Abstract
Changes in gene transcription in the dorsal root ganglion (DRG) after nerve trauma contribute to the genesis of neuropathic pain. We report that peripheral nerve trauma caused by chronic constriction injury (CCI) increased the abundance of the transcription factor C/EBPβ (CCAAT/enhancer binding protein β) in the DRG. Blocking this increase mitigated the development and maintenance of CCI-induced mechanical, thermal, and cold pain hypersensitivities without affecting basal responses to acute pain and locomotor activity. Conversely, mimicking this increase produced hypersensitivity to mechanical, thermal, or cold pain. In the ipsilateral DRG, C/EBPβ promoted a decrease in the abundance of the voltage-gated potassium channel subunit Kv1.2 and µ opioid receptor (MOR) at the mRNA and protein levels, which would be predicted to increase excitability in the ipsilateral DRG neurons and reduce the efficacy of morphine analgesia. These effects required C/EPBβ-mediated transcriptional activation of Ehmt2 (euchromatic histonelysine N-methyltransferase 2), which encodes G9a, an epigenetic silencer of the genes encoding Kv1.2 and MOR. Blocking the increase in C/EBPβ in the DRG improved morphine analgesia after CCI. These results suggest that C/EBPβ is an endogenous initiator of neuropathic pain and could be a potential target for the prevention and treatment of this disorder.
INTRODUCTION
Peripheral nerve trauma–induced neuropathic pain—which is characterized by spontaneous ongoing pain or intermittent burning pain, allodynia, and thermal hyperalgesia—is a worldwide clinical problem (1). About 1/10 of the population in the United States and Europe suffers from neuropathic pain. Of this population, most of the patients do not achieve sufficient pain relief with the use of current medications because most of these medications are nonspecific with regard to the cause of neuropathic pain (2, 3). The estimated cost of neuropathic pain, including direct medical costs and the economic burden on quality of life, is about $600 billion annually. Thus, understanding the causal mechanisms underlying neuropathic pain could provide new and highly efficient therapeutic strategies in neuropathic pain management.
Abnormal ectopic firing and hyperexcitability in the neurons of the dorsal root ganglion (DRG) and neuromas at the injured sites after peripheral nerve trauma are generally considered to contribute to peripheral neuropathic pain genesis (1, 4). This abnormal spontaneous activity in DRG neurons and subsequent enhanced neurotransmitter release from their primary afferents may result from maladaptive changes in gene transcription and translation of receptors, enzymes, and voltage-dependent ion channels in the DRG (5, 6). Kv1.2, an α, pore-forming subunit in the voltage-gated potassium (Kv) channel family, is a key player during neuropathic pain genesis. Kv1.2 is abundant in most DRG neurons and critical for establishing the resting membrane potential and controlling the excitability of DRG neurons (7–10). Rescuing nerve injury–induced reduction in Kv1.2 by blocking the nerve injury–induced increases in euchromatic histone-lysine N-methyltransferase 2 (Ehmt2) mRNA and its gene product EHMT2 (also known as G9a) in the ipsilateral DRG attenuates mechanical, thermal, and cold pain hypersensitivities after peripheral nerve injury (11). Mimicking this reduction by overexpression of G9a in the DRG produces neuropathic pain symptoms (11). In addition, G9a also participates in the nerve injury–induced suppression of genes encoding opioid receptors in the ipsilateral DRG. Blocking the nerve injury–induced G9a increase in the ipsilateral DRG rescues the abundance of μ, κ, and Δ opioid receptors in the DRG; improves morphine analgesia; and impairs neuropathic pain (11–13). Conversely, overexpression of G9a in the DRG decreases the abundance of these three opioid receptors in the DRG and promotes the μ opioid receptor (MOR)–gated release of primary afferent neurotransmitters (12). These lines of evidence suggest that G9a is an endogenous instigator of neuropathic pain.
CCAAT/enhancer binding protein β (C/EBPβ) is a member of a family of transcription factors consisting of six structurally related basic leucine-zipper DNA binding proteins. It is involved in memory formation and synaptic plasticity in the central nervous system (14, 15). The promoter region of the Ehmt2 gene contains a consensus binding motif of C/EBPβ and is transactivated by C/EBPβ in human embryonic kidney (HEK) 293T cells (16). Given that G9a is a key player in neuropathic pain, C/EBPβ may contribute to neuropathic pain genesis by regulating Ehmt2 expression in the DRG.
Here, we showed that peripheral nerve trauma caused by chronic constriction injury (CCI) of the sciatic nerve increases the expression of C/EBPβ in the ipsilateral DRG. This increase contributed to CCI-induced neuropathic induction and maintenance by transcriptionally activating Ehmt2 and subsequently epigenetic silencing of Oprm1 (which encodes MOR) and Kcna2 (which encodes Kv1.2) genes in the ipsilateral DRG. Thus, C/EBPβ may be a potential target in the prevention and treatment of neuropathic pain.
RESULTS
Cebpb abundance is increased at the mRNA and protein levels in the ipsilateral DRG after CCI
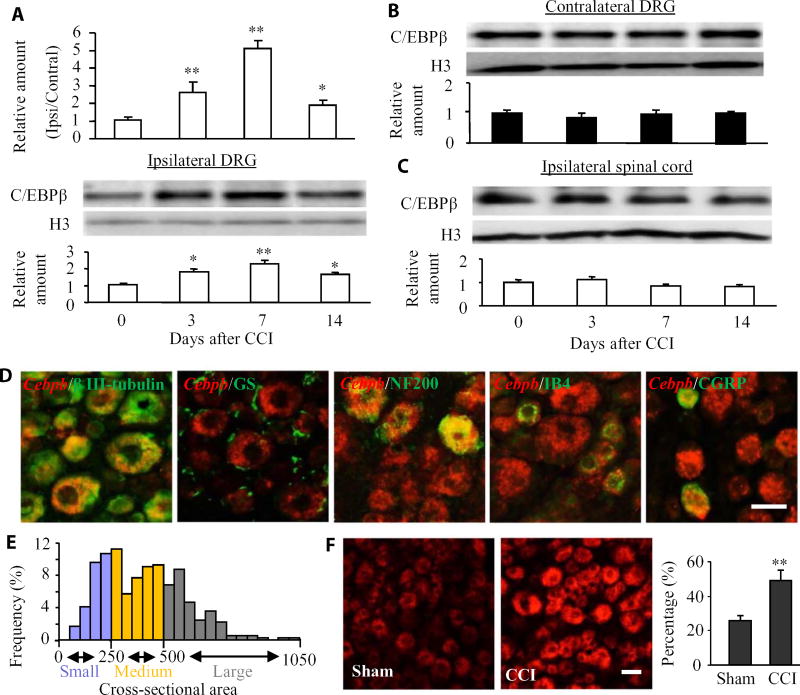
We first examined whether C/EBPβ abundance is altered in two pain-related regions (DRG and spinal cord) after CCI, a preclinical animal model of neuropathic pain that mimics nerve trauma–induced neuropathic pain in the clinic (17). In mice, CCI time-dependently increased the expression of Cebpb mRNA (which encodes C/EBPβ) and protein in the L3/4 DRGs on the ipsilateral side but not the contralateral side (Fig. 1, A and B). The ratios of ipsilateral side to contralateral side of Cebpb mRNA was increased by 2.53-fold on day 3, 4.98-fold on day 7, and 1.84-fold on day 14 after CCI compared to naïve mice (0 day). C/EBPβ protein abundance in the ipsilateral L3/4 DRGs was consistently increased by 1.99-fold on day 3, 2.28-fold on day 7, and 1.67-fold on day 14 after CCI compared to naïve mice. C/EBPβ protein abundance did not significantly change in the ipsilateral L3/4 spinal cord dorsal horn (Fig. 1C). As expected, sham surgery did not produce any changes in the basal abundance of Cebpb mRNA and C/EBPβ protein in the ipsilateral L3/4 DRGs and L3/4 spinal cord dorsal horn (fig. S1, A and B). The significant increase in the expression of C/EBPβ in the ipsilateral DRGs after CCI suggests a possible role of C/EBPβ in neuropathic pain.
Fig. 1. Peripheral nerve injury–induced increases in Cebpb mRNA and C/EBPβ protein in the ipsilateral DRG.
(A) Cebpb mRNA abundance in the ipsilateral (Ipsi) and contralateral (Contral) L3/4 DRGs (top) and C/EBPβ protein expression in the ipsilateral L3/4 DRGs (bottom) after CCI. Unilateral L3/4 DRGs from two mice were pooled together to obtain enough RNA and protein. n = 3 biological replicates (six mice) per time point. One-way analysis of variance (ANOVA) followed by Tukey post hoc test, F3,11 = 26.87 for Cebpb mRNA and F3,11 = 14.04 for C/EBPβ protein. *P < 0.05 or **P < 0.01 compared to the corresponding control group (0 day). H3, histone 3. (B) C/EBPβ protein abundance in the contralateral L3/4 DRGs after CCI. Representative Western blots and a summary of densitometric analysis are shown. n = 3 biological replicates (six mice) per time point. One-way ANOVA followed by Tukey post hoc test, F3,11 = 0.49. (C) C/EBPβ protein abundance in the ipsilateral L3/4 spinal cord after CCI. n = 3 biological replicates (three mice) per time point. One-way ANOVA followed by Tukey post hoc test, F3,11 = 1.99. (D) In situ hybridization histochemistry (ISHH) for Cebpb mRNA and immunohistochemistry of different DRG cell markers: β III tubulin, glutamine synthetase (GS), NF200, IB4, or CGRP in the DRG. n = 3 mice. Scale bar, 25 µm. (E) Distribution of Cebpb mRNA–labeled neuronal somata. Large, 31%; medium, 43%; small, 26%. (F) Neurons labeled by Cebpb mRNA in the ipsilateral L4 DRG on day 7 after CCI or sham surgery. n = 3 mice per group. **P < 0.01 compared to the corresponding sham group by two-tailed paired Student’s t test. Scale bar, 25 µm.
