Abstract
Background
Radon is a known human lung carcinogen, whose underlying carcinogenic mechanism remains unclear. Recently, circular RNA (circRNA), a class of endogenous non-protein coding RNAs that contain a circular loop, was found to exhibit multiple biological effects. In this study, circRNA profiles in mouse lung tissues between control and radon exposure were analyzed.
Methods
Six mice were exposed to radon at concentration of 100,000 Bq/m3, 12 h/d, for up to cumulative doses of 60 working level months (WLM). H&E staining and immunohistochemistry of caspase-3 were used to detect the damages in lung tissue. The lung tissue of control and exposed group were selected for circRNA microarray study. The circRNA/microRNA interaction was analyzed by starBase prediction software. 5 highest expressing circRNAs were selected by real-time PCR to validate the consistency in mouse lung tissue exposed to radon.
Results
Inflammatory reaction was found in mouse lung tissue exposed to radon, and caspase-3 expression was significantly increased. Microarray screening revealed 107 up-regulated and 83 down-regulated circRNAs, among which top 30 circRNAs with the highest fold changes were chosen for further analysis, with 5 microRNAs binding sites listed for each circRNA. Consistency of the top 5 circRNAs with the highest expressions were confirmed in mice exposed with 60WLM of radon.
Conclusion
Mouse lung tissue was severely injured when exposed to radon through pathological diagnosis and immunohistochemical analysis. A series of differentially expressed circRNAs demonstrated that they may play an important role in pulmonary toxicity induced by radon.
Keywords: Radon, Circular RNA, Damage, Mouse, Lung tissue
Background
Radon is the most notorious environmental radioactive gas and has been categorized by the World Health Organization as a carcinogenic substance that causes lung cancer. In the United States, an estimated 21,000 lung cancer deaths annually are attributed to radon [1–4]. Radon decays with the emission of α particles and has a half-life of 3.8 days. When radon is inhaled, α particles could affect lung tissue and may cause lung cancer via DNA and oxidative damages [5, 6]. However the causative effect of radon on lung cancer and its specific mechanisms remain unclear, and one of the carcinogenic mechanisms is thought to be DNA damage through reactive oxygen species produced by α particles [7]. Loss of heterozygosity analysis of the Cdkn2a locus, including the Ink4a and Arf genes, was performed on 33 radon-induced rat lung tumors, and DNA losses were observed in 50% of cases [8]. On the other hand, apoptosis, also known as programmed cell death, can be activated through the intrinsic mitochondrial-dependent pathway, by releasing cytochrome C which binds apoptotic protease activating factor-1 and pro-caspase-9 to create the apoptosome that finally activates the director caspase-3 [9, 10].
Circular RNAs (circRNAs) is a class of RNAs possessing many biological activities, such as sponging miRNA, affecting epithelial-to-mesenchymal transition (EMT)-related cellular functions, regulating RNA polymerase II transcription [11–14]. In addition, studies have shown that circRNAs could act as potential clinical biomarkers in complex disorders of the central nervous system [15], and alteration in the expressions of circRNAs was implicated in basal cell carcinoma [16], pathological hypertrophy and heart failure [17], laryngeal cancer [18].
In this study, we aimed to determine the damages in mouse lung tissue and the changes in circRNAs expression profiles upon exposure to radon, in order to explore their potential functions in radon-induced lung damage.
Methods
Animals and exposure conditions
All animal handling procedures were reviewed and approved by the Animal Care/User Ethical Committee of Soochow University, Suzhou City, P. R. China. 12 male BALB/c mice weighing approximately 15 g were obtained from Shanghai Laboratory Animal Center, and randomly divided into the experimental group and the control group with 6 mice per group. The mice in the experimental group were exposed to radon in a multifunctional radon chamber (Dong-hua University, China), at a concentration of 100,000 Bq/m3, 12 h/d, for up to cumulative doses of 60 working level months (WLM). The radon concentration in the exposure chamber was constantly maintained and continuously measured by scintillation counting. Food and water were available ad libitum during the exposure. Control mice were housed in a same room with a background concentration of radon at 20 Bq/m3 [19].
Sample collection and microarray hybridization
After the last exposure, animals were sacrificed and the left lung tissue was used to extract the total RNA for circRNA analysis. Right lung was used for hematoxylin and eosin (H&E) staining, immunohistochemistry (IHC) analysis and Western blot assay.
Total RNA from each sample was quantified using the NanoDrop ND-1000. The sample preparation and microarray hybridization were performed based on the Arraystar’s standard protocols. Briefly, total RNA from each sample was amplified and transcribed into fluorescent circRNA using random primer according to Arraystar’s Super RNA Labeling protocol (Arraystar Inc.). The labeled circRNAs were hybridized onto the Arraystar Mouse circRNA Array (6x7K, Arraystar). After washing the slides, the arrays were scanned by the Axon GenePix 4000B microarray scanner. Scanned images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Quantile normalization and subsequent data processing were performed using the R software package. Differentially expressed circRNAs with statistical significance between the two groups were identified through Volcano Plot filtering. Differentially expressed circRNAs between two samples were identified through Fold Change filtering. Hierarchical Clustering was performed to show the distinguishable circRNA expression pattern among samples.
