Abstract
Background
This study evaluated the impact of hormonal modulation at the onset of proestrus on ovarian response and uterine gene expression of beef cows.
Methods
A total of 172 anestrous beef cows were assigned to one of four groups according to the treatment with estradiol cypionate (ECP) and/or equine chorionic gonadotropin (eCG) [CON (n = 43), ECP (n = 43), eCG (n = 44) and ECP + eCG (n = 42)].
Results
ECP-treated cows (ECP and ECP + eCG groups) presented greater occurrence of estrus (44.6% vs. 65.4%; P = 0.01) and pregnancy per AI [47.1% vs. 33.3%; P = 0.07], but similar progesterone (P4) concentration at subsequent diestrus than cows not treated with ECP (CON and eCG groups). Nonetheless, eCG-treated cows (eCG and ECP + eCG groups) presented larger follicle at timed AI (12.6 ± 0.3 vs. 13.5 ± 0.3 mm; P = 0.03), greater ovulation rate (96.5% vs. 82.6%; P = 0.008) and greater P4 concentration at d 6 (3.9 ± 0.2 vs. 4.8 ± 0.2 ng/mL; P = 0.001) than cows not treated with eCG (CON and ECP groups). Next, cows with a new corpus luteum 6 d after TAI were submitted to uterine biopsy procedure. Uterine fragments [CON (n = 6), ECP (n = 6)] were analyzed by RNA-Seq and a total of 135 transcripts were differentially expressed between groups (73 genes up-regulated by ECP treatment). Subsequently, uterine samples were analyzed by qPCR (genes associated with cell proliferation). ECP treatment induced greater abundance of PTCH2 (P = 0.07) and COL4A1 (P = 0.02), whereas suppressed EGFR (P = 0.09) expression. Conversely, eCG treatment increased abundance of HB-EGF (P = 0.06), ESR2 (P = 0.09), and ITGB3 (P = 0.05), whereas it reduced transcription of ESR1 (P = 0.05). Collectively, supplementation with ECP or eCG at the onset of proestrous of anestrous beef cows influenced ovarian responses, global and specific endometrial gene expression.
Conclusion
Proestrus estradiol regulate the endometrial transcriptome, particularly stimulating proliferative activity in the endometrium.
Electronic supplementary material
The online version of this article (10.1186/s40104-017-0211-3) contains supplementary material, which is available to authorized users.
Keywords: Cattle, eCG, Endometrium, Estradiol, Transcriptome
Background
Synchronization of estrus and ovulation programs for timed artificial insemination (TAI) has been constantly incorporated on modern reproductive management of beef farms [1, 2]. These programs can induce the first postpartum ovulation and, consequently, hasten the establishment of pregnancy of suckled beef cows [1, 3–5]. However, a significant proportion of ovulated and inseminated cows are detected not-pregnant 30 d after insemination despite the satisfactory ovulation rate (~85%) following protocols for synchronization of ovulation [4, 6, 7]. The uterine environment plays a relevant role among factors that are likely to contribute to the observed failures [8–10].
Early classic studies demonstrated the significant impact of a coordinated and sequential exposure to ovarian steroids on uterine function [11–13]. Gene expression of bovine endometrium changes according to the phase of the estrous cycle and is closely controlled by circulating concentrations of estradiol (E2), progesterone (P4) and the expression ratio of their specific receptors [8, 14–17]. In this regard, proestrus E2 concentration is fundamental in modulation of the uterus for the subsequent luteal phase [8, 14, 18, 19]. This E2 priming may be important for induction of endometrial P4 receptors [20, 21] to avoid premature luteolysis and short cycles in beef cattle [22]. In cyclic dairy heifers, elevated E2 concentrations during proestrus, induce changes in uterine gene expression of E2 and P4 receptors (ESR1 and PGR, respectively), oxytocin receptors, and expression of cyclooxygenase-2, and beta subunit inhibin serpin-14 throughout the subsequent estrus cycle [23]. Also, cyclic beef heifers that are exposed to a longer proestrus period exhibit alterations in the pattern of steroids receptors expression in the uterus and other proteins associated with uterine receptivity to pregnancy [19]. Therefore, it is reasonable to hypothesize that the modulation of E2 concentration during the synchronized proestrus by means of exogenous E2 supplementation could also alter the uterine gene expression of suckled anestrous beef cows.
Two pharmacological strategies to manipulate the proestrus phase have been extensively evaluated in cattle breeding programs; exogenous E2 supplementation or equine chorionic gonadotropin (eCG) administration. Firstly, exogenous E2 supplementation using E2 esters enhances the proportion of cows that display estrus [24–26], increases endometrial thickness in lactating dairy cows [27] and improves the pregnancy success of suckled beef cows [6, 25, 26]. Furthermore, Jinks et al. [28] demonstrated that, recipients beef cows with lower E2 concentration at periovulatory phase, receiving in vivo-produced embryo, presented a dramatic reduction on pregnancy establishment (45% vs. 65% of pregnancy rate). Secondly, administration of eCG at onset of the proestrus is an efficient alternative to increase final follicular growth, ovulation rate and plasma P4 concentration on subsequent diestrus [5, 26, 29, 30]. Such changes may be responsible for the increase in pregnancy rates of anestrous beef cows stimulated with eCG [5, 26, 29, 30]. Altogether, both pharmacological strategies to manipulate the proestrus are capable of altering the periovulatory steroidal endocrine profiles, potentially modulating the expression of genes associated with uterine receptivity and ultimately positively influencing pregnancy establishment of suckled anestrous beef cows.
Therefore, based on the importance of the proestrus hormonal milieu on fertility, we hypothesized that supplementation with estradiol cypionate (ECP) and/or eCG at the onset of proestrus alters the ovarian response and the uterine transcriptome of suckled anestrous beef cows. To assess the above mentioned hypothesis, we chose the following approaches. First, taking a comprehensive approach, RNA extracts from endometrial fragments were submitted to Next Generation RNA sequencing followed by functional enrichment analysis to potentially identify and characterize other ECP-regulated biological and molecular processes and pathways. Secondly, following a candidate gene approach, we tested the effect of ECP and/or eCG supplementation on the expression of selected molecules with relevant biological functions in the context of uterine biology, specifically associated with cell proliferation.
Methods
Animals
Animal procedures were approved by the Ethics and Animal Handling Committee of the Faculdade de Medicina Veterinária e Zootecnia, Universidade de São Paulo (CEUA-FMVZ/USP, No. 2287/2011). This experiment was conducted during the 2012/2013 spring-summer breeding seasons. A total of 172 suckled anestrous Nelore (Bos indicus) beef cows at 30–60 d postpartum from a commercial farm in the state of Parana, Brazil, were enrolled in this study. Cows were maintained on Brachiaria brizantha pasture with water and mineral supplementation ad libitum. Immediately prior to the initiation of the TAI protocol, information about body condition score from each cow were recorded (BCS; range, 1 = emaciated to 5 = obese; with 0.5 scale) [31].
Reproductive management and experimental design
After calving, cows were allocated into breeding groups according to calving date. At 30 to 60 d post-partum, females were synchronized using an E2-plus-P4-based TAI protocol. Briefly, suckled cows received an intravaginal P4-releasing insert previously used for 8 d (1 g of P4; DIB®, MSD Animal Health, São Paulo, Brazil) on D −10 along with an intramuscular (IM) administration of 2 mg estradiol benzoate (EB; Gonadiol®, MSD Animal Health, São Paulo, Brazil; Fig. 1). The P4 insert were removed eight day later (D −2). All cows received an intramuscular administration of 500 mg of cloprostenol (Ciosin®, MSD Animal Health, São Paulo, Brazil) at the moment of the P4 insert removal. At this moment, cows were blocked by BCS, parity (multiparous vs. primiparous) and the diameter of the largest follicle and then randomly assigned into one of four experimental groups [Control (CON): n = 43, Estradiol cypionate (ECP): n = 43, eCG: n = 44, and ECP + eCG: n = 42], in a 2 × 2 factorial arrangement. Cows from ECP group received an IM injection of 1 mg of ECP (E.C.P.; Zoetis, São Paulo, Brazil), cows from eCG group received an IM injection of 400 IU of eCG (Folligon®, MSD Animal Health), while cows from ECP + eCG group received both treatments and cows from CON group did not receive any treatment. In all groups, ovulation was induced by 10 μg of buserelin acetate (GnRH, Sincroforte, Ourofino Saúde Animal, Cravinhos, São Paulo, Brazil) IM administration 48 h after the P4 insert removal (D 0). Cows were artificially inseminated immediately after GnRH treatment. Inseminations were performed by a single technician using frozen-thawed semen from single Angus sire with proven fertility. The sire used had been previously used in TAI programs and had satisfactory (~50%) pregnancy results.
Fig. 1.
