based Gene-Expression Analysis.
Comprehensive mRNA levels were measured essentially as described previously (40). Briefly, RNA was extracted from laser-captured material and used to generate T7 RNA polymerase promoter-linked, oligo(dT)-primed double-stranded cDNA, which was then used as a template to produce T7 transcripts complementary to all mRNAs (cRNA). These T7 transcripts were then subjected to a second round of cDNA synthesis and T7 RNA polymerase-based amplification. The resulting cRNA was hybridized on Affymetrix Mouse Genome430 2.0 microarrays.
Biostatistics and GO.
Robust microarray analysis (RMA)-normalized data were used as input, and analysis was performed using the MeV microarray analysis suite (83). Rank product analysis was performed to compare gene expression between pairs of sample groups. The resulting P values were corrected for multiple analyses using the Benjamini–Hochberg method, which sets the FDR. Statistical cutoffs were set at twofold changes in gene expression and FDR ≤0.05 unless otherwise stated. The GeneCodis program was used for ontology analysis (84–86), using significantly differentially expressed genes as input. All Venn diagrams were created using BioVenn (87).
Coculture Experiments in Early-Passage Cell Strains.
The use of human tissue samples deidentified before receipt was determined to be exempt from the need for Institutional Review Board (IRB) approval and informed consent. Primary human cervical epithelial and fibroblast cell strains were isolated from cervical normal and tumor tissues within 24–48 h of surgical resection. Epithelial cells were cultured in F medium plus 10 μM ROCK inhibitor Y-27632, a Rho kinase inhibitor that extends the life span of keratinocytes (88), with a feeder layer of mitomycin C-treated murine fibroblast J2 3T3 cells. Fibroblasts were cultured in F12 Ham’s medium supplemented with 10% FBS and 1% penicillin/streptomycin (P/S). For all in vitro experiments, derived epithelial cell strains were used before six passages, and fibroblast cell strains were used before 12 passages. hTert-immortalized HDLECs (hTert-HDLECs) have been described previously (89). 3D monocultures or cocultures were generated with fibroblasts embedded in collagen and with epithelial cells layered on top of the collagen. All cultures were maintained in F medium without ROCK inhibitor. At 72 h, CM was assayed for several analytes using a magnetic bead-based multiplex assay.
For more detailed materials and methods, please refer to SI Materials and Methods.
SI Materials and Methods
Animal Care and Estrogen Treatment.
K14E6/K14E7 (referred to as “K14E6/E7” herein) bitransgenic mouse lines maintained on the FVB/N genetic background have been described previously (3, 82). At 4–6 wk of age, nontransgenic FVB/N and K14E6/E7 female mice were anesthetized with 5% isoflurane for s.c. insertion of a continuous-release estrogen (E2) tablet (17β-estradiol; 0.05 mg/60 d) (Innovative Research of America) into the shoulder fat pads. New tablets were inserted every 2 mo as needed. Nontransgenic mice were treated for 1 mo with estrogen to induce hyperplasia, and K14E6/E7 mice were treated for an additional 5 mo to induce cervical cancer (10, 90). Mice were housed and treated in the American Association of Laboratory Animal Care-approved Wisconsin Institute for Medical Research Vivarium of the University of Wisconsin School of Medicine and Public Health (Madison, WI), according to a protocol approved by the University of Wisconsin Institutional Animal Care and Use Committee.
Cryosectioning, Histology, and LCM.
Frozen reproductive tracts embedded in Tissue-Tek optimum cutting temperature (O.C.T.) compound (Sakura) were cryosectioned to provide a series of 7-µm and 14-µm sections. One 7-µm section was H&E stained and used for histopathological analysis to assign lesion location and grade. Adjacent sections were lightly hematoxylin-stained and dehydrated through increasing ethanol concentrations. A PixCell II Laser Capture Microscope (Applied Biosystems/Arcturus) was then used to sample epithelial or stromal compartments.
RNA Extraction, Amplification, Labeling, and Microarray-Based Gene-Expression Analysis.
Comprehensive mRNA levels were measured essentially as described previously (40). Briefly, RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s instructions and was used to generate T7 RNA polymerase promoter-linked, oligo(dT)-primed double-stranded cDNA, which was then used as a template to produce T7 cRNA transcripts. These T7 transcripts were then subjected to a second round of cDNA synthesis and T7 RNA polymerase-based amplification. The resulting cRNA was hybridized on Affymetrix MouseGenome430 2.0 microarrays (Affymetrix).
