ABSTRACT
γδ T cells usually infiltrate many different types of cancer, but it is unclear whether they inhibit or promote tumor progression. Moreover, properties of tumor-infiltrating γδ T cells and those in the corresponding normal tissue remain largely unknown. Here we have studied features of γδ T cells in colorectal cancer, normal colon tissue and peripheral blood, and correlated their levels with clinicopathologic hallmarks. Flow cytometry and transcriptome analyses showed that the tumor comprised a highly variable rate of TILs (5–90%) and 4% γδ T cells on average, with the majority expressing Vδ1. Most Vδ1 and Vδ2 T cells showed a predominant effector memory phenotype and had reduced production of IFN- γ which was likely due to yet unidentified inhibitory molecules present in cancer stem cell secretome. Transcriptome analyses revealed that patients containing abundant γδ T cells had significantly longer 5-year disease free survival rate, suggesting their efficacy in controlling tumor at very early stage.
KEYWORDS: γδ T cells, colon cancer, DFS, IFN-γ, TILs
Introduction
Colorectal cancer (CRC) is one of the top 3 causes of cancer death.1-4 CRC arises from the inner wall of the large intestine and results from the accumulation of diverse genomic aberrations. These include both inherited mutations causing hereditary CRC5 and several chromosomal locations and single nucleotide polymorphisms (SNPs) which confer increased risk for CRC development. Moreover, tumor cells grow in a very complex microenvironment and establish reciprocal interactions with epithelial and mesenchymal cells, vascular and lymphatic vessels, inflammatory and immune cells.6-8
Tumor-infiltrating lymphocytes (TILs) are an immune population composed of different immune cells that have specificity and potential reactivity against the tumor. TILs have been found in a wide variety of solid tumors including CRC.9-10 However, while the functions and anti-CRC activities of CD4 and CD8 T cells within TILs have been extensively studied,11-12 very little is known about γδ T lymphocytes.
γδ T lymphocytes are important effector cells of the immune system that may play a role in the anti-tumor immunosurveillance. Human γδ T cells can be divided into 2 main populations based upon δ chain expression13: γδ T cells expressing the Vδ1 chain are mostly found in mucosal tissues, while γδ T cells expressing the Vδ2 chain (preferentially paired to the Vγ9 chain) predominate in the peripheral blood and secondary lymphoid organs.14 While the ligand(s) recognized by Vδ1 cells remain unknown, Vδ2 T cells recognize non peptidic Ags by a MHC-unrestricted mechanism,15-17 but which involves butyrophilin (BTN) 3A1.18-20 Specifically, Vγ9Vδ2 T cells recognize small unprocessed non peptidic compounds containing phosphate and termed phosphoantigens (PAgs), that are produced through the isoprenoid biosynthesis pathway.15-17 Moreover, these cells can also be activated, through an indirect mechanism, by aminobisphosphonates that inhibit farnesyl pyrophosphate synthase and cause accumulation of downstream endogenous PAgs.21-22 Physiologic levels of PAgs, however, are not stimulatory of Vγ9Vδ2 T cells, but transformed and infected cells would produce increased metabolic intermediates such as PAgs. Upon activation, γδ T lymphocytes undergo a differentiation program resembling that of CD8 T cells and they give rise to both central memory (TCM; CD45RA− CD27+) and effector memory (TEM;CD45RA− CD27−) and terminally differentiated (TEMRA; CD45RA+ CD27−) T cells. γδ TCM cells home to secondary lymphoid organs and lack immediate effector functions, while γδ TEM and TEMRA cells home to sites of inflammation where they display immediate effector functions such as cytokine production and cytotoxicity, respectively.23 Based on their effector properties, Vγ9Vδ2 T lymphocytes are supposed to play an important role in cellular immune responses against tumors.
γδ T cells have been found among TILs in many different types of cancer, but it is unclear if they correlate positively or not with tumor growth, or even fail to correlate with any prognostic feature.24-25 With particular regard to CRC, γδ T cells among TILs are the major source of IL-17 and positively correlate with advanced tumor clinicopathologic features.26 However, a very recent analysis of expression signature from ∼18.000 human tumors with overall survival outcomes across a collection of 39 cancer types, including CRC, revealed intratumoral γδ cells as the most significant favorable prognostic immune population.27 Therefore, there is urgent need to revisit and clarify this issue and also to understand the reciprocal interactions between γδ cells and other components of the tumor microenvironment, because these interactions can influence the function of intratumoral γδ cells and thus the net outcome of their response to tumor.
In this paper, we have studied the frequency, phenotype and functions of γδ T cells infiltrating CRC and correlated levels of intratumoral γδ T cells with clinical outcome. Moreover, we have studied the influence of the tumor microenvironment on the functional responses of γδ T cells.
Results
γδ T cells are present among CRC TILs
To evaluate the composition of tumor-infiltrating leukocytes in human CRC, CRC tissues were freshly obtained from 70 patients undergoing surgery, and analysis of cell surface molecules defining T, B, NK cells and γδ T cells was performed using polychromatic flow cytometry. Cumulative data are shown in Fig. 1a. Immune infiltrates detected with the pan-leukocyte marker CD45 were present in both normal and tumor tissues, but were substantially increased in CRC compared with normal tissue (CRC: median = 68%, range 15–87.8%; Healthy tissue: median = 61%, range 33.9–85%).Lymphocyte subsets were evaluated by the use of cell-surface markers and indicated as percentage of the total number of CD45+ cells in each sample (Fig. 1a). CD3+ T cells represented an average of 20% of the whole leukocyte (CD45+) population within the analyzed primary CRC samples, and consisted mainly of CD4+ and CD8+ T cells, each of which accounted for 8% of the CD45+ population (hence each accounting for 40% of the CD3+ population). Percentages of B and NK lymphocytes were lower than percentages of CD3+ cells (6% for CD3− CD19+ cells and 10% for CD3− CD56+ cells). γδ T cells were present among intratumoral leukocytes and accounted for approximately 4.5% (mean 4.5% ± 4.9%) of the total leukocyte population (Fig. 1a), hence accounting for approximately 20% of the CD3+ population. For comparison, we data mined an independent cohort of 585 CRC transcriptomes28 acquired on Affymetrix U133plus2 microarrays and downloaded from the NCBI-GEO data set repository.29 The algorithmic deconvolution of leucocytes infiltrating these tumors by CIBERSORT–LM730-31 detected on average 4.9% ± 4.4% γδ TILs among the total leucocyte population in these samples. Hence the above FACS results were fully consistent with those from an independent cohort of CRC (Wilcoxon p value = 0.26 for comparison of the 2 series of γδ TIL rates). As shown in Fig. 1a, there was an extremely high variability in percentages of lymphocyte subsets detected among TILs in the tested CRC patients. This was strikingly depicted by both the raw FACS data and the microarray deconvolution for percentages of CD3+ T cells which ranged between 5% to 90% (FACS) and 4% to 70% (microarrays).
Figure 1.
