SUMMARY
In the Drosophila immune response, bacterial derived diaminopimelic acid-type peptidoglycan binds the receptors PGRP-LC and PGRP-LE, which through interaction with the adaptor protein Imd leads to activation of the NF-κB homolog Relish and robust antimicrobial peptide gene expression. PGRP-LC, PGRP-LE, and Imd each contain a motif with some resemblance to the RIP Homotypic Interaction Motif (RHIM), a domain found in mammalian RIPK proteins forming functional amyloids during necroptosis. Here we found that despite sequence divergence, these Drosophila cryptic RHIMs formed amyloid fibrils in vitro and in cells. Amyloid formation was required for signaling downstream of Imd, and in contrast to the mammalian RHIMs, was not associated with cell death. Furthermore, amyloid formation constituted a regulatable step and could be inhibited by Pirk, an endogenous feedback regulator of this pathway. Thus, diverse sequence motifs are capable of forming amyloidal signaling platforms, and the formation of these platforms may present a regulatory point in multiple biological processes.
In Brief

Kleino et al. show that amyloid formation is required for activation of the Drosophila Imd pathway upon recognition of bacterial peptidoglycans. Amyloid formation involves a motif resembling one found in necroptosis-associated mammalian proteins and can be negatively regulated, suggesting that amyloidal signaling platforms may present a regulatory point in multiple biological processes.
INTRODUCTION
The Drosophila humoral innate immune response relies on two NF-κB pathways, the Toll and the Imd pathways, which induce the expression of a battery of potent antimicrobial peptides (AMPs) in response to peptidoglycan (PGN) from bacterial cell wall (Lemaitre and Hoffmann, 2007). PGN is directly sensed by peptidoglycan recognition proteins (PGRPs) (Dziarski and Gupta, 2006). Lys-type peptidoglycan from Gram-positive bacteria is recognized by PGRP-SA, which activates the Toll pathway (Park et al., 2007), while diaminopimelic acid-containing (DAP-type) PGN from Gram-negative bacteria and Gram-positive bacilli specifically trigger the Imd pathway via receptors PGRP-LC and PGRP-LE (Kleino and Silverman, 2014). PGRP-LC is a transmembrane receptor that localizes to the plasma membrane, while PGRP-LE is cytosolic and recognizes DAP-type PGN that is either transported across the plasma membrane or released by cytosolic bacteria (Kaneko et al., 2005, 2006; Paik et al., 2017; Yano et al., 2008).
Imd signaling proceeds from the receptors PGRP-LC and PGRP-LE to the proximal adaptor protein Imd (Lemaitre et al., 1995), which in turn recruits Fadd (Fas-associated death domain) through a homotypic death domain (DD) interface, while Fadd recruits the Caspase-8 homolog Dredd (Death related ced-3/Nedd2-like caspase) via homotypic death effector domain (DED) interaction (Hu and Yang, 2000; Leulier et al., 2000, 2002). Dredd then further transduces the signal by cleaving both Imd and the NF-κB precursor Relish (Kim et al., 2014; Paquette et al., 2010). Imd cleavage at D30 reveals a docking site for the ubiquitin E3 ligase Death-associated inhibitor of apoptosis 2 (Diap2), which together with the E2 enzymes Uev1a, Bendless (Ubc13), and Effete (Ubc5), conjugates the cleaved, active Imd31-273 and other pathway components with K63-polyubiquitin chains (Chen et al., 2017; Meinander et al., 2012; Paquette et al., 2010). Most likely, these ubiquitin chains then recruit Tab2-Tak1 (TAK1-associated binding protein 2, and TGF-β activated kinase 1, respectively) and IKK (IκB kinase) complexes, leading to phosphorylation and activation of the Drosophila IKK complex and the NF-κB transcription factor Relish. Full activation of Relish requires both IKK phosphorylation and Dredd-mediated cleavage (Ertüurk-Hasdemir et al., 2009; Leulier et al., 2000; Silverman et al., 2000; Stöven et al., 2000, 2003).
The PGRP receptors and the adaptor protein Imd do not have orthologs in the mammalian NF-κB signaling. However, they do harbor homologous domains, in particular DDs, which are also present in mammalian proteins such as Receptor Interacting Serine/Threonine Kinase 1 (RIPK1) (Georgel et al., 2001) and FADD (Naitza et al., 2002). Previously, through deletion and site-directed mutagenesis, we identified a conserved sequence motif in the long N-terminal signaling domains of PGRP-LC and PGRP-LE that is required to trigger Imd signaling (Kaneko et al., 2006). The four-residue VxxG core sequence appeared to share similarity to the RIP homotypic interaction motifs (RHIMs), which was first described as a short motif of IQIG or VQVG mediating the interaction of mammalian RIPK1 and RIPK3 proteins (Sun et al., 2002), and is therefore denoted here as cryptic RHIM (cRHIM). RHIMs in RIPK1 and RIPK3 have been shown to form a functional amyloid complex to mediate kinase activation and necroptosis induction (Li et al., 2012) and have been identified in other necroptosis signaling proteins including TRIF (TIR-domain-containing adaptor-inducing interferon-β) (Kaiser and Offermann, 2005) and DAI (DNA-dependent activator of IFN-regulatory factors) (Kaiser et al., 2008; Rebsamen et al., 2009), as well as in viral proteins interfering with necroptosis, such as herpes simplex virus ICP6, ICP10, and murine cytomegalovirus (MCMV) M45 (Guo et al., 2015; Huang et al., 2015; Kaiser et al., 2008; Wang et al., 2014).
cRHIMs are also present in other proteins involved in the Drosophila Imd signaling pathway, including PGRP-LA, Imd, and Pirk, which is a negative regulator of this pathway (Gendrin et al., 2013; Kajava et al., 2014; Kaneko et al., 2006). However, cRHIMs are distinct from the mammalian RHIMs; only the Gly residue in the 4th position is absolutely conserved, and the amyloidogenic Gln of RHIMs is absent in cRHIMs. Therefore, although intact cRHIM domains are required for Imd signaling (Kaneko et al., 2006), the molecular mechanisms underlying this requirement are unclear.
Here we found that the cRHIMs of PGRP-LC, PGRP-LE, and Imd formed amyloid fibrils in vitro and in cells, despite the lack of the amyloidogenic Gln, as evidenced by amyloid dye binding assays, electron microscopy (EM), and X-ray diffraction. Amyloid formation and subsequent Imd signaling were specifically inhibited in cultured cells and in flies by the amyloid-binding dye Thioflavin T (ThT), which binds and interferes with amyloid fibril polymerization. Moreover, we found that Pirk inhibited Imd amyloid fibril polymerization in vitro and in cells, revealing Pirk as an endogenous inhibitor of RHIM-mediated amyloid formation in animals, providing insight into the molecular function of this inhibitor of the Imd pathway. Collectively, our results demonstrate that amyloid assembly is an evolutionarily conserved signaling mechanism that functions in innate immune NF-κB signaling, as well as in necroptosis, and further suggest that amyloid assembly may be a common regulatory mechanism in signaling pathways, mediated by diverse sequences from RHIMs to cRHIMs and beyond.
