Abstract
The current work aims to identify patients with atopic dermatitis for inclusion in genome-wide association studies (GWAS). Here we describe a machine learning-based phenotype algorithm. Using the electronic health record (EHR), we combined coded information with information extracted from encounter notes as features in a lasso logistic regression. Our algorithm achieves high positive predictive value (PPV) and sensitivity, improving on previous algorithms with low sensitivity. These results demonstrate the utility of natural language processing (NLP) and machine learning for EHR-based phenotyping.
I. Introduction
Atopic dermatitis (AD) is a chronic inflammatory disease associated with intense itch and skin hyper-reactivity to environmental triggers that are innocuous in non-atopic individuals [1]. Although AD usually presents in early infancy and childhood, it commonly persists into or begins in adulthood. Recent population-based studies estimated that AD and/or eczema affect 7.2%–10.2% of adults living in the United States [2], [3]. AD has a complex etiology and considerable heterogeneity with respect to skin lesion distribution (e.g., flexural, extensor, head and neck areas, generalized) and morphology (e.g., oozing, scaling, lichenification, prurigo nodules, ill-demarcated, psoriasiform), intensity and time-course (e.g., intermittent, chronic persistent disease, seasonal variation, episodic flares) and associated symptom burden and comorbidities. This heterogeneity can pose diagnostic challenges, and misdiagnoses are common due to the multi-dimensional clinical presentation.
The complexity of this phenotype hinders the identification of large-scale cohorts necessary to undertake clinical, epidemiological, and genetics research. Genetics studies, such as genome-wide association studies (GWAS) contribute to our understanding of the genetic bases of particular phenotypes, a principle goal of the NIH-funded Electronic Medical Records and Genomics (eMERGE) network [4]. In the current work, we used a registry of AD patients as a gold standard to develop a EHR-based phenotype algorithm, which can be used to identify AD cases for clinical research and recruitment. Our goal is to apply the algorithm across eMERGE network sites to identify potential cases for inclusion in GWAS.
To develop the algorithm, we utilized data collected from the Northwestern Medicine Enterprise Data Warehouse (NMEDW), which houses the electronic health records (EHR) of roughly 6 million patients undergoing treatment or involved in research at Northwestern Medicine [5]. The EHR contains two types of information: structured, coded data and unstructured, clinical narratives (such as notes from clinical encounters). Coded data about patients, including demographics, International Classification of Disease (ICD-9 or ICD-10) codes, medication prescriptions, and laboratory results provide substantial information about patients that is useful for clinical research. Previous phenotype algorithms for eMERGE projects have relied exclusively on coded data [4], [6]. However, narrative data (e.g., free-form text in encounter notes written by physicians) is an additional rich source of information about disease history (e.g., symptoms, age of onset, longitudinal course) and clinician assessment (e.g., physical exam, diagnosis, and severity assessment). This information is not readily accessible without the use of natural language processing (NLP). A growing number of phenotype algorithms make use of both structured and unstructured sources of information [7].
Another development in EHR-based phenotyping has been a move towards machine learning to create predictive models. While rule-based phenotype algorithms are still commonly used successfully [8], machine learning can be more appropriate for complex phenotypes, when it is not straightforward to combine criteria using manually constructed Boolean operators over EHR-based criteria. The heterogeneity of AD, and the corresponding diversity of relevant AD data in the EHR, make it a good candidate for a machine learning approach.
II. Related Work
A previous study developed a phenotype algorithm based only on billing (ICD-9) codes for primary diagnoses of AD or contact dermatitis (CD)/eczema and secondary diagnoses of asthma, hay fever, and food allergy [9]. The algorithm achieved a high positive predictive value (PPV) of 95.2% when combining primary and all secondary diagnoses in the algorithm. However, this algorithm severely limited the number of patients that could be included in the cohort, which limits the algorithms effectiveness for GWAS cohort selection. The sensitivity of this best performing algorithm was extremely poor (8.7%). These results motivate the development of an improved and more sophisticated algorithm incorporating features derived from structured data and NLP of encounter notes, as well as machine learning of a predictive model over these features.
A growing number of studies have utilized machine learning and NLP to develop algorithms for phenotypes such as rheumatoid arthritis (RA) [10], [11], bipolar disorder (BD) [12], and cerebral aneurysms (CA) [13]. A study of RA applied lasso logistic regression as a technique for detecting cases based on a combination of features from structured and unstructured sources [10]. As a starting point for developing the algorithm, Liao et al. obtained an enriched set of potential RA patients by searching the EHR for patients with at least one occurrence of the ICD-9 code for RA or related diseases or had been checked for anti-cyclic citrullinated peptide. Using a randomly selected set of patients from this enriched cohort, the phenotype algorithm was trained using a gold standard diagnosis assigned by two rheumatologists following a chart review. The algorithm included features from both structured and unstructured sources. To extract information from unstructured sources (e.g., provider notes, radiology reports, discharge summaries), an NLP tool identified relevant terms, such as mentions of diagnoses of RA (and its comorbidities), RA medications, and relevant laboratory test information. Due to high dimensionality of the set of NLP-derived features, clinicians collapsed similar mentions into broader categories. Dimensionality was reduced further following training of the lasso regression model, which preserves only the most informative features in the final model. When combining these NLP-derived features with features from unstructured sources (e.g., diagnosis codes), the phenotype algorithm outperformed algorithms with only one or the other type of features (5–6% difference in PPV). Furthermore, the regression algorithm drastically outperformed traditional code-based algorithms (38–49% difference in PPV). A subsequent study demonstrated that the RA machine learning-based algorithm generalized well to data from other institutions [11]. Such cross-institutional validation is a critical step in the process of developing high quality phenotype algorithms and GWAS within the eMERGE network.