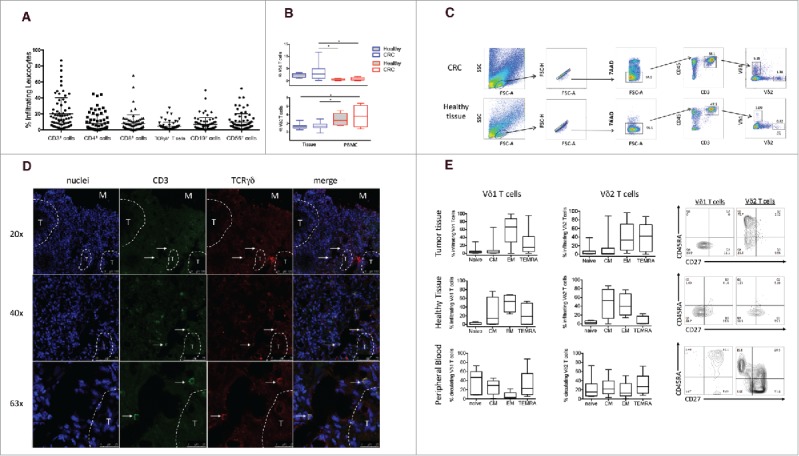
Frequency of infiltrating and circulating γδ T cells expressing either Vδ1 or Vδ2 TCR δ chains in HD and CRC patients. (A) Cumulative analysis of immune infiltrates of 70 colon cancer specimens. Lymphomonocyte populations were evaluated by the use of cell-surface markers and indicated as percentage of the total number of CD45+ cells in each sample. (B) Box plot of percentages of Vδ1 or Vδ2 γδ T cells subsets in healthy tissue, tumor tissue and peripheral blood of CRC patients and peripheral blood of HD subjects. Boxes represent 25th to 75th percentiles; middle bar identifies median; whiskers show minimum and maximum. *p<0.05 performed by nonparametric Mann-Whitney test, unpaired and 2-tailed with confidential interval 95%. (C) Representative dot plots of the gating strategy used to define Vδ1 and Vδ2 T cells from healthy and tumor tissues. The following gating strategy was used to detect γδ T lymphocytes: FSC/SSC, single cells, live cells CD45/CD3, Vδ1 and Vδ2 T cells. (D) Sections from CRC patients were stained with anti-human pan-γδ TCR (red) and anti-CD3 (green) for immunofluorescent (IF) staining. Right panel is a magnified view and the arrows display the colocalization of γδ TCR and CD3. Nuclei were contrasted with DAPI. One of 3 independent experiments is shown. (E) Phenotypical analysis of Vδ1 and Vδ2 T cells among healthy and tumor tissues and PBMC of CRC patients, upon staining with mAbs to CD45RA and CD27, and gating on CD3+ Vδ1+ or CD3+ Vδ2+ T cells. Beside, flow cytometry panels of a representative dot plot. Isotype-matched mAbs were used as controls. Viable lymphocytes were gated by forward and side scatter, and analysis was performed on 100,000 acquired events by using FlowJo. PBMC were stained with anti-CD3, anti-Vδ2, anti-CD45RA and CD27 mAbs.
γδ T cells variably infiltrate several human cancers, but the current data on the prognostic value of intratumoral γδ T cells have shown marked variability.27 To study whether this was dependent on the prevalence of a given γδ subset among TILs, we evaluated Vδ1 and Vδ2 T cells from CRC and adjacent non tumor colon tissue to determine their frequencies and composition. Fig. 1b shows cumulative data from 70 CRC patients, while Fig. 1c shows primary data from one representative sample per each group. As compared with adjacent non tumor colon tissue, intratumoral γδ T cells did not exhibit a distinct prevalence and distribution of Vδ1 and Vδ2 T cell subsets, despite a slightly and not significantly increased abundance of both subsets (Fig. 1b). As expected, the majority of γδ T cells in both CRC and adjacent normal tissues expressed Vδ1, and this pattern was observed in multiple patients despite the frequencies of Vδ1 and Vδ2 T cells among tumor-infiltrating leukocytes varied widely. This TCR bias could not be investigated likewise using the microarray data set which lacks TRDV1 gene, and in which the correlated levels of TRGV9 and TRDV2 genes indicated presence of TCR Vγ9Vδ2 T lymphocytes (data not shown).
Because previous papers11-12 have emphasized the importance of immune cell localization, within distinct tumor regions, related to the risk of tumor recurrence, we also visualized intratumoral γδ T cells by immunofluorescence analysis on frozen sections. In our analysis, γδ T cells were consistently detected in the tumor border/stroma, but only very rarely in the intratumor tissue (Fig. 1d).
Most Vδ1 T cells in tumor tissues were of effector memory phenotype (TEM), whereas TEMRA, TNaive and TCM cells accounted for 15%, 3% and 3.5% of the total γδ population, respectively (Fig. 1e). Conversely, intratumoral Vδ2 T cells had a more heterogeneous phenotype with TEM and TEMRA cells almost equally well represented (33% and 42% respectively) and TCM and TNaive phenotypes accounting for only 3% and 2% of the total γδ population, respectively.
To understand if the predominance of γδ T cells with effector phenotypes in TILs was due to the tumor microenvironment or simply reflected an overall bias in colon cancer patients, we compared the phenotype distribution of Vδ1 and Vδ2 T cells in the tumor tissue with that in adjacent non tumor colon tissue and in peripheral blood. The cell-surface differentiation patterns of CRC-resident Vδ1 and Vδ2 T cells were very similar to those of Vδ1 and Vδ2 T cells residing in adjacent non tumor colon tissue, but distinct from those of the corresponding cells from the peripheral blood (Fig. 1e). This indicates that residence in a non-lymphoid tissue, regardless of whether it is normal or has undergone tumor transformation, serves as a major determinant of the phenotypic characteristics of tumor and tissue γδ T cells.
Functional features of CRC-infiltrating γδ T cells
To further elucidate the functional state of infiltrating γδ T cells, we analyzed the production of IL-17, IFN-γ and TNF-α by tumor-infiltrating Vδ1 and Vδ2 T cells, upon in vitro stimulation with ionomycin and PMA. Fig. 2a shows cumulative data from 20 CRC patients and Fig. 2b shows primary data from one representative sample per each group.
Figure 2.
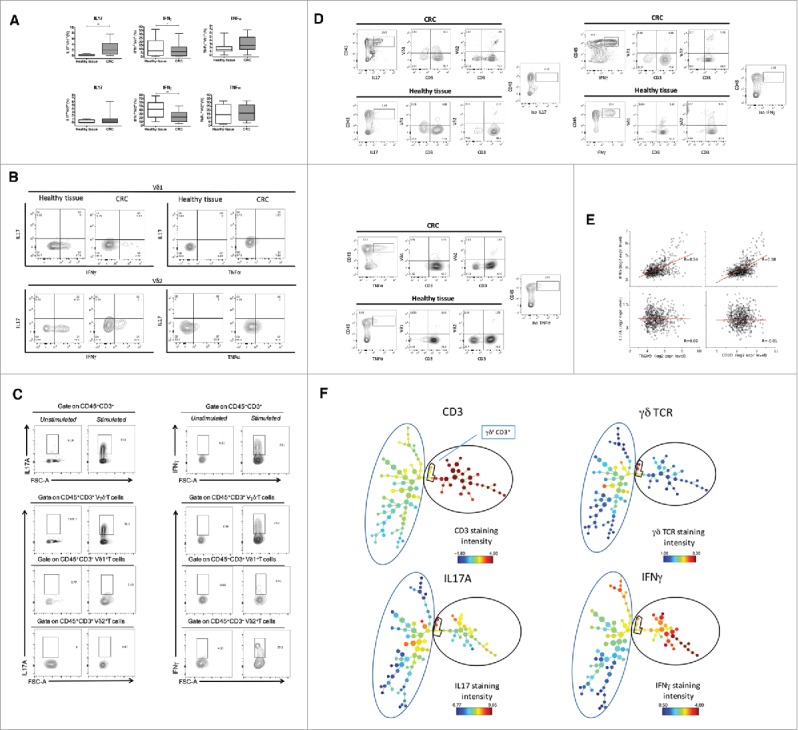
Cytokine production of tumor infiltrating γδ T cells. (A) Box plots of cumulative data of healthy tissue and tumor tissue samples from 20 CRC patients. Cells were stimulated in vitro as described in Materials and Methods and were stained with mAbs to IFN-γ, IL-17 and TNF-α. *p<0.05 and **p<0.01 performed by nonparametric Mann-Whitney test, unpaired and 2-tailed with confidential interval 95%. (B) Flow cytometry analysis of healthy and tumor tissue from one representative CRC patient. (C) Representative dot plots to define IL-17 or IFN-γ producing γδ, Vδ1 and Vδ2 T cells gated separately on CD45+ CD3+γδ−, CD45+ CD3+ Vδ1+ or CD45+ CD3+ Vδ2+ T cells. (D) Representative dot plots to define cells making IL-17 or IFN-γ upon gating on CD45+ IL-17+ or CD45+ IFN-γ+ cells, of healthy and tumor tissue. (E) Pearson correlation of TCR, IFNG and IL17A gene expression levels in n = 585 CRC tumor samples.**p<0.01. (F) CRC-infiltrating CD45+ single cells were used to generate the SPADE tree, and were grouped in 2 different populations, CD3− and CD3+(black outer circles). The distribution of the major populations is showed for one representative sample. The branching tree is based on the number of cells included in each node and the legend indicates the range of cell per node according to relative median fluorescence intensity.
