Abstract
Purpose: Sumoylation plays a critical role in gene regulation and tumorigenesis, and is hypothesized to correlate with the development of various cancers. So far, there has been no reported association between sumoylation-related genes and the risk of gastric cancer (GC).
Methods: A total of 17 tagging single-nucleotide polymorphisms (tag-SNPs) in 5 sumoylation-related genes were selected and genotyped by SNaPshot in a case-control study, including 1021 GC patients and 1304 controls. Odds ratio (OR) and 95% confidential interval (CI) were computed to evaluate the genetic association of the onset of GC.
Results: We demonstrated that CBX4 rs77447679 polymorphism was significantly associated with GC risk (P= 0.017; adjusted OR: 1.71; 95% CI: 1.10-2.66). The patients with CC genotype had a lower risk of GC (CC vs. CA+AA, P= 0.017; adjusted OR: 1.24; 95% CI: 1.04-1.49).
Conclusion: This study revealed that CBX4 rs77447679 polymorphism was positively associated with GC, and individuals with CC genotype had less risk of GC. The risky effects and functional effect of this polymorphism in GC require further investigation.
Keywords: sumoylation, haploview, polymorphism, gastric cancer, susceptibility.
Introduction
Gastric cancer (GC) is the fourth most common cancer diagnosed in men and the fifth in women globally 1. Multiple pathogenic factors, such as unhealthy lifestyle, and Helicobacter pylori infection, contribute to the etiology of GC 2. Consistent with multifactorial pathogenesis, clinical outcomes and GC prevalence might result from environmental factors. Besides, the influence of host-specific factors, especially host genetic variation, could impose a remarkable effect on the susceptibility of GC in a population with high incidence rate. 3.
Post-translational modifications (PTMs) include enzymatic changes such as addition of chemical group adducts, including several ubiquitin, small ubiquitin-like modifiers (SUMO) and phosphor. These changes play an essential role in gene regulation, cellular function, tissue development, and metabolism, and they are closely relevant to cancer occurrence 4, 5. Understanding the influence of sumoylation on gene regulation and protein homeostasis could provide valuable insights of early diagnosis and treatment for cancer 6. Sumoylation refers to the post-translational process that a SUMO protein moiety is enzymatically conjugated to a substrate protein 7, 8. Sumoylation requires the following series of enzymatic events: A. SUMO-activation by enzyme E1, including SUMO-activation enzyme E1 (SAE1) and ubiquitin-activating enzyme 2 (UBA2); B. SUMO-conjugation by enzyme E2, such as UBC9 and ubiquitin-conjugating enzyme 2I (UBE2I); C. SUMO-ligation by enzyme E3, such as ran-binding protein 2 (RANBP2) and polycomb chromobox homolog 4 (CBX4) 4, 9-12. The sumoylation pathway, one of the most important PTMs, was closely related with pathogenesis of human diseases through regulation of genome stability, gene expression, nuclear functions, and protein-protein interactions 13-15. Besides, sumoylation is also relevant in various cancers, including prostate cancer, bladder cancer, brain cancer, breast cancer, and cervical carcinoma, through the mediation of some key oncogenes and tumor suppressors 9, 16-19. Recently, some current GC-association studies for sumoylation have existed 20, 21, but knowledge of the detailed mechanism of GC carcinogenesis is still unclear. So far, there is no study focusing on the influence of sumoylation-related genes on the risk of GC.
This study focuses on the single nucleotide polymorphisms (SNPs) located in the functional region of sumoylation-related genes, and we aim to provide a new perspective on the mechanism of GC. We performed a case-control study of tag-SNPs in 5 potential sumoylation-related genes (RANBP2, SAE1, UBE2I, UBA2 and CBX4) in a Chinese population, and CBX4 rs77447679 has been identified significantly associated with GC.
Methods and Materials
Study participants
A total of 1021 histopathologically confirmed GC patients and 1304 GC-free participants without a family history of GC (as control group) were enrolled in this study. All participants were genetically unrelated Han Chinese. These GC patients had been referred to the Department of Oncology in the Second Affiliated Hospital of Nanjing Medical University, Shanghai Tenth People's Hospital, and Tongji University School of Medicine in China, between 2011 and 2014. The disease-free controls were enrolled from those sought routinely physical examination in the outpatient department of these hospitals. A standardized questionnaire was developed and used to obtain basic demographic information and clinical characteristics of the participants. This study was approved by each of the participating institutes' ethics committee, in accordance with the principles of the Helsinki Declaration (Approval Number: SHSY-IEC-pap-16-15), and written informed consent was obtained from all participants in this study (both patients and control).