Other studies have demonstrated similar success building phenotype algorithms using lasso logistic regression. Similar to the RA algorithm described above, the development of an algorithm for identification of patients with cerebral aneurysms (CA) began by selecting an enriched set of patients with ICD-9 codes for various aneurysm subtypes, relevant procedure and CPT codes, or mentions of the term aneurysm in close proximity to terms such as cerebral, cranial, brain, among others [13]. Following chart review of a random subset of the enriched set, an adaptive lasso logistic regression was trained using gold standard diagnoses. NLP features were derived by extracting a set of clinically relevant terms from clinical notes. Dimensionality of the NLP-derived feature set was initially reduced by including only features present for at least 10% of the patients, and further reduced during training. As was the case for the RA algorithm, the best performing model for selecting a CA cohort from EHR data included both NLP and coded features.
Finally, lasso logistic regression has been applied successfully to algorithms for psychiatric disorders, such as bipolar disorder (BD) [12]. Development of the BD algorithm followed similar procedures as for RA and CA, with initial identification of an enriched set of patients and chart review of a subset of those patients to create a gold standard. Clinicians identified relevant terms in the clinical narratives during chart review, which were then systematically extracted from notes using NLP. No model-independent dimensionality reduction was attempted for the set of NLP-derived features. Consistent with other phenotype algorithms, the BD algorithm incorporating NLP-derived features outperformed code-based algorithms (6–35% difference in PPV).
For the AD phenotype algorithm, we adopted methodologies similar to those utilized for developing phenotype algorithms for RA [10], [11]. We used the same cohort of patients used for developing a previous code-only AD algorithm [9]. For the current study, we developed a machine learning approach to EHR-based AD identification relying on NLP-derived features. Our goal is to match the high PPV of the existing algorithm, but to do so while improving its peformance with respect to sensitivity.
III. Methods
A. Data Source
We utilized an existing registry of potential AD cases created for a previous study [9]. To create this registry, patients with at least one occurrence of an ICD-9 code for AD or CD/eczema between January 2001 and November 2014 (end point of [9], for which the registry was originally created) were drawn from the set of patients available in the NMEDW (N > 2.5 million). From that set of patients (AD: n = 41,162; CD/eczema: 2,106), 577 cases who had a visit documented in the last year were selected. The final set of cases included 562 patients who had encounter notes of the appropriate type within the desired time window (through December 2015; see below). Figure 1 summarizes the cohort creation procedure.
Fig. 1.
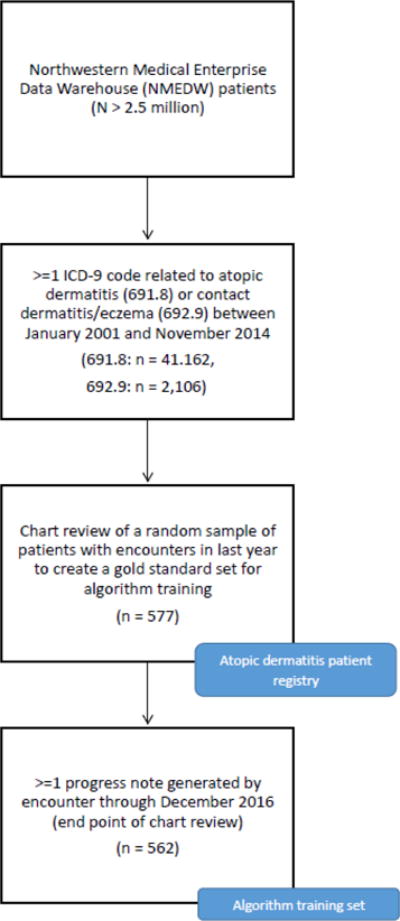
Study flow diagram.
As part of the previous study [9], the entire EHR of these patients were reviewed by four independent reviewers (dermatologists). During this chart review (completed December 2015), the reviewers documented the presence of each of the Hanifin and Rajka (HR) major and minor criteria [14] and UK Working Party (UKWP) criteria [15] (see below), along with other information not relevant to the current study.
