Abstract
While cerebral underdevelopment is a hallmark of fetal alcohol spectrum disorders (FASD), the mechanism(s) guiding the broad cortical neurodevelopmental deficits are not clear. DNA methylation is known to regulate early development and tissue specification through gene regulation. Here, we examined DNA methylation in the onset of alcohol-induced cortical thinning in a mouse model of FASD. C57BL/6 (B6) mice were administered a 4% alcohol (v/v) liquid diet from embryonic (E) days 7–16, and their embryos were harvested at E17, along with isocaloric liquid diet and lab chow controls. Cortical neuroanatomy, neural phenotypes, and epigenetic markers of methylation were assessed using immunohistochemistry, Western blot, and methyl-DNA assays. We report that cortical thickness, neuroepithelial proliferation, and neuronal migration and maturity were found to be deterred by alcohol at E17. Simultaneously, DNA methylation, including 5-methylcytosine (5mC) and 5-hydroxcylmethylcytosine (5hmC), which progresses as an intrinsic program guiding normal embryonic cortical development, was severely affected by in utero alcohol exposure. The intricate relationship between cortical thinning and this DNA methylation program disruption is detailed and illustrated. DNA methylation, dynamic across the multiple cortical layers during the late embryonic stage, is highly disrupted by fetal alcohol exposure; this disruption occurs in tandem with characteristic developmental abnormalities, ranging from structural to molecular. Finally, our findings point to a significant question for future exploration: whether epigenetics guides neurodevelopment or whether developmental conditions dictate epigenetic dynamics in the context of alcohol-induced cortical teratogenesis.
Keywords: Epigenetics, Neurodevelopmental deficit, Neuroepigenetics, Neurogenesis, Fetal Alcohol Spectrum Disorders (FASD)
Introduction
Children with Fetal Alcohol Spectrum Disorders (FASD) have been reported to suffer cognitive and neurological deficits, including learning disabilities, intellectual disabilities, and impairments of expressive and receptive language (Green, 2007; Jacobson, Jacobson, Stanton, Meintjes, & Molteno, 2011; Jones et al., 2010; Lebel et al., 2012). Some of the underlying brain abnormalities of FASD include reduced brain volume (microcephaly), reduced grey matter (Nardelli, Lebel, Rasmussen, Andrew, & Beaulieu, 2011), and reduced corpus callosum (Yang, Phillips, et al., 2012). Cross-sectional neuroimaging studies have also recently revealed that children and adolescents suffering from FASD exhibit abnormalities in the thickness of different regions of the cerebral cortex, as compared to healthy controls (Robertson et al., 2016; Sowell et al., 2008; Yang, Roussotte, et al., 2012). While several observations have been made regarding the fundamental hindrance of alcohol on cortical development, e.g., apoptosis (Lebedeva et al., 2015), deficiency of neurotrophic factors, prevention of cell migration (Aronne, Guadagnoli, Fontanet, Evrard, & Brusco, 2011; Chikhladze, Ramishvili, Tsagareli, & Kikalishvili, 2011; Riar, Narasimhan, Rathinam, Henderson, & Mahimainathan, 2016), and abnormal somatic morphologies of cortical neurons (Chikhladze et al., 2011; Lawrence, Otero, & Kelly, 2012), the mechanism underlying the structural abnormality systemically occurring in relationship to alcohol exposure is not clear.
Recently, alcohol has emerged as a key chemical player, which can reach nuclear chromatin and alter the core functions of DNA (for review see Resendiz, Lo, Badin, Chiu, & Zhou, 2016). We and other investigators (Perkins, Lehmann, Lawrence, & Kelly, 2013) have recently found that DNA methylation, an important regulator of gene expression, progresses in the developing nervous system as a program (Zhou, 2012), and is disturbed by alcohol in many aspects across neural tube (Zhou, Chen, & Love, 2011) and hippocampal development (Chen, Öztürk, & Zhou, 2013; Otero, Thomas, Saski, Xia, & Kelly, 2012). Given the known intricacies of epigenetic mechanisms such as DNA methylation in gene regulation and cellular specification, we sought to characterize the epigenetic and phenotypic changes of chronic, moderate prenatal alcohol exposure in utero in the developing stages of the cortex. In this study, we report that beyond neural tube formation, the formation of the cortices adopt a systemic DNA methylation program (DMP) (including DNA methylation and its binding proteins), by which neuroepithelial cells (NEs) differentiate through the formation of cortical layers in a precise spatiotemporal manner. Aside from confirming cortical phenotypes of FASD, we also demonstrated novel deficiencies. These processes, together with global and cellular epigenetic mechanisms, may drive the consequential dysmorphology of the developing cortex. In this study, we demonstrate how alcohol interferes with the DMP, in parallel with cortical thinning and other abnormalities. Understanding the molecular drivers of alcohol-induced alteration of the highly ordered developmental cortical program is paramount toward uncovering how fetal environmental insults are established, maintained, and manifested into lasting cognitive and behavioral deficits.
Materials and methods
Overview of experimental prenatal alcohol exposure
In this study, alcohol was administered via liquid diet according to the paradigm illustrated in Fig. 1A. The time course and types of analysis are summarized in Fig. 1B. Mice were conditioned to receive the liquid diet prior to mating. After conception, the liquid diet was re-introduced and alcohol was administered from E7–E16 (corresponding to brain development in the late first and second human trimester equivalent). The 4% alcohol liquid diet (v/v) administered in this paradigm has been reported in previous and parallel studies to produce a range of blood alcohol concentrations (BAC) of 100–200 mg/dL (Anthony et al., 2010; Chen et al., 2013). Briefiy, six non-pregnant females receiving 4% v/v alcohol as in the above paradigm were used for BAC analysis. Blood samples were harvested through the tail vein 2 h or 6 h after introducing the fresh alcohol-PMI diet at 10:00 a.m. during the dark cycle, on days 2, 4 and 6 during treatment. Adequate volume of blood (15 μL) was collected in heparinized tubes, and plasma was isolated through centrifugation and stored at −80 °C prior to analysis with a gas chromatograph (GC, Agilent Technologies; model 6890). Each sample was analyzed in duplicate.
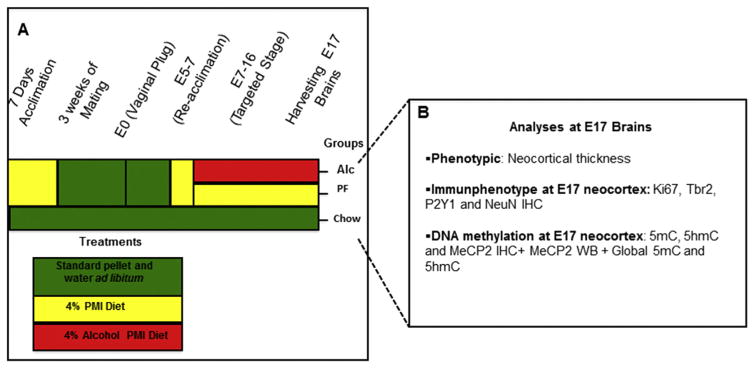
Fig. 1.
Summary of experimental procedure. (A) C57BL/6 (B6) females were conditioned to receive the liquid diet devoid of alcohol for 7 days preceding mating. After conception, the liquid diet was re-introduced at E5 and either an alcohol diet or an isocaloric pair-fed diet was administered from E7–E16 (equivalent to the late first and second human trimesters). Each color in the schema represents a specific treatment: green (standard pellet and water ad libitum), yellow (alcohol-free PMI liquid diet), or red (4% v/v alcohol PMI liquid diet ad libitum). (B) At E17, brains from each litter across the three groups were processed for either immunophenotypic or molecular assessments. Alc (alcohol); IHC (immunohistochemistry); PF (pair-fed). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
While there was no overall gestational weight difference, the Alc and PF groups did exhibit lower gestational weights on E15–E16 (Chen et al., 2013). While our previous studies were aimed at the early neural tube (E10) and early brain primodium (E15), the current study focused on the stage prior to birth, at the peak of rodent cortical layer formation (E17) and alcohol-induced thinning.
Animals and treatments
All mice were used in accordance with National Institute of Health and Indiana University Animal Care and Use (IACUC) guidelines. The protocol was approved by the Laboratory Animal Resource Center (LARC) animal ethics committee of Indiana University. C57BL/6 (B6) (10–14 weeks old, ~20 g body weight) nulliparous female mice (Harlan, Inc., Indianapolis, IN) were used in the study. Mouse breeders were individually housed upon arrival and acclimated for at least one week before mating. The mice were maintained on a 12-h reverse light-dark cycle (lights on: 10:00 p.m.–10:00 a.m.) and were provided laboratory chow and water ad libitum. Mice were then randomly assigned to three treatment groups: N = Chow (7), PF (5), Alc (7). Each litter was considered N = 1; the littermates of each dam were distributed for the analyses described in the following sections. The PF and Alc groups were pre-treated with liquid diet (see below) for 7 days before mating. Females were bred with male breeders for a 2-h period (10:00 a.m. to 12:00 noon). All animals were mated daily over a period of no more than 3 weeks, during which time all animals were on ad libitum chow and water diets. The presence of a vaginal plug at the end of the 2-h mating session was considered as indicative of conceptus, and that hour was designated as hour 0 and embryonic day (E) 0. A liquid-diet paradigm was carried out as previously detailed (Chen et al., 2013). Briefly, all alcohol treatment groups received 4% alcohol v/v in liquid diet (Purina Micro-Stabilized Diet [PMI], Purina Mills Inc., Richmond, Indiana) as instructed by supplier with 4% w/v sucrose added, and administered using a 35-mL drinking tube (Dyets Inc., NY). The PF group was given the PMI diet mixture with the addition of maltose dextran (MD) (to substitute alcohol calories). The volume of the PF diet was restricted to that of a matched dam from the alcohol group throughout the course of treatment. The Chow group was maintained on a standard chow diet and water ad libitum throughout gestation. On E5, pregnant dams in PF and Alc groups were placed on an unrestricted PF liquid diet for acclimation. Either 4% v/v alcohol (Alc group) or restricted volume isocaloric liquid diet (PF group) was initiated on E7 through the end of E16. On E17, dams from all three groups were euthanized for embryo harvest. In addition, E16 embryos from Chow groups (N = 4) were specifically harvested for developmental stage comparison.