We also examined the distribution pattern of C/EBPβ in the DRG. Cebpb mRNA colocalized with β III tubulin (a specific neuronal marker) but not with glutamine synthetase (a marker for satellite glial cells) (Fig. 1D), indicating that Cebpb was expressed exclusively in DRG neurons. Moreover, our analysis showed that about 35% of Cebpb mRNA–labeled neurons were positive for neurofilament 200 (NF200) (a marker for medium/large cells and myelinated Aβ fibers), 32% were positive for isolectin B4 (IB4) (a marker for small nonpeptidergic neurons), and 27% were positive for calcitonin gene-related peptide (CGRP) (a marker for small peptidergic neurons) (Fig. 1D). A cross-sectional area analysis of neuronal somata indicated that about 31% of Cebpb mRNA–labeled neurons were large (>500 µm2 in area), 43% were medium (250 to 500 µm2 in area), and 26% were small (<250 µm2 in area) (Fig. 1E). As expected, the number of Cebpb mRNA–labeled neurons in the ipsilateral L4 DRG on day 7 after CCI increased by 1.94-fold compared with the corresponding sham group (Fig. 1F). On day 7 after CCI, 80.2% of Cebpb mRNA–labeled neurons were positive for activating transcription factor 3 (ATF3), a marker of injury (fig. S2). These findings suggest that most of the Cebpb mRNA–labeled neurons in the ipsilateral DRG are in response to peripheral nerve injury.
Blocking the increase in C/EBPβ abundance in the ipsilateral DRG impairs CCI-induced neuropathic pain
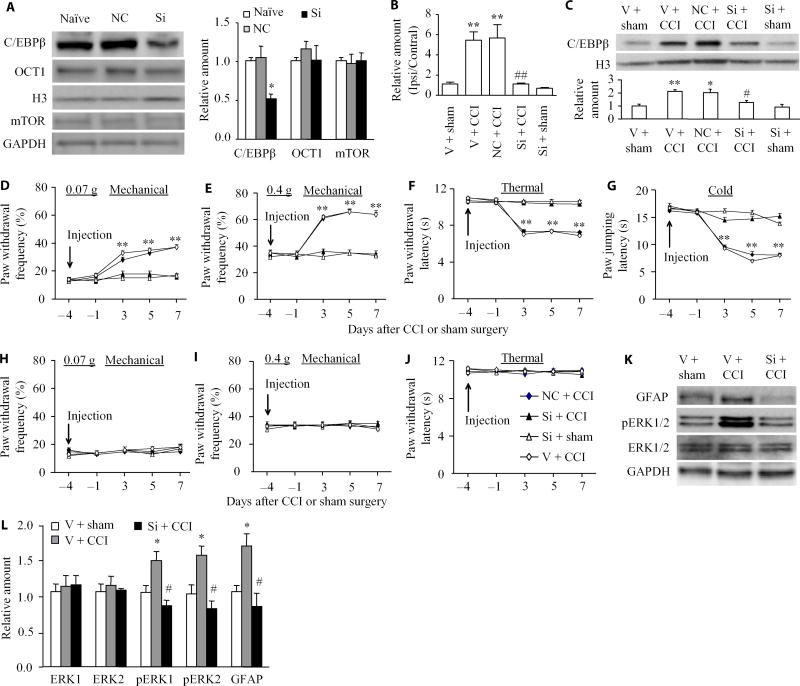
Next, we examined whether blocking the CCI-induced increase in DRG C/EBPβ through microinjection of its small interfering RNA (siRNA) into the ipsilateral DRGs affected CCI-induced pain hypersensitivity (Fig. 2A). Neither Cebpb siRNA nor negative control siRNA altered the basal abundance of another transcription factor, octamer binding factor 1 (OCT1), or that of an intracellular protein, mammalian target of rapamycin (mTOR) (Fig. 2A). As expected, the abundance of C/EBPβ mRNA and protein was significantly increased in the ipsilateral L3/4 DRGs on day 7 after CCI in mice pre-microinjected with vehicle or negative control siRNA as compared to sham mice pre-microinjected with vehicle (Fig. 2, B and C). These increases were not seen in the CCI mice pre-microinjected with Cebpb siRNA (Fig. 2, B and C). No significant decreases in the basal amounts of Cebpb mRNA and protein were observed in the ipsilateral L3/4 DRGs of the sham mice pre-microinjected with Cebpb siRNA (Fig. 2, B and C). This evidence suggests that pre-microinjection of C/EBPβ siRNA could block the CCI-induced increase in C/EBPβ in the ipsilateral DRG.
Fig. 2. Effect of DRG pre-microinjection of Cebpb siRNA on CCI-induced development of pain hypersensitivity and dorsal horn central sensitization.
(A) The abundance of C/EBPβ, OCT1, and mTOR after transfection of Cebpb or negative control siRNA (NC) into cultured DRG neurons. Representative Western blots and a summary of densitometric analysis are shown. n = 3 biological replicates per treatment. One-way ANOVA followed by Tukey post hoc test, F2,8 = 9.647 for C/EBPβ, F2,8 = 0.497 for OCT1, and F2,8 = 0.037 for mTOR. *P < 0.05 compared to naïve group. GAPDH, glyceraldhyde-3-phosphate dehydrogenase. (B and C) Effect of pre-microinjection of Cebpb siRNA (Si) into the ipsilateral L3/4 DRGs on basal or CCI-induced increases in Cebpb mRNA expression (B) and C/EBPβ protein abundance (C) on day 7 after CCI or post-sham surgery in the ipsilateral L3/4 DRGs. Unilateral L3/4 DRGs from two mice were pooled together. n = 6 to 8 mice per group. One-way ANOVA followed by Tukey post hoc test, F4,16 = 17.041 in (B) and F4,19 = 9.614 in (C). **P < 0.01 compared to the vehicle (V) plus sham group. #P < 0.05 or ##P < 0.01 compared to the vehicle plus CCI group. (D to J) Effect of pre-microinjection of Cebpb siRNA, vehicle, or negative control siRNA into the ipsilateral L3/4 DRGs on paw withdrawal frequencies to mechanical stimuli (D and E), paw withdrawal latency to thermal stimulation (F), and paw jumping latency to cold stimulation (G) on the ipsilateral side and on basal paw withdrawal responses to mechanical (H and I) and thermal (J) stimuli on the contralateral side at different days after CCI or sham surgery. n = 10 mice per group. Two-way ANOVA followed by Tukey post hoc test, F4,199 = 37.159 in (D), F4,199 = 27.312 in (E), F4,199 = 37.336 in (F), F4,199 = 56.651 in (G), F4,199 = 2.071 in (H), F4,199 = 0.472 in (I), and F4,199 = 0.681 in (J). **P < 0.01 compared to the corresponding baseline (day –4). (K and L) Effect of pre-microinjection of Cebpb siRNA into the ipsilateral L3/4 DRGs on CCI-induced increases in the phosphorylation of ERK1/2 (p-ERK1/2) and abundance of GFAP in the ipsilateral L3/4 dorsal horn on day 5 post–spinal nerve ligation (SNL). Representative Western blots (K) and a summary of densitometric analysis (L) are shown. n = 6 to 8 mice per group. One-way ANOVA followed by Tukey post hoc test, F2,9 = 9.003 for p-ERK1, F2,9 = 8.899 for p-ERK2, F2,11 = 0.156 for ERK1, F2,11 = 0.192 for ERK2, and F2,11 = 8.163 for GFAP. *P < 0.05 compared to the corresponding vehicle plus sham group. #P < 0.05 compared to the corresponding vehicle plus CCI group.
We inquired whether blocking the CCI-induced increase in C/EBPβ in the DRG would affect the development of CCI-induced pain hypersensitivities. Consistent with previous studies (8, 17), CCI produced long-term mechanical allodynia, thermal hyperalgesia, and cold allodynia on the ipsilateral side in the vehicle-injected mice (Fig. 2, D to G). The paw withdrawal frequencies in response to mechanical stimuli applied to the ipsilateral hindpaw were significantly increased on days 3, 5, and 7 after CCI as compared with preinjury baseline values (Fig. 2, D and E). The paw withdrawal latency and jump latency of the ipsilateral hindpaw in response to heat and cold, respectively, were markedly reduced on days 3, 5, and 7 after CCI as compared to the corresponding baseline (Fig. 2, F and G). Preinjection of Cebpb siRNA did not change basal paw responses to mechanical, heat, or cold stimuli on the ipsilateral side of sham mice (Fig. 2, D to G) but abolished CCI-induced mechanical allodynia, thermal hyperalgesia, and cold allodynia (Fig. 2, D to G). Compared with the baseline values, there were no significant changes in paw withdrawal frequencies and latencies and paw jumping latencies on the ipsilateral side of the Cebpb siRNA–injected mice (Fig. 2, D to G). As expected, preinjection of negative control siRNA did not affect CCI-induced mechanical allodynia, thermal hyperalgesia, and cold allodynia on the ipsilateral side during the observation period (Fig. 2, D to G). There were no marked differences in paw responses between the negative control siRNA–injected and vehicle-injected groups (Fig. 2, D to G). Basal paw responses on the contralateral side(Fig. 2, H to J) and locomotor function (Table 1) were not altered by preinjection of either siRNA or vehicle.
Table 1. Mean changes in locomotor function.
SEM given in parentheses. n = 10 mice per group; five trials.
| Treatment groups | Locomotor functional test | ||
|---|---|---|---|
| Placing | Grasping | Righting | |
| Vehicle + CCI | 5(0) | 5(0) | 5(0) |
| Cebpb siRNA + CCI | 5(0) | 5(0) | 5(0) |
| Cebpb siRNA + sham | 5(0) | 5(0) | 5(0) |
| Negative control siRNA + CCI | 5(0) | 5(0) | 5(0) |
| AAV5-EGFP | 5(0) | 5(0) | 5(0) |
| AAV5-C/EBPβ | 5(0) | 5(0) | 5(0) |
| Saline (5 µl) | 5(0) | 5(0) | 5(0) |
| Lidocaine (0.8%, 5 µl) | 5(0) | 5(0) | 5(0) |
We also examined whether preinjection of Cebpb siRNA affected CCI-induced dorsal horn central sensitization as indicated by increases in the phosphorylation of extracellular signal–regulated kinase 1/2 (ERK1/2) and glial fibrillary acidic protein (GFAP) in the dorsal horn (18, 19). In line with previous studies in a spinal nerve injury–induced neuropathic pain model (19, 20), the phosphorylation of ERK1/2 (but not total ERK1/2) and GFAP was significantly increased in the ipsi-lateral L3/4 dorsal horn in mice subjected to CCI but not in those that received sham surgery and vehicle (Fig. 2, K and L). These increases were not seen in Cebpb siRNA–injected mice (Fig. 2, K and L).
