Abstract
Cancer stem cells (CSCs) can regenerate all facets of a tumour as a result of their stem cell-like capacity to self-renew, survive and become dormant in protective microenvironments. CSCs evolve during tumour progression in a manner that conforms to Charles Darwin’s principle of natural selection. Although somatic DNA mutations and epigenetic alterations promote evolution, post-transcriptional RNA modifications together with RNA binding protein activity (the ‘epitranscriptome’) might also contribute to clonal evolution through dynamic determination of RNA function and gene expression diversity in response to environmental stimuli. Deregulation of these epitranscriptomic events contributes to CSC generation and maintenance, which governs cancer progression and drug resistance. In this Review, we discuss the role of malignant RNA processing in CSC generation and maintenance, including mechanisms of RNA methylation, RNA editing and RNA splicing, and the functional consequences of their aberrant regulation in human malignancies. Finally, we highlight the potential of these events as novel CSC biomarkers as well as therapeutic targets.
With regard to evolution in specific ecological niches, Darwin stated that “the same species, also, often have a somewhat monstrous character…they often differ in an extreme degree in some one part, both when compared one with another, and more especially when compared with all the species in nature to which they are nearest allied” (REF. 1). In a similar sense, cancer cells that develop in specific microenvironments represent a monstrous caricature of normal development. Cancer stem cells (CSCs) are typically rare cells within tumours that exploit stem cell properties, thereby enabling them to become dormant, survive and regenerate in protective microenvironments, albeit in a deregulated manner (BOX 1). These CSCs represent a reservoir of self-sustaining cells that give rise to many types of cancer cell. In contrast to most cells in a tumour, CSCs have the capacity to form self-renewing cells and differentiated cells that comprise the bulk tumour population2. The prevalence of CSCs varies between tumour types and between individual patients3. Research efforts have focused on identifying and understanding the key genetic and epigenetic mechanisms that govern CSC evolution. Recent data suggest that the capacity of CSCs to respond rapidly to environmental changes is predicated, at least in part, on changes in RNA processing. The recently coined term ‘epitranscriptome’ (REF. 4) describes myriad post-transcriptional RNA modifications that bring about functionally relevant changes to the transcriptome. Analogous to the better-defined DNA and protein modifications collectively known as the ‘epigenome’ (REF. 5) and ‘epiproteome’ (REF. 6), epitranscriptomic modifications include several important RNA processing events, including RNA editing, methylation and splicing (FIG. 1).
Box 1 | Discovery of cancer stem cells.
The first evidence supporting the existence of cancer stem cells (CSCs) was reported in acute myeloid leukaemia (AML) in 1994 (REF. 139). A population of primary patient-derived leukaemia cells capable of initiating tumours in immunocompromised mice139, termed leukaemia stem cells (LSCs), were shown to possess cell surface markers (CD34+CD38−) and differentiation capacity similar to those of normal haematopoietic stem cells (HSCs). Serial transplantation into secondary recipient mice resulted in engraftment of human cells with similar morphology and cell surface markers to the original leukaemia, thus establishing the gold standard test for assessing CSC self-renewal capacity. Following the initial discovery of AML LSCs, CSCs were discovered in various other blood cancers, such as chronic myeloid leukaemia (CML). In CML, activation of the BCR-ABL1 fusion oncogene-derived protein tyrosine kinase, P210, was shown to occur at the level of HSCs whereas blast crisis transformation was fuelled by progenitors that had co-opted stem cell self-renewal and survival properties that rendered them impervious to tyrosine kinase inhibitors7,40,55,99,103,104. Similar complexity in the CSC hierarchy was reported in solid tumours. For example, breast CSCs were found to be enriched in the CD44+CD24− population140. Since then, CSC populations have been detected in brain141, lung142, colon143, prostate144 and ovarian cancers145. Whereas these breakthrough studies identified DNA mutations and cell surface phenotypes of relatively rare tumour-initiating cell types, recent research efforts have focused on identifying and understanding the key epigenetic and epitranscriptomic mechanisms that govern CSC evolution.
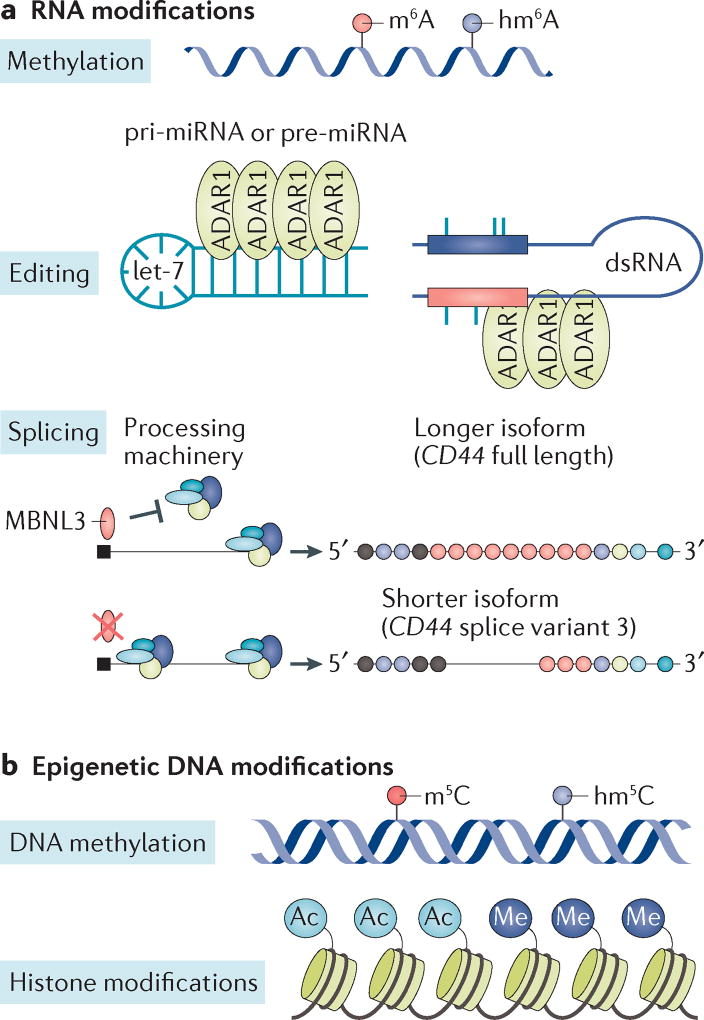
Figure 1. Epitranscriptome regulation contributes to cancer stem cell generation.
Normal haematopoietic stem cells (HSCs) accumulate genetic (DNA) and epitranscriptomic (RNA) changes that promote the emergence of pre-leukaemic clones that have gained survival and/or proliferative advantages. a | Deregulation of epitranscriptomic events include post-transcriptional RNA modifications such as RNA methylation (N6-methyladenosine (m6A) and N6-hydroxymethyladenosine (hm6A)) and adenosine-to-inosine (A-to-I) double-stranded RNA (dsRNA) editing (for example, adenosine deaminase acting on dsRNA 1 (ADAR1) editing of primary microRNA (pri-miRNA) and precursor miRNA (pre-miRNA) such as let-7). Alu element-containing dsRNA is the main target of A-to-I editing, which can result in changes to mRNA, secondary structure, stability and cellular localization. Splicing and the activity of RNA binding protein splicing factors such as muscleblind-like 3 (MBNL3) are additional RNA modifications. These events add additional layers of dynamic regulation that can all contribute to both cancer initiation and cancer progression. b | Epigenetic DNA modifications, such as the formation of 5-methylcytosine (m5C) and 5-hydroxymethylcytosine (hm5C), fulfil important roles in regulating cell differentiation and development. Genes with high levels of hm5C in their promoter regions are typically transcriptionally silent. This addition of methyl groups onto DNA will therefore alter the function of the DNA. For example, if a developmentally regulated gene has a high level of hm5C it may fail to induce differentiation and can potentially contribute to cancer initiation and progression. Histone modifications (methylation (Me) and acetylation (Ac)) are known to have important roles in cell differentiation and development with potential carcinogenic outcomes.
The mechanisms governing human transcriptome diversity have been fine-tuned throughout evolution and involve various regulatory steps in RNA processing such as 5′ processing (capping), 3′ processing (cleavage and polyadenylation), and RNA methylation, editing and splicing. Although these are all important for the phenotypic variability of our species, this Review focuses on RNA editing and splicing and RNA methylation, with an emphasis on the crosstalk between RNA editing and these other RNA processing events. Nascent transcripts are susceptible to RNA sequence modification by RNA editases, such as adenosine deaminases acting on double-stranded RNA (dsRNA) (ADARs). Among these, the activity of ADAR1 has been implicated in the oncogenic transformation of pre-malignant progenitors that harbour clonal self-renewal and survival capacity7. As another key mechanism in RNA processing regulation, precursor mRNA (pre-mRNA) splicing activity dramatically influences RNA and protein diversity in mammals. Alternative splicing occurs in up to 95% of human multi-exon genes during development and ageing8,9. Aberrant RNA splicing has been linked to CSC generation in leukaemia and is being investigated as a source of transcriptomic diversity in other CSC populations10. Finally, although DNA methylation is an important epigenetic arbiter of normal tissue development, which, when disrupted, serves as a driver of cancer evolution, emerging data suggest that methylation of RNA at N6-methyladenosine (m6A)11,12 also has a central role in stem cell fate determination and cancer development. Understanding cell type- and context-specific differences in RNA processing is therefore key to deciphering differences between normal stem cell development and CSCs, which can promote therapeutic resistance and cancer progression.
