Abstract
FAM83B (family with sequence similarity 83, member B) seems to emerge as a new class of players involved in the development of a variety of malignant tumors. Yet the molecular mechanisms are not well understood. The present study is intended to investigate the expression and function of FAM83B in pancreatic ductal adenocarcinoma (PDAC). In this study, we found that the expression of FAM83B was significantly increased both in PDAC cell lines and PDAC tumor tissues. FAM83B expression was positively related with advanced clinical stage and poor vital status. Higher FAM83B expression predicted shorter overall survival in PDAC patients, regardless of lymphatic metastasis status and histological differentiation. Actually, FAM83B may act as an independent prognostic indicator as well. What's more, down-regulation of FAM83B in PDAC cells contributed to G0/G1 phase arrest and inhibition of cell proliferation. Finally, a subcutaneous xenograft model indicated that knockdown of FAM83B significantly reduced the tumor volume in vivo. Our findings have provided supporting evidence for the potential molecular biomarker role of FAM83B in PDAC. It's of great interest and broad significance to target FAM83B in PDAC, which may conduce to develop a meaningful and effective strategy in the diagnosis and treatment of PDAC.
Keywords: FAM83B, pancreatic ductal adenocarcinoma, GSEA, prognosis, cell cycle arrest, cell proliferation, tumor growth.
Introduction
There is no doubt that pancreatic cancer is one of the most devastating malignancies worldwide nowadays. It's reported that pancreatic cancer has been the fourth most leading cause of cancer-related death in developed countries in recent years 1-3. Pancreatic ductal adenocarcinoma (PDAC) is the major type of pancreatic cancer. Patients with advanced PDAC have a relatively poor prognosis. Despite of great efforts made in past decades to improve the diagnosis and treatment of PDAC, little success was achieved in improving the 5-year survival rates of patients 4. Difficulty in early detection, absence of early symptoms, distant and vascular metastasis, lack of specific prognostic indicators, resistance to both chemotherapy and radiation and high recurrence rates cooperatively contribute to poor prognosis of PDAC patients 4-9. Therefore, it's of utmost urgency to explore novel and noninvasive prognostic indicators to detect early cancer, reflect an individual's cancer risk, facilitate the treatment of PDAC and bring benefit to PDAC patients.
Family with sequence similarity 83, member B (FAM83B) has been identified as an oncogene able to promote the transformation of immortalized human mammary epithelial cells with the VBIM strategy 10. It has been reported that FAM83B is vitally involved in EGFR/RAS/MAPK signaling and able to hyperactivate PI3K/AKT/mTOR signaling pathway in breast cancer and thus promotes cell proliferation, anchorage-independent growth (AIG) and tumorigenicity 10, 11. Meanwhile, emerging evidence has indicated that FAM83B is significantly elevated in several malignant tumors, such as breast, bladder, lung, thyroid, cervix, ovary and testis cancer, typically resulting in aberrant expression of gene products that contribute to the progression of varieties of human malignant tumors 10, 12. Nevertheless, to date, the role of FAM83B in PDAC remains largely unknown. Therefore, we examined the expression status of FAM83B in PDAC cell lines, tumor tissues and matched normal tissues, analyzed the correlation between FAM83B expression and clinicopathological characteristics and prognosis in PDAC patients, explored the biological mechanism involved in PDAC progression and detected the biological function of FAM83B in PDAC cell cycle, cell proliferation and tumor growth. In one word, we hope to search for an avenue in the pursuit of novel therapies for PDAC aimed at targeting FAM83B.
Materials and Methods
Clinical patient specimen collection
Freshly-frozen PDAC tumor tissues and matched corresponding non-cancerous tissues (Renji Cohort 1) were recruited from 36 patients who underwent surgical resections at Department of Surgery, Renji Hospital Affiliated to Shanghai Jiao Tong University School of Medicine from January 2012 to December 2013.Human PDAC tissues used for microarray analysis (Renji Cohort 2) were all obtained from Department of Surgery, Shanghai Renji Hospital between June 2009 and July 2013. Renji cohort 2 contains 130 cases of PDAC and adjacent non-cancerous pancreas tissues. The clinicopathological traits consisted of age, gender, tumor size, AJCC stage, T classification, lymph node metastasis, distant metastasis, vascular invasion, histological differentiation, CA199 levels and vital status. The study was approved by the Ethics Committee of Shanghai Jiao Tong University School of Medicine, Renji Hospital. Written informed consent was obtained from all participants before enrollment in this study. None of the participants had received pre-operative tumor-specific therapy including radiotherapy, chemotherapy or hormone therapy. Follow-up was conducted by office visit, telephone call, or outpatient clinic visit, and overall survival information was acquired from all patients in Renji cohort 1 and 130 patients in Renji cohort 2. The last follow-up visit deadline was November 17, 2015. Overall survival was defined as the time from the date of surgery until death from any cause or last follow-up visit deadline.
Bioinformatics analysis
PDAC microarray datasets GSE28735 (Affymetrix Human Gene 1.0 ST Array), GSE71729 (Agilent-014850 Whole Human Genome Microarray 4x44K), TCGA-PAAD (UNC_IlluminaHiSeq_RNASeqV2, level 3) and their corresponding clinical data in this study were directly downloaded from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) and the open access tiers of the Cancer Genome Atlas (TCGA) data portal (https://gdc-portal.nci.nih.gov/). GSE28735 included 45 PDAC samples and 45 pared adjacent non-tumor tissue samples. GSE71729 analysis used data from 145 primary PDAC tumors and 46 pancreas tissues. Among 145 PDAC cases, 130 cases with clinical information were left for further analysis and 124 cases with overall survival information were left for Kaplan-Meier analysis. TCGA-PAAD RNA-Seq included 166 cases with overall survival information. Expression data extraction was performed with R 3.2.5 software. In order to gain insight into the biological mechanism involved in PDAC progression through FAM83B, gene set enrichment analysis (GSEA) was performed in tumor samples of GSE71729 datasets using the GO genesets database (c2.all.v4.0.symbols.gmt) from the Molecular Signatures Database-MsigDB.
Cell culture and treatment
Human PDAC cell lines PANC-1, AsPC-1, Capan-2 and SW1990 were purchased from the Cell Resource Center, Shanghai Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences and the normal control pancreatic ductal epithelial cell line hTERT-HPNE was obtained from American Type Culture Collection (ATCC). All cell lines were cultured as recommended in growth medium supplemented with 10% fetal bovine serum (FBS) and incubated at 37 °C with a humidified atmosphere of 5 % CO2.