RESULTS
Amyloidal Activity of the cRHIMs of PGRP-LC, PGRP-LE, and Imd
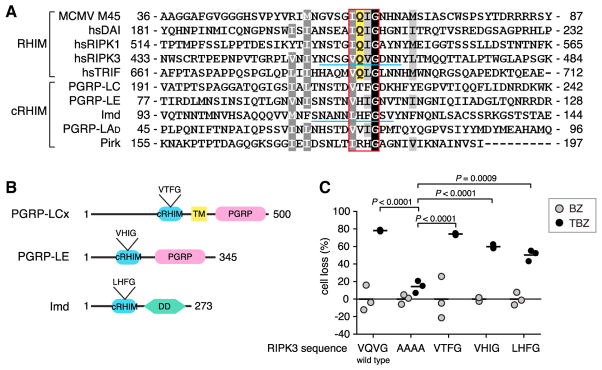
Necroptosis is an inflammatory form of cell death that is mediated by RIPK3 and mixed lineage kinase domain-like (MLKL) in the absence of Caspase-8 activation. Tumor necrosis factor (TNF)-induced RIPK3/MLKL activation also requires RIPK1, the upstream activator of RIPK3. In mammals, the RHIM motifs of RIPK1 and RIPK3 are required for necroptosis signaling and to form functional amyloid fibrils (Cho et al., 2009; Li et al., 2012). The mammalian RHIMs share a well-conserved tetrapeptide core motif marked by an invariant Gln in the P2 position (Figure 1A). Gln residues are also critical for amyloid fibril formation by the yeast prion protein Sup35 and by some pathologic amyloids such as Tau (Sawaya et al., 2007). This is in contrast to the insect cRHIMs, which display variability in the P2 as well as the P3 positions (Figures 1A and S1). This sequence divergence raised the question of whether the Drosophila cRHIMs can form amyloid, as seen with mammalian RHIMs. To test this, we generated chimeric RIPK3 constructs in which the core RHIM sequence VQVG was replaced with the corresponding sequence from PGRP-LC, PGRP-LE, or Imd (Figures 1A and 1B). HeLa cells lack endogenous RIPK3 expression and are normally refractory to necroptosis (Li et al., 2012). When HeLa cells were transfected with wild-type RIPK3, necroptosis induced by TNF, the SMAC mimetic BV6, and the caspase inhibitor zVAD-fmk (TBZ) was restored (Figure 1C). Expression of a non-amyloidal RIPK3 mutant in which the VQVG core motif was replaced with AAAA significantly impaired TBZ-induced necroptosis, as expected. Strikingly, chimeras with the VTFG motif from PGRP-LC completely restored TBZ-induced cell death. In addition, chimeras with VHIG and LHFG, core motifs from PGRP-LE and Imd, respectively, also showed significant restoration of cell death. These results indicate that despite the sequence variability and the lack of a Gln at P2, the Drosophila cRHIMs can functionally substitute for the RHIM motif in RIPK3. These results imply that the Drosophila cRHIMs can also mediate oligomeric complex formation that is required for necroptosis signaling.
Figure 1. Conserved Function of Cryptic RHIM Motifs in the DrosophilaImd Pathway.
(A) cRHIMs of D. melanogaster proteins aligned with human and MCMV RHIM-sequences. The four core amino acids are boxed with red. The amyloidogenic Q that is highly conserved in mammalian sequences but missing in Drosophila cRHIM is highlighted with yellow. Shading represents conservation according to Blosum 62 scoring matrix. Region underlined with blue represents the sequence in RIPK3 that was replaced with the underlined sequence from Imd in Figure 6H.
(B) Schematic illustration of PGRP-LCx, PGRP-LE, and Imd domain composition. Abbreviations are as follows: TM, transmembrane domain; PGRP, PGRP domain; DD, death domain.
(C) Effects of RIPK3 substitution mutants on TNF-induced necroptosis in HeLa cells. Cells were transfected with wild-type or chimeric RIPK3 where the VQVG core motif was substituted with AAAA, VTFG (PGRP-LC), VHIG (PGRP-LE), or LHFG (Imd), and necroptosis activated with combined TNF (T), BV6 (B), and zVAD-fmk (Z) treatment. The graph shows individual data points (N = 3) with mean indicated as a line. BZ indicates no TNF stimulation (gray dots); TBZ indicates BV6 and zVAD-fmk-treated cells stimulated with TNFα (black dots).
See also alignment of selected insect cRHIMs in Figure S1.
cRHIMs of PGRP-LC, PGRP-LE, and Imd Form Amyloidal Aggregates In Vitro
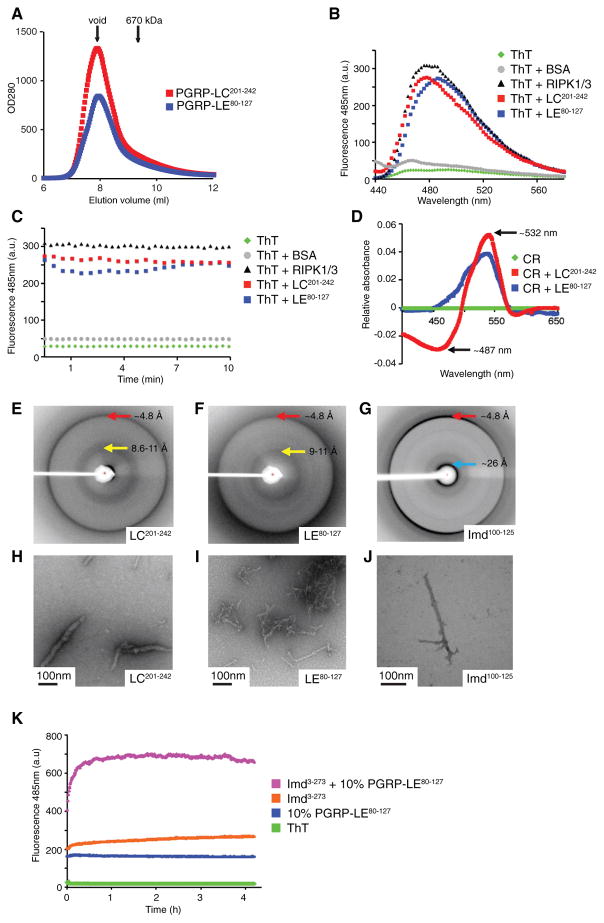
Because RHIMs mediate oligomerization by amyloid formation (Cho et al., 2009; Li et al., 2012), we asked whether the cRHIMs, which lack amyloidogenic residues, also formed cross β-amyloids. To investigate the PGRP-LC and PGRP-LE cRHIMs, we expressed and purified PGRP-LC201-242 and PGRP-LE80-127 as 6xHis-Sumo fusion proteins from Escherichia coli and studied their ability to aggregate in vitro. As expected for amyloids, PGRP-LC and PGRP-LE fusion proteins containing the cRHIM motifs formed large aggregates and fractioned in the void during gel filtration (Figure 2A).
Figure 2. cRHIM Motifs of PGRP-LC, PGRP-LE, and Imd Form Amyloidal AggregatesIn Vitro.
(A) Superimposed gel filtration profiles of PGRP-LC and PGRP-LE cRHIM motifs. The Sumo-tagged fusion proteins eluted in void fractions.