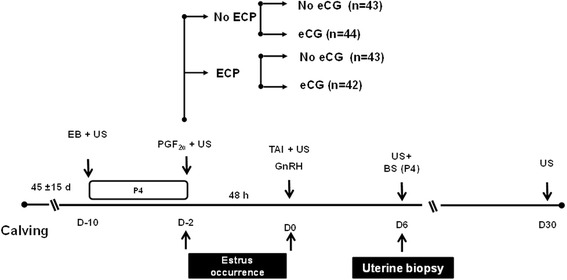
Schematic diagram of the synchronization of ovulation protocol in suckled anestrous beef cows. EB = 2 mg of estradiol benzoate; P4 = progesterone; P4 insert = previously used intravaginal P4 insert containing 1.0 g of P4; GnRH = 100 mg of gonadorelin; ECP = 1 mg of estradiol cypionate; eCG = 400 IU of equine chorionic gonadotropin; PGF2α = 0.25 mg of cloprostenol; US = ultrasound examination; BS = blood sample. Cows from ECP group received ECP and cows from eCG group received eCG, while cows from CON did not receive any further treatment and cows from ECP + ECG received both treatments
Estrus was determined based on the tail-head mark. At the time of the removal of the P4 insert, the tail-head was marked with chalk (Raidl-Maxi, RAIDEX GmbH, Dettingen/Erms, Germany). Estrus was deemed to have occurred in cattle without a tail-head mark at TAI.
Cows presenting a corpus luteum (CL) on D 6 (6 d after GnRH treatment) had the body of the uterus biopsied as previously described [32]. Fragments obtained from uterine biopsies were individually allocated in cryotubes and immediately immersed into liquid nitrogen. Day 6 was strategically selected as the moment in which an early embryo is expected to have recently accessed the uterine environments. Pregnancy was diagnosed by transrectal ultrasonography through the detection of a viable embryo (presence of heartbeat) on d 42 post-AI.
Blood sampling and hormone measurements
Blood sampling for determination of P4 concentrations was performed on D 6, concurrently with the uterine biopsy. Blood samples were collected by coccygeal venipuncture using evacuated tubes containing EDTA (BD, São Paulo, SP, Brazil) and immediately stored in ice. Plasma was separated by centrifugation at room temperature, 1,500 × g for 15 min, and stored at −20 °C. Progesterone concentrations were measured in all samples using a solid-phase radioimmunoassay (Coat-a-count, Siemens, Los Angeles, USA), as validated previously [33]. The P4 assay sensitivity was 0.08 ng/mL and the intra-assay coefficient of variation was 8.7%.
Ultrasound examinations
Transrectal ultrasound examinations were carried out on D −10, D −2, D 0 and D 6 to assess cyclic status, growth of the dominant follicle (DF), ovulation, and the presence of CL. Ultrasonography was performed with the aid of a B-mode (gray-scale) ultrasound instrument (8100, Chison Medical Imaging, Co, China), equipped with a multi-frequency linear-array transducer. The anestrous status was defined as the absence of CL in two consecutive ultrasound examinations performed on D − 10 and D −2. Ovulation was defined as the presence of a recently formed CL on D 6 on the same ovary that the DF was observed on D −2 and D 0. The diameter of the DF at the time of P4 insert removal and at TAI, in addition to the diameter of new CL formed, was calculated as the average between measurements of two perpendicular axes of each structure.
Tissue processing, RNA isolation and cDNA synthesis
Approximately 30 mg of endometrial tissue was macerated in liquid nitrogen using a stainless steel mortar and pestle and immediately mixed with buffer RLT from the PureLink®, RNA Mini kit (Thermo Fisher Scientific, São Paulo, SP, Brazil), as per manufacturer’s instructions. To maximize lysis, tissue suspension was passed at least ten times through a 21-ga needle, and centrifuged at 13,000 × g for 3 min for removal of debris, prior to supernatant loading and processing on RNeasy columns. Columns were eluted with 40 μL of RNase free water. Concentration of total RNA on extracts was measured by a spectrophotometer (Nanovue™ Plus, Spectrophotometer, GE Healthcare, UK, by the absorbance at 260 nm). Subsequently, samples were treated with 80 μL of DNAse I solution (Life Technologies, São Paulo, SP, Brazil) for 15 min at room temperature during RNA extraction protocol, according to manufacturer’s instructions. RNA samples were stored at −80 °C until cDNA synthesis. The cDNA was synthesized by reverse-transcription using High Capacity cDNA Reverse Transcription Kit (Life Technologies) according to manufacturer’s instructions. Briefly, 10 μL of master mix containing RT buffer, dNTP mix, random primers, RNase inhibitor and reverse transcriptase were mixed to 1 μg of total RNA and final volume of the reaction was adjusted to 20 μL. Immediately, reactions were incubated at 25 °C for 10 min, followed by incubation at 37 °C for 2 h, and reverse-transcriptase inactivation at 85 °C for 5 min. Samples were stored at −20 °C.
RNAseq
Prior to the RNA-seq analyses, 12 samples (n = 6/group; ECP and CON) were selected according to previously established criteria by ovarian, occurrence of estrus, pregnancy and endocrine responses. Cows having similar DF diameter at the time of P4 insert removal [ECP (12.1 ± 0.7 mm) and CON (12.1 ± 0.6 mm)] and similar circulating P4 concentration at the time of uterine biopsy [ECP (3.8 ± 0.2 ng/mL) and CON (3.6 ± 0.2 ng/mL)] were considered suitable to further analysis. Additionally, cows were also selected based on pregnancy status 30 d after TAI in order to have both pregnant and non-pregnant cows represented in both experimental groups. Finally, only cows displaying estrus were selected in ECP treated group, whereas only cows that did not display estrus were chosen in the control group. The latter criterion was applied aiming to increase the distinction between two different E2 pre-ovulatory endocrine environments, as cows that display estrus present greater E2 concentration than those not displaying estrus [42].
Integrity of total RNA extracts was assessed using the Agilent RNA 6000 Nano chip (Bioanalyzer, Agilent Technologies). RNA Integrity Number (RIN) of extracts submitted to RNA sequencing analysis ranged from 8.3 to 8.7. Next, 4 μg of RNA were used with the TruSeq RNA Sample Preparation kit (Illumina, San Diego, CA) to prepare the libraries for RNA-Seq. The insert sizes were estimated through the Agilent DNA 1000 chip (Agilent Technologies) and the libraries concentration were measured through Quantitative Real-Time PCR (qPCR) with a KAPA Library Quantification kit (KAPA Biosystems). Samples were diluted, pooled in equimolar amounts and then sequenced at the Centro Genômico Funcional Aplicado a Agropecuária e Agroenergia at the University of São Paulo using a HiScanSQ sequencer (Illumina, San Diego, CA).
Bioinformatics analyses
Raw sequences were trimmed for adaptors and low quality using SeqyClean v1.3.12. (https://github.com/ibest/seqyclean) using 26 Phred quality parameter for maximum average error and a fasta file with contaminant sequences from the Univec database (https://www.ncbi.nlm.nih.gov/tools/vecscreen/univec/). Only high quality paired-end sequences were kept for further analyses The reads were mapped with Bowtie2 v2.1.0 [34] on the masked bovine genome assembly (Bos taurus UMD 3.1, NCBI). The mapping file was sorted using SAMTools v 0.1.18 [35] and read counts were obtained using the script from HTSeq-count v0.5.4p2 (http://htseq.readthedocs.io/en/release_0.9.1/). The differential expression analysis was performed with package DESeq2 [36] from R [37]. Using the function estimateSizeFactors, the normalized counts were obtained (baseMean values, which are the number of reads divided by the size factor or normalization constant). The standard deviation along the baseMean values was also calculated for each gene. In order to avoid artifacts caused by low expression profiles and high expression variance, only transcripts that had an average of baseMean >5 and the mean greater than the standard variation were analyzed. The threshold for evaluating significance was obtained by applying an alpha ≤0.10, considering the FDR-Benjamini-Hochberg P-value [38]. Integrated analysis of different functional databases was done using the functional annotation tool of the Database for Annotation, Visualization, and Integrated Discovery using as background the genes (DAVID) [39] using as background the set of genes that passed through the differential expression analysis filter.
qPCR
The samples employed in qPCR analysis were selected mirroring the general results obtained in regard to ovarian and endocrine responses. Cows receiving ECP should present greater occurrence of estrus, while cows from eCG treatment group should present greater circulation of P4 concentration at the moment of the uterine biopsy. Step-One Plus thermocycler (Life Technologies, Carlsbad, CA) and SYBR Green chemistry were used for quantitative PCR analysis. Primers were designed based on the mRNA sequence of target genes obtained from the RefSeq database, on Genbank (http://www.ncbi.nlm.nih.gov/genbank/). Sequences were masked to remove repetitive sequences with RepeatMasker (http://www.repeatmasker.org/) [40] and then, the masked sequences were used for primer design using the PrimerQuest software (IDT1, http://www.idtdna.com/primerquest/Home/Index). The characteristics of the primers were checked in Oligo Analyzer 3.1 software (IDT1, http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/), while the specificity was compared by BLAST (NCBI, http://blast.ncbi.nlm.nih.gov). The qPCR products obtained from reactions performed with primers not previously validated were submitted to agarose gel electrophoresis and SANGER-DNA sequencing, and identities of target genes were confirmed. Details of primers are provided on Table 1. In order to select reference genes, the GeNorm Microsoft Excel applet was used, as this applet provides a measure of gene expression stability (M) [41]. The Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), Actin, Beta (ACTB) and Ribosomal Protein S18 (RPS18) were the most stable genes and were, therefore, selected as reference genes. Determination of qPCR efficiency and Cq (quantification cycle) values per sample were performed with LinRegPCR software (V2014.2; http://www.hartfaalcentrum.nl/index.php?main=files&fileName=LinRegPCR.zip&description=LinRegPCR:%20qPCR%20data%20analysis&sub=LinRegPCR). Quantification was obtained after normalization of the target genes expression values (Cq values) by the geometric mean of the endogenous control expression values. The following genes, associated with regulation of cell proliferation in the uterus, were selected: ovarian steroid receptors [Estrogen Receptor alpha (ESR1), Estrogen Receptor beta (ESR2), P4 receptor (PGR)], growth factors that regulate cellular proliferation [epidermal growth factor receptor (EGFR), heparin-binding EGF-like growth factor (HB-EGF) and patched homolog 2 (PTCH2)], and extracellular matrix [collagen, type IV, alpha 1 (COL4A1) and integrin, beta 3 (ITGB3)].