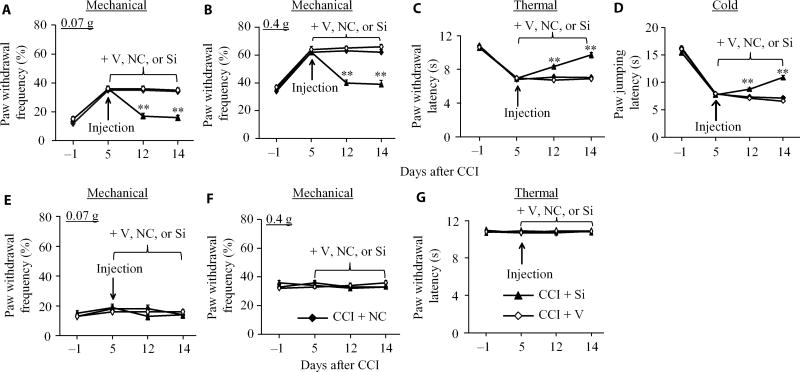
To further examine the role of C/EBPβ in the DRG in the maintenance of CCI-induced pain hypersensitivities, we carried out behavior tests in mice subjected to CCI and injected or not with siRNA. Post-injection of Cebpb siRNA but not negative control siRNA attenuated CCI-induced mechanical allodynia, thermal hyperalgesia, and cold allodynia on the ipsilateral side during the maintenance period (Fig. 3, A to D). At 12 and 14 days after CCI, paw withdrawal frequencies in response to mechanical stimuli markedly decreased (Fig. 3, A and B), paw withdrawal latency in response to heat stimuli greatly increased (Fig. 3C), and paw jumping latency in response to cold stimuli significantly increased (Fig. 3D), compared with the corresponding vehicle-injected group. As expected, post-injection of vehicle, Cebpb siRNA, or negative control siRNA did not alter basal paw withdrawal responses to mechanical, heat, and cold stimuli applied to the contralateral hindpaw during the maintenance period (Fig. 3, E to G).
Fig. 3. Effect of DRG post-microinjection of Cebpb siRNA into the ipsilateral L3/4 DRGs on CCI-induced maintenance of pain hypersensitivity.
(A to G) Effect of DRG microinjection of Cebpb siRNA, vehicle, or negative control siRNA starting on day 5 after CCI on paw withdrawal frequencies to mechanical stimuli (A and B), paw withdrawal latency to thermal stimulation (C), and paw jumping latency to cold stimulation (D) on the ipsilateral side and on basal paw withdrawal responses to mechanical (E and F) and thermal (G) stimuli on the contralateral side on days 12 and 14 after CCI. n = 10 mice per group. Two-way ANOVA followed by Tukey post hoc test, F2,119 = 26.777 in (A), F2,119 = 31.439 in (B), F2,119 = 15.275 in (C), F2,119 = 18.089 in (D), F2,119 = 0.093 in (E), F2,119 < 0.001 in (F), and F2,119 = 0.157 in (G). **P < 0.01 compared to the vehicle plus CCI group at the corresponding time points.
Mimicking the CCI-induced increase in C/EBPβ in the DRG leads to neuropathic pain symptoms
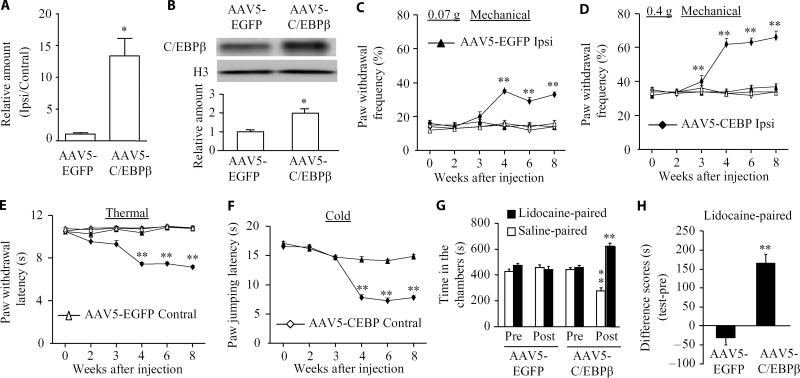
We then determined whether mimicking the CCI-induced increase in C/EBPβ abundance in the DRG through microinjection of adeno-associated virus 5 (AAV5)–C/EBPβ into unilateral L3/4 DRGs altered nociceptive thresholds in naïve mice (Fig. 4, A and B). Injection of AAV5-C/EBPβ but not AAV5–enhanced green fluorescent protein (EGFP) produced mechanical allodynia, heat hyperalgesia, and cold allodynia as demonstrated by ipsilateral increases in paw withdrawal frequency in response to mechanical stimuli (Fig. 4, C and D) and by ipsilateral decreases in paw withdrawal latencies in response to heat (Fig. 4E) and paw jumping latencies in response to cold (Fig. 4F). These pain hypersensitivities developed between 3 and 4 weeks and persisted for at least 8 weeks (Fig. 4, C to F), consistent with the 3- to 4-week lag period for AAV5 expression, which lasts for at least 3 months (8, 10). Neither AAV5-C/EBPβ nor AAV5-EGFP changed basal paw responses to mechanical, heat, and cold stimuli on the contralateral side (Fig. 2, C to E) and locomotor function (Table 1).
Fig. 4. Effect of DRG C/EBPβ overexpression on nociceptive thresholds in naïve mice.
(A and B) Cebpb mRNA expression (A) and C/EBPβ protein abundance (B) in the ipsilateral L3/4 DRGs 8 weeks after microinjection of AAV5-C/EBPβ or control AAV5-EGFP. n = 6 mice per group. Two-tailed, independent Student’s t test, *P < 0.05 compared to the AAV5-EGFP group. (C to F) Effect of microinjection of AAV5-C/EBPβ or AAV5-EGFP into the unilateral L3/4 DRGs on paw withdrawal frequencies to mechanical stimuli (C and D), paw withdrawal latency to thermal stimulation (E), and paw jumping latency to cold stimulation (F) on the ipsilateral and contralateral sides at the different weeks after microinjection. n = 10 mice per group. Two-way ANOVA followed by Tukey post hoc test, F3,239 = 42.76 in (C), F3,239 = 39.60 in (D), F3,239 = 59.08 in (E), and F1,119 = 49.879 in (F). **P < 0.01 compared to the control AAV5-EGFP group on the ipsilateral side at the corresponding time points. (G and H) Effect of microinjection of AAV5-C/EBPβ or AAV5-EGFP into the unilateral L3/4 DRGs on spontaneous ongoing pain as assessed by CPP paradigm. n = 16 mice per group. **P < 0.01 compared to the corresponding preconditioning (G) or the AAV5-EGFP group (H) by two-tailed, independent Student’s t test.
In addition to stimulation-induced evoked pain hypersensitivities, we also examined whether mimicking the CCI-induced increase in C/EBPβ in the DRG led to spontaneous ongoing nociceptive responses using a conditional place preference (CPP) paradigm. The AAV5-C/EBPβ–injected mice displayed an obvious preference (that is, they spent more time) for the lidocaine-paired chamber (Fig. 4, G and H), indicating stimulation-independent spontaneous nociceptive responses. In contrast, the AAV5-EGFP–injected mice did not exhibit significant preference toward either the saline- or lidocaine-paired chamber (Fig. 4, G and H), demonstrating no marked spontaneous nociceptive responses. Together, these findings indicate that mimicking the CCI-induced increase in DRG C/EBPβ leads to both spontaneous and evoked pain hypersensitivity, typical symptoms of neuropathic pain in the clinic.
C/EBPβ transactivates the Ehmt2 gene in the ipsilateral DRG after CCI
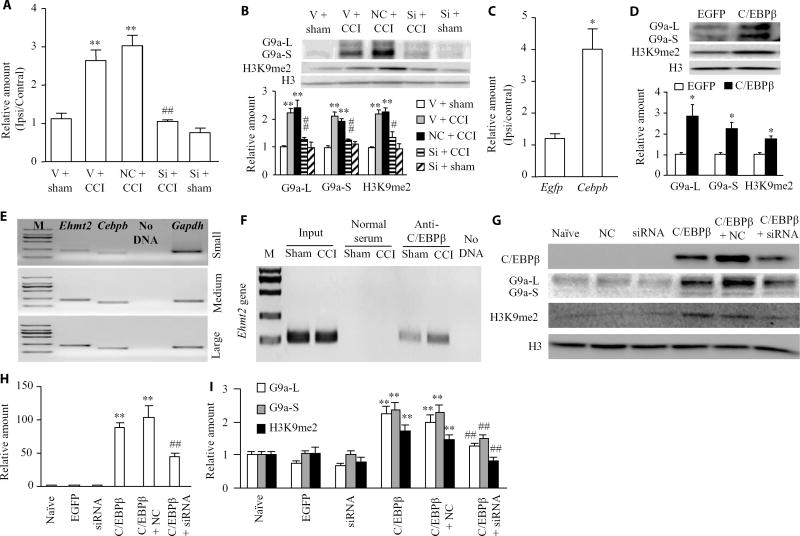
How does C/EBPβ as a transcription factor in the DRG participate in neuropathic pain? Prediction by TFSEARCH ( http://diyhpl.us/~bryan/irc/protocol-online/protocol-cache/TFSEARCH.html) indicates a consensus C/EBPβ binding site (−4770/attgcgcagt/−4760) at the distal promoter region of the Ehmt2 gene. Moreover, C/EBPβ increases the activity of this promoter in HEK-293T cells (16). Given that nerve injury–induced increases in Ehmt2 mRNA and G9a in the ipsilateral DRG are required for neuropathic pain genesis (11–13, 21), we predicted that C/EBPβ contributed to neuropathic pain by transactivating the Ehmt2 gene in the ipsilateral DRG. Preinjection of Cebpb siRNA but not negative control siRNA significantly blocked CCI-induced increases in the abundance of Ehmt2 mRNA, G9a’s long and short protein isoforms, and a G9a-catalyzed repressive marker, dimethyl K9 of histone 3 (H3K9me2), in the ipsilateral DRG on day 7 after CCI (Fig. 5, A and B). Preinjection of Cebpb siRNA did not markedly reduce the basal amounts of Ehmt2 mRNA, G9a’s two protein isoforms, and H3K9me2 in the injected DRG on day 7 post-sham surgery (Fig. 5, A and B). However, microinjection of AAV5-C/EBPβ but not AAV5-EGFP into the unilateral L3/4 DRGs increased the amounts of Ehmt2 mRNA (Fig. 5C), G9a’s two protein isoforms, and H3K9me2 (Fig. 5D) in the injected DRG 5 weeks after injection. Single-cell reverse transcription polymerase chain reaction (RT-PCR) analysis revealed coexpression of Cebpb with Ehmt2 in individual small, medium, or large DRG neurons (Fig. 5E). These results suggest that C/EBPβ may contribute to the increase in G9a induced by nerve injury in the DRG that has been reported previously (11–13, 21).
