Abstract
The emergence of Escherichia coli sequence type 131 (ST131) as a multidrug-resistant and virulent pathogen represents a major challenge to public health globally. Recently, the O25b/ST131 E. coli producing CTX-M-15 with high virulence potential has been reported worldwide, but has received little attention in Iran. This study is the first in Iran to specifically determine the spread of the O25b/ST131 clone producing CTX-M-15 among E. coli isolates belonging to the B2 phylogenetic group. ST131 clone in phylogenetic group B2 was detected based on PCR detection of ST131-specific single-nucleotide polymorphisms in mdh and gyrB. O25b/ST131 E. coli clone was confirmed utilizing O25b/ST131 clone allele-specific PCR for the pabB gene. All group B2 E. coli isolates were characterized based on antibiotic susceptibility, extended-spectrum β-lactamase (ESBL) enzymes, and virulence traits. Our results demonstrated that 38 out of the 154 B2 group isolates (24.7%) were identified as belonging to the ST131 clone. Furthermore, of these, 28 isolates (73.6%) were detected as O25b/ST131 clone. Antibiotic resistance of ST131 E. coli isolates to ciprofloxacin, gentamicin, cefotaxime, and aztreonam was significantly higher than non-ST131 isolates. Almost all of the O25b/ST131 isolates with the ability for ESBL production were reported as CTX-M-15 producing (95.5%). Our results showed that the most prevalent virulence trait in ST131 clone was ompT (94.7%). This study is the first to report the prevalence of the CTX-M-15-producing O25b/ST131 E. coli in Iran. Our findings reinforce the surveillance of dissemination of ST131 E. coli clone as a major drug-resistant pathogen and an important new public health threat.
Keywords: : O25b/ST131 E. coli, extended-spectrum beta-lactamases, CTX-M-15, B2 phylogenetic group, antibiotic resistance
Introduction
Escherichia coli is a major component of the human intestinal flora and certain strains can cause a wide spectrum of intestinal and extraintestinal diseases both in the community and healthcare settings.1 Human extraintestinal E. coli diseases are important causes of morbidity, mortality, and contribute to increased healthcare costs. Management of the infections is complicated by the increasing prevalence and spectrum of antibiotic resistance.2,3
E. coli sequence type 131 (ST131) is the predominant extraintestinal pathogenic E. coli that recently emerged as a global epidemic and multidrug-resistant clone. E. coli ST131 clone belongs to the highly virulent phylogenetic group B2 and its strains are mostly of serotype O25:H4, with a specific O25 type, O25b. In 2008, E. coli O25b/ST131 was identified in three continents.4–6 Emergence of high rates of antimicrobial resistance in E. coli ST131 has become a public health issue because of the limited therapeutic options for management of infections caused by this pathogen.7,8 Of particular concern is the increasing emergence of extended-spectrum β-lactamase (ESBL)-producing E coli. The ST131 clone is strongly associated with ESBLs, predominantly the CTX-M-15 type.9,10 In addition, E. coli ST131 clone occurred in both inpatients and outpatients globally, which represents its widespread dissemination. So, today, E. coli ST131 is a pathogen of significant clinical concern.11,12
Given the ability to withstand antimicrobial treatment, possession of high numbers of virulence factors and widespread dissemination, the E. coli ST131 clone posed a significant threat to human health. This study aims to evaluate the prevalence of E. coli O25b/ST131 clone producing CTX-M-15 in extraintestinal infections among inpatients and outpatients.
Materials and Methods
Bacterial isolates
The study was carried out with a total of 384 nonduplicate clinical E. coli isolates obtained from outpatients and inpatients hospitalized at Imam Reza Hospital, Birjand University of Medical Sciences. The clinical samples contained urine, wound swab, blood, and sputum. These samples were collected over a period of 12 months from September 2012 to September 2013. The E. coli isolates were identified utilizing conventional microbiological methods and biochemical testing.