In contrast to previous reviews, which focus on somatic DNA mutations2,13,14, DNA-based epigenetic modifications15 and transcriptional regulation16–18 (all of which remain crucial areas of cancer research), this Review discusses the emerging role of the deregulation of RNA processing in human malignancies and the importance of examining aberrant RNA editing, methylation and splicing in the orchestration of complex CSC functions, such as self-renewal, dormancy and survival. How these processes are regulated by intrinsic (cell-autonomous) and extrinsic (non-cell-autonomous) factors will be addressed, and innovative CSC eradication strategies targeting RNA processing in various human malignancies will be proposed.
RNA methylation in CSCs
The term epitranscriptome derives from a transcriptome-wide mapping study of the sites of an evolutionarily conserved RNA modification, m6A, first discovered in 1974 (REFS 4,19,20) (FIG. 1). Epitranscriptomic networks have important roles in maintaining the pluripotent state of human embryonic stem cells (hESCs), as well as being a major player in somatic reprogramming21,22. However, how RNA modifications influence the balance between pluripotent and differentiated states remains to be elucidated. Since they were first discovered, m6A modifications have been identified in humans11,12, viruses23 and mice11,12, occurring in mRNAs24, ribosomal RNAs (rRNAs)25 and in long intergenic non-coding RNAs (lincRNAs)26, and have been linked to several diseases, including Alzheimer disease27.
Adenosine methylation is catalysed by the m6A methyltransferase complex containing the methyltransferase-like 3 (METTL3) enzyme. The formation of m6A on mRNAs is influenced directly by micro-RNAs (miRNAs), by modulating the binding capacity of METTL3 to the miRNA targeting sequences via a sequence pairing mechanism28.
Given the important role of m6A in pluripotency and differentiation29, its association with cancer development is not surprising. Described as “An old modification with a novel epigenetic function” (REF. 30), m6A promotes the translation of several oncogene products. For example, a recent study showed that the m6A demethylase FTO has a crucial role in acute myeloid leukaemia (AML) by enhancing leukaemogenesis31. This is achieved by reducing m6A levels in the mRNA transcripts of targets such as ankyrin repeat and SOCS box-containing 2 (ASB2) and retinoic acid receptor-α (RARA). In addition, this RNA modification promotes translation of gene products important for cancer pathogenesis (such as epidermal growth factor receptor (EGFR) and transcriptional co-activator with PDZ-binding motif (TAZ, also known as WWTR1)) by association with ribosomes32. More recently, m6A modifications have been linked to breast CSC generation33–37. Conversely, the loss of m6A can promote translation of NANOG, which in turn enables cancer cells to revert to a primitive functional state akin to that of pluripotent stem cells37. Therefore, m6A methylation signatures could be tested as clinical biomarkers for CSC generation and cancer progression.
A-to-I editing in CSCs
Comprehensive RNA sequencing has provided insights into the regulation of the human transcriptome by RNA editing38 (FIG. 2). In the primate transcriptome, the most frequent type of RNA editing is adenosine-to-inosine (A-to-I), which involves hydrolytic deamination of adenosine by the ADAR family of RNA editases38 (ADAR1, ADAR2 (also known as ADARB1) and ADAR3 (also known as ADARB2)). The resulting inosine bases are subsequently read as guanosines, thus inducing A-to-G post-transcriptional changes that contribute to transcriptome diversity. Although the precise mechanisms governing RNA editing events in cancer are currently the subject of intense investigation, copy number amplification at chromosome 1q39 — the locus of the human ADAR1 gene — and inflammation7,40 are two primary mechanisms that may contribute to induction of RNA editing in cancer. Normally, ADAR1 transcription is activated by RNA viruses and represents an essential component of the innate antiviral immune response41. RNA editases likely evolved to protect cells from retroviral integration by inducing an interferon response, thereby enabling stem cells to proliferate in response to virus-induced tissue injury. Recently, ADAR1-dependent A-to-I editing has been implicated as an essential component of innate immunity. Specifically, in a knock-in mouse model, ADAR1 distinguishes pathogenic dsRNA from host ‘self’ dsRNA by A-to-I RNA editing of en dogenous dsRNA, thus preventing recognition of endogenous transcripts by the cytosolic sensor of dsRNA, interferon-induced helicase C domain-containing protein 1 (IFIH1, also known as MDA5)42. Unedited transcripts may be sensed by IFIH1 as non-self, thereby leading to innate immune system activation.
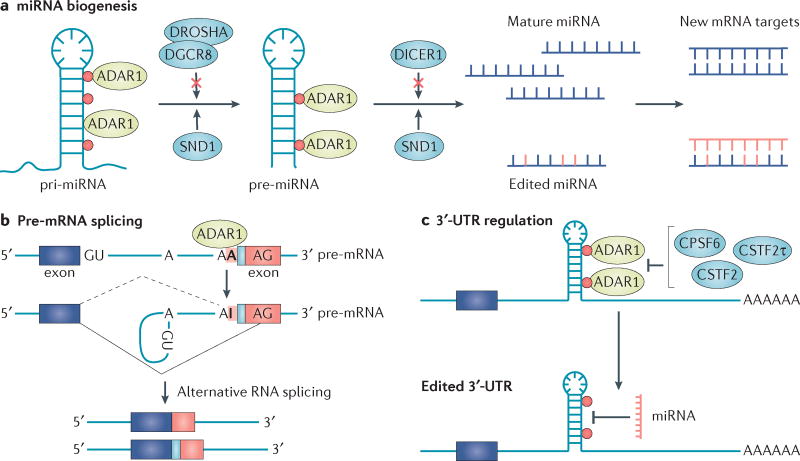
Figure 2. Consequences of RNA editing by ADAR.
a | Adenosine-to-inosine (A-to-I) editing-dependent regulation of microRNA (miRNA) biogenesis and targeting occurs at multiple stages. Editing (depicted by red dots) of primary miRNAs (pri-miRNAs) by adenosine deaminase acting on double-stranded RNA 1 (ADAR1) and ADAR2 prevents processing by the microprocessor complex composed of DROSHA and DGCR8, which results in decreased production of mature miRNA. Similarly, RNA editing of precursor miRNA (pre-miRNA) affects the DICER1 cleavage process. Both edited pri-miRNAs and edited pre-miRNAs are specifically recognized and degraded by the ribonuclease staphylococcal nuclease domain-containing 1 (SND1). Finally, editing within the seed region of mature miRNAs potentially changes the specific mRNA target compared with the unedited version. b | RNA editing at alternative 3′-acceptor sites converts the intronic AA into AI dinucleotides, which mimic the conserved AG sequences normally found at 3′-splicing sites. This introduction of a new splicing acceptor site results in an alternatively spliced mRNA with an insertion (light blue). The dashed line represents an alternative splicing event generated by A-to-I editing. c | ADAR1 competes with canonical 3′-untranslated region (UTR) processing factors (cleavage stimulation factor subunit 2 (CSTF2), CSTF2τ and cleavage and polyadenylation-specific factor 6 (CPSF6)) by direct binding to 3′-UTRs shortly after they are transcribed. As miRNAs predominately target mRNA 3′-UTR sites, RNA editing changes within the miRNA target sequences may prevent miRNA binding and therefore downregulation of the mRNA.
Of the ADAR family members, ADAR1 and ADAR2 are ubiquitously expressed in eukaryotes and participate in various cellular functions through enzymatic RNA editing activity, whereas ADAR3 is specifically expressed in brain tissue and its function remains unknown. Most RNA editing sites occur in non-coding sequences, such as 5′-untranslated regions (UTRs) and 3′-UTRs, and intronic retrotransposable sequences, including Alu elements (a type of short interspersed nuclear element (SINE)) and long interspersed nuclear elements (LINEs). In particular, A-to-I editing events commonly occur in close proximity to inverted Alu elements, which form extended dsRNA structures and represent major ADAR targets. Notably, Alu elements are involved in chromosomal translocations by double-strand or homologous recombination43–45, as well as the generation of aberrant circular RNAs that can contribute to tumour formation and therapeutic resistance46. Notably, ADAR1 depletion in hESCs abrogates pluripotency and induces expression of transcripts that govern differentiation47. Conversely, enforced ADAR1 expression in a human embryonic kidney cell line (HEK293T) directly regulates the expression of more than 300 proteins involved in protein translation and cell cycle regulation48. Together, these studies suggest that the functional impact of changes in the RNA editome may be far-reaching, but this requires further investigation in the context of primary human cell types and tissue-specific stem cells.
Given the important role of RNA editing as a post-transcriptional regulatory mechanism, it is not surprising that aberrant activation of ADAR-mediated RNA editing has emerged as a driver of cancer progression. Cumulative whole-transcriptome RNA sequencing (RNA-seq) analyses have uncovered inflammatory cytokine networks that activate ADAR1 during relapse or progression of lobular breast49,50, hepatocellular51 and oesophageal squamous cell52 carcinomas and in the most highly studied myeloproliferative neoplasm (MPN), chronic myeloid leukaemia (CML)7. Genetic ablation of ADAR1 editase activity in mice leads to embryonic lethality owing to severe defects in erythropoiesis53, and conditional deletion in the haematopoietic system impairs haematopoietic stem cell (HSC) maintenance54, indicative of key roles for ADAR1 in both normal cell fate specification and self-renewal. In human CML CSCs, ADAR1 activation promotes glycogen synthase kinase 3β (GSK3B) missplicing, which prevents degradation of β-catenin, a self-renewal agonist7,55. Thus, activation of ADAR1 may have a significant role in malignancies that have acquired aberrant stem cell self-renewal capacity.