Three small interfering RNAs (siRNAs) against human FAM83B were transfected into human PDAC cells using the DharmaFECT 1 siRNA transfection reagent (Thermo Scientific Dharmacon lnc. USA) according to the manufacturer's instruction, while nonspecific siRNA was applied as a negative control. Real-time PCR was performed to measure the knockdown efficiency of the three FAM83B-siRNAs, and the most efficient one was chosen for further experiment. All the experiments were conducted in the log phase of cell growth. All the siRNAs were designed and synthesized at Genepharm Technologies (Shanghai, China). The sequences targeting FAM83B were listed as follows:
5'- CUCGAGGAGUAUCUGUUUATT-3';
5'- GCCAUCUGAUAGUCUCAGUTT-3';
5'- GGGUCUCAGAAGUUAAGGUTT-3'.
Immunohistochemical staining
Human PDAC tissue microarray sections (Renji cohort 2) were rehydrated and treated with hydrogen peroxide for 15 min. Antigen retrieval was performed by microwave. After blocked with 10 % normal goat serum for 30 min, the tissue microarray sections were incubated with FAM83B antibodies (1:200 dilution, Abcam, UK) on a humidified box at 4˚C overnight. The next day, the sections were incubated with corresponding peroxidase-labeled secondary antibody for 30 min at room temperature and washed with PBS for 3 times. At last, Diaminobenzidine tetrahydrochloride (DAB; Maixin Biotech, China) was used for the color-reaction and hematoxylin was use for nucleus counterstaining. The immunohistochemically stained sections were observed under light microscopy.
Protein expression was assessed according to the intensity and extent of staining. The intensity of staining was evaluated on a scale of 0-3: 0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining. The extent of FAM83B positive cells was assessed on a scale of 0-4: 0, 0-5%; 1, 6-25%; 2, 26-50%; 3, 51-75%; 4, 76-100%; A final score was obtained by using grades of the intensity staining × grades of extent. The tissues with a final score < 6 were sorted into “FAM83B low expression” and those with a final score ≥ 6 were classified as “FAM83B high expression”.
Total RNA extraction and quantitative real-time PCR
Total RNA was extracted from PDAC cells, primary PDAC tissues and adjacent non-cancerous tissues using Trizol reagent (Takara, Japan). 1μg of total RNA was reverse-transcribed using the PrimeScript RT Reagent Kit (Takara, Japan), and quantitative real-time PCR was performed using SYBR Premix Ex Taq Ⅱ (Takara, Japan) by the StepOne real-time PCR system according to the manufacturer's instruction.
The relative mRNA expression levels were quantified with the 2-ΔΔCt method 13-15 and the amplified transcript level of each specific gene was normalized to the expression of the internal control GAPDH. Primer sequences used in this study were listed as follows: FAM83B, forward, 5'-AGGCGAAGTTTCCCGTTATT-3'; reverse, 5'-TCTTTGGGCTTTTCTGGTTG-3'. GAPDH, forward, 5'-GCATTGCCCTCAACGACCAC-3'; reverse, 5'-CCACCACCCTGTTGCTGTAG-3'.
Western blotting
Total protein was extracted from PDAC cells using a total protein extraction buffer (Beyotime, China) containing a protease inhibitor mixture (protease inhibitors; phosphatase inhibitors; PMSF; KangChen, Shanghai, China). BCA Protein Assay Kit (Pierce Biotechnology) was used to measure the concentration of protein. Proteins were separated by 10-12% SDS-polyacrylamide gels and transferred to PVDF membranes (Millipore, Bedford, MA, USA). After blocked in 5 % BSA at room temperature for 1.5 hours, the membranes were incubated overnight with primary rabbit anti-FAM83B (1:1000 dilution, Abcam, UK), rabbit anti-p21Cip1 (1:1000 dilution, CST, USA), rabbit anti-Cyclin D1 (1:1000 dilution, CST, USA), rabbit anti-CDK4 (1:1000 dilution, CST, USA), mouse anti-CDK6 (1:1000 dilution, CST, USA), GAPDH (1:1000 dilution, KangChen, Shanghai, China) antibodies at 4 ℃, and then washed with TBST for five times and incubated with species-specific secondary antibodies for one hour at room temperature the next day. At last, the ECL detection system was used for visualization. Antibodies against GAPDH acted as an internal control.
Cell cycle analysis by flow cytometry
PDAC cell lines PANC-1 and AsPC-1 were planted at an amount of 3× 105 cells per well in 6-well plates. Transfection with si-RNA was conducted when cells reached approximately 30 % confluency. Cells were harvested 48 hours later, washed twice with pre-cooling PBS and then fixed with 70 % ice-old ethanol at -20 °C overnight. Then the cells were resuspended with PBS containing 1mg/ml RNAase and incubated at 37 °C for 30 min. At last, cells were stained with 50μg/ml PI in the dark for 15 min and analyzed using a fluorescence-activated cell-sorting (FACS) flow cytometer (BD Biosciences, San Jose, CA, USA). The fluorescence profiles represented the DNA contents of PI-positive cells and the populations of G0/G1, S, and G2/M phases were analyzed by FlowJo V10 software.
Cell proliferation assay
PDAC cells were seeded at a density of 1500 cells/well of each group in 96-well plates in 100 μL of medium and transfected with corresponding si-RNAs the next day. Cells were treated with Cell Counting Kit-8 (CCK8, Dojindo Molecular Technologies, Kyushu, Japan) at 37 ℃ for 2 hours at various time points of 24 h, 48 h, 72 h, 96 h and 120 h after the initial seeding. The absorbance was detected at wavelength 450 nm (450 OD) using a microplate reader. The experiment was performed in six replicate wells for each group and all the experiments were conducted in triplicate. The ultimate results were calculated by mean value and SEM.
Transwell invasion assay
PANC-1 cells were transfected with control-siRNA and FAM83B-siRNA, and the cells were collected 48 hours later and suspended in serum-free medium. Transwell chambers were coated with fresh matrigel 3 hours before 2*105 cells were seeded into the upper chamber. At the same time, 600ul culture medium containing 20% FBS was added into the lower chamber. After incubated in 37 ℃ for 48 hours, cells on the upper chamber were removed. Then, cells on the lower chamber were washed with PBS softly, fixed with 4% paraformaldehyde for 20 minutes, stained with 0.1% crystal violet for 30 minutes and washed with PBS for 3 times. At last, the invasion cells were counted under a light microscope.
Apoptosis analysis by flow cytometry
PANC-1 cells were transfected with control-siRNA and FAM83B-siRNA. 24 hours later, the cells were collected and centrifuged at 1500 rpm for 5 minutes. Then, the cells were washed with 1x binding buffer, stained with Annexin V-FITC for 30 minutes and propidium iodide (PI) for 5 minutes. Finally, the samples were analyzed using a fluorescence-activated cell-sorting (FACS) flow cytometer (BD Biosciences, San Jose, CA, USA).
