Abstract
Eleven viral isolates derived mostly in Aedes cells from mosquito pools collected in Southeast Asia and the Americas between 1966 and 2014 contained particles with electron microscopy morphology typical of reoviruses. Metagenomics analysis yielded the near complete genomes of three novel reoviruses, Big Cypress orbivirus, Ninarumi virus, and High Island virus and a new tetravirus, Sarawak virus. Strains of previously characterized Sathuvarachi, Yunnan, Banna and Parry’s Lagoon viruses (Reoviridae), Bontang virus (Mesoniviridae), and Culex theileri flavivirus (Flaviviridae) were also characterized. The availability of these mosquito virus genomes will facilitate their detection by metagenomics or PCR to begin to determine their geographic range, extent of host tropism, and possible association with arthropod or vertebrate disease.
Keywords: Metagenomics, Reoviridae, Viral discovery, Mosquito, Deep sequencing, Orbivirus
Introduction
During the past decade, advances in DNA sequencing technology have allowed the identification of a wide range of novel viruses associated with mosquitoes (Aguiar et al., 2015; Auguste et al., 2014; Bolling et al., 2015; Carissimo et al., 2016; Chandler et al., 2015; Charles et al., 2016; Cholleti et al., 2016; Coffey et al., 2014; Contreras-Gutierrez et al., 2017; Fauver et al., 2016; Frey et al., 2016; Li et al., 2015a; Li et al., 2015b; Nasar et al., 2012; Ng et al., 2011; Palacios et al., 2015; Pauvolid-Correa et al., 2016; Shi et al., 2015; Simmonds et al., 2017; Vasilakis et al., 2014a; Wang et al., 2015). We examine here, by electron microscopy and metagenomics analysis, 11 mosquito derived viral isolates from the extensive collection of the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) (https://www.niaid.nih.gov/research/wreceva), whose morphology indicated the likely presence of multi-segmented reoviruses.
Materials and Methods
Viral isolates
Culture supernatants of each virus were obtained from the WRCEVA at the University of Texas Medical Branch (UTMB). Table 1 lists the eleven viruses in this study, including mosquito source, geographic locality and date of collection, and cell lines tested. All of the viruses were initially identified through cytopathic effect in mosquito cell cultures (Aedes albopictus C6/36 line or Aedes pseudoscutellaris AP-61 line). Four of the viruses were isolated at UTMB from mosquito pools sent for testing; and seven of the viruses were isolated elsewhere and submitted to the WRCEVA for identification and characterization.
Table 1.
Origins of 11 viral isolates.
| Species name | Strain | Family Subfamily Genus | Collection date and location | Source of isolate, cell line used1 | Segment/s | GenBank accession numbers |
|---|---|---|---|---|---|---|
| Big Cypress orbivirus (BCPOV) | BCNP 2-151 |
Reoviridae Sedoreovirinae Orbivirus |
7/7/2014 USA, FLORIDA Sawgrass, Big Cypress |
Pool of 50 Psorophora columbiae, C6/36 |
VP1-7 NS1-3 |
MF094109-MF094118 |
| Ninarumi virus (NRUV) | LO-041 |
Reoviridae Sedoreovirinae Orbivirus |
2/22/2009 PERU, LORETO Puerto Almendra |
Pool of 41 Ochlerotatus fulvus, C6/36 |
VP1 VP3-7 NS1-3 |
MF094119-MF094127 |
| High Island virus (HISLV) | HI-BSC-18 |
Reoviridae Spinareovirinae Idnoreovirus |
7/25/2013 USA, TEXAS High Island. Galveston Co. |
Pool of 5 Psorophora ciliate, C6/36, Vero Neg. |
s1, s2, s3, s4, s6, s8 | MF094128-MF094133 |
| Sarawak virus (SWKV) | SWK-M26 |
Tetraviridae Betatetravirus |
10/17/2013 MALAYSIA |
Pool of males Aedes albopictus, C6/36 |
ORF1-3 | MF094134 |
| Yunnan orbivirus (YOUV) | JKT-8650 |
Reoviridae Sedoreovirinae Orbivirus |
1/21/1981 INDONESIA, BALI, Tag-Tag |
Anopheles vagus, C6/36, Vero Neg. |
VP1-7 NS1-3 |
MF152977-MF152987 |
| Yunnan orbivirus (YOUV) | JKT-10087 |
Reoviridae Sedoreovirinae Orbivirus |
12/29-30/1981 INDONESIA,JAVA,CILACAP, Karang Sari |
Mansonia uniformis, AP-61, Vero Neg. |
VP1-7 NS1-3 |
MF152988-MF152998 |
| Banna virus (BAV) | JKT-6957 |
Reoviridae Sedoreovirinae Orbivirus |
6/9/1981 INDONESIA, CENTRAL JAVA, Yogyakarta |
Culex fuscocephala, C6/36, Vero Neg. |
VP1-12 | ND |
| Sathuvachai virus (SVIV) | CG LT 392 |
Reoviridae Sedoreovirinae Orbivirus |
1966 or before VIETNAM |
Unknown source C6/36, BHK | VP1-7 NS1-3 |
MF152967-MF152976 |
| Parry’s Lagoon virus (PLV) | KP84-0156 |
Reoviridae Sedoreovirinae Orbivirus |
1984 Thailand Kamphaeng Phet |
unknown source C6/36, BHK Neg. |
VP1-12 | ND |
| Bontang Baru virus (BBaV) | JKT-7815 | Mesoniviridae | 2-3/26-10/1981 INDONESIA, EAST KALIMANTAN, Bontang Baru |
Pool of 50 Culex vishnui, C6/36, Vero Neg. |
NA | MF158348 |
| Culex theileri flavivirus (CTFV) | JKT-8650 | Flaviviridae | 1/21/1981 INDONESIA, BALI, Tag-Tag |
Anopheles vagus, C6/36, Vero Neg. |
NA | MF153378 |
All inoculated cell lines are listed and were CPE positive unless followed by Neg.
Viral cultures
Pools of mosquitoes tested at UTMB were homogenized in 1.0 ml of phosphate-buffered saline, pH 7.4, with 25% fetal bovine serum, using a TissueLyser (Qiagen, Valencia, CA) and 3 mm stainless steel beads at a frequency of 1,260 oscillations/minute for 1 minute at ambient temperature (25°C). After centrifugation at 10,000 rpm in a microcentrifuge for 10 min., 100 μl of the supernatant were inoculated into two 12.5 cm2 flasks with monolayer cultures of C6/36 and Vero E6 or baby hamster kidney (BHK) cells. After inoculation, the C6/36 cells were maintained at 28°C for 6–7 days and were examined every 2 days for evidence of viral cytopathic effect (CPE) (Igarashi, 1978). Vero and BHK cell cultures were incubated at 37°C and examined for CPE for two weeks. All JKT labeled isolates were inoculated intra-cranially into newborn mice and none developed signs of diseases. The Sathuvachai virus (SVIV) isolate was also inoculated and was pathogenic for newborn mice. None of the other isolates were inoculated into newborn mice.