B. Gold Standard Classification
Several criteria have been developed and validated for clinical diagnosis of AD. The Hanifin and Rajka (HR) criteria [14] is comprised of a detailed set of signs, symptoms, and comorbidities present in patients with AD. Major criteria include pruritus, flexural dermatitis, chronic or relapsing dermatitis, and a personal or family history of atopic disease (e.g., asthma, hay fever). HR minor criteria include facial symptoms or diseases (e.g., infraorbital darkening, conjunctivitis), environmental or emotional triggers (e.g., anxiety, depression, wool intolerance), common complications (e.g., skin infections, cataracts), and other symptoms and diseases (e.g., dry skin, ichthyosis, white dermatographism). Patients meet the diagnostic criteria for AD if they possess at least three major and three minor criteria.
The UK Working Party (UKWP) criteria [15] is an abridged modification of the HR criteria. Patients must have a documented itchy skin condition and least three of the following symptoms: skin crease involvement, personal history of atopic disease, dry skin, flexural dermatitis, or an early age of onset.
To create a gold standard for algorithm development in the current study, each patient was assigned definite, probable, or negative status according to the HR and UKWP diagnostic criteria and information documented during chart review (Table I). For the HR criteria, patients with at least three major and minor HR criteria were definite for AD. Patients with at least two major and minor criteria were classified as probable for AD. For the UKWP criteria, patients with pruritus and at least three minor criteria were definite for AD, while those with pruritus but only two minor criteria were probable for AD. Patients who met fewer criteria were classified as not having AD. Patients with definite and probable classifications were grouped to a single AD-positive class, while negative classifications comprised the AD-negative class.
TABLE I.
Gold standard classification distributions
| AD Criteria | Classifications
|
|||
|---|---|---|---|---|
| Definite | Probable | Positive | Negative | |
| HR | 150 | 128 | 278 | 284 |
| UKWP | 164 | 63 | 226 | 335 |
C. Coded EHR Data
We included counts of the following structured codes from the EHR as features for the phenotype algorithm: ICD-9 and ICD-10 codes for AD, eczema, and AD comorbidities (Table II), laboratory values, coded medications, and demographic information (age, sex, race). We imposed no restriction on provider type for coded data in the EHR. All structured codes were de-duplicated so that any given value was only counted once per day (regardless of the number of encounters on that day).
TABLE II.
Diagnosis codes included in algorithm
| Phenotype | Diagnosis Codes
|
|
|---|---|---|
| ICD-9 | ICD-10 | |
| Atopic dermatitis | 691.8 | L20.x |
| Contact dermatitis/eczema | 692.9 | L30.9 |
| Asthma | 493.x | J45 |
| Hay fever | 477.x | J30 |
| Food allergy | 995.3, 995.6, 995.7, 693.1 | – |
Counts of structured codes associated with a patient were transformed in a number of ways for inclusion in the machine learning algorithm. Diagnosis code counts were normalized in two ways: dividing by the total number of (de-duplicated) codes in the EHR for each patient and natural-log transformation. Laboratory values were included as a binary variable, indicating whether any laboratory test relevant to AD diagnosis was performed (e.g., skin prick test). AD-relevant medications were also included as a single, grouped category and represented as a binary variable. These medications include topical corticosteroids, topical calcineurin inhibitors, prescription emollients, antihistamines, oral corticosteroids, phototherapy, cyclosporine A, interferon gamma, doxepin, azathioprine, methotrexate, and mycophenolate mofetil [14], [16]. Patient ages were calculated by subtracting date of birth from March 15, 2017. Patient sex was dummy coded with female equal to 1 and male equal to 0. Finally, due to many missing values and indications not to disclose race, race was broadly grouped and dummy coded as white (1) vs. non-white (0).
D. Narrative EHR Data and NLP
Many types of narrative data exist in the EHR, some with more useful information than others for developing phenotype algorithms. For the current purposes, we focused on progress notes generated by both inpatient and outpatient encounters with physicians (with no restriction on provider type). We excluded telephone encounter notes, procedural notes, and nursing notes, among others. We queried the NMEDW for progress notes generated by encounters through December 2015 (the end point of chart review). Of our cohort of 577 AD cases, only 562 patients had encounter notes of the appropriate type within the specified time window. Our analyses, therefore, included only these 562 patients.
Encounter notes contain mentions of many concepts (e.g., diagnoses, signs, symptoms, medications) that are irrelevant for the target phenotype. To focus our NLP efforts, we identified a set of target AD concepts with the help of an AD domain expert clinician (JS). We curated a set of key terms based on the clinicians experience with AD patients and the specific HR and UKWP criteria. These terms were then mapped to Unified Medical Language System (UMLS) [17] concept unique identifiers (CUIs) and were used to create a custom dictionary of concepts. Alternate phrasings of each term were included based on UMLS entries for CUIs associated with the term. For example, for the target term pruritus (CUI: C0033774), there were several alternate phrasings in the UMLS entry, including itching (pruritus), itchy skin, and pruritic dermatitis. These alternate phrasings ensure the named entity recognition (NER) component can cope with variations in how concepts are expressed. Common misspellings (e.g., ichthyosis icthyosis and pruritus pruritis) were added to the dictionary manually based on the guidance of our AD domain expert (JS). Finally, the same set of medications targeted in the coded data were included in the dictionary. Our final dictionary contained 437 unique concepts.
