Abstract
Premise of the study:
The use of genome skimming allows systematists to quickly generate large data sets, particularly of sequences in high abundance (e.g., plastomes); however, researchers may be overlooking data in low abundance that could be used for phylogenetic or evo-devo studies. Here, we present a bioinformatics approach that explores the low-abundance portion of genome-skimming next-generation sequencing libraries in the fan-flowered Goodeniaceae.
Methods:
Twenty-four previously constructed Goodeniaceae genome-skimming Illumina libraries were examined for their utility in mining low-copy nuclear genes involved in floral symmetry, specifically the CYCLOIDEA (CYC)-like genes. De novo assemblies were generated using multiple assemblers, and BLAST searches were performed for CYC1, CYC2, and CYC3 genes.
Results:
Overall Trinity, SOAPdenovo-Trans, and SOAPdenovo implementing lower k-mer values uncovered the most data, although no assembler consistently outperformed the others. Using SOAPdenovo-Trans across all 24 data sets, we recovered four CYC-like gene groups (CYC1, CYC2, CYC3A, and CYC3B) from a majority of the species. Alignments of the fragments included the entire coding sequence as well as upstream and downstream regions.
Discussion:
Genome-skimming data sets can provide a significant source of low-copy nuclear gene sequence data that may be used for multiple downstream applications.
Keywords: CYCLOIDEA, floral symmetry, genome skimming, Goodeniaceae
The use of low-coverage and cost-effective genome skimming, also known as whole-genome shotgun sequencing (WGS), has become increasingly prevalent in plant phylogenomics and has allowed for the creation of large data sets, many of which have made possible the resolution of long-standing phylogenetic questions. The premise of the technique is to characterize high-copy fractions of total genomic DNA (i.e., organellar genomes, nuclear ribosomal DNA, and other multicopy elements) through random shearing and inexpensive multiplexing (Steele et al., 2012; Straub et al., 2012). Recent uses of genome skimming include determining the origin of Jerusalem artichoke (Bock et al., 2013); ultra-barcoding accessions of cultivated cacao (Kane et al., 2012); and resolving phylogenetic relationships at deep levels in palms and other commelinid monocots (Barrett et al., 2015), generic-level relationships in Chrysobalanaceae (Malé et al., 2014) and Goodeniaceae (Gardner et al., 2016a), and shallow species-level relationships in Oreocarya Greene (Ripma et al., 2014).
Although genome-skimming data generated in prior studies have been primarily targeted for their high-copy fraction (i.e., plastomes), additional uses of WGS data have been suggested (Godden et al., 2012; Straub et al., 2012; Soltis et al., 2013). For instance, one potential use is mining the data sets for candidate low-copy nuclear genes of interest in nonmodel organisms to use in phylogeny reconstruction or downstream evo-devo experiments (Straub et al., 2011, 2012, 2014; Blischak et al., 2014; Ripma et al., 2014). The inclusion of nuclear genes in phylogenetic reconstruction often relies on genes with at least two conserved domains from distantly related taxa and the use of degenerate primers (e.g., ADH, GAPDH, CYCLOIDEA [Strand et al., 1997; Hileman, 2003]), which limits the amount of known coding sequence (see Howarth and Donoghue, 2005) and does not include upstream or downstream regulatory elements. Here we present an approach to obtain low-copy nuclear genes that could be used for detailed evo-devo experiments. We target candidate genes involved in establishing bilateral symmetry in flowers, specifically the CYCLOIDEA (CYC)-like genes in the “ECE” clade (Howarth and Donoghue, 2006).
The CYC-like genes are a plant-specific clade of TCP transcription factors identified by a characteristic TCP DNA-binding domain (177 bp), a conserved R domain (51 bp), and, in some copies, by a short conserved motif referred to as the “ECE” motif (Cubas et al., 1999; Howarth and Donoghue, 2006). CYC-like genes have undergone at least two duplication events with three main paralogs found across Pentapetalae (CYC1, CYC2, and CYC3; Howarth and Donoghue, 2006). Full-length coding regions of these paralogs vary from 807 to 1245 bp in Helianthus L. (Chapman et al., 2008), Gerbera L. (Broholm et al., 2008), and Antirrhinum L. (Luo et al., 1996, 1999). Many studies of floral symmetry have used the portion of the gene easily obtained with Sanger sequencing, specifically the roughly 300–400 bp spanning the TCP domain through to the R domain (the two areas in which primers can be generated across large clades) (Howarth and Donoghue, 2005, 2006; Zhang et al., 2010; Howarth et al., 2011; Citerne et al., 2013; W. Zhang et al., 2013; Berger et al., 2016).
Among the three clades of paralogs, CYC2-like genes are the most studied and are known to be involved in regulating the development and position of dorsal petals and stamens across Pentapetalae (Hileman, 2014; Specht and Howarth, 2014). However, CYC3-like genes are also asymmetrically expressed across the dorsoventral flower axis and may also be involved in patterning symmetry (Berger et al., 2016). Previous studies of CYC-like genes have primarily focused on the most common form of bilateral flower symmetry in which two petals are positioned dorsally and three petals ventrally (see Fig. 1D, E) (Hileman, 2014). One exception to this basic pattern of dorsoventral asymmetry is the highly derived ray florets of Asteraceae, in which novel expression patterns have been observed (Tahtiharju et al., 2012; Juntheikki-Palovaara et al., 2014). The paraphyletic grade subtending Asteraceae includes two radially symmetrical groups (Menyanthaceae and Calyceraceae) and the distinctive bilaterally symmetrical Goodeniaceae (Tank and Donoghue, 2010). With the exception of the monotypic Brunonia australis R. Br. (Jabaily et al., 2012; Gardner et al., 2016a), Goodeniaceae have a large dorsal slit down their corolla tube, with dorsal petals varying in their placement around the slit (Gardner et al., 2016b). In the most extreme cases, typified by Scaevola L. species, all of the petals are arranged in the ventral region of the flower, making the flowers resemble individual ray florets of Asteraceae with unfused petal lobes (Fig. 1). The expression and function of the CYC-like genes in Goodeniaceae is of particular interest for understanding the evolution of complex bilateral symmetry.
Fig. 1.
Sample images from Core Goodeniaceae showing diversity in floral symmetry, including lacking dorsal slit (A); fan flowered (B, E, F); and bilabiate, with two dorsal petals (C, D). (A) Brunonia australis R. Br. (K. A. Shepherd & S. R. Willis KS 1512), (B) Scaevola porocarya F. Muell. (K. A. Shepherd & S. R. Willis KS 1518), (C) Diaspasis filifolia R. Br. (K. A. Shepherd s.n.), (D) Goodenia ovata Sm. (K. A. Shepherd & S. R. Willis KS 1530), (E) Velleia rosea S. Moore (K. A. Shepherd & S. R. Willis KS 1509), (F) Goodenia decursiva W. Fitzg. (R. Meissner s.n.). Images A–C, E, F by K. A. Shepherd; D by S. R. Willis.