(B–D) PGRP-LC and PGRP-LE cRHIMs bind am-yloid binding dyes Thioflavin T (ThT) and Congo Red (CR).
(B and C) Fluorescent emission spectra of ThT in the absence (green, no protein; gray, BSA) and presence (black, RIPK1/RIPK3; red, PGRP-LC201-242; blue, PGRP-LE80-127) of RHIM proteins.
(D) Absorption difference spectra of CR in the presence of PGRP-LC201-242 (red) or PGRP-LE80-127 (blue) compared to CR alone (green).
(E–J) PGRP-LC, PGRP-LE, and Imd filaments are classical amyloid fibrils. Scale bar: 100 nm.
(E–G) X-ray diffraction images of PGRP-LC (E), PGRP-LE (F), and Imd (G) cRHIM regions. The resolutions of dominant diffractions are labeled.
(H–J) Negative staining EM images of PGRP-LC (H), PGRP-LE (I), and Imd (J) cRHIM regions.
(K) ThT fluorescent emission spectra of Imd3-273 in the presence and absence of preformed PGRP-LE80-127 fibrils.
Amyloids bind aromatic, cross β-specific dyes such as Thioflavin T (ThT) and Congo Red (CR), changing their spectral properties (Klunk et al., 1989; LeVine, 1999). To test whether PGRP-LC and PGRP-LE cRHIM aggregates were classical amyloids, we added ThT to either PGRP-LC201-242 or PGRP-LE80-127 and measured fluorescence after excitation at 430 nm. Like RIPK1-RIPK3 RHIM amyloids used as a positive control, ThT fluorescence spectra displayed an emission peak around 485 nm upon binding with PGRP-LC and PGRP-LE cRHIMs (Figures 2B and 2C). Similarly, CR showed a characteristic red shift from an absorption maximum of ~487 nm to ~532 nm after incubation with either PGRP-LC or PGRP-LE cRHIMs (Figure 2D).
Amyloids form cross-β quaternary structures, which produce characteristic X-ray diffraction patterns (Sunde et al., 1997). When cRHIMs of PGRP-LC and PGRP-LE were analyzed by X-ray diffraction, dominant diffractions at around 4.8 Å were observed, corresponding to the inter-β strand spacing within a β sheet along the length of a fibril that is present in all β-amyloids (Figures 2E and 2F). Similar analysis of the cRHIM from the adaptor protein Imd, Imd100-125, also showed diffraction at the characteristic ~4.8 Å inter-β strand spacing (Figure 2G), suggesting that the cRHIMs of PGRP-LC, PGRP-LE, and Imd all formed cross-β amyloid structures. Diffractions that most likely represent inter-β sheet packing distances in equatorial directions perpendicular to the fiber axis were broader for the three cRHIM samples, at ~8.6–11 Å and ~9–11 Å for PGRP-LC and PGRP-LE, respectively (Figures 2E and 2F), and at ~11–26 Å with an intense ~26 Å reflection for Imd (Figure 2G). These equatorial reflections are known to be more sensitive to amyloid sequences and to protofilament structures (Sunde et al., 1997). When visualizing the aggregated cRHIMs of PGRP-LC, PGRP-LE, and Imd, we found that all the proteins showed filamentous morphology on negative staining EM, consistent with the nature of amyloids (Figures 2H–2J).
PGRP-LC and PGRP-LE are PGN-sensing receptors while Imd is a critical intracellular signaling molecule in the Imd pathway, all of which formed amyloid fibrils in vitro. Thus, we hypothesized that amyloids formed by the PGRP receptors may nucleate the formation of Imd cRHIM fibrils. To test this idea, Imd-cRHIM amyloid transformation was monitored by ThT fluorescence, in the absence or presence of sub-stoichiometric amounts of preformed PGRP-LE cRHIM amyloids (Figure 2K). In this assay we studied a longer version of Imd, Imd3-273, which was easier to solubilize than the highly aggregation-prone Imd100-125 studied above (Figures 2G and 2J). When measured separately, Imd3-273 and the preformed PGRP-LE80-127 amyloids both showed modest ThT fluorescence, similar to that in Figure 2C. However, combining the preformed PGRP-LE80-127 cRHIM amyloids with Imd3-273 further increased ThT fluorescence compared to the Imd alone sample. This experiment indicates that the receptors of the Imd pathway form an amyloidal nucleus that enhances the formation of Imd fibrils. Altogether, these in vitro data suggest that not only do the cRHIMs of PGRP-LC, PGRP-LE, and Imd form β-amyloid, but also that the receptor amyloid aggregates can nucleate enhanced Imd-cRHIM amyloid fibril formation.
Mutations Disrupt the Formation of cRHIM Filaments In Vitro
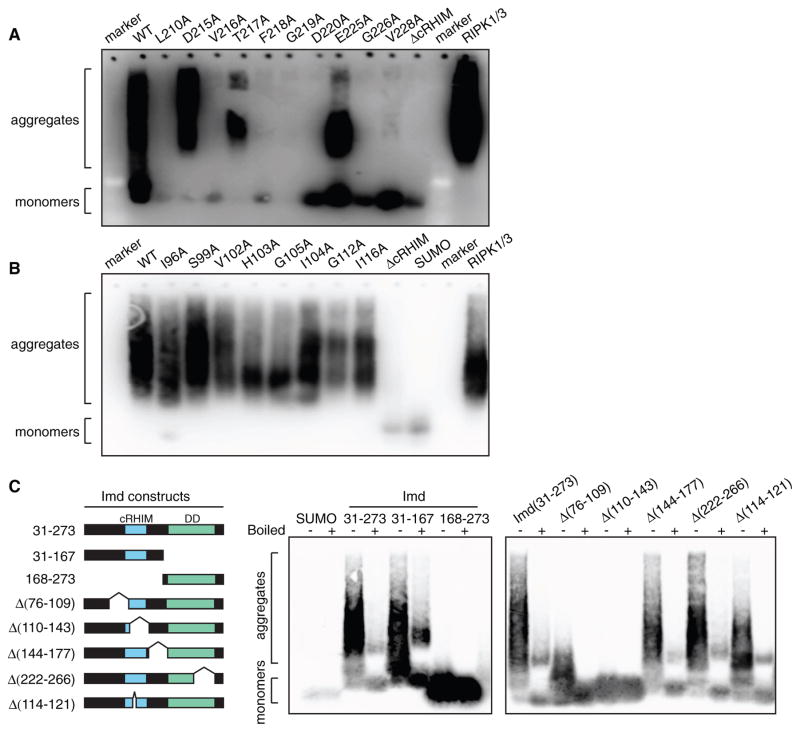
To begin to link PGRP-LC and PGRP-LE cRHIM fibril formation with the biological function of these receptors, we first characterized deletion mutants lacking the cRHIM. We previously reported that cRHIM-deficient PGRP-LC or PGRP-LE fail to support Imd signaling (Kaneko et al., 2006). Wild-type and mutant proteins were analyzed by semi-denaturing detergent agarose gel electrophoresis (SDD-AGE), a technique that identifies detergent-resistant aggregates, such as amyloid fibrils (Halfmann and Lindquist, 2008). In this assay, RIPK1/3 amyloid fibrils were used as a positive control throughout. Wild-type PGRP-LC201-242 and PGRP-LE80-127 both formed large aggregates, while the cRHIM deletion mutants, lacking residues 209–225 or 98–113, respectively, were monomeric (Figures 3A and 3B). These data further support the amyloidal structure of these cRHIMs indicated by the biophysical analysis in Figure 2.