PDAC subcutaneous xenograft animal model
Short hairpin RNA (shRNA) targeting FAM83B was packaged into lenti-virus. PANC-1 cells were infected with sh-FAM83B lenti-virus and sh-control lenti-virus to construct PANC-1-FAM83B knockdown and PANC-1-control stable cell lines. The stable cell lines were selected. PANC-1 cells were collected and resuspended in PBS. A total of 5×106 resuspended cells were injected subcutaneously into the right flank of 5-week-old male BALB/c nude mice. 3 weeks later, the tumors were removed after the mice were euthanized. The volumes were calculated by the equation: V= (length x width2)/2, (V, volume). All the animal experiments were approved by the Institutional Animal Care and Use Committee.
Statistical analysis
All statistical analyses were carried out using SPSS 19.0 software (SPSS Inc, Chicago, IL, USA). Correlation between FAM83B expression and clinicopathologic parameters in patients with PDAC was examined by chi-square test or Fisher's exact test. Overall survival was evaluated by Kaplan-Meier survival curve and analyzed by the log-rank test. Cox proportional hazards model was performed to identify the prognostic factors by univariable and multivariable Cox regression analysis. Data from at least 3 independent experiments conducted in triplicates were presented as the means ± SD. Differences were considered to be significant with a value of p < 0.05.
Results
The expression of FAM83B was significantly increased in PDAC cell lines and tumor tissues
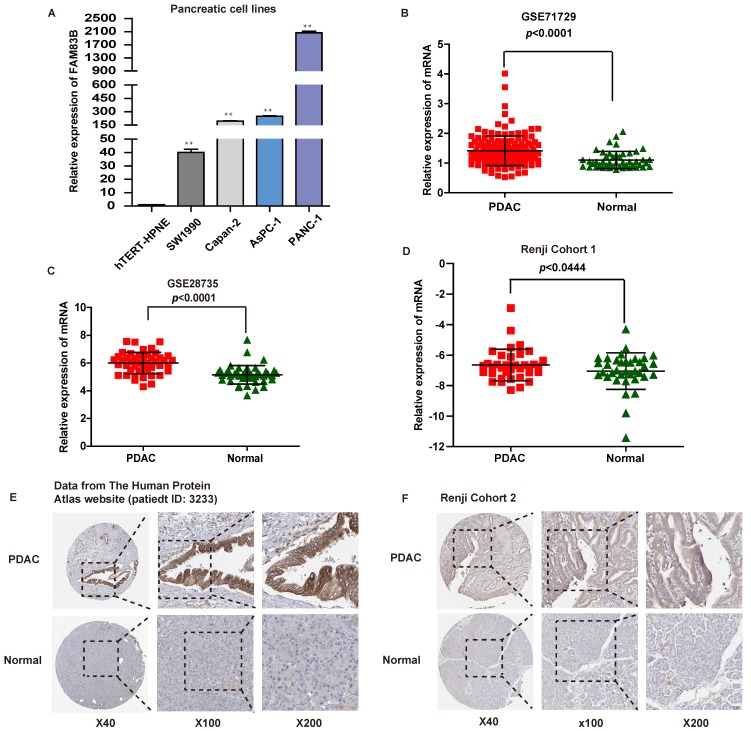
First of all, the expression of FAM83B was measured in four human PDAC cell lines and one normal pancreatic cell line by real-time PCR. The four PDAC cell lines were listed as follows: PANC-1, AsPC-1, Capan-2 and SW1990 cells, and the normal pancreatic cell line was hTERT-HPNE. It turned out that FAM83B expression was increased in PDAC cell lines to a remarkable extent compared with normal pancreatic cell line (p <0.01, Figure 1A).
Figure 1.
The expression of FAM83B in PDAC cell lines and tissues. A. The expression of FAM83B was measured by real-time PCR in PDAC cell lines. ** p<0.01. B-D. The relative expression of FAM83B was detected in PDAC tissues and adjacent non-tumor pancreas tissues in dataset GSE71729, GSE28735 and Renji Cohort 1 patients. Error bars in the scatter plots represent SD. E-F. FAM83B protein level was measured by immunohistochemistry in normal pancreas and PDAC tissue from The Human Protein Atlas website and Renji cohort 2. FAM83B: Family with sequence similarity 83, member B; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; PDAC: pancreatic ductal adenocarcinoma.
What's more, the mRNA expression levels of FAM83B in PDAC tissues were evaluated in two independent microarray datasets (GSE71729 and GSE28735) from GEO database 16, 17. As expected, PDAC tissues displayed higher mRNA expression level of FAM83B compared with paired normal pancreatic tissues in the above two independent microarray datasets (p <0.0001, Figure 1B-1C). In addition, in order to validate microarray data, real-time PCR was performed to quantify the mRNA expression levels of FAM83B in fresh PDAC tissues and adjacent non-tumor pancreas tissues from patients enrolled in Renji Hospital (n=36, Renji Cohort 1). Obviously, the expression of FAM83B was remarkably increased in PDAC tissues compared to adjacent normal tissues (p =0.0444, Figure 1D). Furthermore, the protein expression of FAM83B was also over-expressed in most PDAC tissues, which could be summarized from the data in the Human Protein Atlas website (patient ID: 3233, female, age 56, Figure 1E). Also, we detected FAM83B protein expression in 130 pairs of clinical PDAC samples by immunohistochemistry (n = 130, Renji Cohort 2). The results confirmed the above consequence that FAM83B displayed a remarkable higher expression level in PDAC tissues compared with matched corresponding non-cancerous tissues (p <0.0001, Table 1, Figure 1F). In one word, the expression of FAM83B was significantly up-regulated in PDAC tissues.
Table 1.
FAM83B protein levels in normal pancreas and PDAC tissues from Renji cohort 2
| Tissue | p value (x2 test) | ||
|---|---|---|---|
| PDAC (n=130, %) |
Normal (n=130, %) |
||
| Immunohistochemical grade | <0.001 | ||
| High FAM83B expression | 103 (79.2) | 45 (34.6) | |
| Low FAM83B expression | 27 (20.8) | 85 (65.4) | |
FAM83B was significantly related with advanced clinical stage and poor vital status in PDAC patients
In order to assess the pathologic and clinical significance of FAM83B in PDAC, we investigated the association between the expression of FAM83B and clinicopathological features of PDAC in Renji Cohort 2. The clinicopathological characteristics included: age, gender, tumor size, T classification, lymph node metastasis, distant metastasis, vascular invasion, AJCC stage, histological differentiation, CA199 levels and vital status. As illustrated in Table 2, the expression of FAM83B was significantly correlated with clinical stage (stage Ⅲ/Ⅳ vs. stage Ⅰ/Ⅱ, p =0.021) and vital status (dead vs. alive, p =0.005). However, there was no apparent relevance between the expression of FAM83B with age, gender, tumor size, T classification, lymph node metastasis, distant metastasis, vascular invasion, histological differentiation and CA199 levels. All in all, FAM83B was clinically relevant in PDAC patients.