Goodeniaceae is a moderately sized family of flowering plants (>420 species, 12 genera) almost entirely endemic to Australia. The family is divided into two clades: LAD and Core Goodeniaceae (Jabaily et al., 2012; Gardner et al., 2016a). Core Goodeniaceae has roughly 330 species (>80% of the family) and is divided into three groups: Brunonia Sm. ex R. Br., Goodenia Sm. s.l., and Scaevola s.l. (Fig. 1). Monotypic Brunonia, sister to the other two groups, is the only radially symmetrical species of Goodeniaceae, lacking the dorsal slit that typifies the family. Scaevola s.l. are primarily fan-flowered with all petals arranged on the lower side of the flower and all five lobes uniform in morphology. Diaspasis filifolia R. Br., embedded within Scaevola s.l., develops two upper dorsal petals, but those petals are identical morphologically to the ventral petals. Goodenia s.l. displays more diversity in floral symmetry, although they are primarily bilabiate, having two upper dorsal petals that differ in morphology from the lower three petal lobes. Within Goodenia s.l., the location and morphology of the dorsal petal lobes are very labile and include multiple shifts to a fan-flowered morphology (Gardner et al., 2016b).
Analysis of the evolution of genetic changes across a diverse clade requires generating sequence data from many independent species. Here we show the utility of genome-skimming data to generate sequence information for morphologically important candidate genes. We show that previously generated high-copy fractions of WGS data, which were used to resolve the backbone of Core Goodeniaceae (Gardner et al., 2016a), can also be mined to recover portions of the low-copy fraction, including full-length coding sequences of CYC-like genes, and in some cases, even upstream promoter regions.
MATERIALS AND METHODS
Taxon sampling
Sampling of 24 species was based on data generated to resolve the phylogeny of the Core Goodeniaceae clade (Gardner et al., 2016a). Vouchers and silica-dried tissue samples for all species collected in the field or obtained from cultivated specimens were deposited at the Western Australian Herbarium (PERTH). Genomic DNA was extracted for WGS with a QIAGEN DNeasy Plant Mini Kit (Valencia, California, USA) following the manufacturer’s protocol, but eluting the DNA in 30 μL of elution buffer to maximize concentration. All Goodeniaceae Illumina libraries were generated using the Nextera DNA Sample Preparation Kit (Illumina, San Diego, California, USA). Barcodes were added and 500–600-bp fragments were size-selected after quality assessment using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, California, USA) and qPCR. Multiplexing using 150-bp paired-end chemistry was performed in two lanes on a HiSeq 2500 (Gardner et al., 2016a). Reads passing Illumina’s standard quality filters were used for assemblies. All assembly-generated scaffolds were mined to identify putative full-length and/or partial CYCLOIDEA-like sequences (see Appendix S1 (54.8KB, xlsx) ). These 24 taxa spanned the Core Goodeniaceae clade, encompassing much of the floral shape variation found across the group.
NGS data sets and de novo assemblies
Because a reference genome is currently unavailable for Goodeniaceae, we first compared the ability of four de novo assembly software packages (SPAdes version 3.9.0 [Bankevich et al., 2012], SOAPdenovo version 2.04 [Luo et al., 2012], SOAPdenovo-Trans version 1.03 [Xie et al., 2014], and Trinity version 2.2.0 [Grabherr et al., 2011; Haas et al., 2013]) to generate scaffolds from paired-end (PE) (2 × 150-bp) read data sets. Assemblies were performed for a subset of taxa (Brunonia australis, Coopernookia polygalacea (de Vriese) Carolin, Goodenia decursiva W. Fitzg., and Scaevola porocarya F. Muell.) using (1) the complete PE read data set for each taxon and (2) a reduced data set composed only of ‘Merged Paired Reads’ created in Geneious version 9.1.6 (Kearse et al., 2012) using the default parameters (minimum overlap: 10; maximum overlap: 65; maximum mismatch density: 0.25) of the FLASH version 1.2.9 plugin (Magoč and Salzberg, 2011). Merging PE reads prior to genome assembly has been shown to increase N50 values of both contigs and scaffolds, while decreasing mis-assemblies (Magoč and Salzberg, 2011). Assemblies were generated in SPAdes using the –careful command to limit the number of mismatches; 8 threads (-t 8); k-mer sizes of 21, 33, and 55 (-k 21, 33, 55); and the –cov-cutoff auto command to allow SPAdes to determine conservative read coverage cutoff value. SOAPdenovo assemblies were performed using the SOAPdenovo-63mer source code with the following commands: all, -K 25 (also assembled with -K 31 and -K 63), -F, -f, -R, -r, -e 2, -L 100, and -p 8. Additional parameters in the configuration file for SOAPdenovo included a max read length of 100 bp (max_rd_len = 100), an average insert size of 200 bp (avg_ins = 200), specifying that sequences should not be complementary reversed (reverse_seq = 0), indicating reads should be used in construction of both contigs and assemblies (asm_flags = 3), and designating a cutoff value of pair number reliably connecting contigs and/or scaffolds (pair_num_cutoff = 3). SOAPdenovo-Trans assemblies were run using the SOAPdenovo-Trans-31mer source code and the same parameters as SOAPdenovo; however, only -K 25 was analyzed based on preliminary data. Trinity assemblies were performed with default parameters plus the implementation of the –trimmomatic and –full_cleanup commands. All assemblies were conducted on the high-performance computing cluster (HiPerGator 2.0) at the University of Florida.
To estimate genomic coverage, we mapped each PE read to the Helianthus annuus L. chloroplast (NC_007977) and mitochondrion (NC_023337) genomes in Geneious version 9.1.6 (Kearse et al., 2012). The total number of reads mapping to those genomes was subtracted from the total number of PE reads obtained for each taxon. Genomic coverage was then calculated using the reduced read pool and the nuclear DNA estimate of Goodenia mimuloides S. Moore (C-value = 0.52 pg; Hanson, 2001).