Concepts were extracted from encounter notes using the dictionary look-up NER component of the clinical Text Analysis and Knowledge Extraction System (cTAKES) [18] with our custom dictionary of AD-relevant concepts. In cases where cTAKES assigned multiple CUIs to a single mention (approximately 3.5% of mentions), we selected the first CUI assigned. cTAKES assertion annotation modules were also included in the NLP pipeline in order to capture the subject of concepts (i.e., referring to the patient or a family member), the status of concepts (e.g., relation to patient history), and the polarity of concepts (asserted or negated). Therefore, a concept such as pruritus (C0033774) could appear in the feature set multiple times (e.g., negated mention of pruritus regarding patient history vs. positive mention of pruritus for patient in present). Extracted concepts were converted into features by calculating the number of mentions of each concept for each patient. These concepts were then grouped into broader categories to reduce the dimensionality of the feature set. These broader categories were based on the HR and UKWP criteria from which the concepts were originally derived. Following this grouping, there were 56 narrative-based features remaining for inclusion in the machine learning algorithm. Each feature was natural log transformed. Concepts related to medications and laboratory tests described within the clinical narrative were transformed to binary variables to mirror their coded counterparts.
E. Lasso Logistic Regression
We used lasso logistic regression to develop an algorithm to predict whether patients had AD. The lasso logistic regression technique has been used for previous phenotype algorithms [10]–[13] due to its ability to eliminate non-influential features from the model during training and generalizability to unseen data from novel sources. The result is an easily interpretable model with a relatively small set of influential features.
Prior to training, we created three non-overlapping data subsets stratified by class label: training (60%), validation (20%), testing (20%). The training set was used for model selection and fitting, using the glmet implementation of lasso logistic regression in R [19], which includes a cross-validation (CV) training function that selects the optimal value of the tuning parameter lambda. We fit the model using 10-fold CV, and used the optimal lambda value to evaluate the model on the held-out validation set. We do not report here results on the held-out test set, as we plan further improvements to the NLP pipeline and machine learning approach (see below).
F. Evaluation Measures
The primary goal of phenotype algorithms within eMERGE projects is to identify a cohort of patients for which there is high confidence the selected patients represent true cases of the phenotype. The secondary goal is ensure the high precision cohort of patients is large enough to perform GWAS. To achieve these goals, we evaluated the performance of our phenotype algorithm in terms of PPV, sensitivity, and F-measure. PPV was calculated as the total number of positive cases accurately identified by the algorithm (true positives) divided by the total number of predicted positive cases (true positives and false positives). Sensitivity was calculated as the total number of true positives divided by the total number of positive cases (true positives and false negatives). F-measure was calculated as the harmonic mean of PPV and sensitivity.
IV. Results
We report two sets of results based on each of the two gold standards, the HR and UKWP criteria. As shown in Table I, our gold standard classification procedure using the HR criteria identified 278 AD-positive patients and 284 AD-negative patients. Characteristics of the HR training set are summarized in Table III. The HR training set is quite balanced across classes (AD positive vs. AD negative) for each of our demographic variables (age, sex, and race) and for the mean number of diagnosis codes in the EHR (a proxy measure of the approximate size of each patients health record).
TABLE III.
Characteristics of Training Set (n = 562)
| Patient Feature | HR
|
UKWP
|
||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| Total | 278 | 284 | 227 | 335 |
| Gender (female) | 173 | 180 | 145 | 208 |
| Race (white) | 144 | 148 | 107 | 185 |
| Age (mean) | 43.8 | 46.4 | 43.6 | 46.2 |
| Age (sd) | 15.1 | 15.1 | 14.6 | 15.4 |
| Dx Code (mean) | 138.4 | 139.4 | 153.9 | 128.8 |
| Dx Code (sd) | 193.8 | 234.2 | 249 | 187.5 |
HR model performance on the validation set is summarized in Table IV. While there was little difference between the PPV achieved by models including NLP-based features (code+NLP and NLP-only), these models outperformed the model including only coded data from the EHR (10.5–10.8% difference). Similarly, models with NLP-based features had comparable levels of sensitivity, both were substantially higher than the code-only model (19.6–21.4% difference). The highest performing model, taking all evaluation measures into account, is the model including both structured and unstructured sources, although both models including NLP-based features performed quite well. These models provide reasonably good PPV, but crucially a close to 10-fold improvement on sensitivity compared to a previous AD phenotype algorithm based solely on ICD-9 code counts using the same set of patients [9].
TABLE IV.