Biostatistics.
RMA was performed on the resulting Affymetrix data. RMA-normalized data were used as input, and analysis was performed using MeV (mev.tm4.org/). Rank product analysis was performed to compare gene expression between pairs of sample groups. The resulting P values were corrected for multiple analyses using the Benjamini–Hochberg method, which sets the FDR. Statistical cutoffs were set at twofold changes in gene expression and a FDR ≤0.05 unless otherwise stated.
Data Access.
A complete set of Affymetrix CEL files and GC-RMA–normalized gene-expression values for specimens was deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession no. GSE102232. Individual sample accession numbers are listed in Table S1.
GO.
The GeneCodis program was used for ontology analysis (84–86). Significantly differentially expressed genes (fold change >2, corrected P value ≤ 0.05) as determined by rank product analysis were used as input, and up-regulated and down-regulated genes were analyzed separately to gauge directional GO associations. Results were parsed to include only GO terms with at least five genes and P value ≤ 0.05.
Gene-Expression Measurement by Real-Time qPCR.
Additional cryosections were used to independently validate the Affymetrix microarray-based mRNA-profiling data by real-time qPCR for a small subset of genes. Using the criteria and procedures describe above, RNA was extracted from laser-captured cells and used for cDNA synthesis, T7 RNA polymerase-based linear amplification, and subsequent second-round cDNA synthesis. Using TaqMan technology-based real-time qPCR with a Bio-Rad CFX96 real-time qPCR thermocycler and SsoFast Probes Supermix reagents according to the manufacturer’s instructions, we used 1-µL aliquots of 20-µL final cDNA preparations to determine the cDNA levels of 10 mRNAs: KRT14, CXCL1, CXCL2, CXCL3, CXCL5, IL1A, IL1B, S100A8, S100A9, and the HPVE6/E7/KRT14 virus-mouse chimeric transcript. Levels of ACTB and GAPDH mRNAs were measured for normalization. Reference measurements to generate standard curves were obtained using serial dilutions of synthetic DNA fragments (gBLOCKs; Integrated DNA Technologies) containing primer/probe set target sequences for each of the mRNAs. The following primers were used in analysis: K14E6/E7 transcript (F: 5′-ACCTGTTAATGGGCACACTAG-3′ and R: 5′-TCCAGCTGTGAAGTGCTTG-3′); CXCL1 (F: 5′-GAGACCACTAAGTGTCAACCAC-3′ and R: 5′-CACACATGTCCTCACCCTAATAC-3′); CXCL2 (F: 5′-GGTTTCAGT GTTTGTAAACTGTATG-3′ and R: 5′-GTGCCTTACGAGGAAGACATAA-3′); CXCL3 (F: 5′-TTTGGTGGCAGCTGTGATAG-3′ and R: 5′-TGTGACACCGTAAGACCATTTC-3′); CXCL5 (F: 5′-CCTGGGCTGGCATATAACTT-3′ and R: 5′-AGATGAGAACACATTGATCCTTCTA-3′); IL1A (F: 5′-CCACGAAGCTCTCTGTACATTC-3′ and R: 5′-GCTTTAAGGATGGGAGGGAAA-3′); IL1B (F: 5′-GGTACATCAGCACCTCACAA-3′ and R: 5′-TTAGAAACAGTCCAGCCCATAC-3′); S100A8 (F: 5′-GTGACAATGCCGTCTGAACT-3′ and R: 5′-GGGCATGGTGATTTCCTTGTA-3′); and S100A9 (F: 5′-GAGGAGTGTATGATGCTGATGG-3′ and R: 5′-GTCACATGGCTGACCTCTTAAT-3′).
Early-Passage Cell Strains and Cell Culture.