Multiple assembler approach and alignments
Assemblies were read into Geneious version 9.1.6 (Kearse et al., 2012) as new databases. Helianthus annuus sequences (EU088366–EU088375; Chapman et al., 2008) were BLASTed against the databases (TBLASTX, extended region with annotation, E-value = 1e−10, word size = 3, matrix = BLOSUM62). Because few BLAST hits were obtained in Brunonia australis using H. annuus sequences, we also BLASTed CYC1, CYC2, CYC3A, and CYC3B BLAST hits obtained from the other 23 Goodeniaceae taxa against the B. australis database. BLAST hits from each assembly were verified in the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov; accessed 6 February 2017). Confirmed CYC-like gene hits were separated by species and copy (e.g., those that BLASTed to CYC2 were organized together) and initially aligned using MAFFT version 7.222 (Katoh and Standley, 2013) as implemented in Geneious. BLAST hits with the same overlapping sequence were aligned and joined into a single consensus sequence. Consensus sequences of each CYC-like copy were then aligned for all taxa. Manual adjustments to the consensus alignment were made by hand based on amino acid translation. Fragments of copies that were recovered for a single taxon were amalgamated into a single consensus sequence with N’s inserted for regions missing coverage.
In many hits within a species, there were overlapping regions that allowed them to be combined. These overlapping regions were identical to each other and so it was logical to assess them as from the same genomic region. In a few cases, duplications were hypothesized within a species because the overlapping regions were clearly distinct from each other (i.e., had multiple mismatches).
RESULTS
De novo genome-skimming coverage and assemblies
Within Goodeniaceae, only a single species, Goodenia mimuloides, currently has a measurement of nuclear DNA content (C-value = 0.52 pg; Hanson, 2001). Given this value, we estimate a 509-Mb genome size. Most Core Goodeniaceae have similar chromosome numbers of N = 7, 8, or 9 (Carolin et al., 1992), including G. mimuloides; therefore, we hypothesize C-values to be similar across the group, excluding the occurrence of polyploidization. After determining the percentage of reads mapping to the chloroplast (∼3%) and mitochondrion (∼0.5%), total remaining PE reads ranged between 7–12 million reads (Gardner et al., 2016a), providing between 1.05 and 1.8 billion base pairs of coverage. Dividing base pair coverage by the 509-Mb estimated genome size, we roughly estimate 2× to 3.5× coverage across these species.
Scaffold number, the longest scaffold generated, and N50 values varied based on k-mer size and which de novo assembler was used (see Tables 1, 2; Appendix S1 (54.8KB, xlsx) ). The number of scaffolds obtained for the four-taxa subset ranged from 30,955 in Brunonia australis (SOAPdenovo, k-mer 63) up to 3,344,837 in B. australis (SOAPdenovo, k-mer 31). Scaffold length varied from 1259 bp in Coopernookia polygalacea (SOAPdenovo, k-mer 25) up to 103,842 bp in C. polygalacea (SPAdes). N50 values, which are a common metric to determine contiguity of an assembly, ranged from 100 in B. australis (SOAPdenovo, k-mer 25, k-mer 31) up to 1820 in Goodenia decursiva (SPAdes).
Table 1.
Table 1. Comparison of assembly data for four taxa: Brunonia australis (B. aust.), Coopernookia polygalacea (C. poly.), Diaspasis filifolia (D. fili.), and Goodenia decursiva (G. decu.).
| Taxon | SPAdes merged | SOAPdenovo25 | SOAPdenovo31 | SOAPdenovo63 | SOAPdenovo-Trans | Trinity | ||||||||||||
| No. of scaffolds | Longest scaffold | N50 | No. of scaffolds | Longest scaffold | N50 | No. of scaffolds | Longest scaffold | N50 | No. of scaffolds | Longest scaffold | N50 | No. of scaffolds | Longest scaffold | N50 | No. of scaffolds | Longest scaffold | N50 | |
| B. aust. | 41,029 | 35,892 | 1781 | 2,026,187 | 1298 | 100 | 3,344,847 | 1803 | 100 | 30,955 | 7575 | 153 | 164,715 | 43,894 | 134 | 294,509 | 22,268 | 725 |
| C. poly. | 65,404 | 103,842 | 1449 | 1,125,793 | 1259 | 116 | 1,581,193 | 3989 | 100 | 74,904 | 32,134 | 157 | 354,963 | 51,989 | 179 | 352,727 | 16,926 | 774 |
| D. fili. | 189,002 | 56,220 | 875 | 1,008,445 | 1977 | 119 | 1,391,445 | 1718 | 100 | 105,937 | 28,394 | 146 | 331,448 | 85,261 | 199 | 327,239 | 21,987 | 813 |
| G. decu. | 52,324 | 36,308 | 1810 | 1,236,654 | 2863 | 165 | 1,632,591 | 2741 | 149 | 235,467 | 11,760 | 208 | 621,984 | 81,414 | 322 | 409,522 | 27,649 | 1065 |
Table 2.
BLAST sequence results from different assemblers in four taxa: Brunonia australis, Coopernookia polygalacea, Diaspasis filifolia, and Goodenia decursiva. Results from four gene clades: CYC1 - CYC2 - CYC3A - CYC3B.a
| Taxon | Trinity | SOAPdenovo-Trans | SOAPdenovo-25 | SOAPdenovo-31 | SOAPdenovo-63 | SPAdes merged |
| Brunonia australis | 0 - 0 - 0 - 0 | 0 - 0 - 0 - 0 | 0 - 100 - 100 - 100 | 0 −100 - 100 −100 | 0 - 0 - 0 - 0 | NA |
| Coopernookia polygalacea | 800 - 719(2) - 289 - 466 | 0 - 234 - 100 - 0 | 553 - 501(2) - 0 - 0 | 411 - 193 - 100 - 0 | 196 - 0 - 0 - 0 | 527 - 357 - 0 - 0 |
| Diaspasis filifolia | 715(2) - 859 - 529;289 - 449 | 895 - 130 - 0 - 0 | 100 - 100 - 187 - 194 | 100;476 - 130 - 118 - 194 | 0 - 0 - 0 - 0 | 0 - 0 - 0 - 0 |
| Goodenia decursiva | 1384 - 989 - 669 - 1348 | 876 - 775 - 144 - 894;429 | 349 - 274 - 0 - 657;429 | 666(2) - 293(2) - 0 - 268;764 | 0 - 0 - 0 - 281 | 0 - 0 - 0 - 0 |
Note: NA = not available.
Numbers represent sequence lengths recovered in BLAST searches. Semicolons separate multiple gene copies from a single gene group. Parentheses note that the sequence was broken between nonoverlapping sequences.
None of the four assemblers consistently outperformed any of the others, although overall Trinity, SOAPdenovo-Trans, and SOAPdenovo implementing lower k-mer values uncovered the most sequences and the longest scaffolds. When comparing BLAST results, the assemblies from B. australis overall yielded the least amount of CYC-like data while those assemblies from G. decursiva yielded the greatest amount of CYC-like data (Table 2). This is correlated with the length of the scaffolds and N50 values determined for each species and assembler (Table 1). Overall, on average, SOAPdenovo-Trans combined the fastest assembly times with the longest scaffolds, and was, therefore, our assembler of choice to compare all of the genome-skimming data sets quickly and efficiently.