Table 1.
Gene name, accession number, forward and reverse primer sequences used for qPCR analysis
| Gene Name | Gene ID | Sequence ID | Forward primer sequence (5′–3′) | Reverse sequence (5′–3′) | Primer efficiency, % | Amplicon length, bp |
|---|---|---|---|---|---|---|
| Progesterone receptor | PGR | NM_001205356.1 | GCCGCAGGTCTACCAGCCCTA | GTTATGCTGTCCTTCCATTGCCCTT | 96.9 | 199 |
| Estrogen receptor 1 | ESR1 | NM_001001443.1 | CAGGCACATGAGCAACAAAG | TCCAGCAGCAGGTCGTAGAG | 99.1 | 82 |
| Estrogen receptor 2 | ESR2 | NM_174051.3 | TCACGTCAGGCACGCCAGTAAC | CACCAGGTTGCGCTCAGACCC | 99.5 | 155 |
| Patched 2a | PTCH2 | XM_005197904.1 | CATCCTGCTGCTGTGTACTT | ATCGCCAGGACCAGTACTAT | 99.9 | 87 |
| Epidermal growth factor receptor | EGFR | XM_002696890.3 | ATGCTCTATGACCCTACCAC | TTCCGTTACAAACTTTGCCA | 97.6 | 178 |
| Heparin-binding EGF-like growth factor | HB-EGF | NM_001144090.1 | CATCCACGGAGAATGCAAATAC | CAGCAGACAGACGGATGATAG | 98.6 | 181 |
| Collagen, type IV, alpha 1 | COL4A1 | NM_001166511.1 | CACGGCTACTCTTTGCTCTAC | GAAGGGCATGGTACTGAACTT | 96.48 | 102 |
| Integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | ITGB3 | NM_001206490.1 | GGGAGAGTGCTATGGTTAGA | CTTCACAAGACACCCAAGAG | 92.09 | 142 |
| Actin Beta | ACTB | NM_173979.3 | GGATGAGGCTCAGAGCAAGAGA | TCGTCCCAGTTGGTGACGAT | 93.7 | 77 |
| Glyceraldehyde-3-Phosphate Dehydrogenase | GAPDH | NM_001034034.2 | GCCATCAATGACCCCTTCAT | TGCCGTGGGTGGAATCA | 99.99 | 69 |
| Ribosomal Protein S18 | RPS18 | AY786141.1 | TGGAGAGTATTGCGCCTTCTC | CACAAGTTCCACCACACTATTGG | 97.9 | 79 |
aTranscript variants X1 to X7
Statistical analyses from ovarian, endocrine and gene expression responses
The statistical analyses for ovarian responses were performed using the PROC GLIMMIX of SAS for Windows (SAS 9.3 Institute Inc., Cary, NC, USA, 2003). Continuous variables were presented as mean ± standard error of the mean (mean ± SEM) and percentage (%) for frequency of occurrence for binomial variables. The continuous response variables were subjected to response scaling test through the solution Guided Data Analysis of SAS. Variables that did not follow these assumptions were transformed accordingly. Binomial variables (i.e. occurrence of estrus and ovulation rate) were analyzed by logistic regression using the SAS GLIMMIX procedure with models fitted to binomial distributions. The explanatory variables considered for inclusion in the models were the treatment with ECP, eCG and interaction of ECP and eCG. The effect of cow within each replicate was included as a random effect.
The qPCR data were tested for normality of residuals and homogeneity of variances followed by ANOVA using the GLIMMIX procedure of SAS fitting log normal distribution. The explanatory variables considered for inclusion in the models were the treatment with ECP, eCG and interaction between ECP and eCG. Final results are presented in natural log (Ln) scale (because of the log normal distribution considered) as normalized values of a specific gene transcript by the mean level of the transcript from Control (No-ECP and No-eCG treated animals). Down-regulation of expression in a specific experimental group may be represented by negative values relative to control because of Ln scale. To avoid negative values, the mean used for data normalization was divided by the fifth negative exponent. All data were compared with the relative mean expression level of the control group.
Statistical difference was considered when P < 0.10. Graphs were plotted with Sigmaplot (version 11.0; Systat Software, Inc. San Jose, CA, USA).
Results
Ovarian, pregnancy and endocrine responses
Animals receiving different hormonal therapies at the proestrus presented different rates of occurrence of estrus between P4-releasing device removal and TAI, final follicular growth, ovulatory responses and subsequent CL function (Table 2). There were no interactions between ECP and eCG treatment on response variables, except for the CL diameter 6 d after the TAI (P = 0.06). Larger CLs were observed in cows treated with eCG, especially in cows not treated with ECP. The ECP treated cows presented a greater frequency of occurrence of estrus [ECP = 64.7% (55/85) vs. No-ECP = 44.8% (39/87); P = 0.008] and presented greater pregnancy per TAI [ECP = 47.1% (40/85) vs. No-ECP = 33.3% (29/87); P = 0.07]. Cows treated with eCG presented greater rate of final follicular growth [eCG = 1.2 ± 0.1 mm/d vs. No-eCG = 0.9 ± 0.1 mm/d; P = 0.01], resulting in a greater DF diameter at TAI [eCG = 13.5 ± 0.3 mm vs. No-eCG = 12.6 ± 0.3 mm; P = 0.03]. Also, a greater proportion of cows receiving eCG displayed estrus [eCG = 62.8% (54/86) vs. No-eCG = 46.5% (40/86); P = 0.03] and ovulated [eCG = 96.5% (83/85) vs. No-eCG = 82.6% (71/86); P = 0.008]. A greater P4 concentration at the moment of uterine biopsy (D 6) was observed in cows receiving eCG at the onset of the proestrus [eCG = 4.8 ± 0.2 ng/mL vs. No-eCG = 3.9 ± 0.2 ng/mL; P = 0.001]. However, there was no influence of eCG treatment on the pregnancy per TAI [eCG = 43.0% (37/86) vs. No-eCG = 37.2% (32/86); P = 0.42].
Table 2.