Electron microscopy
Infected cell monolayers showing CPE were fixed in a mixture of 2.5% formaldehyde and 0.1% glutaraldehyde in 0.05 M cacodylate buffer pH 7.2 containing 0.03% trinitrophenol and 0.03% CaCl2 for at least 1 h at room temperature. Fixed cells were kept in the fixative solution at 4°C until further processing. After washing in 0.1 M cadodylate buffer, cells were scraped off the plastic, pelleted and processed further as a pellet. The pellets were postfixed in 1% OsO4 in 0.1 M cacodylate buffer pH 7.2, en bloc stained with 2% aqueous uranyl acetate for 20 min. at 60°C, dehydrated in graded series of ethanol, and embedded in Poly/Bed 812 (Polysciences, Warrington, PA). Ultrathin sections were cut on a Leica-Reichart, Ultracut S ultramicrotome, (Vienna, Austria) stained with lead citrate, and examined in a Philips (Eindhoven, Netherlands) CM-100 electron microscope at 60 KV (Kim et al., 2009).
Deep Sequencing and sequence analysis
Supernatants of cell cultures showing cytopathic effects were filtered (0.45-μm Millipore) and treated with nucleases (Sadeghi et al., 2017; Zhang et al., 2016), prior to nucleic acid extraction (MagMAX Viral RNA Isolation Kit, Ambion, Inc., Austin, Tx, USA). Following random RT-PCR and Nextera reagents (Illumina), doubly tagged DNA was generated from each supernatant for sequencing using a MiSeq instrument (250 bases PE) (Li et al., 2015c; Sadeghi et al., 2017; Zhang et al., 2016). Library preparation and computational analysis were performed as previously described (Li et al., 2015c; Sadeghi et al., 2017; Zhang et al., 2016). Sequence reads were assembled de novo, using the Ensemble program and compared to all viral protein sequences in RefSeq in GenBank (Deng et al., 2015).
Results
Complete or partial protein coding sequences (CDS) were generated from three novel uncharacterized reoviruses, and one novel tetravirus. Seven viral genomes, closely related to those of previously characterized viruses were also sequenced (Table 1). All eleven viruses produced CPE in mosquito cells and one also caused CPE in vertebrate cells (BHK) and was lethal to newborn mice (Sathuvachai virus-CGLT 392).
Big Cypress orbivirus (BCPOV)
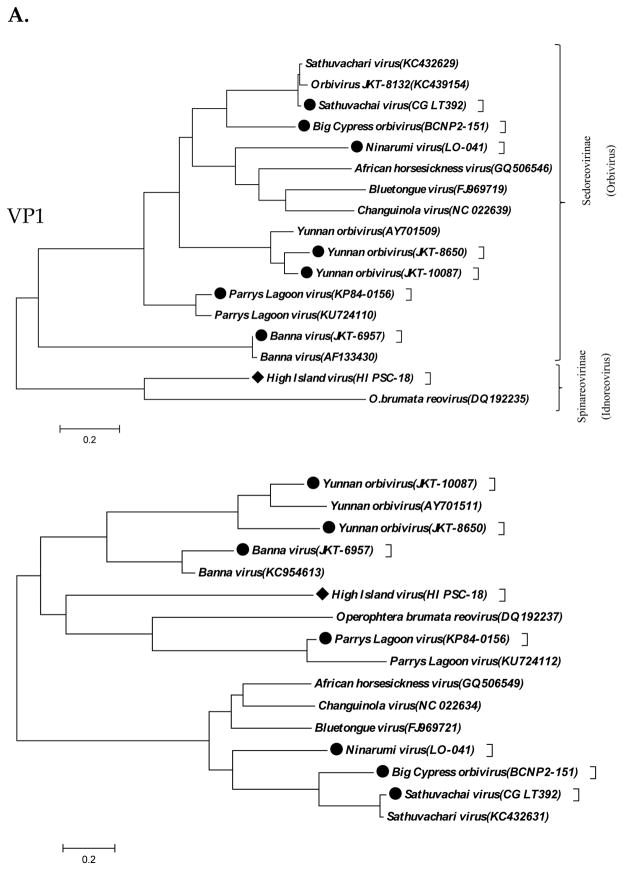
Ten segments (VP1–VP7 and NS1–NS3) were assembled from a new orbivirus species isolated from a pool of 50 Psorophora columbia mosquitoes collected in Big Cypress National Preserve, South Florida on July 7, 2014 (GenBank MF094109- MF094118). The VP1 and VP3 encoding RNA dependent RNA polymerase (RdRp) and outer capsid proteins showed 63% and 74% amino acid identifies to those of the recently sequenced Sathuvachari virus (SVIV) (Kapoor et al., 2013). We named this agent Big Cypress orbivirus, strain BCNP 2-151. Amino acid identity with other related reoviruses is given in Table 2. Results of phylogenetic studies are shown in Figures 1–2.
Table 2.
Amino acid identities of reoviruses to closest known relatives.