When RNA editing occurs in coding sequences of pre-mRNA transcripts, functional changes in the protein can promote cancer progression56. A pioneering RNA-seq study in oestrogen receptor-α-positive metastatic lobular breast cancer revealed that ADAR1 is among the top 5% of genes expressed during molecular evolution. Specifically, ADAR1 activation was linked to high-frequency editing of component of oligomeric Golgi complex 3 (COG3) and signal recognition particle 9 (SRP9) transcripts, although the functional consequences of RNA editing were not specifically evaluated49. Another well-studied example of RNA editing is the ADAR1-induced coding sequence change in antizyme inhibitor 1 (AZIN1) in hepatocellular carcinoma (HCC)56. The A-to-I editing level was elevated in HCC specimens compared with benign liver samples. Hyperediting was associated with tumour recurrence and lower disease-free survival rates. The A-to-I editing of AZIN1 results in a serine-to-glycine substitution at residue 367 that induces a protein conformational change leading to relocalization of AZIN1 from the cytoplasm to the nucleus. Notably, edited AZIN1 (AZIN1-S367G) exhibits gain-of-function phenotypes typified by enhanced cell-invasive capacity, proliferation and tumour initiation56.
A seminal RNA-seq study involving more than 6,000 patient samples from 17 cancer types revealed clinically relevant differences in A-to-I RNA editing profiles between tumour and normal control tissues57. Both edited COG3 (COG3-I635V) and AZIN1-S367G could be detected in multiple cancer types, including breast cancer, head and neck squamous cell carcinoma and lung adenocarcinoma57. Moreover, this study found that RNA editing events can act as ‘driver’ mutations and contribute to therapeutic response, suggesting that ADAR activation may be a crucial contributor to therapy-resistant CSC generation. Future studies should focus on the functional characterization of these RNA editing-induced mutations in primary patient CSCs, which may provide novel biomarkers as well as therapeutic targets in CSCs.
RNA editing disrupts miRNA biogenesis in CSCs
Some of the first insights into the functional effects of A-to-I RNA editing came from early studies demonstrating that aberrant A-to-I RNA editing in primary microRNAs (pri-miRNAs) and precursor microRNAs (pre-miRNAs) directly interfered with RNA interference (RNAi) pathways58–60 (FIG. 2a). Editing of pri-miRNAs and pre-miRNAs can impair mature miRNA biogenesis with potentially wide-ranging effects on stem cell regulatory gene expression in hESCs and CSCs61–64. Because of dsRNA stem-loop structure formation, both pri-miRNAs and pre-miRNAs are targets of A-to-I editing by ADAR1, resulting in reduced production of mature miRNAs60,65–67. The best-studied example is the haematopoietic-specific pri-miR-142 (REF. 68). Editing of pri-miR-142 at +4 and +5 sites by ADARs suppresses cleavage by the ribonuclease III DROSHA, and the edited transcripts are degraded by staphylococcal nuclease domain-containing 1 (SND1, also known as Tudor-SN), a ribonuclease specific to inosine-containing dsRNA60. Notably, mature miR-142 is highly expressed in human breast CSCs, activating the canonical WNT signalling pathway69. Moreover, inhibition of endogenous miR-142 suppresses organoid formation and prevents tumour initiation by breast CSCs69. Interestingly, ADARs also stimulate RNAi machinery by interacting directly with DICER1 to promote pre-miRNA splicing62, thereby demonstrating that ADARs have a profound impact on miRNA regulation.
Altered miRNA expression is associated with CSC-specific properties of self-renewal, metastasis and drug resistance70. Recently, we demonstrated that the self-renewal capacity of CSCs derived from patients with blast crisis (BC) CML is associated with impaired biogenesis of the let-7 miRNA family owing to ADAR1 activation mediated by inflammatory cytokine signalling and the Janus kinase 2 (JAK2)–signal transducer and activator of transcription (STAT) pathway40. Overexpression of wild-type ADAR1 also reduced mature let-7 family miRNA levels by promoting A-to-I editing at DROSHA cleavage sites in pri-let-7 transcripts, which in turn increased expression of the pluripotency gene LIN28B, a target of let-7 miRNA. LIN28B is an important RNA binding protein (RBP) that promotes progression and metastasis in various human malignancies71. Indeed, the disruption of the let-7-LIN28B axis induced malignant self-renewal of BC CML progenitors40. Interestingly, deregulation of miRNA expression and processing is associated with human cancer72, raising the interesting question of whether inflammation-induced ADAR1 activation is linked to impaired miRNA biogenesis in a broad array of recalcitrant malignancies, and whether this effect is therapeutically targetable.
A-to-I editing of the 3′-UTR landscape regulates gene expression
According to the DARNED human RNA editome database (see Further information), approximately 23% of all editing events occur in 3′-UTR and 2.6% in 5′-UTR sequences73. This difference might be explained by the prevalence of Alu elements. Approximately 5% of all cDNAs contain Alu elements. Although the 5′-UTR has a few Alu elements, most Alu elements (82%) are located in 3′-UTRs74. Although the significance of hyperediting of 3′-UTR Alu elements remains largely unknown in cancer biology, several lines of evidence suggest diverse roles, including nuclear retention by the inosine-specific RBP, non-POU domain-containing, octamer binding (NONO, also known as p54nrb)75, shortening of the 3′-UTR by the inosine-selective nuclease SND1 (REF. 76) and interference with miRNA accessibility77,78.
A comparison of tumour and normal controls from lymphoma, neuroblastoma and head and neck sarcoma showed that an estimated 2,000 genes have at least one differentially edited site in the 3′-UTR78. Increased A-to-I changes were observed in the 3′-UTR of several cancer-associated genes, including BRCA2, TP53 (which encodes p53), ataxia telangiectasia mutated (ATM ) and MDM2. The editing events in MDM2 are located within miR-200b or miR-200c binding sites, thereby disrupting miRNA-mediated repression and stabilizing MDM2 expression, which leads to disruption of p53 (REF. 78). Conversely, hyperediting within the 3′-UTR can create new miRNA binding sites in mRNAs77. The dual role of 3′-UTR editing in miRNA-mediated differential gene expression is indicative of a tightly orchestrated and complex regulatory role of ADAR1 in CSC maintenance, which should provoke future studies of 3′-UTR editing in CSC populations (FIG. 2c).
To add another layer of complexity, ADAR1 also interferes with the 3′-UTR shortening process79 (FIG. 2c). Regulation of 3′-UTR length via alternative cleavage and polyadenylation introduces shortened 3′-UTRs that are more stably expressed than their full-length counterparts80. The loss of 3′-UTR sequences results in differential mRNA nuclear export and stability, and evasion of suppression by miRNAs. This epigenetic mechanism is often used by cancer cells to promote proto-oncogene activation, ultimately resulting in enhanced proliferation and tumorigenesis80. Crosslinking immunoprecipitation (CLIP)-sequencing in human U87MG glioma cells revealed that ADAR1 alters 3′-UTR length by RNA editing-dependent and -independent mechanisms79. ADAR1 prohibits access of cleavage and polyadenylation factors, such as cleavage stimulation factor subunit 2 (CSTF2), CSTF2τ and cleavage and polyadenylation-specific factor 6 (CPSF6), to the 3′-UTR by competition for binding substrates or induction of structural alterations by introducing A-to-I changes79. This interesting aspect of ADAR1-mediated 3′-UTR control and whether it has a role in CSC generation will require further exploration.
RBP deregulation in CSCs
RBPs are highly versatile post-transcriptional modulators involved in gene expression, including maturation, nuclear transport, stability, degradation and translational control of RNAs. Individual RBPs have hundreds of potential RNA targets that form complex and dynamic ‘RNA regulons’ (REF. 81). In addition to ADAR1, many RBPs have emerged as key pluripotency determinants in progenitor cells82 and regulators of CSC generation83. In particular, RNA binding motif protein 15 (RBM15), a spliceosomal component identified as a 5′ fusion partner of megakaryoblastic leukaemia 1 (MKL1) in acute megakaryoblastic leukaemia84, is required for HSC proliferation under stress conditions85. As noted above, the RBPs LIN28 and LIN28B are important pluripotency factors that exhibit oncogenic effects in human malignancies71. Along with suppression of let-7 miRNAs, LIN28 also enhances the post-transcriptional expression of CSC markers such as leucine-rich repeat-containing G-protein-coupled receptor 5 (LGR5) and prominin 1 (PROM1)86. The role of LIN28 and LIN28B in CSC maintenance was confirmed in prostate cancer, ovarian cancer, non-small-cell lung cancer, colon cancer and CML40,86–89.