SNP selection
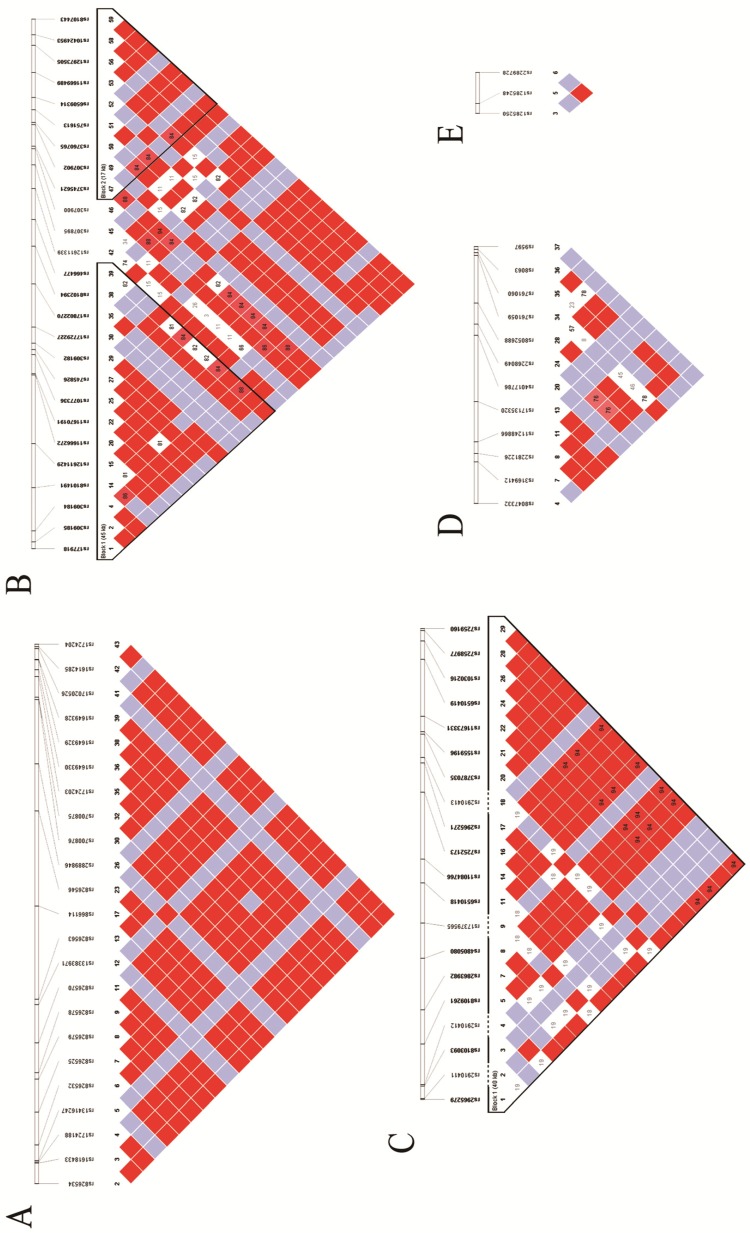
The potential SNPs were screened and selected from unrelated Han Chinese genotype data from the 1000 Genome Project database. Identification of the tag-SNPs was performed in the Haploview 4.2 software (BROAD institute, Cambridge, MA, UK) with pairwise option, using r2=0.8 as the threshold. Besides, we also scanned these tag-SNPs' corresponding minor allele frequencies (MAFs) in the NCBI SNP database, and excluded those without reports in Chinese population or MAF < 0.05. The identification number of the final candidate tag-SNPs, their relative positions in the genome, and the linkage disequilibrium (LD) of these tag-SNPs are shown in Fig 1.
Figure 1.
Hapmaps of Sumoylation-related genes: A. Hapmap of RANBP2; B. Hapmap of UBE2I; C. Hapmap of CBX4; D. Hapmap of UBA2 and E. Hapmap of SAE.