SOAPdenovo-Trans assemblies for all taxa
We used SOAPdenovo-Trans to assemble the genome-skimming data sets for all 24 species. SOAPdenovo-Trans–based assemblies (see Appendix S1 (54.8KB, xlsx) ) generated varying numbers of scaffolds ranging from 164,715 in Brunonia australis to 621,984 in Goodenia decursiva, with an average number of 459,996 scaffolds. The longest scaffold of each assembly ranged from 23,907 bp in G. ovata Sm. to 123,122 bp in G. pinifolia de Vriese, with the average longest scaffold across all samples being 57,646 bp. N50 values varied across all taxa from 134 in B. australis to 322 in G. decursiva, with the average N50 across all samples being 228. Total scaffold size varied from 24,779,056 bp in B. australis to 158,074,932 bp in G. decursiva, with the average total scaffold size being 95,657,739 bp. Mean contig length varied from 150 bp in B. australis to 254 bp in G. decursiva, with the average mean contig length across all taxa being 203 bp. The total number of contigs incorporated into the scaffolds ranged from 159,656 in B. australis to 575,138 in G. decursiva, with the average number of contigs included in scaffolds across all taxa being 437,830. With regard to overall contig length across those included in scaffolds, B. australis had the fewest number of contigs of at least 100 bp and is the only species having less than 80% of scaffolds greater than 100 bp. Goodenia decursiva had the highest percentage of scaffolds greater than 100 bp (93.83%), as well as the largest number of scaffolds greater than 500 bp and 1000 bp.
CYC-like gene BLAST results and alignments
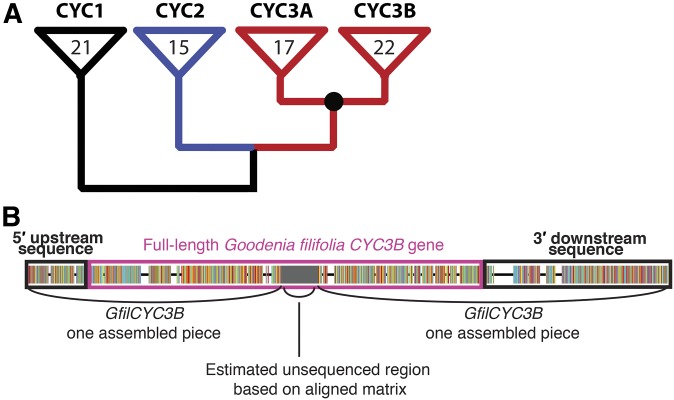
CYC-like gene BLAST hits were found in 23 of the 24 (all but B. australis) genome-skimming SOAPdenovo-Trans–assembled data sets and are deposited in Dryad (http://dx.doi.org/10.5061/dryad.0500c; Berger et al., 2017). Sequence fragments from each of the three core eudicot CYC-like clades (CYC1, CYC2, and CYC3) were confirmed in NCBI. Additionally, by aligning the CYC3-like data sets with other Goodeniaceae and Helianthus sequences, two separate clades of CYC3 emerged: CYC3A and CYC3B. There were, therefore, four separate gene clades and resulting alignments in total: CYC1, CYC2, CYC3A, and CYC3B (Fig. 2). In some cases, multiple BLAST hits from a single species did not overlap with each other; therefore, a single species could have two different scaffolds in different parts of the same genic region, leading to more scaffolds than numbers of species.
Fig. 2.
CYCLOIDEA-like gene clades and gene annotation. (A) CYC-like genes could be separated into four gene clades based on sequence diversity. CYC1, CYC2, and CYC3 clades occur across core eudicots. Additionally, a duplication of CYC3 found in Asteraceae is shared with Goodeniaceae, resulting in two CYC3 clades. Numbers inside the triangles represent the number of scaffolds obtained for each CYC-like copy across the 24 taxa. (B) A single example of an annotated CYC-like gene from the genome-skimming alignment. Black boxes note sequence regions upstream and downstream of the coding sequence. Pink box indicates the hypothesized coding gene sequence. Gaps are from alignment with CYC-like genes from other species. Gray internal box indicates the hypothesized missing region between two separately sequenced BLAST results.
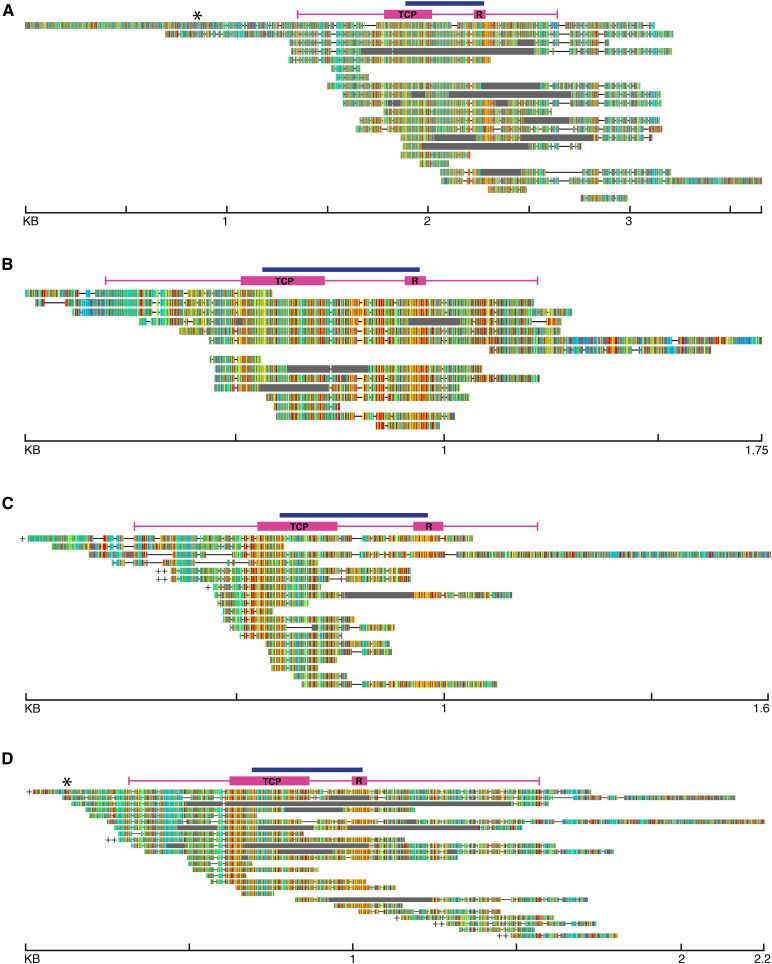
Thirty-five partial to full-length CYC1-like fragments were found in the genome-skimming data sets of 21 species (Fig. 3A, Table 3, Appendix S2 (30.7KB, docx) ). The total aligned matrix was 3656 bp (Fig. 3A). BLAST hits from Goodenia filiformis R. Br. and G. decursiva pulled out sequence approximately 500 bp upstream of the methionine start codon and included the hypothesized TATA box of the promoter (with a sequence of CTATAWAWA; Shahmuradov, 2003). Thirteen species had sequence upstream of the TCP domain, and five species, all in Goodenia s.l., included sequence of the protein start codon. Fifteen species contained sequence downstream of the R domain, with 14 of these including the hypothesized stop codon. There was no evidence that any of the CYC1-like genes had multiple copies in any of the 21 species in which sequences were obtained.