Phylogenetic group analysis and detection of virulence determinants
For determination of major E. coli phylogenetic group (A, B1, B2, and D), the chuA and yjaA genes and TspE4.C2 fragments of DNA were examined by triplex PCR assay.13 In addition, all B2 group isolates were screened for three genes encoding putative virulence factors (cnf1, ompT, and iha) by PCR with primers as previously described.14
O25b/ST131 screening
ST131 clone in phylogenetic group B2 was detected on the basis of PCR detection of ST131-specific single-nucleotide polymorphisms (SNPs) in mdh and gyrB.15 The PCR was carried out in the total volume of 20 μl (10 μl of 2× Hot Star Taq Master Mix, 1 μl of the DNA template, 1 μl of each primer [20 pmol], and 7 μl of ddH2O) utilizing the Hot Star Taq Master Mix kit (SinaClon). DNA amplification was carried out in a thermocycler (Eppendorf) with an initial denaturation step at 95°C for 10 min, 35 amplification cycles each with 30 sec at 95°C; 30 sec at different temperatures for the various genes (Table 1); and 50 sec at 72°C, followed by an additional extension step of 7 min at 72°C. The amplified products were electrophoresed on 1% gel agarose containing 1× GelRed DNA stain (Biotium, Inc.). Moreover, in this study, detection of O25b/ST131 clone was confirmed by PCR using primers O25pabBspe described by Clermont et al.16
Table 1.
Polymerase Chain Reaction Primers and Annealing Temperatures Used for the Various Genes
| Primer | Sequence (5′-3′) | Products sizes (bp) | Annealing (°C) |
|---|---|---|---|
| blaTEM | Fw- AGATTTCATCTTTGATTCTTGG | 868 | 45 |
| Rv- AATTGATTCTTTAGCATCTGG | |||
| blaSHV | Fw- CCAGCCAACTATGGCGGAATC | 822 | 53 |
| Rv- CCTGTCGCAAGATCGACTGTA | |||
| blaCTX-M | Fw- ATGTGCAGCACCAGTAAAGT | 542 | 53 |
| Rv- ACCGCGATATCGTTGGTGG | |||
| blaCTX-M-15 | Fw- AAAAATGATTCGGCTCCAGAA | 483 | 54 |
| Rv- TGCCAGATTCGCTCTCAAAG | |||
| Mdh | Fw- GTTTAACGTTAACGCCGGT | 275 | 65 |
| Rv- GGTAACACCAGAGTGACCA | |||
| gyrB | Fw- CGCGATAAGCGCGAC | 132 | 65 |
| Rv- ACCGTCTTTTTCGGTGGAA | |||
| O25pabBspe | Fw- TCCAGCAGGTGCTGGATCGT | 347 | 56 |
| Rv- GCGAAATTTTTCGCCGTACTGT |
Antibiotic susceptibility testing
The antibiotic resistance profile of all group B2 isolates was determined by the Kirby-Bauer disk diffusion method, and the results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines.17 The antimicrobial agents (MAST Co.) tested in this study included Amikacin (30 μg), Amoxicillin–Clavulanic Acid (20/10 μg), Aztreonam (30 μg), Cefepime (30 μg), Ceftazidime (30 μg), Cefotaxime (30 μg), Ciprofloxacin (5 μg), Gentamicin (10 μg), Imipenem (10 μg), Meropenem (10 μg), Tetracycline (30 μg), Trimethoprim–Sulfamethoxazole (1.25/23.75 μg). E. coli ATCC 25922 was utilized as a standard strain.
ESBL screening test
ESBL production was detected utilizing combination disc test method based on CLSI recommendations.17 Briefly, phenotypic confirmatory test was performed by comparing the inhibition zone of disks containing cefotaxime and ceftazidime with and without clavulanic acid. Zones of inhibition were measured after overnight incubation at 37°C. An increase in the inhibition zone diameter of ≥5 mm for a combination disc versus ceftazidime disc alone confirmed ESBL production. E. coli ATCC 25922 (Negative control) and Klebsiella pneumoniae ATCC 700603 (Positive control) were utilized as reference strains.
Identification of bla genes
In this study, the blaTEM, blaSHV, blaCTX-M, and blaCTX-M-15 genes responsible for the ESBL activity in the B2 group ESBL-producing isolates were detected employing PCR and primers described in Table 1. The PCR was performed on the final volume of 20 μl using HotStar Taq Master Mix kit (SinaClon) containing 10 μl of 2× HotStar Taq Master Mix, 1 μl of the DNA template, 1 μl of each primer (20 pmol), and 7 μl of ddH2O. DNA amplification was conducted in a thermocycler (Eppendorf) with an initial denaturation step at 95°C for 5 min, 35 amplification cycles each with 1 min at 95°C; 30 sec at different temperatures for different genes (Table 1); and 50 sec at 72°C, followed by an additional extension step of 10 min at 72°C. The amplified products were electrophoresed on 1% agarose gel containing 1 × GelRed DNA stain (Biotium, Inc.).