Table 2.
The association between the expression of FAM83B and clinicopathological features of PDAC in Renji Cohort 2.
| Clinicopathological feature | Total 130 |
Expression of FAM83B | p Value (x2 test) |
|
|---|---|---|---|---|
| High (n=103, 79.2%) |
Low (n=27, 20.8%) |
|||
| Age (years) | ||||
| <65 | 60 | 45 (75.0) | 15 (25.0) | 0.271 |
| ≥65 | 70 | 58 (82.9) | 12 (17.1) | |
| Gender | ||||
| Male | 74 | 60 (81.1) | 14 (18.9) | 0.550 |
| Female | 56 | 43 (76.8) | 13 (23.2) | |
| Tumor size | ||||
| ≤3cm | 48 | 36 (75.0) | 12 (25.0) | 0.363 |
| >3cm | 82 | 67 (81.7) | 15 (18.3) | |
| Clinical stage (AJCC) | ||||
| Stage I / II | 105 | 79 (75.2) | 26 (24.8) | 0.021 |
| Stage III / IV | 25 | 24 (96.0) | 1 (4.0) | |
| T classification | ||||
| T1/T2 | 27 | 20 (74.1) | 7 (25.9) | 0.458 |
| T3/T4 | 103 | 83(80.6) | 20 (19.4) | |
| Lymph node metastasis | ||||
| Absent | 85 | 66 (77.6) | 19 (22.4) | 0.541 |
| Present | 45 | 37 (82.2) | 8 (17.8) | |
| Distant metastasis | ||||
| Absent | 118 | 92 (78.0) | 26 (22.0) | 0.265 |
| Present | 12 | 11 (91.7) | 1 (8.3) | |
| Vascular invasion | ||||
| Absent | 118 | 93 (78.8) | 25 (21.2) | 0.713 |
| Present | 12 | 10 (83.3) | 2 (16.7) | |
| Histological differentiation | ||||
| Well/Moderate | 80 | 63 (78.7) | 17(21.3) | 0.864 |
| Poor | 50 | 40 (80.0) | 10 (20.0) | |
| CA199 level (U/ml) | ||||
| ≤35 | 18 | 16(88.9) | 2 (11.1) | 0.371 |
| >35 | 69 | 55 (79.7) | 14 (20.3) | |
| Missing | 43 | |||
| Vital status | ||||
| Alive | 17 | 9 (52.9) | 8 (47.1) | 0.005 |
| Dead | 95 | 79 (83.2) | 16 (16.8) | |
The bold number represents p<0.05.
Up-regulated FAM83B predicted poor prognosis in PDAC patients
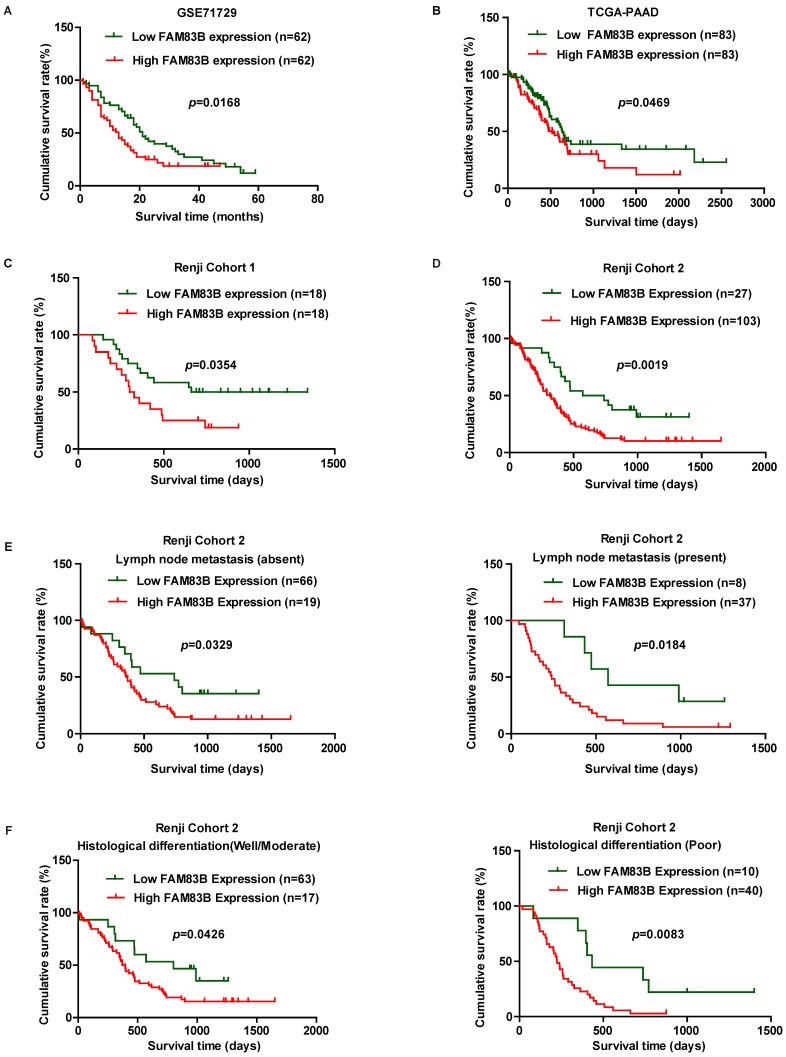
In order to validate whether FAM83B could predict clinical outcome in PDAC patients, Kaplan-Meier analysis and log-rank test were used to assess the association between FAM83B expression and overall survival in GSE71729, TCGA-PAAD, Renji Cohort 1 and Renji Cohort 2. First, we analyzed the prognostic value of FAM83B at mRNA level in GSE71729, TCGA-PAAD dataset and Renji Cohort 1. The results demonstrated that overall survival time was dramatically shorter in patients with high expression of FAM83B than in those with low expression of FAM83B in GSE71729 (p =0.0168), TCGA-PAAD (p =0.0469) and Renji Cohort 1 (p =0.0354) (Figure 2A-2C). Next, we analyzed the predictive value for clinical outcome of FAM83B at protein level in Renji Cohort 2. As displayed in Figure 2D, patients with higher FAM83B level had notably decreased overall survival time compared with patients with lower FAM83B levels (p =0.0019). Moreover, the correlation between FAM83B expression and overall survival in PDAC patients was assessed in different sub-groups according to lymphatic metastasis status and histological differentiation in Renji Cohort 2. Kaplan-Meier analysis demonstrated that high expression of FAM83B predicted poor prognosis in PDAC patients, regardless of lymphatic metastasis status and histological differentiation (Figure 2E-2F).