| Virus/Segment | S1/VP1 | S2/VP2 | S3/VP3 | S4/VP4 | S5/VP5 | S6/VP6 | S7/VP7 | S8/NS1 | S9/NS2 | S10/NS3 | S11/NS4 | S12/NS5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Big Cypress orbivirus (BCPOV) | VP1 | VP2 | VP3 | VP4 | VP5 | VP6 | VP7 | NS1 | NS2 | NS3 | --- | --- |
| 1 SVIV | SVIV | SVIV | SVIV | SVIV | SVIV | SVIV | SVIV | SVIV | SVIV | |||
| (63%) | (32%) | (74%) | (60%) | (62%) | (41%) | (75%) | (42%) | (57%) | (60%) | |||
|
| ||||||||||||
| Ninarumi virus (NRUV) | VP1 | --- | VP3 | VP4 | VP5 | VP6 | VP7 | NS1 | NS2 | NS3 | --- | --- |
| 2 AHSV | 3 CGLV | 4 BTV | BTV | CGLV | 5 TILV | CGLV | 6 LEBV | SVIV | ||||
| (53%) | (49%) | (53%) | (41%) | (32%) | (37%) | (28%) | (34%) | (35%) | ||||
|
| ||||||||||||
| High Island virus (HISLV) | S1 | S2 | S3 | S4 | --- | S6 | --- | S8 | --- | --- | --- | --- |
| 7 OpbuRV | OpbuRV | OpbuRV | OpbuRV | OpbuRV | OpbuRV | |||||||
| (34%) | (29%) | (23%) | (36%) | (23%) | (27%) | |||||||
|
| ||||||||||||
|
Yunnan orbivirus (YOUV) JKT-8650 |
VP1 | VP2 | VP3 | VP4 | VP5 | VP6 | VP7 | NS1 | NS2 | NS3 | NS4 | --- |
| 8 YNV | YNV | 9 MPO | YNV | YNV | YNV | YNV | YNV | YNV | YNV | YNV | ||
| (94%) | (99%) | (97%) | (97%) | (99%) | (99%) | (97%) | (92%) | (97%) | (94%) | (98%) | ||
|
| ||||||||||||
|
Yunnan orbivirus (YOUV) JKT-10087 |
VP1 | VP2 | VP3 | VP4 | VP5 | VP6 | VP7 | NS1 | NS2 | NS3 | NS4 | --- |
| YNV | YNV | YNV | YNV | YNV | YNV | YNV | YNV | YNV | YNV | YNV | ||
| (94%) | (98%) | (86%) | (98%) | (98%) | (95%) | (97%) | (92%) | (96%) | (94%) | (93%) | ||
|
| ||||||||||||
| Banna virus (BAV) | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 | S12 |
| 10 BAV | BAV | BAV | BAV | BAV | BAV | BAV | BAV | BAV | BAV | BAV | BAV | |
| (99%) | (90%) | (89%) | (86%) | (96%) | (93%) | (89%) | (98%) | (97%) | (97%) | (95%) | (93%) | |
|
| ||||||||||||
| Sathuvachai virus (SVIV) | VP1 | VP2 | VP3 | VP4 | VP5 | VP6 | VP7 | NS1 | NS2 | VP3 | --- | --- |
| SVIV | SVIV | SVIV | SVIV | SVIV | SVIV | SVIV | SVIV | SVIV | SVIV | |||
| (98%) | (88%) | (99%) | (97%) | (97%) | (97%) | (99%) | (99%) | (98%) | (99%) | |||
|
| ||||||||||||
| Parry’s Lagoon virus (PLV) | VP1 | VP2 | VP3 | VP4 | VP5 | --- | --- | NS1 | NS3 | --- | --- | |
| PLV | PLV | PLV | PLV | PLV | PLV | PLV | ||||||
| (91%) | (92%) | (73%) | (89%) | (90%) | (96%) | (79%) | ||||||
Sathuvachari virus (SVIV)
African horse sickness virus (AHSV)
Changuinola virus (CGLV)
Bluetongue virus (BTV)
Tilligerry virus (TILV)
Lebombo virus (LEBV)
Operophtera brumata reovirus (OpbuRV) (MPO)
Yunnan virus (YNV)
Middle Point orbivirus
Banna virus
Figure 1.
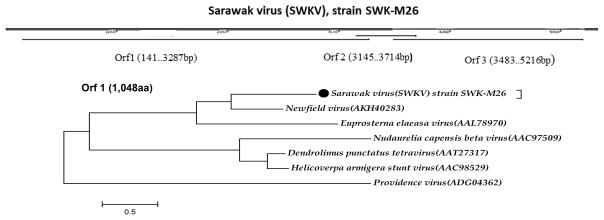
A. Phylogeny of the Orbivirus and Idnoreovirus genera based on the RdRp gene (VP1). B. Phylogeny of Orbiviruses based on innermost protein capsid (VP3). C. Tetravirus genome open reading frames and phylogeny of ORF1 protein.
Figure 2.
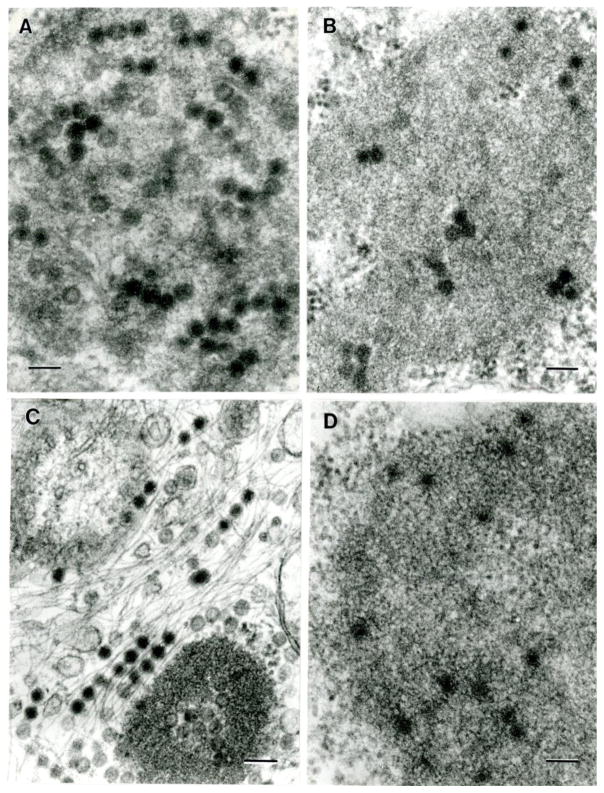
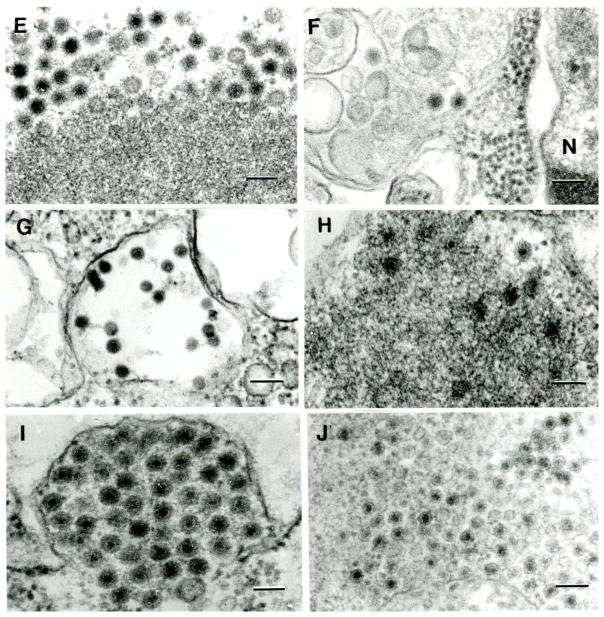
Electron microscopy of ten viral isolates included in this study. A. Big Cypress virus; B. Ninarumi virus; C. Parry’s Lagoon virus; D. Yunnan virus (JKT-10087); E. Banna virus; F. High Island orbivirus; G. Bontang virus; H. Yunnan virus (JKT-8650); I. Sathuvachari virus; J. Sarawak tetravirus. Bars = 100 nm
Ninarumi virus (NRUV)
Nine segments (VP1, VP3–VP7 and NS1–NS3) could be assembled from a novel orbivirus isolated in C6/36 cells at UTMB from a pool of 41 Aedes (Ochlerotatus) fulvus mosquitoes collected near Iquitos, Loreto department, Peru on February 22, 2009. The VP1 and VP3 of NRUV showed 53% and 49% amino acid identities to those of African horse sickness virus (AHSV) and Changuinola virus (CGLV), respectively. This isolate was named Ninarumi virus, strain LO-041 after the neighborhood from which it originated. AHSV is a species of the genus Orbivirus and currently consists of 9 different serotypes (Attoui and Mohd Jaafar, 2015) which are endemic in sub-Saharan Africa. AHSV is a vector-borne orbivirus that is transmitted primarily by Culicoides midges (Attoui and Mohd Jaafar, 2015). Changuinola virus is made up of at least twelve known serotypes that have been isolated mainly from phlebotomine sandflies (Attoui and Mohd Jaafar, 2015). The phylogenetic analyses of VP1 and VP2 of Ninarumi virus are shown in Figure 1 and 2.