Primary human cervical epithelial and fibroblast cell strains were isolated from normal tissue no. 1 (WICVX-1), normal tissue no. 2 (WICVX-3), and cancer tissue no. 1 (HPV16+ WICVX-2). Human cervical tissue samples were kindly provided by J.S.R. Cancer tissue no. 2 (no. 6204; HPV18+ cervical adenocarcinoma) and samples from which three primary fibroblast strains (no. 5559, no. 5602, and no. 6204) were derived were procured from the National Disease Research Interchange. Within 24–48 h of surgical resection, tissues were incubated overnight at 4 °C with P/S, gentamycin, fungizone, and 1 unit/mL dispase. Epithelial layers were peeled from digested tissue, and the remaining tissue was used for fibroblast isolation. Both cell preparations were minced; then the epithelial portion was treated with 0.25% trypsin at 37 °C, and the stromal portion was treated with 0.35% collagenase at 37 °C. The epithelial cell preparation was cultured in F medium + ROCK inhibitor, composed of three parts F12 Ham’s medium to one part DMEM supplemented with 5% FBS, 24 μg/mL adenine, 8.4 ng/mL cholera toxin, 10 ng/mL epidermal growth factor, 2.4 μg/mL hydrocortisone, 5 μg/mL insulin, P/S, and 10 μM ROCK inhibitor Y-27632, a Rho kinase inhibitor that extends the life span of keratinocytes (no. S1049; Selleck Chemicals) (88). The fibroblast cell preparation was resuspended in F12 Ham’s medium supplemented with 10% FBS and 1% P/S. Epithelial cell colonies were pooled and cultured with a feeder layer of mitomycin C-treated murine fibroblast J2 3T3 cells. For all in vitro experiments, derived epithelial cell strains were used before six passages, and fibroblast cell strains were used before 12 passages. hTert-HDLECs have been described previously (89).
Immunohistochemistry.
Portions of primary human tissue samples were fixed in 4% paraformaldehyde and were paraffin-embedded, and 5-μM sections were cut from the tissue blocks. Sections were deparaffinized and rehydrated with xylenes and graded ethanol, respectively. Endogenous peroxidase activity was quenched with 3% H2O2 in methanol followed by heat-induced antigen retrieval in 10 mM citrate buffer (pH 6.0). Immunohistochemistry used anti-p16INK4A (p16) mouse monoclonal antibody (CINtec p16, clone E6H4; Ventana Medical Systems, Inc.), and biotinylated horse anti-mouse IgG (Vector Laboratories) as a secondary antibody. Proteins were visualized using 3,3′-diaminobenzidine (Vector Laboratories), and tissues were counterstained with hematoxylin. Images were acquired on a Zeiss Axio Imager M2 microscope using AxioVision software version 4.8.2.
Immunoblotting.
Protein concentrations of lysates from primary cell strains were determined using Bio-Rad Protein Assay reagent (Bio-Rad). Equivalent amounts of protein were resolved in precast Mini-PROTEAN TGX Any kD or Mini-PROTEAN TGX 4–20% gradient gels (Bio-Rad) and were transferred to a 0.45-μM nitrocellulose membrane (Whatman Protran BA85; GE Healthcare). Following transfer, membranes were blocked with 5% nonfat dry milk in TBS-BGT (Tris-buffered saline containing BSA and glycine supplemented with 0.1% Tween-20). Proteins were detected using anti–β-actin (1:5,000 dilution) (Sigma) and anti-HPV16 E7 (1:500 dilution) (Santa Cruz Biotechnology). Horseradish peroxidase-conjugated secondary antibodies (1:10,000) (Jackson ImmunoResearch) and chemiluminescent substrates (Clarity ECL Substrates; Bio-Rad) were used to visualize immune complexes on a Bio-Rad ChemiDoc Imaging System.
HPV Genotyping.
DNA was isolated using a Qiagen DNeasy Blood and Tissue kit (Qiagen). Equivalent amounts of DNA were analyzed by PCR for high-risk mucosotropic HPV types using primers specific for HPV16 (709-1: 5′-CCCGGATCCTACCTGCAGGATCAGCCATG-3′ and 709-4: 5′-GGCGGATCCTTTTATGCACCAAAAGAGAACTG-3′), HPV18 (MG#3: 5′-CGGTTGCAGCACGAATGGC-3′ and HPV18E6F: 5′-GGTCGGGACCGAAAACGG-3′), and HPV31 (HPV31E6F: 5′-CTGCCAAGGTTGTGTCATGC-3′ and HPV31E6R: 5′-CCTCCTCATCTGAGCTGTCG-3′). PCR products were resolved on a 1% agarose gel and visualized with ethidium bromide.