Upon activation, Vδ1 T cells from adjacent non tumor colon tissue produced IFN-γ, but very poor, if any, IL-17 and TNF-α. In contrast, Vδ1 T cells from CRC tissues produced significantly less IFN-γ compared with adjacent non tumor colon tissue (8.1% versus 1.2%), but expressed more IL-17 (2.7% vs. 0.2%) and TNF-α (3% vs. 1.9%). Similarly, upon activation Vδ2 T cells from adjacent non tumor colon tissue expressed both IFN-γ and TNF-α, but very low levels IL-17, while Vδ2 T cells from CRC tissues expressed significantly less IFN-γ (23.5% vs. 41.2%) and had similar TNF-α (16.9% vs. 14.6%) and IL-17 expression (1% vs. 0.4%). These results were confirmed by the measurement of IFN-γ, IL-17 and TNF-α concentrations in culture supernatants of Vδ1 and Vδ2 T cells by ELISA (data not shown). Moreover, the very poor IL-17 production by CRC infiltrating γδ T cells was confirmed when we differently gated through more stringent leukocyte gates (CD45+ CD3+γδ−, CD45+ CD3+ Vδ1+ or CD45+ CD3+ Vδ2+ T cells): in fact, 5% Vδ1+ and 0.87% Vδ2+ cells expressed IL-17, and 10% Vδ1+ and 27.6% Vδ2+ cells expressed IFN-γ (Fig. 2c).
The above finding was unexpected, because a previous study has shown that γδ T cells are the major cellular source of IL-17 in human CRC.26 Given that, and to exclude a general failure of IL-17 production in our experimental settings, we used another different strategy: we gated first on IL-17-producing leukocytes (CD45+ IL-17+) and then checked the phenotype of cells making IL-17 among tumor-infiltrating leukocytes. As shown in Fig. 2d the majority of CD45+ IL-17+ cells both in CRC and in adjacent non tumor colon tissues were CD3+ but did not express either Vδ1 or Vδ2, suggesting they may be typical Th17 or Tc17 αβ T cells. Moreover, a discrete fraction of CD45+ IL-17+ cells (ranging from 10% to 50% in different samples), which was increased in CRC tissues as compared with adjacent non tumor colon tissue, did not express CD3 and probably corresponds to type 3 innate lymphoid cells (ILC3) or other leukocyte populations. Using this same gating strategy, we also show that most of cells expressing IFN-γ and TNF-α were CD3+ but Vδ1− and Vδ2− T cells, both in CRC and adjacent non tumor colon tissues.
In the CRC microarray data set as well, expression of TRGV9, TRDV2 or CD3D genes was correlated with expression of IFNG gene, suggesting that TILs in general and TCRVγ9Vδ2 cells in particular are involved in producing this cytokine in CRC samples. By contrast such correlations were not found with expression of IL17A gene, suggesting that IL-17 production arises from more diverse cell sources than IFNG (Fig. 2e). This possibility was also supported by SPADE (Spanning-Tree Progression Analysis of Density-normalized Events) algorithm, which distinguishes cell subsets by clustering, based on surface antigen expression denoted by a color gradient: as shown in Fig. 2f, IFN-γ had diverse maps compared with those of IL-17.
Together, these results indicate that Vδ1 and Vδ2 T cells in CRC and adjacent normal tissues preferentially produce IFN-γ, but very low IL-17, almost as all the other CD3+ TILs in human CRC. Moreover, IFN-γ production by both Vδ1 and Vδ2 T cells is significantly reduced in CRC tissues, as compared with adjacent normal colon tissue samples.
CRC-infiltrating γδ T cells are largely shaped by the local tumor microenvironment
Next we sought to explore whether the tumor microenvironment imparts distinct functional features on γδ T cells. In particular, it is conceivable that tissue resident γδ T cells are functionally affected locally or it is also possible that γδ T cells are recruited into the tumor from draining lymph nodes or peripheral blood and then undergo functional changes in response to the local microenvironment.
To test the influence of the tumor environment on γδ T cells, we initially isolated cancer stem cells (CSC) and cancer-associated fibroblasts (CAF) from 15 CRC patients and tested the effect of the 48-hrs culture supernatants from CSC and CAF on polyclonal γδ T cell lines (containing both Vδ1 and Vδ2 T cells) obtained from peripheral blood of healthy donors. As shown in Fig. 3a, supernatants from CSC, but not supernatants from CAF significantly impaired proliferation of polyclonal γδ T cells to PHA. Supernatants from CSC also inhibited proliferation of polyclonal γδ T cells stimulated with anti-CD3 and anti-CD28, or proliferation of Vδ2 T cells from peripheral blood stimulated with zoledronate + IL-2 in a 7-day culture setting. (data not shown).
Figure 3.
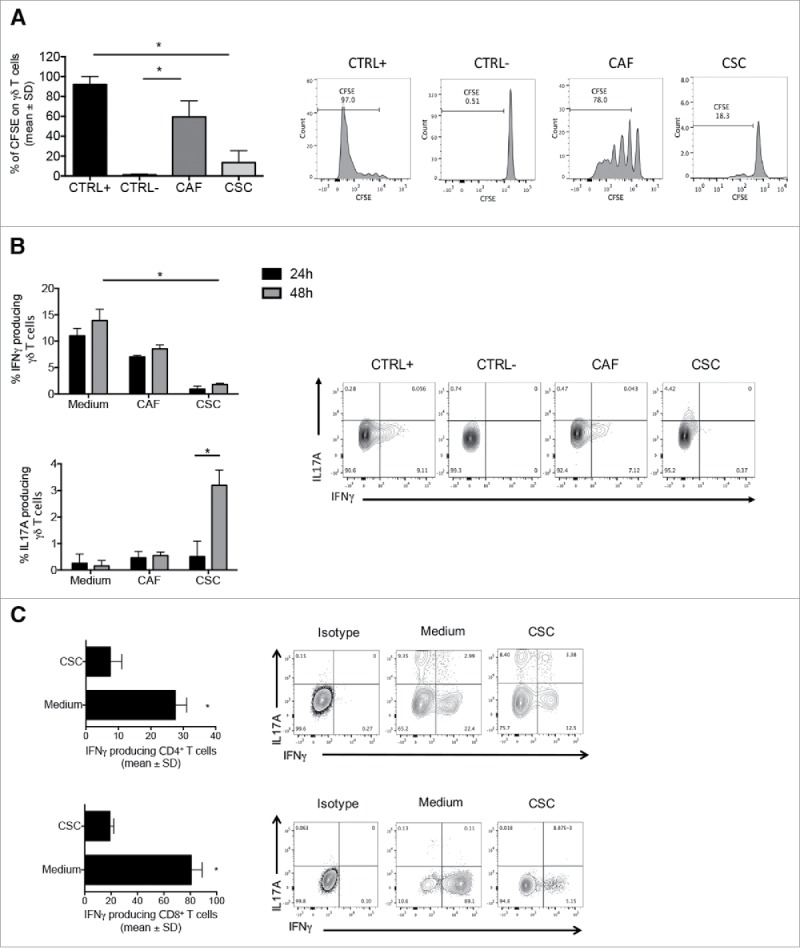
CSC supernatants inhibit proliferation and IFN-γproduction by γδ, CD4 and CD8 T cells. (A) Cumulative data (n = 5) from proliferation assay. Positive control (CTRL+) and negative control (CTRL-) refer to cells stimulated with PHA and unstimulated cells, respectively. Data are mean percentage of positive cells ± SD. Shown also histogram plots of proliferation of γδ T cells upon culture with PHA and in the presence of CAF and CSC supernatants. (B) Frequency of IL-17- or IFN-γ-producing γδ T cells upon incubation for 24 or 48 hrs with PHA in the presence of CAF or CSC supernatant. Histograms show cumulative data from 5 different experiments. Error bars indicate SD. Shown are also representative dot plots.(C) Cumulative and flow cytometry analysis of IFN-γ and IL-17 production by CD4 and CD8 T cells upon incubation for 48 hrs with PHA in the presence of CAF or CSC supernatant.*p<0.05.