Statistical analysis
Comparisons of proportions were analyzed using the Fisher's exact test in SPSS v.21 software. The level of statistical significance was set at p < 0.05.
Results
Source of isolates and phylogenetic group analysis
In this study, a total of 384 E. coli isolates were isolated from urine (97.3%), wound (1.6%), blood (0.3%), and sputum (0.8%) samples and these isolates were obtained from outpatients (94%) and inpatients (6%). Our analysis revealed that the most prevalent phylogenetic group among E. coli isolates was B2 (154 isolates, 40.1%), followed by group D (149 isolates, 38.8%), group A (75 isolates, 19.5%), and group B1 (six isolates, 1.6%).
Prevalence of the O25b/ST131 clone
In our study, 154 E. coli isolates belonging to group B2 were analyzed for detection of ST131 clone. The results demonstrated that 38 out of 154 isolates of group B2 (24.7%) were identified as belonging to the ST131 clone using PCR-based assay for specific SNPs in mdh and gyrB genes. Prevalence of ST131 clone among outpatients and inpatients was reported as 22.8% and 55.6%, respectively (p value = 0.024) (Table 2). In addition, the O25b/ST131 clone was identified by PCR using primers O25pabBspe. Accordingly, of the 38 ST131 strains, 28 strains (73.6%) were detected as the O25b/ST131 clone (Fig. 1).
Table 2.
Prevalence of ST131 Clone in B2 Group Escherichia coli Isolates Among Outpatients and Inpatients
| E. coli phylogenetic group B2 (n = 154) | E. coli ST131 (n = 38) | ||||
|---|---|---|---|---|---|
| Patients | Non-ST131 (%) | ST131 (%) | Non-O25b (%) | O25b (%) | p-Value* |
| Inpatients | 4 (44.4) | 5 (55.6) | 1 (20) | 4 (80) | 0.024 |
| Outpatients | 112 (77.2) | 33 (22.8) | 9 (27.3) | 24 (72.7) | |
p-Values (ST131 vs. non-ST131), by Fisher's exact test.
ST131, sequence type 131.
FIG. 1.
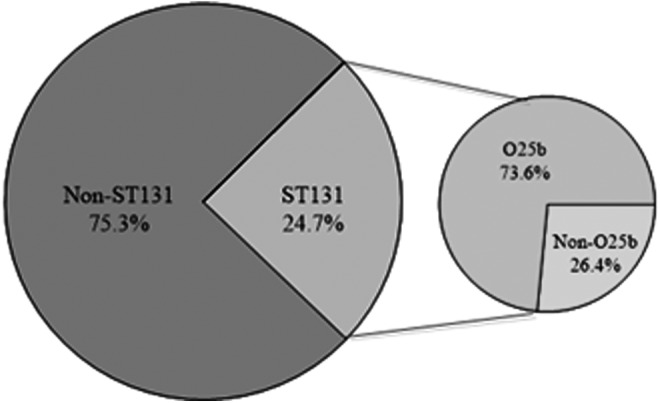
Distribution of ST131 and O25b/ST131 clone among 154 B2 group Escherichia coli isolates.