Figure 2.
Kaplan-Meier analysis of overall survival in PDAC patients. A-D. Survival curves were generated using the Kaplan-Meier method and the log-rank test was used to evaluate the statistical significance of differences. Higher FAM83B expression predicted shorter overall survival of PDAC patients in GSE71729, TCGA-PAAD, Renji Cohort 1 and Renji Cohort 2. E-F. FAM83B expression was related with poor prognosis in PDAC patients, regardless of lymphatic metastasis status and histological differentiation
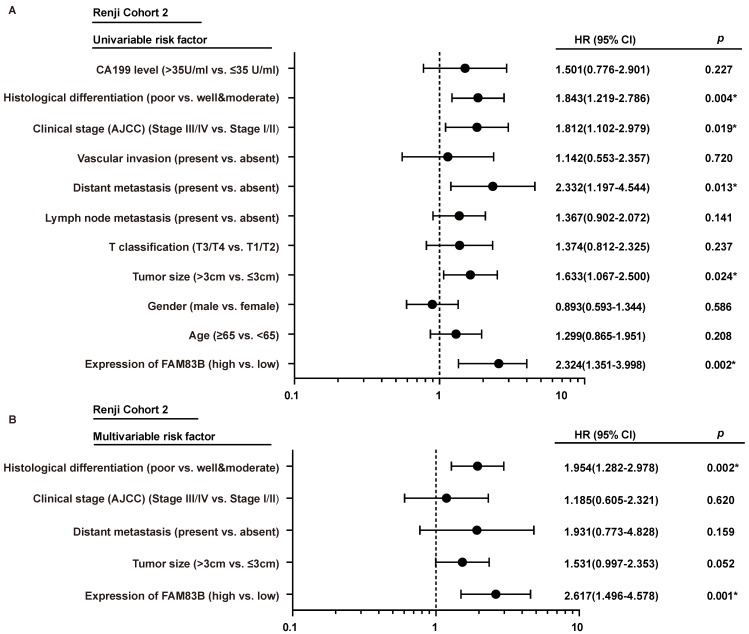
In order to gain further insight into the risk factors of prognosis in PDAC patients, Cox univariable and multivariable analyses were performed to evaluate the function of FAM83B expression and other ten prognostic parameters in Renji Cohort 2. In univariable analysis, higher FAM83B expression (HR, 2.324; 95% CI, 1.351-3.998; p=0.002), larger tumor size (HR, 1.633; 95% CI, 1.067-2.500; p=0.024), distant metastasis (HR, 2.332; 95% CI, 1.197-4.544; p =0.013), advanced AJCC clinical stage (HR, 1.812; 95% CI, 1.102-2.979; p =0.019) and poor histological differentiation (HR, 1.843; 95% CI, 1.219-2.786; p =0.004) were significant risk factors for overall survival of PDAC patients (Table 3, Figure 3A). Multivariable Cox regression analysis confirmed that expression of FAM83B (HR, 2.617; 95% CI, 1.496-4.578; p =0.001) and histological differentiation (HR, 1.954; 95% CI, 1.282-2.978; p =0.002) were two predictors of overall survival in patients with PDAC, independent of the other clinical covariates (Table 3, Figure 3B). To sum up, increased FAM83B expression emerged as an independent predictor of poor prognosis of PDAC patients.
Table 3.
Cox univariable and multivariable analyses of prognostic parameters including FAM83B expression of PDAC patients in Renji Cohort 2
| Prognostic Parameter | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Expression of FAM83B (high vs. low) | 2.324 | 1.351-3.998 | 0.002 | 2.617 | 1.496-4.578 | 0.001 | |
| Age (≥65 vs. <65) | 1.299 | 0.865-1.951 | 0.208 | ||||
| Gender (male vs. female) | 0.893 | 0.593-1.344 | 0.586 | ||||
| Tumor size (>3cm vs. ≤3cm) | 1.633 | 1.067-2.500 | 0.024 | 1.531 | 0.997-2.353 | 0.052 | |
| T classification (T3/T4 vs. T1/T2) | 1.374 | 0.812-2.325 | 0.237 | ||||
| Lymph node metastasis (present vs. absent) | 1.367 | 0.902-2.072 | 0.141 | ||||
| Distant metastasis (present vs. absent) | 2.332 | 1.197-4.544 | 0.013 | 1.931 | 0.773-4.828 | 0.159 | |
| Vascular invasion (present vs. absent) | 1.142 | 0.553-2.357 | 0.720 | ||||
| Clinical stage (AJCC) (Stage I / II vs. Stage III / IV) | 1.812 | 1.102-2.979 | 0.019 | 1.185 | 0.605-2.321 | 0.620 | |
| Histological differentiation (poor vs. well/moderate) | 1.843 | 1.219-2.786 | 0.004 | 1.954 | 1.282-2.978 | 0.002 | |
| CA199 level (>35U/ml vs. ≤35 U/ml) | 1.501 | 0.776-2.901 | 0.227 | ||||
HR: Hazard Ratio; CI: Confidence Interval. The bold number represents p<0.05.
Figure 3.
The forest plot of univariable and multivariable analyses of prognostic parameters for survival of PDAC patients in Renji cohort 2. A-B. The forest plot demonstrated the relation between overall survival and FAM83B expression as well as other clinical characteristics in PDAC patients using the univariable and multivariable analysis.