Supernatants from CSC were also capable to significantly inhibit IFN-γ production by polyclonal γδ T cell lines and promoted production of IL-17, while supernatants from CAF only minimally inhibited IFN-γ production and did not induce IL-17 production by γδ T cell lines (Fig. 3b). Overall, these results indicate that cells in the tumor microenvironment, and particularly CSC, produce immunomodulatory molecules capable to inhibit proliferation and IFN-γ production by γδ T cells and to promote their IL-17 production.
To test whether the effect of CSC supernatants was restricted to γδ T cells or was rather a more general phenomenon, we generated polyclonal CD4 and CD8 αβ T cell lines and tested the capability of 48-hrs CSC supernatants to inhibit IFN-γ production. As shown in Fig. 3c, supernatants from CSC significantly inhibited IFN-γ production by both CD4 and CD8 T cells lines, thus indicating that inhibitory molecules produced by CSC have a profound effect on several components of both adaptive (αβ) and innate-like (γδ)T cell immune response.
The above reported data clearly demonstrate the presence of biologically important immunomodulatory molecules in CSC secretome. Therefore, we performed a comparative analysis of levels of 50 different cytokines in supernatants of CSC and CAF by the Luminex platform. As shown in Fig. 4a, CSC produced remarkably elevated levels of several cytokines, while CAF had a more limited cytokine producing capabilities, with the exception of TGF-β which was preferentially produced by CAF. We then checked at cytokines which were differentially expressed in the inhibitory CSC secretome, but not in the non inhibitory CAF secretome. As shown in Fig. 4b, although there were several molecules unique to each CSC or CAF secretome, there were only 8 common molecules to every CSC secretome (FGF, GM-CSF, VEGF, Groα, IL-3, IL-8, IL-12 and IL-12p40), and 9 common molecules to every CAF secretome (FGF, GM-CSF, Groα, TGF-β, SDF1, HGF, OPN, IL-3 and IL-12p40). When analysis was restricted to cytokines differentially expressed by the CSC and CAF secretomes, there were only 3 cytokines uniquely expressed by the inhibitory CSC secretome, but absent (or produced at very low levels) in the non inhibitory CAF secretome, namely IL-8, IL-12 and VEGF. Among these 3 top overexpressed cytokines, IL-12 does not inhibit T cell proliferation and IFN-γ production but instead induces differentiation to IFN-γ production. Therefore, IL-8 and VEGF remain as potential candidates of the immunosuppressive activities of the CSC secretome.
Figure 4.
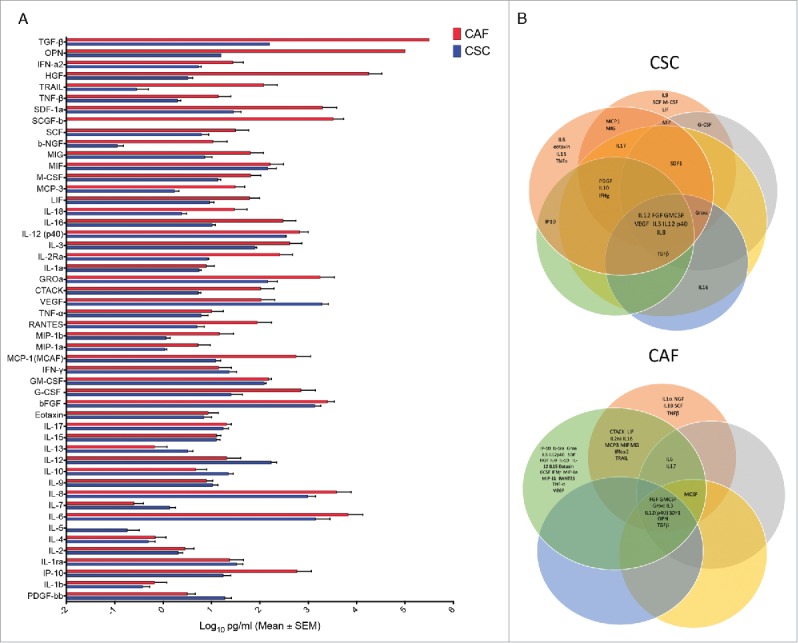
Comparative analysis of 50 different cytokines in CSC and CAF secretome (A) Levels of 50 different cytokines in 48 hrs supernatants of CSC and CAF by the Luminex platform. (B) Cytokine grouping in 6 CRC and 5 CAF samples as represented by Venn diagrams.
Intratumoral γδ T cells correlate with CRC outcome
Previous studies on the prognostic value of tumor-infiltrating γδ T cells have shown marked variability, with positive, negative, or even no correlation.22-23 This is strikingly depicted by results obtained in CRC in which γδ T cells making IL-17 positively correlated with advanced tumor clinicopathologic features in one study,24 but emerged as the most significant favorable prognostic population in another study.25
We initially investigated whether intratumoral γδ T cells had clinical relevance, by data mining transcriptomes and clinical files from the larger cohort of n = 585 CRC samples mentioned above. The leucocyte deconvolution of this data set evidenced a link between molecular markers of CRC and their abundance of TILs, whether of the γδ or αβ TCR subtype. The KRAS mutation status made no difference for these criteria. By contrast, the abundance of both subsets of TILs was significantly higher in mismatch repair-deficient (MMR-D) than -proficient (MMR-P) tumors, in BRAFmutated vs. BRAFwt, and in TP53wt vs. TP53mutated tumors. A higher content of TILs was also apparent in tumors positive for the CPG island methylator phenotype (CIMP) vs. their negative counterparts and in tumors negative for the chromosomal instability phenotype (CIN) vs. their positive counterparts. Importantly, each of these prognostic factors influenced in the same direction the abundance of γδ TILs and of αβ TILs (Fig. 5).
Figure 5.
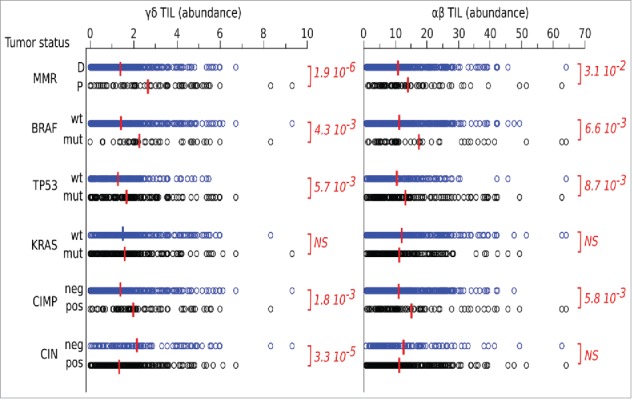
Data mining transcriptomes and abundance of TILs. Deconvolution of γδ TIL and αβ TIL abundances in CRC tumors according to their molecular and clinical hallmarks. Red bar indicate group means. Student's p values (2-sided) are indicated.
We then correlated frequencies of total γδ T cells with clinical outcome by analyzing the independent data set mentioned previously for n = 557 CRC patients for whom follow-up was available. Across the whole cohort, those patients with more abundant γδ TILs had a better DFS, as confirmed by stratifying patients according to expression of the TCRGV9-encoding gene. Patients with higher expression of IFNG also presented a higher DFS, whereas those with higher expression of IL17A had significantly reduced DFS. This γδ TIL and IFNG-dependent favorable pattern was conserved when reducing the whole cohort to those patients without lymph node invasion. By contrast, the IL17-dependent unfavorable pattern was rather observed in patients with invaded lymph nodes (Fig. 6a). Hence, stratifying the entire cohort according to both γδ TIL abundance and IFNG expression produced groups with strikingly different DFS, and demonstrated most notably that γδ TIL abundance overweighted IFNG expression in contributing to this outcome. The same conclusions hold true for γδ TIL abundance with regard to IL17A expression (Fig. 6b).
Figure 6.