Embryo isolation and tissue preparation
After deep CO2 euthanasia, embryos were harvested from dams at E17 by removal from the embryonic sack. Each embryo was either immersion-fixed in 20 mL of fixative prepared from 4% paraformaldehyde (PFA) for immunohistochemistry or immediately dissected for brain tissue and snap-frozen and stored in a −80 °C freezer until Western blot or global methylation analysis. Fixed embryos were subsequently weighed, dissected for brains, gelatin-blocked, and post-fixed for at least 24 h at 4 °C before sectioning was performed for immunocytochemistry (average N = Chow (5), PF (4), Alc (5); animal number for each staining is shown in Results).
Immunocytochemistry analysis
One Alc and either one PF or Chow brain were embedded in a single 10% gelatin block with careful rostrocaudal and dorsoventral alignments. Gelatin blocks were fixed with 4% PFA and sectioned in 40-μm thick coronal sections on a floating vibratome (Leica Microsystems; Buffalo Grove, IL). The section pairs (Alc-PF or Alc-Chow) were processed equally in all immunocytochemical procedures. The section pairs were then cleared of endogenous per-oxidases using 10% H2O2 in phosphate-buffered saline (PBS) for 10 min and permeabilized with 1% TritonX-100 in PBS for 30 min before incubation with a primary antibody diluted in goat kit (1.5% goat serum, 0.1% TritonX-100 in PBS) for 18 h at room temperature. Epigenetic antibodies used in this study were: 5mC (1:2000, mouse monoclonal; Eurogenetec, Fremont, CA), 5hmC (1:3000, rabbit monoclonal; Active Motif, Carlsbad, CA), and MeCP2 (a DNA methylation-binding protein; 1:1000, rabbit monoclonal; Cell Signaling, Danvers, MA). Stage differentiation markers used were: Ki67 (a marker for cell proliferation; 1:500, rabbit polyclonal, Millipore, Billerica, MA), NeuN (a marker for mature neuron; 1:500, mouse monoclonal, Cell Signaling, Danvers, MA), Tbr2 (T-box brain protein 2, a marker for intermediate neural progenitors, 1:500, rabbit polyclonal, Millipore, Billerica, MA), and P2Y1 (metabotropic G-protein P2 receptor, a marker for specialized neural cells capable of communication; 1:1000, rabbit polyclonal, Millipore, Billerica, MA). The section pairs were then incubated for 90 min in goat anti-rabbit IgG or goat anti-mouse secondary antibodies conjugated with biotin (Jackson ImmunoResearch, West Grove, PA) followed by Streptavidin-AP (1:500, Jackson ImmunoResearch, West Grove, PA) for 90 min. The immunostaining was visualized by incubation in 0.05% 3,3′-diaminobenzidine (DAB) and 0.003% H2O2 over an average of 3–8 min, followed by counterstaining with methyl green. All stainings were photographed under light microscopy for cellular analysis (Leitz Orthoplan 2 microscope; Ernst Leitz GMBH, Wetzlar, Germany).
Densitometry analysis and cortical thickness assessment
Upon observing epigenetic immunostainings under a light microscope, the immunoreactive nuclei (based on evidence of the brown-color DAB reactions) apparently exhibited a differential staining profile within different subcortical regions. In order to reflect this differential expression, we employed H scoring for nuclear densitometry analysis (Chen et al., 2013; Goulding et al., 1995; Singh, Shiue, Schomberg, & Zhou, 2009) of each cell nucleus within each individual selected subcortical region (VZ + SVZ, SP, and CP).
For the analysis, all immunostained pictures were taken using a Leitz Orthoplan 2 microscope with a Spot RT color camera (Diagnostic Instruments, Inc., Sterling Heights, MI). Bright-field images were taken with consistent setup and exposure time for each antibody staining. Immunostained images were converted to the 16-bit color format, and staining intensity was measured using Image J (National Institutes of Health, Bethesda, MD). Calibration was set based on 256 levels of the gray scale. To measure the subcortical regions of prefrontal neocortex, a rectangular box of equal dimensions (150 μm in width) was selected at the same rostrocaudal level of E17 coronal brain sections. Lateral ventricle and corpus callosum were considered as landmarks of the pre-frontal cortex. The staining intensities of marks were defined based on the optical density (OD) values of the nuclei in each subregion of neocortex as follows: Absent – 0 – (OD = 90–120); Weak – 1 – (OD = 120–150); Moderate – 2 – (OD = 150–180); and High – 3 – (OD = 180–210). Overall, the immunohistochemical H score of each subcortical region was obtained by the following formula: 3 × percentage of highly stained nuclei +2 × percentage of moderately stained nuclei + percentage of weakly stained nuclei, giving a range of 0–400. A Kruskal-Wallis test was used for non-parametric statistical analysis of nuclear intensity to address differences between the three groups (N = Chow (5), PF (4), Alc (5)), while Conover post hoc testing was used to identify differences between each of the groups. Statistical analysis was performed using MedCalc software.
Western blot of the methylation-binding protein MeCP2
Western blotting was carried out to confirm MeCP2 protein expression differences at E17 between groups, which were initially observed in MeCP2 immunostainings. From the preliminary MeCP2 staining, we noticed that MeCP2 was unilaterally upregulated by alcohol across all cortical layers and in various other brain regions, such as the hippocampus and cerebellum. As such, Western blots of the entire E17 fetal brain were used (N = 4 each) following a standard protocol (Anthony, Zhou, Ogawa, Goodlett, & Ruiz, 2008; Mason et al., 2012; Zhou, Patel, Swartz, Xu, & Kelley, 1999). Nuclear protein was isolated from tissue lysates using NE-PER nuclear and cytoplasmic reagents (Thermo Fischer Scientific, Waltham, MA), and sample concentrations were evaluated against a BSA standard curve at OD595. All samples were run in triplicate on two independent gels for each protein examined. Immunoreactive blots were detected using ECL Western Blotting Detection Kit (Thermo Fisher Scientific, Rockford, IL, USA; catalog# RPN2108) and exposed to a biomolecular imaging system (ImageQuant,LAS 4000). Densitometric comparisons were made with Image J software. GAPDH density measurements were used as loading controls. All changes in protein expression were reported as a percentage change compared to Chow and PF groups, with a minimum of four samples/treatment group. Statistical analysis was performed by one-way ANOVA on MedCalc software.
Global DNA methylation analysis
Fetal brains were isolated and microdissected under a dissection microscope (Leica MZ6, Leica Microsystems). Neocortical brain tissues were separated from subcortical brain tissue using the borders of the nascent internal capsule as a visual guide. DNA extraction and purification were subsequently performed using silica-based spin-column purification (DNeasy Blood and Tissue kit, Qiagen) according to manufacturer’s instructions. Purified DNA was quantified by spectrophotometric absorption at 230, 260, and 280 nm, and the quality and concentration were calculated as the A260/A230 and A260/A280 ratio (Nanodrop 2000, Thermo Scientific). An average of 100–200 ng of genomic DNA was used for DNA global methylation analysis performed with the MethylFlash Methylated DNA Quantification Kit and MethylFlash Hydroxymethylated DNA Quantification Kit (Colorimetric; Epigentek Group) according to the manufacturer’s instructions. OD values were determined using a PHERAstar FSX microplate reader and MARS Data Analysis Software (BMG Labtech, Cary, NC). Methylation levels were estimated using a standard curve of methylated DNA standards provided by the manufacturer. Values are presented as methylation percent relative to the control group. Statistical analysis was performed by non-parametric Kruskal-Wallis test followed by Conover post hoc test for multiple comparisons using MedCalc software.
Results
First, we report the parallel development of the phenotypes and cellular features of normal corticogenesis alongside DNA methylation markers and their binding proteins layer-by-layer, revealing the cortical DNA methylation program of differentiating neuroepithelial cells into mature neurons. Subsequently, alcohol-induced aberrations of phenotypic and epigenetic features are demonstrably associated. Finally, a global analysis of the average cortical DNA methylation and MeCP2 protein are summarized and quantitatively assessed.