DNA extraction and genotyping
Approximately 10 ml whole blood from each participant was extracted and transferred into an ACD tube. Genomic DNA was extracted from peripheral blood samples by proteinase K (Roche Applied Science, IN, USA) digestion and phenol-chloroform extraction. DNA extraction was performed in the laminar flow hood using sterile techniques. Genotypes of the 33 tag-SNPs were determined by SNaPshot (Applied Biosystems, CA, USA). The primers to amplify different fragments, including each tag-SNP, were designed with the Primer 5 software (PREMIER Biosoft, Palo Alto, CA, USA). Amplifications were performed in the ABI3130 genetic analyzer (Applied Biosystems, CA, USA), and allelic discrimination was completed using the GeneMapperTM software (Applied Biosystems, CA, USA), using manufacturers' instruction and protocols. Positive controls through sequencing and negative controls without DNA samples were contained in each plate to ensure the accuracy of genotyping. The genotyping procedure in this study was conducted in a double-blinded way. Approximately 10% of the total samples were randomly selected for duplicated assays, and the results were 100% concordant.
Statistical analysis
χ2 test and Student's t-test were performed to quantify the differences of demographic characteristics, selected variables, genotype frequencies and allele distributions between the case (GC-patient) and control groups. Association between genetic variation and the risk of GC was estimated by odds ratio (OR) and corresponding 95% confidence intervals (CI) from an unconditional logistic regression, adjusted for age and sex. Hardy-Weinberg equilibrium (HWE) for each SNP in the case and control groups were checked using χ2 test. P< 0.05 was considered as statistically significant in this study. Statistical analyses were performed in SPSS 23.0 software (SPSS, Inc., IL, USA).
Results
Characteristics of the study cases and controls
1021 patients (case group) with histopathologically confirmed GC (806 patients, comprising 79% of all cases were non-cardia, and the remaining 21% were cardia) and 1304 healthy participants (control group) were enrolled in this study. There was no significant difference of age or sex between the case and control groups. 470 patients (46%) had clinical stage I/II GC, and the other 551 (54%) were in stage III/IV GC. The demographic information and clinical characteristics of the participants are shown in Table 1.
Table 1.
Demographic and clinical characteristics of GC patient (cases) and control groups.
| Variables | Cases N = 1021 | Control N = 1304 | P | |
|---|---|---|---|---|
| age | 0.364 | |||
| ≤60 | 459 (0.45) | 612 (0.47) | ||
| >60 | 562 (0.55) | 692 (0.53) | ||
| sex | 0.321 | |||
| male | 715 (0.70) | 887 (0.68) | ||
| female | 306 (0.30) | 417 (0.32) | ||
| Tumor site | ||||
| Non-carida | 806 (0.79) | |||
| Cardia | 214 (0.21) | |||
| TNM stage | ||||
| I/II | 470 (0.46) | |||
| III/IV | 551 (0.54) |
Association between SUMO gene polymorphisms and the risk of GC
19 potential SNPs were selected from RANBP2, UBE2I, CBX4, UBA2 and SAE1 genes as the candidate sumoylation-related tag-SNPs. The characteristics of these 19 tag-SNPs, as well as the allele frequencies of the polymorphisms among the case and control groups are reported in Table 2. After SNP genotyping, 5 SNPs (UBE2I rs8062881, rs75998799 and rs12925270, CBX4 rs190846371, and SAE1 rs6509314) were excluded because of their low MAF (P < 0.05). The observed genotype frequencies of the SUMO genes were consistent with the HWE. Of the remaining 14 SNPs, there was significant difference between the case and control group for rs77447679 in CBX4 gene, and consequently strong association between SNP rs77447679 and the risk of GC (P = 0.017; adjusted OR: 1.24; 95% CI: 1.07-1.44).
Table 2.