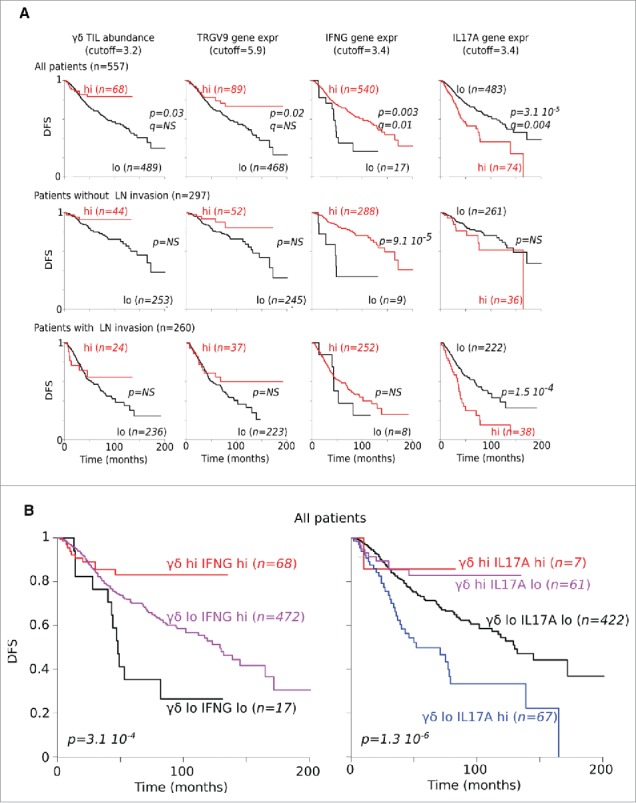
Correlation between gene expression and DFS. (A) DFS of CRC patients according to abundance of γδ TILs as well as of TCRGV9, IFNG and IL17A gene expression levels. (B) DFS of CRC patients according to γδ TILs abundance and IFNG or IL17A gene expression.
Discussion
Accumulating evidences that high densities of mature T cells, particularly with a Th1 and cytotoxic orientation, in different locations of a primary tumor, correlate with favorable prognosis both in terms of disease-free and overall survival, strongly support the fact that a natural immune reaction controls tumor cell growth and smoothens cancer aggressiveness.10-11,32 The presence of tumor infiltrating lymphocytes in CRC is associated with a favorable prognosis but is not sufficient to overcome inhibitory changes within the tumor microenvironment over a prolonged period of time. Migration of lymphocytes from the circulation to the tumor site implies that the host immune system is capable of initiating an anti-tumor response. Unfortunately, changes that occur as a result of mutations within tumor cells eventually create an immunosuppressive tumor microenvironment that prevents tumor eradication by TILs.33-34
Tumor immunoevasion mechanisms are common and include the downregulation of tumor associated antigens, of MHC, and of costimulatory molecules. By contrast to αβ T cells, γδ T cells are not MHC restricted and show less dependence on costimulators such as CD28. Moreover, γδ T cells in humans display potent MHC unrestricted cytotoxic activity in vitro against various tumors and for example they are fully capable to kill colon cancer stem cells that had been sensitized in vitro by zoledronate or low dose chemotherapy.35-37
In this paper, we have studied CRC-infiltrating γδ T cells and correlated their levels with clinical outcome. Moreover, we have studied the influence of the tumor microenvironment on the functional responses of γδ T cells.
Results herewith reported show that γδ T cells are present among intratumoral leukocytes but they are a minor T cell population accounting for approximately 20% of total CD3+ cells. Immunofluorescence analysis additional revealed that γδ T cells were mainly detected in the tumor border/stroma, but very rarely in the intratumor tissue.
T cells expressing Vδ1 were the dominant γδ subset in CRC tissue and also in adjacent normal tissues, but Vδ1 and Vδ2 T cell subsets were not significantly increased in tumor tissue. Phenotypic analysis showed that most of CRC-infiltrating Vδ1 and Vδ2 T cells had TEM and TEMRA phenotypes, similar to the phenotype of Vδ1 and Vδ2 T cells residing in adjacent normal colon tissue, but distinct from that of the corresponding cells from the peripheral blood, suggesting that residence in a non-lymphoid peripheral tissue, regardless of whether it is normal or has undergone tumor transformation, may be a major determinant of the phenotypic characteristics of tumor and tissue γδ T cells.
Functional analysis of tumor-infiltrating γδ T cells demonstrate that both Vδ1 and Vδ2 T cells in CRC and adjacent normal tissues preferentially produce IFN-γ, but very low IL-17. Moreover, IFN-γ production by both Vδ1 and Vδ2 T cells is significantly reduced in CRC tissues, as compared with adjacent normal colon tissue samples. These finding are surprising in light of a previous study showing that γδ T cells are the major cellular source of IL-17 in human CRC.26 Using 3 different FACS gating strategies we were able to confirm the majority of IL-17-producing leukocytes both in CRC and in adjacent normal tissues were in fact CD3+ but not Vδ1 or Vδ2, suggesting they are Th17 or Tc17 αβ T cells, and a sizeable fraction of IL-17 was expressed by CD3− cells, probably corresponding to ILC3. We also demonstrated that CD3+ but Vδ1− and Vδ2− T cells were the major source of TNF-α and IFN-γ in both CRC and adjacent normal tissues.
Current data on the prognostic value of CRC-infiltrating γδ T cells show marked variability: while one initial study found that γδ17 T cells (expressing either Vδ1 or Vδ2) positively correlate with advanced tumor clinicopathologic features,26 2 most recent studies27,31 of expression signature from CRC with overall survival outcomes, revealed intratumoral γδ cells,27 and particularly the Vγ9Vδ2 subset,31 as the most significant favorable prognostic immune population. Results of data mining transcriptomes and clinical files from a large cohort of CRC samples revealed that 5-year DFS probability was significantly higher in CRC patients with high number of tumor infiltrating γδ T cells and IFN-γ positive cells.
Thus, it is the maintenance of IFN-γ production, that positively associates with better patient outcome, and it is largely influenced by the tumor microenvironment. Accordingly, supernatants from colon CSCs significantly inhibited proliferation and IFN-γ production by γδ T cells and promoted production of IL-17. Supernatants from other components of the tumor tissue microenvironment such as CAF had limited suppressive ability and did not promote production of IL-17. We also found that colon CSC supernatants significantly inhibited IFN-γ production by both CD4 and CD8 T cells, clearly indicating that inhibitory molecules produced by colon CSC have a profound effect on several components of both adaptive (αβ) and innate-like (γδ) T cell immune response. We do not have clear evidence on which molecule(s) is made by colon CSC which is responsible for their immunosuppressive activities: there were only 3 cytokines differentially expressed by the inhibitory CSC secretome, but absent in the non inhibitory CAF secretome, namely IL-8, IL-12 and VEGF. IL-12 does not have inhibitory activity on T cell proliferation and IFN-γ production, which leaves IL-8 and VEGF as potential candidates of the immunosuppressive activities of the colon CSC secretome. While both these 2 molecules have the capability to suppress T cell responses, this is not due to a direct on T cells but is rather mediated by other cell types like dendritic cells, myeloid-derived suppressor cells, M2 macrophages and Treg cells.38-39 Additionally, it is also possible that immunosuppressive elements like prostaglandins,40 kynurenins41 or potassium42 may be responsible. Therefore, additional studies are needed to find out the molecule responsible for the immunosuppressive activities of the colon CSC secretome on αβ and γδ T cells.
In conclusion, our results clearly show that γδ T cells are a minor population among colon cancer-infiltrating leukocytes, have an effector phenotype but reduced capacity to produce IFN-γ, when compared with γδ T cells from adjacent normal colon tissue and peripheral blood, but do not produce IL-17. Moreover, they are correlated with clinical outcome indicating they are probably involved in controlling tumor growth at a early stage of disease.
Materials and methods
Characteristics of sample cohort
Colon cancer tissues and adjacent normal colon tissues were obtained from the Department of Surgery at the University Hospital of Palermo. We enrolled 70 patients (52 males, 18 females, median age 62 years, age range 42–82 years) undergoing a colon resection for colon adenocarcinoma and diagnosis of CRC was histologically confirmed.