Histological assessment and immunohistochemistry
H&E staining was conducted to examine histological pathology of lung tissue under a light microscope. Digital images were collected by a conventional light microscope in 5 visual fields/per section with 400× magnification under bright-field viewing. IHC stains were performed to examine the caspase-3 expression level in tissues from mice exposed to radon. The paraffin-embedded sections were stained with antibody against caspase-3. Tissue array slides were deparaffinized 3 times with Xylene, each for 5 min. The slides were rehydrated by a sequential treatment with 100% ethanol twice for 5 min each; 95% ethanol twice for 5 min each; and 80% ethanol twice for 5 min. Antigen retrieval was performed by microwaving in 0.01 M sodium citrate buffer (pH 6.0) at 96–100 °C for 15 min. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide at room temperature for 25 min. After rinsing with PBS (pH 7.4) three times each for 5 min, the sections were then incubated with rabbit polyclonal antibody against caspase-3 (1:100 dilution, Abcam, USA) overnight at 4 °C. Following extensive washing with PBS, anti-rabbit secondary antibody (KPL, USA) was applied with 1:2000 dilutions for 1 h. The slides were then visualized using 3,3-diaminobenzidine (DAB) chromogen (LabVision Corp., Fremont, CA, USA) and counterstained with H&E. The sections were washed in PBS, dried, mounted in neutral gum, and observed at 400× magnification with a Nikon TE200 inverted microscope (Nikon Co, Japan). The sections for which the primary antibody was omitted were used as negative controls. 5 non-repeat high-power field images were randomly selected in which the brown particles were considered as positive areas.
Western blot analysis
Total protein was extracted from lung tissues. The protein concentrations of individual samples were assessed using a standard bicinchoninic acid assay (Beyotime, Nantong, China). For each sample, 30 μg of protein was loaded on 10% SDS-PAGE gel (Bio-Rad, USA), transferred onto a polyvinylidene difluoride membrane and blocked with 5% skimmed milk and 0.1% Tris-buffered saline-Tween 20 (TBST) at room temperature for 1.5 h. The membranes were incubated overnight at 4 °C with caspase-3 antibody (1:1,000 dilution, Abcam, USA), mouse anti-GAPDH and mouse anti-β-Actin antibodies (1/8,000 dilution, Beyotime, Nantong, China). The membranes were then washed with TBST and incubated with anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology, Boston, MA). The protein expression was evaluated using chemiluminescence and exposure to Kodak film (Kodak, Rochester, NY).
RNA Isolation and real-time PCR
Five highest expressing circRNAs were chosen to be examined in the mice of the 60 WLM group and the control group. The total RNA was extracted with TRIzol and reversely transcribed using SuperScriptTM III Reverse Transcriptase(Invitrogen, USA). The primers used in this assay are listed in Table 1. The following PCR conditions were used: 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s, 62 °C for 30 s. All RNA samples were analyzed in triplicate for each tested transcript and normalized to GAPDH. Expression fold changes were calculated using the 2−ΔΔCt methods for quantitative analysis.
Table 1.
Primer sequences
| Gene | Sequence (5’-3’) |
|---|---|
| GAPDH | F:5’GTTGTCTCCTGCGACTTCA3’ R:5’GCCCCTCCTGTTATTATGG3’ |
| mmu_circ_0001028 | F:5’ TATTCTTCTGGGTGAGGATGGC3’ R:5’ TTGATCCGCTTTATGGCTACG3’ |
| mmu_circ_0001311 | F:5’ CCCCAGGATCTTCGTAGGTTA3’ R:5’ TCTTTCTTCCTTCAGCCACTTC3’ |
| mmu_circ_0001154 | F:5’ TCTGGAACATTGCGATTTGGA 3’ R:5’ TTGTTGACCGATGCCCACTC3’ |
| mmu_circ_0001052 | F:5’ CCGTACAGGGTTAAAGTGATAG3’ R:5’ TTTGATAAACATACGGTGGGT3’ |
| mmu_circ_0001796 | F:5’ GTCCGTACCCGATCAGTTGG 3’ R:5’ CGTTCTCAGACCTGCCTCCT 3’ |
Statistical analysis
The SPSS 17.0 software was used for the statistical analysis. The data are presented as mean ± SEM. Group comparisons were evaluated by one-way ANOVA to determine statistical significance. Differences were considered significant when P<0.05.
Results
Alteration in histological pathology of mouse lung tissue
The H&E stainings of lung tissues from mice exposed to 60 WLM radon displayed inflammatory cells in the alveolus interval (Fig. 1A. b: black arrow) and some alveolar septal fracture (Fig. 1A. b: red arrow). Immunohistochemistry results showed that the brown represented positive cells. The expression of apoptotic activator protein caspase-3 was significantly increased in 60 WLM mice (Fig. 1A. d: black arrow). Western blot result showed that caspase-3 protein expression was also significantly increased compared with the control group (Fig. 1B).
Fig. 1.