High Island virus (HISLV)
Six segments (s1, s2, s3, s4, s6, s8) were assembled from a novel reovirus that was isolated at UTMB from a pool of 5 Psorophora ciliate mosquitos collected on High Island, Galveston County, Texas on July 25, 2013. Segment 1 (RNA-dependent RNA polymerase) and segment 3 (T2 subcore shell) showed 34% and 23% amino acid identities, respectively, to those of Operophtera brumata reovirus (ObRV), genus Idnoreovirus (Graham et al., 2006). We named this isolate High Island virus, strain HI PSC-18. Additionally, we assembled another potential segment (2,357nt) from this library, without measurable amino acid identities to known viruses, possibly reflecting one of the missing segments. ObRV was isolated from O. brumata and its parasitoid wasp Phobocampe tempestiva and has a genome composed of 10 segments of double stranded RNA (Graham et al., 2006; Graham et al., 2008; Graham et al., 2007).
A novel tetravirus: Sarawak virus (SWKV)
A novel genome from the family Alphatetraviridae was derived from an isolate obtained from a pool of male Aedes albopictus mosquitoes collected in Sarawak, (island of Borneo), Malaysia on October 17, 2013. The large ORF (1,048aa) showed 38% identity with putative replicase of recently described Newfield virus, associated with Drosophila melanogaster (Webster et al., 2015). The second ORF (577aa) and small ORF (189aa) showed no significant similarity to any protein in GenBank. We named this isolate Sarawak virus (SWKV), strain SWK-M26.
Previously described orbivirus genomes
Yunnan viruses
Two new strains of Yunnan orbivirus (YOUV) were identified (Table 1). Strain JKT-8650 was isolated from a pool of Anopheles vagus mosquitoes collected in Tag-Tag, Bali, Indonesia on January 21, 1981. Strain JKT-10087 was isolated from a pool of Mansonia uniformis mosquitoes collected in Karang Sari, Java, Indonesia on December 29 and 30, 1981. All eleven segments (VP1 to VP7 and NS1 to NS4) were sequenced from both strains. The VP1 of the two isolates showed >90% amino acid identity to the prototype YOUV strain isolated from Culex tritaeniorhynchus mosquitoes collected in Yunnan province of China (Attoui et al., 2005). Subsequent phylogenetic studies have shown that Yunnan orbivirus is a species complex that includes three closely related serotypes: Yunnan virus, isolated from mosquitoes in China and Indonesia; Rioja virus, isolated from cattle, sheep, donkeys and a dog in Peru; and Middle Point virus, isolated from a cow in Australia (Attoui et al., 2009).
Banna virus (BAV)
Twelve partial segments (VP1 to VP12) of a new BAV strain, designated JKT-6957, were obtained. Strain JKT-6957 was isolated from a pool of Culex fuscocephala mosquitoes collected on June 9, 1981 in Yogyakarta, Java, Indonesia. The 12 segments of JKT-6957 showed >90% amino acid identity to VP1-VP12 of Banna virus strain JKT-6423 (Attoui et al., 2000). BAV was originally isolated from persons with encephalitis and fever in Yunnan province, China (Xu et al., 1990). Subsequently, it has been obtained from pigs and cattle as well as from ticks and mosquitoes in China, Indonesia and Vietnam (Liu et al., 2010).
Sathuvachai virus (SVIV)
Ten segments (VP1 – VP7) and NS1 and NS3) of a new isolate of SVIV, designated CG LT392, were generated de novo. CG LT 92 showed 98% and 99% amino acid identity to the VP1 and VP3 of the prototype SVIV strain I An 66411 (Figures 1 and 2). The prototype SVIV strain was isolated from a starling (Brahminy myna) collected in Vellore, India in 1963 (Kapoor et al., 2013). Our isolate, CG LT92, was received on March 2, 1966 from a U.S. Army Laboratory in Saigon, Vietnam. The sample was sent for study as infected newborn mouse brain following pathogenic intra-cranial inoculation, along with two other isolates of Sindbis and Bagaza viruses from South Vietnam. No other information accompanied the samples, but we assume they were all mosquito isolates.
In addition to its identity with the Indian prototype strain of SVIV, based on comparative genetic analysis, the other nearest genetic relative of CG LT92 is Tagtag virus (TGV, strain JKT-8132) TGV was isolated from a pool of Culex vishnui mosquitoes collected in Tagtag, Bali, Indonesia in 1980 (Kapoor et al., 2013). In view of their close phylogenetic relationship, it was proposed that SVIV and TGV represent a single species within the genus Orbivirus (Kapoor et al., 2013).
Parry’s Lagoon virus (PLV)
PLV was isolated from a pool of unidentified mosquitoes collected in Kamphaeng Phet, Thailand in 1984. The pool, designated KP 84-0156, contained three different mosquito viruses: Kamphaeng Phet virus (KPhV), a mesonivirus reported previously (Vasilakis et al., 2014b); PLV, the virus described here; and a novel Tymovirus (to be described). We obtained 7 partial segments of the PLV-KP84-0156 with nearly 100% identity at the amino acid level with PLV. The prototype PLV strain was isolated from mosquitoes in the Kimberley region of Western Australia and shares sequence similarities to Corriparta virus (CORV) (Harrison et al., 2016). PLV is classified as an Orbivirus (subfamily Sedoreovirinae).
Previously described mesonivirus and insect-specific flavivirus
Bontang Baru virus (BBaV)
An isolate of Bontang Baru virus (BBaV), designated JKT 7815, was recovered from a pool of Culex vishuui mosquitoes, collected at Bontang Baru, East Kalimantan, Indonesia on February 10, 1981. We generated a 20,736 nt contig from JKT-7815, with 99% nucleotide identity to the BBaV prototype, strain JKT 7774. Organization of the genome was similar to that described for other mesoniviruses (Lauber et al., 2012; Vasilakis et al., 2014b) with a 5′-untranslated region (5′-UTR) of 359 to 370 nt, four major long open reading frames (ORFs), and a long terminal region of 1780 to 1804 nt preceding the poly [A] tail. Four previous isolates of BBaV were reported from Culex mosquitoes captured on the Indonesian islands of Java and Kalimantan (Vasilakis et al., 2014b).