Overall occurrence and effects of treatment with estradiol cypionate (ECP) and/or equine chorionic gonadotropin (eCG) at onset of the proestrus on follicular and luteal development in an estradiol/progesterone-based synchronization protocol on anestrous suckled beef cows
| Itens | Treatments1 | Pvalue | |||||
|---|---|---|---|---|---|---|---|
| No ECP | ECP | ||||||
| No eCG | eCG | No eCG | eCG | ECP | eCG | ECP × eCG | |
| Number of cows | 43 | 44 | 43 | 42 | – | – | – |
| BCS at onset of the synchronization2 | 3.1 ± 0.1 | 3.0 ± 0.1 | 2.9 ± 0.1 | 3.0 ± 0.1 | 0.12 | 0.71 | 0.43 |
| DF diameter at insert removal, mm3 | 11.0 ± 0.4 | 11.2 ± 0.4 | 10.8 ± 0.4 | 11.3 ± 0.4 | 0.90 | 0.38 | 0.77 |
| DF diameter at TAI, mm4 | 12.6 ± 0.4 | 13.6 ± 0.4 | 12.7 ± 0.4 | 13.4 ± 0.4 | 0.90 | 0.03 | 0.68 |
| Daily DF growth, mm/d5 | 0.9 ± 0.1 | 1.3 ± 0.1 | 0.9 ± 0.1 | 1.1 ± 0.1 | 0.52 | 0.01 | 0.25 |
| Occurrence of estrus, %6 | 37.2 | 52.3 | 55.8 | 73.8 | 0.008 | 0.03 | 0.77 |
| Ovulation rate, %7 | 81.4 | 95.5 | 83.7 | 97.6 | 0.54 | 0.008 | 0.71 |
| CL diameter at d 6 after TAI, mm | 17.8 ± 0.6b | 20.1 ± 0.5a | 18.6 ± 0.6ab | 18.7 ± 0.6ab | 0.54 | 0.04 | 0.06 |
| Plasma P4 at d 6 after TAI, ng/mL | 3.8 ± 0.3 | 5.1 ± 0.3 | 4.1 ± 0.3 | 4.6 ± 0.3 | 0.94 | 0.001 | 0.18 |
| Pregnancy per TAI, % | 30.2 | 36.4 | 44.2 | 50.0 | 0.07 | 0.42 | 0.95 |
1Suckled anestrous beef cows received an previously used intravaginal insert containing 1.0 g of progesterone (P4) and 2.0 mg of estradiol benzoate on the first day of the estrus/ovulation synchronization protocol (D −10). The P4 insert was removed eight days later (D −2), and cows from ECP group received an IM treatment of 1 mg of ECP, cows from eCG group received an IM injection of 400 IU of eCG, while cows from ECP + ECG received both treatments and cows from CON did not receive any treatment at this moment. All cows received GnRH IM and were timed artificially inseminated (TAI) 48 h after the P4 insert removal (D 0). Different letters within the same row indicate the presence of difference between groups (P <0.05) when an interaction between eCG and ECP was observed
2BCS = Body condition score collected at insertion of the P4 insert
3DF = Dominant follicle
4TAI = timed artificial insemination
5DF growth between the P4 insert removal and TAI divided by two
6Estrus determined based on the tail-head mark
7Number of cows with a new CL formed 6 d after the TAI divided by the number of animal synchronized
RNA-seq
RNA sequencing produced a total of ~334 million reads with an average of 27.5 million reads for each group. Six biological replicates were analyzed for each phenotype (please see Statistical Analyses section above) with the reads ranging from 17 to 26 million per sample after filtering (Additional file 1: Table S1). Approximately ~65% of the total reads uniquely mapped to the UMD 3.1 reference genome (https://www.ncbi.nlm.nih.gov/genome?term=bos%20taurus). Only the uniquely mapped reads were considered in the analysis. From the remaining, approximately 20% of the reads were not uniquely mapped, and 15% unmapped reads. After applying the variance and minimal value of baseMean filtering, a total of 15,161 genes were included on the differential expression analysis. A total of 310 out of the 15,161 analyzed genes showed differential expression (adjusted P-value <0.1), of which 73 and 62 were upregulated in the endometrium of ECP and CON samples, respectively (see Volcano plot, Fig. 2 and Additional file 2: Table S2). Differentially expressed genes (DEG) with the greatest expression profiles were RPS2 [ribosomal protein S2], GABARAP [GABA (A) receptor-associated protein], up-regulated in the CON endometrium, and PEPD [peptidase D], SG100g [calcium binding protein G] and CEACAM1 [carcinoembryonic antigen-related cell adhesion molecule 1], up-regulated in the ECP group. Heatmap on Fig. 3 shows the 50 genes with the lowest p-adjusted values. It is possible to observe the similarity of gene expression patterns among individuals within each group, as indicated by the shades of green (for low expression) or red color (high expression).
Fig. 2.
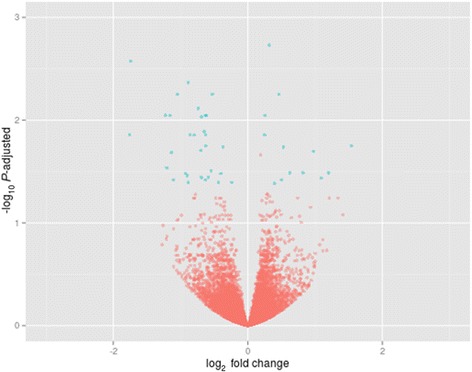
Volcano plot obtained from DESeq analysis. Volcano plot shows that the vertical lines axe is log2-fold change and the horizontal axis is the statistical significance (P value ≤ 0.10). Genes with P value ≤0.10 are marked with blue dots
Fig. 3.
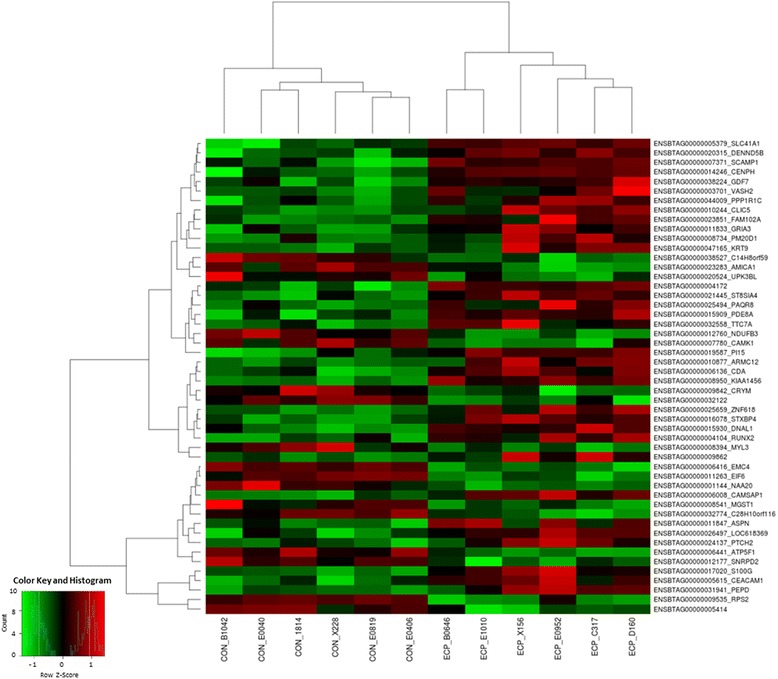
Heat map obtained from DESeq2 analysis. Each column represent one sample showing the intensity of expression profile per gene. The colors in the map display the relative standing of the reads count data; GREEN indicates a count value that is lower than the mean value of the row while red indicates higher than the mean. The shades of the color indicate distance from each data point to the mean value of the row
Sequences of all reads were deposited in the Sequence Read Archive (SRA) of the NCBI (http://www.ncbi.nlm.nih.gov/sra/; Additional file 3: Table S3) and, an overview of these data has been deposited in NCBI’s Gene Expression Omnibus (GEO) and is accessible through GEO Series accession number GSE67807.
Functional enrichment analysis of RNA-seq data - DAVID results
KEGG pathway and Gene ontology (GO) term analyses were performed with DAVID (Table 3). Functional enrichment analysis using DAVID revealed two KEGG pathways overrepresented by the ECP-upregulated transcripts: pathways in cancer (5 genes; P < 0.01) and small cell lung cancer (3 genes; P < 0.05). On the other hand, ECP downregulated transcripts indicated the enrichment of three pathways: Parkinson’s disease (3 genes; P = 0.06), oxidative phosphorylation (3 genes; P = 0.06) and Alzheimer’s disease (3 genes; P = 0.09). More specifically, ECP-upregulated transcripts associated with pathways in cancer were [gene symbol (fold change; adjusted P value on RNA-seq); respectively]: LAMC3 (1.55; P = 0.10), PTCH1 (1.51; P = 0.09), PTCH2 (1.52; P = 0.03), PIK3R3 (1.22; P = 0.10), and PIAS1 (1.18; P = 0.09), whereas ECP downregulated transcripts associated with oxidative phosphorylation were ATP5F1 (1.18; P = 0.01), ATP5J (1.24; P = 0.06), and NDUFB3 (1.37; P = 0.01). Additionally, analysis of GO terms identified that ECP upregulated transcripts over represented epidermis development [ADAM9 and ENSBTAG00000017455 (uncharacterized protein)]. On the other hand, ECP downregulated GO terms indicated the enrichment of five biological processes: generation of metabolic precursors and energy (GPI, NDUFB3, ATP5F1, IDH3B, ATP5J), Translation (RPS2, EEF1D, ENSBTAG00000013866, ENSBTAG00000011263), and mRNA processing, mRNA metabolic process and RNA splicing with 3 common genes (GEMIN7, SNRPD2, STRAP).
Table 3.
KEGG Pathways and Gene ontologies (GO category) of mRNA transcripts differentially expressed in cows treated with estradiol cypionate (ECP).
| Category | Term | Count | P value | Genes |
|---|---|---|---|---|
| Upregulated in ECP | ||||
| KEGG PATHWAY | Pathways in cancer | 5 | 0.008 | PIK3R3, PTCH1, PTCH2, LAMC3, PIAS1 |
| KEGG PATHWAY | Small cell lung cancer | 3 | 0.020 | PIK3R3, LAMC3, PIAS1 |
| GO TERM BP_FAT | Epidermis development | 2 | 0.095 | ADAM9, ENSBTAG00000017455 |
| Downregulated in ECP | ||||
| KEGG PATHWAY | Parkinson’s disease | 3 | 0.062 | NDUFB3, ATP5F1, ATP5J |
| KEGG PATHWAY | Oxidative phosphorylation | 3 | 0.064 | NDUFB3, ATP5F1, ATP5J |
| KEGG PATHWAY | Alzheimer’s disease | 3 | 0.090 | NDUFB3, ATP5F1, ATP5J |
| GO TERM BP_FAT | Generation of precursor metabolites and energy | 5 | 0.004 | GPI, NDUFB3, ATP5F1, IDH3B, ATP5J |
| GO TERM BP_FAT | Translation | 4 | 0.059 | RPS2, EEF1D, ENSBTAG00000013866, ENSBTAG00000011263 |
| GO TERM BP_FAT | mRNA processing | 3 | 0.071 | GEMIN7, SNRPD2, STRAP |
| GO TERM BP_FAT | mRNA metabolic process | 3 | 0.087 | GEMIN7, SNRPD2, STRAP |
| GO TERM BP_FAT | RNA splicing | 3 | 0.041 | GEMIN7, SNRPD2, STRAP |
Enrichment analysis was performed with DAVID tools (https://david.ncifcrf.gov/tools.jsp)
Cell proliferation-related gene expression
According to selection criteria described previously, uterine tissue used in the qPCR analysis derived from cows that presented different estrus responses [CON (10.1%), ECP (90.9%), eCG (66.7%) and ECP + ECG (83.3%)] and P4 concentration at uterine biopsy [CON (3.4 ± 0.2 ng/mL), ECP (3.7 ± 0.2 ng/mL), eCG (5.3 ± 0.4 ng/mL) and ECP + ECG (5.0 ± 0.6 ng/mL)].