Analyte Measurements in Coculture CM.
For monocultures, fibroblasts were embedded in 3 mg/mL rat-tail collagen (Wako Chemicals), and epithelial cells were cultured on top of collagen. For cocultures, cells were cultured at a fibroblast:epithelial cell ratio of 4,000:20,000 with fibroblasts embedded in collagen and epithelial cells layered on top of the collagen. All cultures were maintained in F medium without ROCK inhibitor. At 72 h, CM from three replicate cultures were pooled. Undiluted, clarified CM (50 μL) was used to assay several analytes using a magnetic bead-based multiplex assay (R&D Systems) following the manufacturer’s protocol. Prepared samples were analyzed in a 96-well plate using a MagPix instrument (Luminex Corporation) and xPONENT software (Luminex Corporation). To compare analyte concentrations in epithelial or fibroblast monocultures with those in cocultured epithelial cells and fibroblasts, at least three independent biological replicates were compared using a two-sided multiple-experiment permutation test. To compare chemokine concentrations in normal epithelial cells cocultured with fibroblasts versus tumor epithelial cells cocultured with fibroblasts, a two-sided Wilcoxon rank-sum test was performed. Results were considered significant when P values were ≤0.05. Statistical analyses used the MSTAT statistical software version 6.3.1 (https://mcardle.wisc.edu/mstat/; last accessed August 11, 2017).
Supplementary Material
Acknowledgments
We thank Craig Woodworth for providing technical guidance with primary cell isolation. We acknowledge the use of tissues procured by the National Disease Research Interchange (NDRI) with support from NIH Grant U42OD11158. This study was supported by NIH Grants CA022443 (to P.F.L.) and CA211246 (to M.E.S.), University of Wisconsin Carbone Cancer Center Cancer Center Support Grant P30 CA014520 (to D.J.B.), a Morgridge Institute for Research Postdoctoral Fellowship (to O.F.), and a Wisconsin Partnership Program Collaborative Health Sciences grant. P.A. is an investigator of the Howard Hughes Medical Institute and the Morgridge Institute for Research.
Footnotes
Conflict of interest statement: D.J.B. holds equity in Bellbrook Labs LLC, Tasso Inc., Stacks to the Future LLC, Lynx Biosciences LLC, Onexio Biosystems LLC, and Salus Discovery LLC. D.D. is a member of the external advisory board for the University of Wisconsin Cancer Center, for which he receives an honorarium. He is not an active collaborator with any of the authors.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (accession no. GSE102232).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712018114/-/DCSupplemental.
References
- 1.de Martel C, et al. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 2.zur Hausen H. Papillomaviruses in the causation of human cancers–A brief historical account. Virology. 2009;384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 3.Herber R, Liem A, Pitot H, Lambert PF. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J Virol. 1996;70:1873–1881. doi: 10.1128/jvi.70.3.1873-1881.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song S, Pitot HC, Lambert PF. The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J Virol. 1999;73:5887–5893. doi: 10.1128/jvi.73.7.5887-5893.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert PF, et al. Epidermal cancer associated with expression of human papillomavirus type 16 E6 and E7 oncogenes in the skin of transgenic mice. Proc Natl Acad Sci USA. 1993;90:5583–5587. doi: 10.1073/pnas.90.12.5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stelzer MK, et al. A mouse model for human anal cancer. Cancer Prev Res (Phila) 2010;3:1534–1541. doi: 10.1158/1940-6207.CAPR-10-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jabbar S, Strati K, Shin MK, Pitot HC, Lambert PF. Human papillomavirus type 16 E6 and E7 oncoproteins act synergistically to cause head and neck cancer in mice. Virology. 2010;407:60–67. doi: 10.1016/j.virol.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strati K, Pitot HC, Lambert PF. Identification of biomarkers that distinguish human papillomavirus (HPV)-positive versus HPV-negative head and neck cancers in a mouse model. Proc Natl Acad Sci USA. 2006;103:14152–14157. doi: 10.1073/pnas.0606698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brake T, Lambert PF. Estrogen contributes to the onset, persistence, and malignant progression of cervical cancer in a human papillomavirus-transgenic mouse model. Proc Natl Acad Sci USA. 2005;102:2490–2495. doi: 10.1073/pnas.0409883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riley RR, et al. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res. 2003;63:4862–4871. [PubMed] [Google Scholar]