Antibiotic susceptibility testing
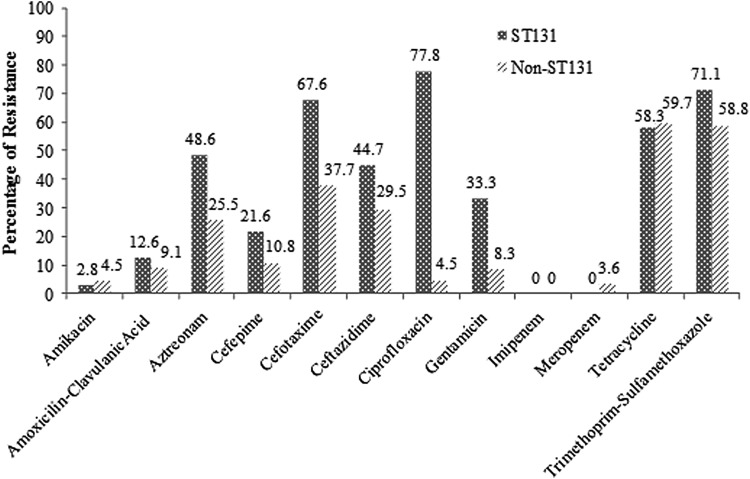
Antimicrobial resistance pattern was studied in 154 ST131 and non-ST131 E. coli isolates belonging to group B2 (Fig. 2). High resistance in ST131 E. coli isolates was reported to ciprofloxacin, trimethoprim–sulfamethoxazole, cefotaxime, and tetracycline (77.8%, 71.1%, 67.6%, and 58.3%, respectively). Moreover, resistance to these antibiotics (84.6%, 75%, 79.2%, and 57.9%, respectively) was most prevalent in O25b/ST131 isolates. It is noteworthy that both tetracycline and trimethoprim/sulfamethoxazole resistances were highly prevalent in both ST131 and non-ST131 isolates. Statistical analysis using Pearson's chi-Square test showed that antibiotic resistance of ST131 E. coli isolates to ciprofloxacin (p = 0.0000), gentamicin (p = 0.001), cefotaxime (p = 0.007), and aztreonam (p = 0.04) was significantly higher than non-ST131 E. coli isolates.
FIG. 2.
Antimicrobial resistance pattern of 154 ST131 and non-ST131 E. coli isolates.
Distribution of ESBL enzymes
ESBL production and the blaTEM, blaSHV, blaCTX-M, and blaCTX-M-15 genes responsible for the ESBL activity were evaluated in the B2 group E. coli isolates. In our study, 56 (46.3%) E. coli isolates belonging to group B2 had the ability of ESBL production. The prevalence of ESBL-producing strains among ST131 and non-ST131 isolates was reported for 65.7% and 26.7%, respectively. In addition, of the 28 total O25b/ST131 isolates, 22 isolates (78.5%) produced ESBL enzymes (Table 3). The results revealed that from the 25 ST131 ESBL-producing E. coli isolates, 17 (68%) were positive for blaTEM, 8 (32%) for blaSHV, 24 (96%) for blaCTX-M variants, and 22 (88%) for blaCTX-M-15. The blaTEM, blaSHV, blaCTX-M variants, and blaCTX-M-15 genes were detected in 35.5%, 38.7%, 64.5%, and 54.8% of non-ST131 ESBL-producing E. coli isolates, respectively. Moreover, almost all of the O25b/ST131 isolates with the ability for ESBL production were reported as CTX-M-15 producing (95.5%). Statistical analysis indicated that compared with non-ST131 isolates, ESBL-producing ST131 isolates were associated positively with CTX-M-15 (p = 0.007), CTX-M variants (p = 0.004), and TEM (p = 0.015) (Table 4).
Table 3.
Phylogenetic and Clonal Grouping of Extended-Spectrum β-Lactamase-Producing E. coli Isolates
| Phylogenetic/clonal group | ESBL positive (%) | ESBL negative (%) |
|---|---|---|
| Group A (n = 75) | 19 (25.3) | 56 (74.7) |
| Group B1 (n = 6) | 1 (16.6) | 5 (83.4) |
| Group B2 (n = 154) | 56 (36.4) | 98 (63.6) |
| Group D (n = 149) | 45 (30.2) | 104 (69.8) |
| ST131 (n = 38) | 25 (65.7) | 13 (34.3) |
| O25b/ST131 (n = 28) | 22 (78.5) | 6 (21.5) |
ESBL, extended-spectrum β-lactamase.
Table 4.
Extended-Spectrum β-Lactamase-Encoding Genes Among Fifty-Six ST131 and Non-ST131 Extended-Spectrum β-Lactamase-Producing E. coli Isolates
| Prevalence [no. (%)] of ESBL genotype in E. coli isolates | ||||
|---|---|---|---|---|
| ESBL genotype | Non-ST131 (31) | ST131 (25) | O25b/ST131 (22) | p-Value* |
| TEM | 11 (35.5) | 17 (68) | 15 (68.2) | 0.015 |
| SHV | 12 (38.7) | 8 (32) | 5 (22.7) | 0.406 |
| CTX-M | 20 (64.5) | 24 (96) | 22 (100) | 0.004 |
| CTX-M-15 | 17 (54.8) | 22 (88) | 21 (95.5) | 0.007 |
p-Values (ST131 vs. non-ST131), by Fisher's exact test.