Fig. 5. C/EBPβ-triggered transcriptional activation of Ehmt2 in the ipsilateral DRG after CCI.
(A and B) Ehmt2 mRNA expression (A), the abundance of G9a’s two protein isoforms (B), and the amount of H3K9me2-marked protein (B) in the ipsilateral L3/4 DRGs 7 days after CCI or sham surgery in the mice pre-microinjected with vehicle, negative control siRNA, or Cebpb siRNA into the ipsilateral L3/4 DRGs. Unilateral L3/4 DRGs from two mice were pooled together. n = 6 to 8 mice per group. One-way ANOVA followed by Tukey post hoc test, F4,16 = 33.704 for Ehmt2 mRNA, F4,19 = 16.879 for G9a-L, F4,19 = 28.072 for G9a-S, and F4,14 = 19.035 for H3K9me2. **P < 0.01 compared to the corresponding vehicle plus sham group; #P < 0.05 or ##P < 0.01 compared to the corresponding vehicle plus CCI group. L, long isoform; S, short isoform. (C and D) The amounts of Ehmt2 mRNA (C), G9a’s two protein isoforms (D), and H3K9me2 protein (D) in the injected L3/4 DRGs 8 weeks after microinjection of AAV5-C/EBPβ or control AAV5-EGFP. n = 6 to 8 mice per group. Two-tailed, independent Student’s t test, *P < 0.05 compared to the corresponding AAV5-EGFP group. (E) Coexpression of Cebpb mRNA with Ehmt2 mRNA in small, medium, and large DRG neurons. Gapdh mRNA was used a positive control. n = 3 biological replicates. M, ladder marker. (F) ChIP analysis of C/EBPβ binding to a fragment of the Ehmt2 gene distal promoter in the ipsilateral L3/4 DRGs on day 7 after CCI or sham surgery. Input, total purified fragments. Ipsilateral L3/4 DRGs from 3 mice were pooled together. n = 9 mice per group. (G to I) The abundance of C/EBPβ, G9a’s two protein isoforms, and H3K9me2 in mouse cultured DRG neurons transduced as indicated. Representative Western blots (G) and a summary of densitometric analysis [C/EBPβ (H) and G9a-L, G9a-S, and H3K9me2 (I)] are shown. n = 5 biological replicates per treatment. One-way ANOVA followed by Tukey post hoc test, F5,29 = 51.42 for C/EBPβ, F5,29 = 19.66 for G9a-L, F5,29 = 27.95 for G9a-S, and F5,29 = 10.16 for H3K9me2. **P < 0.01 compared to the corresponding naïve group; ##P < 0.01 compared to the corresponding C/EBPβ-treated group.
Chromatin immunoprecipitation (ChIP) assays revealed that a fragment within the Ehmt2 promoter including the C/EBPβ binding site was amplified from the complex immunoprecipitated with an anti-C/EBPβ antibody (but not normal serum) in nuclear fractions from DRGs in the sham group (Fig. 5F), suggesting that C/EBPβ specifically bound to the Ehmt2 gene in the DRG. The binding activity of C/EBPβ to the Ehmt2 gene was significantly increased after CCI, as shown by a fourfold increase in band density in the ipsilateral L3/4 DRGs from CCI mice compared to those from sham mice on day 7 after CCI (Fig. 5F). This increased binding of C/EBPβ may be due to the increase of C/EBPβ protein in the ipsilateral L3/4 DRGs on day 7 after CCI (Fig. 1A).
These findings suggest that C/EBPβ may directly regulate G9a expression in the DRG after CCI. Neurons transduced with AAV5-C/EBPβ but not with AAV5-EGFP showed significant increases in C/EBPβ, G9a’s two isoforms, and H3K9me2 proteins (Fig. 5, G to I). These increases were attenuated in the neurons with cotreatment with Cebpb siRNA but not with negative control siRNA (Fig.5, G to I), suggesting that the induction of G9a and H3K9me2 was specific in responses to C/EBPβ. Collectively, our findings suggest that the nerve injury–induced increase in C/EBPβ protein enhances its binding to the Ehmt2 promoter, resulting in increased Ehmt2 mRNA transcription and translation in the ipsilateral DRG.
Participation of C/EBPβ in CCI-induced suppression of Oprm1 and Kcan2 genes requires G9a in the ipsilateral DRG
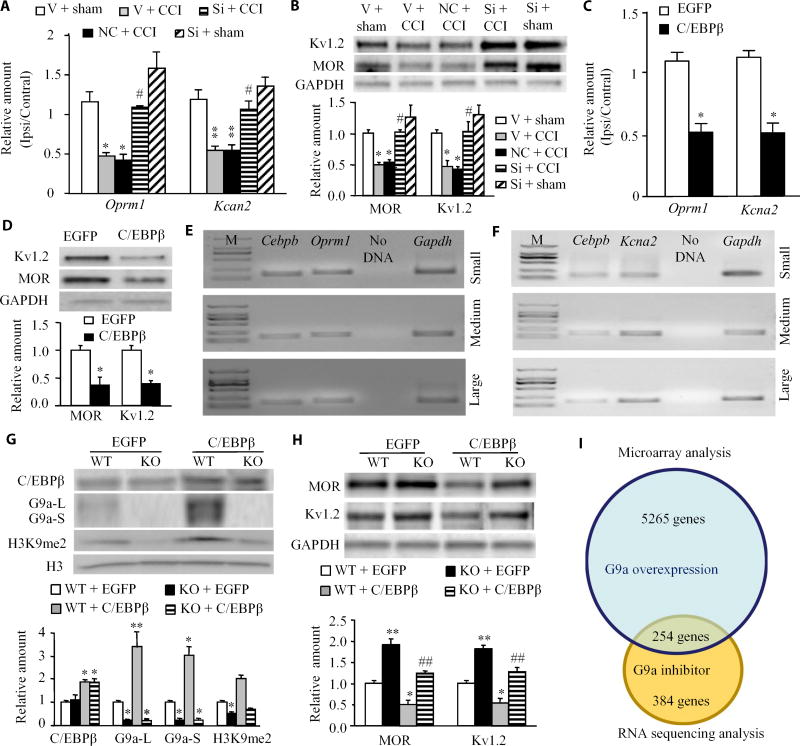
The increase in G9a in the DRG induced by nerve injury contributes to neuropathic pain through epigenetic silencing of Oprm1 (which encodes MOR) and Kcna2 (which encodes Kv1.2) (11–13). Consistent with the previous studies (11–13), CCI decreased the abundance of MOR and Kv1.2 at the mRNA and protein levels in the ipsilateral DRG on day 7 after CCI in vehicle-injected mice, effects that were rescued by injection of Cebpb siRNA but not negative control siRNA (Fig. 6, A and B). Injection of Cebpb siRNA did not significantly alter the basal amounts of MOR and Kv1.2 at the mRNA and protein levels in the injected DRG of sham mice (Fig. 6, A and B). We also found that injection of AAV5-C/EBPβ, but not AAV5-EGFP, decreased the abundance of MOR and Kv1.2 at the mRNA and protein levels 5 weeks after injection (Fig. 6, C and D). Single-cell RT-PCR analysis revealed coexpression of Cebpb with Oprm1 or Kcna2 in individual small, medium, or large DRG neurons (Fig. 6, E and F). These data indicate that the nerve injury increase of C/EBPβ may participate in nerve injury–induced suppression of MOR and Kv1.2 abundance in the ipsilateral DRG.
Fig. 6. G9a is required for the effect of C/EBPβ in CCI-induced suppression of MOR and Kv1.2 in the ipsilateral DRGs.
(A and B) Oprm1 and Kcan2 mRNA expression (A) and protein abundance (B) in the ipsilateral L3/4 DRGs 7 days after sham surgery or CCI in the mice pre-microinjected with vehicle, negative control siRNA, or Cebpb siRNA into the ipsilateral L3/4 DRGs. Unilateral L3/4 DRGs were pooled together. n = 6 to 8 mice per group. One-way ANOVA followed by Tukey post hoc test, F4,16 = 13.249 for Oprm1 mRNA, F4,16 = 14.464 for Kcna2 mRNA, F4,19 = 12.658 for MOR protein, and F4,19 = 11.820 for Kv1.2 protein. *P < 0.05 or **P < 0.01 compared to the corresponding vehicle plus sham group; #P < 0.05 compared to the corresponding vehicle plus CCI group. (C and D) Oprm1 and Kcna2 mRNA expression (C) and MOR and Kv1.2 protein abundance (D) in the injected L3/4 DRGs 8 weeks after microinjection of AAV5-C/EBPβ or control AAV5-EGFP. n = 6 to 8 mice per group. Two-tailed, independent Student’s t test, *P < 0.05 compared to the corresponding AAV5-EGFP group. (E and F) Coexpression of Cebpb mRNA with Oprm1 mRNA (E) or Kcna2 mRNA (F) in small, medium, and large DRG neurons. Gapdh mRNA was used as a positive control. n = 3 biological replicates per gene. (G and H) The abundance of C/EBPβ protein, G9a’s two protein isoforms, and H3K9me2 protein (G) and MOR and Kv1.2 protein (H) in cultured DRG neurons from adult wild-type (WT) (G9afl/fl mice) or G9a conditional knockout (KO) mice. The cultured neurons were collected 2 days after they were transduced with AAV5-EGFP or AAV5-C/EBPβ. n = 3 biological replicates per group. One-way ANOVA with Tukey post hoc test, F3,11 = 10.206 for C/EBPβ, F3,11 = 51.332 for G9a-L, F3,11 = 38.517 for G9a-S, F3,11 = 52.955 for H3K9me2, F3,11 = 27.503 for MOR, and F3,11 = 30.199 for Kv1.2. *P < 0.05 or **P < 0.01 compared to the corresponding AAV5-EGFP–treated WT mice; ##P < 0.01 compared to the corresponding the AAV5-C/EBPβ–treated WT mice. (I) Venn diagram of genes from microarray analysis of the DRG in the mice that overexpressed Ehmt2 (11) and from RNA sequencing data of the injured DRG in a neuropathic pain mouse model treated with a G9a inhibitor (21). About 254 genes overlapped between these two databases.