A blood drawing was taken before the surgical excision. The study received authorisation by the local ethical committee and was performed in accordance to the principles of the Helsinki declaration. All individuals gave written informed consent to participate.
Isolation of tumor-infiltrating and circulating immune cells and flow cytometry analysis
Colon cancer specimens and adjacent normal colon tissues were freshly obtained at the time of primary surgery and transported to the laboratory for processing. Tissue was minced into small pieces followed digestion with Collagenase type IV and DNAse (Sigma, St Louis, MO) for 2 hrs at 37°C 5% CO2. After digestion, the cells extracted were washed twice in incomplete medium (RPMI 1640, Gibco, Grand Island, NY). Whole blood samples were obtained from the same patients recruited for the collection of tissue specimens before the surgical procedure, and used for the comparative analysis between peripheral blood and cancer tissue. The peripheral blood mononuclear cells (PBMCs) were separated from whole blood by density gradient centrifugation using Ficoll-Hypaque (Pharmacia Biotech, Uppsala, Sweden).
Both PBMC and tumor infiltrating cells were stained for live/dead discrimination using Invitrogen Live/Dead fixable violet dead cell stain kit (Invitrogen, Carlsbad, CA). Fc receptor blocking was performed with human immunoglobulin (Sigma, 3 μg/ml final concentration) followed by surface staining with different fluorochrome-conjugated antibodies to study the composition of the different subpopulations. The fluorescein isothiocyanate (FITC)-, phycoerythrin (PE)-, PE-Cy5-, allophycocyanin (APC)-, phycoerythrin-Cy7 (PECy7)-, allophycocyanin-Cy7 (APC-Cy7)-conjugated monoclonal antibodies (mAbs) used to characterize the entire population were the following: anti-CD3 (Cat 45–0037 eBioscience, Cat 300412 and Cat 300420 Biolegend), anti-CD45 (Cat 560274 BD Bioscience), anti-pan γδ TCR (Cat 555717 BD), anti-Vδ1 (Cat PG196007 ThermoFisher), anti-Vδ2 (Cat 331408 Biolegend), anti-CD4, (Cat 17–0279 eBioscience), anti-CD45RA (Cat 25–0458 eBioscience), anti-CD19 (Cat 302228 Biolegend), anti-CD4, (Cat 130–094–158 Miltenyi, Cat 348809 BD), anti CD8 (Cat 555367 BD) and anti-CD56 (Cat 341027 BD).
Expression of surface markers was determined by flow cytometry on a FACSCanto II Flow Cytometer with the use of the FlowJo software (BD Biosciences). The gating strategy involved progressively measuring total cells; viable cells only; lymphomonocytes and specific cell types. For every sample 100.000 nucleated cells were acquired and values are expressed as percentage of viable lymphomonocytes, as gated by forward and side scatter.
To study intracellular IFN-γ, IL-17 and TNF-α, cells from tumor and adjacent normal colon tissues were stimulated with Ionomycin and PMA in the presence of monensin for 4 hrs at 37°C in 5% CO2. The cells were harvested, washed twice in PBS with 1% FCS and fixed with PBS containing 4% paraformaldehyde overnight at 4°C. Fixation was followed by permeabilization with PBS containing 1% FCS, 0.3% saponin, and 0.1% Na azide for 15 min at 4°C. Staining of intracellular cytokines was performed by incubation of fixed permeabilized cells with FITC-labeled anti-IFN-γ (Cat 502506 Biolegend), APC-labeled anti-IL-17A (Cat 130–096–748 Miltenyi) and PeVio770 anti-TNF-a (Cat 130–096–748 Miltenyi). After 2 more washes in PBS containing 1% FCS, the cells were analyzed by Facs Canto II flow cytometer (BD Bioscience). Viable lymphocytes were gated by forward and side scatter, and analysis was performed on 100,000 acquired events for each sample by using FlowJo and the following gating strategy to detect lymphocytes from FSC/SSC, single cells, double positive CD45+ CD3+, and Vδ1 and Vδ2 positive T cells.
Polyclonal γδ T cell lines were prepared as described previously,43-44 were labeled with CFSE (Molecular Probes, Eugene, USA) and 5 × 105 cells were incubated with CAF and CSC supernatants in 24-well plates (Costar, Cambridge, MA) for 7 d at 37°C, 5% CO2, in addition to PHA or anti CD3/CD28. Proliferation was assessed after 7 d of culture according to loss of CFSE labeling.
To obtain CD4 and CD8 polyclonal αβ T cell lines, CD4 and CD8 T cells were sorted from PBMC of healthy donors using MACS cell separation kit, and incubated for 2 weeks with PHA, IL-2 and Beads (Dynabeads, ThermoFisher). Cell lines were incubated with CAF and CSC supernatants for 48 hrs and then stained for IL-17 and IFN-γ content upon Ionomycin and PMA stimulation.
Preparation of CAF and CSC conditioned medium, Luminex and ELISA analysis
Primary cancer associated fibroblasts (CAFs) and colon cancer stem cells (CSCs) were obtained from 15 surgical resection of CRC subjected to mechanical and enzymatic digestion with collagenase (0.6 mg/ml, Gibco) and hyaluronidase (10μg/ml, Sigma). Cell suspension was cultured in 10% fetal bovine serum (FBS) Dulbecco's modified Eagle's medium (DMEM) in adhesion flasks, to obtain CAFs, or in low-adhesion conditions and in serum-free medium supplemented with EGF and β-FGF, which allows the selective growth of colon CSCs.45-46 Cells were plated and incubated in their specific medium for 48 hrs; the medium was then collected and used for luminex assay.
Fourthy-eight cytokines (IL-1α, IL-1β, IL-1R antagonist, IL-2, IL-2Rα, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-12, IL-12 (p40), IL-13, IL-15, IL-16, IL-17, IL-18, TNF-α, TNF-β, IFN-α2, IFN-γ, G-CSF, GM-CSF,M-CSF, FGF-β, VEGF, PDGF, MIF, MIG, HGF, LIF, NGF-β,SCF, SCGF-β, SDF-1α, TRAIL, Eotaxin, IP-10, IL-8, MIP-1α, MIP-1β, MCP-1, RANTES, CTACK, GRO-α, MCP-3) were analyzed in CAF and CSC conditioned medium by xMAP multiplex technology on the Luminex platform (Luminex, Austin, TX), using Bio-Rad reagents (Bio-Plex Pro™ Human Cytokine 27-plex Assay #M500KCAF0Y and Bio-Plex Pro™ Human Cytokine 21-plex Assay #MF0005KMII, Bio-Rad, Hercules, CA) acquired and analyzed with the Bioplex Manager Software (Bio-Rad). Responses were scored positive if the value was 2-fold over the negative control. TGF-β and OPN were measured by ELISA according to the manufacturer's instructions (R&D Systems).46
Immunfluorescence analysis
Fresh frozen tissue samples were incubated with primary antibodies to the pan-γδ TCR and CD3 and in a subsequent secondary step, FITC-conjugated goat anti-rabbit and Rhodamine B200-conjugated goat anti-mouse IgG were used to detect them. Irrelevant isotype-matched primary mAbs were used to control for nonspecific staining. Analysis was performed with a confocal laser scanning microscopy equipped with 20x, 40x and 63x objectives. The tumor border configuration was diagnosed according to the method proposed by Jass et al.47 at low magnification. Briefly, the tumor margins were identified as infiltrating when there was no recognizable margin of growth and a “streaming dissection” between the normal structures of the bowel wall was present. Margins were considered pushing when they were reasonably well circumscribed, and they often were associated with a well-developed inflammatory lamina.
Transcriptome analysis
Public raw data of 585 colon cancers transcriptomes using Affymetrix HGU133 Plus 2.0 microarrays were downloaded from the NCBI-GEO data set repository (GSE3958229, https://www.ncbi.nlm.nih.gov/geo), normalized together and collapsed to HUGO gene symbols using chipset definition files available from the NCBI gene expression omnibus. The Pearson correlation coefficient between IFNG, IL17A, TRGV9 and CD3D gene expressions were calculated.