There were no interactions (P > 0.10) between ECP and eCG on the expression of the transcripts evaluated. ECP treatment induced greater endometrial abundance of PTCH2 (P = 0.07) and COL4A1 (P = 0.02) genes, whereas it reduced EGFR (P = 0.09) gene expression (Figs. 4 and 5). The ECP treatment did not affect gene expression of ESR1 (P = 0.90), ESR2 (P = 0.61), HB-EGF (P = 0.80) and ITGB3 (P = 0.57). On the other hand, eCG treatment induced greater endometrial abundance of HB-EGF (P = 0.06), ESR2 (P = 0.09), and ITGB3 (P = 0.05) genes, whereas reduced the gene expression of ESR1 (P = 0.05). Supplementation with eCG did not alter expression of EGFR (P = 0.34), PTCH2 (P = 0.31) and COL4A1 (P = 0.19). Additionally, expression of PGR was not altered by either ECP (P = 0.51) or eCG (P = 0.25) treatments.
Fig. 4.
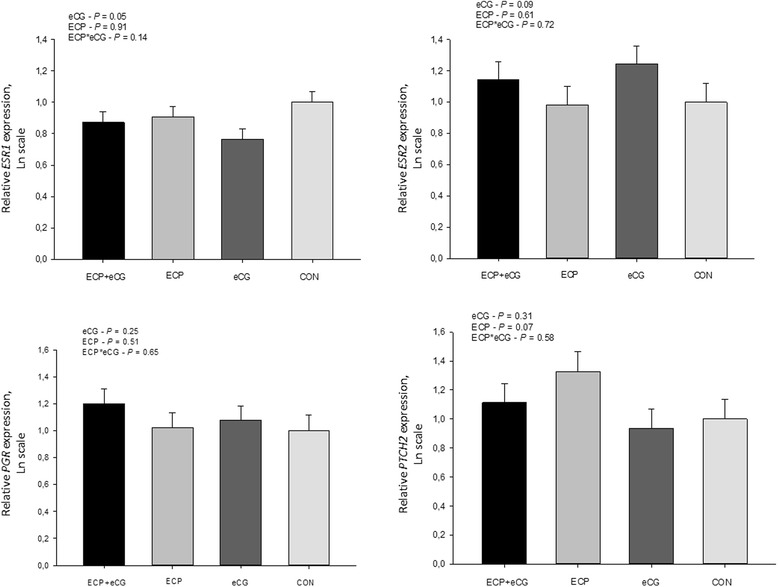
Comparison of gene expression between suckled anestrous beef cows receiving 1 mg of estradiol cipionate (ECP) and/or 400 IU of equine chorionic gonadotropin (eCG) at onset of the proestrus [CON (n = 11), ECP (n = 11), eCG (n = 12) and ECP + ECG (n = 11)]. The amounts of ESR1, ESR2, PGR, and PTCH2 transcripts are expressed in relation to control (CON) untreated cows. Expression values were normalized by the geometric mean of GAPDH, ACTB, and RPS18. The P values refer to comparisons made for each gene between groups (effects of ECP, eCG and interaction between ECP and eCG)
Fig. 5.
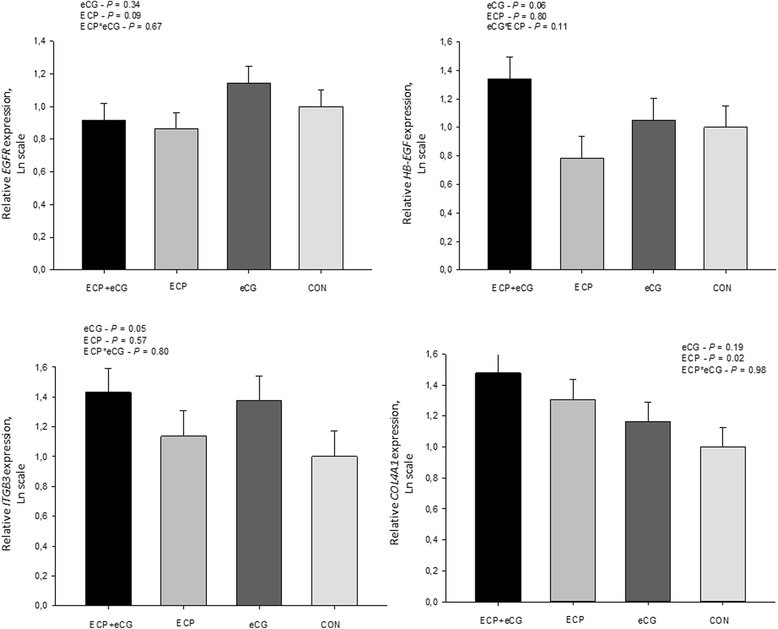
Comparison of gene expression between suckled anestrous beef cows receiving 1 mg of estradiol cipionate (ECP) and/or 400 IU of equine chorionic gonadotropin (eCG) at onset of the proestrus [CON (n = 11), ECP (n = 11), eCG (n = 12) and ECP + ECG (n = 11)]. The amounts of EGFR, HBEGF, ITGB3, and COL4A1 transcripts are expressed in relation to control (CON) untreated cows. Expression values were normalized by the geometric mean of GAPDH, ACTB, and RPS18. The P values refer to comparisons made for each gene between groups (effects of ECP, eCG and interaction between ECP and eCG)
Discussion
The present study investigated the impact of hormonal manipulation of proestrus on ovarian response and on uterine transcriptome 6 d post-TAI. The most relevant observations from this study are: 1) ECP treatment improves occurrence of estrus and pregnancy per AI, whereas eCG treatment enhances final follicular growth, size of ovulatory follicle, ovulation rate and subsequent P4 concentration, 2) the endometrial transcriptional profile is regulated by ECP supplementation and cell proliferation was one of the overrepresented gene ontology terms; 3) selected candidate genes with altered expression further support an ECP effect on cellular proliferation and tissue morphology.
Synchronized cows displaying estrus before TAI exhibited larger dominant follicles, greater E2 concentration during the proestrus/estrus, greater luteal function on the subsequent estrus cycle, and greater conception rate when compared to cows that did not display estrus [6, 25, 42–44]. In agreement, exogenous ECP treatment at the onset of proestrus improved the proportion of suckled beef cows displaying estrus, determining greater pregnancy outcomes following TAI than non-ECP treated cows [25, 26], similar to what was observed in the present study. Furthermore, the eCG treatment at onset of the proestrus was effective to increase conception rates in suckled beef cows [1, 5, 29, 30]. Also corroborating with the present results, eCG-treated cows presented greater final follicular growth, follicular diameter at TAI, ovulation rate and plasma P4 concentration on subsequent diestrus [5, 26, 29, 30]. Therefore, the hormonal therapies established in the present study may be considered a pro-fertility model for suckled anestrous beef cows and potentially allow the establishment of two distinct periovulatory endocrine milieus, that are associated with an uterine environment of better receptivity. Specifically, it was expected that ECP-treated cows present greater periovulatory E2 concentration due to the exogenous estradiol administration. Additionally, those cows treated with eCG also presented greater concentrations of E2 during proestrus/estrus due to endogenous estradiol from a healthy larger DF at TAI, in addition to presenting greater concentrations of P4 during early diestrus.
Unexpectedly, transcriptome analysis of D 6 endometrium from cows treated or not with ECP did not reveal dramatic differences of gene expression patterns. Our model was unique in selecting for cows displaying estrus behavior in ECP-treated group versus not displaying estrus behavior in the control group. Estrus behavior is associated with higher pregnancy rates [6, 25, 43, 44]. Global transcriptome analysis of D 14 endometrium from high fertility heifers compared to low fertility ones did not reveal substantial differences [45]. Another study using a similar criterion for high and low fertility revealed that D 7 endometrium presented 417 DEG, however, most of the DEG exhibited fold change between 1.0 and 2.0 [46]. These results are in agreement with our data showing that endometrial gene expression is not dramatically different between groups with contrasting fertility; however, it is important to point out that half of the samples came from pregnant animals, whereas the other half came from non-pregnant cows in both ECP or control groups.
Estradiol levels are higher after ECP administration [28, 42, 47] and estrus behavior is correlated with estradiol levels [42, 48]; however, we did not quantify estradiol plasma concentrations. It was observed in ovariectomized cows that estradiol benzoate injection alters global gene expression of the endometrium when compared to a control group or progesterone treatment; whereas a combined estradiol and progesterone group shows data closer to estradiol treatment, suggesting that estradiol counteracts progesterone effects [49]. In our model, progesterone is the dominant steroid hormone at the time of sample collection; however its impact on gene expression is likely influenced by the previous exposure to estradiol.