Figure 3. Mutations within the cRHIM Motif Disrupt the Formation of Amyloidal Aggregates.
(A and B) SDD-AGE profiles of wild-type (WT), deletion mutant, and alanine substitution mutants of PGPR-LC201-242 (A) and PGRP-LE80-127 (B). ΔcRHIM represents deletion of amino acids 209–225 in PGRP-LC201-242 and of amino acids 98–113 in PGRP- LE80-127. RIPK1/3-RHIM complex was loaded as a positive control.
(C) Schematic representation of recombinant IMD31-273, deletion mutants, and their SDD-AGE profiles.
In addition to these cRHIM deletion mutants, we used SDD-AGE to analyze a series of alanine substitution mutants we previously characterized in both the PGRP-LC and PGRP-LE cRHIM regions (Kaneko et al., 2006). In particular, substitutions of core cRHIM residues (VTFG) in PGRP-LC caused a dramatic loss of signal transduction, as measured by AMP gene induction (Kaneko et al., 2006). Remarkably, single alanine substitutions on core residues caused a marked reduction in aggregation of PGRP-LC201-242, as monitored by SDD-AGE (Figure 3A). In PGRP-LE, the core cRHIM residues (VHIG) were similarly critical for signal transduction and aggregation, with the exception of I104, which had a partial effect in both signaling and aggregation assays (Figure 3B; Kaneko et al., 2006). Substitution of residues flanking the core cRHIM showed similar correlations between activity and aggregation. L210A, G226A, and V228A mutants of PGRP-LC as well as G112A of PGRP-LE, which are known to prevent signal transduction (Kaneko et al., 2006), failed to aggregate in SDD-AGE, while D215A or E225A of PGRP-LC as well as I96A, S99A, or I116A of PGRP-LE could be mutated with little effect on aggregation or signaling (Figures 3A and 3B; Kaneko et al., 2006). These data suggest that single residue substitutions of critical amino acids are sufficient to disrupt the aggregation and assembly of PGRP-LC and PGRP-LE cRHIMs into amyloid fibrils as well as subsequent signal transduction (Kaneko et al., 2006). Moreover, our results indicate that flanking sequences around the core tetrapeptide contribute to amyloid formation.
PGRP-LC and PGRP-LE directly interact with the adaptor protein Imd, which also contains a cRHIM, as well as a C-terminal DD. DDs can potentially form large, very stable aggregates (Kagan et al., 2014; Lin et al., 2010). To determine whether Imd could aggregate in a cRHIM-dependent manner in SDD-AGE, as suggested by the studies with Imd100-125, we produced recombinant mature Imd31-273 as well as two truncated proteins: a cRHIM-containing and DD-deficient form (Imd31-167) and a DD-containing and cRHIM-deficient form (Imd168-273). Like Imd31-273, Imd31-167 formed large aggregates on SDD-AGE, while the DD-only Imd168-273 migrated as monomer (Figure 3C). To further confirm the importance of the cRHIM region for Imd aggregation, we also tested smaller deletion mutants of Imd within the 31–273 region. A small deletion of the cRHIM core motif LHFG and the adjacent residues, Δ114–121, resulted in reduced aggregate size but did not totally abolish aggregation. Deletion of neighboring regions, amino acids 76–109, reduced aggregate size, while deletion of a slightly larger region encompassing most of the cRHIM (Δ110–143), totally abolished aggregation. Deletions outside the predicted cRHIM region, Δ144–177 and Δ178–221, had no effect on aggregation (Figure 3C). Altogether, these data indicate that while the cRHIM region in Imd is necessary and sufficient for amyloid formation in vitro, the flanking sequences further facilitate aggregation.
cRHIM-Containing Proteins Form Amyloidal Aggregates in S2* Cells
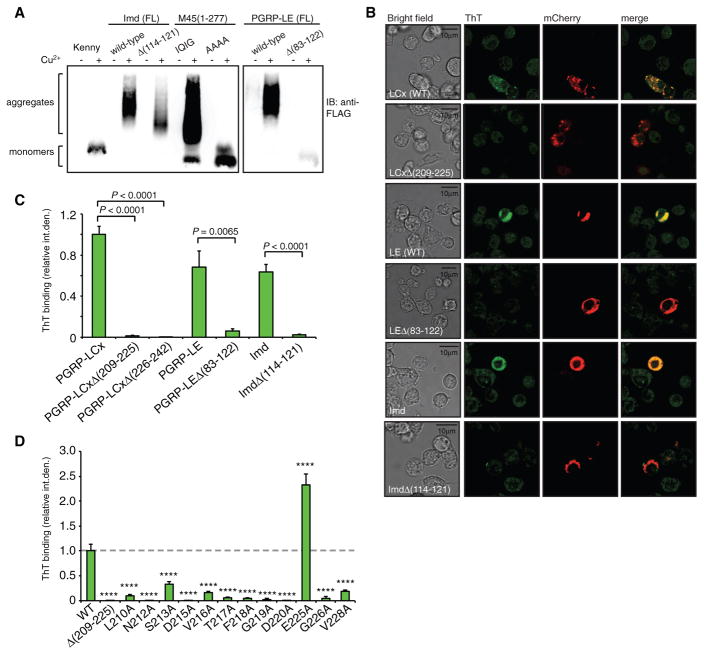
To examine the amyloidal properties of full-length cRHIM-containing proteins in a more physiological environment, we generated constructs expressing FLAG-tagged PGRP-LE and Imd in Drosophila cells. Expression of either of these proteins from the copper-inducible metallothionein promoter is sufficient to induce Imd signaling and AMP gene expression (Choe et al., 2005; Takehana et al., 2002). Wild-type MCMV protein M451-277, which is known to form amyloidal aggregates, was used as a positive control, while a RHIM tetra-alanine substitution and the Imd pathway component IKKγ (Kenny), which lacks a RHIM, were used as negative controls. Drosophila hemocyte-like S2* cells were transfected with either wild-type or RHIM deletion/substitution constructs, and whole cell lysates were analyzed by SDD-AGE. As expected, M45 displayed RHIM-dependent aggregation, as did wild-type full-length PGRP-LE and Imd. Deletion of residues 83–122 in PGRP-LE rendered the protein monomeric, while deletion of residues 114–121 in Imd reduced the size and intensity of aggregates but did not totally abolish aggregation (Figure 4A). This is consistent with the aggregation properties of in vitro produced proteins and protein fragments (Figures 3B and 3C). Note that we were unable to produce detectable quantities of PGRP-LC for similar assays.
Figure 4. PGRP-LC, PGRP-LE, and Imd Form Amyloidal Aggregates in S2* Cells.
(A) SDD-AGE profiles of S2* cell lysates expressing WT or mutant forms of PGRP-LE and Imd. MCMV protein M45(1-277) was used as positive control and Kenny and M45(1-277) IQIG/AAAA mutant as negative controls.