Other developmentally regulated RBPs are the Musashi (MSI) family, which were originally discovered in 1994 as crucial determinants of cell fate in Drosophila90. Like other important self-renewal regulators that are co-opted by CSCs, MSI proteins have been implicated in CSC generation and progression of haematological malignancies and solid tumours91–95 (FIG. 3). In vertebrates, there are two highly conserved homologues of Musashi genes, Musashi 1 (MSI1) and Musashi 2 (MSI2), which arose from a gene duplication event. The encoded proteins appear to be functionally redundant in some cell types96. Normally, the main impact of MSI activity is the inhibition of protein translation by binding to mRNA. A well-known example is the inhibition of translational control of NUMB, a repressor of the Notch intracellular signalling pathway, which MSI inhibits by binding to the 3′-UTR of the mRNA transcript92,97.
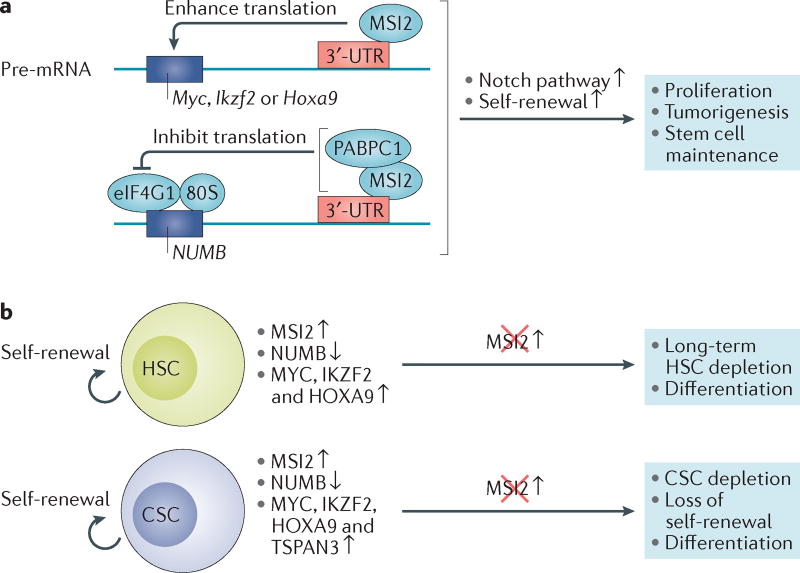
Figure 3. Translational control by MSI2 directs normal and leukaemic stem cell function.
a | Musashi 2 (MSI2) protein inhibits NUMB translation by binding directly to the 3′-untranslated region (UTR) of NUMB precursor-mRNA (pre-mRNA) and also competing with eukaryotic translation initiation factor 4 γ1 (eIF4G1) for binding to poly(A) binding protein cytoplasmic 1 (PABPC1). MSI2 also enhances the translation of crucial transcription factor genes such as Myc, IKAROS family zinc finger 2 (Ikzf2) and homeobox A9 (Hoxa9), by binding directly to the mRNA transcripts. The resulting NUMB inhibition and enhanced MYC, IKZF2 and HOXA9 protein expression promotes Notch signalling pathway activation and self-renewal programmes that are required for haematopoietic stem cell (HSC) maintenance and cancer stem cell (CSC) regeneration. b | MSI2 upregulation is observed in self-renewing normal HSCs and malignant progenitors (CSCs) of blast crisis chronic myeloid leukaemia (CML) and acute myeloid leukaemia (AML). In cooperation with BCR–ABL1 oncogene activation, which induces a chronic phase-like CML phenotype, MSI2 represents a second hit required to enhance CSC self-renewal capacity. Similar to the role for the MSI2–NUMB pathway in aggressive leukaemia, blocking MSI2 restores NUMB expression and inhibits CSC propagation, thus preventing leukaemic initiation in CML and AML. TSPAN3, tetraspanin-3.
The important function of MSI2 in normal haematopoiesis was demonstrated by decreased engraftment potential following Msi2 knockdown in LIN−SCA1+KIT+ cells, and increased long-term HSC proliferation and an expansion of committed progenitors driven by ectopic Msi2 overexpression98. In CML, progression is typified by expansion of CSCs in the immature granulocyte–macrophage progenitor compartment99. Notably, MSI2 is highly expressed in these blast cells, suggesting that MSI2 may be a marker for CSC generation92. In cooperation with the CML oncogene BCR–ABL1, MSI2 overexpression is associated with progression to accelerated phase CML or BC CML98. BC transformation of chronic phase CML is accompanied by increased MSI2 expression and decreased NUMB expression, whereas genetic ablation of MSI2 restores NUMB expression and impairs BC CML propagation92. In addition, high expression of MSI2 and reduced NUMB expression were found in AML, and elevated MSI2 expression directly correlated with decreased survival and poor prognosis in both malignancies92,98. Recently, the role of MSI2 in AML was directly linked to maintenance of mixed-lineage leukaemia (MLL, also known as KMT2A) self-renewal programmes94. Here, instead of suppression of translation, increased MSI2 expression supports the translational efficiency of transcripts involved in self-renewal such as Myc, IKAROS family zinc finger 2 (Ikzf2) and homeobox A9 (Hoxa9), thus supporting a positive feedback mechanism for efficient CSC maintenance94. The inhibitory effect of MSI on translation is achieved by inhibiting formation of the 80S ribosome complex and competing with eukaryotic translation initiation factor 4 γ1 (eIF4G1) for binding to poly(A) binding protein cytoplasmic 1 (PABPC1), which is required for ribosome recruitment and translation initiation100. The mechanism of enhanced translation by MSI is not well understood, but likely occurs through interaction with other cellular components of the translational machinery.
Taken together, these studies highlight the importance of RBP regulatory pathways such as MSI in leukaemogenesis and identify MSI as a potential prognostic factor and therapeutic target in human malignancies. A recent preclinical report indicates that MSI targeting with an antisense oligonucleotide (ASO) strategy efficiently inhibits pancreatic tumour colony formation, as well as tumour growth in patient cell line-derived xenografts of pancreatic adenocarcinoma93. In the future, the ASO strategy and other MSI antagonists should be developed and examined in cancers with high MSI expression with the goal of preventing CSC generation and cancer progression and relapse.
Splicing deregulation in CSCs
Recent whole-transcriptome analyses revealed widespread splice isoform alterations in splicing factor-mutated and non-mutated cancers101,102 and in MPN progenitors55,103,104. Notably, various stem cell regulatory transcripts, such as CD44 and those encoding members of the BCL2 family of apoptosis regulatory genes, have been implicated as functionally relevant targets displaying pro-survival pre-mRNA splicing patterns in CSCs7,10,55,103–105. For example, CD44v3, a splice variant of the cell adhesion glycoprotein CD44, which is typically expressed in hESCs104, along with other truncated isoforms of CD44 (REF. 10) and long, pro-survival isoforms of the BCL2 family, including BCL2, MCL1, BCL-2-like 1 (BCL2L1) and BFL1 (also known as BCL2A1)103, are enriched in CSCs during leukaemic transformation. In contrast, during normal HSC and progenitor cell ageing, pro-survival BCL2 family splice isoforms are depleted10. This is consistent with the alterations observed in ageing human bone marrow10, typified by stem cell exhaustion and myeloid differentiation106, and highlights the functional relevance of splice isoform switching in malignant versus normal stem cell populations (FIG. 4). Furthermore, preclinical studies support the utility of combination treatment strategies, including inhibitors of pro-survival BCL-2 family activity, in eradicating dormant CSCs and preventing cancer relapse103. In AML, whole-transcriptome-based splice isoform signatures of CSCs revealed overexpression of several alternatively spliced signal transduction (for example, protein tyrosine phosphatase, non-receptor type 6 (PTPN6), protein tyrosine kinase 2β (PTK2B)) and cell adhesion (for example, integrin subunit β2 (ITGB2)) transcripts compared with normal age-matched control progenitors10. Other myelodysplastic syndrome (MDS)107 and AML108 gene expression studies demonstrate differential exon usage of epigenetic modifier (for example, enhancer of zeste 1 (EZH1)) and tumour suppressor (for example, TP53) transcripts, underscoring the importance of pre-mRNA splicing in the evolution of haematopoietic malignancy. Together, these studies shed light on the key functional networks that may be affected by alternative splicing in CSCs.
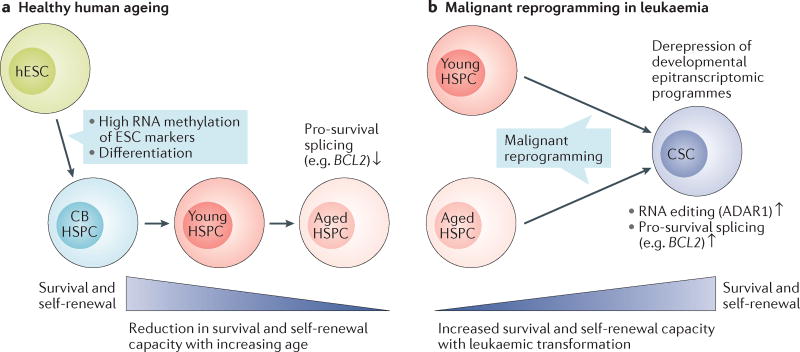
Figure 4. RNA processing in normal and malignant haematopoiesis.