- 11.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 12.Walboomers JM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 13.Bronowicka-Klys DE, Lianeri M, Jagodzinski PP. The role and impact of estrogens and xenoestrogen on the development of cervical cancer. Biomed Pharmacother. 2016;84:1945–1953. doi: 10.1016/j.biopha.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Chung SH, Franceschi S, Lambert PF. Estrogen and ERalpha: Culprits in cervical cancer? Trends Endocrinol Metab. 2010;21:504–511. doi: 10.1016/j.tem.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinaldi S, et al. Endogenous sex steroids and risk of cervical carcinoma: Results from the EPIC study. Cancer Epidemiol Biomarkers Prev. 2011;20:2532–2540. doi: 10.1158/1055-9965.EPI-11-0753. [DOI] [PubMed] [Google Scholar]
- 16.Salazar EL, Mercado E, Sojo I, Salcedo M. Relationship between estradiol 16 alpha-hydroxylation and human papillomavirus infection in cervical cell transformation. Gynecol Endocrinol. 2001;15:335–340. [PubMed] [Google Scholar]
- 17.Appleby P, et al. International Collaboration of Epidemiological Studies of Cervical Cancer Cervical cancer and hormonal contraceptives: Collaborative reanalysis of individual data for 16,573 women with cervical cancer and 35,509 women without cervical cancer from 24 epidemiological studies. Lancet. 2007;370:1609–1621. doi: 10.1016/S0140-6736(07)61684-5. [DOI] [PubMed] [Google Scholar]
- 18.Luhn P, et al. The role of co-factors in the progression from human papillomavirus infection to cervical cancer. Gynecol Oncol. 2013;128:265–270. doi: 10.1016/j.ygyno.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreno V, et al. International Agency for Research on Cancer Multicentric Cervical Cancer Study Group Effect of oral contraceptives on risk of cervical cancer in women with human papillomavirus infection: The IARC multicentric case-control study. Lancet. 2002;359:1085–1092. doi: 10.1016/S0140-6736(02)08150-3. [DOI] [PubMed] [Google Scholar]
- 20.Muñoz N, et al. International Agency for Research on Cancer Multicentric Cervical Cancer Study Group Role of parity and human papillomavirus in cervical cancer: The IARC multicentric case-control study. Lancet. 2002;359:1093–1101. doi: 10.1016/S0140-6736(02)08151-5. [DOI] [PubMed] [Google Scholar]
- 21.Arbeit JM, Howley PM, Hanahan D. Chronic estrogen-induced cervical and vaginal squamous carcinogenesis in human papillomavirus type 16 transgenic mice. Proc Natl Acad Sci USA. 1996;93:2930–2935. doi: 10.1073/pnas.93.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung SH, Wiedmeyer K, Shai A, Korach KS, Lambert PF. Requirement for estrogen receptor alpha in a mouse model for human papillomavirus-associated cervical cancer. Cancer Res. 2008;68:9928–9934. doi: 10.1158/0008-5472.CAN-08-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung SH, Lambert PF. Prevention and treatment of cervical cancer in mice using estrogen receptor antagonists. Proc Natl Acad Sci USA. 2009;106:19467–19472. doi: 10.1073/pnas.0911436106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo YA, et al. Progesterone signaling inhibits cervical carcinogenesis in mice. Am J Pathol. 2013;183:1679–1687. doi: 10.1016/j.ajpath.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castle PE. Do selective estrogen receptor modulators treat cervical precancer and cancer? Time to pool data from relevant trials. Int J Cancer. 2011;128:997–998. doi: 10.1002/ijc.25393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh CJ, Hong MK, Chen PC, Wang JH, Chu TY. Antiestrogen use reduces risk of cervical neoplasia in breast cancer patients: A population-based study. Oncotarget. 2017;8:29361–29369. doi: 10.18632/oncotarget.12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung SH, Shin MK, Korach KS, Lambert PF. Requirement for stromal estrogen receptor alpha in cervical neoplasia. Horm Cancer. 2013;4:50–59. doi: 10.1007/s12672-012-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.den Boon JA, et al. Molecular transitions from papillomavirus infection to cervical precancer and cancer: Role of stromal estrogen receptor signaling. Proc Natl Acad Sci USA. 2015;112:E3255–E3264. doi: 10.1073/pnas.1509322112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar MM, et al. Role of estrogen receptor alpha in human cervical cancer-associated fibroblasts: A transcriptomic study. Tumour Biol. 2016;37:4409–4420. doi: 10.1007/s13277-015-4257-6. [DOI] [PubMed] [Google Scholar]