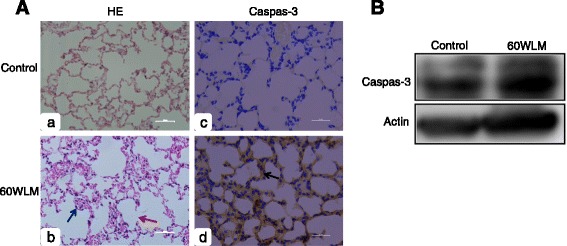
H&E and immunohistochemistry in mouse lung tissue exposed to radon. A Representative H&E and immunohistochemistry staining images of caspase-3 in normal mouse lung tissues and exposure samples. Black arrow indicates inflammatory cell in the alveolus interval, red arrow indicates alveolar septal fracture, black arrow indicates positive cells. B Western blot analysis of caspase-3 expression in mouse lung tissue
CircRNA expression profiles in control and 60WLM group
Three mice from the control group and 60 WLM group, respectively, were chosen to assess the circRNA profiles using the circRNA microarray. A box view was used to compare the distribution of expression values for the samples after normalization. The distribution of log2 ratios was similar in all the tested samples as shown in Fig. 2.
Fig. 2.
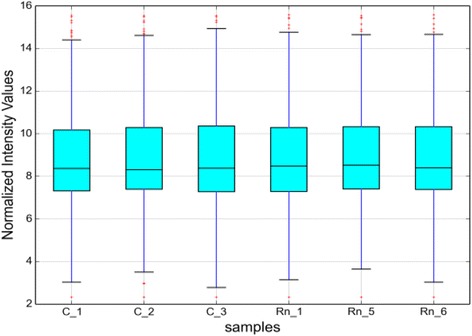
Box plot. X-axis: 6 tissue samples; Y-axis: normalized intensity values. All the 6 samples in the databases were normalized. The distribution of circRNAs was similar in all samples
Hierarchical clustering is one of the simplest and widely used clustering techniques for gene expression data analysis. Cluster analysis arranges are based on their expression levels, which allows us to hypothesize on the relationships between two groups. The dendrogram shows the relationships of the expression levels between the two groups. In this study, hierarchical clustering was performed based on “All Targets Value - CircRNAs”. The result showed that a distinguishable mRNA expression profiling between the control and radon-exposed groups as shown in Fig. 3.
Fig. 3.
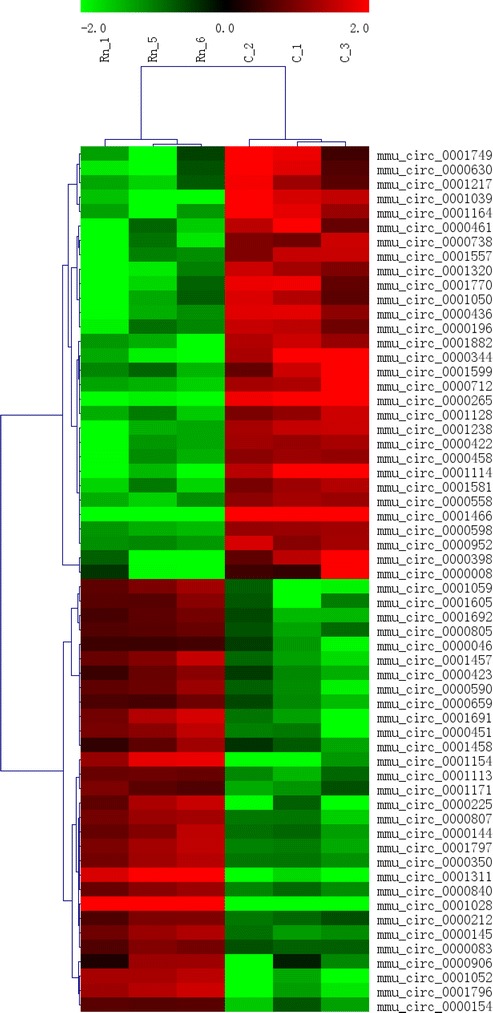
Top 30 up-regulated and down-regulated circRNAs were chose for analysis. Hierarchical cluster analysis diagram of the control and 60 WLM group (n=3) versus the sham-irradiated control (n=3). The hierarchical clustering is displayed as color saturation, which is directly proportional to the measured expression. The color saturation represents the log2 value of the normalized intensity of each sample (ranges from 7 to 11; P value cut-off 0.05). Rows indicate the individual probe set, and the columns represent the experimental sample
The Scatter plot was used to assess the variations in circRNA expressions between the lung tissues of the control and exposure groups. The values of X and Y axes in the Scatter plot are the normalized signal values of the samples (log2 scaled) or the averaged normalized signal values of groups of samples (log2 scaled). The green lines are fold change lines. The circRNAs above the top green line and below the green line on the bottom indicated more than 2 fold change in circRNAs between the two groups as shown in Fig. 4.
Fig. 4.
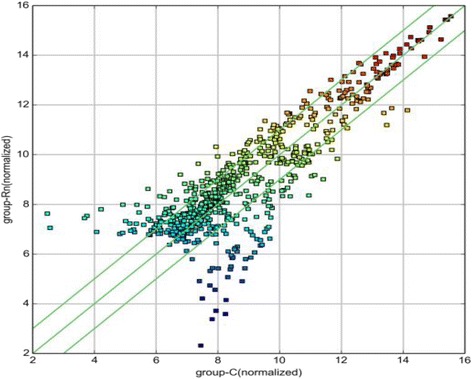
Scatter plot. X-axis: adjacent non-tumor tissues (normalized), Yaxis: LSCC tissues (normalized). The green lines represent fold change. The circRNAs above the top green line and below the bottom green line indicate more than 2 fold change in circRNA expressions between the two groups of samples
The Volcano plot was performed to visualize the differential expressions between the control and exposed groups. The significant differences (fold change >2.0, P < 0.05) between the two different conditions are illustrated in the Volcano plot, and the red points in the plot represents the differentially expressed circRNAs with statistical significance as shown in Fig. 5.