Culex theileri flavivirus (CTFV)
Mosquito pool JKT-8650 (Anopheles vagus collected in Tag-Tag, Bali, Indonesia on January 21, 1981) also contained a second virus in addition to the Yunnan orbivirus described above. The full genome of an insect-specific flavivirus (ISV) was generated with a genome length of at least 10,280 nucleotides with 91% identity to that of two Culex theileri flavivirus isolates from Portugal (153 and 178) (Parreira et al., 2012). Like other flaviviruses, it encoded a single 3357 residue polyprotein. CTFV was also recently isolated from mosquitoes collected in Turkey (Ergunay et al., 2016).
Transmission electron microscopy
Figure 2 show the ultrastructure of 10 of the viruses in C6/36 or BHK cells. Fig 1A–E and IH show fragments of viroplasm (virus factories and viral inclusion bodies) in the cytoplasm of cells forming subviral particles 55–65 nm in diameter. Figure 1F and 1I show reovirus particles ~65 nm in diameter in the cytoplasm of C6/36 cells. Figure 1G shows mesonivirus (Bontang virus) particles, 55–65 nm in diameter, in C6/36 cell. 1J shows Sarawak tetravirus particles, 45 nm in diameter, in the cytoplasm of C6/36 cell.
Discussion
Metagenomic sequencing of viral nucleic acids was used to characterize 11 viral genomes generated from 10 mosquito pools that produced CPE in mosquito cell cultures and where tested failed to cause CPE in Vero or BHK cells or kill newborn mice (except for Sathuvachai virus). Initial attempts to identify these agents by classical serologic methods (hemagglutination-inhibition, complement fixation and immunofluorescence) were unsuccessful, as specific antibodies were not available. Subsequent transmission electron microscopy at UTMB on infected C6/36 or vertebrate cell cultures demonstrated the presence of virus-like structures. Metagenomics sequencing was therefore undertaken and identified members of four virus families: Reoviridae, Tetraviridae, Mesoniviridae and Flaviviridae.
The family Reoviridae currently consists of 15 genera (Attoui H et al., 2012). The genus Orbivirus currently contains 22 recognized species, plus 10 additional probable but still unapproved species. Orbiviruses typically contain 10 to 11 dsRNA segments, encoding seven structural proteins (Virion proteins, VP1–VP7) and three to four non-structural proteins (NS1–NS4) (Belhouchet et al., 2011). The Orbivirus RNA-dependent RNA polymerases (RdRp), encoded by the Seg1 gene (VP1), is an important marker for classification. Amino acid identity >30% with other characterized orbiviruses is required to be considered a member of the genus (Attoui H et al., 2012). In addition, the sub-core protein (T2) of orbiviruses is used to classify species within genera. Orbiviruses within a single species group show greater than 91% identity with respect to the T2 protein sequence (Attoui H et al., 2012). Orbiviruses are transmitted primarily by arthropod vectors such as culicoid midges, phlebotomine sandflies, mosquitoes or ticks, and can infect a wide variety of vertebrate hosts including humans (Drolet et al., 2015). Using ICTV taxonomic criteria, the two novel orbiviruses described here (Big Cypress and Ninarumi) qualify as new species.
The genus Idnoreovirus is a seperate member of the family Reoviridae. Idnoreovirus genomes are composed of 10 dsRNA segments (Attoui H et al., 2012). Insects serve as natural hosts for idnoreoviruses. There are currently five species in the genus, which are designated Idnoreovirus 1–5. Two additional viruses isolated from Drosophila simulans (Drosophila S virus) and from populations of the winter moth Operophtera brumata (Operophtera brumata idnoreovirus) are still unclassified members of the genus (Attoui H et al., 2012; Graham et al., 2006; Lopez-Ferber et al., 1989). The addition of the genome of another Idnoreovirus species from Psorophora ciliata mosquitoes adds to our understanding of the host range and diversity of that genus.
The family Tetraviridae is restricted mainly to insects in the order Lepidoptera (butterflies and moths). Viruses in this family have positive-sense ssRNA genomes. Viral particles are about 40 nm in diameter and composed of 240 copies of two proteins of approximately 60kDa (L) and 8 kDa (S) (Dorrington et al., 2012). The structures of Nudaurelia capensis β virus (NβV) and Nudaurelia capensis ω virus (NωV) have been solved and reveal an unusual T = 4 icosahedral capsid architecture that distinguishes the family from other non-enveloped viruses (Jiwaji et al., 2016). Eleven viruses are recognized in this family including NβV, NωV and Helicoperva armigera stunt virus (HaSV). Viruses in this family are classified into two genera: Betatetravirus and Omegatetravirus according to their viral replicases (Dorrington et al., 2012). Betatetraviruses, such as NβV, have monopartite genomes; with a 6.5 kb genomic RNA encoding the replicase and capsid precursor genes. Omegatetraviruses, such as NωV and HaSV, have bipartite ssRNA+ genomes of approximately 5.2 and 2.5 kb separately encoding the replicase and capsid precursor genes, respectively. Our isolate (SWK M26) has a monopartite betatetravirus genome encoding two large ORFs with the 5′ ORF encoding RdRp showing 38% identity to that of Newfield virus. The second ORF did not show similarity to any protein currently in GenBank. Since SWK M26 was derived from a pool of Aedes albopictus mosquitoes and replicated in C6/36 cells, the tropism of this viral family likely extends beyond that of lepidopterans. The C6/36 cell isolate is also the second reported member of this family to be amplified in cell culture. Providence virus (PrV) was previously shown to replicate in both insect and human HeLa cells (Pringle et al., 2003).
The viral family Mesoniviridae (order Nidovirales) consists of ([+]ssRNA) insect viruses with ~20-kb genome size (Lauber et al., 2012). Members of the family Mesoniviridae replicate in mosquitoes (Lauber et al., 2012; Nga et al., 2011; Zirkel et al., 2011). Cavally virus (CavV) and Nam Dinh virus (NDiV) were the first mesoniviruses to be characterized (Nga et al., 2011; Zirkel et al., 2011). These two viruses are closely related and belong to the same species, Alphamesonivirus 1, which is the prototype species of the genus Alphamesonivirus (Lauber et al., 2012). Other phylogenetically diverse mesoniviruses have been identified in recent studies of viruses isolated from a wide range of mosquito species and geographic locations (Vasilakis et al., 2014b). The latter remain to be assigned to existing or yet-to-be-established taxa within the family Mesoniviridae (Lauber et al., 2012; Thuy et al., 2013). Mesoniviruses have genomes of approximately 20 kb and have been proposed to provide an evolutionary link between small (13 to 16 kb) and large (26 to 32 kb) nidoviruses (Lauber et al., 2012; Nga et al., 2011; Zirkel et al., 2011).