We have observed previously that the endometrial tissue of cows ovulating larger follicles expressed markers of proliferative activity earlier than cows ovulating smaller follicles [8]. Similarly, gene expression changes suggesting reduction of proliferative activity and transition to a biosynthetic phenotype were also hastened in cows with larger ovulatory follicles. Larger follicles led to increased estradiol concentrations during proestrus and greater progesterone concentrations during early diestrus [8, 14]. Functional enrichment analysis using DAVID identified gene ontology terms associated with regulation of cell proliferation such as pathways in cancer and small cell lung cancer. Similarly, endometrial gene expression at D 7 in one estrous cycle prior to embryo transfer revealed enrichment of GO-terms cell cycle and anti-apoptosis in cows that successfully established pregnancy [50]. It is noteworthy that the above mentioned studies obtained samples from non-lactating cyclic cows [8] or heifers [50], whereas in the present study all cows were lactating and in anestrus. Additionally, assessment of the expression of proliferation-related candidate genes showed that PTCH2 and COL4A1 were induced by ECP treatment, whereas EGFR expression was suppressed. PTCH2 is a membrane receptor, member of the Hedgehog signaling pathway [51], and has been associated with proliferation-related disorders such as endometriosis and ovarian carcinoma [52], playing a role as a tumor suppressor gene [53]. COL4A1 encodes a type IV collagen protein that is an integral component of basement membranes [54]. In the endometrium, the breakdown of the basement membrane as well as increased expression of COL4A1 have been related with inhibition of angiogenesis and reduced tumor growth [55]. In addition, the oncogene EGFR, which is associated with growth of placental tissue [56], was downregulated by ECP-treatment suggesting a suppression of the endometrial ability to respond to mitogenic stimuli. The collective interpretation of these data is that estrogenic stimulus given by ECP induced a non-proliferative status on D6 endometrium. Such findings are consistent with our previous report, in which ovulation of a larger follicle (associated with greater proestrus and estrus plasma concentrations of estradiol) inhibited proliferation in both luminal and glandular epithelial cells on D 7 endometrium [8]. Importantly, such regulation occurred despite similar plasma concentrations of P4 between animals that received an did not receive ECP.
The most remarkable eCG-induced changes in gene expression were associated to E2 signaling. Indeed, transcript abundance was greater for ESR2 and lesser for ESR1 in eCG-treated cows than No eCG-treated cows, suggesting the establishment of a transition phase, from proliferative to secretory. The recognized proliferative role of estrogens in the female reproductive tract appears to be mediated by ESR1 [57]. After estrus, ESR1 abundance decreases and reaches nadir endometrium concentrations during the mid-luteal phase of the estrous cycle [58]. In contrast, uterine ESR2 expression is positively associated with the increasing P4 concentration observed from early to mid diestrus. The greater abundance of ESR2 expression found in eCG treated cows could be justified by the positive effect of eCG on P4 concentration during early diestrus. Altogether, these results suggest that the endometrium of suckled anestrous cows at Day 6 receiving either ECP or eCG is still transitioning from a proliferative to a secretory state, as previously reported [8].
Conclusions
Supplementation with ECP or eCG at onset of the synchronized proestrus of suckled anestrous beef cows significantly influence the ovarian responses; however, the impact on global uterine gene expression is discrete, presenting few DEG that are associated with ceasing cell proliferation. Such phenotype is consistent with the beginning of the secretory phase of the endometrium, required to support conceptus growth and survival.
Additional files
Number of reads from all samples from suckled cows receiving (ECP) or not (CON) 1 mg of ECP at the onset of the proestrous (DOCX 14 kb)
Differential gene expression results. BaseMean is the average of all samples expression profile after normalization; lfcSE – standard error from log2FoldChange; padj – P-value adjusted after correction of BH-FDR for multiple tests (DOCX 26 kb)
Bio-samples and Experiment accession numbers of the Raw reads resulted from the RNAseq of endometrial biopsis in the SRA data base (DOCX 20 kb)
Acknowledgements
We would like to thank the Empyreo Farm (Jacarezinho – PR) for allowing the use of their animals and facilities during the trial. These experiments were supported by Firmasa-Pecuária com Tecnologia. This research was funded, in part, by São Paulo Research Foundation (FAPESP; 2012/14731-4). We are thankful to Professor Marcos Roberto Chiaratti from Departamento de Genética e Evolução, CCBS, Universidade Federal de São Carlos - UFSCar for the statistical analysis of qPCR data. We also want to thank Bruno Moura Monteiro, Julia G. Soares and Milena L. Oliveira for technical support.
Funding
FAPESP (2012/14731–4) to MFSF.
CAPES PEC-PG 15068–12-9 to AMGD.
CNPq 142,387–2015-0 to MS.
CNPq 481,199/2012–8 and FAPESP- 2011/03226–4 to MB.
The funding bodies had no participation on the study, collection, analysis, interpretation of data or in writing the manuscript.
Availability of data and materials
Sequences of all reads generated in the RNAseq were deposited in the Sequence Read Archive (SRA) of the NCBI (http://www.ncbi.nlm.nih.gov/sra/; Additional file 3: Table S3) and, an overview of these data has been deposited in NCBI’s Gene Expression Omnibus (GEO) and is accessible through GEO Series accession number GSE67807.
Abbreviations
- ACTB
actin, Beta
- ADAM9
ADAM metallopeptidase domain 9
- ANOVA
Analysis of variance
- ATP5F1
ATP Synthase, H+ Transporting, Mitochondrial Fo Complex Subunit B1
- ATP5J
ATP Synthase, H+ Transporting, Mitochondrial Fo Complex Subunit F6
- BCS
Body contition score
- CEACAM1
Carcinoembryonic antigen-related cell adhesion molecule 1
- CL
Corpus luteum
- COL4A1
Collagen, type IV, alpha 1
- CON
Control
- Cq
Quantification cycle
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- DEG
Differentially expressed genes
- DF
Dominant follicle
- E2
Estradiol
- eCG
Equine chorionic gonadotropin
- ECP
Estradiol cypionate
- EDTA
Ethylenediaminetetraacetic acid
- EEF1D
Eukaryotic translation elongation factor 1 delta
- EGFR
Epidermal growth factor receptor
- ESR1
Estrogen receptor alpha
- ESR2
Estrogen receptor beta
- GABARAP
GABA (A) receptor-associated protein
- GAPDH
Glyceraldehyde-3-Phosphate Dehydrogenase
- GEMIN7
Gem nuclear organelle associated protein 7
- GEO
NCBI’s gene expression omnibus
- GnRH
Gonadotropin-releasing hormone
- GO
Gene ontology
- GPI
Glucose-6-Phosphate Isomerase
- HB-EGF
Heparin-binding EGF-like growth factor
- IDH3B
Isocitrate dehydrogenase 3 (NAD(+)) Beta
- IM
Intramuscular
- ITGB3
Integrin, beta 3
- LAMC3
Laminin Subunit Gamma 3
- LN
Natural logarithm
- NDUFB3
NADH:Ubiquinone Oxidoreductase Subunit B3
- P4
Progesterone
- PEPD
Peptidase D
- PGR
Progesterone receptor
- PIAS1
Protein Inhibitor Of Activated STAT 1
- PIK3R3
Phosphoinositide-3-Kinase regulatory subunit 3
- PTCH1
Patched 1
- PTCH2
Patched homolog 2
- qPCR
Quantitative real time PCR
- RIN
RNA integrity number
- RNAseq
RNA sequencing
- RPS18
Ribosomal Protein S18
- RPS2
Ribosomal protein S2
- SEM
Standard error of the mean
- SG100G
Calcium binding protein G
- SNRPD2
Small nuclear ribonucleoprotein D2 polypeptide
- SRA
Sequence read archive
- STRAP
Serine/Threonine Kinase Receptor Associated Protein
- TAI
Timed artificial insemination
Authors’ contributions
MFSF contributed to experimental design, animal management, samples preparation for RNAseq and qPCR analysis, statistical analysis of transcripts and was a major contributor in writing the manuscript. RSR, and MFM contributed to animal management. AMGD, GP, and MS performed samples preparation for RNAseq and qPCR analysis, performed qPCR analysis, and contributed in writing the manuscript. SSCA, GG, and LLC performed the RNAseq and the bioinformatics analysis. MDG, FSM, and PSB contributed to experiment design and writing the manuscript. MB was the PI and contributed to experiment design and writing the manuscript. All authors read and approved the final manuscript.
Ethics approval
Animal procedures were approved by the Ethics and Animal Handling Committee of the Faculdade de Medicina Veterinária e Zootecnia, Universidade de São Paulo (CEUA-FMVZ/USP, No. 2287/2011).
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s40104-017-0211-3) contains supplementary material, which is available to authorized users.
Contributor Information
Manoel Francisco de Sá Filho, Email: mfsvt@yahoo.com.br.
Angela Maria Gonella-Diaza, Email: angelamvz@gmail.com.