Fig. 5.
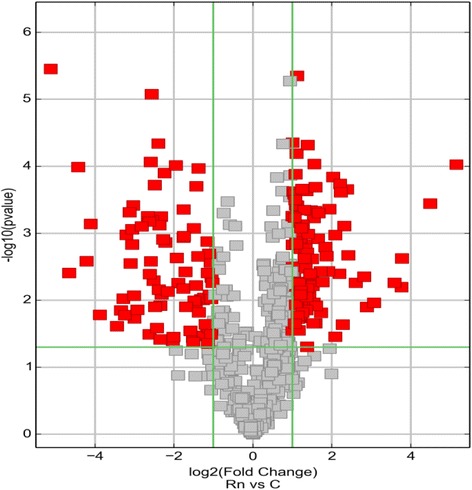
Volcano plot. X-axis: log2 (fold change); Y-axis: −log10 (P-value). The vertical green lines represent 2 fold up-regulation (left) and down-regulation (right), and the horizontal green line represents a p-value of 0.05. The red points in the plot represent circRNAs expressed differentially with statistical significance
CircRNA expression profile and function prediction
Microarray screening revealed 107 up-regulated and 83 down-regulated circRNAs, among which the top 30 circRNAs with the most fold changes were chosen for further analysis, with 5 microRNA binding sites were listed for each circRNA (Tables 2 and 3).
Table 2.
Fold change of top 30 circRNAs up-regulation
| CircBase ID | Fold Change | best_transcript | GeneSymbol | miRNA Binding Sites 1 | miRNA Binding Sites 2 | miRNA Binding Sites3 | miRNA Binding Sites4 | miRNA Binding Sites5 |
|---|---|---|---|---|---|---|---|---|
| mmu_circ_0001028 | 35.63598 | NM_016866 | Stk39 | mmu-miR-703 | mmu-miR-879-3p | mmu-miR-3058-5p | mmu-miR-693-3p | mmu-miR-7015-5p |
| mmu_circ_0001311 | 22.46768 | NM_001003909 | Ankib1 | mmu-miR-103-3p | mmu-miR-107-3p | mmu-miR-7092-3p | mmu-miR-712-5p | mmu-miR-6935-3p |
| mmu_circ_0001154 | 13.69381 | NM_133869 | Cept1 | mmu-miR-6946-3p | mmu-miR-22-5p | mmu-miR-1955-5p | mmu-miR-298-5p | mmu-miR-500-5p |
| mmu_circ_0001052 | 13.58701 | NM_001145824 | Hipk3 | mmu-miR-7222-5p | mmu-miR-1929-5p | mmu-miR-338-3p | mmu-miR-6922-3p | mmu-miR-143-5p |
| mmu_circ_0001796 | 12.02444 | NM_172536 | Zfp609 | mmu-miR-7009-3p | mmu-miR-3971 | mmu-miR-6974-3p | mmu-miR-6975-3p | mmu-miR-344 g-3p |
| mmu_circ_0001691 | 8.370497 | NM_030113 | Arhgap10 | mmu-miR-6898-3p | mmu-miR-7221-5p | mmu-miR-201-3p | mmu-miR-210-5p | mmu-miR-326-3p |
| mmu_circ_0001059 | 7.40114 | NM_029617 | Casc5 | mmu-miR-6344 | mmu-miR-7035-5p | mmu-miR-28a-5p | mmu-miR-7233-3p | mmu-miR-28c |
| mmu_circ_0000225 | 7.036664 | mmu-miR-8092 | mmu-miR-875-3p | mmu-miR-7679-3p | mmu-miR-532-5p | mmu-miR-6481 | ||
| mmu_circ_0000451 | 6.093728 | NM_001123386 | Cdyl | mmu-miR-7083-5p | mmu-miR-6902-5p | mmu-miR-185-3p | mmu-miR-3085-3p | mmu-miR-3064-5p |
| mmu_circ_0001457 | 5.334011 | NM_013791 | Mkln1 | mmu-miR-30d-3p | mmu-miR-3071-3p | mmu-miR-7214-5p | mmu-miR-5615-5p | mmu-miR-7b-5p |
| mmu_circ_0001797 | 5.213906 | NM_172536 | Zfp609 | mmu-miR-7009-3p | mmu-miR-3971 | mmu-miR-6974-3p | mmu-miR-6975-3p | mmu-miR-6961-3p |
| mmu_circ_0000807 | 4.974568 | NM_001025572 | Ankrd12 | mmu-miR-7092-3p | mmu-miR-683 | mmu-miR-6944-3p | mmu-miR-7684-3p | mmu-miR-6899-3p |