Fig. 3.
Alignments of BLAST results from all genome-skimming data sets for each CYC-like clade. Each line is the BLAST results from a single species. Colors are based on nucleotide sequence, generated in Geneious. Hypothesized coding sequence is indicated by the pink bar, with conserved TCP and R domains labeled. The blue bar shows the length of previously generated data with degenerate primers and Sanger sequencing. Total length in kilo base pairs is labeled on the bottom of each alignment. (A) CYC1 alignment, with sequences from 21 species; (B) CYC2 alignment, with sequences from 15 species; (C) CYC3A alignment, with sequences from 17 species with 19 total sequences (two copies are found in each of two species); (D) CYC3B alignment with sequences from 22 species with 25 total sequences (two or three duplicate sequences found in each of two species). * indicate putative TATA box position upstream of the start codon, while + and ++ highlight possible duplicate copies.
Table 3.
CYC-like gene sequence lengths found in genomic skimming data sets.a
| Taxon | CYC1 | CYC2 | CYC3A | CYC3B |
| Brunonia australis R. Br. | None | None | None | None |
| Coopernookia polygalacea (de Vriese) Carolin | 800 | 730 | 289 | 479† |
| Coopernookia strophiolata (F. Muell.) Carolin | None | 534† | 506† | 469 |
| Diaspasis filifolia R. Br. | 1419* | 859* | 531; 507 | 449 |
| Goodenia decursiva W. Fitzg. | 2229†* | 1146†* | 127 | 1602†*; 433* |
| Goodenia drummondii Carolin | 1293(2)†* | 1082† | 932; 244† | 842†; 395*; 316* |
| Goodenia filiformis R. Br. | 2862†* | 1258* | 1436†* | 1677(2)†* |
| Goodenia hassallii F. Muell. | 1302(3)* | 785(3)* | 302 | 516† |
| Goodenia helmsii Carolin | 1183(2)* | 491* | 109 | 203 |
| Goodenia micrantha (F. Muell. ex K. Krause) Hemsl. ex Carolin | 225* | 454 | 109 | 105 |
| Goodenia ovata Sm. | 151 | 155 | 529(2) | 541(4)† |
| Goodenia phillipsiae Carolin | 1140(3)* | 391 | None | 787(3)* |
| Goodenia pinifolia de Vriese | 1033(2)†* | None | 434 | 1171(3)* |
| Goodenia tripartita Carolin | 274(2)* | None | None | 207 |
| Goodenia viscida R. Br. | None | None | 211 | 758(2) |
| Scaevola collaris F. Muell. | 336 | 114 | None | None |
| Scaevola phlebopetala F. Muell. | 133 | 382 | 229 | 100 |
| Scaevola porocarya F. Muell. | 143 | 146 | None | 502(2)* |
| Scaevola tomentosa Gaudich. | 665(2)* | None | 158 | 437(2)†* |
| Selliera radicans Cav. | 927† | None | 306 | 364 |
| Velleia discophora F. Muell. | 624(3)* | None | None | 376 |
| Velleia foliosa K. Krause | 1244(2)* | None | 309† | 1752(2)†* |
| Velleia rosea S. Moore | 187 | None | None | 168 |
| Verreauxia reinwardtii Benth. | 840(3)* | 438(2) | 267 | 806(2)† |
Numbers in parentheses indicate that sequence was split between multiple regions. Semicolons separate sequences that could not be concatenated because sequences differed. †’s include sequence upstream of the start codon and *’s include sequence downstream of the stop codon.
For CYC2-like sequences, 18 BLAST hits from 15 species were obtained. The total alignment of the matrix was 1749 bp. The longest upstream sequence from the start codon was 160 bp, with no evidence that the promoter was reached (Fig. 3B). BLAST hits from 10 species included sequence upstream of the TCP domain and three species included the protein start codon (Fig. 3B, Table 3). Twelve species contained sequence downstream of the R domain, with five species including sequence downstream of the hypothesized stop codon. There was no evidence that any of the CYC2-like genes had duplicated in any of the 15 species in which sequence reads were found.
Twenty partial CYC3A-like fragments were found across 17 species (Table 3). The total aligned length of the matrix was 1788 bp (Fig. 3C). Four species contained the protein start codon, with the longest upstream sequences being 201 bp. Four species contained sequence downstream of the R domain, with a single species (Goodenia filiformis) containing the hypothesized downstream stop codon plus 485 bp past the stop codon. Two species (G. drummondii Carolin and D. filifolia) contained evidence of a duplication in the CYC3A clade, meaning that there were separate BLAST hits with overlapping regions not identical to each other. In the case of G. drummondii, one duplicate closely matched the sequences of CYC3A from other species, while the second copy contained a stop codon with additional divergent nucleotides in the reading frame. This pattern of divergence suggests that this second copy could be a pseudogene. Diaspasis filifolia similarly had two separate copies; however, one copy was most similar to Scaevola phlebopetala F. Muell., while the other copy shared more sequence similarity with the rest of the Core Goodeniaceae sequences.
Thirty-eight CYC3B-like gene fragments were identified across 22 species (Table 3). The total aligned length of the matrix was 2227 bp (Fig. 3D). Nine species contained the hypothesized protein start codon, with the longest upstream region containing 286 bp. There is a putative TATA box roughly 192 bp upstream of the translational start codon found in two species, G. filiformis and G. decursiva. Fourteen species contained sequence downstream of the R domain, with 12 of those sequences continuing past the hypothesized stop codon. The longest downstream sequence extended 786 bp beyond the hypothesized stop codon. Closely related G. drummondii and G. decursiva each contained an extra, shared sequence, suggesting a duplication in this clade of Goodenia. Additionally, there was an upstream region of G. drummondii that did not overlap with either of the other sequence reads and therefore could not be joined to either copy.