(B) S2* cells were transiently transfected with wild-type mCherry-tagged PGRP-LCx, PGRP-LE, or Imd, or respective cRHIM deletion mutants, and amyloidal protein aggregates were visualized by ThT fluorescence. Scale bar: 10 μm.
(C) Quantification of ThT fluorescence in cells expressing mCherry-tagged PGRP-LCx, PGRP-LE, and Imd, wild-type, and cRHIM deletion mutants. Data shown are mean ± SEM of at least 15 cells pooled from three independent experiments.
(D) Quantification of ThT fluorescence in S2* cells expressing single alanine substitution mutants of mCherry-PGRP-LCx. Columns represent the mean ± SEM of at least 20 cells pooled from three independent experiments. ****p < 0.0001.
To examine amyloid formation in cells, we exploited the fluorescent properties of ThT combined with mCherry-tagged PGRP-LC, PGRP-LE, or Imd. PGRP-LC encodes three isoforms, which all share the same cRHIM-containing cytoplasmic domain; assays were performed with PGRP-LCx as it is involved in the recognition of both polymeric and monomeric PGN (Kleino and Silverman, 2014). S2* cells were transfected with constructs encoding full-length mCherry-tagged wild-type proteins or cRHIM deletion mutants. Fixed cells were then stained with ThT and analyzed by confocal microscopy. The expression levels of wild-type versus cRHIM deletion mutant constructs were comparable based on the strength of the mCherry signal, but only the wild-type PGRP-LCx, PGRP-LE, or Imd showed strong ThT fluorescence, which overlapped with the mCherry signal in all cases. These results demonstrate cRHIM-mediated amyloid fibrils formation in cells (representative images in Figure 4B and quantitation in Figure 4C).
Using the same confocal microscopy-based assay, we next tested the full-length alanine substitution mutants of PGRP-LCx (Figure 4D), corresponding to the fragments analyzed by SDD-AGE in Figure 3A. While E225A showed enhanced ThT fluorescence consistent with its strong signaling activity (Kaneko et al., 2006), all other substitution mutants showed strikingly reduced ThT staining compared to wild-type PGRP-LCx. In conclusion, PGRP-LC mutants that fail to signal properly were also defective in amyloid formation in vitro and in cells. Hence, PGRP-LC, PGRP-LE, and Imd signal through cRHIM-dependent amyloidal fibrils in Drosophila cells.
Amyloid Formation Is Required for Imd Pathway Signaling
ThT not only binds amyloid but also inhibits further amyloid polymerization (Alavez et al., 2011). We therefore hypothesized that if amyloid formation were required for Imd pathway activation, an amyloid inhibitor such as ThT would suppress the signaling output, i.e., AMP gene induction. To test this, S2* cells were treated with ThT, and 30 min later Imd signaling was activated by adding DAP-type PGN. After 6 hr, the induction of the AMP gene Diptericin (Dpt) was quantified by qRT-PCR. ThT potently inhibited Dpt induction in a dose-dependent manner (Figure 5A). Note, this dose range of ThT (0.1–1.0 μM) had no effect on cell viability. ThT reduced Dpt induction also at 2 hr and 24 hr after PGN addition (Figure S2A). Other Imd signaling-induced AMPs were also affected: 1 μM ThT significantly reduced Attacin A (AttA) mRNA levels at 2 hr and 6 hr time points and Cecropin A1 (CecA1) mRNA levels at 2 hr post-induction (Figures S2B and S2C). To determine whether the effect of ThT on Imd signaling was specific or affecting general cellular processes or signal transduction, we also analyzed the induction of the AMP gene Drosomycin (Drs) via the Toll pathway activated with recombinant Spätzle (Spz-C106) (Weber et al., 2003). ThT did not interfere with Toll signaling (Figure 5B). These data demonstrate that ThT specifically inhibits Imd signaling, strongly arguing that amyloid formation is uniquely required for this signaling pathway.
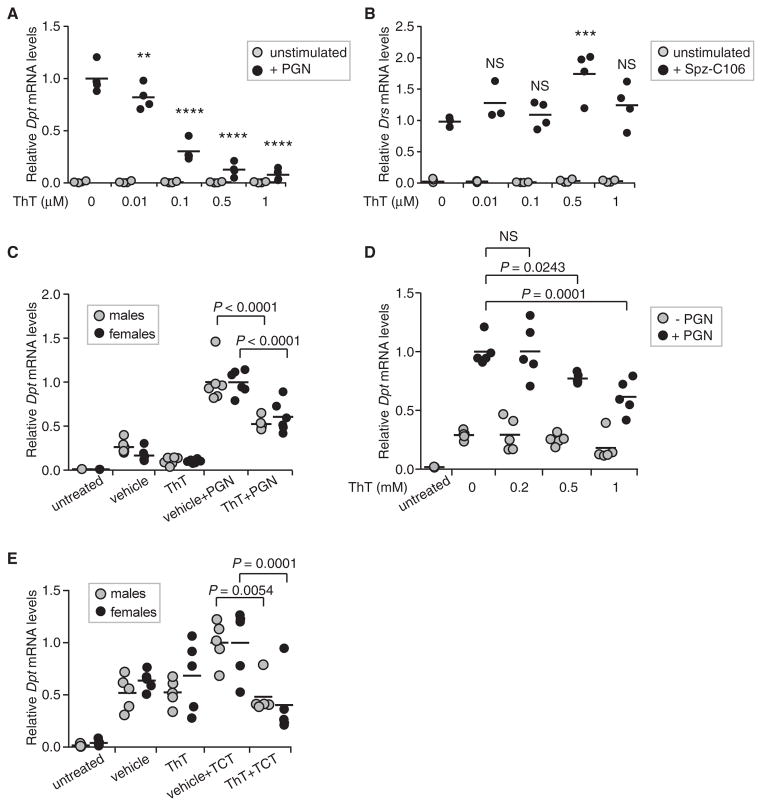
Figure 5. Dose-Dependent Inhibition of Imd Signaling by ThT in Cells and in Flies.
(A and B) S2* cells were treated with ThT before triggering the activation of the Imd pathway with DAP-type PGN (A), or the Toll pathway with recombinant cleaved Spätzle, Spz-C106 (B). Transcript levels of the target genes for the Imd and Toll pathways, Diptericin and Drosomycin, respectively, were measured by qRT-PCR and normalized to Rp49 expression. Graph shows individual data points from 3 or 4 independent experiments, the line indicating the mean.
(C and D) ThT suppresses Diptericin induction in flies. w1118 male and female flies were co-injected with vehicle (5% DMSO in sterile PBS), and 1 mM ThT ± 0.5 mg/mL PGN (C) or 40 μM TCT (D), and harvested 1 hr after injection. Diptericin transcript levels were measured by qRT-PCR and normalized to Rp49 values. Data shown are individual data points of six (C) and five (D) biological replicates, the bar indicating the mean.