- 30.Cooke PS, et al. Stromal estrogen receptors mediate mitogenic effects of estradiol on uterine epithelium. Proc Natl Acad Sci USA. 1997;94:6535–6540. doi: 10.1073/pnas.94.12.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cunha GR. Stromal induction and specification of morphogenesis and cytodifferentiation of the epithelia of the mullerian ducts and urogenital sinus during development of the uterus and vagina in mice. J Exp Zool. 1976;196:361–370. doi: 10.1002/jez.1401960310. [DOI] [PubMed] [Google Scholar]
- 32.Cunha GR, Cooke PS, Kurita T. Role of stromal-epithelial interactions in hormonal responses. Arch Histol Cytol. 2004;67:417–434. doi: 10.1679/aohc.67.417. [DOI] [PubMed] [Google Scholar]
- 33.Kurita T, Cooke PS, Cunha GR. Epithelial-stromal tissue interaction in paramesonephric (müllerian) epithelial differentiation. Dev Biol. 2001;240:194–211. doi: 10.1006/dbio.2001.0458. [DOI] [PubMed] [Google Scholar]
- 34.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srikrishna G. S100A8 and S100A9: New insights into their roles in malignancy. J Innate Immun. 2012;4:31–40. doi: 10.1159/000330095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chow MT, Luster AD. Chemokines in cancer. Cancer Immunol Res. 2014;2:1125–1131. doi: 10.1158/2326-6066.CIR-14-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLaughlin-Drubin ME, Münger K. Oncogenic activities of human papillomaviruses. Virus Res. 2009;143:195–208. doi: 10.1016/j.virusres.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibarra Sierra E, et al. Differential gene expression between skin and cervix induced by the E7 oncoprotein in a transgenic mouse model. Virology. 2012;433:337–345. doi: 10.1016/j.virol.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendoza-Villanueva D, et al. Gene expression profile of cervical and skin tissues from human papillomavirus type 16 E6 transgenic mice. BMC Cancer. 2008;8:347. doi: 10.1186/1471-2407-8-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pyeon D, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell. 2010;17:135–147, and erratum (2010) 17:523. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 42.Gius D, et al. Profiling microdissected epithelium and stroma to model genomic signatures for cervical carcinogenesis accommodating for covariates. Cancer Res. 2007;67:7113–7123. doi: 10.1158/0008-5472.CAN-07-0260. [DOI] [PubMed] [Google Scholar]
- 43.Pietras K, Pahler J, Bergers G, Hanahan D. Functions of paracrine PDGF signaling in the proangiogenic tumor stroma revealed by pharmacological targeting. PLoS Med. 2008;5:e19. doi: 10.1371/journal.pmed.0050019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pickard A, et al. HPV16 down-regulates the insulin-like growth factor binding protein 2 to promote epithelial invasion in organotypic cultures. PLoS Pathog. 2015;11:e1004988. doi: 10.1371/journal.ppat.1004988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 46.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schorey JS, Harding CV. Extracellular vesicles and infectious diseases: New complexity to an old story. J Clin Invest. 2016;126:1181–1189. doi: 10.1172/JCI81132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meckes DG, Jr, et al. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci USA. 2010;107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meckes DG, Jr, et al. Modulation of B-cell exosome proteins by gamma herpesvirus infection. Proc Natl Acad Sci USA. 2013;110:E2925–E2933. doi: 10.1073/pnas.1303906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Honegger A, Leitz J, Bulkescher J, Hoppe-Seyler K, Hoppe-Seyler F. Silencing of human papillomavirus (HPV) E6/E7 oncogene expression affects both the contents and the amounts of extracellular microvesicles released from HPV-positive cancer cells. Int J Cancer. 2013;133:1631–1642. doi: 10.1002/ijc.28164. [DOI] [PubMed] [Google Scholar]