Prevalence of virulence determinants
In this study, three virulence genes, including cnf1 (Cytotoxic necrotizing factor), iha (adhesin-siderophore), and ompT (outer membrane protease T), were studied in 154 ST131 and non-ST131 E. coli isolates belonging to group B2. Our results showed that the most prevalent virulence trait in ST131 E. coli isolates was ompT (94.7%). Furthermore, prevalence of cnf1 and iha factors in these isolates was reported as 39.5%. Compared with non-ST131 isolates, the ST131 isolates exhibited significantly greater prevalence of the iha gene (Table 5).
Table 5.
Prevalence of Virulence Traits of One Hundred Fifty-Four ST131 and Non-ST131 Escherichia coli Isolates
| Virulence traits | Non-ST131 (%) (n = 116) | ST131 (%) (n = 38) | O25b/ST131 (%) (n = 28) | p-Value* |
|---|---|---|---|---|
| cnf1 | 42 (36.2) | 15 (39.5) | 11 (39.3) | 0.43 |
| ompT | 97 (83.6) | 36 (94.7) | 27 (96.4) | 0.08 |
| Iha | 22 (19) | 15 (39.5) | 12 (42.9) | 0.01 |
p values (ST131 vs. non-ST131), by Fisher's exact test.
cnf1, cytotoxic necrotizing factor; iha, adhesin-siderophore; ompT, outer membrane protease T.
Discussion
E. coli ST131 clone belongs to the B2 phylogenetic group, which is known as the highly virulent and antimicrobial resistance group. In recent years, public health concern is that ST131 with CTX-M-15 is increasingly found in E. coli isolates with the ability to produce ESBL.18–20 Recently, the O25b/ST131 E. coli producing CTX-M-15 with high virulence potential has been reported worldwide.21 This study is the first in Iran to specifically determine the spread of the O25b/ST131 clone producing CTX-M-15 among E. coli.
In this study, 38 out of 154 E. coli isolates of group B2 (24.7%) were identified as belonging to the ST131 clone. Furthermore, of the 38 ST131 strains, 28 strains (73.6%) were detected as the O25b/ST131 clone. This study is the first report of prevalence of ST131 E. coli from Iran. Other countries such as Canada (78%),22 United States (47%),2 Japan (41%),23 and Denmark (38%)5 have also reported the prevalence of ST131 clone. This difference could be attributed to the geographical location and in some studies, the ST131 clone has been detected among all B2 group E. coli isolates, or among ESBL-producing isolates only.
Our results demonstrated that the highest resistance rate of ST131 and O25b/ST131 E. coli isolates was seen to ciprofloxacin, trimethoprim–sulfamethoxazole, cefotaxime, and tetracycline. Statistical analysis showed that antibiotic resistance of ST131 E. coli isolates to ciprofloxacin, gentamicin, cefotaxime, and aztreonam was significantly higher than non-ST131 E. coli isolates. In the study of Olesen et al., of the 115 ESBL study isolates, 44 (38%) represented the ST131 clonal group, all from phylogenetic group B2. They stated that antibiotic resistance of ST131 E. coli isolates to ciprofloxacin, florfenicol, gentamicin, nalidixic acid, and neomycin was significantly higher than non-ST131 E. coli isolates.5 The high levels of trimethoprim–sulfamethoxazole and ciprofloxacin resistance in E. coli isolates in this and other study have implications regarding empiric therapy for urinary tract infections since these frequently used antibiotics are less effective for such pathogens.
Recently, ESBL production in E. coli primarily due to the spread of CTX-M types has increased significantly. In addition, significant spread of O25b/ST131 E. coli producing CTX-M-15 represents a major public health problem globally.16,21 Our study demonstrated that blaCTX-M gene was most common in ST131 and O25b/ST131 ESBL-producing E. coli (96% and 100%, respectively), which confirms the importance of spread of CTX-M types in the ESBL production. In addition, almost all of the O25b/ST131 isolates with the ability for ESBL production were reported as CTX-M-15 producing (95.5%). Statistical analysis indicated that compared with non-ST131 isolates, ESBL-producing ST131 isolates were positively associated with CTX-M-15, CTX-M variants, and TEM genes. The high prevalence of the CTX-M-15-producing O25b/ST131 E. coli clone has been reported in other studies such as Markovska et al. (96%),24 Blanco et al. (74%),21 and Coelho et al. (63%).25 The presence of blaCTX-M-15 gene in E. coli isolates is significantly associated with antibiotics resistance to first-choice agents for empiric therapy (β-lactams) and may favor a selection pressure for the CTX-M-15-producing strains. So, the identification of high prevalence of CTX-M-15-producing ST131 E. coli clone may help in predicting the increasing rate of antibiotic treatment failure, which is proportional to the increasing rate of ST131 among other E. coli clones.