HR Algorithm Results
| Model | Performance Metric
|
||
|---|---|---|---|
| PPV | Sensitivity | F-measure | |
|
|
|||
| Code+NLP | 84.0% | 75.0% | 79.2% |
| Code–only | 73.2% | 53.6% | 61.9% |
| NLP–only | 83.7% | 73.2% | 78.1% |
The final model contained 26 features, with features from both data sources. The top ten features with highest predictive value are summarized with regression coefficients in Table V. This set of best features are noteworthy for a number of reasons. First, the normalized count of AD diagnosis codes coded in the EHR has the highest predictive power in the model, indicating that AD diagnosis codes contribute important information to the model. The high predictive power of this diagnosis code likely contributes to the competitive PPV of the code-only algorithm. However, previous work has shown that diagnosis codes alone are not enough to achieve high sensitivity with the same cohort of patients [9], a finding we replicate here. Eight of the remaining top performing features are derived from applying NLP to the free text of clinicians encounter notes, which demonstrates the necessity of NLP for achieving high sensitivity in an AD phenotype algorithm.
TABLE V.
Top 10 Features in Best HR Model
| Feature | Coefficient |
|---|---|
|
Positive predictors
| |
| Coded normalized count of AD diagnosis codes | 3.253 |
| NLP pytiriasis alba (patient, positive) | 1.112 |
| NLP atopic disease (patient, negative) | 1.099 |
| NLP white dermatographism (patient, positive) | 0.448 |
| Coded laboratory tests | 0.328 |
| NLP ichthyosis (patient, positive) | 0.318 |
| NLP AD/eczema (patient history, positive) | 0.289 |
| NLP contact dermatitis (patient, negative) | 0.249 |
|
Negative predictors | |
| NLP white dermatographism (patient, negative) | −0.929 |
| NLP perifollicular accentuation (patient, negative) | −0.404 |
A somewhat different pattern of results was observed for models trained with the UKWP gold standard labels. Characteristics of patients in the UKWP training set are summarized in Table III and results are summarized in Table VI. As with the HR models, there was little difference in PPV between the code+NLP model and the NLP-only model (3% difference). However, the PPV for the code-only model outperformed either model including NLP-based features (5.6–8.6% difference). Similar to the HR model, the UKWP models including NLP-derived features out-performed the code-only model in terms of sensitivity (12.2–14.7% difference). The code+NLP and NLP-only models had the same F-measure, which was higher than the code-only F-measure (5.8% difference). Although the code-only model had the highest PPV in Table VI, with respect to F-measure (the harmonic mean of PPV and sensitivity) the best model was the full model that included features from both structured and unstructured sources.
TABLE VI.
UKWP Algorithm Results
| Model | Performance Metric
|
||
|---|---|---|---|
| PPV | Sensitivity | F-measure | |
|
|
|||
| Code+NLP | 72.2% | 63.4% | 67.5% |
| Code–only | 77.8% | 51.2% | 61.7% |
| NLP–only | 69.2% | 65.9% | 67.5% |
Therefore, the highest performing model included features from both structured and unstructured sources. The final UKWP model included 20 features from both data sources. Table VII shows the ten features with the highest predictive value with their regression coefficients. As in the best HR model, the feature with the highest predictive power in the UKWP model was AD diagnosis codes, again a likely contributor to the high PPV of the code-only model. There is notable overlap in the features selected for the HR and UKWP models, such as mention within the clinicians encounter notes of pytiriasis alba, patient history of AD/eczema and ichthyosis.
TABLE VII.
Top 10 Features in Best UKWP Model
| Feature | Coefficient |
|---|---|
|
Positive predictors
| |
| Coded normalized count of AD diagnosis codes | 3.311 |
| NLP pytiriasis alba (patient, negative) | 0.515 |
| NLP medications (positive) | 0.464 |
| NLP AD/eczema (patient history, positive) | 0.345 |
| NLP atopic disease (patient history, positive) | 0.199 |
| NLP ichthyosis (patient, positive) | 0.175 |
|
Negative predictors | |
| NLP keratoconus (patient, positive) | −0.210 |
| NLP facial erythema (patient, negative) | −0.181 |
| Race | −0.096 |
| NLP perifollicular accentuation (patient, positive) | −0.093 |
V. Discussion
The results of the current study indicate that the development of the AD phenotype algorithm benefited from including information from both structured and unstructured sources within the EHR, and from the use of a machine learning approach. Using NLP, we derived a set of features representing important information for identifying patients with this phenotype that was not represented by coded information. By including these NLP-derived features, we were able to achieve a near 10-fold improvement in the sensitivity reported with the same cohort of patients using an algorithm based solely on diagnosis codes [9]. Our NLP-derived features were able to capture information about patient history (e.g., history of atopic disease, such as asthma and hay fever) and conditions (e.g., pytiriasis alba) used by physicians to diagnose AD in clinical settings.