Mariana Sponchiado, Email: msponchiado@usp.br.
Marcio Ferreira Mendanha, Email: Marciofm@dallasolucoes.com.br.
Guilherme Pugliesi, Email: pugliesi_vet@hotmail.com.
Roney dos Santos Ramos, Email: roney.ramos@yahoo.com.br.
Sónia Cristina da Silva Andrade, Email: soniacsandrade@gmail.com.
Gustavo Gasparin, Email: ggasparin@illumina.com.
Luiz Lehmann Coutinho, Email: llcoutinho@usp.br.
Marcelo Demarchi Goissis, Email: marcelo.febem@gmail.com.
Fernando Silveira Mesquita, Email: Fernando.Mesquita@unipampa.edu.br.
Pietro Sampaio Baruselli, Email: barusell@usp.br.
Mario Binelli, Phone: +55 (19) 3565-4220, Email: binelli@usp.br.
References
- 1.Baruselli PS, Reis EL, Marques MO, Nasser LF, Bó GA. The use of hormonal treatments to improve reproductive performance of anestrous beef cattle in tropical climates. Anim Reprod Sci. 2004;82–83:479–486. doi: 10.1016/j.anireprosci.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 2.Baruselli PS, Sales JNS, Sala RV, Vieira LM, Sa Filho MF. History, evolution and perspectives of timed artificial insemination programs in Brazil. Anim Reprod. 2012;9(3):139–152. [Google Scholar]
- 3.Sa Filho MF, Penteado L, Reis EL, Reis TANPS, Galvao KN, Baruselli PS. Timed artificial insemination early in the breeding season improves the reproductive performance of suckled beef cows. Theriogenology. 2013;79(4):625–632. doi: 10.1016/j.theriogenology.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 4.Meneghetti M, Sa Filho OG, Peres RFG, Lamb GC, Vasconcelos JLM. Fixed-time artificial insemination with estradiol and progesterone for Bos Indicus cows I: basis for development of protocols. Theriogenology. 2009;72(2):179–189. doi: 10.1016/j.theriogenology.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Sa Filho OG, Meneghetti M, Peres RFG, Lamb GC, Vasconcelos JLM. Fixed-time artificial insemination with estradiol and progesterone for Bos Indicus cows II: strategies and factors affecting fertility. Theriogenology. 2009;72(2):210–218. doi: 10.1016/j.theriogenology.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Sa Filho MF, Crespilho AM, Santos JEP, Perry GA, Baruselli PS. Ovarian follicle diameter at timed insemination and estrous response influence likelihood of ovulation and pregnancy after estrous synchronization with progesterone or progestin-based protocols in suckled Bos Indicus cows. Anim Reprod Sci. 2010;120(1–4):23–30. doi: 10.1016/j.anireprosci.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Peres RFG, Claro Junior I, Sa Filho OG, Nogueira GP, Vasconcelos JLM. Strategies to improve fertility in Bos Indicus postpubertal heifers and nonlactating cows submitted to fixed-time artificial insemination. Theriogenology. 2009;72(5):681–689. doi: 10.1016/j.theriogenology.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 8.Mesquita FS, Ramos RS, Pugliesi G, Andrade SC, Van Hoeck V, Langbeen A, et al. The receptive endometrial Transcriptomic signature indicates an earlier shift from proliferation to metabolism at early Diestrus in the cow. Biol Reprod. 2015;93(2):52. doi: 10.1095/biolreprod.115.129031. [DOI] [PubMed] [Google Scholar]
- 9.Satterfield MC, Song G, Kochan KJ, Riggs PK, Simmons RM, Elsik CG, et al. Discovery of candidate genes and pathways in the endometrium regulating ovine blastocyst growth and conceptus elongation. Physiol Genomics. 2009;39(2):85–99. doi: 10.1152/physiolgenomics.00001.2009. [DOI] [PubMed] [Google Scholar]
- 10.Bauersachs S, Ulbrich SE, Gross K, Schmidt SEM, Meyer HHD, Einspanier R, et al. Gene expression profiling of bovine endometrium during the oestrous cycle: detection of molecular pathways involved in functional changes. J Mol Endocrinol. 2005;34(3):889–908. doi: 10.1677/jme.1.01799. [DOI] [PubMed] [Google Scholar]
- 11.Miller BG, Moore NW. Effects of progesterone and oestradiol on RNA and protein metabolism in the genital tract and on survival of embryos in the ovariectomized ewe. Aust J Biol Sci. 1976;29(5–6):565–573. doi: 10.1071/BI9760565. [DOI] [PubMed] [Google Scholar]
- 12.Moore NW. The use of embryo transfer and steroid-hormone replacement therapy in the study of prenatal mortality. Theriogenology. 1985;23(1):121–128. doi: 10.1016/0093-691X(85)90077-9. [DOI] [Google Scholar]
- 13.Wilmut I, Sales DI, Ashworth CJ. Maternal and embryonic factors associated with prenatal loss in mammals. J Reprod Fertil. 1986;76(2):851–864. doi: 10.1530/jrf.0.0760851. [DOI] [PubMed] [Google Scholar]
- 14.Mesquita FS, Pugliesi G, Scolari SC, Franca MR, Ramos RS, Oliveira M, et al. Manipulation of the periovulatory sex steroidal milieu affects endometrial but not luteal gene expression in early diestrus Nelore cows. Theriogenology. 2014;81(6):861–869. doi: 10.1016/j.theriogenology.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 15.Forde N, Beltman ME, Duffy GB, Duffy P, Mehta JP, O'Gaora P, et al. Changes in the endometrial Transcriptome during the bovine estrous cycle: effect of low circulating progesterone and consequences for Conceptus elongation. Biol Reprod. 2011;84(2):266–278. doi: 10.1095/biolreprod.110.085910. [DOI] [PubMed] [Google Scholar]
- 16.Forde N, Carter F, Fair T, Crowe MA, Evans ACO, Spencer TE, et al. Progesterone-regulated changes in endometrial gene expression contribute to advanced Conceptus development in cattle. Biol Reprod. 2009;81(4):784–794. doi: 10.1095/biolreprod.108.074336. [DOI] [PubMed] [Google Scholar]
- 17.Okumu LA, Forde N, Fahey AG, Fitzpatrick E, Roche JF, Crowe MA, et al. The effect of elevated progesterone and pregnancy status on mRNA expression and localisation of progesterone and oestrogen receptors in the bovine uterus. Reproduction. 2010;140(1):143–153. doi: 10.1530/REP-10-0113. [DOI] [PubMed] [Google Scholar]
- 18.Mann GE, Lamming GE. The role of sub-optimal preovulatory oestradiol secretion in the aetiology of premature luteolysis during the short oestrous cycle in the cow. Anim Reprod Sci. 2000;64(3–4):171–180. doi: 10.1016/S0378-4320(00)00205-0. [DOI] [PubMed] [Google Scholar]
- 19.Bridges GA, Mussard ML, Pate JL, Ott TL, Hansen TR, Day ML. Impact of preovulatory estradiol concentrations on conceptus development and uterine gene expression. Anim Reprod Sci. 2012;133(1–2):16–26. doi: 10.1016/j.anireprosci.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Lamming GE, Mann GE. Control of endometrial oxytocin receptors and prostaglandin F2 alpha production in cows by progesterone and oestradiol. J Reprod Fertil. 1995;103(1):69–73. doi: 10.1530/jrf.0.1030069. [DOI] [PubMed] [Google Scholar]
- 21.Robinson RS, Mann GE, Lamming GE, Wathes DC. Expression of oxytocin, oestrogen and progesterone receptors in uterine biopsy samples throughout the oestrous cycle and early pregnancy in cows. Reproduction. 2001;122(6):965–979. doi: 10.1530/rep.0.1220965. [DOI] [PubMed] [Google Scholar]
- 22.Kieborz-Loos KR, Garverick HA, Keisler DH, Hamilton SA, Salfen BE, Youngquist RS, et al. Oxytocin-induced secretion of prostaglandin F2alpha in postpartum beef cows: effects of progesterone and estradiol-17beta treatment. J Anim Sci. 2003;81(7):1830–1836. doi: 10.2527/2003.8171830x. [DOI] [PubMed] [Google Scholar]
- 23.Ulbrich SE, Frohlich T, Schulke K, Englberger E, Waldschmitt N, Arnold GJ, et al. Evidence for estrogen-dependent uterine Serpin (SERPINA14) expression during estrus in the bovine endometrial glandular epithelium and lumen. Biol Reprod. 2009;81(4):795–805. doi: 10.1095/biolreprod.108.075184. [DOI] [PubMed] [Google Scholar]
- 24.Hillegass J, Lima FS, Filho MFS, Santos JEP. Effect of time of artificial insemination and supplemental estradiol on reproduction of lactating dairy cows. J Dairy Sci. 2008;91(11):4226–4237. doi: 10.3168/jds.2007-0901. [DOI] [PubMed] [Google Scholar]
- 25.Sa Filho MF, Santos JEP, Ferreira RM, Sales JNS, Baruselli PS. Importance of estrus on pregnancy per insemination in suckled Bos Indicus cows submitted to estradiol/progesterone-based timed insemination protocols. Theriogenology. 2011;76(3):455–463. doi: 10.1016/j.theriogenology.2011.02.022. [DOI] [PubMed] [Google Scholar]