Seven of the viruses identified in this study are viruses that have been previously described. Nonetheless, results of the present study provide additional sequences of variant strains and expand the known host range and geographic distribution of these agents. For example, isolates of Sathuvachari virus (SVIV) were previously reported from a bird in India and mosquitoes in Bali, Indonesia (Kapoor et al., 2013). Our identification of an isolate of SVIV from Vietnam suggests that this virus may occur throughout southern India and Southeast Asia. Likewise, the identification of Culex theileri flavivirus (CTFV) from Anopheles mosquitoes in Indonesia, along with the previously reported isolations of CTFV from Portugal and Turkey, indicate that this virus has a much wider geographic distribution and is not restricted to a single mosquito genus or species. The identification of Parry’s Lagoon virus (PLV) from northern Thailand indicates that this virus also has a wider geographic distribution than Australia. Banna virus (BAV) has been isolated before in Indonesia (Attoui et al., 2000); but its wide geographic distribution in Asia (China, Indonesia and Vietnam) and its reported association with human illness indicates that this virus deserves further study as a regional public health threat.
Viruses identified in this study (YOUV, PLV and CTFV) were obtained from cultures of mosquito pools containing more than one virus. Traditionally, arbovirologists used newborn mice, embryonated eggs or various vertebrate cell lines when attempting to isolate viruses from pools of mosquitoes and other hematophagous arthropods (Work, 1964). However, following the development of arthropod cell lines, virologists also began to use mosquito and tick cell lines for primary isolation of viruses from hematophagous arthropods (Jozan, 1987). Two of the most widely used are the C6/36 clone of Aedes albopictus cells (Igarashi, 1978) and the AP-61 line of Aedes pseudoscutellaris cells (Varma et al., 1974). These two mosquito cell lines have proved to be more sensitive for the isolation of some arboviruses, such as dengue, yellow fever and Zika viruses, than newborn mice or vertebrate cell lines (Igarashi, 1987; Tesh, 1979; R.B. Tesh, unpublished data). But these mosquito cell lines also support the growth of mosquito-specific or mosquito only viruses. Consequently, when assaying mosquito pools for arboviruses in C6/36 or AP-61 cells, it is not uncommon to isolate two or more mosquito-associated viruses from a single pool. In the past, some of these mosquito-specific viruses remained genetically un-characterized because many were novel and no antibodies or other reagents were available to identify them. Deep sequencing now allows the genetic characterization of a plethora of novel virus genomes present in cell ultures of mosquito pools.
All of the isolates produced CPE in mosquito (C6/36) cells; but of the subset inoculated into vertebrate (Vero or BHK) cells only SVIV resulted in CPE. For the subset of isolates inoculated into newborn mice their inability to cause signs of disease (except for SVIV) also support the likelihood that the virus(es) in these isolates are restricted to insect host(s). Further inoculations using a wider range of cells will be required to determine the full range of susceptible hosts.
One of the disadvantages of testing insects in pools of is that it does not provide the true frequency of infection. For example, if 50 or 100 insects are assayed in a single pool, one does not know if a single mosquito is infected or if multiple insects are. Likewise, if two or more viruses are isolated, one is uncertain if they came from a single mosquito or from several different infected insects. The latter information is important in understanding the interaction of viruses in the mosquito host and their potential effects on the insect’s immune system as well as its vector competence, longevity and fecundity. Ideally, one would like to test individual mosquitoes; but at present, the cost of deep sequencing is still prohibitive and testing mosquitoes in pools is the only feasible option.
Highlights.
We genetically characterized viral isolates derived from different mosquito species in insect cells and identified novel species of reoviruses and tetravirus.
The known geographic range and mosquito species tropism of some mosquito viruses was expanded.
Understanding virus diversity in mosquitoes vector may be important for control strategies.
Acknowledgments
Blood Systems Research Institute for support to MS, TGP, and ED. The authors are indebted to the following individuals and institutions for providing the viruses or mosquito specimens used in this study: James D. Converse (deceased), Ashley Wilson, Fanny Castro, Katherine Young, Kathryn Hanley, David Perera, Durland Fish and the Armed Forces Research Institute of Medical Science (AFRIMS), Bankok, Thailand.
Work at UTMB was supported by NIH contract HHSN272701000040I/HHSN2720004/D04, NIH grant R24 AI 120942 and a pilot grant by the Institute for Human Infections and Immunity.
Abbreviations
- BCPOV
Big Cypress orbivirus
- NRUV
Ninarumi virus
- HISLV
High Island virus
- SWKV
Sarawak virus
- YOUV
Yunnan orbivirus
- BAV
Banna virus
- SVIV
Sathuvachari virus
- AHSV
African horse sickness virus
- CGLV
Changuinola virus
- BTV
Bluetongue virus
- TILV
Tilligerry virus
- LEBV
Lebombo virus
- OpbuRV
Operophtera brumata reovirus
- YNV
Yunnan virus
- MPO
Middle Point orbivirus
- BBaV
Bontang Baru virus
- CTFV
Culex theileri flavivirus
Footnotes
Competing interests
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguiar ER, Olmo RP, Paro S, Ferreira FV, de Faria IJ, Todjro YM, Lobo FP, Kroon EG, Meignin C, Gatherer D, Imler JL, Marques JT. Sequence-independent characterization of viruses based on the pattern of viral small RNAs produced by the host. Nucleic acids research. 2015;43(13):6191–6206. doi: 10.1093/nar/gkv587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attoui H, Mertens PPC, Becnel J, Belaganahalli S, Bergoin M, Brussaard CP, Chappell JD, Ciarlet M, del Vas M, TSD . Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. x. Academic Press; London; Waltham, MA: 2012. p. 1327. [Google Scholar]
- Attoui H, Billoir F, Biagini P, de Micco P, de Lamballerie X. Complete sequence determination and genetic analysis of Banna virus and Kadipiro virus: proposal for assignment to a new genus (Seadornavirus) within the family Reoviridae. The Journal of general virology. 2000;81(Pt 6):1507–1515. doi: 10.1099/0022-1317-81-6-1507. [DOI] [PubMed] [Google Scholar]