As mentioned earlier, the ST131 E. coli clone significantly exceeded the non-ST131 isolates for extent of virulence profiles. Our results indicated that the most prevalent virulence trait in ST131 E. coli isolates was ompT (94.7%). Compared with non-ST131 isolates, the ST131 isolates exhibited significantly greater prevalence of the iha gene. Other studies have also found that various virulence genes were significantly more prevalent among ST131 than non- ST131 isolates.3,25 This may represent the importance of global dissemination of E. coli ST131 as a major public health threat.
Conclusion
This study is the first to report the prevalence of CTX-M-15-producing O25b/ST131 E. coli in Iran. We found that a relatively high prevalence of O25b/ST131 E. coli possess high virulence factors and antimicrobial resistance. Most of these isolates had the ability to produce ESBL and were harboring gene of CTX-M-15. Our findings reinforce the surveillance of dissemination of E. coli ST131 as a major drug-resistant pathogen and an important new public health threat.
Acknowledgment
This research was supported by Birjand University of Medical Sciences, Birjand, Iran (Grant number: 1080).
Disclosure Statement
No competing financial interests exist.
References
- 1.Qureshi Z.A., and Doi Y. 2014. Escherichia coli sequence type 131: epidemiology and challenges in treatment. Expert Rev. Anti Infect. Ther. 12:597–609 [DOI] [PubMed] [Google Scholar]
- 2.Johnson J.R., Urban C., Weissman S.J., Jorgensen J.H., Lewis J.S., Hansen G., Edelstein P., Robicsek A., Cleary T., and Adachi J. 2012. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25: H4) and blaCTX-M-15 among extended-spectrum β-lactamase-producing E. coli from the United States (2000–2009). Antimicrob. Agents Chemother. 56:2364–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson J.R., Johnston B., Clabots C., Kuskowski M.A., and Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294 [DOI] [PubMed] [Google Scholar]
- 4.Rogers B.A., Sidjabat H.E., and Paterson D.L. 2010. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14 [DOI] [PubMed] [Google Scholar]
- 5.Olesen B., Hansen D.S., Nilsson F., Frimodt-Møller J., Leihof R.F., Struve C., Scheutz F., Johnston B., Krogfelt K.A., and Johnson J.R. 2013. Prevalence and characteristics of the epidemic multi-resistant Escherichia coli ST131 clonal group among extended-spectrum β-lactamase (ESBL)-producing E. coli in Copenhagen. J. Clin. Microbiol. 51:1779–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolas-Chanoine M.H., Bertrand X., and Madec J.Y. 2014. Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27:543–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alqasim A., Emes R., Clark G., Newcombe J., La Ragione R., and McNally A., 2014. Phenotypic microarrays suggest Escherichia coli ST131 is not a metabolically distinct lineage of extra-intestinal pathogenic E. coli. PLoS One 9:e88374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platell J.L., Cobbold R.N., Johnson J.R., Heisig A., Heisig P., Clabots C., Kuskowski M.A., and Trott D.J. 2011. Commonality among fluoroquinolone-resistant sequence type ST131 extraintestinal Escherichia coli isolates from humans and companion animals in Australia. Antimicrob. Agents Chemother. 55:3782–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Can F., Azap O.K., Seref C., Ispir P., Arslan H., and Ergonul O. 2014. Emerging Escherichia coli O25b/ST131 clone predicts treatment failure in urinary tract infections. Clin. Infect. Dis. 60:523–527 [DOI] [PubMed] [Google Scholar]
- 10.Dhanji H., Doumith M., Clermont O., Denamur E., Hope R., Livermore D.M., and Woodford N. 2010. Real-time PCR for detection of the O25b-ST131 clone of Escherichia coli and its CTX-M-15-like extended-spectrum β-lactamases. Int. J. Antimicrob. Agents 36:355–358 [DOI] [PubMed] [Google Scholar]