Down-regulation of FAM83B in PDAC cells leaded to G0/G1 phase arrest and inhibition of cell proliferation and tumor growth
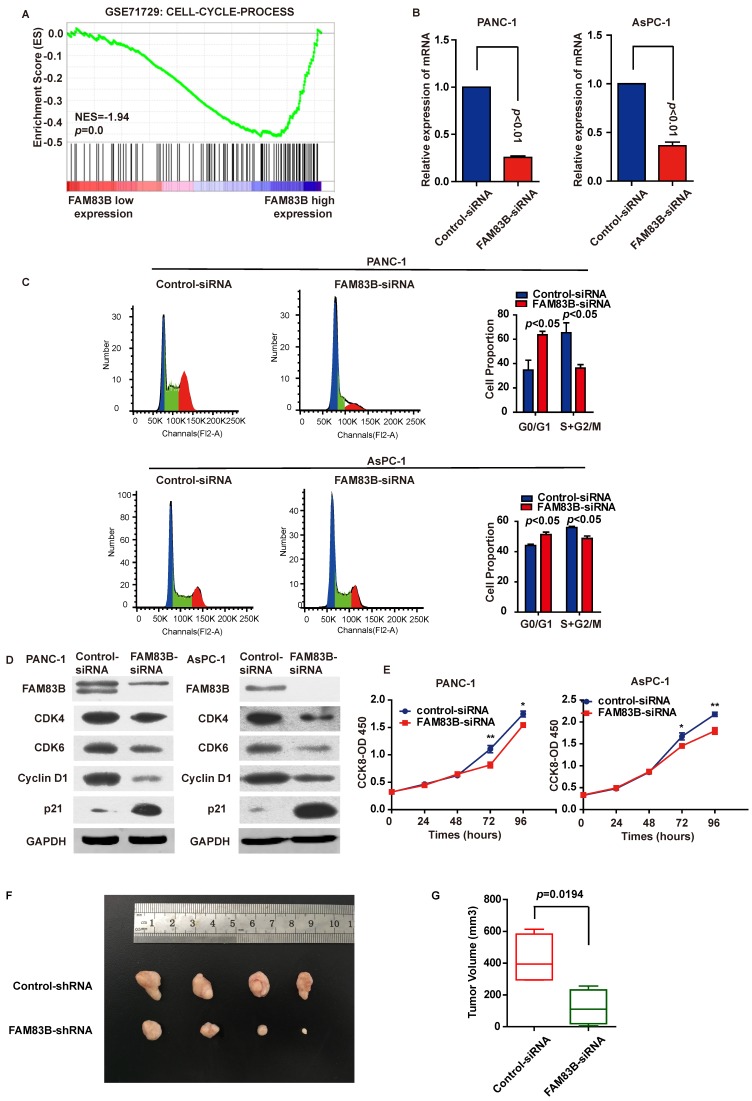
To explore the biological pathways involved in PDAC progression through FAM83B, gene set enrichment analysis (GSEA) was performed for further study. As a result, GSEA performed in GSE71729 illustrated that 'CELL-CYCLE-PROCESS' pathway was enriched in patients with FAM83B-higher expression versus patients with FAM83B-lower expression (Figure 4A). Therefore, cell cycle analysis and cell proliferation assay were carried out to clarify the oncogenic properties and effects of FAM83B on PDAC. First and foremost, PANC-1 and AsPC-1 cells were transfected with FAM83B-siRNAs. The transfection efficiency was confirmed by real-time PCR (Figure 4B). Flow cytometry for cell cycle analysis showed that the percentage of cells in G0/G1 (2N) phase increased from 34.7% to 63.7% in PANC cells. On the other hand, the percentage of cells containing 2N-4N DNA (in the S or G2 phase) decreased from 65.3% to 36.3% (p<0.05, Figure 4C). Meanwhile, the same phenomenon was observed in AsPC-1 cells. The percentage of cells in G0/G1 (2N) phase increased from 44.1% to 51.3%, while the percentage of cells containing 2N-4N DNA (in the S or G2 phase) decreased from 55.9% to 48.7% (p<0.05, Figure 4C). Consistently, down-regulation of FAM83B reduced the protein expression of G1-S transition promoter cyclin D1, CDK4 and CDK6. At the same time, p21Cip1 was enhanced in response to the down-regulation of FAM83B in PANC-1 and AsPC-1 cells (Figure 4D). These results suggested that FAM83B inhibition acted on the G1/S checkpoint to prohibit the cell cycle in PDAC cells. What's more, proliferation assay demonstrated that cell proliferation was suppressed when FAM83B was reduced in PANC-1 and AsPC-1 cells (Figure 4E). However, down-regulation of FAM83B had no significant effect on cell invasion and apoptosis in PDAC cell lines (Supplementary Figure 1).
Figure 4.
Down-regulation of FAM83B in PDAC cells leads to G1/S phase arrest and inhibition of cell proliferation and tumor growth. A. GSEA in GSE71729 tumor samples (median split according to FAM83B expression) indicated that 'CELL-CYCLE-PROCESS' pathway was enriched in patients with FAM83B-higher expression versus patients with FAM83B-lower expression. B. Knockdown efficiency was confirmed by real-time PCR in PANC-1 and AsPC-1 cell line. C. Down-regulation of FAM83B increased cell population at G0/G1 phase but decreased at S/G2 phase by flow cytometry analysis in PANC-1 and AsPC-1 cells. D. Western blot indicated down-regulation of FAM83B reduced the protein expression of CDK4, CDK6, cyclin D1, and enhanced the expression of p21Cip1. GAPDH was used as a loading control. E. The cell counting kit-8 assay indicated cell proliferation was blocked when FAM83B was reduced in PANC-1 and ASPC-1 cells.*p<0.05; ** p<0.01. The experiment was performed in six replicate wells for each group and all the experiments were conducted in triplicate. Error bars represented SEM. F-G. Down-regulation of FAM83B significantly reduced the PDAC tumor volume in the subcutaneous xenograft model (p=0.0194). Error bars represented SEM.
In order to verify the effect of FAM83B on tumor growth, we used a subcutaneous xenograft model in vivo. The PANC-1-FAM83B knockdown and PANC-1-control stable cells were subcutaneously injected into the right flank of 5-week-old nude mice. The tumor volume was measured and the tumors were removed 3 weeks later. The results showed that knockdown of FAM83B significantly reduced the tumor volume (Figure 4F-4G, p=0.0194). Combined with the in vitro experiments, we could draw a conclusion that FAM83B promoted cell proliferation and tumor growth.
In conclusion, down-regulation of FAM83B in PDAC cells leaded to G1 phase arrest and inhibition of cell proliferation and tumor growth.
Discussion
Pancreatic ductal adenocarcinoma is one of the most malignant cancers worldwide with increasing morbidity and mortality 18-20. Despite the progress made in targeted anticancer therapies in recent years, challenges remain. FAM83B is one of eight members of FAM83 protein family, which is sufficient and necessary to drive transformation 21. It's proved to be significantly elevated in several malignant tumors including breast, bladder, lung, thyroid, cervix, ovary and testis cancer. FAM83B is emerging as a prominent layer of oncogene. In previous study, FAM83B has emerged as a critical intermediary in EGFR/RAS and PI3K/AKT signaling pathway 10, 11. Grant S 22 also illustrated that FAM83B served as a candidate oncogene and TKI resistance mediator. Elevated FAM83B expression has been proved to increase Phospholipase D (PLD) activity, which was engaged in hyperactivation of EGFR. Thus, ablation of FAM83B suppressed PLD activity and MAPK pathway in breast cancer 23. It's also mentioned that FAM83B can interact with both Axin-1 and APC, and is likely involved in Wnt signaling pathway 24. Moreover, previous studies have illustrated an association between increased FAM83B expression and elevated tumor grade, increased recurrence rates and decreased overall survival time in breast cancer 10, 11, 23. The expression of FAM83B is significantly elevated in lung squamous cell carcinoma (SSC) compared with normal lung or adenocarcinoma. Thus, it may serve as a reliable diagnostic and prognostic biomarker for SCC 12. However, little investigation was made to evaluate the clinical significance in PDAC. Hence the current study was focused on the clinical significance and biological functions in PDAC.