RNA processing alteration influence human haematopoietic stem and progenitor cell (HSPC) development, ageing and disease. a | In human embryonic stem cells (hESCs), RNA methylation represses pluripotency markers, promoting differentiation along distinct lineages when coupled with derepression of pathway-specific regulatory transcripts. During healthy human development and ageing, precise regulation of stem cell regulatory RNA processing activities such as alternative splicing of pro-survival gene families (for example, BCL2) and RNA binding protein activities (including splicing factors) are required to maintain HSPC survival and self-renewal from fetal stages (cord blood (CB)) through adulthood and ageing. b | In age-related malignancies such as chronic myeloid leukaemia (CML) and acute myeloid leukaemia (AML), aberrant RNA editing and preferential expression of pro-survival splice isoforms (for example, long isoforms of BCL2 and BCL2L1 gene products) promotes malignant reprogramming of progenitors and supports leukaemia cancer stem cell (CSC) survival and self-renewal through derepression of developmental epitranscriptomic programmes. ADAR1, adenosine deaminase acting on double-stranded RNA 1.
Aberrant splicing activity in CSCs as opposed to normal stem cell populations is regulated by various cellular components and mechanisms. Selected RBPs, spliceosome-associated transcripts and proteins, transcription factors and RNA editing activity have all been highlighted in recent studies investigating the mechanisms governing alternative splicing regulation in CSCs. With regard to RBP-mediated regulation of alternative splicing in CSC generation, comparative RNA sequencing analyses of pluripotent cells, including ESCs and induced pluripotent stem cells (iPSCs), and differentiated cells from both humans and mice identified striking species-specific differences in alternative splicing events that govern pluripotency109. Dozens of alternative splicing events differed between pluripotent and differentiated cells and this was governed, at least in part, by muscleblind-like 1 (MBNL1) and MBNL2 splicing factors, which are expressed at very low levels in pluripotent cells and upregulated in differentiated cells. Of 1,348 RBP genes analysed from 11 solid tumour types, MBNL1 was shown to be the main factor controlling the altered splicing patterns; this provides a strong correlation between reversion to pluripotency and solid tumour progression110. Downregulation of MBNL3 promotes generation of CSCs in CML, and this population of self-renewing cells harbours enriched ADAR1 expression and activity, similar to hESCs and in contrast to differentiated cells104, thus rendering CSC-specific transcripts susceptible to an additional layer of RNA processing deregulation through aberrant RNA editing-mediated modulation of splicing.
To date, most cancer-associated mutations affecting key spliceosome components such as splicing factor 3B subunit 1 (SF3B1), serine/arginine-rich splicing factor 2 (SRSF2) and U2 small nuclear RNA auxiliary factor 1 (U2AF1), among others, have been identified in haematopoietic malignancies, in particular MPNs, MDS and chronic lymphocytic leukaemia (CLL)111–118. Although initial studies focused primarily on the presence and clinical significance of splicing factor mutations and their correlation with disease outcome, more recent studies have begun to elucidate the functional consequences of splicing factor mutations in HSC maintenance and tumorigenic potential10. Additionally, specific alternative splicing events can recapitulate cancer phenotypes associated with splicing factor mutation or overexpression, and key splicing networks involving deregulation of RBPs contribute to cancer development and progression110, highlighting the need to evaluate these splicing alterations in drug-resistant CSC populations.
Interestingly, in support of a functional role for the spliceosome components SRSF2 and SF3B1 in human leukaemia progression, preclinical studies demonstrated that small-molecule splicing modulator compounds reduced leukaemia cell engraftment in mouse models of AML119, and impaired CSC self-renewal in humanized AML models10 (BOX 2). Notably, AML CSCs were exquisitely sensitive to splicing modulator treatment and short hairpin RNA (shRNA)-mediated SF3B1 knockdown, whereas the survival and self-renewal capacity of normal haematopoietic stem and progenitor cells were relatively unimpaired10.
Box 2 | Challenges in therapeutic targeting of cancer stem cells.
Although cancer stem cells (CSCs) are typically a relatively rare subset of the entire tumour cell population, selectively targeting CSCs on the basis of activation of specific functional pathways is proving to have significant clinical potential146. Dormant CSCs residing in fibrotic tissue or protective niches can be difficult to fully eradicate with conventional cytotoxic treatments147. Thus, targeting stem cell regulatory pathways such as Notch, Hedgehog, WNT and BCL-2 (REFS 103,146) that are either inactive or less active in non-CSC tumour cells and healthy normal stem cells provides an important strategy to target CSCs while sparing normal healthy cell types. Demonstration of a favourable therapeutic index against CSCs compared with normal counterpart tissue-specific stem cells facilitates highly selective targeting of CSCs10,103. With regard to targeting RNA processing for CSC eradication, a growing body of evidence suggests that CSCs10 as well as bulk tumour cells in leukaemia119, breast cancer123 and melanoma125 display sensitivity to splicing modulation, thus providing a potential strategy to clear dividing malignant cells along with CSCs in various recalcitrant human cancers.
In addition to the ongoing studies investigating the functional consequences of splicing factor mutations in haematopoietic malignancies, other reports have implicated non-mutation-driven mechanisms leading to disruption of similar components of the spliceosome in the pathogenesis of solid tumours110,120–122. Specifically, the spliceosome has been implicated as a target of oncogenic stress in MYC-driven cancers123. Increased RNA synthesis through MYC-dependent transcriptional activation of its downstream target genes places a high burden of transcript stress on the spliceosome, the activity of which is dependent on the spliceosome component BUD31 (REF. 123). Although this study did not focus specifically on CSC populations, because MYC is a vital stem cell pluripotency and reprogramming factor, these findings highlight the complex relationship that exists between transcriptional regulation and spliceosome activity in normal as opposed to malignant stem cells. First, monitoring splicing activity may be relevant to predicting reprogramming efficiency during iPSC generation. Second, MYC-driven cancers are therapeutically vulnerable to splicing modulation, thereby warranting further evaluation of this mechanism in purified CSCs, as well as the role of non-mutation-driven mechanisms of splicing alteration in leukaemia. Although MYC was not differentially expressed in AML compared with normal age-matched haematopoietic progenitors, decreased expression of tumour suppressor genes such as TP53 and interferon regulatory factor 8 (IRF8)10 could lead to widespread deregulation of transcription. However, the consequences of tumour suppressor loss on spliceosome function are currently unknown. Together, these data implicate spliceosome disruption as a therapeutic vulnerability in a growing array of human malignancies123–125, and in AML this may drive splicing alterations of stem cell regulatory genes contributing to CSC generation10.
Splicing and RNA editing crosstalk
Deregulated RNA editing and splicing activities have the potential to synergize with each other, thus potentially exacerbating malignant reprogramming events in human cancers (FIG. 2b). Enzymatically active ADAR1 and ADAR2 localize to large nuclear ribonucleoprotein (lnRNP) particles126 that contain myriad pre-mRNA processing factors, together known as the supraspliceosome, comprising protein splicing factors and U small nuclear RNPs (snRNPs)127. Thus, it is likely that editing and splicing occur in close succession or at overlapping times along single transcripts and that the local RNP landscape has a key role in mediating RNA processing events. Additionally, RNA editing can directly influence alternative splicing through at least two distinct mechanisms: first, in cis by directly creating or destroying splice sites128 leading to intron retention129 through a process termed ‘exonization’ (REF. 130), and second, in trans by interfering with splicing factor binding61. Intriguingly a handful of functional splicing alterations have been reported to result from A-to-I RNA editing events that directly target splice sites. For example, RNA hyperediting of a putative branch site in PTPN6 leads to retention of intron 3, which is expressed at high levels at diagnosis in patients with AML129. A broader analysis revealed that A-to-I editing sites were rarely detected within canonical 5′- or 3′-splice sites61. This is likely related to the preference of ADAR enzymes for a G at the +1 position downstream of the editing locus, which is not present in the human consensus branch site sequence. Bioinformatics approaches, RNA-seq and exon-specific microarray analyses of ADAR1 knockdown cells demonstrated that RNA editing events more frequently target splicing regulatory elements (SREs) contained within exons, thus modulating trans-acting factors involved in the splicing machinery61.
Adding another layer of molecular complexity, RNA editing of rat Adar2 mRNAs can result in alternative splicing of this RNA editase128; however, these results have yet to be confirmed in human cells. Providing evidence that aberrant RNA editing-mediated alternative splicing events have functional relevance to CSC maintenance, RNA editing-induced missplicing of GSK3B promoted survival and self-renewal of dormant, therapy-resistant CSCs7,55,103,105. Conversely, RNA splicing efficiency can control the extent of RNA editing of transcripts when editing is guided by intronic elements (for example, Alu elements)131,132. Moreover, RBPs such as ribosomal protein S14 (RPS14), the splicing factor SRSF9 and DEAH-box helicase 15 (DHX15) act as site-specific repressors of ADAR2-mediated RNA editing133, highlighting the crosstalk between these essential RNA processing pathways. In future studies, it will be important to consider RNA processing alterations in CSCs in the broader context of all activities occurring from transcription to translation.
Targeting RNA splicing and editing in CSCs
Fundamental differences in pre-mRNA processing between humans and mice9 have hampered efforts to develop RNA splicing and RNA editing targeted thera-pies134. A growing body of evidence supports the potential utility of RNA splicing modulation for the treatment of MYC-driven solid tumours and splicing factor-deregulated haematological malignancies119,123,134. As shown in preclinical AML models, treatment with a small-molecule, pladienolide-derived, splicing modulatory agent, 17S-FD-895, significantly impaired AML CSC survival and self-renewal at doses that spared normal HSCs10. This favourable therapeutic index suggests that RNA splicing modulation could be used clinically as a potent and selective method to eradicate CSCs in drug-resistant leukaemia and other recalcitrant malignancies (BOX 2).