Phenotypic and epigenetic features of normally developing neocortex
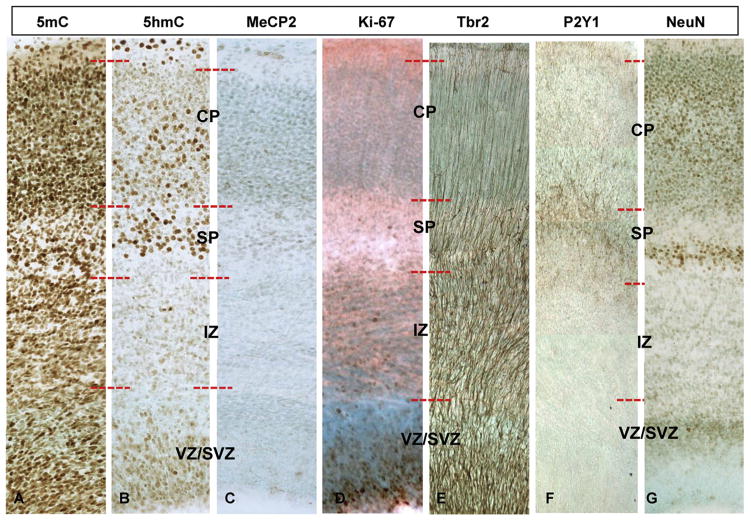
During normal development, cellular DNA methylation progresses in an orderly manner in differentiating NEs parallel to neocortical maturation from ventricular zone (VZ) to cortical plate (CP). This simultaneous developmental and epigenetic progression has been similarly described in neural tube formation previously (Zhou, 2012; Zhou, Chen, et al., 2011). In the E17 Chow cortices, the VZ (Fig. 2A–C), the neurogenic layer of the developing neocortex, exhibited strong proliferative activity as demonstrated by Ki67-im (Fig. 2D) and dense Tbr2-im cells (indicative of intermediate neural progenitors (INPs) detaching from the ventricular surface and migrating into the upper layers along radial glia fibers (Fig. 2E). We found a full expression of 5-methylcytosine (5mC) (Fig. 2A), followed by emerging (lightly stained) 5-hydroxymethylcytosine (5hmC) (Fig. 2B) at this highly proliferative zone. Meanwhile, the DNA methyl-binding protein MeCP2 was almost completely absent (Fig. 2C). 5mC consistently escalated prior to (~1 day in rodent) and in far greater amounts than 5hmC in this layer, marking the beginning of differentiation.
Fig. 2.
Comparative phenotypic and DNA methylation dynamics in the embryonic neocortex. (A–C) Representative cortical columns from the Chow E17 frontal neocortex immunostained with DMP markers (5mC, 5hmC, MeCP2) and (D–G) phenotypic neural markers (Ki67, Tbr2, P2Y1, and NeuN) are presented for comparison of the DMP dynamics along the radially progressing corticogenesis of the E17 brain. SVZ/VZ (Subventricular Zone/Ventricular Zone); IZ (Intermediate Zone); SP (Subplate); CP (Cortical plate).
At the subventricular zone (SVZ), the secondary proliferative compartment of the developing cortex, Ki67-im-positive proliferative cells were less apparent as compared to the VZ (Fig. 2D), but contained dense Tbr2-im fibers extending from the VZ (Fig. 2E). Similar 5mC and 5hmC distribution was observed in the SVZ as in the VZ, with only a slight reduction of 5mC (Fig. 2A and B). MeCP2 was also absent in the SVZ layers, similar to the VZ (Fig. 2C).
The intermediate zone (IZ) at E17 features the vertical projection fibers of Tbr2-im-positive intermediate neural progenitors that originated in the SZ and VZ. In the IZ, the Ki67-im (Fig. 2D and E) dwindled. DNA methylation marks (DMMs) at this region appeared either unchanged or in a transitional state, entering a new cycle of increasing 5mC and a lagging increase of 5hmC as compared to the SVZ (Fig. 2A and B).
The subplate (SP) featured the first appearance of round-shaped (representing a more mature state than the ellipsoidal shape nuclei; compare the cells in VZ and SP) and highly NeuN-impositive neurons, occupying the middle layer of the SP (Fig. 2G). While being a proliferation-free (devoid of Ki67-im) zone (Fig. 2D), actively migrating INPs were detected by Tbr2-im (cell bodies and fibers) and P2Y1-im (membrane component of cell bodies and dendritic profiles) (Fig. 2F and G4). In the SP, 5mC is present in both round and ellipsoidal-shaped nuclei (Fig. 2A). In contrast, the intense 5hmC-im distinctly occupied only the round-shaped (more mature) nuclei (Fig. 2B). The 5hmC-im-positive neurons are highly correlated with the NeuN-im cells (Fig. 2G). Further, MeCP2-im appeared only in few cells of the SP (Fig. 2C).
The cortical plate (CP) contains maturing neurons at E17, which will make up future cortical layers II–VI. Here, proliferation was absent (devoid of Ki67-im), while radially extending Tbr2-im fibers were tapered compared to the lower layers of the cortex (Fig. 2E). Interestingly, mature NeuN-im neurons appeared to be distributed into three distinct sublayers within the CP (Fig. 2G). In the CP, late-arrived cells densely populate the top NeuN-im layer (Angevine & Sidman, 1961; Hatanaka, Hisanaga, Heizmann, & Murakami, 2004), while the middle NeuN-im is, based on cellular density, likely made up of more mature and scattered layer V neurons undergoing arborization (Meyer et al., 2010). Finally, the lower NeuN-im CP layer contains a mix of upward-migrating immature cells (dense) and the smaller, denser layer VI neurons. Concordant with the three CP layers, 5mC-im also appeared distributed in the three sublayers with an immuno-intensity gradient inversely proportional to that of NeuN-im (middle < top and bottom) (Fig. 2A). The distribution of 5hmC-im is, in contrast, well-aligned with the NeuN-im pattern (middle > top and bottom) (Fig. 2B). Similar to the SP layer, 5hmC appeared to only occupy the mature, round-shaped nuclei, whereas 5mC was more widely expressed. Lastly, MeCP2-im patterns similarly reflect those observed in the SP (Fig. 2C).
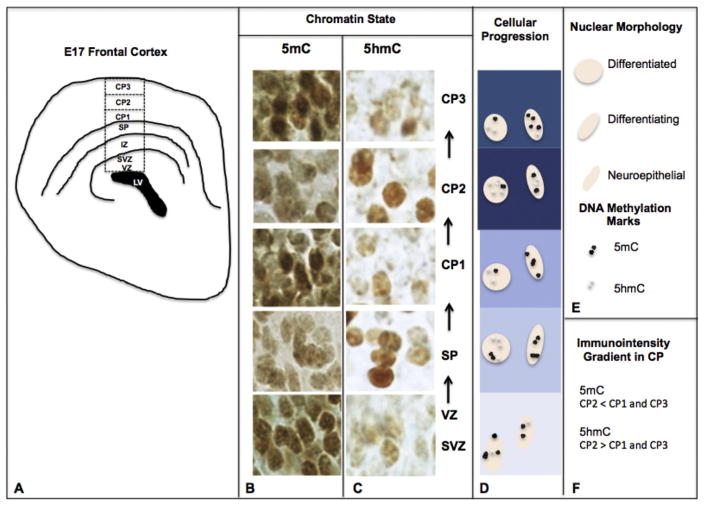
The complex laminar organization of the mouse cortex results from the tightly regulated temporal and spatial transcriptional cues, which direct progenitors from the VZ to their target regions in the upper cortex. During this course, our epigenetic evaluation has demonstrated a patterned presentation along the cortical developmental trajectory, which indicated that 5mC precedes 5hmC at the ventricular zone and is non-discriminant throughout the SP and CP (Fig. 3B). 5hmC, though less abundant than 5mC, demonstrated a cellular restriction toward neurons exhibiting a more mature morphology in the SP and CP (Fig. 3C).
Fig. 3.
The DNA methylation program of the embryonic cortex. (A) At E17, the embryonic cortex develops in distinct layers progressing from the roof of the lateral ventricle (LV). Neuroepithelial cells (NEs) sequentially migrate through the proliferative ventricular zone (VZ) to the uppermost cortical superficial layers. (B, C) During this developmental progression, cells of the layers are diverse in their maturational states and simultaneously unique in their chromatic distribution of DNA methylation markers. Specifically, NECs of the proliferative VZ exhibit strong 5mC (B) followed by a weaker 5hmC signal (C). (B–D) As these cells undergo differentiation and radial migration into the subplate layer (SP), cellular morphology changes from ellipsoidal to larger, rounded nuclei. During this transition, a characteristic rise in 5hmC is observed, in contrast to a weakening of 5mC. (E) Legend for nuclear morphology and DNA methylation mark. (B–D, F) As cells reach their target layers within the cortical plate (CP), the distribution (immunointensity gradient) of 5mC/5hmC shows an opposite trend within three sublayers of the CP (CP1/2/3). SVZ (Subventricular Zone).