Tag-SNPs results for GC in sumolytion-related genes.
| SNP | Gene | CHR | position | Major allele | Minor allele | MAF in control | OR (95% CI)a | Pa |
|---|---|---|---|---|---|---|---|---|
| rs12614691 | RANBP2 | 2 | 109362565 | A | G | 0.092 | 1.08 (0.88-1.33) | 0.465 |
| rs8062881 | UBE2I | 16 | 1361651 | C | G | 0.036 | 0.98 (0.71-1.34) | 0.885 |
| rs12920764 | UBE2I | 16 | 1362187 | T | C | 0.073 | 0.94 (0.75-1.18) | 0.605 |
| rs75998799 | UBE2I | 16 | 1366079 | G | A | 0.020 | 1.14 (0.72-1.79) | 0.576 |
| rs761059 | UBE2I | 16 | 1374524 | G | A | 0.106 | 0.92 (0.72-1.16) | 0.473 |
| rs12925270 | UBE2I | 16 | 1375937 | C | A | 0.049 | 0.86 (0.61-1.21) | 0.372 |
| rs1285251 | CBX4 | 17 | 77809823 | C | T | 0.343 | 1.10 (0.97-1.25) | 0.153 |
| rs1285249 | CBX4 | 17 | 77810143 | G | C | 0.156 | 1.03 (0.87-1.21) | 0.756 |
| rs73422123 | CBX4 | 17 | 77810391 | T | C | 0.289 | 0.97 (0.85-1.10) | 0.606 |
| rs77447679 | CBX4 | 17 | 77810474 | C | A | 0.166 | 1.24 (1.04-1.49) | 0.017 |
| rs2289728 | CBX4 | 17 | 77811854 | A | G | 0.488 | 1.06 (0.94-1.19) | 0.316 |
| rs1285243 | CBX4 | 17 | 77814106 | T | C | 0.177 | 0.96 (0.82-1.13) | 0.641 |
| rs12980229 | UBA2 | 19 | 34914144 | A | G | 0.126 | 0.98 (0.82-1.18) | 0.851 |
| rs177918 | SAE1 | 19 | 47634313 | G | T | 0.187 | 0.92 (0.79-1.07) | 0.272 |
| rs309184 | SAE1 | 19 | 47636827 | T | G | 0.403 | 1.01 (0.90-1.14) | 0.848 |
| rs55801246 | SAE1 | 19 | 47650489 | C | T | 0.080 | 0.90 (0.72-1.12) | 0.337 |
| rs12611339 | SAE1 | 19 | 47684084 | G | A | 0.171 | 1.02 (0.87-1.21) | 0.786 |
| rs3745621 | SAE1 | 19 | 47689921 | C | G | 0.275 | 0.89 (0.78-1.02) | 0.091 |
| rs6509314 | SAE1 | 19 | 47696626 | T | C | 0.042 | 1.00 (0.73-1.35) | 0.983 |
a adjusted for age and sex in logistic regression
Detailed allele distributions of rs77447679 in the case and control groups are shown in Table 3. Individuals with the CA genotype had significantly higher risk of GC, (1.71-fold compared to the baseline genotype CC, P= 0.018; adjusted OR: 1.71; 95% CI: 1.10-2.66); besides, recessive model also had a significantly increased risk of GC (CC vs. CA+AA, P= 0.017; adjusted OR: 1.24; 95% CI: 1.04-1.49). However, there was no statistically significant difference of AA genotype between the patient and control groups (P= 0.065; adjusted OR: 1.19; 95% CI: 0.99-1.44).
Table 3.
Genotype frequencies of CBX4 rs77447679 polymorphism in GC patients and controls.
| rs77447679 genotype | Controls | OR (95% CI)a | Pa | |
|---|---|---|---|---|
| C>A | Cases | |||
| CC | 648 | 890 | 1.00 (Reference) | |
| CA | 296 | 341 | 1.71 (1.10-2.66) | 0.018 |
| AA | 46 | 40 | 1.19 (0.99-1.44) | 0.065 |
| CA+AA | 342 | 381 | 1.24 (1.04-1.49) | 0.017 |
| CC+CA | 944 | 1231 | 1.00 (Reference) | |
| AA | 46 | 40 | 1.63 (1.05-2.52) | 0.030 |
a Adjustment for age and sex in logistic regression
Discussion
In this study, we have identified 17 tag-SNPs in 5 sumoylation-related genes (RANBP2, SAE1, UBE2I, UBA2 and CBX4) and quantified their association with GC susceptibility in a Chinese population. We demonstrated that CBX4 rs77447679 was significantly associated with the risk of GC, and individuals with CA genotype had a significantly higher risk of GC, comparing to the baseline CC genotype. This is a pioneer investigation to quantify the relationship between the sumoylation-related gene polymorphism and the risk of GC in Chinese population.