Transfection of AAV5-C/EBPβ but not AAV5-EGFP in cultured DRG neurons from wild-type (G9afl/fi) mice resulted in increased abundance of C/EBPβ protein, G9a’s two protein isoforms, and H3K9me2 and in a decrease in MOR and Kv1.2 protein abundance (Fig. 6, G and H). In contrast, these decreases were absent in cultured DRG neurons from Ehmt2 knockout mice transfected with AAV5-C/EBPβ (Fig. 6, G and H). In agreement with the previous reports (11, 12), Ehmt2 knockout increased the basal abundance of MOR and Kv1.2 proteins in cultured DRG neurons transfected with AAV5-EGFP (Fig. 6, G and H). Combined with previous reports (11, 12), our findings indicate that C/EBPβ-triggered Ehmt2 gene activation participates in epigenetic silencing of Oprm1 and Kcna2 genes in the ipsilateral DRG neurons under neuropathic pain conditions.
In addition to Oprm1 and Kcna2 genes, C/EBPβ-triggered Ehmt2 gene activation might be involved in other changes in gene expression in the DRG under neuropathic pain conditions. We compared our previous microarray data from the DRG in the mice that overexpressed Ehmt2 (11) with RNA sequencing data from the injured DRG in a neuropathic pain mouse model treated with the G9a inhibitor UNC0638 (21). About 5519 genes were affected by Ehmt2 overexpression in the DRG (11), whereas about 638 genes that were in association with SNL could be normalized by UNC0638 (21). We found that about 254 genes including Kcna2 and Oprm1 were found in both databases (Fig. 6I and table S1). These genes could be regulated by C/EBPβ through its transcriptional activation of Ehmt2 in the ipsilateral DRG after peripheral nerve injury.
Blocking the increase in C/EBPβ in the DRG improves morphine analgesia after CCI
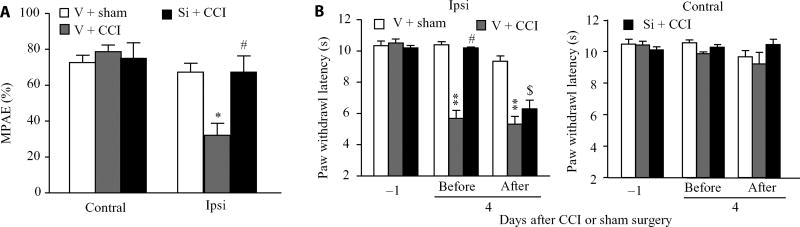
Because blocking the CCI-induced increase in C/EBPβ restored MOR abundance in the ipsilateral DRG, we questioned whether this blockade improved morphine analgesia. Consistent with previous studies (19, 22, 23), morphine analgesia on day 3 after CCI significantly declined compared to that on day 3 post-sham surgery on the ipsilateral side of the vehicle-injected group (Fig. 7A). This reduction was rescued on the ipsilateral side of the Cebpb siRNA–injected group (Fig. 7A). As expected, morphine produced marked analgesia on the contralateral side of all treated groups (Fig. 7A). Cebpb siRNA–injected mice exhibited the complete attenuation of CCI-induced decreases in paw withdrawal latency to heat stimulation on the ipsilateral side before intraperitoneal methylnaltrexone injection (Fig. 7B). However, this effect was significantly reduced 30 min after intraperitoneal methylnaltrexone administration (Fig. 7B). Methylnaltrexone at the dose used did not alter the CCI-induced decrease in paw withdrawal latency to heat stimulation on the ipsilateral side of the vehicle-injected CCI mice and did not affect basal paw responses on the ipsilateral side of the vehicle-injected sham mice and on the contralateral side of all treated mice (Fig. 7B). These findings provide further evidence for the rescue of MOR abundance after C/EBPβ knockdown in the ipsilateral DRG of CCI mice, implicating MOR in the DRG in the antinociceptive effect of C/EBPβ knockdown under neuropathic pain conditions.
Fig. 7. Blocking CCI-induced increase in C/EBPβ in the DRG improves MOR-mediated analgesia under CCI-induced neuropathic pain conditions.
(A) Effect of pre-microinjection of Cebpb siRNA or vehicle into the ipsilateral DRG on morphine (1 mg/kg, subcutaneously) analgesia on the ipsilateral side 3 days after CCI. n = 9 mice per group. One-way ANOVA with Tukey post hoc test, F2,26 = 7.833 on the ipsilateral side and F2,26 = 0.969 on the contralateral side. *P < 0.05 compared to the corresponding vehicle plus sham group. #P < 0.01 compared to the corresponding vehicle plus CCI group. MPAE, maximal possible analgesic effect. (B) Effect of intraperitoneal injection with methylnaltrexone (2 mg/kg) on the Cebpb siRNA–produced antinociceptive effect on day 4 after CCI on the ipsilateral side of the Cebpb siRNA–treated group. Paw withdrawal latency to thermal stimulation was measured before and 30 min after drug administration on day 4 after CCI or sham surgery. n = 9 mice per group. Two-way ANOVA with Tukey post hoc test, F2,80 = 109.908 on the ipsilateral side and F2,80 = 0.718 on the contralateral side. **P < 0.01 compared to the corresponding vehicle plus sham group −1 day after CCI or sham surgery. #P < 0.05 compared to the vehicle plus CCI group before drug administration on day 4 after CCI or sham surgery. $P < 0.05 compared to the Cebpb siRNA plus CCI group before drug administration on day 4 after CCI or sham surgery.
DISCUSSION
Peripheral nerve injury caused by CCI leads to persistent pain hypersensitivities including spontaneous pain, mechanical allodynia, thermal hyperalgesia, and cold allodynia in a preclinical rat model, which mimics trauma-induced neuropathic pain in the clinic. Investigating how CCI produces nociceptive hypersensitivity may open new avenues to manage neuropathic pain. Here, we report that C/EBPβ was required for the induction and maintenance of CCI-induced neuropathic pain through suppressing the G9a-mediated increase in MOR and Kv1.2 abundance in the ipsilateral DRG. Thus, C/EBPβ may be a potential target for therapeutic treatment of peripheral neuropathic pain.
The abundance of the transcription factor C/EBPβ, like that of other transcription factors (such as myeloid zinc finger 1) and non–transcription factor proteins (such as G9a) (8, 11, 12), can be regulated after peripheral nerve injury. We demonstrated that CCI led to an increase of C/EBPβ at both mRNA and protein levels in the ipsilateral DRG, suggesting activation of Cebpb gene transcription under neuropathic pain conditions. How CCI causes an increase in Cebpb mRNA in the DRG is still unclear, but this increase is likely triggered by other transcription factors and/or caused by epigenetic modifications or increases in RNA stability.
To examine the role of C/EBPβ in the DRG in neuropathic pain, we used Cebpb siRNA, which did not alter locomotor activity and basal responses to mechanical, thermal, and cold stimuli, suggesting the specificity and selectivity of Cebpb siRNA effects. Unexpectedly, microinjection of Cebpb siRNA into the DRG did not markedly affect the basal amount C/EBPβ protein in the sham group, although this siRNA was effective in cultured DRG neurons. Why Cebpb siRNA did not affect basal C/EBPβ abundance in vivo is unclear but could be because of its low abundance in the DRG under normal conditions. In addition, the remaining Cebpb mRNA after its knockdown may have a high translational efficacy, and thus, the normal amount of basal C/EBPβ protein was maintained in the injected DRG of the sham group.
The increased C/EBPβ in the ipsilateral DRG could participate in the induction and maintenance of CCI-induced pain hypersensitivity by triggering Ehmt2 gene transcription in the ipsilateral DRG. The histone methyltransferase G9a dimethylates histone H3 at Lys9 (H3K9me2), resulting in condensed chromatin and gene transcription repression (24, 25). Ehmt2 mRNA and G9a protein are required for nerve injury–induced suppression of several potassium channel encoding genes (such as Kcna2) and opioid receptor encoding genes in the ipsilateral DRG (11–13, 21). Because the suppression of these genes contributes to neuropathic pain genesis (7, 8, 10, 19), the increase in G9a abundance in the DRG is considered as an endogenous instigator of neuropathic pain (11, 21). We further revealed that C/EBPβ triggered the transcriptional activation of Ehmt2 in the DRG after CCI, resulting in increases in G9a’s two isoform proteins and H3K9me2 and in the decreases in Kcna2 and Oprm1 mRNAs and their proteins in the ipsilateral DRG. Thus, the antihyperalgesic effect caused by blocking the CCI-induced increase in C/EBPβ in the ipsilateral DRG may result from the failure to transcriptionally activate Ehmt2 in the ipsilateral DRG after CCI. Without the increase in G9a protein abundance, no changes would occur in Kcna2 and Oprm1 mRNA expression, their proteins and total Kv current, and neuronal excitability in the ipsilateral DRG neurons and central sensitization in the dorsal horn. Consistent with this conclusion, blocking the CCI-induced increase of C/EBPβ in the ipsilateral DRG attenuated the CCI-induced increases in markers for central sensitization in the ipsilateral L3/4 dorsal horn and pain hypersensitivity. Moreover, the antihyperalgesic effect caused by blocking-increased DRG C/EBPβ on CCI-induced neuropathic pain was diminished by antagonizing peripheral MOR. We also demonstrated that C/EBPβ did not directly affect MOR and Kv1.2 abundance because the down-regulation of Kv1.2 and MOR caused by C/EBPβ overexpression disappeared in the absence of G9a in DRG neurons. Together, our findings suggest that DRG C/EBPβ contributes to neuropathic pain induction and maintenance through epigenetic silencing of Kv1.2 and MOR caused by C/EBPβ-triggered transcriptional activation of the Ehmt2 gene in the ipsilateral DRG. However, other potential mechanisms of C/EBPβ involvement in neuropathic pain cannot be excluded. After comparing microarray data from the DRG in the mice that overexpressed Ehmt2 (11) with RNA sequencing data from the injured DRG in a neuropathic pain mouse model treated with a G9a inhibitor (21), we found that the expression of about 252 genes appear to be controlled by G9a in the ipsilateral DRG under neuropathic pain conditions. In addition, C/EBPβ as a transcription factor may target some genes that are not affected by G9a in the ipsilateral DRG after peripheral nerve injury (26, 27).