Phenotypic and epigenetic features of alcohol-pervaded neocortex
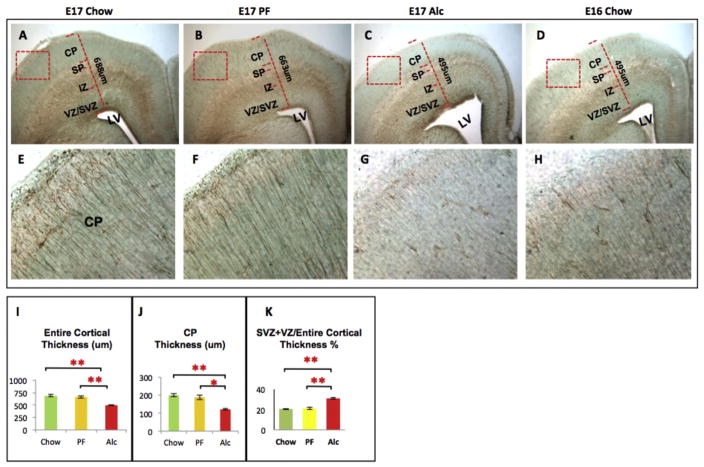
We observed that while inducing neuroanatomical and phenotypic dysregulation, alcohol closely disrupted several aspects of the cortical DMP. A reduction of thickness was characterized throughout the neocortices and appears to be a continuous cortical deficit consistent with our previous observations at E15 (Zhou, Sari, & Powrozek, 2005; Zhou, Sari, Powrozek, Goodlett, & Li, 2003). At E17, a primary feature of the experimental group was a significant reduction in the CP size (Fig. 4J), in addition to a reduction of the entire frontal neocortex compared to Chow and PF control groups (Fig. 4I). Second, a marked increase in the proportion of the VZ and SVZ to the total cortical length was observed in the Alc group compared to the PF and Chow control groups (Fig. 4K). Further evidence of neocortical thinning was demonstrated by abnormal expansion of lateral ventricles in the Alc group compared to the control groups (Fig. 4A–C). In addition to the anatomical abnormalities observed in the alcohol-treated cortex, several phenotype markers described in normal development (above) were compared alongside with DNA methylation markers.
Fig. 4.
Tbr2-im at the E17 frontal cortex across experimental groups. Structural abnormalities were observed during immunophenotypic investigation. Notably, the thickness of cortical plate (J) as well as the entire thickness of frontal cortex (I) were markedly reduced in Alcohol group frontal cortex as compared to their Chow and PF control cortices (A–C). Fetal alcohol exposure also increased the proportion of SVZ + VZ/entire cortical thickness (K) as compared to controls. Lateral Ventricle (LV) expansion was also observed in E17 alcohol cortices (A–C). (G) Finally, Tbr2-im (a marker for neural progenitor migration) was normally observed as a radially extending fiber ascending from the base of the lateral ventricle up to the pial surface. Alcohol noticeably reduced the Tbr2 immunoreactivity in the CP layer. E16 Chow brains were used as developmental stage controls and more closely resembled the E17 alcohol developmental state than E17 Chow (C–D, G–H). Quantitative measurements among the three groups were analyzed by one-way ANOVA, and the difference between paired groups were compared by Student t-test *p < 0.05, **p < 0.005. N (structural analysis) = Chow (5), PF (5), Alc (5). N (Tbr2-im analysis) = Chow (3), PF (3), Alc (3), E16 Chow (3). SVZ/VZ (Subventricular Zone/Ventricular Zone); IZ (Intermediate Zone); SP (Subplate); CP (Cortical plate).
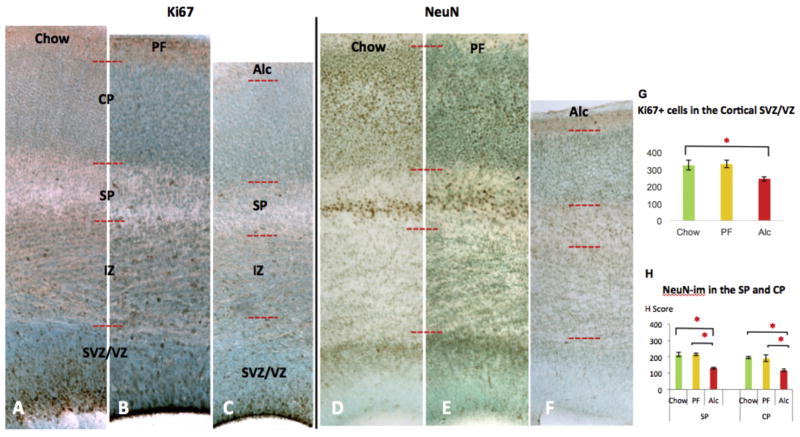
In the VZ/SVZ, a significant reduction of Ki67-im (+) cells (p < 0.05; Kruskal-Wallis test statistics [KW] = 8.61) (Fig. 5A–C) was demonstrated compared to Chow controls, though the decrease was not significantly lower than the PF controls (p > 0.05). Further, a notable reduction of Tbr2 immunoreactivity was evident in the E17 Alc group compared to E17 Chow and PF control groups (Fig. 4A–C). When further compared to E16 Chow stage controls, the E17 alcohol group was anatomically and phenotypically reminiscent of E16 Chow controls (Fig. 4C and D). Epigenetic marks showed that although changes in the 5mC-im were less apparent (p > 0.05; KW = 0.86) (Fig. 6) in the VZ/SVZ, a conspicuous reduction of 5hmC was observed (p < 0.05; KW = 7.71) (Fig. 7D). Interestingly, a marked increase of MeCP2-im in the Alc group was observed in the neurogenic VZ/SVZ compared to controls (p < 0.05; KW = 8.18) (Fig. 8A–C, F).
Fig. 5.
Alcohol-induced changes of Ki67-im and NeuN-im across different groups of E17 cortices. Representative cortical column of E17 Chow (A, D), PF (B, E), and (C, F) Alc group coronal sections for Ki67 and NeuN immunostaining. Fetal alcohol-induced reduction of Ki67 immunoreactivity was observed mainly in the SVZ/VZ zone, the neuroepithelial cellular zones. Quantitative assessment of Ki67-im (+) cells further confirmed an alcohol-related reduction in the SVZ/VZ zone (G); N = Chow (5), PF (4), Alc (7). Alcohol reduced NeuN-im throughout cortical SP and CP layers (F) compared to Chow (D) and PF (E). No significant change was observed between Chow and PF groups. Quantitative assessment of NeuN-im was further quantified by single-cell density analysis (H Scoring) across the three groups (H). *p < 0.05. Data are mean ± SEM. VZ/SVZ (Ventricular Zone/Subventricular Zone); IZ (Intermediate Zone); SP (Subplate); CP (Cortical plate).
Fig. 6.
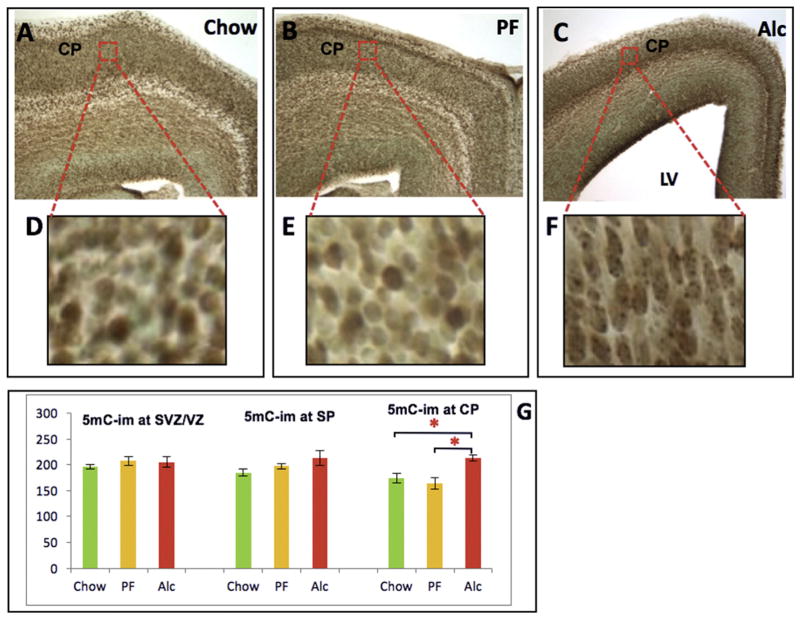
Developmental 5mC-im in the E17 frontal cortex and alcohol-induced developmental delay. (A–C) E17 frontal cortex across three groups (Chow, PF, and Alc). Red-boxed areas in CP (A–C) were enlarged in all D–F. While no change in 5mC-im was detected across the groups in the SVZ/VZ or SP, 5mC was significantly increased in the Alcohol group CP (G). Enlarged CP areas further demonstrated that alcohol induced a morphological delay of CP neurons (as observed by their ellipsoidal shape and granular intranuclear 5mC-im distribution) compared to the mature roundedness of Chow and PF CP neurons (D–F). *p < 0.05. N = Chow (5), PF (4), Alc (5). LV (Lateral Ventricle); SVZ/VZ (Subventricular Zone/Ventricular Zone); IZ (Intermediate Zone); SP (Subplate); CP (Cortical plate); MZ (Marginal Zone). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
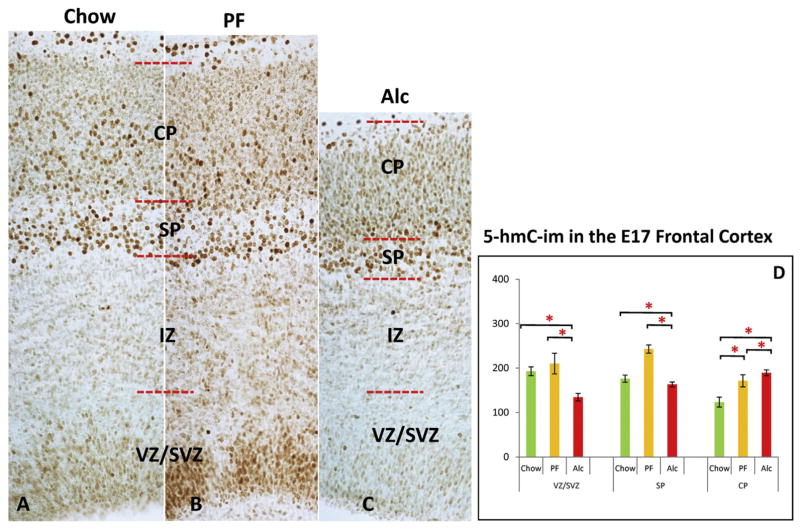
5hmC-im in the E17 frontal cortex across groups. Alcohol reduced 5hmC-im in the cortical SVZ/VZ layers (C) compared to Chow (A) and PF controls (B), while no significant change was observed between controls (D). At the SP cortical layer, the only significant alteration detected was an increase in the PF group as compared to both Chow and Alc groups. (A–D) A marked increase of 5hmC-im was observed at the CP region in both the PF and Alc groups as compared to the Chow group, while a significant increment was also evident in the Alc group CP as compared to the PF group. *p < 0.05. N = Chow (5), PF (4), Alc (5). CP (cortical plate); MZ (marginal zone); SP (subplate); SVZ (subventricular zone); VZ (Ventricular zone).