First reported in 1996, sumoylation modifies various eukaryotic proteins 7. Sumoylation is governed by a conserved cascade consisting an E1-activating enzyme complex, an E2-conjugating enzyme and an E3 ligase 22. The 5 sumoylation-related genes involved in this study (RANBP2, SAE1, UBE2I, UBA2, and CBX4) are responsible for gene regulation of the E1 enzyme complex, E2 or multiple E3 ligases. In this study, we have demonstrated that CBX4 polymorphism had strong association with GC development. CBX4 gene is a critical component of the polycomb repressive complex 1 (PRC1), which synergizes with PRC2 to silence gene expression by specifically modifying nucleosomal histones 23-25. Additionally, CBX4 is a SUMO E3 ligase, which could upregulate the sumoylation of a more limited repertoire of substrate involved in tumorigenesis 26, 27. Several recent studies have reported that CBX4 is closely related with the proliferation and differentiation of cloned embryos, thymic epithelial cells, and transarterial chemoembolization in hepatocellular carcinoma (HCC) 28-31. Furthermore, CBX4 expression has been reported to upregulate tumor tissues, and higher CBX4 expression is correlated with the α-fetoprotein (AFP) level in serum, tumor size, pathological differentiation, and the TNM (tumor, node, metastasis) stages in HCC 32. In addition, Jiao HK et al. demonstrated that CBX4 could promote angiogenesis and metastasis in HCC by the sumoylation pathway dependent on the hypoxia-inducible factor-1α protein and could be an independent prognostic factor for HCC patients 30. The amplification and biological roles of the CBX4 gene have not yet been reported in GC. Regarding its well-known role in HCC, we hypothesize that CBX4 gene is involved in the pathogenesis of GC. However, subsequent studies on different populations are needed to confirm our finding.
This study has suggested that SNPs in gene's functional regions could influence gene expression and functions 33-35. Thus, 7 selected tag-SNPs (rs1285251, rs1285249, rs73422123, rs77447679, rs2289728, rs190846371, and rs1285243) of the CBX4 genes, located in the functional regions of CBX4, were examined for their potential association with GC. CBX4 rs77447679 variant could be involved in the development of GC and leading to abnormal expression of CBX4 proteins; moreover, patients with CC genotype had a lower risk of GC, which suggested that the C to A substitution of CBX4 rs77447679 could increase the transcription and expression of CBX4, and contribute to the development of GC (P= 0.018; adjusted OR: 1.71; 95% CI: 1.10-2.66). Thus, CBX4 rs77447679 polymorphism is a biologically plausible factor in GC. The association between CBX4 gene polymorphism and cancer risk has been reported in previous studies 36. In a large-scale study of esophageal squamous cell carcinoma (ESCC), it was demonstrated that CBX4 gene was substantially amplified and upregulated in a Chinese population and could act as oncogenes and potential therapeutic targets in ESCC. Recurrent focal CNAs, including amplified chromosomal segments containing CBX4, were validated in ESCC, which implied that CBX4 amplification and the corresponding protein upregulation contributed to the development of ESCC 36. Our findings on CBX4 rs77447679 polymorphism were consistent with previous studies of ESCC, which together could provide a novel interventional target for GC patients and people with certain susceptible genotypes (e.g., CC comparing to CA) and thus with a higher GC risk. We expect a better understanding of sumoylation in the occurrence and development of GC.
There were, however, some limitations in this study. First, multiple factors, such as age, sex, smoking status, drinking status, and dietary habits, might influence the development of GC, and stratification would be recommended. Besides, the expression of CBX4 proteins in patients' tumor tissues and controls, as well as the underlying mechanisms of this polymorphism in GC-related genes require further investigation. Finally, this case-control study was based on hospital participants, so there was inherent possibility of selection bias.
Conclusion
We have identified that the CBX4 rs77447679 polymorphism was statistically significantly associated with GC, and individuals with CC genotype had lower GC risk. More comprehensive investigation is necessary to evaluate the interactions between the genetic and environmental factors, as well as the underlying mechanisms of the association between rs77447679 polymorphism and the risk of GC.