In addition, CSC-specific splice isoform biomarkers open up novel avenues to predict and prevent disease relapse and monitor response to CSC-targeted and RNA splicing modulatory therapies10. Future preclinical studies will be necessary to fully elucidate whether splicing modulation can eradicate CSCs in other human malignancies. Because splicing is an essential cellular function that is active in all known cell types across human tissues, development of splicing modulators with favourable therapeutic indices will be vital for the development of safe and effective RNA processing-targeted therapies. Sensitization of CSCs compared with normal stem cell populations in patients who have developed therapeutic resistance may represent one of the most compelling splicing modulator-based strategies aimed at preventing cancer relapse10.
Modulation of aberrant ADAR1-mediated RNA editing of mRNA and miRNA target transcripts also represents a relatively unexplored avenue for CSC-directed therapeutic strategies (BOX 2). Previously we showed in a humanized mouse model of CML that shRNA-mediated knockdown of ADAR1 inhibits serial transplantation of malignant progenitors that promote leukaemic transformation7. This suggests that therapeutic inhibition of ADAR1 expression or activity might reduce self-renewal of CSCs responsible for disease relapse. Although a few studies have shown that locked nucleic acid oligonucleotides135 or substrate-mimic peptides136 have the potential to block RNA editase activity in a substrate-selective manner, these agents have not been investigated in a therapeutic context, and small-molecule inhibitors of ADAR1 enzymatic activity have yet to be identified. Thus, the molecular regulators that contribute to ADAR1 activation provide a valuable means to probe the therapeutic efficacy of blocking aberrant RNA editing activity in CSC maintenance. Consistent with a role for inflammation-associated, JAK2-driven induction of ADAR1 expression and activity, treatment with a JAK2 inhibitor in combination with a BCR–ABL1-targeted tyrosine kinase inhibitor significantly reduces the self-renewal capacity of BC CML CSCs40. Moreover, in stromal co-culture CSC models, a small-molecule tool compound interfered with the effect of ADAR1 on CSC self-renewal and restored let-7 biogenesis40. Together, these results suggest that therapeutic strategies aimed at antagonizing the widespread transcriptome reprogramming effects of aberrant ADAR1 activity may have the potential to selectively reduce the self-renewal capacity of CSC in malignancies typified by aberrant RNA editing activity.
Conclusions and future perspectives
The discovery of widespread deregulated RNA processing events in a plethora of human cancers indicates that transcriptome remodelling and translational deregulation are hallmarks of malignant transformation and therapeutic resistance. Importantly, the rapidly advancing sensitivity of sequencing technologies has facilitated the detection of rare but functionally relevant transcripts in human primary cells. However, further improvements in sequencing sensitivity and functional validation methods will be required to characterize the full complement of rare edited or spliced RNA variants that contribute to human cancer initiation and progression. Importantly, alternatively spliced isoforms and edited RNA species that are translated into proteins may represent tumour-specific antigens and thus represent novel potential diagnostic and immunotherapeutic targets.
In the case of RNA editing, a close examination of the human transcriptome reveals that the efficiency of A-to-I editing is cell type and context specific132. Thus, additional mechanisms may shape editome signatures in a given cell population and remain to be elucidated in future studies. Moreover, the activation of oncoproteins such as BCR–ABL1 and MYC may profoundly influence the RNA processing machinery in CSCs and bulk tumour cell populations. To enhance existing transcriptome editing databases, sequential whole-genome and whole-transcriptome sequencing of the same patients and cell types is required to accurately identify malignant RNA editing and splicing events. RNA editing activity can also be detected by alignment of RNA-seq data alone to existing reference genome databases by single nucleotide comparison at known RNA edited loci or by RNA editing site-specific quantitative PCR (RESSqPCR) in rare CSC populations137. Currently, there are two comprehensive RNA editing databases, DARNED73 and RADAR138 (see Further information), which enable identification of A-to-I editing in particular transcripts. In primary CSCs, detection of A-to-I editing of less abundant transcripts and transcripts subject to degradation may require more sensitive methods such as CLIP-sequencing. Future RNA editome and transcriptome studies to investigate the precise mechanisms by which CSCs gain stem cell-like properties will provide key insights into biomarkers of cancer progression and novel therapeutic targets. Because RNA processing activities are important for normal tissue development and stem cell self-renewal, development of small-molecule inhibitors to target CSCs will require careful examination to ensure that normal stem cell function remains intact. Future research aimed at deciphering the malignant epitranscriptome should pave the way for reversing the ‘monstrous’ evolutionary potential of cancer.
Acknowledgments
The authors acknowledge the inspiration and support of their patients and their professional colleagues. This work was supported by the Moores Foundation, California Institute for Regenerative Medicine (CIRM) grants (RN2-00910-1, DR1-01430 and RS1-00228-1); CIRM Training Grant (TG2-01154); National Institutes of Health (NIH) National Institute of General Medical Sciences grant 5K12GM068524; NIH National Cancer Institute (NCI) grant 2P30CA023100-28; NIH NCI grants R01CA205944, R21CA189705 and R21CA194679; the Leukemia & Lymphoma Society (0754-14); the Sanford Stem Cell Clinical Center; the San Diego Foundation; the Ratner Family Foundation; and the Swedish Childhood Cancer Foundation (Barncancerfonden).
Glossary
- 5′-Untranslated regions
(UTRs). Located upstream of the translation initiation codon, the 5′-UTRs are important for translational regulation of mRNA transcripts.
- 3′-UTRs
(Untranslated regions). The 3′-UTRs have an important role in regulation of gene expression by controlling RNA degradation, cellular localization and translation.
- Alu elements
A class of SINE elements Alu elements comprise approximately 10% of the human genome, Inverted Alu elements are favourable targets of adenosine deaminase acting on double-stranded RNA (ADAR)-mediated RNA editing as much as 90% of adenosine-to-inosine editing in the human transcriptome occurs within Alu elements.
- Short interspersed nuclear element
(SINE). Presented at high frequency in the eukaryotic genome SINEs are short (<700 bp) non-coding DNA sequences that retrotranspose themselves by a copy and paste mechanism.
- Long interspersed nuclear elements
(LINEs). Similar to SINEs, LINEs are a class of retrotransposons (~6 kb) comprising approximately 17%. of the human genome. They consist of a 5′-untranslated region (UTR), two open reading frames (ORF1 and ORF2) and a 3′-UTR. Misregulation of LINEs has been linked to tumorigenesis by retrotransposition-dependent and -independent functions.
- Primary microRNAs
(Pri-miRNAs). The miRNA genes are transcribed by RNA polymerase II and cleaved to large pri-miRNA transcripts that are subsequently cleaved by DROSHA to form the precursor miRNA (pre-miRNA) transcripts.
- Precursor micro RNAs
(Pre-miRNAs). Pre-miRNA transcripts are exported from the nucleus by exportin 5 (XPO5), to be processed by DICER1 to form the mature miRNAs.
- Blast crisis (BC) CML
BC CML is characterized by the elevated numbers of self-renewing cancer stem cells residing in the granulocyte–. macrophage progenitor compartment which express higher levels of the BCR–ABL1 oncogene and nuclear β-catenin. Patients with BC live an average of 3–6 months.
- Crosslinking immunoprecipitation
(CLIP). CLIP is UV crosslinking followed by immunoprecipitation to examine the interactions between RNA transcripts and RNA binding proteins and location of RNA modifications. The isolated RNA is reverse transcribed for PCR, microarray or sequencing analysis.
- LIN−SCA1+KIT+ cells
These cells are lineage-negative (LIN−), stem-cell antigen 1 positive (SCA1+) and KIT positive (KIT+) and make up a population of mouse bone marrow cells (~0.5%) with long-term multi-lineage repopulation capacity.
- Accelerated phase CML
(AP CML). In this phase of chronic myeloid leukaemia (CML), the cancer stem cells often acquire new genetic mutations, causing more severe symptoms and poor response to treatment.
- Chronic phase CML
(CP CML). The beginning phase of chronic myeloid leukaemia (CML) in which the patients have acquired the BCR–ABL1 oncogene, which induces abnormal production of myeloid cells. The standard treatment for CP CML is tyrosine kinase inhibitors such as imatinib or dasatinib. However, CP CML can progress slowly to an accelerated phase and later a blastic phase (blast crisis) over several years.
- Branch site
Also called branch point;. these occur predominantly at adenosine, highly conserved and closely localized to the 3′-splice site of an intron. The consensus sequence for an intron branch site (in IUPAC nucleic acid notation) is Y-U-R-A-C (20–50 nucleotides upstream of the acceptor site). Intronic RNA editing events, point mutations in the underlying DNA or errors during transcription have the potential to either destroy a branch site or activate a cryptic splice site in part of the transcript that is usually not spliced.
Footnotes
Competing interests statement
The authors declare competing interests: see Web version for details.