Other C/EBPβ-independent mechanisms are involved in the nerve injury–induced suppression of Kcna2 and Oprm1 in the ipsilateral DRG, such as transcriptional activation of Kcna2 antisense RNA by myeloid zinc finger protein 1 (7, 8, 10). We have shown that DNA methyltransferase 3a (DNMT3a)–induced de novo DNA methylation in the promoters and 5′ untranslated regions of the Kcna2 and Oprm1 genes is crucial in nerve injury–induced silencing of these genes in the ipsilateral DRG (28, 29). It appears that these mechanisms do not compensate for each other, but how these mechanisms work together to regulate Kv1.2 and MOR abundance and whether they interact with or affect each other after peripheral nerve injury remain to be further studied.
In summary, we demonstrated that blocking the CCI-induced increase in C/EBPβ abundance in the DRG alleviated CCI-induced pain hypersensitivity without altering basal or acute nociceptive responses and locomotor functions. C/EBPβ promoted the G9a-mediated epigenetic silencing of Kcna2 and Oprm1 in the ipsilateral DRG. Because MOR and Kv1.2 are key players in neuropathic pain initiation and maintenance (30), C/EBPβ may be an endogenous initiator of neuropathic pain and could be a potential target for the prevention and treatment of this disorder.
MATERIALS AND METHODS
Animal preparation
Adult male CD-1 mice, aged 7 to 8 weeks, were purchased from Charles River Laboratories. G9afl/fl mice (provided by E. J. Nestler, Icahn School of Medicine at Mount Sinai, New York, NY) were fully backcrossed to C57BL/6J mice and were homozygous for a floxed G9a allele. Male AvilCre/+ mice (provided by F. Wang, Duke University Medical Center, Durham, NC) were crossed with G9afl/fl mice to obtain G9a conditional knockout mice (that is, Ehmt2 knockout mice). All animals were housed under a standard 12-hour light/dark cycle, with water and food pellets available ad libitum. All animal procedures were conducted in strict accordance with the Animal Care and Use Committee at New Jersey Medical School and were consistent with the ethical guidelines of the National Institutes of Health (NIH) and the International Association for the Study of Pain. All efforts were made to minimize animal suffering and the number of animals used. All experimenters were blind to treatment condition.
DRG microinjection
DRG microinjection was carried out as described with minor modifications (7, 8, 10). Briefly, a midline incision was performed in the lower lumbar back region and then L3/4 DRGs were exposed. Viral solution (1 µl, 4 × 1012) or siRNA solution (1 µl, 20 µM) was injected into the unilateral L3/4 DRGs with the use of a glass micropipette connected to a Hamilton syringe. After each injection, a 10-min pipette retention was used before the glass pipette was removed. After injection, the surgical field was irrigated with sterile saline, and the skin incision was closed with wound clips. Mice showing abnormal locomotor activities were excluded from the experiment.
For siRNA injection, Cebpb siRNA (catalog no. sc-29862) and its negative control siRNA (catalog no. sc-44230) were purchased from Santa Cruz Biotechnology. To improve delivery and prevent degeneration of siRNA, we used TurboFect in vivo transfection reagent (Thermo Fisher Scientific) as a delivery vehicle as described (7, 8, 10). For viral injection, the pcDNA3.1(−) plasmid harboring mouse full-length Cebpb gene was provided by X. Li (Fudan University, Shanghai, China) (16). After digestion with XbaI/BamHI, full-length Cebpb cDNA was gel-purified and ligated into proviral plasmids. The resulting plasmid expressed C/EBPβ under the control of the cytomegalovirus promoter. The plasmid expressing EGFP was used as a control. Recombinant AAV5 viral particles carrying these two cDNAs were produced at the UNC Vector Core (Chapel Hill, NC).
CCI model
The CCI-induced neuropathic pain model was carried out with minor modification, as described previously (8). Briefly, mice were placed under anesthesia with isoflurane. The sciatic nerve was exposed and loosely ligated with 7-0 silk thread at four sites with an interval of about 1 mm proximal to trifurcation of the sciatic nerve. Sham animals received an identical surgery but without the ligation.
Behavioral testing
All mice were habituated 1 to 2 hours every day for 2 to 3 days before basal behavioral testing. Behavioral testing including mechanical, thermal, and cold tests was carried out in order with 1-hour intervals. CPP test was performed at the seventh week after viral injection. Locomotor functional testing was carried out after all behavioral tests described above were done.
Paw withdrawal thresholds in response to mechanical stimuli were measured with two calibrated von Frey filaments (0.07 and 0.4 g, Stoelting Co.), as described previously (31, 32). Briefly, mice were placed in a Plexiglas chamber on an elevated mesh screen and allowed to habituate for 30 min. Each von Frey filament was applied to the plantar sides of both hindpaws for 10 times. A quick withdrawal of the paw was regarded as a positive response. The amount of positive responses of 10 stimulations was recorded as percentage withdrawal frequency [(number of paw withdrawals/10 trials) × 100 = %response frequency].
Paw withdrawal latencies in response to noxious heat stimuli were examined as described (31, 32). Briefly, mice were placed in a Plexiglas chamber on a glass plate. The heat stimuli were performed with a Model 336 Analgesic Meter (IITC Life Science Inc.). A beam of light was emitted from a hole of the light box and applied on the middle of the plantar surface of each hindpaw. A quick lift of the hindpaw was regarded as a signal to turn off the light. The length of time between the start and the stop of the light beam was defined as the paw withdrawal latency. For each side, five trials with 5-min intervals were carried out. A cutoff time of 20 s was used to avoid tissue damage to the hindpaw.
Paw withdrawal latencies to noxious cold (0°C) were measured with a cold aluminum plate, the temperature of which was monitored continuously by a thermometer as described (7, 8). Each mouse was placed in a Plexiglas chamber on the cold aluminum plate. The length of time between the placement of the hindpaw on the plate and the sign of mouse paw jumping was defined as the paw jumping latency. Each trial was repeated three times at 10-min intervals for the paw on the ipsilateral side. A cutoff time of 20 s was used to avoid tissue damage.
CPP test was carried out as described with minor modification (33). Briefly, an apparatus with two Plexiglas chambers connected with an internal door (Med Associates Inc.) was used. One of the chambers was made of rough floor and walls with black and white horizontal stripes, and another one was composed of a smooth floor and walls with black and white vertical stripes. Movement of the mice and time spent in each chamber were monitored by photobeam detectors installed along the chamber walls and automatically recorded in MED-PC IV CPP software. Mice were first preconditioned for 30 min with full access to two chambers to habituate them to the environment. At the end of the preconditioning phase, the basal duration time spent in each chamber was recorded within 15 min to check whether mice had a preexisting chamber bias. Animals spending more than 80% or less than 20% of the total time in any chamber were excluded from further testing. The conditioning protocol was performed for the following 3 days with the internal door closed. The mice first received intrathecal injection of saline (5 µl) specifically paired with one conditioning chamber in the morning. Six hours later, lidocaine (0.8 % in 5 µl of saline) was given intrathecally paired with the opposite conditioning chamber in the afternoon. Lidocaine at this dosage did not affect motor function (Table 1). Injection order of saline and lidocaine every day was switched. On the test day (at least 20 hours later) after the conditioning, the mice were placed in one chamber with free access to both chambers. The duration of time that each mouse spent in each chamber was recorded for 15 min. Difference scores were calculated as test time-preconditioning time spent in the lidocaine chamber.
Locomotor function was tested as previously described (32). Three reflex tests were carried out as follows: To test the placing reflex, we held the mouse with the hindlimbs slightly lower than the forelimbs and brought the dorsal surfaces of the hindpaws into contact with the edge of a table. The experimenter recorded whether the hindpaws were placed on the table surface reflexively. To test the grasping reflex, we placed the mouse on a wire grid; the experimenter recorded whether the hindpaws grasped the wire on contact. To test the righting reflex, we placed the mouse on its back on a flat surface; the experimenter noted whether it immediately assumed the normal upright position. Scores for placing, grasping, and righting reflexes were based on the counts of each normal reflex exhibited in five trials.
Morphine-induced analgesia
Morphine-induced analgesia was carried out as previously described (31, 34). Briefly, morphine (1 mg/kg; West-Ward Pharmaceuticals) was given subcutaneously on day 3 after CCI or sham surgery. In addition, methylnaltrexone bromide (a peripheral MOR antagonist) (2 mg/kg; dissolved in saline, MedChemExpress) or saline was intraperitoneally injected on day 4 after CCI or sham surgery. Paw withdrawal latencies in response to noxious heat stimuli (see above) were examined before surgery and 20 min after drug injection. Maximal possible analgesic effect was calculated as follows: [(final paw withdrawal latency − baseline paw withdrawal latency)/(cutoff time − baseline paw withdrawal latency)] × 100%.
Cell culture and transfection
HEK-293T cell cultures and DRG neuronal cultures were prepared according to previously described methods (10, 32). Briefly, HEK-293T cells were cultured in Dulbecco’s modified Eagle’s medium/high glucose medium (Gibco/Thermo Fisher Scientific) containing 10% fetal bovine serum (FBS) and 1% antibiotics. DRGs from adult mice were collected in ice-cold Neurobasal medium (Life Technologies) with 10% FBS (J R Scientific) and penicillin (100 U/ml) and streptomycin (100 µg/ml) (Quality Biological) and then digested with enzyme solution [dispase (5 mg/ml) and collagenase type I (1 mg/ml)] in Hanks’ balanced salt solution without Ca2+ and Mg2+ (Life Technologies) at 37°C for 20 to 25 min. After trituration and centrifugation, the dissociated neurons were resuspended in mixed Neurobasal medium and plated into six-well plates coated with poly-d-lysine (50 µg/ml) (Sigma) at a density of 1.5 × 105 to 4 × 105 cells. The neurons were incubated in a humidified 95% O2 and 5% CO2 atmosphere at 37°C. For viral infection, 2 µl of AAV5 virus (titer ≥ 1 × 1012/ml) was added to each 2-ml well after 24 hours of incubation. For siRNA transfection, siRNA was diluted to the concentration of 100 nM and transfected to cultured cells by Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instruction. Cells were collected 2 or 3 days later for Western blot analysis.