| mmu_circ_0001605 | 4.840995 | NM_175388 | Rnf169 | mmu-miR-3095-5p | mmu-miR-693-3p | mmu-miR-207 | mmu-miR-6344 | mmu-miR-7038-3p |
| mmu_circ_0000145 | 4.680084 | NM_011682 | Utrn | mmu-miR-6958-3p | mmu-miR-1968-5p | mmu-miR-22-3p | mmu-miR-19b-2-5p | mmu-miR-450b-5p |
| mmu_circ_0000350 | 4.620216 | NM_001162465 | Dtnb | mmu-miR-3066-5p | mmu-miR-6938-3p | mmu-miR-6929-5p | mmu-miR-103-1-5p | mmu-miR-103-2-5p |
| mmu_circ_0000590 | 4.569337 | NM_018745 | Azin1 | mmu-miR-216a-5p | mmu-miR-7657-3p | mmu-miR-489-5p | mmu-miR-203-5p | mmu-miR-5625-3p |
| mmu_circ_0000144 | 4.247965 | NM_011682 | Utrn | mmu-miR-6958-3p | mmu-miR-1968-5p | mmu-miR-22-3p | mmu-miR-328-3p | mmu-miR-19b-2-5p |
| mmu_circ_0000906 | 4.241155 | ENSMUST00000182122 | Zfp236 | mmu-miR-1930-3p | mmu-miR-143-5p | mmu-miR-669d-5p | mmu-miR-1187 | mmu-miR-466i-5p |
| mmu_circ_0000840 | 4.08919 | NM_001039692 | Arhgap12 | mmu-miR-30c-1-3p | mmu-miR-450a-5p | mmu-miR-30c-2-3p | mmu-miR-592-5p | mmu-miR-320-5p |
| mmu_circ_0001113 | 3.831634 | NM_173182 | Fndc3b | mmu-miR-93-3p | mmu-miR-7231-3p | mmu-miR-412-3p | mmu-miR-6998-3p | mmu-miR-298-5p |
| mmu_circ_0001692 | 3.745827 | NM_030113 | Arhgap10 | mmu-miR-6898-3p | mmu-miR-201-3p | mmu-miR-326-3p | mmu-miR-7011-3p | mmu-miR-7b-5p |
| mmu_circ_0000154 | 3.65036 | NM_018747 | Akap7 | mmu-miR-1903 | mmu-miR-135b-5p | mmu-miR-135a-5p | mmu-miR-106a-5p | mmu-miR-93-5p |
| mmu_circ_0001171 | 3.562626 | NM_146141 | Ppa2 | mmu-miR-7092-3p | mmu-miR-6349 | mmu-miR-206-3p | mmu-miR-6382 | mmu-miR-1a-3p |
| mmu_circ_0000423 | 3.392009 | NM_011625 | Ppp1r13b | mmu-miR-7649-3p | mmu-miR-6946-3p | mmu-miR-7083-3p | mmu-miR-3473c | mmu-miR-1903 |
| mmu_circ_0000046 | 3.298026 | NM_009952 | Creb1 | mmu-miR-5623-5p | mmu-miR-6911-5p | mmu-miR-7688-5p | mmu-miR-7242-3p | mmu-miR-216a-3p |
| mmu_circ_0000659 | 3.281563 | NM_029582 | Txndc11 | mmu-miR-21a-3p | mmu-miR-5110 | mmu-miR-7038-5p | mmu-miR-331-3p | mmu-miR-3064-3p |
| mmu_circ_0000212 | 3.168443 | NM_001037846 | Cnot2 | mmu-miR-6902-5p | mmu-miR-6914-3p | mmu-miR-7052-5p | mmu-miR-7214-3p | mmu-miR-6972-5p |
| mmu_circ_0000805 | 3.13632 | NM_013933 | Vapa | mmu-miR-1298-5p | mmu-miR-672-3p | mmu-miR-19b-1-5p | mmu-miR-7212-3p | mmu-miR-19b-2-5p |
| mmu_circ_0001458 | 3.029251 | NM_013791 | Mkln1 | mmu-miR-296-5p | mmu-miR-8103 | mmu-miR-135a-5p | mmu-miR-135b-5p | mmu-miR-30d-3p |
| mmu_circ_0000083 | 2.999915 | NM_001166501 | Dennd1b | mmu-miR-145a-5p | mmu-miR-145b | mmu-miR-122-5p | mmu-miR-103-3p | mmu-miR-107-3p |
Table 3.