Feature design and gold standard creation relied upon two diagnostic criteria utilized in clinical settings to diagnose patients with AD [14], [15]. Our algorithm achieves superior performance when gold standard labels were assigned via the HR criteria compared to the UKWP criteria (more than 10% higher PPV than UKWP criteria). As discussed previously, the UKWP criteria is an abridged modification of the HR criteria, which assigns diagnoses based on fewer signs and symptoms than the HR criteria. Our results suggest that the more detailed set of signs, symptoms, and comorbidities included in the HR criteria lead to more accurate diagnoses. These findings are consistent with the clinical experience and intutions of our domain expert (JS). While the high level of detail of the HR criteria may create burdens in a clinical setting, it is critical for high performance in our phenotype algorithm.
These algorithm results are a promising start towards our ultimate eMERGE consortium goal of using EHR data to assemble a large cohort of patients for GWAS. Once this cohort has been created, the genetic bases of AD can be explored using linked biobank samples from each participating institution. Our AD phenotype algorithm has further applications for the identification of patients for epidemiological research and for inclusion in clinical trials for AD treatment. Treatment of AD is challenging, and large-scale clinical trials with AD patients are necessary to develop effective treatments.
The success of our phenotyping approach may not be unique to AD. Rather, it may be useful for other complex phenotypes that cannot be adequately characterized with coded data alone. Many autoimmune disorders fall into this category. For example, systemic lupus erythematosus (SLE) is a systemic autoimmune disease characterized by pronounced inflammation that is often difficult to diagnose because of the diverse manifestations that occur over time [20]. Another complex phenotype is polycystic ovary syndrome (PCOS), a common endocrine system disorder. As is the case with AD, PCOS has high prevalence but is commonly misdiagnosed. Further, physicians rely on multiple diagnostic criteria to identify patients with PCOS [21]–[24], complicating the process of creating a gold standard cohort of patients. Finally, many PCOS symptoms, which are critical to diagnosis, may not be codified in the EHR, highlighting the need for NLP.
VI. Future Work
There are a number of clear directions for future work. A preliminary error analysis revealed that cTAKES generally succeeded in identifying the concepts of interest. However, a few clear improvements can be made to the NLP pipeline. Currently, the dictionary look-up algorithm does not handle misspellings. Misspelled terms are not currently identified unless the misspelling was manually added to the input dictionary. While a number of common misspellings have been manually added, it is infeasible to predict all possible misspellings in the encounter notes. We are currently exploring the performance impact of combining dictionary look-up with an implementation of minimum edit distance, a sequence alignment technique that calculates the minimum number of changes (insertions, deletions, substitutions) needed to map from one dictionary entry to a target string.
The NLP pipeline also currently cannot detect relations among concepts. A number of potentially key concepts for the current phenotype involve symptoms localized to a particular body region (e.g., dermatitis on the antecubital and popliteal fossae), which are not contained in the UMLS Metathesaurus. Therefore, such concepts cannot be identified by the cTAKES dictionary look-up algorithm. Temporal relations are also not currently captured by the NLP pipeline. The time-course of AD contributes to its heterogeneity, as patients experience intermittent, chronic persistent, seasonal, or episodic flares. Accurate capture of time-course information could improve the performance of our algorithm, and shed light on possible subphenotypes of AD. Other temporal information, such as age of disease onset, plays an important role in the clinical diagnosis of AD due to its inclusion in the HR and UKWP criteria. Modifications to the NLP pipeline could be made to handle each of these kinds of relations.
Finally, the cTAKES NER module does not handle word sense disambiguation (i.e., assigning the most contextually appropriate CUI for terms with more than one possible CUI). The ambiguity in the current dataset is quite low (approximately 3.5% of mentions), in large part due to the creation of a dictionary customized to the current phenotype with few overlapping concepts. However, planned extensions of the current work (the identification of subphenotypes of AD) will require extraction of a broader set of clinical concepts outside of the set of known symptoms and comorbidities of AD. Preliminary explorations of our dataset suggest that up to 30% of all concepts identified using the standard cTAKES dictionary are ambiguous. To ensure the success of our subphenotyping efforts, resolution of this ambiguity is required.
While certain improvements can still be made to the NLP pipeline, we can also make a number of changes to the machine learning approaches to our phenotype algorithm. The NLP-derived features included in the logistic regression were extracted from encounter notes generated by clinicians with many different specialties. Therefore, much of the content of some notes was not relevant to the current phenotype (e.g., treatment for injury or other illness by a primary care physician). Furthermore, the quality of phenotype-specific information recorded by clinicians may vary based on specialty; documentation by dermatologists and allergists may contain more and better information than documentation by a primary care physician or hospitalist. Future work could index the note source of each concept (e.g., dermatologist/allergist vs. other) within the machine learning algorithm to account for this possible source of variation across notes.
We could also improve our approach by experimenting with other machine learning algorithms. We opted for lasso logistic regression given previous research with other phenotype algorithms [10]–[13]. However, other classification algorithms could achieve even higher performance. Ensemble methods, such as random forest, in particular are a promising avenue for future experiments. Aside from higher performance, a benefit of random forest is the potential to interpret the fit model in terms of a series of human-readable rules. Ongoing work is exploring this option.