(E) Inhibition of the Imd pathway activation by ThT is dose dependent. Male flies were injected with vehicle (5% DMSO in sterile PBS), 0.2 mM, 0.5 mM, or 1 mM ThT ± 0.5 mg/mL PGN and harvested 1 hr after injection. Diptericin transcript levels were measured by qPCR and normalized to Rp49 values. Data shown are individual data points of five biological replicates, the bar indicating the mean.
NS = not significant, **p < 0.01, ***p < 0.001, ****p < 0.0001. See further analysis of ThT inhibitor effects on Imd signaling in Figures S2 and S3.
S2* cells are derived from embryonic hemocytes and resemble plasmatocytes, phagocytes capable of immune signaling and AMP release (Lemaitre and Hoffmann, 2007). However, the main site of AMP production upon septic injury is the fat body, and this prompted us to ask whether amyloid was also involved in AMP induction in this tissue in the adult fly. To address this, flies were injected with 1 mM ThT along with monomeric or polymeric DAP-type PGN (TCT or PGN, respectively), and Dpt induction was analyzed 1 hr after injection. Of note, we tested both higher concentrations of ThT and longer time points in our preliminary assays, but noticed that ThT concentrations above 2 mM transiently altered fly behavior, with full recovery in less than 2 hr, suggesting that ThT was quickly metabolized and/or inactivated. Thus, our assays were limited to 1 hr after injection, with simultaneous ThT and PGN delivery. As expected for this early time point, injection of vehicle alone caused Dpt induction (Lemaitre et al., 1997); nevertheless, injection with polymeric PGN further increased Dpt induction by ~4-fold in males and ~6-fold in females (Figure 5C). Co-injection with ThT significantly suppressed Dpt induction in both males and in females (48% in males and 39% in females). The slightly stronger effect of ThT in males was reproducible and probably was due to the smaller size and volume of the male flies and the subsequently higher final concentration of ThT in tissues. We therefore focused on males when testing the dose response of ThT in adult flies. Dpt expression in flies co-injected with polymeric PGN and 0.2 mM ThT was similar to control flies, but co-injection of 0.5 mM or 1 mM ThT clearly interfered with Dpt induction in a dose-dependent manner (Figure 5D). The effect of ThT was similar when the flies were challenged with monomeric PGN, TCT. Co-injection of 1 mM ThT with 40 μM TCT suppressed Dpt, AttA, and CecA1 mRNA levels (Figures 5E, S3A, and S3B). However, Dpt induction at this dose and time point was only 2-fold when compared to vehicle-only-injected flies. We therefore repeated this assay with 10-fold more TCT. Unexpectedly, males did not recover well from injections with this high dose of TCT plus ThT, so females were analyzed. As previously reported, higher concentration of TCT resulted in stronger Dpt induction (Neyen et al., 2016). At both TCT concentrations, 1 mM ThT significantly suppressed Dpt induction by 45%–60%. These data demonstrate that ThT inhibits Imd signaling in vivo, further indicating that amyloid fibrils are critical for this innate immune signaling pathway in adult animals and consistent with our earlier analysis of cRHIM-deficient PGPR-LCx alleles (Kaneko et al., 2006).
Pirk Acts as a Regulator of Signaling Amyloid Formation
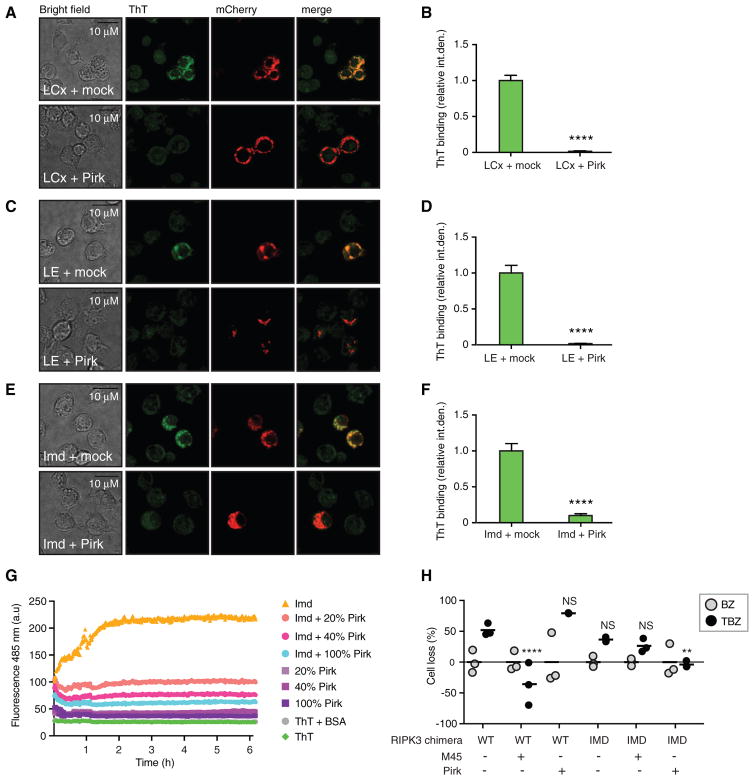
Unlike mammalian RHIM-mediated necroptosis signaling, Imd signaling is transient and does not lead to cell death. This suggests that fly cells possess mechanisms to limit the potential cytotoxic effects of signaling amyloids. To investigate these mechanisms, we focused on the Imd pathway negative regulator Pirk. Pirk is a potent inhibitor of Imd signaling that was originally identified as directly interacting with the cRHIM of PGRP-LC, PGRP-LE, and Imd (Aggarwal et al., 2008; Kleino et al., 2008). In fact, our earlier work argued that Pirk interferes with the complex formed by PGRP-LC and Imd. Moreover, Pirk contains a putative cRHIM (Kajava et al., 2014) within a predicted β sheet-rich region, which is sufficient to block Imd signaling (Kleino et al., 2008). Together, these earlier findings led us to hypothesize that Pirk might block Imd signaling by interfering with the formation of the PGRP-Imd amyloid fibrils. To test this idea, FLAG-tagged Pirk was expressed along with mCherry-tagged PGRP-LCx, PGRP-LE, or Imd in S2* cells, and cells were stained with ThT to detect amyloid (Figures 6A–6F). PGRP-LCx, PGRP-LE, and Imd expression, as assayed by mCherry fluorescence, was similar in both control and Pirk-expressing cells. However, in Pirk-expressing cells, these proteins no longer bound ThT (representative confocal microscopy images in Figures 6A, 6C, and 6E, and quantitation of all the images in Figures 6B, 6D, and 6F), suggesting that Pirk prevents the formation of PGRP-LC, PGRP-LE, and Imd amyloidal aggregates in cells.
Figure 6. Pirk Inhibits Amyloid Formation.
(A–F) ThT fluorescence in Drosophila S2* cells expressing mCherry-fusions of PGRP-LCx (A, B), PGRP-LE (C, D), or Imd (E,F), and either an empty vector (mock) or FLAG-Pirk. Representative images (A, C, E) and quantification (B, D, F) showing mean ± SEM of at least 39 cells pooled from three independent experiments. Scale bar: 10 μm. ****p < 0.0001.
(G) The inhibition of Imd amyloidal polymerization by Pirk, assayed by ThT fluorescence.