Deconvolution of immune population are calculated using the CIBERSORT software with 500 Monte Carlo iterations30 (https://cibersort.stanford.edu/) associated with the LM7 matrix.31 Sample Enrichment Scores (SES)48 of immune genes were computed using the open source software AutoCompare-SES (https://sites.google.com/site/fredsoftwares/products/autocompare_ses) with normalized settings. αβ and γδ TIL abundance were automatically calculated from the deconvolution result and SES using the open source software DeepTIL (https://sites.google.com/site/fredsoftwares/products/deeptil).31
SPADE analysis
Spanning-tree progression analysis of density-normalized events (SPADE)49 clustering algorithm on the Cytobank.org platform was performed to visualize single cells, among live CD45+ lymphocytes from 6 subjects. The nodes of the tree reproduce clusters of cells that are similar in marker expression. SPADE uses the size and color of each node to signify the number of cells and median marker expression, respectively.
Statistics
Data were analyzed for statistical significance using Mann-Whitney test for 2 groups and Kruskal-Wallis test for more than 2 groups. Differences between groups with a probability of ≤ 0.05 were regarded as significant. All data were analyzed using GraphPad Prism version 6.0e (GraphPad, San Diego, CA). All values are expressed as mean ± SD. Comparison of TIL abundance in clinical groups were done using unpaired Student t-test.
For Kaplan-Meier plots, optimal cutoffs were determined with the survival R package50 and the Log-Rank p values were corrected using the Benjamini-Hochberg method.51
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Achim Jungbluth, Department of Pathology Memorial Sloan-Kettering Cancer Center New York, for helping with immunofluorescence analysis of colon cancer tissue samples and Adrian Hayday for reading the manuscript and for training Elena Lo Presti at King's College London.
Funding
This work was supported in part by grants from the Ministry of Health “Ricerca Finalizzata 2007” to F.D and by Associazione Italiana per la Ricerca sul Cancro (AIRC) to M.T. (AIRC IG 14415).
References
- 1.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet 2010; 375(9719):1030-47; PMID:20304247; https://doi.org/ 10.1016/S0140-6736(10)60353-4 [DOI] [PubMed] [Google Scholar]
- 2.Aune D, Lau R, Chan DS, Vieira R, Greenwood DC, Kampman E, Norat T. Dairy products and colorectal cancer risk: A systematic review and meta-analysis of cohort studies. Ann Oncol 2012; 23(1):37-45; PMID:21617020; https://doi.org/ 10.1093/annonc/mdr269 [DOI] [PubMed] [Google Scholar]
- 3.Kennedy DA, Stern SJ, Moretti M, Matok I, Sarkar M, Nickel C, Koren G. Folate intake and the risk of colorectal cancer: A systematic review and meta-analysis. Cancer Epidemiol 2011; 35(1):2-10; PMID:21177150; https://doi.org/ 10.1016/j.canep.2010.11.004 [DOI] [PubMed] [Google Scholar]
- 4.Ma Y, Zhang P, Wang F, Yang J, Liu Z, Qin H. Association between vitamin D and risk of colorectal cancer: A systematic review of prospective studies. J Clin Oncol 2011; 29(28):3775-82; PMID:21876081; https://doi.org/ 10.1200/JCO.2011.35.7566 [DOI] [PubMed] [Google Scholar]
- 5.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology 2010; 138(6):2044-58; PMID:20420945; https://doi.org/ 10.1053/j.gastro.2010.01.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coussens LM, Werb Z. Inflammation and cancer. Nature 2002; 420:860-7; PMID:12490959; https://doi.org/ 10.1038/nature01322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hannahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell 2011; 144(5):646-74; PMID:21376230; https://doi.org/ 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 8.Fridman WH, Dieu-Nosjean MC, Pagès F, Cremer I, Damotte D, Sautès-Fridman C, Galon J. The immune microenvironment of human tumors: General significance and clinical impact. Cancer Microenviron 2013; 6(2):117-22; PMID:23108700; https://doi.org/ 10.1007/s12307-012-0124-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasada T, Suekane S. Variation of tumor-infiltrating lymphocytes in human cancers: Controversy on clinical significance. Immunotherapy 2011; 3(10):1235-51; PMID:21995574; https://doi.org/ 10.2217/imt.11.106 [DOI] [PubMed] [Google Scholar]
- 10.Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: A systematic review with meta-analysis. Br J Cancer 2011; 105(1):93-103; PMID:21629244; https://doi.org/ 10.1038/bjc.2011.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, et al.. Type, density, and location of immune cells within human colorectal tumors predicts clinical outcome. Science 2006; 313(5795):1960-4; PMID:17008531; https://doi.org/ 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 12.Pagès F, Kirilovsky A, Mlecnik B, Asslaber M, Tosolini M, Bindea G, Lagorce C, Wind P, Marliot F, Bruneval P, et al.. In situ cytotoxic and memory T cells predict outcome in early-stage colorectal cancer patients. J Clin Oncol 2009; 27(35):5944-51; PMID:19858404; https://doi.org/ 10.1200/JCO.2008.19.6147 [DOI] [PubMed] [Google Scholar]
- 13.Groh V, Porcelli S, Fabbi M, Lanier LL, Picker LJ, Anderson T, Warnke RA, Bhan AK, Strominger JL, Brenner MB. Human lymphocytes bearing T cell receptor γδ are phenotypically diverse and evenly distributed throughout the lymphoid system. J Exp Med 1989; 169(4):1277-94; PMID:2564416; https://doi.org/ 10.1084/jem.169.4.1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonneville M, O'Brien RL, Born WK. γδ T cell effector functions: A blend of innate programming and acquired plasticity. Nat Rev Immunol 2010; 10(7):467-78; PMID:20539306; https://doi.org/ 10.1038/nri2781 [DOI] [PubMed] [Google Scholar]
- 15.Constant P, Davodeau F, Peyrat MA, Poquet Y, Puzo G, Bonneville M, Fournié JJ. Stimulation of human γδ T cells by nonpeptidic mycobacterial ligands. Science 1994; 264(5156):267-70; PMID:8146660; https://doi.org/ 10.1126/science.8146660 [DOI] [PubMed] [Google Scholar]
- 16.Eberl M, Hintz M, Reichenberg A, Kollas AK, Wiesner J, Jomaa H. Microbial isoprenoid biosynthesis and human γδ T cell activation. FEBS Lett 2003; 544(1-3):4-10; PMID:12782281; https://doi.org/ 10.1016/S0014-5793(03)00483-6 [DOI] [PubMed] [Google Scholar]
- 17.Tanaka Y, Morita CT, Nieves E, Brenner MB, Bloom BR. Natural and synthetic non-peptide antigens recognized by human γδ T cells. Nature 1995; 375(6527):155-8; PMID:7753173; https://doi.org/ 10.1038/375155a0 [DOI] [PubMed] [Google Scholar]
- 18.Vavassori S, Kumar A, Wan GS, Ramanjaneyulu GS, Cavallari M, El Daker S, Beddoe T, Theodossis A, Williams NK, Gostick E, et al.. Butyrophilin 3A1 binds phosphorylated antigens and stimulates human γδ T cells. Nat Immunol 2013; 14(9):908-16; PMID:23872678; https://doi.org/ 10.1038/ni.2665 [DOI] [PubMed] [Google Scholar]
- 19.Harly C, Guillaume Y, Nedellec S, Peigné CM, Mönkkönen H, Mönkkönen J, Li J, Kuball J, Adams EJ, Netzer S, et al.. Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human γδT-cell subset. Blood 2012; 120(11):2269-79; PMID:22767497; https://doi.org/ 10.1182/blood-2012-05-430470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandstrom A, Peigné CM, Léger A, Crooks JE, Konczak F, Gesnel MC, Breathnach R, Bonneville M, Scotet E, Adams EJ. The intracellular B30.2 domain of butyrophilin 3A1 binds phosphoantigens to mediate activation of human Vγ9Vδ2 T cells. Immunity 2014; 40(4):490-500; PMID:24703779; https://doi.org/ 10.1016/j.immuni.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gober H, Kistowska M, Angman L, Jenö P, Mori L, De Libero G. Human T cell receptor γδ cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med 2003; 197(2):163-8; PMID:12538656; https://doi.org/ 10.1084/jem.20021500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dieli F, Gebbia N, Poccia F, Caccamo N, Montesano C, Fulfaro F, Arcara C, Valerio MR, Meraviglia S, Di Sano C, et al.. Induction of γδ T lymphocyte effector functions by bisphosphonate zoledronic acid in cancer patients in vivo. Blood 2003; 102(6):2310-1; PMID:12959943; https://doi.org/ 10.1182/blood-2003-05-1655 [DOI] [PubMed] [Google Scholar]
- 23.Dieli F, Poccia F, Lipp M, Sireci G, Caccamo N, Di Sano C, Salerno A. Differentiation of effector/memory Vδ2 T cells and migratory routes in lymph nodes or inflammatory sites. J Exp Med 2003; 198(3):391-7; PMID:12900516; https://doi.org/ 10.1084/jem.20030235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo Presti E, Dieli F, Meraviglia S. Tumor-infiltrating γδ T lymphocytes: Pathogenic role, clinical significance, and differential programming in the tumor microenvironment. Front Immunol 2014; 5:607; PMID:25505472; https://doi.org/ 10.3389/fimmu.2014.00607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva-Santos B, Serre K, Norell H. γδT cells in cancer. Nat Rev Immunol 2015; 15(11):683-91; PMID:26449179; https://doi.org/ 10.1038/nri3904 [DOI] [PubMed] [Google Scholar]
- 26.Wu P, Wu D, Ni C, Ye J, Chen W, Hu G, Wang Z, Wang C, Zhang Z, Xia W, et al.. γδT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity 2014; 40:785-800; PMID:24816404; https://doi.org/ 10.1016/j.immuni.2014.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al.. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015; 21(8):938-45; PMID:26193342; https://doi.org/ 10.1038/nm.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marisa L, Reyniès LA, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D, Ayadi M, et al.. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 2013; 10:e1001453; PMID:23700391; https://doi.org/ 10.1371/journal.pmed.1001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GSE39582, https://www.ncbi.nlm.nih.gov/geo.