- Attoui H, Mendez-Lopez MR, Rao S, Hurtado-Alendes A, Lizaraso-Caparo F, Jaafar FM, Samuel AR, Belhouchet M, Pritchard LI, Melville L, Weir RP, Hyatt AD, Davis SS, Lunt R, Calisher CH, Tesh RB, Fujita R, Mertens PP. Peruvian horse sickness virus and Yunnan orbivirus, isolated from vertebrates and mosquitoes in Peru and Australia. Virology. 2009;394(2):298–310. doi: 10.1016/j.virol.2009.08.032. [DOI] [PubMed] [Google Scholar]
- Attoui H, Mohd Jaafar F. Zoonotic and emerging orbivirus infections. Rev Sci Tech. 2015;34(2):353–361. doi: 10.20506/rst.34.2.2362. [DOI] [PubMed] [Google Scholar]
- Attoui H, Mohd Jaafar F, Belhouchet M, Aldrovandi N, Tao S, Chen B, Liang G, Tesh RB, de Micco P, de Lamballerie X. Yunnan orbivirus, a new orbivirus species isolated from Culex tritaeniorhynchus mosquitoes in China. The Journal of general virology. 2005;86(Pt 12):3409–3417. doi: 10.1099/vir.0.81258-0. [DOI] [PubMed] [Google Scholar]
- Auguste AJ, Carrington CV, Forrester NL, Popov VL, Guzman H, Widen SG, Wood TG, Weaver SC, Tesh RB. Characterization of a novel Negevirus and a novel Bunyavirus isolated from Culex (Culex) declarator mosquitoes in Trinidad. The Journal of general virology. 2014;95(Pt 2):481–485. doi: 10.1099/vir.0.058412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhouchet M, Mohd Jaafar F, Firth AE, Grimes JM, Mertens PP, Attoui H. Detection of a fourth orbivirus non-structural protein. PloS one. 2011;6(10):e25697. doi: 10.1371/journal.pone.0025697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolling BG, Weaver SC, Tesh RB, Vasilakis N. Insect-Specific Virus Discovery: Significance for the Arbovirus Community. Viruses. 2015;7(9):4911–4928. doi: 10.3390/v7092851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carissimo G, Eiglmeier K, Reveillaud J, Holm I, Diallo M, Diallo D, Vantaux A, Kim S, Menard D, Siv S, Belda E, Bischoff E, Antoniewski C, Vernick KD. Identification and Characterization of Two Novel RNA Viruses from Anopheles gambiae Species Complex Mosquitoes. PloS one. 2016;11(5):e0153881. doi: 10.1371/journal.pone.0153881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JA, Liu RM, Bennett SN. RNA shotgun metagenomic sequencing of northern California (USA) mosquitoes uncovers viruses, bacteria, and fungi. Frontiers in microbiology. 2015;6:185. doi: 10.3389/fmicb.2015.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles J, Firth AE, Lorono-Pino MA, Garcia-Rejon JE, Farfan-Ale JA, Lipkin WI, Blitvich BJ, Briese T. Merida virus, a putative novel rhabdovirus discovered in Culex and Ochlerotatus spp. mosquitoes in the Yucatan Peninsula of Mexico. The Journal of general virology. 2016;97(4):977–987. doi: 10.1099/jgv.0.000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholleti H, Hayer J, Abilio AP, Mulandane FC, Verner-Carlsson J, Falk KI, Fafetine JM, Berg M, Blomstrom AL. Discovery of Novel Viruses in Mosquitoes from the Zambezi Valley of Mozambique. PloS one. 2016;11(9):e0162751. doi: 10.1371/journal.pone.0162751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey LL, Page BL, Greninger AL, Herring BL, Russell RC, Doggett SL, Haniotis J, Wang C, Deng X, Delwart EL. Enhanced arbovirus surveillance with deep sequencing: Identification of novel rhabdoviruses and bunyaviruses in Australian mosquitoes. Virology. 2014;448:146–158. doi: 10.1016/j.virol.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Gutierrez MA, Nunes MR, Guzman H, Uribe S, Gallego Gomez JC, Suaza Vasco JD, Cardoso JF, Popov VL, Widen SG, Wood TG, Vasilakis N, Tesh RB. Sinu virus, a novel and divergent orthomyxovirus related to members of the genus Thogotovirus isolated from mosquitoes in Colombia. Virology. 2017;501:166–175. doi: 10.1016/j.virol.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, Delwart EL. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic acids research. 2015;43(7):e46. doi: 10.1093/nar/gkv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet BS, van Rijn P, Howerth EW, Beer M, Mertens PP. A Review of Knowledge Gaps and Tools for Orbivirus Research. Vector Borne Zoonotic Dis. 2015;15(6):339–347. doi: 10.1089/vbz.2014.1701. [DOI] [PubMed] [Google Scholar]
- Ergunay K, Litzba N, Brinkmann A, Gunay F, Kar S, Oter K, Orsten S, Sarikaya Y, Alten B, Nitsche A, Linton YM. Isolation and genomic characterization of Culex theileri flaviviruses in field-collected mosquitoes from Turkey. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2016;46:138–147. doi: 10.1016/j.meegid.2016.11.008. [DOI] [PubMed] [Google Scholar]
- Fauver JR, Grubaugh ND, Krajacich BJ, Weger-Lucarelli J, Lakin SM, Fakoli LS, 3rd, Bolay FK, Diclaro JW, 2nd, Dabire KR, Foy BD, Brackney DE, Ebel GD, Stenglein MD. West African Anopheles gambiae mosquitoes harbor a taxonomically diverse virome including new insect-specific flaviviruses, mononegaviruses, and totiviruses. Virology. 2016;498:288–299. doi: 10.1016/j.virol.2016.07.031. [DOI] [PubMed] [Google Scholar]
- Frey KG, Biser T, Hamilton T, Santos CJ, Pimentel G, Mokashi VP, Bishop-Lilly KA. Bioinformatic Characterization of Mosquito Viromes within the Eastern United States and Puerto Rico: Discovery of Novel Viruses. Evolutionary bioinformatics online. 2016;12(Suppl 2):1–12. doi: 10.4137/EBO.S38518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RI, Rao S, Possee RD, Sait SM, Mertens PP, Hails RS. Detection and characterisation of three novel species of reovirus (Reoviridae), isolated from geographically separate populations of the winter moth Operophtera brumata (Lepidoptera: Geometridae) on Orkney. Journal of invertebrate pathology. 2006;91(2):79–87. doi: 10.1016/j.jip.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Graham RI, Rao S, Sait SM, Attoui H, Mertens PP, Hails RS, Possee RD. Sequence analysis of a reovirus isolated from the winter moth Operophtera brumata (Lepidoptera: Geometridae) and its parasitoid wasp Phobocampe tempestiva (Hymenoptera: Ichneumonidae) Virus research. 2008;135(1):42–47. doi: 10.1016/j.virusres.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RI, Rao S, Sait SM, Mertens PP, Hails RS, Possee RD. Characterisation and partial sequence analysis of two novel cypoviruses isolated from the winter moth Operophtera brumata (Lepidoptera: Geometridae) Virus genes. 2007;35(2):463–471. doi: 10.1007/s11262-007-0113-0. [DOI] [PubMed] [Google Scholar]
- Harrison JJ, Warrilow D, McLean BJ, Watterson D, O’Brien CA, Colmant AM, Johansen CA, Barnard RT, Hall-Mendelin S, Davis SS, Hall RA, Hobson-Peters J. A New Orbivirus Isolated from Mosquitoes in North-Western Australia Shows Antigenic and Genetic Similarity to Corriparta Virus but Does Not Replicate in Vertebrate Cells. Viruses. 2016;8(5) doi: 10.3390/v8050141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiwaji M, Short JR, Dorrington RA. Expanding the host range of small insect RNA viruses: Providence virus (Carmotetraviridae) infects and replicates in a human tissue culture cell line. The Journal of general virology. 2016;97(10):2763–2768. doi: 10.1099/jgv.0.000578. [DOI] [PubMed] [Google Scholar]