(H) Effect of Pirk on TNF-induced necroptosis in HeLa cells expressing wild-type RIPK3 or RIPK3/Imd chimera on TNF-induced necroptosis in HeLa cells. Cells were co-transfected with M45 or Pirk with WT or chimeric Imd-RIPK3. Necroptosis was activated with combined TNF, BV6, and zVAD-fmk treatment. The graph shows individual data points (N = 3) with mean indicated as a bar. Abbreviations: BZ, no TNF stimulation (gray dots); TBZ-, BV6-, and zVAD-fmk-treated cells stimulated with TNFα (black dots).
See also Figure S4 for a model of amyloid signaling and Pirk inhibition.
Next we tested whether full-length Pirk could interfere with the amyloidal aggregation of Imd in vitro. In this assay, denatured, monomeric Imd60-273 was mixed with denatured, monomeric Pirk at different ratios. The proteins were then diluted in assay buffer containing ThT, and the amyloid polymerization was measured as an increase in the ThT fluorescence, as in Figures 2C and 2K. Imd alone readily polymerized into ThT binding aggregates (Figure 6G). However, increasing concentrations of Pirk dose-dependently interfered with ThT binding to Imd, showing that Pirk inhibits amyloidal polymerization of Imd.
The viral necroptosis inhibitor MCMV M45 harbors a RHIM motif and possibly functions by interfering with the RHIM-mediated interaction of RIPK1 and RIPK3. To test whether the function of Pirk was analogous to M45, we triggered necroptosis in HeLa cells as in Figure 1C, co-expressing either M45 or Pirk with RIPK3 (Figure 6H). In these conditions, M45 efficiently blocked TNF-induced necroptosis, while expression of Pirk had no protective effect. We hypothesized that this could be due to the significant sequence diversity of RHIMs versus cRHIMs. We therefore tested whether Pirk could interfere with a chimeric RIPK3, in which 11 amino acids within the RHIM were replaced by 11 amino acids from the cRHIM of Imd. Similar to the four amino acid RHIM substitutions tested in Figure 1C, this longer substitution supported necroptosis signaling but failed to interact with M45, demonstrated by the lack of protection from necroptosis when co-expressed with M45. However, co-expression of Pirk significantly reduced necroptosis in cells expressing the chimeric RIPK3, suggesting that the 11 amino acids within the Imd cRHIM were necessary and sufficient for Pirk-mediated interference. Altogether, these data demonstrate that Pirk specifically interferes with the formation of Imd pathway signaling amyloids.
DISCUSSION
Proteins forming amyloidal aggregates have been associated with several neurodegenerative and systemic pathologies (Knowles et al., 2014). This has fueled research focusing on how amyloid is formed and which proteins are prone to form amyloidal aggregates. A recent discovery has been that some proteins, which are not misfolded, form amyloid fibrils in their active and functional states, so-called functional amyloids (Fowler et al., 2007).
Amyloid fibrils are highly organized structures that grow quickly into very large detergent-resistant polymers featuring a cross β sheet fold. Shearing these fibrils creates new ends that can seed new fibrils by promoting conformational change of monomeric proteins and thereby accelerate the fibril polymerization into exponential growth. These prion-like conformational changes have also been found to promote the formation of innate immune signaling complexes in mammals. The CARD (Caspase activation and recruitment domain) and PYRIN domains of the inflammasome adaptor protein ASC (apoptosis-associated speck-like protein containing a CARD) polymerize into a prion-like core filament, around which the other inflammasome proteins are assembled (Cai et al., 2014; Franklin et al., 2014; Lu and Wu, 2015; Yin et al., 2015). Likewise, the adaptor MAVS (mitochondrial antiviral signaling protein) forms prion-like filaments upon activation by RIG-I (retinoic acid-inducible gene I) or MDA5 (melanoma differentiation associated factor 5). Upon necroptotic signaling, the RHIMs of RIPK1 and RIPK3 polymerize to form functional amyloid, while the DAI-RIPK1 RHIM complex also forms filamentous aggregates (Li et al., 2012). Another RHIM-containing innate immune signaling protein, TRIF, was recently reported to form detergent-resistant aggregates in cells (Gentle et al., 2017), but the amyloidal nature of the aggregates, and whether this aggregation is RHIM or TIR mediated, remains elusive.
The current model for the formation of prion or amyloid platforms in innate immune signaling suggests that aggregation of a receptor protein forms the nucleus that seeds fibril formation by receptor-interacting adaptor proteins (Kagan et al., 2014). In the case of Imd signaling, the receptors PGRP-LC and PGRP-LE are known to oligomerize or cluster in a ligand-dependent manner, via their C-terminal PGN binding domains. This may bring their N-terminal signaling domains into close proximity and promote conversion of their cRHIMs into a proto-amyloid oligomer. The receptor cRHIM oligomer, in turn, could recruit the adaptor protein Imd and seed the propagation of amyloid fibrils, as depicted in our model (Figure S4). Relative proportions of the receptor and adaptor proteins in Drosophila cells support this hypothesis: endogenous PGRP-LC is barely detectable on the plasma membrane (Neyen et al., 2012), while Imd is abundant in the cytoplasm and in the nucleus (Boyer et al., 2011). Thus, the amyloid seed would be formed by the limiting amounts of PGRP-LC or PGRP-LE, following their ligand-induced clustering and aggregation of their cRHIMs. The abundant Imd protein would then elongate a fibril via its centrally positioned cRHIM. This architecture would allow the C-terminal DD and N-terminal cleavage site, with the Iap binding motif (IBM) of Imd, to be positioned on the exterior of the amyloid fibril and would support the interaction between the Imd DD and Fadd and enable Fadd-mediated recruitment of the caspase Dredd. Interestingly, Dredd cleaves Imd at D30, creating a neo-N-terminal recognition site for the E3 ligase Diap2. This cleavage site near the N terminus is likely positioned on the exterior of the fibril, enabling Dredd-catalyzed cleavage and subsequent interaction with the ubiquitin ligase Diap2. Together with the E2 ligases Effete, Bendless, and Uev1a, Diap2 links K63 polyubiquitin chains to lysine 137 and 153 of Imd (Chen et al., 2017), which are outside the predicted core amyloid (cRHIM) region, and thus also likely to be exterior to the fibril. With multiple monomers, each with a potential site of K63-ubiquitination, the Imd amyloid fibril would create a macromolecular signaling platform potentially decorated with many K63-ubiquitin chains and capable of driving rapid and robust NF-κB activation and the observed 100- to 1,000-fold activation of target gene expression.
In addition to PGRP-LC and PGRP-LE, cRHIM-like motifs have been predicted to be present in two other Drosophila proteins, PGRP-LA and Pirk (Kajava et al., 2014). PGRP-LA has been associated with Imd signaling, but its function is unclear. PGRP-LA is mostly expressed in the barrier epithelia, is upregulated upon infection, and encodes three isoforms, one of which (PGRP-LAD) includes an N-terminal cRHIM (Gendrin et al., 2013; Werner et al., 2000). Ubiquitous overexpression of PGRP-LAD, but not the cRHIM-lacking isoforms, is sufficient to trigger expression of the Imd pathway target gene Dpt, suggesting that PGRP-LA is a positive regulator of the Imd pathway via its cRHIM (Gendrin et al., 2013). Presumably, this also involves Imd amyloid fibrils.