Acknowledgments
This study was supported by the Fundamental Research Funds for the Central Universities (grant no. 2016KJ048), the National Science Foundation of China for Young Scientists (grant no. 81602944 and 81602431), the excellent young teachers program of school of basic medical sciences in Nanjing Medical University (grant no. 12JC014), and the Key Project supported by Medical Science and technology development Foundation, Nanjing Department of Health (grant no. YKK16216).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.O'Connor A, Gisbert JP, O'Morain C, Ladas S. Treatment of Helicobacter pylori Infection 2015. Helicobacter. 2015;20(Suppl 1):54–61. doi: 10.1111/hel.12258. [DOI] [PubMed] [Google Scholar]
- 3.Chiurillo MA. Role of gene polymorphisms in gastric cancer and its precursor lesions: current knowledge and perspectives in Latin American countries. World J Gastroenterol. 2014;20(16):4503–4515. doi: 10.3748/wjg.v20.i16.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Lu W. Roles of ubiquitination and SUMOylation on prostate cancer: mechanisms and clinical implications. Int J Mol Sci. 2015;16(3):4560–4580. doi: 10.3390/ijms16034560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JH, Choi HJ, Kim B, Kim MH, Lee JM, Kim IS. et al. Roles of sumoylation of a reptin chromatin-remodelling complex in cancer metastasis. Nat Cell Biol. 2006;8(6):631–639. doi: 10.1038/ncb1415. [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135(6 Pt 1):1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez A, Pangas SA. Regulation of germ cell function by SUMOylation. Cell Tissue Res; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattoscio D, Casadio C, Fumagalli M, Sideri M, Chiocca S. The SUMO conjugating enzyme UBC9 as a biomarker for cervical HPV infections. Ecancermedicalscience. 2015;9:534. doi: 10.3332/ecancer.2015.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delfino DV, Spinicelli S, Pozzesi N, Pierangeli S, Velardi E, Bruscoli S. et al. Glucocorticoid-induced activation of caspase-8 protects the glucocorticoid-induced protein Gilz from proteasomal degradation and induces its binding to SUMO-1 in murine thymocytes. Cell Death Differ. 2011;18(1):183–190. doi: 10.1038/cdd.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B, Zhou J, Liu P, Hu J, Jin H, Shimono Y, Takahashi M, Xu G. Polycomb protein Cbx4 promotes SUMO modification of de novo DNA methyltransferase Dnmt3a. Biochem J. 2007;405(2):369–378. doi: 10.1042/BJ20061873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer LM, Koonin EV, Aravind L. Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle. 2004;3(11):1440–1450. doi: 10.4161/cc.3.11.1206. [DOI] [PubMed] [Google Scholar]
- 13.Yang XJ, Chiang CM. Sumoylation in gene regulation, human disease, and therapeutic action. F1000Prime Rep. 2013;5:45. doi: 10.12703/P5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J. Sumoylation regulates diverse biological processes. Cell Mol Life Sci. 2007;64(23):3017–3033. doi: 10.1007/s00018-007-7137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cashman R, Cohen H, Ben-Hamo R, Zilberberg A, Efroni S. SENP5 mediates breast cancer invasion via a TGFbetaRI SUMOylation cascade. Oncotarget. 2014;5(4):1071–1082. doi: 10.18632/oncotarget.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan M, Gong H, Wang J, Tao L, Xu D, Bao E, Liu Z, Qiu J. SENP2 regulates MMP13 expression in a bladder cancer cell line through SUMOylation of TBL1/TBLR1. Sci Rep. 2015;5:13996. doi: 10.1038/srep13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivenugopal KS, Rawat A, Niture SK, Paranjpe A, Velu C, Venugopal SN, Madala HR, Basak D, Punganuru SR. Posttranslational Regulation of O6-Methylguanine-DNA Methyltransferase (MGMT) and new opportunities for treatment of brain cancers. Mini Rev Med Chem; 2015. [DOI] [PubMed] [Google Scholar]