- 30.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Meth 2015; 12:453-7; PMID:25822800; https://doi.org/ 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tosolini M, Pont F, Poupot M, Vergez F, Nicolau-Travers ML, Vermijlen D, Sarry JE, Dieli F, Fournie JJ. Assessment of tumor-infiltrating TCRVγ9Vδ2 lymphocyte abundance by deconvolution of human cancers microarrays. Oncoimmunology 2017; 6:e1284723; PMID:28405516; https://doi.org/ 10.1080/2162402X.2017.1284723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smyth MJ, Dunn GP, Schreiber RD. Cancer immunosurveillance and immunoediting: The roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Adv Immunol 2006; 90:1-50; PMID:16730260; https://doi.org/ 10.1016/S0065-2776(06)90001-7 [DOI] [PubMed] [Google Scholar]
- 33.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer 2012; 12(4):298-306; PMID:22419253; https://doi.org/ 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- 34.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013; 14:1014-22; PMID:24048123; https://doi.org/ 10.1038/ni.2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todaro M, D'Asaro M, Caccamo N, Iovino F, Francipane MG, Meraviglia S, Orlando V, La Mendola C, Gulotta G, Salerno A, et al.. Efficient killing of human colon cancer stem cells by γδ T lymphocytes. J Immunol 2009; 182(11):7287-96; PMID:19454726; https://doi.org/ 10.4049/jimmunol.0804288 [DOI] [PubMed] [Google Scholar]
- 36.Todaro M, Orlando V, Cicero G, Caccamo N, Meraviglia S, Stassi G, Dieli F. Chemotherapy sensitizes colon cancer initiating cells to Vγ9Vδ2 T cell-mediated cytotoxicity. PLoS One 2013; 8(6):e65145; PMID:23762301; https://doi.org/ 10.1371/journal.pone.0065145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Todaro M, Meraviglia S, Caccamo N, Stassi G, Dieli F. Combining conventional chemotherapy and γδ T cell-based immunotherapy to target cancer-initiating cells. Oncoimmunology 2013; 2(9):e25821; PMID:24244907; https://doi.org/ 10.4161/onci.25821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T, Chen Y, Han Y, Wu K. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev 2016; 31:61-71; PMID:27578214; https://doi.org/ 10.1016/j.cytogfr.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voron T, Marcheteau E, Pernot S, Colussi O, Tartour E, Taieb J, Terme M. Control of the immune response by pro-angiogenic factors. Front Oncol 2014; 4:70; PMID:24765614; https://doi.org/ 10.3389/fonc.2014.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basingab FS, Ahmadi M, Morgan DJ. IFN-γ-dependent interactions between ICAM-1 and LFA-1 counteract prostaglandin E2-mediated Inhibition of antitumor CTL responses. Cancer Immunol Res 2016; 4(5):400-11; PMID:26928462; https://doi.org/ 10.1158/2326-6066.CIR-15-0146 [DOI] [PubMed] [Google Scholar]
- 41.Zhai L, Spranger S, Binder DC, Gritsina G, Lauing KL, Giles FJ, Wainwright DA. Molecular pathways: Targeting IDO1 and other tryptophan dioxygenases for cancer immunotherapy. Clin Cancer Res 2015; 21(24):5427-33; PMID:26519060; https://doi.org/ 10.1158/1078-0432.CCR-15-0420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eil R, Vodnala SK, Clever D, Klebanoff CA, Sukumar M, Pan JH, Palmer DC, Gros A, Yamamoto TN, Patel SJ, et al.. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 2016; 537(7621):539-43; PMID:27626381; https://doi.org/ 10.1038/nature19364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sireci G, Espinosa E, Di Sano C, Dieli F, Fournié JJ, Salerno A. Differential activation of human γδ cells by non peptide phosphoantigens. Eur J Immunol 2001; 31(5):1628-35; PMID:11465120; https://doi.org/ 10.1002/1521-4141(200105)31:5%3c1628::AID-IMMU1628%3e3.0.CO;2-T [DOI] [PubMed] [Google Scholar]
- 44.Sireci G, Champagne E, Fourniè JJ, Dieli F, Salerno A. Patterns of phosphoantigen stimulation of human Vγ9Vδ2 T cell clones include Th0 cytokines. Hum Immunol 1997; 58(2):70-82; PMID:9475336; https://doi.org/ 10.1016/S0198-8859(97)00211-5 [DOI] [PubMed] [Google Scholar]
- 45.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007; 445(7123):111-5; PMID:17122771; https://doi.org/ 10.1038/nature05384 [DOI] [PubMed] [Google Scholar]
- 46.Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G, Medema JP, et al.. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 2007; 1(4):389-402; PMID:18371377; https://doi.org/ 10.1016/j.stem.2007.08.001 [DOI] [PubMed] [Google Scholar]
- 47.Shinto E, Mochizuki H, Ueno H, Matsubara O, Jass JR. A novel classification of tumour budding in colorectal cancer based on the presence of cytoplasmic pseudo-fragments around budding foci. Histopathology 2005; 47(1):25-31; PMID:15982320; https://doi.org/ 10.1111/j.1365-2559.2005.02162.x [DOI] [PubMed] [Google Scholar]
- 48.Tosolini M, Algans C, Pont F, Ycart B, Fournie JJ. Large scale microarray profiling reveals four stages of immune escape in non-Hodgkin's lymphomas. Oncoimmunology 2016; 5(7):e1188246; PMID:27622044; https://doi.org/ 10.1080/2162402X.2016.1188246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu P, Simonds EF, Bendall SC, Gibbs KD Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 2011; 29(10):886-91; PMID:21964415; https://doi.org/ 10.1038/nbt.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Therneau TM, Grambsch PM. Modeling survival data: Extending the cox model. New York: Springer; (2000). [Google Scholar]
- 51.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med 1990; 9(7):811-8; PMID:2218183; https://doi.org/ 10.1002/sim.4780090710 [DOI] [PubMed] [Google Scholar]