DISCUSSION
Genome skimming and phylogenetics
The bulk of studies utilizing genome-skimming data have generally focused on improving resolution and support for phylogenetic relationships in groups where single or multigene phylogenies have not yielded well-supported backbones (Gardner et al., 2016a) or resolved relationships among recently diverged lineages (Ripma et al., 2014). The target(s), in most cases, are the high-copy elements such as the plastid and mitochondrial genomes, nuclear ribosomal repeats, and additional repetitive elements (Straub et al., 2011, 2012; Steele et al., 2012; Bock et al., 2013; Ripma et al., 2014; Dodsworth et al., 2016; Gardner et al., 2016a). While these markers are easily obtainable because of their high abundance and are capable of being assembled against a growing number of references, the data being used represent only a small fraction of the total data obtained (e.g., plastome data represent only 3% of the data sequenced in Core Goodeniaceae; Gardner et al., 2016a).
While genome skimming provides low-coverage sequencing depth across the genome, there are a growing number of studies developing pipelines for mining low-copy nuclear genes for phylogenetic purposes. For example, conserved orthologous gene (COS; Fulton et al., 2002), single-copy conserved orthologous gene (COSII; Wu et al., 2006), shared single-copy gene (SSC; Duarte et al., 2010), and/or pentatricopeptide repeat (PPR; Yuan et al., 2009, 2010) databases have been used to identify phylogenetic markers or regions of those genes for primer development in Oreocarya (Boraginaceae) (Ripma et al., 2014), Asclepias L. (Apocynaceae) (Straub et al., 2011, 2012, 2014), and Penstemon Schmidel (Plantaginaceae) (Blischak et al., 2014). Additionally, genome-skimming data are now being used in conjunction with transcriptome data to develop novel probes for targeting low-copy nuclear genes via Hyb-Seq in taxa such as Asclepias (Apocynaceae) (Weitemier et al., 2014) and Oxalis L. (Oxalidaceae) (Schmickl et al., 2015).
Genome-skimming data and de novo assemblies
While genome skimming allows for quick, relatively inexpensive acquisition of large amounts of data, particularly those elements in high abundance, the bigger question is how to use and make sense of the low-copy fraction and the gene(s) of interest that can potentially be mined. Although no clear-cut protocol can be assigned for all data sets given the variability in depth and quality of the sequencing, we found that when performing de novo assemblies to search for a gene(s) of interest, BLASTing scaffolds generated by multiple assemblers using smaller k-mer values (i.e., k-mer of 25 to 31) yielded the highest number of hits in our data sets. Similar findings supporting the use of multiple assemblers were recently reported when generating complete de novo transcriptome assemblies of Lonicera japonica Thunb. (Caprifoliaceae), as well as Pinus patula Schltdl. & Cham. (Pinaceae) using deep RNA sequencing (Visser et al., 2015; Rai et al., 2016). The de novo assemblies were compared using multiple assemblers, and it was found that none consistently outperformed the others. Additionally, as observed in our study, the number and length of contigs assembled varied greatly both within and between assemblers. Our data suggest that using multiple assemblers maximizes the variability of assembly variants, and therefore the amount of available data that could be mined.
Model clades for evo-devo
Understanding the specific DNA sequence changes that underlie morphological diversification is an exciting frontier given the speed of new data being generated. Growing numbers of quickly generated genome-skimming data sets can provide the necessary backbone to develop well-sampled model clades to examine the genetic underpinnings of trait changes. Specifically, evo-devo studies have previously used a few disparately related species to uncover major organ patterning genes such as HOX and MADS-box genes (Coen and Meyerowitz, 1991; Gehring, 1998) and have used those to determine the nature of single trait changes, usually by comparing two species. However, data provided by genome skimming could open the door for gene expression and functional studies across multiple species simultaneously. This would allow for a more “model-clade” approach, which could include comparisons of multiple trait gains, losses, or modifications (Specht and Howarth, 2014; Howarth and Dunn, 2016). Comparing gene expression and function across a clade can determine if losses or gains of a trait occur via changes to the same regulatory region or through different mechanisms (Prud’homme et al., 2006; R. Zhang et al., 2013; W. Zhang et al., 2013). For instance, it has recently been argued that much of species diversification is likely due to a change in the timing or location of gene expression, either through cis- or trans- regulatory changes (Hoekstra and Coyne, 2007; Specht and Howarth, 2014). Only by analyzing entire clades will we be able to discern major patterns in how genome dynamics shape phenotypic diversity.
In this article, we demonstrate that a genome-skimming data set, which was originally aimed at obtaining full plastome sequences of 24 Core Goodeniaceae species for phylogenetic reconstruction, has provided enough nuclear genome data to create an alignment of all four CYC-like genes in Core Goodeniaceae. Single contiguous or multiple noncontiguous portions from each CYC-like copy were successfully identified from genome-skimming data in 21 species (CYC1), 15 species (CYC2), 17 species (CYC3A), and 22 species (CYC3B). Although there was limited sequence information from any single species, examining all sampled species together allowed for the alignment of detailed matrices for each gene clade encompassing entire gene lengths and regulatory regions. All the sequence fragments were easily alignable within each copy for the entire coding sequence of the gene. Even the 5′ and 3′ ends of the coding region were alignable through much of the sequence.
CYC-like genes are involved in floral symmetry and are generally more highly expressed in dorsal (upper) regions of the corolla and androecium (Hileman, 2014) to regulate dorsoventral asymmetry. It has been suggested that the amount of disparity of gene expression of CYC-like genes across the dorsoventral axis is directly correlated with the degree of zygomorphy when examining two different floret types in a single species (Berger et al., 2016). We aim to use this data set to expand sampling for gene tree reconstruction and as a stepping-stone to examine expression and regulatory sequence differences among CYC-like genes in Goodeniaceae, encompassing significant variation in corolla symmetry.
Using this approach, we now have enough information to use a target enrichment approach to batch-sequence CYC-like genes from a large number of Core Goodeniaceae species, including upstream and downstream regulatory regions. With these sequences, we can align and analyze coding gene changes as well as changes in regulatory regions. Databases such as PlantPAN 2.0 (Chow et al., 2016) can be used to scan regulatory regions for changes in possible enhancers. Therefore, genome skimming could provide a cost-effective way to generate enough genomic data to create baits for efficient and thorough sequencing of coding and regulatory regions of candidate genes from across clades. Additionally, previously sequenced genome-skimming data sets can likely be mined for important candidate genes for traits of interest.