- 51.Honegger A, et al. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog. 2015;11:e1004712. doi: 10.1371/journal.ppat.1004712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiantore MV, et al. Human papillomavirus E6 and E7 oncoproteins affect the expression of cancer-related microRNAs: Additional evidence in HPV-induced tumorigenesis. J Cancer Res Clin Oncol. 2016;142:1751–1763. doi: 10.1007/s00432-016-2189-1. [DOI] [PubMed] [Google Scholar]
- 53.Johung K, Goodwin EC, DiMaio D. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J Virol. 2007;81:2102–2116. doi: 10.1128/JVI.02348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: A central regulator of cancer progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eiró N, et al. Potential therapeutic effect of the secretome from human uterine cervical stem cells against both cancer and stromal cells compared with adipose tissue stem cells. Oncotarget. 2014;5:10692–10708. doi: 10.18632/oncotarget.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valencia C, et al. Human papillomavirus E6/E7 oncogenes promote mouse ear regeneration by increasing the rate of wound re-epithelization and epidermal growth. J Invest Dermatol. 2008;128:2894–2903. doi: 10.1038/jid.2008.156. [DOI] [PubMed] [Google Scholar]
- 58.Stappenbeck TS, Miyoshi H. The role of stromal stem cells in tissue regeneration and wound repair. Science. 2009;324:1666–1669. doi: 10.1126/science.1172687. [DOI] [PubMed] [Google Scholar]
- 59.Coussens LM, Hanahan D, Arbeit JM. Genetic predisposition and parameters of malignant progression in K14-HPV16 transgenic mice. Am J Pathol. 1996;149:1899–1917. [PMC free article] [PubMed] [Google Scholar]
- 60.Rey O, Lee S, Park NH. Human papillomavirus type 16 E7 oncoprotein represses transcription of human fibronectin. J Virol. 2000;74:4912–4918. doi: 10.1128/jvi.74.10.4912-4918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toussaint-Smith E, Donner DB, Roman A. Expression of human papillomavirus type 16 E6 and E7 oncoproteins in primary foreskin keratinocytes is sufficient to alter the expression of angiogenic factors. Oncogene. 2004;23:2988–2995. doi: 10.1038/sj.onc.1207442. [DOI] [PubMed] [Google Scholar]
- 62.Bornstein P. Thrombospondins function as regulators of angiogenesis. J Cell Commun Signal. 2009;3:189–200. doi: 10.1007/s12079-009-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sahebali S, et al. Stromal issues in cervical cancer: A review of the role and function of basement membrane, stroma, immune response and angiogenesis in cervical cancer development. Eur J Cancer Prev. 2010;19:204–215. doi: 10.1097/CEJ.0b013e32833720de. [DOI] [PubMed] [Google Scholar]
- 64.Keeley EC, Mehrad B, Strieter RM. Chemokines as mediators of tumor angiogenesis and neovascularization. Exp Cell Res. 2011;317:685–690. doi: 10.1016/j.yexcr.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Begley LA, et al. CXCL5 promotes prostate cancer progression. Neoplasia. 2008;10:244–254. doi: 10.1593/neo.07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolitho C, Hahn MA, Baxter RC, Marsh DJ. The chemokine CXCL1 induces proliferation in epithelial ovarian cancer cells by transactivation of the epidermal growth factor receptor. Endocr Relat Cancer. 2010;17:929–940. doi: 10.1677/ERC-10-0107. [DOI] [PubMed] [Google Scholar]
- 67.Curry JM, et al. Tumor microenvironment in head and neck squamous cell carcinoma. Semin Oncol. 2014;41:217–234. doi: 10.1053/j.seminoncol.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 68.Gao Y, et al. CXCL5/CXCR2 axis promotes bladder cancer cell migration and invasion by activating PI3K/AKT-induced upregulation of MMP2/MMP9. Int J Oncol. 2015;47:690–700. doi: 10.3892/ijo.2015.3041. [DOI] [PubMed] [Google Scholar]
- 69.Li A, et al. Overexpression of CXCL5 is associated with poor survival in patients with pancreatic cancer. Am J Pathol. 2011;178:1340–1349. doi: 10.1016/j.ajpath.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou SL, et al. CXCR2/CXCL5 axis contributes to epithelial-mesenchymal transition of HCC cells through activating PI3K/Akt/GSK-3β/Snail signaling. Cancer Lett. 2015;358:124–135. doi: 10.1016/j.canlet.2014.11.044. [DOI] [PubMed] [Google Scholar]