Western blot analysis
Protein extraction and Western blot analysis were carried out as described (8, 10). Briefly, the bilateral L3/4 DRGs and L3/4 spinal cords were harvested because the mouse sciatic nerve originates mainly from the L3/4 spinal nerve as previously described (35). The tissues were homogenized in lysis buffer (10 mM tris, 1 mM phenylmethylsulfonyl fluoride, 5 mM MgCl2, 5 mM EGTA, 1 mM EDTA, 1 mM dithiothreitol, 40 µM leupeptin, 250 mM sucrose). After centrifugation at 4°C for 15 min at 1000g, the supernatant was collected for cytosolic proteins, and the pellet was collected for nuclear proteins. The pellet was dissolved in lysis buffer plus 2% SDS and 0.1% Triton X-100 (SDS lysis buffer) and then sonicated on ice. The samples from the cultured neurons were directly dispersed in lysis buffer, centrifuged, and sonicated. After protein concentration was measured, the samples were heated at 99°C for 5 min and loaded onto a 4 to 15% stacking/7.5% separating SDS–polyacrylamide gel (Bio-Rad Laboratories). The proteins were then electrophoretically transferred onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories). After being blocked with 3% nonfat milk in tris-buffered saline containing 0.1% Tween 20 for 2 hours, the membranes were then incubated at 4°C overnight with the following antibodies: mouse anti-Kv1.2 (1:200, NeuroMab), rabbit anti-MOR (1:500, Neuromics), rabbit anti-GAPDH (1:1000, Sigma), rabbit anti-C/EBPβ (1:200, Santa Cruz Biotechnology), rabbit anti-G9a (1:1000, Cell Signaling Technology), rabbit anti-H3K9me2 (1:500, EMD Millipore), rabbit anti-OCT1 (1:1000, Abcam), rabbit anti-mTOR (1:1000, Cell Signaling Technology), rabbit anti–phospho-ERK1/2 (Thr202/Tyr204; 1:1000, Cell Signaling Technology), rabbit anti-ERK1/2 (1:1000, Cell Signaling Technology), mouse anti-GFAP (1:1000, Cell Signaling Technology), and rabbit anti–histone H3 (1:1000, Cell Signaling Technology). The proteins were detected by horseradish peroxidase–conjugated anti-rabbit or anti-mouse secondary antibody (1:3000, Bio-Rad Laboratories) and visualized by chemiluminescence reagent (enhanced chemiluminescence, Bio-Rad Laboratories). Images were generated using ChemiDoc XRS System with Image Lab software (Bio-Rad Laboratories). Band intensities were quantified with densitometry by System with Image Lab software (Bio-Rad Laboratories). Band intensities for nuclear proteins were normalized to total histone H3, and those for cytosol proteins were normalized to GAPDH.
Quantitative real-time RT-PCR
Total RNA extraction and quantitative real-time RT-PCR were carried out as described (7, 8, 10). Briefly, unilateral L3/4 DRGs from two adult mice were collected rapidly and pooled together to achieve enough RNA. Total RNA was extracted by TRIzol-chloroform methods (Invitrogen), treated with an overdose of deoxyribonuclease I (New England Biolabs), and reverse-transcribed with the ThermoScript Reverse Transcriptase (Invitrogen/Thermo Fisher Scientific) and oligo(dT) primers (Invitrogen/Thermo Fisher Scientific) or specific RT primers. Template (4 µl) was amplified in a Bio-Rad CFX96 real-time PCR system by using the primers listed in table S2. Each sample was run in triplicate in a 20-µl reaction volume containing 250 nM forward and reverse primers, 10 µl of SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories), and 20 ng of cDNA. The PCR amplification consisted of 30 s at 95°C, 30 s at 60°C, 30 s at 72°C, and 5 min at 72°C for 39 cycles. Ratios of ipsilateral-side mRNA levels to contralateral-side mRNA levels were calculated using the ΔCt method (2−ΔΔCt). All data were normalized to tuba1a mRNA, which was stable even after peripheral nerve injury insult in mice as shown in our pilot study.
For single-cell RT-PCR, freshly dissociated mouse DRG neurons from adult mice were first prepared as described previously (8, 10). Briefly, 4 hours after plating, the diameter of a single living DRG neuron was first measured under an inverted microscope, and the cell collected by using a glass micropipette fit was measured with a micromanipulator and micro-injector. Each cell was placed in a PCR tube with 5 to 10 µl of cell lysis buffer (Signosis). After incubation on ice for 10 min and then centrifugation at 4°C for 2 min, the supernatants were collected and used as the template for RT. The remaining real-time RT-PCR procedure was carried out according to the manufacturer’s instructions with the single-cell real-time RT-PCR assay kit (Signosis). Primers are listed in table S2.
In situ hybridization histochemistry
ISHH was carried out as described previously (10). Briefly, after mice were anesthetized with isoflurane, they were perfused with 50 to 80 ml of 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4). Bilateral L3/4 DRGs were harvested for the double labeling of Cebpb mRNA ISHH with DRG marker immunohistochemistry from naïve mice. Five sets of 20-µm sections were collected from each DRG by grouping every sixth section (one to two sections per set per DRG), for a total of four to eight sections per set per mouse. The ipsilateral L3/4 DRGs to label Cebpb mRNA through ISHH and/or ATF3 through immunohistochemistry were harvested from CCI and sham mice on day 7 after CCI or sham surgery. Two sets of 20-µm sections were collected from each DRG by grouping every third section (three to four sections per set per DRG), for a total of six to eight sections per set per mouse. Cebpb complementary RNA (cRNA) probe (0.6 kb) were prepared by in vitro transcription and labeled with digoxigenin–deoxyuridine triphosphate (dUTP) according to the manufacturer’s instructions (Roche Diagnostics). Primers are listed in table S2. After treatment of proteinase K (25 µg/ml) and prehybridization, the sections were hybridized with digoxigenin-dUTP-labeled Cebpb cRNA probe (1 to 2 ng/µl) at 62°C overnight. After washing, sections were incubated with alkaline phosphatase (AP)–conjugated anti-digoxigenin antibody (1:500) overnight. Fluorescent signals were developed with Fast Red (5 mg/ml) per 1 ml of detection buffer (filtered through 0.2-µm nylon filter) for 30 min. Slides were rinsed in phosphate-buffered saline (PBS) and mounted with Fluoromount (Southern Biotech). For fluorescence in situ hybridization combined with immunofluorescence, protocols before blocking with PBS with Tween 20 (PBT) containing 20% lamb serum were similar to single in situ hybridization. Sections were incubated with AP-conjugated anti-digoxigenin (1:500, Roche) and chicken anti-β III tubulin (1:1000, EMD Millipore), rabbit anti-glutamine synthetase (1:10000, Sigma-Aldrich), rabbit anti-NF200 (1:100, Sigma), biotinylated IB4 (1:100, Sigma), or mouse anti-CGRP (1:50, Abcam) at 4°C overnight in 20% lamb serum blocking solution. After washing in PBT, slides were incubated in species-appropriate fluorescein isothiocyanate (FITC) or Cy2-conjugated secondary antibody (1:500 in 5% lamb serum in PBT) for 1 hour at room temperature. HNPP (2-hydroxy-3-naphthoic acid-2′-phenylanilide phosphate)/Fast Red detection was then developed for 30 min, and slides were mounted after washing in PBS.
Immunohistochemistry
Immunohistochemistry was carried out as previously described (10, 11, 29, 34, 36). Briefly, after being blocked for 1 hour at 37°C in PBS containing 10% lamb serum and 0.3% Triton X-100, five sets of sections from naïve mice were incubated with chicken anti-β III tubulin (1:1000, EMD Millipore), rabbit anti-glutamine synthetase (1:10,000, Sigma-Aldrich), rabbit anti-NF200 (1:100, Sigma), biotinylated IB4 (1:100, Sigma), and mouse anti-CGRP (1:50, Abcam) at 4°C overnight. One set of sections from CCI mice was incubated with rabbit anti-ATF3 (1:200, Sigma) at 4°C overnight. Sections were incubated with species-appropriate Cy2-conjugated secondary antibody (1:500, Jackson ImmunoResearch) or with FITC-labeled Avidin D (1:200, Sigma) for 1 hour at room temperature. Images were taken with a Leica DMI4000 fluorescence microscope (Leica) with DFC365 FX camera (Leica) and analyzed using NIH ImageJ software.
ChIP assay
ChIP assays were conducted using the EZ-ChIP kit (Upstate/EMD Millipore) as described (10). The homogenization solution from DRGs was cross-linked with 1% formaldehyde for 10 min at room temperature. The reaction was terminated by the addition of 0.25 M glycine. After centrifugation, the collected pellet was lysed by SDS lysis buffer with protease inhibitor cocktail and sonicated until the DNA was broken into fragments with a mean length of 200 to 1000 base pairs. After the samples were precleaned with protein G–agarose, they were subjected to immunoprecipitation with 2 µg of rabbit antibodies against C/EBPβ (Abcam) or with 2 µg of normal rabbit serum at 4°C overnight. Input (10 to 20% of the sample for immunoprecipitation) was used as a positive control. The DNA fragments were purified and identified using PCR/real-time PCR with the primers in table S2.
Statistical analysis
All data were presented as means ± SEM. The data were statistically analyzed with two-tailed, independent Student’s t test and a one- or two-way ANOVA. When ANOVA showed a significant difference, pairwise comparisons between means were tested by the post hoc Tukey method (SigmaPlot 12.5). P < 0.05 was considered statistically significant in all analyses.
Supplementary Material
Fig. S1. Changes in Cebpb mRNA expression and C/EBPβ protein abundance in the DRG and spinal cord after sham surgery.
Fig. S2. Colocalization of Cebpb mRNA with ATF3 in DRG neurons.
Table S1. List of 254 overlapping genes from microarray and RNA sequencing databases.
Table S2. Primer sequences.
Acknowledgments
We thank X. Li (Fudan University, Shanghai, China) for providing mouse full-length Cebpb plasmid, E. J. Nestler (Icahn School of Medicine at Mount Sinai, New York, NY) for providing the G9afl/fl mice, and F. Wang for providing AvilCre/+ mice (Duke University Medical Center, Durham, NC).
Funding: This work was supported by the NIH (grants R01NS094664, R01NS094224, and R01DA033390 to Y.-X.T.) and by the National Natural Science Foundation of China [81571082 to W.Z. and 81671094 to Z.L.].
Footnotes
Author contributions: Y.-X.T. and W.Z. conceived the project and supervised all experiments. Z.L., Y.M., L.L., W.Z., and Y.-X.T. designed the project. Z.L., Y.M., L.L., S.W., J.Y., K.M., W.C., Q.M., and J.C. performed molecular, biochemical, and behavioral experiments. Z.L., Y.M., L.L., S.W., A.B., W.Z., and Y.-X.T. analyzed the data. Y.-X.T. wrote the manuscript. All authors read and discussed the manuscript.
Competing interests: The authors declare that they have no competing interests.