Supplementary Material
LITERATURE CITED
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., Lesin V. M., et al. 2012. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett C. F., Baker W. J., Comer J. R., Conran J. G., Lahmeyer S. C., Leebens-Mack J. H., Li J., et al. 2015. Plastid genomes reveal support for deep phylogenetic relationships and extensive rate variation among palms and other commelinid monocots. New Phytologist 209: 855–870. [DOI] [PubMed] [Google Scholar]
- Berger B. A., Thompson V., Lim A., Ricigliano V., Howarth D. G. 2016. Elaboration of bilateral symmetry across Knautia macedonica capitula related to changes in ventral petal expression of CYCLOIDEA-like genes. EvoDevo 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B. A., Han J., Sessa E. B., Gardner A. G., Shepherd K. A., Ricigliano V. A., Jabaily R. S., Howarth D. G. 2017. Data from: The unexpected depths of genome-skimming data: A case study examining Goodeniaceae floral symmetry genes. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.0500c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blischak P. D., Wenzel A. J., Wolfe A. D. 2014. Gene prediction and annotation in Penstemon (Plantaginaceae): A workflow for marker development from extremely low-coverage genome sequencing. Applications in Plant Sciences 2: 1400044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock D. G., Kane N. C., Ebert D. P., Rieseberg L. H. 2013. Genome skimming reveals the origin of the Jerusalem Artichoke tuber crop species: Neither from Jerusalem nor an artichoke. New Phytologist 201: 1021–1030. [DOI] [PubMed] [Google Scholar]
- Broholm S. K., Tahtiharju S., Laitinen R. A. E., Albert V. A., Teeri T. H., Elomaa P. 2008. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proceedings of the National Academy of Sciences, USA 105: 9117–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carolin R. C., Rajput M. T. M., Morrison P. 1992. Goodeniaceae. In: A. S. George , Flora of Australia, vol. 35, 4–300. Australian Government Publishing Service, Canberra, Australia. [Google Scholar]
- Chapman M. A., Leebens-Mack J. H., Burke J. M. 2008. Positive selection and expression divergence following gene duplication in the sunflower CYCLOIDEA gene family. Molecular Biology and Evolution 25: 1260–1273. [DOI] [PubMed] [Google Scholar]
- Chow C.-N., Zheng H.-Q., Wu N.-Y., Chien C.-H., Huang H.-D., Lee T.-Y., Chiang-Hsieh Y.-F., et al. 2016. PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Research 44: D1154–D1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citerne H. L., Le Guilloux M., Sannier J., Nadot S., Damerval C. 2013. Combining phylogenetic and syntenic analyses for understanding the evolution of TCP ECE genes in eudicots. PLoS ONE 8: e74803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E., Meyerowitz E. 1991. The war of the whorls: Genetic interactions controlling flower development. Nature 353: 31–37. [DOI] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. 1999. The TCP domain: A motif found in proteins regulating plant growth and development. Plant Journal 18: 215–222. [DOI] [PubMed] [Google Scholar]
- Dodsworth S., Chase M. W., Särkinen T., Knapp S., Leitch A. R. 2016. Using genomic repeats for phylogenomics: A case study in wild tomatoes (Solanum section Lycopersicon: Solanaceae). Biological Journal of the Linnean Society. Linnean Society of London 117: 96–105. [Google Scholar]
- Duarte J. M., Wall P. K., Edger P. P., Landherr L. L., Ma H., Pires P. K., Leebens-Mack J., dePamphilis C. W. 2010. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evolutionary Biology 10: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton T. M., Van der Hoeven R., Eannetta N. T., Tanksley S. D. 2002. Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 14: 1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner A. G., Sessa E. B., Michener P., Johnson E., Shepherd K. A., Howarth D. G., Jabaily R. S. 2016a. Utilizing next-generation sequencing to resolve the backbone of the Core Goodeniaceae and inform future taxonomic and floral form studies. Molecular Phylogenetics and Evolution 94: 605–617. [DOI] [PubMed] [Google Scholar]
- Gardner A. G., Fitz Gerald J. N., Menz J., Shepherd K. A., Howarth D. G., Jabaily R. S. 2016b. Characterizing floral symmetry in the Core Goodeniaceae with geometric morphometrics. PLoS ONE 11: e0154736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring W. J. 1998. Master control genes in development and evolution: The homeobox story. Yale University Press, New Haven, Connecticut, USA. [Google Scholar]
- Godden G. T., Jordon-Thaden I. E., Chamala S., Crowl A. A., García N., Germain-Aubrey C. C., Heaney J. M., et al. 2012. Making next-generation sequencing work for you: Approaches and practical considerations for marker development and phylogenetics. Plant Ecology & Diversity 5: 427–450. [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nature Biotechnology 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Couger M. B., et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson L. 2001. First nuclear DNA C-values for another 25 angiosperm families. Annals of Botany 88: 851–858. [DOI] [PubMed] [Google Scholar]
- Hileman L. C. 2003. Why do paralogs persist? Molecular evolution of CYCLOIDEA and related floral symmetry genes in Antirrhineae (Veronicaceae). Molecular Biology and Evolution 20: 591–600. [DOI] [PubMed] [Google Scholar]
- Hileman L. C. 2014. Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 369: 20130348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra H. E., Coyne J. A. 2007. The locus of evolution: Evo devo and the genetics of adaption. Evolution 61: 995–1016. [DOI] [PubMed] [Google Scholar]
- Howarth D. G., Donoghue M. J. 2005. Duplications in CYC-like genes from Dipsacales correlate with floral form. International Journal of Plant Sciences 166: 357–370. [Google Scholar]
- Howarth D. G., Donoghue M. J. 2006. Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proceedings of the National Academy of Sciences, USA 103: 9101–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth D. G., Dunn M. P. 2016. Phylogenetic approach to studying developmental evolution: A model clade approach. In R. M. Kliman , Encyclopedia of evolutionary biology, 246–253. Academic Press, Waltham, Massachusetts, USA. [Google Scholar]
- Howarth D. G., Martins T., Chimney E., Donoghue M. J. 2011. Diversification of CYCLOIDEA expression in the evolution of bilateral flower symmetry in Caprifoliaceae and Lonicera (Dipsacales). Annals of Botany 107: 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabaily R. S., Shepherd K. A., Gustafsson M. H. G., Sage L. W., Krauss S. L., Howarth D. G., Motley T. J. 2012. Systematics of the Austral-Pacific family Goodeniaceae: Establishing a taxonomic and evolutionary framework. Taxon 61: 419–436. [Google Scholar]
- Juntheikki-Palovaara I., Tähtiharju S., Lan T., Broholm S. K., Rijpkema A. S., Ruonala R., Kale L., et al. 2014. Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae). Plant Journal 79: 783–796. [DOI] [PubMed] [Google Scholar]