- Kapoor A, Tesh RB, Duraisamy R, Popov VL, Travassos da Rosa AP, Lipkin WI. A novel mosquito-borne Orbivirus species found in South-east Asia. The Journal of general virology. 2013;94(Pt 5):1051–1057. doi: 10.1099/vir.0.046748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C, Ziebuhr J, Junglen S, Drosten C, Zirkel F, Nga PT, Morita K, Snijder EJ, Gorbalenya AE. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Archives of virology. 2012;157(8):1623–1628. doi: 10.1007/s00705-012-1295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CX, Shi M, Tian JH, Lin XD, Kang YJ, Chen LJ, Qin XC, Xu J, Holmes EC, Zhang YZ. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife. 2015a:4. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015b;31(10):1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Li L, Giannitti F, Low J, Keyes C, Ullmann LS, Deng X, Aleman M, Pesavento PA, Pusterla N, Delwart E. Exploring the virome of diseased horses. The Journal of general virology. 2015c;96(9):2721–2733. doi: 10.1099/vir.0.000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li MH, Zhai YG, Meng WS, Sun XH, Cao YX, Fu SH, Wang HY, Xu LH, Tang Q, Liang GD. Banna virus, China, 1987–2007. Emerging infectious diseases. 2010;16(3):514–517. doi: 10.3201/eid1603.091160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Ferber M, Veyrunes JC, Croizier L. Drosophila S virus is a member of the Reoviridae family. Journal of virology. 1989;63(2):1007–1009. doi: 10.1128/jvi.63.2.1007-1009.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasar F, Palacios G, Gorchakov RV, Guzman H, Da Rosa AP, Savji N, Popov VL, Sherman MB, Lipkin WI, Tesh RB, Weaver SC. Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(36):14622–14627. doi: 10.1073/pnas.1204787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TF, Willner DL, Lim YW, Schmieder R, Chau B, Nilsson C, Anthony S, Ruan Y, Rohwer F, Breitbart M. Broad surveys of DNA viral diversity obtained through viral metagenomics of mosquitoes. PloS one. 2011;6(6):e20579. doi: 10.1371/journal.pone.0020579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nga PT, del Parquet MC, Lauber C, Parida M, Nabeshima T, Yu F, Thuy NT, Inoue S, Ito T, Okamoto K, Ichinose A, Snijder EJ, Morita K, Gorbalenya AE. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS pathogens. 2011;7(9):e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G, Wiley MR, Travassos da Rosa AP, Guzman H, Quiroz E, Savji N, Carrera JP, Bussetti AV, Ladner JT, Lipkin WI, Tesh RB. Characterization of the Punta Toro species complex (genus Phlebovirus, family Bunyaviridae) The Journal of general virology. 2015;96(8):2079–2085. doi: 10.1099/vir.0.000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauvolid-Correa A, Solberg O, Couto-Lima D, Nogueira RM, Langevin S, Komar N. Novel Viruses Isolated from Mosquitoes in Pantanal, Brazil. Genome announcements. 2016;4(6) doi: 10.1128/genomeA.01195-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle FM, Johnson KN, Goodman CL, McIntosh AH, Ball LA. Providence virus: a new member of the Tetraviridae that infects cultured insect cells. Virology. 2003;306(2):359–370. doi: 10.1016/s0042-6822(02)00052-1. [DOI] [PubMed] [Google Scholar]
- Sadeghi M, Kapusinszky B, Yugo DM, Phan TG, Deng X, Kanevsky I, Opriessnig T, Woolums AR, Hurley DJ, Meng XJ, Delwart E. Virome of US bovine calf serum. Biologicals: journal of the International Association of Biological Standardization. 2017;46:64–67. doi: 10.1016/j.biologicals.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Liu Y, Hu X, Xiong J, Zhang B, Yuan Z. A metagenomic survey of viral abundance and diversity in mosquitoes from Hubei province. PloS one. 2015;10(6):e0129845. doi: 10.1371/journal.pone.0129845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P, Adams MJ, Benko M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B, Hull R, King AM, Koonin EV, Krupovic M, Kuhn JH, Lefkowitz EJ, Nibert ML, Orton R, Roossinck MJ, Sabanadzovic S, Sullivan MB, Suttle CA, Tesh RB, van der Vlugt RA, Varsani A, Zerbini FM. Consensus statement: Virus taxonomy in the age of metagenomics. Nature reviews Microbiology. 2017;15(3):161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- Thuy NT, Huy TQ, Nga PT, Morita K, Dunia I, Benedetti L. A new nidovirus (NamDinh virus NDiV): Its ultrastructural characterization in the C6/36 mosquito cell line. Virology. 2013;444(1–2):337–342. doi: 10.1016/j.virol.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Castro-Llanos F, Widen SG, Aguilar PV, Guzman H, Guevara C, Fernandez R, Auguste AJ, Wood TG, Popov V, Mundal K, Ghedin E, Kochel TJ, Holmes EC, Walker PJ, Tesh RB. Arboretum and Puerto Almendras viruses: two novel rhabdoviruses isolated from mosquitoes in Peru. The Journal of general virology. 2014a;95(Pt 4):787–792. doi: 10.1099/vir.0.058685-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Guzman H, Firth C, Forrester NL, Widen SG, Wood TG, Rossi SL, Ghedin E, Popov V, Blasdell KR, Walker PJ, Tesh RB. Mesoniviruses are mosquito-specific viruses with extensive geographic distribution and host range. Virology journal. 2014b;11:97. doi: 10.1186/1743-422X-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li H, He Y, Zhou Y, Meng J, Zhu W, Chen H, Liao D, Man Y. Isolation and Genetic Characterization of Mangshi Virus: A Newly Discovered Seadornavirus of the Reoviridae Family Found in Yunnan Province, China. PloS one. 2015;10(12):e0143601. doi: 10.1371/journal.pone.0143601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster CL, Waldron FM, Robertson S, Crowson D, Ferrari G, Quintana JF, Brouqui JM, Bayne EH, Longdon B, Buck AH, Lazzaro BP, Akorli J, Haddrill PR, Obbard DJ. The Discovery, Distribution, and Evolution of Viruses Associated with Drosophila melanogaster. PLoS biology. 2015;13(7):e1002210. doi: 10.1371/journal.pbio.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li L, Deng X, Blumel J, Nubling CM, Hunfeld A, Baylis SA, Delwart E. Viral nucleic acids in human plasma pools. Transfusion. 2016;56(9):2248–2255. doi: 10.1111/trf.13692. [DOI] [PubMed] [Google Scholar]
- Zirkel F, Kurth A, Quan PL, Briese T, Ellerbrok H, Pauli G, Leendertz FH, Lipkin WI, Ziebuhr J, Drosten C, Junglen S. An insect nidovirus emerging from a primary tropical rainforest. mBio. 2011;2(3):e00077–00011. doi: 10.1128/mBio.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]