Fig. 8.
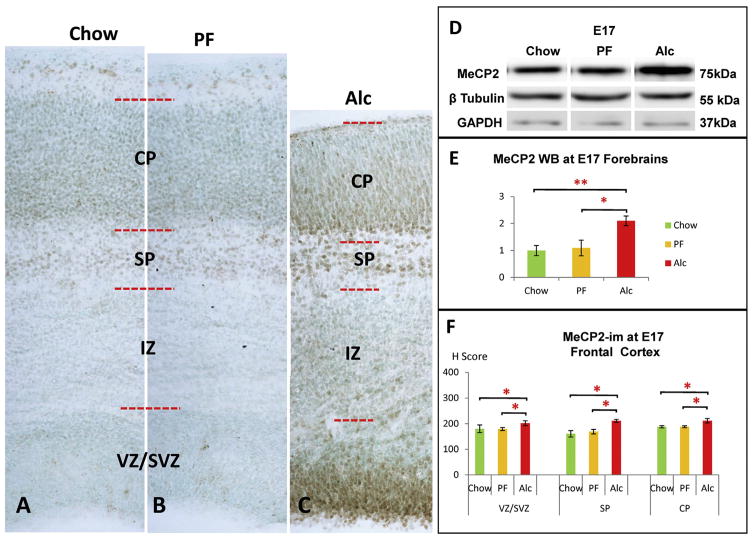
Alcohol-induced MeCP2 increase in the E17 cortex. (A–C) Representative columns from the E17 frontal cortex across the three groups. (C, F) Alcohol increased MeCP2-im throughout cortical SVZ/VZ, SP, and CP layers compared to controls. No significant change was observed between Chow and PF groups. *p < 0.05. N = Chow (5), PF (4), Alc (5). (D–E) Densitometry of MeCP2 whole-brain Western blot (WB) showed a significant increase of MeCP2 expression at E17 in the Alc group compared to its counterparts. (One-way ANOVA: F = 6.95, p < 0.05). Post hoc analysis showed no significant difference between PF and Chow groups. Western blot band intensity was normalized to GAPDH as an internal control. N = Chow (4), PF (3), Alc (4) mean ± SEM *p < 0.05 **p < 0.005. WB (Western blot).
In the SP, a significant reduction of NeuN-im neurons was found in the E17 Alc group as compared to the Chow and PF groups (p < 0.05; KW = 8.07) (Fig. 5D–F). The only significant difference in 5hmC at the SP layer was seen as an increment of the PF (p < 0.05; KW = 8.18) group as compared to both Chow and Alc groups, whereas those two did not significantly differ from each other (p > 0.05) (Fig. 7D). Meanwhile, the 5mC was not different among the groups, though a marked increase of MeCP2-im (p < 0.05; KW = 8.06) was observed in the Alc group as compared to Chow and PF groups (Fig. 8F).
In the CP, a key morphological feature imposed by alcohol is that the cortical cells arrive in an immature form (ellipsoidal shape (rather than round/mature) (Fig. 6D–F) and with shortened distance between cells, presumably due to decreased arborization between cells. In the densely packed CPs, both 5mC-im (p < 0.05; KW = 9.64) (Fig. 6G) and 5hmC-im (p < 0.05; KW = 10.01) (Fig. 7D) were up-regulated by alcohol exposure. Similarly, a marked increase of MeCP2-im was also observed in the alcohol group (p < 0.05; KW = 7.98) (Fig. 8F). Another major alcohol effect was a differential intranuclear 5mC-im chromatic distribution between control and experimental groups. In the thinned (alcohol-treated) CP, the immature ellipsoidal-shaped nuclei contained highly granular (punctate) 5mC (Fig. 6F), reminiscent of nascent 5mC distributions in less mature cells. Alternatively, a euchromatic, more heterogeneous 5mC-im pattern was observed alongside the matured CP neurons characteristic in E17 Chow and PF (Fig. 6D and E). In lieu of these findings, the comparisons of 5mC-im and 5hmC-im positive cells in the CP are presumably entangled with intra-nuclear distribution and cellular density incongruities that occur in the alcohol-exposed group.
In general, there were no major structural or phenotypic differences detected between Chow and PF groups. However, epigenetically, 5hmC-im did demonstrate some sensitivity to the PF treatment, particularly in the CP where increases could be detected (Fig. 7D).
Global 5mC and 5hmC analyses, and MeCP2 protein analysis
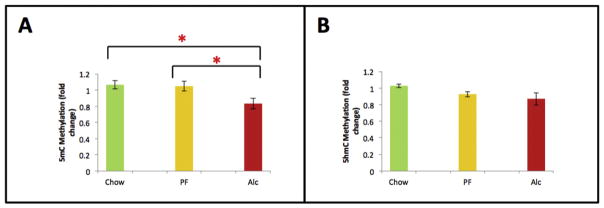
In addition to cellular and cortical layer-specific DNA methylation analyses by immunocytochemistry, an independent molecular 5mC and 5hmC analysis was performed in the E17 neocortex. 5mC analysis demonstrated that alcohol induced a global reduction in DNA methylation compared to Chow and PF animals (p < 0.05; KW = 6.03) (Fig. 9). This analysis represents an average of overall genomic levels of 5mC and 5hmC, in contrast to the layer-specific observations. In contrast, no treatment-specific differences were detected by the global 5hmC analysis (p = 0.08; KW = 4.87). This may be due to the relatively lower abundance of 5hmC in the brain compared to 5mC, or due the heterogeneity of cells that was represented in the tissue sample compared to the more cell-specific, immunohistochemical analysis (Fig. 7).
Fig. 9.
Global quantitative DNA methylation (5mC and 5-hmC) of E17 cortex across experimental groups. (A) Global DNA methylation (5mC) was significantly decreased in the neocortex at E17. (B) Global DNA hydroxylmethylation (5hmC) was not significantly decreased in the neocortex in response to fetal alcohol exposure. Means of the three groups were compared by non-parametric Kruskal-Wallis test followed by Conover post hoc test for multiple comparisons. *p < 0.05. Data are mean ± SEM. N = Chow (6), PF (6), Alc (6).
Global MeCP2 protein expression was further analyzed via Western blot analysis, which confirmed that alcohol significantly increased MeCP2 expression in the forebrain as compared to the controls (F = 6.95, Chow/Alc, p < 0.005 and PF/Alc, p < 0.05). No MeCP2 protein differences were observed between Chow and PF groups (p > 0.05) (Fig. 8D and E).
Discussion
Fetal alcohol exposure has been associated with lasting cortical deficits through various molecular constructs and functional outcomes in human and rodent models of FASD (Abbott, Kozanian, Kanaan, Wendel, & Huffman, 2016; El Shawa, Abbott, & Huffman, 2013; Robertson et al., 2016; Zhou, Lebel, et al., 2011). To date, however, the underpinnings of cortical thinning reported in human and animal models of FASD have not been clearly defined. We have previously demonstrated that an orderly progression of DNA methylation marks occurs parallel to the progression of spatiotemporal neurodevelopment during early neurulation and early hippocampal formation. Alteration of the program with the DNMT inhibitor AZA has been previously observed to lead to abnormal neural tube development (Zhou, Chen, et al., 2011). Additionally, we have described that neuro-developmental courses are disrupted by prenatal alcohol at the cellular and DNA methylation level (Chen et al., 2013; Zhou, Chen, et al., 2011). The DNA methylation program occurs spatiotemporally parallel with differentiation of neural stem cells observed in the neural tube and hippocampus. This process is not restricted to just those regions or those ages, but occurs throughout the brain wherever and whenever neuroprogenitor differentiation occurs, including the cortex presented here. This progression is best demonstrated in the cortical layers, which simultaneously feature distinct stages of neuronal development layer-by-layer at E17. This cortical model also provides an excellent illustration of the alcohol interference of the neuroepigenetic program during development. Examined together, the normal and alcohol-disrupted cortical models help to corroborate the participation of DNA methylation in cortical cellular differentiation and its disruption in cortical thinning.