- Kane N., Sveinsson S., Dempewolf H., Yang J. Y., Zhang D., Engels J. M. M., Cronk Q. 2012. Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. American Journal of Botany 99: 320–329. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. 2013. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., et al. 2012. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England) 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L., Coen E. 1996. Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799. [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Copsey L., Vincent C., Clark J., Coen E. 1999. Control of organ asymmetry in flowers of Antirrhinum. Cell 99: 367–376. [DOI] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., et al. 2012. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T., Salzberg S. L. 2011. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics (Oxford, England) 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malé P.-J. G., Bardon L., Besnard G., Coissac E., Delsuc F., Engel J., Lhuillier E., et al. 2014. Genome skimming by shotgun sequencing helps resolve the phylogeny of a pantropical tree family. Molecular Ecology Resources 14: 966–975. [DOI] [PubMed] [Google Scholar]
- Prud’homme B., Gompel N., Rokas A., Kassner V. A., Williams T. M., Yeh S.-D., True J. R., Carroll S. B. 2006. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440: 1050–1053. [DOI] [PubMed] [Google Scholar]
- Rai A., Kamochi H., Suzuki H., Nakamura M., Takahashi H., Hatada T., Saito K., Yamazaki M. 2016. De novo transcriptome assembly and characterization of nine tissues of Lonicera japonica to identify potential candidate genes involved in chlorogenic acid, luteolosides, and secoiridoid biosynthesis pathways. Journal of Natural Medicines 71: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripma L. A., Simpson M. G., Hasenstab-Lehman K. 2014. Geneious! Simplified genome skimming methods for phylogenetic systematic studies: A case study in Oreocarya (Boraginaceae). Applications in Plant Sciences 2: 1400062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickl R., Liston A., Zeisek V., Oberlander K., Weitemier K., Straub S. C. K., Cronn R. C., et al. 2015. Phylogenetic marker development for target enrichment from transcriptome and genome skim data: The pipeline and its application in southern African Oxalis (Oxalidaceae). Molecular Ecology Resources 16: 1124–1135. [DOI] [PubMed] [Google Scholar]
- Shahmuradov I. A. 2003. PlantProm: A database of plant promoter sequences. Nucleic Acids Research 31: 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D. E., Gitzendanner M. A., Stull G., Chester M. 2013. The potential of genomics in plant systematics. Taxon 62: 886–898. [Google Scholar]
- Specht C. D., Howarth D. G. 2014. Adaptation in flower form: A comparative evodevo approach. New Phytologist 206: 74–90. [DOI] [PubMed] [Google Scholar]
- Steele P. R., Hertweck K. L., Mayfield D., McKain M. R., Leebens-Mack J., Pires J. C. 2012. Quality and quantity of data recovered from massively parallel sequencing: Examples in Asparagales and Poaceae. American Journal of Botany 99: 330–348. [DOI] [PubMed] [Google Scholar]
- Strand A. E., Leebens-Mack J., Milligan B. G. 1997. Nuclear DNA-based markers for plant evolutionary biology. Molecular Ecology 6: 113–118. [DOI] [PubMed] [Google Scholar]
- Straub S. C. K., Fishbein M., Livshultz T., Foster Z., Parks M., Weitemier K., Cronn R. C., Liston A. 2011. Building a model: Developing genomic resources for common milkweed (Asclepias syriaca) with low coverage genome sequencing. BMC Genomics 12: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub S. C. K., Parks M., Weitemier K., Fishbein M., Cronn R. C., Liston A. 2012. Navigating the tip of the genomic iceberg: Next-generation sequencing for plant systematics. American Journal of Botany 99: 349–364. [DOI] [PubMed] [Google Scholar]
- Straub S. C. K., Moore M. J., Soltis P. S., Soltis D. E., Liston A., Livshultz T. 2014. Phylogenetic signal detection from an ancient rapid radiation: Effects of noise reduction, long-branch attraction, and model selection in crown clade Apocynaceae. Molecular Phylogenetics and Evolution 80: 169–185. [DOI] [PubMed] [Google Scholar]
- Tähtiharju S., Rijpkema A. S., Vetterli A., Albert V. A., Teeri T. H., Elomaa P. 2012. Evolution and diversification of the CYC/TB1 gene family in Asteraceae—A comparative study in Gerbera (Mutisieae) and sunflower (Heliantheae). Molecular Biology and Evolution 29: 1155–1166. [DOI] [PubMed] [Google Scholar]
- Tank D., Donoghue M. J. 2010. Phylogeny and phylogenetic nomenclature of the Campanulidae based on an expanded sample of genes and taxa. Systematic Botany 35: 425–441. [Google Scholar]
- Visser E. A., Wegrzyn J. L., Steenkmap E. T., Myburg A. A., Naidoo S. 2015. Combined de novo and genome guided assembly and annotation of the Pinus patula juvenile shoot transcriptome. BMC Genomics 16: 1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitemier K., Straub S. C. K., Cronn R. C., Fishbein M., Schmickl R., McDonnell A., Liston A. 2014. Hyb-Seq: Combining target enrichment and genome skimming for plant phylogenomics. Applications in Plant Sciences 2: 1400042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Mueller L. A., Crouzillat D., Pétiard V., Tanksley S. D. 2006. Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: A test case in the euasterid plant clade. Genetics 174: 1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Wu G., Tang J., Luo R., Patterson J., Liu S., Huang W., et al. 2014. SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-seq reads. Bioinformatics (Oxford, England) 30: 1660–1666. [DOI] [PubMed] [Google Scholar]
- Yuan Y.-W., Liu C., Marx H. E., Olmstead R. G. 2009. The pentatricopeptide repeat (PPR) gene family, a tremendous resource for plant phylogenetic studies. New Phytologist 182: 272–283. [DOI] [PubMed] [Google Scholar]
- Yuan Y.-W., Liu C., Marx H. E., Olmstead R. G. 2010. An empirical demonstration of using pentatricopeptide repeat (PPR) genes as plant phylogenetic tools: Phylogeny of Verbenaceae and the Verbena complex. Molecular Phylogenetics and Evolution 54: 23–35. [DOI] [PubMed] [Google Scholar]
- Zhang R., Guo C., Zhang W., Wang P., Li L., Duan X., Du Q., et al. 2013. Disruption of the petal identity gene APETALA3-3 is highly correlated with loss of petals within the buttercup family (Ranunculaceae). Proceedings of the National Academy of Sciences, USA 110: 5074–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Kramer E. M., Davis C. C. 2010. Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proceedings of the National Academy of Sciences, USA 107: 6388–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Steinmann V. W., Nikolov L., Kramer E. M., Davis C. C. 2013. Divergent genetic mechanisms underlie reversals to radial floral symmetry from diverse zygomorphic flowered ancestors. Frontiers in Plant Science 4: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.