In this study, we found that FAM83B may act as a tumor-promoting gene in PDAC progression. It boiled down to three main points. First of all, FAM83B displayed a remarkable trend of increasing expression levels from adjacent normal pancreatic tissues to carcinoma tissues in two microarray datasets as well as two independent cohorts in Renji Hospital. Secondly, FAM83B is positively associated with elevated tumor grade, which revealed a similar phenomenon with previous studies 11, 23. What's more, Kaplan-Meier analysis demonstrated that overall survival time was dramatically shorter in patients with high expression of FAM83B than in those with low expression of FAM83B. Univariable Cox regression analysis illustrated that high expression of FAM83B was a significant risk factor for overall survival of PDAC patients. At the same time, multivariable Cox regression analysis confirmed that high expression of FAM83B was an independent predictor of shorter overall survival in patients with PDAC. Thirdly, GSEA data indicated that 'CELL-CYCLE-PROCESS' pathway was enriched in patients with FAM83B-higher expression. Our experiments also confirmed that down-regulation of FAM83B inhibited cell proliferation and cell cycle.
It's well known that cell proliferation plays a critical role in tumorigenesis and tumor progression 25. Obviously, inhibition of FAM83B prohibited the growth of pancreatic cancer cells, which suggested that FAM83B might be responsible for the enhanced growth of PDAC and subsequently facilitated the development of PDAC. Our result was consistent with the phenomenon previously observed in breast cancer that FAM83B promoted cancer cell growth and anchorage-independent growth (AIG) 11.
To determine whether inhibition of FAM83B suppresses cell proliferation through cell cycle distribution, we performed cell cycle analysis by flow cytometry. The cell cycle is a continuous progress, which is divided into G0/G1, S, G2 and M phases 26. A great number of anti-tumor drugs have been reported to induce cell death by arresting the cell cycle at different phases 27-31. In particular, Cyclin D1 and p21Cip1 are main regulatory proteins that control the G1 checkpoint in the cell cycle. Cyclin D1, a G1 cyclin, acts as a G1-S transition promoter. Simultaneously, p21Cip1, a key CDK inhibitor, serves as a G1 gatekeeper and can arrest cell cycle progression at the G1 phase 32, 33. Cyclin D1 binds with CDK4/6 and activates CDK4/6, which subsequently phosphorylates tumor suppressor protein Rb and allows the cell cycle to progress through G1 into S phase 34-36. In this study, down-regulation of FAM83B induced cell cycle arrest at G0/G1 phase after 48-h treatment when analyzed by flow cytometry. The data of cell cycle arrest was similar to that observed in western blot analysis. Western blot indicated that FAM83B inhibition enhanced p21 protein expression and conversely reduced the protein expression of CDK4, CDK6 and cyclin D1 in both PANC-1 and AsPC-1 cells. Hence, we could draw a conclusion that down regulation of FAM83B could arrest cell cycle. However, knockdown of FAM83B had no significant effect on cell invasion and apoptosis in PDAC cell lines. Therefore, the impaired PDAC cell proliferation due to FAM83B inhibition could be accounted, at least in part, for the arrest of cell cycle. Finally, the subcutaneous xenograft model further verified that FAM83B promoted tumor growth in vivo.
Even so, the mechanism between FAM83B and cell proliferation and tumor growth remains unknown. Previous study reported that elevated FAM83B expression was engaged in hyperactivation of EGFR (epidermal growth factor receptor) and PLD (Phospholipase D) in breast cancer. We hypothesize that FAM83B may promote EGFR expression in PDAC, and then may active downstream MAPK and mTOR signaling and eventually promote tumor growth. Nevertheless, the specific mechanism and the hypothesis needed our further exploration and verification in future study.
Taken together, FAM83B is significantly up-regulated in PDAC tissues and negatively related with prognosis, which can act as an independent prognostic factor among PDAC patients. Moreover, FAM83B inhibition plays an anti-proliferative role through arrest of cell cycle in PDAC cells. The exact mechanism remains to be further elucidated in our future study. Therefore, our study reveals that FAM83B may be a potential biomarker of PDAC and open a new avenue for targeted PDAC therapy.
Supplementary Material
Supplementary figure 1.
Supplementary table 1.
Supplementary table 2.
Supplementary table 3.
Supplementary table 4.
Supplementary table 5.
Acknowledgments
The authors are grateful to Dr Jie Hong, Hao-Yan Chen, Dr Jing-yuan Fang and Dr Yong-wei Sun for their assistances. This work was supported by grants from the National Natural Science Foundation (No.31271366, 81522008) to JH, Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (No.20151512), Chenxing Project of Shanghai Jiao-Tong University to JH, and the Doctoral Innovation Fund Projects from Shanghai Jiaotong University School of Medicine to YTT (BXJ201720).