Fold change of top 30 circRNAs down-regulation
| CircBase ID | Fold Change | best_transcript | GeneSymbol | miRNA Binding Sites 1 | miRNA Binding Sites 2 | miRNA Binding Sites3 | miRNA Binding Sites4 | miRNA Binding Sites5 |
|---|---|---|---|---|---|---|---|---|
| mmu_circ_0001466 | 34.8883803 | NM_194061 | D630045J12Rik | mmu-miR-6390 | mmu-miR-468-3p | mmu-miR-7232-3p | mmu-miR-29b-2-5p | mmu-miR-146a-3p |
| mmu_circ_0001039 | 25.2731871 | NM_016965 | Nckap1 | mmu-miR-7065-3p | mmu-miR-5619-5p | mmu-miR-7010-3p | mmu-miR-143-5p | mmu-miR-6344 |
| mmu_circ_0000265 | 21.5734915 | NM_173753 | Fnip1 | mmu-miR-221-5p | mmu-miR-145a-3p | mmu-miR-146a-3p | mmu-miR-504-5p | mmu-miR-3113-5p |
| mmu_circ_0000344 | 18.5911228 | NM_029878 | Tbcd | mmu-miR-3474 | mmu-miR-7239-3p | mmu-miR-6945-5p | mmu-miR-1982-5p | mmu-miR-204-3p |
| mmu_circ_0001114 | 17.2864639 | NM_001163007 | Tnik | mmu-miR-384-5p | mmu-miR-199a-3p | mmu-miR-199b-3p | mmu-miR-543-3p | mmu-miR-30e-5p |
| mmu_circ_0000398 | 14.6857941 | NM_001167920 | Slc8a3 | mmu-miR-8113 | mmu-miR-6900-5p | mmu-miR-6957-5p | mmu-miR-3552 | mmu-miR-7050-5p |
| mmu_circ_0000738 | 10.8968513 | NM_027060 | Btbd9 | mmu-miR-15b-5p | mmu-miR-15a-5p | mmu-miR-322-5p | mmu-miR-16-5p | mmu-miR-195b |
| mmu_circ_0000461 | 10.1374001 | NM_011078 | Phf2 | mmu-miR-29b-2-5p | mmu-miR-29b-1-5p | mmu-miR-191-3p | mmu-miR-337-3p | mmu-miR-673-3p |
| mmu_circ_0001770 | 9.7481856 | NM_008775 | Pafah1b2 | mmu-miR-1224-3p | mmu-miR-683 | mmu-miR-6935-3p | mmu-miR-3092-5p | mmu-miR-6342 |
| mmu_circ_0001557 | 9.492165 | NM_016682 | Uba2 | mmu-miR-7649-3p | mmu-miR-6399 | mmu-miR-7226-5p | mmu-miR-380-5p | mmu-miR-3090-5p |
| mmu_circ_0001164 | 9.234634 | TCONS_00021080 | XLOC_016164 | mmu-miR-7062-5p | mmu-miR-7116-3p | mmu-miR-106a-3p | mmu-miR-691 | mmu-miR-7093-3p |
| mmu_circ_0001238 | 8.7123832 | NM_001080926 | Lrp8 | mmu-miR-7032-5p | mmu-miR-7094b-2-5p | mmu-miR-6990-5p | mmu-miR-674-5p | mmu-miR-1956 |
| mmu_circ_0001320 | 8.5860481 | NM_010305 | Gnai1 | mmu-miR-7011-3p | mmu-miR-6360 | mmu-miR-1291 | mmu-miR-324-3p | mmu-miR-3105-3p |
| mmu_circ_0000436 | 8.3430278 | NM_172120 | Vps41 | mmu-miR-7b-5p | mmu-miR-670-3p | mmu-miR-7034-3p | mmu-miR-7117-3p | mmu-miR-7093-3p |
| mmu_circ_0001882 | 8.3381778 | NM_001160403 | Il1rapl1 | mmu-miR-337-3p | mmu-miR-7018-5p | mmu-miR-8095 | mmu-miR-195b | mmu-miR-6975-3p |
| mmu_circ_0000712 | 8.1893468 | NM_139145 | Hlcs | mmu-miR-5133 | mmu-miR-6938-5p | mmu-miR-7118-5p | mmu-miR-6911-5p | mmu-miR-7672-5p |
| mmu_circ_0000630 | 8.0822088 | NM_172610 | Mpped1 | mmu-miR-7037-5p | mmu-miR-5120 | mmu-miR-7062-5p | mmu-miR-5132-5p | mmu-miR-6897-5p |
| mmu_circ_0001749 | 8.0148488 | NM_145610 | Ppan | mmu-miR-6954-5p | mmu-miR-6938-3p | mmu-miR-6914-5p | mmu-miR-486b-3p | mmu-miR-486a-3p |
| mmu_circ_0001050 | 7.5953109 | NM_010163 | Ext2 | mmu-miR-7019-5p | mmu-miR-30b-3p | mmu-miR-3547-5p | mmu-miR-6982-5p | mmu-miR-6930-5p |
| mmu_circ_0000422 | 7.0486583 | NM_021516 | Mark3 | mmu-miR-466f | mmu-miR-1187 | mmu-miR-574-5p | mmu-miR-709 | mmu-miR-297a-5p |
| mmu_circ_0000458 | 6.3303882 | NM_019930 | Ranbp9 | mmu-miR-3109-5p | mmu-miR-145b | mmu-miR-490-3p | mmu-miR-7007-5p | mmu-miR-5124a |
| mmu_circ_0001581 | 6.1968327 | NR_028143 | Lrrc28 | mmu-miR-3087-5p | mmu-miR-1231-5p | mmu-miR-6998-5p | mmu-miR-1912-5p | mmu-miR-34a-5p |
| mmu_circ_0000008 | 6.143468 | NM_026493 | Cspp1 | mmu-miR-1903 | mmu-miR-3154 | mmu-miR-6340 | mmu-miR-203-5p | mmu-miR-6946-3p |
| mmu_circ_0001599 | 6.1137112 | NM_011035 | Pak1 | mmu-miR-1903 | mmu-miR-6516-5p | mmu-miR-107-5p | mmu-miR-181d-5p | mmu-miR-6946-3p |
| mmu_circ_0000558 | 6.0158681 | NM_001164503 | Akap11 | mmu-miR-452-5p | mmu-miR-6925-5p | mmu-miR-6999-5p | mmu-miR-6976-5p | mmu-miR-7241-5p |
| mmu_circ_0000196 | 6.0061817 | NM_001277188 | Ano4 | mmu-miR-691 | mmu-miR-5622-5p | mmu-miR-302a-3p | mmu-miR-5620-3p | mmu-miR-20b-5p |
| mmu_circ_0000598 | 5.9125661 | NM_001113554 | Nudcd1 | mmu-miR-494-5p | mmu-miR-6946-3p | mmu-miR-7054-5p | mmu-miR-6935-3p | mmu-miR-376b-5p |
| mmu_circ_0001128 | 5.9008556 | NM_001198766 | Postn | mmu-miR-29b-2-5p | mmu-miR-29b-1-5p | mmu-miR-23a-5p | mmu-miR-344d-2-5p | |
| mmu_circ_0001217 | 5.7224481 | NM_001163732 | Frmd3 | mmu-miR-7024-3p | mmu-miR-6939-5p | mmu-miR-7663-5p | mmu-miR-1967 | mmu-miR-450a-1-3p |
| mmu_circ_0000952 | 5.6172869 | NM_145501 | Pi4k2a | mmu-miR-1907 | mmu-miR-133c | mmu-miR-133a-3p | mmu-miR-133b-3p | mmu-miR-6989-3p |
Identification of circRNAs
Five circRNAs with the highest expressions were selected to validate the data from microarray in mice exposed to 60 WLM of radon as shown in Fig. 6. The result confirmed the consistency between data from microarray and real-time PCR.