References
- 1.Campbell JN, Meyer RA. Mechanisms of neuropathic pain. Neuron. 2006;52:77–92. doi: 10.1016/j.neuron.2006.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vorobeychik Y, Gordin V, Mao J, Chen L. Combination therapy for neuropathic pain: A review of current evidence. CNS Drugs. 2011;25:1023–1034. doi: 10.2165/11596280-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 3.Mao J, Gold MS, Backonja MM. Combination drug therapy for chronic pain: A call for more clinical studies. J. Pain. 2011;12:157–166. doi: 10.1016/j.jpain.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costigan M, Scholz J, Woolf CJ. Neuropathic pain: A maladaptive response of the nervous system to damage. Annu. Rev. Neurosci. 2009;32:1–32. doi: 10.1146/annurev.neuro.051508.135531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung JM, Chung K. Importance of hyperexcitability of DRG neurons in neuropathic pain. Pain Pract. 2002;2:87–97. doi: 10.1046/j.1533-2500.2002.02011.x. [DOI] [PubMed] [Google Scholar]
- 6.Devor M. Ectopic discharge in Aβ afferents as a source of neuropathic pain. Exp. Brain Res. 2009;196:115–128. doi: 10.1007/s00221-009-1724-6. [DOI] [PubMed] [Google Scholar]
- 7.Fan L, Guan X, Wang W, Zhao J-Y, Zhang H, Tiwari V, Hoffman PN, Li M, Tao Y-X. Impaired neuropathic pain and preserved acute pain in rats overexpressing voltage-gated potassium channel subunit Kv1.2 in primary afferent neurons. Mol. Pain. 2014;10:8. doi: 10.1186/1744-8069-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Gu X, Sun L, Wu S, Liang L, Cao J, Lutz BM, Bekker A, Zhang W, Tao Y-X. Dorsal root ganglion myeloid zinc finger protein 1 contributes to neuropathic pain after peripheral nerve trauma. Pain. 2015;156:711–721. doi: 10.1097/j.pain.0000000000000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasband MN, Park EW, Vanderah TW, Lai J, Porreca F, Trimmer JS. Distinct potassium channels on pain-sensing neurons. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13373–13378. doi: 10.1073/pnas.231376298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao J-Y, Liang L, Wang W, Guan X, Kao S-C, Tiwari V, Gao Y-J, Hoffman PN, Cui H, Li M, Dong X, Tao Y-X. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat. Neurosci. 2013;16:1024–1031. doi: 10.1038/nn.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang L, Gu X, Zhao JY, Wu S, Miao X, Xiao J, Mo K, Zhang J, Lutz BM, Bekker A, Tao YX. G9a participates in nerve injury-induced Kcna2 downregulation in primary sensory neurons. Sci. Rep. 2016;6:37704. doi: 10.1038/srep37704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang L, Zhao J-Y, Gu X, Wu S, Mo K, Xiong M, Bekker A, Tao Y-X. G9a inhibits CREB-triggered expression of mu opioid receptor in primary sensory neurons following peripheral nerve injury. Mol. Pain. 2016;12 doi: 10.1177/1744806916682242. 1744806916682242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Chen S-R, Laumet G, Chen H, Pan H-L. Nerve injury diminishes opioid analgesia through lysine methyltransferase-mediated transcriptional repression of μ-opioid receptors in primary sensory neurons. J. Biol. Chem. 2016;291:8475–8485. doi: 10.1074/jbc.M115.711812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taubenfeld SM, Milekic MH, Monti B, Alberini CM. The consolidation of new but not reactivated memory requires hippocampal C/EBPβ. Nat. Neurosci. 2001;4:813–818. doi: 10.1038/90520. [DOI] [PubMed] [Google Scholar]
- 15.Pulido-Salgado M, Vidal-Taboada JM, Saura J. C/EBPβ and C/EBPδ transcription factors: Basic biology and roles in the CNS. Prog. Neurobiol. 2015;132:1–33. doi: 10.1016/j.pneurobio.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Li S-F, Guo L, Qian S-W, Liu Y, Zhang Y-Y, Zhang Z-C, Zhao Y, Shou J-Y, Tang Q-Q, Li X. G9a is transactivated by C/EBPβ to facilitate mitotic clonal expansion during 3T3-L1 preadipocyte differentiation. Am. J. Physiol. Endocrinol. Metab. 2013;304:E990–E998. doi: 10.1152/ajpendo.00608.2012. [DOI] [PubMed] [Google Scholar]
- 17.Bennett GJ, Xie Y-K. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain. 1988;33:87–107. doi: 10.1016/0304-3959(88)90209-6. [DOI] [PubMed] [Google Scholar]
- 18.Latremoliere A, Woolf CJ. Central sensitization: A generator of pain hypersensitivity by central neural plasticity. J. Pain. 2009;10:895–926. doi: 10.1016/j.jpain.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Liang L, Miao X, Wu S, Cao J, Tao B, Mao Q, Mo K, Xiong M, Lutz BM, Bekker A, Tao Y-X. Contribution of the suppressor of variegation 3–9 homolog 1 in dorsal root ganglia and spinal cord dorsal horn to nerve injury-induced nociceptive hypersensitivity. Anesthesiology. 2016;125:765–778. doi: 10.1097/ALN.0000000000001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang Z-Y, Gerner P, Woolf CJ, Ji R-R. ERK is sequentially activated in neurons, microglia, and astrocytes by spinal nerve ligation and contributes to mechanical allodynia in this neuropathic pain model. Pain. 2005;114:149–159. doi: 10.1016/j.pain.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 21.Laumet G, Garriga J, Chen S-R, Zhang Y, Li D-P, Smith TM, Dong Y, Jelinek J, Cesaroni M, Issa J-P, Pan H-L. G9a is essential for epigenetic silencing of K+ channel genes in acute-to-chronic pain transition. Nat. Neurosci. 2015;18:1746–1755. doi: 10.1038/nn.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rashid MH, Inoue M, Toda K, Ueda H. Loss of peripheral morphine analgesia contributes to the reduced effectiveness of systemic morphine in neuropathic pain. J. Pharmacol. Exp. Ther. 2004;309:380–387. doi: 10.1124/jpet.103.060582. [DOI] [PubMed] [Google Scholar]
- 23.Zhou X-L, Yu L-N, Wang Y, Tang L-H, Peng Y-N, Cao J-L, Yan M. Increased methylation of the MOR gene proximal promoter in primary sensory neurons plays a crucial role in the decreased analgesic effect of opioids in neuropathic pain. Mol. Pain. 2014;10:51. doi: 10.1186/1744-8069-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonzheim I, Irmler M, Klier-Richter M, Steinhilber J, Anastasov N, Schäfer S, Adam P, Beckers J, Raffeld M, Fend F, Quintanilla-Martinez L. Identification of C/EBPβ target genes in ALK+ anaplastic large cell lymphoma (ALCL) by gene expression profiling and chromatin immunoprecipitation. PLOS ONE. 2013;8:e64544. doi: 10.1371/journal.pone.0064544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Guo F, Ni Y, Zhao R. LPS-induced inflammation in the chicken is associated with CCAAT/enhancer binding protein beta-mediated fat mass and obesity associated gene down-regulation in the liver but not hypothalamus. BMC Vet. Res. 2013;9:257. doi: 10.1186/1746-6148-9-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun L, Zhao J-Y, Gu X, Liang L, Wu S, Mo K, Feng J, Guo W, Zhang J, Bekker A, Zhao X, Nestler EJ, Tao Y-X. Nerve injury-induced epigenetic silencing of opioid receptors controlled by DNMT3a in primary afferent neurons. Pain. 2017;158:1153–1165. doi: 10.1097/j.pain.0000000000000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J-Y, Liang L, Gu X, Li Z, Wu S, Sun L, Atianjoh FE, Feng J, Mo K, Jia S, Lutz BM, Bekker A, Nestler EJ, Tao Y-X. DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat. Commun. 2017;8:14712. doi: 10.1038/ncomms14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang L, Lutz BM, Bekker A, Tao Y-X. Epigenetic regulation of chronic pain. Epigenomics. 2015;7:235–245. doi: 10.2217/epi.14.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liaw WJ, Zhu XG, Yaster M, Johns RA, Gauda EB, Tao YX. Distinct expression of synaptic NR2A and NR2B in the central nervous system and impaired morphine tolerance and physical dependence in mice deficient in postsynaptic density-93 protein. Mol. Pain. 2008;4:45. doi: 10.1186/1744-8069-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao Y-X, Rumbaugh G, Wang G-D, Petralia RS, Zhao C, Kauer FW, Tao F, Zhuo M, Wenthold RJ, Raja SN, Huganir RL, Bredt DS, Johns RA. Impaired NMDA receptor-mediated postsynaptic function and blunted NMDA receptor-dependent persistent pain in mice lacking postsynaptic density-93 protein. J. Neurosci. 2003;23:6703–6712. doi: 10.1523/JNEUROSCI.23-17-06703.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Y, Tian X, Hu X, Porreca F, Wang ZJ. Negative reinforcement reveals non-evoked ongoing pain in mice with tissue or nerve injury. J. Pain. 2012;13:598–607. doi: 10.1016/j.jpain.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J-T, Zhao J-Y, Zhou X, Zhao X, Ligons D, Tiwari V, Lee C-Y, Atianjoh FE, Liang L, Zang W, Njoku D, Raja SN, Yaster M, Tao Y-X. Opioid receptor-triggered spinal mTORC1 activation contributes to morphine tolerance and hyperalgesia. J. Clin. Invest. 2014;124:592–603. doi: 10.1172/JCI70236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rigaud M, Gemes G, Barabas M-E, Chernoff DI, Abram SE, Stucky CL, Hogan QH. Species and strain differences in rodent sciatic nerve anatomy: Implications for studies of neuropathic pain. Pain. 2008;136:188–201. doi: 10.1016/j.pain.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang L, Fan L, Tao B, Yaster M, Tao Y-X. Protein kinase B/Akt is required for complete Freund’s adjuvant-induced upregulation of Nav1.7 and Nav1.8 in primary sensory neurons. J. Pain. 2013;14:638–647. doi: 10.1016/j.jpain.2013.01.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Changes in Cebpb mRNA expression and C/EBPβ protein abundance in the DRG and spinal cord after sham surgery.
Fig. S2. Colocalization of Cebpb mRNA with ATF3 in DRG neurons.
Table S1. List of 254 overlapping genes from microarray and RNA sequencing databases.
Table S2. Primer sequences.