- 26.Pitaluga PCSF, Sa Filho MF, Sales JNS, Baruselli PS, Vincenti L. Manipulation of the proestrous by exogenous gonadotropin and estradiol during a timed artificial insemination protocol in suckled Bos Indicus beef cows. Livest Sci. 2013;154(1–3):229–234. doi: 10.1016/j.livsci.2013.02.023. [DOI] [Google Scholar]
- 27.Souza AH, Gumen A, Silva EP, Cunha AP, Guenther JN, Peto CM, et al. Supplementation with estradiol-17beta before the last gonadotropin-releasing hormone injection of the Ovsynch protocol in lactating dairy cows. J Dairy Sci. 2007;90(10):4623–4634. doi: 10.3168/jds.2007-0172. [DOI] [PubMed] [Google Scholar]
- 28.Jinks EM, Smith MF, Atkins JA, Pohler KG, Perry GA, MacNeil MD, et al. Preovulatory estradiol and the establishment and maintenance of pregnancy in suckled beef cows. J Anim Sci. 2013;91(3):1176–1185. doi: 10.2527/jas.2012-5611. [DOI] [PubMed] [Google Scholar]
- 29.Sa Filho MF, Ayres H, Ferreira RM, Marques MO, Reis EL, Silva RCP, et al. Equine chorionic gonadotropin and gonadotropin-releasing hormone enhance fertility in a norgestomet-based, timed artificial insemination protocol in suckled Nelore (Bos Indicus) cows. Theriogenology. 2010;73(5):651–658. doi: 10.1016/j.theriogenology.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Sales JNS, Crepaldi GA, Girotto RW, Souza AH, Baruselli PS. Fixed-time AI protocols replacing eCG with a single dose of FSH were less effective in stimulating follicular growth, ovulation, and fertility in suckled-anestrus Nelore beef cows. Anim Reprod Sci. 2011;124(1–2):12–18. doi: 10.1016/j.anireprosci.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Ayres H, Ferreira RM, de Souza Torres-Junior JR, Borges Demetrio CG, de Lima CG, Baruselli PS. Validation of body condition score as a predictor of subcutaneous fat in Nelore (Bos Indicus) cows. Livest Sci. 2009;123(2–3):175–179. doi: 10.1016/j.livsci.2008.11.004. [DOI] [Google Scholar]
- 32.Pugliesi G, Scolari SC, Mesquita FS, Maturana Filho M, Araujo ER, Cardoso D, et al. Impact of probing the reproductive tract during early pregnancy on fertility of beef cows. Reprod Domestic Anim. 2014;49(4):E35–EE9. doi: 10.1111/rda.12345. [DOI] [PubMed] [Google Scholar]
- 33.Garbarino EJ, Hernandez JA, Shearer JK, Risco CA, Thatcher WW. Effect of lameness on ovarian activity in postpartum Holstein cows. J Dairy Sci. 2004;87(12):4123–4131. doi: 10.3168/jds.S0022-0302(04)73555-9. [DOI] [PubMed] [Google Scholar]
- 34.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. [DOI] [PMC free article] [PubMed]
- 35.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed]
- 37.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. [DOI] [PMC free article] [PubMed]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57(1):289–300. [Google Scholar]
- 39.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 40.Smit A, Hubley R, Green P. RepeatMasker Open-3.0. http://www.repeatmasker.org/.
- 41.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7)RESEARCH 0034.1-0034-12. [DOI] [PMC free article] [PubMed]
- 42.Perry GA, Swanson OL, Larimore EL, Perry BL, Djira GD, Cushman RA. Relationship of follicle size and concentrations of estradiol among cows exhibiting or not exhibiting estrus during a fixed-time AI protocol. Domest Anim Endocrinol. 2014;48:15–20. doi: 10.1016/j.domaniend.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Fields SD, Gebhart KL, Perry BL, Gonda MG, Wright CL, Bott RC, et al. Influence of standing estrus before an injection of GnRH during a beef cattle fixed-time AT protocol on LH release, subsequent concentrations of progesterone, and steriodogenic enzyme expression. Domest Anim Endocrinol. 2012;42(1):11–19. doi: 10.1016/j.domaniend.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Perry GA, Smith MF, Roberts AJ, MacNeil MD, Geary TW. Relationship between size of the ovulatory follicle and pregnancy success in beef heifers. J Anim Sci. 2007;85(3):684–689. doi: 10.2527/jas.2006-519. [DOI] [PubMed] [Google Scholar]
- 45.Minten MA, Bilby TR, Bruno RGS, Allen CC, Madsen CA, Wang Z, et al. Effects of Fertility on Gene Expression and Function of the Bovine Endometrium. Plos One. 2013;8(8)E69944. [DOI] [PMC free article] [PubMed]
- 46.Killeen AP, Morris DG, Kenny DA, Mullen MP, Diskin MG, Waters SM. Global gene expression in endometrium of high and low fertility heifers during the mid-luteal phase of the estrous cycle. BMC Genomics. 2014;15:234. [DOI] [PMC free article] [PubMed]
- 47.Perry GA, Perry BL. Effect of preovulatory concentrations of estradiol and initiation of standing estrus on uterine pH in beef cows. Domest Anim Endocrinol. 2008;34(3):333–338. doi: 10.1016/j.domaniend.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Allrich RD. Endocrine and neural control of estrus in dairy-cows. J Dairy Sci. 1994;77(9):2738–2744. doi: 10.3168/jds.S0022-0302(94)77216-7. [DOI] [PubMed] [Google Scholar]
- 49.Shimizu T, Krebs S, Bauersachs S, Blum H, Wolf E, Miyamoto A. Actions and interactions of progesterone and estrogen on transcriptome profiles of the bovine endometrium. Physiol Genomics. 2010;42A(4):290–300. doi: 10.1152/physiolgenomics.00107.2010. [DOI] [PubMed] [Google Scholar]
- 50.Salilew-Wondim D, Hoelker M, Rings F, Ghanem N, Ulas-Cinar M, Peippo J, et al. Bovine pretransfer endometrium and embryo transcriptome fingerprints as predictors of pregnancy success after embryo transfer. Physiol Genomics. 2010;42(2):201–218. doi: 10.1152/physiolgenomics.00047.2010. [DOI] [PubMed] [Google Scholar]
- 51.Toftgard R. Hedgehog signalling in cancer. Cell Mol Life Sci. 2000;57(12):1720–1731. doi: 10.1007/PL00000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Worley MJ, Jr, Liu S, Hua Y, Kwok JS-L, Samuel A, Hou L, et al. Molecular changes in endometriosis-associated ovarian clear cell carcinoma. Eur J Cancer. 2015;51(13):1831–1842. doi: 10.1016/j.ejca.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhulyn O, Nieuwenhuis E, Liu YC, Angers S, Hui C-C. Ptch2 Shares overlapping functions with Ptch1 in Smo regulation and limb development. Dev Biol. 2015;397(2):191–202. doi: 10.1016/j.ydbio.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 54.Kuhn K. Basement membrane (type IV) collagen. Matrix Biol. 1995;14(6):439–445. doi: 10.1016/0945-053X(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 55.Rogers PAW, Lederman F, Taylor N. Endometrial microvascular growth in normal and dysfunctional states. Hum Reprod Update. 1998;4(5):503–508. doi: 10.1093/humupd/4.5.503. [DOI] [PubMed] [Google Scholar]
- 56.Hu T, Li C. Convergence between Wnt-beta-catenin and EGFR signaling in cancer. Mol Cancer. 2010;9:236. [DOI] [PMC free article] [PubMed]
- 57.Wang H, Masironi B, Eriksson H, Sahlin L. A comparative study of estrogen receptors alpha and beta in the rat uterus. Biol Reprod. 1999;61(4):955–964. doi: 10.1095/biolreprod61.4.955. [DOI] [PubMed] [Google Scholar]
- 58.Kurita T, Young P, Brody JR, Lydon JP, O'Malley BW, Cunha GR. Stromal progesterone receptors mediate the inhibitory effects of progesterone on estrogen-induced uterine epithelial cell deoxyribonucleic acid synthesis. Endocrinology. 1998;139(11):4708–4713. doi: 10.1210/endo.139.11.6317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Number of reads from all samples from suckled cows receiving (ECP) or not (CON) 1 mg of ECP at the onset of the proestrous (DOCX 14 kb)
Differential gene expression results. BaseMean is the average of all samples expression profile after normalization; lfcSE – standard error from log2FoldChange; padj – P-value adjusted after correction of BH-FDR for multiple tests (DOCX 26 kb)
Bio-samples and Experiment accession numbers of the Raw reads resulted from the RNAseq of endometrial biopsis in the SRA data base (DOCX 20 kb)
Data Availability Statement
Sequences of all reads generated in the RNAseq were deposited in the Sequence Read Archive (SRA) of the NCBI (http://www.ncbi.nlm.nih.gov/sra/; Additional file 3: Table S3) and, an overview of these data has been deposited in NCBI’s Gene Expression Omnibus (GEO) and is accessible through GEO Series accession number GSE67807.