Abbreviations
- PDAC
pancreatic ductal adenocarcinoma
- FAM83B
family with sequence similarity 83, member B
- CI
Confidence interval
- HR
Hazard ratio
- EGFR
epidermal growth factor receptor
- PLD
Phospholipase D.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA: a cancer journal for clinicians. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.The Lancet O. Pancreatic cancer in the spotlight. The Lancet Oncology. 2014;15:241. doi: 10.1016/S1470-2045(14)70097-X. [DOI] [PubMed] [Google Scholar]
- 4.Carr RM, Fernandez-Zapico ME. Pancreatic cancer microenvironment, to target or not to target? EMBO molecular medicine. 2016;8:80–2. doi: 10.15252/emmm.201505948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuramitsu Y, Taba K, Ryozawa S, Yoshida K, Zhang X, Tanaka T. et al. Identification of up- and down-regulated proteins in gemcitabine-resistant pancreatic cancer cells using two-dimensional gel electrophoresis and mass spectrometry. Anticancer research. 2010;30:3367–72. [PubMed] [Google Scholar]
- 6.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet (London, England) 2011;378:607–20. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nature reviews Clinical oncology. 2010;7:163–72. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 8.Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK. et al. Recent progress in pancreatic cancer. CA: a cancer journal for clinicians. 2013;63:318–48. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remmers N, Bailey JM, Mohr AM, Hollingsworth MA. Molecular pathology of early pancreatic cancer. Cancer biomarkers: section A of Disease markers. 2010;9:421–40. doi: 10.3233/CBM-2011-0168. [DOI] [PubMed] [Google Scholar]
- 10.Cipriano R, Graham J, Miskimen KL, Bryson BL, Bruntz RC, Scott SA. et al. FAM83B mediates EGFR- and RAS-driven oncogenic transformation. The Journal of clinical investigation. 2012;122:3197–210. doi: 10.1172/JCI60517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW. FAM83B-mediated activation of PI3K/AKT and MAPK signaling cooperates to promote epithelial cell transformation and resistance to targeted therapies. Oncotarget. 2013;4:729–38. doi: 10.18632/oncotarget.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okabe N, Ezaki J, Yamaura T, Muto S, Osugi J, Tamura H. et al. FAM83B is a novel biomarker for diagnosis and prognosis of lung squamous cell carcinoma. International journal of oncology. 2015;46:999–1006. doi: 10.3892/ijo.2015.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pu J, Yuan A, Shan P, Gao E, Wang X, Wang Y. et al. Cardiomyocyte-expressed farnesoid-X-receptor is a novel apoptosis mediator and contributes to myocardial ischaemia/reperfusion injury. European heart journal. 2013;34:1834–45. doi: 10.1093/eurheartj/ehs011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corona-Meraz FI, Navarro-Hernandez RE, Ruiz-Quezada SL, Madrigal-Ruiz PM, Castro-Albarran J, Chavarria-Avila E. et al. Inverse Relationship of the CMKLR1 Relative Expression and Chemerin Serum Levels in Obesity with Dysmetabolic Phenotype and Insulin Resistance. Mediators of inflammation. 2016;2016:3085390. doi: 10.1155/2016/3085390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan TT, Fu XL, Li J, Bian YN, Liu DJ, Hua R. et al. Downregulation of RPL15 may predict poor survival and associate with tumor progression in pancreatic ductal adenocarcinoma. Oncotarget. 2015;6:37028–42. doi: 10.18632/oncotarget.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA. et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nature genetics. 2015;47:1168–78. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang G, He P, Tan H, Budhu A, Gaedcke J, Ghadimi BM. et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:4983–93. doi: 10.1158/1078-0432.CCR-13-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H. et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. European journal of cancer (Oxford, England: 1990) 2013;49:1374–403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 19.Hessmann E, Johnsen SA, Siveke JT, Ellenrieder V. Epigenetic treatment of pancreatic cancer: is there a therapeutic perspective on the horizon? Gut; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW. Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Molecular cancer research: MCR. 2014;12:1156–65. doi: 10.1158/1541-7786.MCR-13-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant S. FAM83A and FAM83B: candidate oncogenes and TKI resistance mediators. The Journal of clinical investigation. 2012;122:3048–51. doi: 10.1172/JCI64412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cipriano R, Bryson BL, Miskimen KL, Bartel CA, Hernandez-Sanchez W, Bruntz RC. et al. Hyperactivation of EGFR and downstream effector phospholipase D1 by oncogenic FAM83B. Oncogene. 2014;33:3298–306. doi: 10.1038/onc.2013.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilger M, Mann M. Triple SILAC to determine stimulus specific interactions in the Wnt pathway. Journal of proteome research. 2012;11:982–94. doi: 10.1021/pr200740a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhi X, Chen W, Xue F, Liang C, Chen BW, Zhou Y. et al. OSI-027 inhibits pancreatic ductal adenocarcinoma cell proliferation and enhances the therapeutic effect of gemcitabine both in vitro and in vivo. Oncotarget. 2015;6:26230–41. doi: 10.18632/oncotarget.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coqueret O. Linking cyclins to transcriptional control. Gene. 2002;299:35–55. doi: 10.1016/s0378-1119(02)01055-7. [DOI] [PubMed] [Google Scholar]
- 27.Aix E, Gutierrez-Gutierrez O, Sanchez-Ferrer C, Aguado T, Flores I. Postnatal telomere dysfunction induces cardiomyocyte cell-cycle arrest through p21 activation. The Journal of cell biology. 2016;213:571–83. doi: 10.1083/jcb.201510091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baan B, Dihal AA, Hoff E, Bos CL, Voorneveld PW, Koelink PJ. et al. 5-Aminosalicylic acid inhibits cell cycle progression in a phospholipase D dependent manner in colorectal cancer. Gut. 2012;61:1708–15. doi: 10.1136/gutjnl-2011-301626. [DOI] [PubMed] [Google Scholar]
- 29.Cannell IG, Merrick KA, Morandell S, Zhu CQ, Braun CJ, Grant RA. et al. A Pleiotropic RNA-Binding Protein Controls Distinct Cell Cycle Checkpoints to Drive Resistance of p53-Defective Tumors to Chemotherapy. Cancer cell. 2015;28:623–37. doi: 10.1016/j.ccell.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao Z, Xue J, Sowin TJ, Zhang H. Differential roles of checkpoint kinase 1, checkpoint kinase 2, and mitogen-activated protein kinase-activated protein kinase 2 in mediating DNA damage-induced cell cycle arrest: implications for cancer therapy. Molecular cancer therapeutics. 2006;5:1935–43. doi: 10.1158/1535-7163.MCT-06-0077. [DOI] [PubMed] [Google Scholar]
- 31.Cully M. Combinations with checkpoint inhibitors at wavefront of cancer immunotherapy. Nature reviews Drug discovery. 2015;14:374–5. doi: 10.1038/nrd4648. [DOI] [PubMed] [Google Scholar]
- 32.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Developmental cell. 2008;14:159–69. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer research. 2005;65:3980–5. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 34.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–47. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 35.Ye D, Luo H, Lai Z, Zou L, Zhu L, Mao J. et al. ClC-3 Chloride Channel Proteins Regulate the Cell Cycle by Up-regulating cyclin D1-CDK4/6 through Suppressing p21/p27 Expression in Nasopharyngeal Carcinoma Cells. Scientific reports. 2016;6:30276. doi: 10.1038/srep30276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landis MW, Pawlyk BS, Li T, Sicinski P, Hinds PW. Cyclin D1-dependent kinase activity in murine development and mammary tumorigenesis. Cancer cell. 2006;9:13–22. doi: 10.1016/j.ccr.2005.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1.
Supplementary table 1.
Supplementary table 2.
Supplementary table 3.
Supplementary table 4.
Supplementary table 5.