FURTHER INFORMATION
DARNED: http://darned.ucc.ie
RADAR: http://rnaedit.com
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Darwin C. On the Origin of Species by Means of Natural Selection. John Murray; 1859. [Google Scholar]
- 2.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 3.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat. Rev. Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 4.Saletore Y, et al. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012;13:175. doi: 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chadwick LH. The NIH Roadmap Epigenomics Program data resource. Epigenomics. 2012;4:317–324. doi: 10.2217/epi.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai B, Rasmussen TP. Global epiproteomic signatures distinguish embryonic stem cells from differentiated cells. Stem Cells. 2007;25:2567–2574. doi: 10.1634/stemcells.2007-0131. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Q, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc. Natl Acad. Sci. USA. 2013;110:1041–1046. doi: 10.1073/pnas.1213021110. This study showed for the first time that A-to-I RNA editing by ADAR1 is associated with leukaemic transformation and is required for CSC self-renewal capacity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 9.Pan Q, et al. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Crews LA, et al. RNA splicing modulation selectively impairs leukemia stem cell maintenance in secondary human AML. Cell Stem Cell. 2016;19:599–612. doi: 10.1016/j.stem.2016.08.003. This study was the first to show that small-molecule splicing modulators can selectively target self-renewing CSCs while sparing normal HSCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. This paper describes that silencing of m6A methyltranferase affects gene expression as well as alternative splicing patterns, resulting in modulation of p53 and indicating a role for m6A in gene expression regulation. [DOI] [PubMed] [Google Scholar]
- 12.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. By uncovering an association between m6A residues and miRNA-binding sites in 3’-UTRs this study provided insight into the epigenetic regulation of the mammalian transcriptome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat. Rev. Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 14.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Los Angeles A, et al. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- 16.Licht K, Jantsch MF. Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications. J. Cell Biol. 2016;213:15–22. doi: 10.1083/jcb.201511041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonda TJ, Ramsay RG. Directly targeting transcriptional dysregulation in cancer. Nat. Rev. Cancer. 2015;15:686–694. doi: 10.1038/nrc4018. [DOI] [PubMed] [Google Scholar]
- 19.Perry RP, Kelley DE. Existence of methylated messenger-RNA in mouse L cells. Cell. 1974;1:37–42. [Google Scholar]
- 20.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basanta-Sanchez M, Temple S, Ansari SA, D’Amico A, Agris PF. Attomole quantification and global profile of RNA modifications: epitranscriptome of human neural stem cells. Nucleic Acids Res. 2016;44:e26. doi: 10.1093/nar/gkv971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batista PJ, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moss B, Gershowitz A, Stringer JR, Holland LE, Wagner EK. 5′-terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J. Virol. 1977;23:234–239. doi: 10.1128/jvi.23.2.234-239.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desrosiers RC, Friderici KH, Rottman FM. Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5′ terminus. Biochemistry. 1975;14:4367–4374. doi: 10.1021/bi00691a004. [DOI] [PubMed] [Google Scholar]
- 25.Iwanami Y, Brown GM. Methylated bases of transfer ribonucleic acid from HeLa and L cells. Arch. Biochem. Biophys. 1968;124:472–482. doi: 10.1016/0003-9861(68)90355-x. [DOI] [PubMed] [Google Scholar]
- 26.Liu N, et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19:1848–1856. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller L, et al. The obesity related gene, FTO, interacts with APOE, and is associated with Alzheimer’s disease risk: a prospective cohort study. J. Alzheimers Dis. 2011;23:461–469. doi: 10.3233/JAD-2010-101068. [DOI] [PubMed] [Google Scholar]
- 28.Chen T, et al. m6A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Geula S, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 30.Niu Y, et al. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11:8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. This is a seminal study of the functional importance of m6A methylation in cancer, its impact on leukaemogenesis and ultimately its role in drug response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. This is a novel report of METTL3 and its involvement in human cancer by promoting translation of oncogenes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu T, et al. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer Res. 2008;68:6533–6540. doi: 10.1158/0008-5472.CAN-07-6642. [DOI] [PubMed] [Google Scholar]
- 35.Yu F, et al. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30:2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leis O, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31:1354–1365. doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- 37.Iv Santaliz-Ruiz LE, Xie X, Old M, Teknos TN, Pan Q. Emerging role of nanog in tumorigenesis and cancer stem cells. Int. J. Cancer. 2014;135:2741–2748. doi: 10.1002/ijc.28690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zipeto MA, Jiang Q, Melese E, Jamieson CH. RNA rewriting, recoding, and rewiring in human disease. Trends Mol. Med. 2015;21:549–559. doi: 10.1016/j.molmed.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Anadon C, et al. Gene amplification-associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis. Oncogene. 2016;35:4407–4413. doi: 10.1038/onc.2015.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zipeto MA, et al. ADAR1 activation drives leukemia stem cell self-renewal by impairing let-7 biogenesis. Cell Stem Cell. 2016;19:177–191. doi: 10.1016/j.stem.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mannion NM, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liddicoat BJ, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strout MP, Marcucci G, Bloomfield CD, Caligiuri MA. The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proc. Natl Acad. Sci. USA. 1998;95:2390–2395. doi: 10.1073/pnas.95.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elliott B, Richardson C, Jasin M. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol. Cell. 2005;17:885–894. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 45.Jeffs AR, Benjes SM, Smith TL, Sowerby SJ, Morris CM. The BCR gene recombines preferentially with Alu elements in complex BCR-ABL translocations of chronic myeloid leukaemia. Hum. Mol. Genet. 1998;7:767–776. doi: 10.1093/hmg/7.5.767. [DOI] [PubMed] [Google Scholar]
- 46.Guarnerio J, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 47.Osenberg S, et al. Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing. PLoS ONE. 2010;5:e11173. doi: 10.1371/journal.pone.0011173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo J, et al. Unraveling molecular effects of ADAR1 overexpression in HEK293T cells by label-free quantitative proteomics. Cell Cycle. 2016;15:1591–1601. doi: 10.1080/15384101.2016.1176657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 50.Fumagalli D, et al. Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep. 2015;13:277–289. doi: 10.1016/j.celrep.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi L, Chan TH, Tenen DG, Chen L. RNA editome imbalance in hepatocellular carcinoma. Cancer Res. 2014;74:1301–1306. doi: 10.1158/0008-5472.CAN-13-3485. [DOI] [PubMed] [Google Scholar]
- 52.Qin YR, et al. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 2014;74:840–851. doi: 10.1158/0008-5472.CAN-13-2545. [DOI] [PubMed] [Google Scholar]
- 53.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 54.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abrahamsson AE, et al. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proc. Natl Acad. Sci. USA. 2009;106:3925–3929. doi: 10.1073/pnas.0900189106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen L, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013;19:209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han L, et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015;28:515–528. doi: 10.1016/j.ccell.2015.08.013. By profiling genome-wide A-to-I RNA editing events in 17 cancer types, the authors report here that nonsynomymous RNA editing is associated with tumour cell viability and drug sensitivity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kawahara Y, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 60.Yang W, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. This article shows that A-to-I editing of pri-miRNA-142 by ADAR1 and ADAR2 suppresses DROSHA processing and mature miRNA production, revealing a role for RNA editing in miRNA biogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Solomon O, et al. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR) RNA. 2013;19:591–604. doi: 10.1261/rna.038042.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ota H, et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen T, et al. ADAR1 is required for differentiation and neural induction by regulating microRNA processing in a catalytically independent manner. Cell Res. 2015;25:459–476. doi: 10.1038/cr.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lechman ER, et al. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell. 2016;29:602–606. doi: 10.1016/j.ccell.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heale BS, et al. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wulff BE, Nishikura K. Modulation of microRNA expression and function by ADARs. Curr. Top. Microbiol. Immunol. 2012;353:91–109. doi: 10.1007/82_2011_151. [DOI] [PubMed] [Google Scholar]
- 67.Nishikura K. A-To-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 69.Isobe T, et al. miR-142 regulates the tumorigenicity of human breast cancer stem cells through the canonical WNT signaling pathway. eLife. 2014;3:e01977. doi: 10.7554/eLife.01977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garofalo M, Croce CM. Role of microRNAs in maintaining cancer stem cells. Adv. Drug Deliv. Rev. 2015;81:53–61. doi: 10.1016/j.addr.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou J, Ng SB, Chng WJ. LIN28/LIN28B: an emerging oncogenic driver in cancer stem cells. Int. J. Biochem. Cell Biol. 2013;45:973–978. doi: 10.1016/j.biocel.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 72.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 73.Kiran A, Baranov PV. DARNED: a database of RNA editing in humans. Bioinformatics. 2010;26:1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 74.Yulug IG, Yulug A, Fisher EM. The frequency and position of Alu repeats in cDNAs, as determined by database searching. genomics. 1995;27:544–548. doi: 10.1006/geno.1995.1090. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54nrb-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 76.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 77.Borchert GM, et al. Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum. Mol. Genet. 2009;18:4801–4807. doi: 10.1093/hmg/ddp443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang L, Yang CS, Varelas X, Monti S. Altered RNA editing in 3’ UTR perturbs microRNA-mediated regulation of oncogenes and tumor-suppressors. Sci. Rep. 2016;6:23226. doi: 10.1038/srep23226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bahn JH, et al. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat. Commun. 2015;6:6355. doi: 10.1038/ncomms7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morris AR, Mukherjee N, Keene JD. Systematic analysis of posttranscriptional gene expression. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010;2:162–180. doi: 10.1002/wsbm.54. [DOI] [PubMed] [Google Scholar]