VII. Conclusions
The current study demonstrates the effectiveness of utilizing NLP and machine learning for developing phenotype algorithms. Future iterations of our algorithm will be used for large-scale clinical, epidemiological, and genomics research, and will further our understanding of the complex phenotype of AD.
Acknowledgments
This project is part of the eMERGE Network funded by the NHGRI, grant number U01HG8673. This project was also made possible with support from the Agency for Healthcare Research and Quality (AHRQ), grant number K12HS023001, and the Dermatology Foundation. We thank the members of the eMERGE team at Northwestern University for useful discussion at various stages of this project.
Contributor Information
Erin Gustafson, Email: erin.gustafson@northwestern.edu.
Jennifer Pacheco, Email: japecheco@northwestern.edu.
Firas Wehbe, Email: firas.wehbe@northwestern.edu.
Jonathan Silverberg, Email: jonathan.silverberg@northwestern.edu.
William Thompson, Email: wkt@northwestern.edu.
References
- 1.Karimkhani Chante, Dellavalle Robert P, Coffeng Luc E, Flohr Carsten, Hay Roderick J, Langan Sinad M, Nsoesie Elaine O, Ferrari Alize J, Erskine Holly E, Silverberg Jonathan I, Vos Theo, Naghavi Mohsen. Global Skin Disease Morbidity and Mortality: An Update From the Global Burden of Disease Study 2013. JAMA Dermatology. 2017 March; doi: 10.1001/jamadermatol.2016.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silverberg Jonathan I, Hanifin Jon M. Adult eczema prevalence and associations with asthma and other health and demographic factors: A US population-based study. Journal of Allergy and Clinical Immunology. 2013 November;132:1132–1138. doi: 10.1016/j.jaci.2013.08.031. [DOI] [PubMed] [Google Scholar]
- 3.Silverberg JI, Garg NK, Paller AS, Fishbein AB, Zee PC. Sleep Disturbances in Adults with Eczema Are Associated with Impaired Overall Health: A US Population-Based Study. Journal of Investigative Dermatology. 2015 January;135:56–66. doi: 10.1038/jid.2014.325. [DOI] [PubMed] [Google Scholar]
- 4.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N, Crane PK, Pathak J, Chute CG, Bielinski SJ, Kullo IJ, Li R, Manolio TA, Chisholm RL, Denny JC. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011 April;3:79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Starren Justin B, Winter Andrew Q, Lloyd-Jones Donald M. Enabling a Learning Health System through a Unified Enterprise Data Warehouse: The Experience of the Northwestern University Clinical and Translational Sciences (NUCATS) Institute. Clinical and Translational Science. 2015 August;8(4):269–271. doi: 10.1111/cts.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, Lai AM. A review of approaches to identifying patient phenotype cohorts using electronic health records. Journal of the American Medical Informatics Association: JAMIA. 2014 April;21:221–30. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao Katherine P, Cai Tianxi, Savova Guergana K, Murphy Shawn N, Karlson Elizabeth W, Ananthakrishnan Ashwin N, Gainer Vivian S, Shaw Stanley Y, Xia Zongqi, Szolovits Peter, Churchill Susanne, Kohane Isaac. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ (Clinical research ed) 2015 April;350:h1885. doi: 10.1136/bmj.h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirby JC, Speltz P, Rasmussen LV, Basford M, Gottesman O, Peissig PL, Pacheco JA, Tromp G, Pathak J, Carrell DS, Ellis SB, Lingren T, Thompson WK, Savova G, Haines J, Roden DM, Harris PA, Denny JC. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. Journal of the American Medical Informatics Association: JAMIA. 2016 November;23:1046–1052. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu DY, Dalal P, Sable KA, Voruganti N, Nardone B, West DP, Silverberg JI. Validation of International Classification of Disease Ninth Revision codes for atopic dermatitis. Allergy. 2016 December; doi: 10.1111/all.13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao Katherine P, Cai Tianxi, Gainer Vivian, Goryachev Sergey, Zeng-treitler Qing, Raychaudhuri Soumya, Szolovits Peter, Churchill Susanne, Murphy Shawn, Kohane Isaac, Karlson Elizabeth W, Plenge Robert M. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care & Research. 2010;62:1120–1127. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll Robert J, Thompson Will K, Eyler Anne E, Mandelin Arthur M, Cai Tianxi, Zink Raquel M, Pacheco Jennifer A, Boomershine Chad S, Lasko Thomas A, Xu Hua, Karlson Elizabeth W, Perez Raul G, Gainer Vivian S, Murphy Shawn N, Ruderman Eric M, Pope Richard M, Plenge Robert M, Kho Abel Ngo, Liao Katherine P, Denny Joshua C. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. Journal of the American Medical Informatics Association: JAMIA. 