- 71.Kogan-Sakin I, et al. Prostate stromal cells produce CXCL-1, CXCL-2, CXCL-3 and IL-8 in response to epithelia-secreted IL-1. Carcinogenesis. 2009;30:698–705. doi: 10.1093/carcin/bgp043. [DOI] [PubMed] [Google Scholar]
- 72.Gallagher PG, et al. Gene expression profiling reveals cross-talk between melanoma and fibroblasts: Implications for host-tumor interactions in metastasis. Cancer Res. 2005;65:4134–4146. doi: 10.1158/0008-5472.CAN-04-0415. [DOI] [PubMed] [Google Scholar]
- 73.Li K, et al. Letrozole-induced functional changes in carcinoma-associated fibroblasts and their influence on breast cancer cell biology. Med Oncol. 2016;33:64. doi: 10.1007/s12032-016-0779-z. [DOI] [PubMed] [Google Scholar]
- 74.Mangino G, Chiantore MV, Iuliano M, Fiorucci G, Romeo G. Inflammatory microenvironment and human papillomavirus-induced carcinogenesis. Cytokine Growth Factor Rev. 2016;30:103–111. doi: 10.1016/j.cytogfr.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 75.Coussens LM, Tinkle CL, Hanahan D, Werb Z. MMP-9 supplied by bone marrow-derived cells contributes to skin carcinogenesis. Cell. 2000;103:481–490. doi: 10.1016/s0092-8674(00)00139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coussens LM, et al. Inflammatory mast cells up-regulate angiogenesis during squamous epithelial carcinogenesis. Genes Dev. 1999;13:1382–1397. doi: 10.1101/gad.13.11.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spurgeon ME, Chung SH, Lambert PF. Recurrence of cervical cancer in mice after selective estrogen receptor modulator therapy. Am J Pathol. 2014;184:530–540. doi: 10.1016/j.ajpath.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woodworth CD, McMullin E, Iglesias M, Plowman GD. Interleukin 1 alpha and tumor necrosis factor alpha stimulate autocrine amphiregulin expression and proliferation of human papillomavirus-immortalized and carcinoma-derived cervical epithelial cells. Proc Natl Acad Sci USA. 1995;92:2840–2844. doi: 10.1073/pnas.92.7.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun C, et al. FGF9 from cancer-associated fibroblasts is a possible mediator of invasion and anti-apoptosis of gastric cancer cells. BMC Cancer. 2015;15:333. doi: 10.1186/s12885-015-1353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fillmore CM, et al. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc Natl Acad Sci USA. 2010;107:21737–21742. doi: 10.1073/pnas.1007863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murata T, et al. HB-EGF and PDGF mediate reciprocal interactions of carcinoma cells with cancer-associated fibroblasts to support progression of uterine cervical cancers. Cancer Res. 2011;71:6633–6642. doi: 10.1158/0008-5472.CAN-11-0034. [DOI] [PubMed] [Google Scholar]
- 82.Song S, Liem A, Miller JA, Lambert PF. Human papillomavirus types 16 E6 and E7 contribute differently to carcinogenesis. Virology. 2000;267:141–150. doi: 10.1006/viro.1999.0106. [DOI] [PubMed] [Google Scholar]
- 83.Saeed AI, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 84.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: A web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nogales-Cadenas R, et al. GeneCodis: Interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–W322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: A non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40:W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hulsen T, de Vlieg J, Alkema W. BioVenn–A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chapman S, Liu X, Meyers C, Schlegel R, McBride AA. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J Clin Invest. 2010;120:2619–2626. doi: 10.1172/JCI42297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nisato RE, et al. Generation and characterization of telomerase-transfected human lymphatic endothelial cells with an extended life span. Am J Pathol. 2004;165:11–24. doi: 10.1016/S0002-9440(10)63271-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Elson DA, et al. Sensitivity of the cervical transformation zone to estrogen-induced squamous carcinogenesis. Cancer Res. 2000;60:1267–1275. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.