Fig. 6.
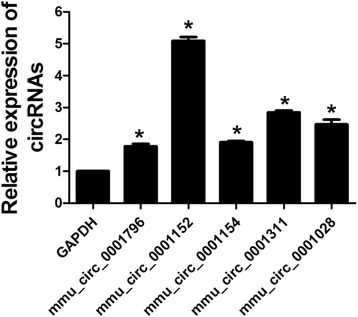
The expressions of 5 circRNAs were examined in mice of 60 WLM group. Real-time PCR was used to determine the expressions of the 5 highest circRNAs in the 60 WLM group. * P<0.05 vs GAPDH
Discussion
Radon (222Rn) gas is usually released from rocks and soils, and rapidly disperses in the atmosphere. When radon is inhaled into the human respiratory system, the entire body could be subjected to internal exposure. Many reports showed that radon causes cancers, including lung cancer and childhood leukemia [1–4]. There have been strong evidences showing a significantly increased risk of lung cancer followed by long-term low radon exposure among Wismut miners [1]. Genome damage, such as genome instability, was found in peripheral blood lymphocytes of lung cancer patients living in a region with high air pollution and increased background radon levels [5, 20]. Moreover, IL-6 promoter variant was associated with lung cancer in uranium miners [21]. Reports show that caspase-3 is involved in cell apoptosis [22, 23]. Our results showed that caspas-3 expression was significantly increased in radon-exposed group, suggesting that the lung tissue was severely injured when exposed to radon.
CircRNA is an abundant, developmentally regulated, tissue and cell-type specific, stable transcriptional product, and is evolutionarily conserved across the eukaryotic tree of life [24, 25]. Studies showed that interactions between circRNAs and miRNAs indicate that circRNAs are potentially associated with many disease, such as gastric cancer [26], colorectal cancer [27] and breat cancer [28]. Since circRNAs are involved in many kinds of diseases and provide new insights for exploring the disease pathological mechanisms. In this study, we have identified 107 up-regulated and 83 down-regulated circRNAs. 5 miRNAs of each circRNA was successfully predicted. These circRNAs and miRNAs could play important and synergistic roles in radon-induced lung damage, even in lung cancer. Further studies need to be conducted to illustrate the functional mechanisms of circRNAs in radon-induced lung damages.
Recent evidence suggests that circRNAs play a crucial role in fine-tuning the level of miRNA-mediated regulation of gene expression by sequestering miRNAs. MiRNA response elements (MREs) were found in some circRNAs, which could sponge miRNAs, such as miR-7, miR-17, and miR-214 [29]. In addition, circRNA could affect miRNA activities and increase the complexity of RNA regulatory networks hence playing a role in gene expression [30]. Evidences indicated that circRNAs play important roles in tumorigenesis. For instance, ciRS-7 was found to inhibit cancer via miR-7 by down-regulating some critical oncogenes [31]. Hsa_circ_002059 expression in gastric cancer was involved in TNM stage, gender and age [32]. Moreover, hsa_circRNA_100855 and hsa_circRNA_104912 were involved in lymph node metastasis, poor differentiated or advanced clinical stages in laryngeal cancer [18]. In summary, these results showed that circRNAs may serve as potential biomarkers in certain diseases such as cancer. However, few circRNA/miRNA interactions have been experimentally validated. Therefore, the functional effect of their interaction will be the focus of our future research.
Conclusion
In summary, our results suggest that mouse lung tissue was severely injured when exposed to radon through and a series of differentially expressed circRNAs were found. It will hopefully to further research the biological effects induced by radon.
Acknowledgements
The circRNAs microarray hybridyzation and bioinformatics analysis were conducted by KangChen Biotech Co Ltd (Shanghai, China).
Funding
This research was supported by National Science Foundation of China (81402626 and 81472920), China Postdoctoral Science Foundation (2015 M571811).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors’ contributions
Text for this manuscript was written through contribution of all authors. All authors have given approval to the final version of the manuscript.
Competing interest
The authors declare that they have no competing interests.
Consent for publication
All authors have approved the manuscript be published.
Ethics approval and consent to participate
All animal handling procedures were reviewed and approved by the Animal Care/User Ethical Committee of Soochow University, Suzhou City, P. R. China.