- 11.Vimont S., Boyd A., Bleibtreu A., Bens M., Goujon J.M., Garry L., Clermont O., Denamur E., Arlet G., and Vandewalle A. 2012. The CTX-M-15-producing Escherichia coli clone O25b: H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS One 7:e46547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ranjan A., Shaik S., Hussain A., Nandanwar N., Semmler T., Jadhav S., Wieler L.H., and Ahmed N. 2015. Genomic and functional portrait of a highly virulent, CTX-M-15-producing H30-Rx subclone of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 59:6087–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clermont O., Bonacorsi S., and Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdi H.A., and Ghalehnoo M.R. 2015. Virulence genes, genetic diversity, antimicrobial susceptibility and phylogenetic background of Escherichia coli isolates. Int. J. Enter. Pathog. 3:25692 [Google Scholar]
- 15.Johnson J.R., Menard M., Johnston B., Kuskowski M.A., Nichol K., and Zhanel G.G. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob. Agents Chemother. 53:2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clermont O., Dhanji H., Upton M., Gibreel T., Fox A., Boyd D., Mulvey M.R., Nordmann P., Ruppé E., and Sarthou J.L. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 64:274–277 [DOI] [PubMed] [Google Scholar]
- 17.Wikler M.A. 2007. Performance standards for antimicrobial susceptibility testing: seventeenth informational supplement. Clinical and Laboratory Standards Institute. 26th Inform Suppl 2016; 36:M100-S25 [Google Scholar]
- 18.Perna N., Glasner J., Burland V., and Plunkett G., III. 2002. Escherichia coli: Virulence Mechanism of a Versatile Pathogen. Academic Press, Elsevier Science, San Diego, CA, pp. 3–53 [Google Scholar]
- 19.Dahbi G., Mora A., López C., Alonso M.P., Mamani R., Marzoa J., Coira A., García-Garrote F., Pita J.M., and Velasco D. 2013. Emergence of new variants of ST131 clonal group among extraintestinal pathogenic Escherichia coli producing extended-spectrum β-lactamases. Int. J. Antimicrob. Agents 42:347–351 [DOI] [PubMed] [Google Scholar]
- 20.Clermont O., Lavollay M., Vimont S., Deschamps C., Forestier C., Branger C., Denamur E., and Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024–1028 [DOI] [PubMed] [Google Scholar]
- 21.Blanco J., Mora A., Mamani R., López C., Blanco M., Dahbi G., Herrera A., Marzoa J., Fernández V., and de la Cruz F. 2013. Four main virotypes among extended-spectrum-β-lactamase-producing isolates of Escherichia coli O25b: H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J. Clin. Microbiol. 51:3358–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peirano G., van der Bij A.K., Gregson D.B., and Pitout J.D. 2012. Molecular epidemiology over an 11-year period (2000 to 2010) of extended-spectrum β-lactamase-producing Escherichia coli causing bacteremia in a centralized Canadian region. J. Clin. Microbiol. 50:294–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumura Y., Yamamoto M., Higuchi T., Komori T., Tsuboi F., Hayashi A., Sugimoto Y., Hotta G., Matsushima A., and Nagao M. 2012. Prevalence of plasmid-mediated AmpC β-lactamase-producing Escherichia coli and spread of the ST131 clone among extended-spectrum β-lactamase-producing E. coli in Japan. Int. J. Antimicrob. Agents 40:158–162 [DOI] [PubMed] [Google Scholar]
- 24.Markovska R., Schneider I., Ivanova D., Keuleyan E., Stoeva T., Sredkova M., Markova B., Bojkova K., Gergova R., and Bauernfeind A. 2012. High prevalence of CTX-M-15-producing O25b-ST131 Escherichia coli clone in Bulgarian hospitals. Microb. Drug Resist. 18:390–395 [DOI] [PubMed] [Google Scholar]
- 25.Coelho A., Mora A., Mamani R., López C., González-López J.J., Larrosa M.N., Quintero-Zarate J.N., Dahbi G., Herrera A., and Blanco J.E. 2010. Spread of Escherichia coli O25b: H4-B2-ST131 producing CTX-M-15 and SHV-12 with high virulence gene content in Barcelona (Spain). J. Antimicrob. Chemother. 66:517–526 [DOI] [PubMed] [Google Scholar]