The DNA methylation program in the developing neocortex
Prior to delving into alcohol-induced epigenetic aberration as an underlying factor of the alcohol-induced cortical phenotype, it is necessary to understand the DNA methylation dynamics of the embryonic cortex. A key finding is that during the formation of multi-layer cytoarchitecture of the cortex, 5mC and 5hmC marks are dynamically established in the progression of neuroprogenitor cell differentiation, which is in many ways similar to our previous observation of neuroepithelial cells (NEs) in neural tube and hippocampus formation. Commonly, the escalation of 5mC in the NEs marks the preparation and initiation of specification toward neural cells, likely occurring to assist in the down-regulation of multi-potent and proliferation genes (Kim et al., 2014; Resendiz, Mason, Lo, & Zhou, 2014). 5mC subsequently declines as migration begins from the upper limits of the VZ and through the IZ and enters a second cycle of up-regulation in the upper cortical layers as maturing neurons prepare for synaptogenesis. 5hmC, which functions as a bivalent or activating epigenetic mark (Chen, Damayanti, Irudayaraj, Dunn, & Zhou, 2014; Diotel et al., 2016; Resendiz, Chen, Öztürk, & Zhou, 2013), appears as differentiation proceeds and peaks when neurons are specified. In the VZ, where cortical proliferation (Ki67+) and neurogenesis progress and in SVZ where neural specification begins (Tbr2+ cell bodies), 5mC is prominent in the cells with mostly ellipsoidal-shaped nuclei, while 5hmC weakly appears in a subpopulation of the VZ and SVZ. These features are reminiscent of the NEs in the VZ of neural tube (at E10), and hippocampal ventral CA and dentate infragranular layers perinatally (Chen et al., 2013; Zhou, Chen, et al., 2011). Cells in the IZ are in a transitional state, as characterized by a changing morphology (transforming from ellipsoidal shape to round), differentiation (migrating intermediate progenitor cells, INPs; also marked by Tbr2-im cell bodies and fibers), and epigenetic landscape (decrease of 5mC-im and maintaining of 5hmC-im).
At the SP and CP, where bona fide neurons appear, 5hmC-im is at peak expression in round-shaped neuronal nuclei, demonstrated by NeuN-im (mature neurons) and P2Y1-im (mature neural cells capable of communication) co-localization. This has been a common observation of NEs throughout their transition to neurons during neural tube, hippocampal, and now cortex formation. Both mid-level and escalated 5mC-im is distributed in the sub-layers of the CP, likely reflecting a mixed population of newly arrived INPs (small, ellipsoid, and lightly 5mC-im) and conformed neurons (round and dark 5mC-im) entering the next phase of differentiation – synaptogenesis. Because the radially migrating cells of the neocortex occur in an “inside-out” fashion (with newly arriving cells settling atop existing neurons), the deep layer of neurons in the CP exhibits a higher (than superficial layer of CP) intensity of NeuN-im and P2Y1 compared to the upper strata of the CP. 5hmC-im is similarly distributed in the lower CP limits where cellular maturity is most advanced.
The methyl-binding protein MeCP2 is barely present in the VZ, SZ, and IZ, instead appearing only in the SP and CP at E17. This is consistent with previous findings indicating that MeCP2 generally appears only in mature neurons (Kishi & Macklis, 2004; Mullaney, Johnston, & Blue, 2004). Though the early role of MeCP2 was thought to be largely repressive (Guy, Cheval, Selfridge, & Bird, 2011), recent evidence has established that MeCP2, through the binding of 5hmC, may actually play dual roles (activation and repression) in dynamic DNA methylation distributions (Chahrour et al., 2008; Mellén, Ayata, Dewell, Kriaucionis, & Heintz, 2012). Additionally, an examination of 5mC and 5hmC co-localization with MeCP2 during corticogenesis indicated that while there is a modest co-localization between 5hmC and MeCP2 which persists from E17 to postnatal day 7, 5mC undergoes a significant transition from weak to strong co-localization with MeCP2 during that same time period (Chen et al., 2014).
Ultimately, the outcomes of our normal cortical analysis illustrated that DNA methylation progresses predictably in tandem with phenotypic landmarks of neuroepithelial differentiation and the formation of mature neurons.
Alcohol-induced cortical thinning
Among a multitude of fetal alcohol-induced structural and phenotypic anomalies, cortical thinning was a hallmark of the dysmorphology, indicated by reduced thickness of the entire cortical length as well as the CP. Our finding of an increased ratio of SVZ + VZ/whole cortical length further explains a stagnant progression of NEs. Disruption of neocortical thickness as a consequence of alcohol (prenatal and perinatal) has been documented in animal studies, including various time courses and doses of alcohol (Table 1). Cortical thinning is apparently a continuous effect occurring over mid-gestation (Aronne, Evrard, Mirochnic, & Brusco, 2008; Zhou, Sari, Powrozek, & Spong, 2004) to postnatal and young adult ages in rodent models. Alcohol-induced cortical abnormalities can be more complex and varied across different cortical regions postnatally (Abbott et al., 2016). Other factors which contribute to this complexity include human versus rodent imaging studies (Sowell et al., 2008), the ratio of neuron to glia or white matter to gray matter, along with various environmental factors which are important considerations in alcohol-derived cortical abnormalities. The current study demonstrated distinct cortical thinning, which allowed for close examination of cellular epigenetic and phenotypic changes unfolding alongside cortical thinning.
Table 1.
Comparative FASD animal models reporting fetal alcohol-induced cortical phenotypes.
| Strain | Animal Model | Dose, intake of EtOH; (v/v) | Period of Exposure | Age of Cortical Analysis | BEC (mg/dL) | Cortical phenotype | Reference |
|---|---|---|---|---|---|---|---|
| Wistar Rat | IP | 20%; 3.5 g/kg/d | E10–18 | E18 | 119–300 | Cortical thinning | Aronne et al., 2008 |
| C57 BL6 Mice | SA of EtOH in PMI liquid diet | 25% | E7–15 | E15 | 40–120 | Cortical thinning | Zhou et al., 2004 |
| CD1 Mice | SA of EtOH in Water | 25%; 6.76 ± 0.25 mL/d | E0.5–19.5 | P0 | 103–138 | Reduced cortical length | El Shawa et al., 2013 |
| CD1 Mice | SA of EtOH in Water | 25%; 6.55 ± 0.2 mL/d | E0.5–19.5 | P0, P20, P50 | 100–135 | Thickening of the frontal, somatosensory and visual cortices; thinning of prelimbic and auditory cortex (P0) | Abbott et al., 2016 |
| C57 BL/6 (B6) Mice | SA of EtOH in PMI liquid diet | 4%; 13 mL/d | E7–16 | E17 | 120–160 | Thinning of the frontal cortex | Current Study |
BEC: blood ethanol content; IP: intraperitoneal injection; SA: self-administered.
Alcohol alters the DNA methylation program
Alcohol exposure has been shown to affect global (Liu, Balaraman, Wang, Nephew, & Zhou, 2009; Zhou, Zhao, et al., 2011) and gene-specific (Govorko, Bekdash, Zhang, & Sarkar, 2012; Ouko et al., 2009) DNA methylation through the alteration of methyl donor metabolism (Resendiz et al., 2016). Here, we exemplify for the first time that the altered neocortical DMP is concomitant with the aberrant laminar patterning of the neocortex as a consequence of embryonic alcohol insult. While a global investigation of DNA methylation markers revealed a cortex-wide reduction following prenatal alcohol exposure, a more detailed examination of the developing cortical laminae revealed that the sub-structural DNA methylation patterning exhibited a far more complex response to alcohol.
In the VZ/SVZ, two seemingly contradictory features were observed – decreased proliferation and increase of layer thickness. Interestingly, one group has reported that alcohol-induced depletion of neural stem cells (and by default, their proliferation) is not due to neuroepithelial cell apoptosis, but rather is due to a premature transformation of the NEs into radial glial-like cells (Camarillo & Miranda, 2008). Another group has proposed that reduced progenitor proliferation may be the result of cytostasis, or cell cycle arrest in the absence of apoptosis (Riar et al., 2016). Whichever the case, the apparent result would denote a higher number of cells remaining in the VZ/SVZ, meaning that these cells do not survive past differentiation or represent a delay in development, given the similarities between the E17 alcohol-exposed cortex and E16 controls. Simultaneously the DMP points to stagnant cellular epigenetic progression. First, the seemingly unaffected 5mC levels of the VZ/SVZ may be the result of a delay in the initial 5mC escalation (seen in control groups) obscured by the inhibited progression of the secondary reduction in the program. In contrast, 5hmC appears delayed in tandem with increased MeCP2 at these neurogenic layers. Since the initial escalation of 5hmC-im marks the initiation of neuroepithelial differentiation (Zhou, Chen, et al., 2011; Zhou, Zhao, et al., 2011), reduced 5hmC-im occurs in parallel with the reduced differentiation of NEs.