Abbreviations
- circRNA
Circular RNA
- EMT
Epithelial-to-mesenchymal transition
- IHC
Immunohistochemistry
- TBST
Tris-buffered saline-Tween
- WLM
Working level months
- MREs
MiRNA response elements
Contributor Information
Jian Tong, Email: tongjian@suda.edu.cn.
Jihua Nie, Email: niejihua@suda.edu.cn.
References
- 1.Kreuzer M, Fenske N, Schnelzer M, Walsh L. Lung cancer risk at low radon exposure rates in German uranium miners. Br J Cancer. 2015;113:1367–1369. doi: 10.1038/bjc.2015.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peckham EC, Scheurer ME, Danysh HE, Lubega J, Langlois PH, Lupo PJ. Residential Radon Exposure and Incidence of Childhood Lymphoma in Texas, 1995–2011. Int J Environ Res Public Health. 2015;12:12110–12126. doi: 10.3390/ijerph121012110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen DH, Worrell J, Gibson PR. Radon Action Month: Why Nurses Should Care About Radon Exposure. Tar Heel Nurse. 2016;78:10. [PubMed] [Google Scholar]
- 4.Schwartz AG, Cote ML. Epidemiology of Lung Cancer. Adv Exp Med Biol. 2016;893:21–41. doi: 10.1007/978-3-319-24223-1_2. [DOI] [PubMed] [Google Scholar]
- 5.Ruano-Ravina A, Aragones N, Perez-Rios M, Lopez-Abente G, Barros-Dios JM. Residential radon exposure and esophageal cancer. An ecological study from an area with high indoor radon concentration (Galicia, Spain) Int J Radiat Biol. 2014;90:299–305. doi: 10.3109/09553002.2014.886792. [DOI] [PubMed] [Google Scholar]
- 6.Meenakshi C, Sivasubramanian K, Venkatraman B. Mutat Res. Nucleoplasmic bridges as a biomarker of DNA damage exposed to radon. Mutat Res. 2017;814:22–8. [DOI] [PubMed]
- 7.Nie JH, Chen ZH, Liu X, Wu YW, Li JX, Cao Y, et al. Oxidative damage in various tissues of rats exposed to radon. J Toxicol Environ Health A. 2012;75:694–699. doi: 10.1080/15287394.2012.690086. [DOI] [PubMed] [Google Scholar]
- 8.Bastide K, Guilly MN, Bernaudin JF, Joubert C, Lectard B, Levalois C, et al. Molecular analysis of the Ink4a/Rb1-Arf/Tp53 pathways in radon-induced rat lung tumors. Lung Cancer. 2009;63:348–353. doi: 10.1016/j.lungcan.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Kanduc D, Mittelman A, Serpico R, Sinigaglia E, Sinha AA, Natale C, et al. Cell death: apoptosis versus necrosis (review) Int J Oncol. 2002;21:165–170. [PubMed] [Google Scholar]
- 10.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 12.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 13.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 15.Lu D, Xu AD. Mini Review: Circular RNAs as Potential Clinical Biomarkers for Disorders in the Central Nervous System. Front Genet. 2016;7:53. doi: 10.3389/fgene.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M, Stockfleth E, Hessam S. Circular RNA expression in basal cell carcinoma. Epigenomics. 2016;8:619–632. doi: 10.2217/epi-2015-0019. [DOI] [PubMed] [Google Scholar]
- 17.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016. [DOI] [PubMed]
- 18.Xuan L, Qu L, Zhou H, Wang P, Yu H, Wu T, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8:932–939. [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z, Wang D, Gu C, Liu X, Pei W, Li J, et al. Down-regulation of let-7 microRNA increased K-ras expression in lung damage induced by radon. Environ Toxicol Pharmacol. 2015;40:541–548. doi: 10.1016/j.etap.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Minina VI, Sinitsky MY, Druzhinin VG, Fucic A, Bakanova ML, Ryzhkova AV, et al. Chromosome aberrations in peripheral blood lymphocytes of lung cancer patients exposed to radon and air pollution. Eur J Cancer Prev. 2016;5:26. doi: 10.1097/CEJ.0000000000000270. [DOI] [PubMed] [Google Scholar]
- 21.Leng S, Thomas CL, Snider AM, Picchi MA, Chen W, Willis DG, et al. Radon Exposure, IL-6 Promoter Variants, and Lung Squamous Cell Carcinoma in Former Uranium Miners. Environ Health Perspect. 2016;124:445–451. doi: 10.1289/ehp.1409437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/S1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 23.Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell. 2008;29:350–361. doi: 10.1016/j.molcel.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng X, Li X, Zhang P, Wang J, Zhou Y, Chen M. Circular RNA: an emerging key player in RNA world. Brief Bioinform. 2016. [DOI] [PubMed]
- 26.Li P, Chen H, Chen S, Mo X, Li T, Xiao B, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;12. doi:10.1038/bjc.2016.451 [DOI] [PMC free article] [PubMed]
- 27.Guo JN, Li J, Zhu CL, Feng WT, Shao JX, Wan L, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451–7458. doi: 10.2147/OTT.S123220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair AA, Niu N, Tang X, Thompson KJ, Wang L, Kocher JP, et al. Circular RNAs and their associations with breast cancer subtypes. Oncotarget. 2016;7(49):80967–80979. doi: 10.18632/oncotarget.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F, Zhang L, Li W, Deng J, Zheng J, An M, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- 32.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