- 18.Puhr M, Hoefer J, Eigentler A, Dietrich D, van Leenders G, Uhl B, PIAS1 is a determinant of poor survival and acts as a positive feedback regulator of AR signaling through enhanced AR stabilization in prostate cancer. Oncogene; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim KI, Baek SH. SUMOylation code in cancer development and metastasis. Mol Cells. 2006;22(3):247–253. [PubMed] [Google Scholar]
- 20.Song JG, Xie HH, Li N, Wu K, Qiu JG, Shen DM, Huang CJ. SUMO-specific protease 6 promotes gastric cancer cell growth via deSUMOylation of FoxM1. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(12):9865–9871. doi: 10.1007/s13277-015-3737-z. [DOI] [PubMed] [Google Scholar]
- 21.Ren YH, Liu KJ, Wang M, Yu YN, Yang K, Chen Q. et al. De-SUMOylation of FOXC2 by SENP3 promotes the epithelial-mesenchymal transition in gastric cancer cells. Oncotarget. 2014;5(16):7093–7104. doi: 10.18632/oncotarget.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8(12):947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 23.Volkel P, Angrand PO. The control of histone lysine methylation in epigenetic regulation. Biochimie. 2007;89(1):1–20. doi: 10.1016/j.biochi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Xu Y, Jiao H, Wang W, Mei Z, Chen G. Sumoylation of hypoxia inducible factor-1alpha and its significance in cancer. Sci China Life Sci. 2014;57(7):657–664. doi: 10.1007/s11427-014-4685-3. [DOI] [PubMed] [Google Scholar]
- 26.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113(1):127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 27.Wotton D, Merrill JC. Pc2 and SUMOylation. Biochem Soc Trans. 2007;35(Pt 6):1401–1404. doi: 10.1042/BST0351401. [DOI] [PubMed] [Google Scholar]
- 28.Hao L, Midic U, Garriga J, Latham KE. Contribution of CBX4 to cumulus oophorus cell phenotype in mice and attendant effects in cumulus cell cloned embryos. Physiol Genomics. 2014;46(2):66–80. doi: 10.1152/physiolgenomics.00071.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B, Liu YF, Du YR, Mardaryev AN, Yang W, Chen H. et al. Cbx4 regulates the proliferation of thymic epithelial cells and thymus function. Development. 2013;140(4):780–788. doi: 10.1242/dev.085035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiao HK, Xu Y, Li J, Wang W, Mei Z, Long XD, Chen GQ. Prognostic significance of Cbx4 expression and its beneficial effect for transarterial chemoembolization in hepatocellular carcinoma. Cell Death Dis. 2015;6:e1689. doi: 10.1038/cddis.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Xu Y, Long XD, Wang W, Jiao HK, Mei Z. et al. Cbx4 governs HIF-1alpha to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell. 2014;25(1):118–131. doi: 10.1016/j.ccr.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Wang B, Tang J, Liao D, Wang G, Zhang M, Sang Y. et al. Chromobox homolog 4 is correlated with prognosis and tumor cell growth in hepatocellular carcinoma. Ann Surg Oncol. 2013;20(Suppl 3):S684–692. doi: 10.1245/s10434-013-3171-7. [DOI] [PubMed] [Google Scholar]
- 33.Xu M, Qiang F, Gao Y, Kang M, Wang M, Tao G. et al. Evaluation of a novel functional single-nucleotide polymorphism (rs35010275 G>C) in MIR196A2 promoter region as a risk factor of gastric cancer in a Chinese population. Medicine (Baltimore) 2014;93(26):e173. doi: 10.1097/MD.0000000000000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Putra AC, Tanimoto K, Arifin M, Hiyama K. Hypoxia-inducible factor-1alpha polymorphisms are associated with genetic aberrations in lung cancer. Respirology. 2011;16(5):796–802. doi: 10.1111/j.1440-1843.2011.01972.x. [DOI] [PubMed] [Google Scholar]
- 35.Bienkiewicz J, Smolarz B, Malinowski A. Association Between Single Nucleotide Polymorphism +276G > T (rs1501299) in ADIPOQ and Endometrial Cancer. Pathol Oncol Res; 2015. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Zhou Y, Cheng C, Cui H, Cheng L, Kong P. et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet. 2015;96(4):597–611. doi: 10.1016/j.ajhg.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]