In the SP, where the earliest mature neurons arrive, 5hmC correspondingly peaks in rounded neurons. Alcohol significantly reduced neuronal maturity (denoted by NeuN-im) alongside a reduction in 5hmC-im and an increase in MeCP2. While we were not able to currently explain the inverse 5hmC/MeCP2 fetal alcohol response, it warrants further examination in future studies. A recent report mimicking alcohol exposure and withdrawal in vitro, during the differentiation of neural stem cells, showed an induction and reduction of both transcript and protein levels of MeCP2 during exposure and withdrawal, respectively. These changes were highly correlated with dynamic changes of 5hmC and 5mC at specific MeCP2 regulatory elements (Liyanage, Zachariah, Davie, & Rastegar, 2015) and constitute some of the early unravelings of the 5mC/5hmC/MeCP2 dynamic.
Overall, the CP demonstrated the most notable alcohol response. In the CP, thickness was reduced in addition to decreased NeuN-im and Tbr2-im. Using E16 Chow stage controls, the radial migration marker Tbr2-im demonstrated an alcohol-induced developmental delay of about one day (Fig. 4). Simultaneously, all three DNA methylation marks (5mC, 5hmC and MeCP2) were increased in the CP of the Alc group. This is in contrast to the DNA methylation levels measured cortex-wide, in which 5mC was decreased relative to controls and no treatment-specific differences were detected in global 5hmC analysis (Fig. 9). Previous cortical analyses have similarly reported reductions in DNA methylation in primordial and postnatal cortical regions (Garro, McBeth, Lima, & Lieber, 1991; Otero et al., 2012). There are a few explanations to the discrepancy between laminar and region-wide DNA methylation analysis. First, as outlined earlier, epigenetic landscapes vary according to cellular states of maturity. As such, analysis of the entire cortical region may oversimplify the complexity of the epigenetic alcohol response that was observed layer-by-layer. Second, the cortex-wide reduction of DNA methylation may be biased toward the lower cortical layers due to inherent cell number. Third, in light of several reports (Prock & Miranda, 2007; Riar et al., 2016) demonstrating a developmental delay of cortical progenitors devoid of apoptosis, it can be presumed that cellular density in the CP may be more compacted than in untreated CPs (see Fig. 6), thereby skewing the immunohisto-chemical abundance of DNA methylation markers. One report observed that prenatal alcohol was associated with an increase in the number of medial ganglionic eminence-derived interneurons in the medial prefrontal cortex (Skorput, Gupta, Yeh, & Yeh, 2015), supporting the likely increased cellular density of the alcohol CP. Finally, as shown in Fig. 6(D–F), differential cellular morphology and intranuclear 5mC distribution in the alcohol CP may factor into perceived alcohol-induced hypermethylation in the CP.
While the role of DNA methylation in alcohol-mediated cortical aberration continues to be explored, important considerations include study variables such as alcohol model, dose, exposure period, age/region of analysis, etc. The appropriate controls should also be evaluated to accurately disseminate the contribution of alcohol in these. Here, for example, the administration of alcohol via a liquid-diet paradigm required the inclusion of an isocaloric liquid-diet control group (PF), which nearly mirrored the Chow brain anatomically, phenotypically, and epigenetically. Some liquid-diet sensitivity (PF compared to Chow) was detectable in the global 5mC analysis and in 5hmC analysis of the CP. Even though the liquid-diet alcohol paradigm allows for control over the alcohol dose and caloric equilibrium in a relatively non-invasive method, some epigenetic differences may be linked to inherent differences between the Chow and PF diet, such as micronutrition, fat content, and stress induced by the yoking of liquid-diet volume to equilibrate caloric content of the alcohol and control group.
Whether epigenetics leads neurodevelopment or whether developmental conditions dictate epigenetic dynamics is a question worthy of future exploration. Our ongoing study using epigenetic editing to alter the DNA methylation of a proneuron gene will shed further light on the functional impact of DNA methylation in neural stem cells. Here, we revealed that there is a unique DMP occurring parallel to the patterning of each cortical layer during brain development. This patterning was not uniformly responsive to alcohol, but instead exhibited a distinctive response to the fetal alcohol insult. We propose that the tightly regulated developmental course of the embryonic cortex is an optimal model for the pursuit of the elusive question of epigenetic governance in development.
Conclusion
Here we report that a dynamic DNA methylation program can be demonstrated throughout the distinct cortical laminae during development. 5mC and 5hmC, the two key methylation marks in the brain, demonstrated a differential distribution consistent with neural maturity. In the presence of fetal alcohol exposure, the DMP in the normal cortical condition was altered, globally, across individual cortical layers, and at a cellular level. More importantly, alcohol-induced alterations of the DMP overlapped with critical facets of neocortical development, such as the gross reduction of cortical thickness, reduced proliferation in the neuroepithelial zones, reduced expression of radial migration markers, and reduced neuronal maturation in the upper layers. Although cortical thinning in FASD may be the result of multiple causes, our findings show that DNA methylation at the cellular level is altered by fetal alcohol, underlying phenotypic abnormality, and cortical thinning.
Acknowledgments
Funding sources
This work was supported by the National Institutes of Health AA024216, AA016698, and P50AA07611 to FCZ. Additional support was provided by the Indiana CTSI Cohort 11 core pilot grant to FCZ and BAP-SBEA 2011-5DR (Mersin University) to HO. NCO is supported in part by the Fulbright Visiting Researcher fellowship. MR is supported by National Institutes of Health training grant T32 AA007462.
References
- Abbott CW, Kozanian OO, Kanaan J, Wendel KM, Huffman KJ. The impact of prenatal ethanol exposure on neuroanatomical and behavioral development in mice. Alcoholism: Clinical and Experimental Research. 2016;40:122–133. doi: 10.1111/acer.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- Anthony B, Vinci-Booher S, Wetherill L, Ward R, Goodlett C, Zhou FC. Alcohol-induced facial dysmorphology in C57BL/6 mouse models of fetal alcohol spectrum disorder. Alcohol. 2010;44:659–671. doi: 10.1016/j.alcohol.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony B, Zhou FC, Ogawa T, Goodlett CR, Ruiz J. Alcohol exposure alters cell cycle and apoptotic events during early neurulation. Alcohol and Alcoholism. 2008;43:261–273. doi: 10.1093/alcalc/agm166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronne MP, Evrard SG, Mirochnic S, Brusco A. Prenatal ethanol exposure reduces the expression of the transcriptional factor Pax6 in the developing rat brain. Annals of the New York Academy of Sciences. 2008;1139:478–498. doi: 10.1196/annals.1432.006. [DOI] [PubMed] [Google Scholar]
- Aronne MP, Guadagnoli T, Fontanet P, Evrard SG, Brusco A. Effects of prenatal ethanol exposure on rat brain radial glia and neuroblast migration. Experimental Neurology. 2011;229:364–371. doi: 10.1016/j.expneurol.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Camarillo C, Miranda RC. Ethanol exposure during neurogenesis induces persistent effects on neural maturation: Evidence from an ex vivo model of fetal cerebral cortical neuroepithelial progenitor maturation. Gene Expression. 2008;14:159–171. [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Damayanti NP, Irudayaraj J, Dunn K, Zhou FC. Diversity of two forms of DNA methylation in the brain. Frontiers in Genetics. 2014;5:46. doi: 10.3389/fgene.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Öztürk NC, Zhou FC. DNA methylation program in developing hippocampus and its alteration by alcohol. PLoS One. 2013;8:e60503. doi: 10.1371/journal.pone.0060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhladze RT, Ramishvili NSh, Tsagareli ZG, Kikalishvili NO. The spectrum of hemispheral cortex lesions in intrauterine alcoholic intoxication. Georgian Medical News. 2011:81–87. [PubMed] [Google Scholar]
- Diotel N, Mérot Y, Coumailleau P, Gueguen MM, Sérandour AA, Salbert G, et al. 5-hydroxymethylcytosine marks postmitotic neural cells in the adult and developing vertebrate central nervous system. The Journal of Comparative Neurology. 2016;525:478–497. doi: 10.1002/cne.24077. [DOI] [PubMed] [Google Scholar]
- El Shawa H, Abbott CW, 3rd, Huffman KJ. Prenatal ethanol exposure disrupts intraneocortical circuitry, cortical gene expression, and behavior in a mouse model of FASD. The Journal of Neuroscience. 2013;33:18893–18905. doi: 10.1523/JNEUROSCI.3721-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: Implications for the fetal alcohol syndrome. Alcoholism: Clinical and Experimental Research. 1991;15:395–398. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- Goulding H, Pinder S, Cannon P, Pearson D, Nicholson R, Snead D, et al. A new immunohistochemical antibody for the assessment of estrogen receptor status on routine formalin-fixed tissue samples. Human Pathology. 1995;26:291–294. doi: 10.1016/0046-8177(95)90060-8. [DOI] [PubMed] [Google Scholar]
- Govorko D, Bekdash RA, Zhang C, Sarkar DK. Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biological Psychiatry. 2012;72:378–388. doi: 10.1016/j.biopsych.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JH. Fetal alcohol spectrum disorders: Understanding the effects of prenatal alcohol exposure and supporting students. The Journal of School Health. 2007;77:103–108. doi: 10.1111/j.1746-1561.2007.00178.x. [DOI] [PubMed] [Google Scholar]
- Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annual Review of Cell and Developmental Biology. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- Hatanaka Y, Hisanaga S, Heizmann CW, Murakami F. Distinct migratory behavior of early- and late-born neurons derived from the cortical ventricular zone. The Journal of Comparative Neurology. 2004;479:1–14. doi: 10.1002/cne.20256. [DOI] [PubMed] [Google Scholar]
- Jacobson SW, Jacobson JL, Stanton ME, Meintjes EM, Molteno CD. Biobehavioral markers of adverse effect in fetal alcohol spectrum disorders. Neuropsychology Review. 2011;21:148–166. doi: 10.1007/s11065-011-9169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KL, Hoyme HE, Robinson LK, Del Campo M, Manning MA, Prewitt LM, et al. Fetal alcohol spectrum disorders: Extending the range of structural defects. American Journal of Medical Genetics. Part A. 2010;152A:2731–2735. doi: 10.1002/ajmg.a.33675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Park YK, Kang TW, Lee SH, Rhee YH, Park JL, et al. Dynamic changes in DNA methylation and hydroxymethylation when hES cells undergo differentiation toward a neuronal lineage. Human Molecular Genetics. 2014;23:657–667. doi: 10.1093/hmg/ddt453. [DOI] [PubMed] [Google Scholar]
- Kishi N, Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Molecular and Cellular Neurosciences. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Lawrence RC, Otero NK, Kelly SJ. Selective effects of perinatal ethanol exposure in medial prefrontal cortex and nucleus accumbens. Neurotoxicology and Teratology. 2012;34:128–135. doi: 10.1016/j.ntt.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva J, Zakharov A, Ogievetsky E, Minlebaeva A, Kurbanov R, Gerasimova E, et al. Inhibition of cortical activity and apoptosis caused by ethanol in neonatal rats in vivo. Cerebral Cortex. 2015:1–15. doi: 10.1093/cercor/bhv293. pii: bhv293. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Lebel C, Mattson SN, Riley EP, Jones KL, Adnams CM, May PA, et al. A longitudinal study of the long-term consequences of drinking during pregnancy: Heavy in utero alcohol exposure disrupts the normal processes of brain development. The Journal of Neuroscience. 2012;32:15243–15251. doi: 10.1523/JNEUROSCI.1161-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics. 2009;4:500–511. doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage VR, Zachariah RM, Davie JR, Rastegar M. Ethanol deregulates Mecp2/MeCP2 in differentiating neural stem cells via interplay between 5-methylcytosine and 5-hydroxymethylcytosine at the Mecp2 regulatory elements. Experimental Neurology. 2015;265:102–117. doi: 10.1016/j.expneurol.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason S, Anthony B, Lai X, Ringham HN, Wang M, Witzmann FA, et al. Ethanol exposure alters protein expression in a mouse model of fetal alcohol spectrum disorders. International Journal of Proteomics. 2012;2012:867141. doi: 10.1155/2012/867141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HS, Wimmer VC, Oberlaender M, de Kock CP, Sakmann B, Helmstaedter M. Number and laminar distribution of neurons in a thalamocortical projection column of rat vibrissal cortex. Cerebral Cortex. 2010;20:2277–2286. doi: 10.1093/cercor/bhq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney BC, Johnston MV, Blue ME. Developmental expression of methyl-CpG binding protein 2 is dynamically regulated in the rodent brain. Neuroscience. 2004;123:939–949. doi: 10.1016/j.neuroscience.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Nardelli A, Lebel C, Rasmussen C, Andrew G, Beaulieu C. Extensive deep gray matter volume reductions in children and adolescents with fetal alcohol spectrum disorders. Alcoholism: Clinical and Experimental Research. 2011;35:1404–1417. doi: 10.1111/j.1530-0277.2011.01476.x. [DOI] [PubMed] [Google Scholar]
- Otero NK, Thomas JD, Saski CA, Xia X, Kelly SJ. Choline supplementation and DNA methylation in the hippocampus and prefrontal cortex of rats exposed to alcohol during development. Alcoholism: Clinical and Experimental Research. 2012;36:1701–1709. doi: 10.1111/j.1530-0277.2012.01784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouko LA, Shantikumar K, Knezovich J, Haycock P, Schnugh DJ, Ramsay M. Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: Implications for fetal alcohol spectrum disorders. Alcoholism: Clinical and Experimental Research. 2009;33:1615–1627. doi: 10.1111/j.1530-0277.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- Perkins A, Lehmann C, Lawrence RC, Kelly SJ. Alcohol exposure during development: Impact on the epigenome. International Journal of Developmental Neuroscience. 2013;31:391–397. doi: 10.1016/j.ijdevneu.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prock TL, Miranda RC. Embryonic cerebral cortical progenitors are resistant to apoptosis, but increase expression of suicide receptor DISC-complex genes and suppress autophagy following ethanol exposure. Alcoholism: Clinical and Experimental Research. 2007;31:694–703. doi: 10.1111/j.1530-0277.2007.00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendiz M, Chen Y, Öztürk NC, Zhou FC. Epigenetic medicine and fetal alcohol spectrum disorders. Epigenomics. 2013;5:73–86. doi: 10.2217/epi.12.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendiz M, Lo C-L, Badin JK, Chiu Y, Zhou FC. Alcohol metabolism and epigenetic methylation and acetylation. In: Patel VB, editor. Molecular aspects of alcohol and nutrition. 1. Waltham, MA: Academic Press; 2016. pp. 287–303. [Google Scholar]
- Resendiz M, Mason S, Lo CL, Zhou FC. Epigenetic regulation of the neural transcriptome and alcohol interference during development. Frontiers in Genetics. 2014;5:285. doi: 10.3389/fgene.2014.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riar AK, Narasimhan M, Rathinam ML, Henderson GI, Mahimainathan L. Ethanol induces cytostasis of cortical basal progenitors. Journal of Biomedical Science. 2016;23:6. doi: 10.1186/s12929-016-0225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson FC, Narr KL, Molteno CD, Jacobson JL, Jacobson SW, Meintjes EM. Prenatal alcohol exposure is associated with regionally thinner cortex during the preadolescent period. Cerebral Cortex. 2016;26:3083–3095. doi: 10.1093/cercor/bhv131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RP, Shiue K, Schomberg D, Zhou FC. Cellular epigenetic modifications of neural stem cell differentiation. Cell Transplant. 2009;18:1197–1211. doi: 10.3727/096368909X12483162197204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorput AG, Gupta VP, Yeh PW, Yeh HH. Persistent interneuronopathy in the prefrontal cortex of young adult offspring exposed to ethanol in utero. The Journal of Neuroscience. 2015;35:10977–10988. doi: 10.1523/JNEUROSCI.1462-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell ER, Mattson SN, Kan E, Thompson PM, Riley EP, Toga AW. Abnormal cortical thickness and brain-behavior correlation patterns in individuals with heavy prenatal alcohol exposure. Cerebral Cortex. 2008;18:136–144. doi: 10.1093/cercor/bhm039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Phillips OR, Kan E, Sulik KK, Mattson SN, Riley EP, et al. Callosal thickness reductions relate to facial dysmorphology in fetal alcohol spectrum disorders. Alcoholism: Clinical and Experimental Research. 2012;36:798–806. doi: 10.1111/j.1530-0277.2011.01679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Roussotte F, Kan E, Sulik KK, Mattson SN, Riley EP, et al. Abnormal cortical thickness alterations in fetal alcohol spectrum disorders and their relationships with facial dysmorphology. Cerebral Cortex. 2012;22:1170–1179. doi: 10.1093/cercor/bhr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC. DNA methylation program during development. Frontiers in Biology, (Beijing) 2012;7:485–494. doi: 10.1007/s11515-012-9246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Patel TD, Swartz D, Xu Y, Kelley MR. Production and characterization of an anti-serotonin 1A receptor antibody which detects functional 5-HT1A binding sites. Brain Research. Molecular Brain Research. 1999;69:186–201. doi: 10.1016/s0169-328x(99)00101-1. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Sari Y, Powrozek T, Goodlett CR, Li TK. Moderate alcohol exposure compromises neural tube midline development in prenatal brain. Brain Research. Developmental Brain Research. 2003;144:43–55. doi: 10.1016/s0165-3806(03)00158-5. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Sari Y, Powrozek TA, Spong CY. A neuroprotective peptide antagonizes fetal alcohol exposure-compromised brain growth. Journal of Molecular Neuroscience. 2004;24:189–199. doi: 10.1385/JMN:24:2:189. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Sari Y, Powrozek TA. Fetal alcohol exposure reduces serotonin innervation and compromises development of the forebrain along the serotonergic pathway. Alcoholism: Clinical and Experimental Research. 2005;29:141–149. doi: 10.1097/01.alc.0000150636.19677.6f. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Chen Y, Love A. Cellular DNA methylation program during neurulation and its alteration by alcohol exposure. Birth Defects Research. A, Clinical and Molecular Teratology. 2011;91:703–715. doi: 10.1002/bdra.20820. [DOI] [PubMed] [Google Scholar]
- Zhou D, Lebel C, Lepage C, Rasmussen C, Evans A, Wyper K, et al. Developmental cortical thinning in fetal alcohol spectrum disorders. Neuro-Image. 2011;58:16–25. doi: 10.1016/j.neuroimage.2011.06.026. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Zhao Q, Liu Y, Goodlett CR, Liang T, McClintick JN, et al. Alteration of gene expression by alcohol exposure at early neurulation. BMC Genomics. 2011;12:124. doi: 10.1186/1471-2164-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]