2012;19:e162–169. doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castro Victor M, Minnier Jessica, Murphy Shawn N, Kohane Isaac, Churchill Susanne E, Gainer Vivian, Cai Tianxi, Hoffnagle Alison G, Dai Yael, Block Stefanie, Weill Sydney R, Nadal-Vicens Mireya, Pollastri Alisha R, Rosenquist J Niels, Goryachev Sergey, Ongur Dost, Sklar Pamela, Perlis Roy H, Smoller Jordan W, Consortium International Cohort Collection for Bipolar Disorder Validation of electronic health record phenotyping of bipolar disorder cases and controls. The American Journal of Psychiatry. 2015;172:363–372. doi: 10.1176/appi.ajp.2014.14030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castro Victor M, Dligach Dmitriy, Finan Sean, Yu Sheng, Can Anil, Abd-El-Barr Muhammad, Gainer Vivian, Shadick Nancy A, Murphy Shawn, Cai Tianxi, Savova Guergana, Weiss Scott T, Du Rose. Large-scale identification of patients with cerebral aneurysms using natural language processing. Neurology. 2017 January;88:164–168. doi: 10.1212/WNL.0000000000003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichenfield Lawrence F, Tom Wynnis L, Berger Timothy G, Krol Alfons, Paller Amy S, Schwarzenberger Kathryn, Bergman James N, Chamlin Sarah L, Cohen David E, Cooper Kevin D, Cordoro Kelly M, Davis Dawn M, Feldman Steven R, Hanifin Jon M, Margolis David J, Silverman Robert A, Simpson Eric L, Williams Hywel C, Elmets Craig A, Block Julie, Harrod Christopher G, Begolka Wendy Smith, Sidbury Robert. Guidelines of care for the management of atopic dermatitis: Section 2. Management and treatment of atopic dermatitis with topical therapies. Journal of the American Academy of Dermatology. 2014;71:116–132. doi: 10.1016/j.jaad.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams Hc, Jburney Pg, Pembroke Ac, Hay Rj, Party Atopic Dermatitis Diagnostic Criteria Working The U.K. Working Party’s Diagnostic Criteria for Atopic Dermatitis. III. Independent hospital validation. British Journal of Dermatology. 1994 September;131:406–416. doi: 10.1111/j.1365-2133.1994.tb08532.x. [DOI] [PubMed] [Google Scholar]
- 16.Sidbury R, Davis DM, Cohen DE, Cordoro KM, Berger TG, Bergman JN, Chamlin SL, Cooper KD, Feldman SR, Hanifin JM, Krol A, Margolis DJ, Paller AS, Schwarzenberger K, Silverman RA, Simpson EL, Tom WL, Williams HC, Elmets CA, Block J, Harrod CG, Begolka WS, Eichenfield LF, Dermatology American Academy of. Guidelines of care for the management of atopic dermatitis: section 3 Management and treatment with phototherapy and systemic agents. J Am Acad Dermatol. 2014 August;71:327–49. doi: 10.1016/j.jaad.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphreys BL, Lindberg DA, Schoolman HM, Barnett GO. The Unified Medical Language System: an informatics research collaboration. Journal of the American Medical Informatics Association: JAMIA. 1998 February;5:1–11. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savova Guergana K, Masanz James J, Ogren Philip V, Zheng Jiaping, Sohn Sunghwan, Kipper-Schuler Karin C, Chute Christopher G. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010 September;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman Jerome, Hastie Trevor, Tibshirani Rob. Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of statistical software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 20.Narain S, Richards HB, Satoh M, Sarmiento M, Davidson R, Shuster J, Sobel E, Hahn P, Reeves WH. Diagnostic accuracy for lupus and other systemic autoimmune diseases in the community setting. Arch Intern Med. 2004 December;164:2435–41. doi: 10.1001/archinte.164.22.2435. [DOI] [PubMed] [Google Scholar]
- 21.Zawadski JK. Diagnostic criteria for polycystic ovary syndrome: towards a rational approach. In: Dunaif A, editor. Polycystic Ovary Syndrome, pages. Blackwell Scientific Publications; Boston: 1994. pp. 377–384. [Google Scholar]
- 22.Eshre Asrm-Sponsored Pcos Consensus Workshop Group Rotterdam. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril. 2004 January;81:19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Eshre Asrm-Sponsored Pcos consensus workshop group Rotterdam. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS) Hum Reprod. 2004 January;19:41–7. doi: 10.1093/humrep/deh098. [DOI] [PubMed] [Google Scholar]
- 24.Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, Janssen OE, Legro RS, Norman RJ, Taylor AE, Witchel SF, Excess Task Force on the Phenotype of the Polycystic Ovary Syndrome of The Androgen, and Pcos Society The Androgen Excess and PCOS Society criteria for the polycystic ovary syndrome: the complete task force report. Fertil Steril. 2009 February;91:456–88. doi: 10.1016/j.fertnstert.2008.06.035. [DOI] [PubMed] [Google Scholar]