- 82.Ye J, Blelloch R. Regulation of pluripotency by RNA binding proteins. Cell Stem Cell. 2014;15:271–280. doi: 10.1016/j.stem.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Okano H, Imai T, Okabe M. Musashi: a translational regulator of cell fate. J. Cell Sci. 2002;115:1355–1359. doi: 10.1242/jcs.115.7.1355. [DOI] [PubMed] [Google Scholar]
- 84.Ma Z, et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 2001;28:220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- 85.Xiao N, et al. Hematopoietic stem cells lacking Ott1 display aspects associated with aging and are unable to maintain quiescence during proliferative stress. Blood. 2012;119:4898–4907. doi: 10.1182/blood-2012-01-403089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.King CE, et al. LIN28B promotes colon cancer progression and metastasis. Cancer Res. 2011;71:4260–4268. doi: 10.1158/0008-5472.CAN-10-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene. 2010;29:2153–2159. doi: 10.1038/onc.2009.500. [DOI] [PubMed] [Google Scholar]
- 88.Zhang WC, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 89.Yang X, et al. Double-negative feedback loop between reprogramming factor LIN28 and microRNA let-7 regulates aldehyde dehydrogenase 1-positive cancer stem cells. Cancer Res. 2010;70:9463–9472. doi: 10.1158/0008-5472.CAN-10-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 91.Fox RG, Park FD, Koechlein CS, Kritzik M, Reya T. Musashi signaling in stem cells and cancer. Annu. Rev. Cell Dev. Biol. 2015;31:249–267. doi: 10.1146/annurev-cellbio-100814-125446. [DOI] [PubMed] [Google Scholar]
- 92.Ito T, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fox RG, et al. Image-based detection and targeting of therapy resistance in pancreatic adenocarcinoma. Nature. 2016;534:407–411. doi: 10.1038/nature17988. These authors showed that MSI2 expression can identify pancreatic CSCs, and that antisense oligonucleotides against MSI effectively block pancreatic cancer invasion and growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park SM, et al. Musashi2 sustains the mixed-lineage leukemia-driven stem cell regulatory program. J. Clin. Invest. 2015;125:1286–1298. doi: 10.1172/JCI78440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kwon HY, et al. Tetraspanin 3 is required for the development and propagation of acute myelogenous leukemia. Cell Stem Cell. 2015;17:152–164. doi: 10.1016/j.stem.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sakakibara S, et al. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc. Natl Acad. Sci. USA. 2002;99:15194–15199. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Imai T, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kharas MG, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 2010;16:903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jamieson CH, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 100.Kawahara H, et al. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 2008;181:639–653. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.DeBoever C, et al. Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLoS Comput. Biol. 2015;11:e1004105. doi: 10.1371/journal.pcbi.1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barrett CL, et al. Systematic transcriptome analysis reveals tumor-specific isoforms for ovarian cancer diagnosis and therapy. Proc. Natl Acad. Sci. USA. 2015;112:E3050–E3057. doi: 10.1073/pnas.1508057112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goff DJ, et al. A Pan-BCL2 inhibitor renders bone-marrow-resident human leukemia stem cells sensitive to tyrosine kinase inhibition. Cell Stem Cell. 2013;12:316–328. doi: 10.1016/j.stem.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Holm F, et al. Reversion to an embryonic alternative splicing program enhances leukemia stem cell self-renewal. Proc. Natl Acad. Sci. USA. 2015;112:15444–15449. doi: 10.1073/pnas.1506943112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Crews LA, Jamieson CH. Selective elimination of leukemia stem cells: hitting a moving target. Cancer Lett. 2013;338:15–22. doi: 10.1016/j.canlet.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 106.Pang WW, et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl Acad. Sci. USA. 2011;108:20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dolatshad H, et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia. 2015;29:1092–1103. doi: 10.1038/leu.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Adamia S, et al. A genome-wide aberrant RNA splicing in patients with acute myeloid leukemia identifies novel potential disease markers and therapeutic targets. Clin. Cancer Res. 2014;20:1135–1145. doi: 10.1158/1078-0432.CCR-13-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Han H, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sebestyen E, et al. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 2016;26:732–744. doi: 10.1101/gr.199935.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lasho TL, et al. SF3B1 mutations in primary myelofibrosis: clinical, histopathology and genetic correlates among 155 patients. Leukemia. 2012;26:1135–1137. doi: 10.1038/leu.2011.320. [DOI] [PubMed] [Google Scholar]
- 112.Wang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. This is one of the first seminal papers identifying DNA mutations occurring in splicing regulatory genes in pre-malignant haematopoietic disorders. [DOI] [PubMed] [Google Scholar]
- 114.Hahn CN, Scott HS. Spliceosome mutations in hematopoietic malignancies. Nat. Genet. 2011;44:9–10. doi: 10.1038/ng.1045. [DOI] [PubMed] [Google Scholar]
- 115.Thol F, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119:3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- 116.Zhang SJ, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119:4480–4485. doi: 10.1182/blood-2011-11-390252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Malcovati L, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118:6239–6246. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee SC, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat. Med. 2016;22:672–678. doi: 10.1038/nm.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Adler AS, et al. An integrative analysis of colon cancer identifies an essential function for PRPF6 in tumor growth. Genes Dev. 2014;28:1068–1084. doi: 10.1101/gad.237206.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jiang L, et al. Genomic landscape survey identifies SRSF1 as a key oncodriver in small cell lung cancer. PLoS Genet. 2016;12:e1005895. doi: 10.1371/journal.pgen.1005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Golan-Gerstl R, et al. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 2011;71:4464–4472. doi: 10.1158/0008-5472.CAN-10-4410. [DOI] [PubMed] [Google Scholar]
- 123.Hsu TY, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–388. doi: 10.1038/nature14985. This work describes a non-mutation-dependent mechanism of spliceosome disruption in solid tumours that functionally sensitizes tumour cells to treatment with small-molecule splicing modulatory compounds. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kashyap MK, et al. Targeting the spliceosome in chronic lymphocytic leukemia with the macrolides FD-895 and pladienolide-B. Haematologica. 2015;100:945–954. doi: 10.3324/haematol.2014.122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Salton M, et al. Inhibition of vemurafenib-resistant melanoma by interference with pre-mRNA splicing. Nat. Commun. 2015;6:7103. doi: 10.1038/ncomms8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Raitskin O, Cho DS, Sperling J, Nishikura K, Sperling R. RNA editing activity is associated with splicing factors in lnRNP particles: the nuclear pre-mRNA processing machinery. Proc. Natl Acad. Sci. USA. 2001;98:6571–6576. doi: 10.1073/pnas.111153798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shefer K, Sperling J, Sperling R. The supraspliceosome — a multi-task machine for regulated pre-mRNA processing in the cell nucleus. Comput. Struct. Biotechnol. J. 2014;11:113–122. doi: 10.1016/j.csbj.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 129.Beghini A, et al. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum. Mol. Genet. 2000;9:2297–2304. doi: 10.1093/oxfordjournals.hmg.a018921. [DOI] [PubMed] [Google Scholar]
- 130.Lev-Maor G, et al. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lev-Maor G, et al. Intronic Alus influence alternative splicing. PLoS Genet. 2008;4:e1000204. doi: 10.1371/journal.pgen.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Licht K, Kapoor U, Mayrhofer E, Jantsch MF. Adenosine to inosine editing frequency controlled by splicing efficiency. Nucleic Acids Res. 2016;44:6398–6408. doi: 10.1093/nar/gkw325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tariq A, et al. RNA-interacting proteins act as site-specific repressors of ADAR2-mediated RNA editing and fluctuate upon neuronal stimulation. Nucleic Acids Res. 2013;41:2581–2593. doi: 10.1093/nar/gks1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mizrahi RA, Schirle NT, Beal PA. Potent and selective inhibition of A-to-I RNA editing with 2’-O-methyl/locked nucleic acid-containing antisense oligoribonucleotides. ACS Chem. Biol. 2013;8:832–839. doi: 10.1021/cb300692k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schirle NT, Goodman RA, Krishnamurthy M, Beal PA. Selective inhibition of ADAR2-catalyzed editing of the serotonin 2c receptor pre-mRNA by a helix-threading peptide. Org. Biomol. Chem. 2010;8:4898–4904. doi: 10.1039/c0ob00309c. [DOI] [PubMed] [Google Scholar]
- 137.Crews LA, et al. An RNA editing fingerprint of cancer stem cell reprogramming. J. Transl Med. 2015;13:52. doi: 10.1186/s12967-014-0370-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014;42:D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. This is the first study to identify a self-renewing CSC population in haematological malignancies. [DOI] [PubMed] [Google Scholar]
- 140.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc. Natl Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hemmati HD, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc. Natl Acad. Sci. USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim CF, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 143.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 144.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 145.Szotek PP, et al. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian inhibiting substance responsiveness. Proc. Natl Acad. Sci. USA. 2006;103:11154–11159. doi: 10.1073/pnas.0603672103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Takebe N, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.LaBarge MA. The difficulty of targeting cancer stem cell niches. Clin. Cancer Res. 2010;16:3121–3129. doi: 10.1158/1078-0432.CCR-09-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]