Pirk is an Imd pathway negative feedback regulator, and its expression is under the transcriptional control of Relish. The mechanism of Pirk-mediated Imd pathway inhibition has remained elusive, but it has been suggested that Pirk traffics PGRP-LC into lysosomes to be degraded (Lhocine et al., 2008), and/or that by binding both receptors and Imd, Pirk prevents their interaction (Aggarwal et al., 2008; Kleino et al., 2008). Here we reported that Pirk interfered with the formation of Imd amyloid fibrils and suppressed Imd signaling. Our data demonstrated that Pirk stops fibril elongation, perhaps by capping the nascent Imd fibrils, similar to the human inhibitor of CARD (INCA), which inhibits inflammasome assembly by terminating Caspase-1 filaments (Lu et al., 2016). It is also conceivable that Pirk may insert into the Imd fibrils and destabilize them, although this hypothesis requires further study. Amyloid inhibition may be mediated either by the predicted C-terminal cRHIM of Pirk (Kajava et al., 2014) or by the multiple repetitive, beta-sheet motifs spanning the conserved region of the protein (Kleino et al., 2008). Further, it is yet unknown whether Pirk also traffics Imd fibrils for destruction, by proteasomal, autophagosomal, or other degradative systems.
Imd signaling is robust yet transient. Thus, Imd amyloid fibrils must be regulated such that they trigger rapid NF-κB activation and robust AMP gene expression, but for only a limited time. Hyperactivation of Imd signaling is highly deleterious; overexpression of Imd leads to spontaneous immune signaling and excessive cell death (Akbar et al., 2016; Cao et al., 2013; Georgel et al., 2001). However, cell death is not observed during experimental immune challenge. S2 cells do not die with immune stimulation; neither cell death nor fat body remodeling is observed following immune challenge of flies. These cRHIM-based amyloid fibrils are involved in a process not obligatorily leading to cell death. Together, these findings argue that Imd amyloids must be tightly regulated so that immune activation is robust yet limited and implies active mechanisms in the turnover of these fibrils. The Drosophila cRHIMs are regulated by Pirk and also lack a Gln at P2 in their core, which is invariant in the mammalian RHIMs. This sequence variation could potentially be related to the unique turnover properties of these amyloids. Polar residues within amyloids, such as the Gln in position P2, have been suggested to promote amyloid oligomerization through hyperpolarized H-bonds between the side chains (Mompeán et al., 2016). cRHIM sequences instead contain other polar residues at P2—His or Thr in Drosophila, and even Lys in other insect species. These residues may form H-bonds to support oligomerization that are weaker or altered in a manner that allows these Imd fibrils to be sensitive to Pirk and to disassemble more readily. In addition, the sequence flanking the tetrapeptide core of the Drosophila cRHIMs, which demonstrably contributed to fibril formation, are also rich with polar residues, such as Q, N, S, and T. It is plausible that the flanking regions further contribute to the unique oligomerization and disassembly properties of the Imd fibrils. In the case of yeast Sup35 prions, disruption of these polar residue-generated H-bonds was shown to cause disassembly of the fibrils (Mompeán et al., 2016). If similar H-bonds contribute to the cRHIM amyloid formation, breaking these bonds could be one way of destabilizing the fibrils and disassembling the Imd signaling complex. Future studies will focus on how cRHIM fibrils support signal transduction, perhaps as macromolecular scaffold, as well as probe the mechanisms limiting amyloid formation, activity, and toxicity.
STAR★METHODS
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-FLAG M2 | Sigma | F3165; RRID: AB_259529 |
| Mouse monoclonal anti-His Tag | Abgent | AM1010a; RRID: AB_352469 |
| Bacterial and Virus Strains | ||
| E. coli DH5α | Thermo Fisher | Cat# 18258012 |
| E. coli BL21 DE3 (RIPL) | Agilent Technologies | Cat# 230280 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| D. melanogaster Spätzle-C106 | Weber et al., 2003 | N/A |
| Human TNF | Invitrogen | 34-8329-85 |
| Smac mimetic BV6 | Genentech | N/A |
| zVAD-fmk | APExBio | A1902 |
| Thioflavin T | Santa Cruz | sc-359849 |
| Polymeric peptidoglycan from E. coli strain 1106 | Kaneko et al., 2004 | N/A |
| Monomeric peptidoglycan (TCT) from Bordetella pertussis | Cookson et al., 1989 | N/A |
| Propidium iodide | Sigma | P4864-10ML |
| Experimental Models: Cell Lines | ||
| D. melanogaster cell line S2* | Samakovlis et al., 1992; Schneider 1972 | N/A |
| Human HeLa cell line | ATCC | CCL-2 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster strain w1118 | N.S. lab stock | N/A |
| Oligonucleotides | ||
| Primer: Dpt forward: CATTGCCGTCGCCTTACTT | This paper | N/A |
| Primer: Dpt reverse: TAGGTGCTTCCCACTTTCCA | This paper | N/A |
| Primer: AttA forward: CAATGTGGTGGGTCAGGTTT | This paper | N/A |
| Primer: AttA reverse: ACCTTGGCATCCAGATTGTG | This paper | N/A |
| Primer: CecA1 forward: TTTCGTCGCTCTCATTCTGG | This paper | N/A |
| Primer: CecA1 reverse: TTGTTGAGCGATTCCCAGTC | This paper | N/A |
| Primer: Drs forward: CTCTTCGCTGTCCTGATGCT | This paper | N/A |
| Primer: Drs reverse: ATCCTTCGCACCAGCACTT | This paper | N/A |
| Primer: Rp49 forward: GCACTCTCTGTTGTCGATACCCTTG | This paper | N/A |
| Primer: Rp49 reverse: AGCGCACCAAGCACTTCATC | This paper | N/A |
| Recombinant DNA | ||
| pEGFP-N1-RIPK3 WT | Cho et al., 2009; Li et al., 2012 | N/A |
| pEGFP-N1-RIPK3 VQVG/AAAA | Cho et al., 2009; Li et al., 2012 | N/A |
| pEGFP-N1-RIPK3 VQVG/VTFG | This paper | N/A |
| pEGFP-N1-RIPK3 VQVG/VHIG | This paper | N/A |
| pEGFP-N1-RIPK3 VQVG/LHFG | This paper | N/A |
| pEGFP-N1-RIPK3/IMD (NCSGVQVGDNN from RIPK3 RHIM replaced with SNANNLHFGSV from Imd cRHIM) | This paper | N/A |
| pSmt3-PGRP-LC(201-242) WT | This paper | N/A |
| pSmt3-PGRP-LC(201-242) L210A | This paper | N/A |
| pSmt3-PGRP-LC(201-242) D215A | This paper | N/A |
| pSmt3-PGRP-LC(201-242) V216A | This paper | N/A |
| pSmt3-PGRP-LC(201-242) T217A | This paper | N/A |
| pSmt3-PGRP-LC(201-242) F218A | This paper | N/A |
| pSmt3-PGRP-LC(201-242) G219A | This paper | N/A |
| pSmt3-PGRP-LC(201-242) D220A | This paper | N/A |
| pSmt3-PGRP-LC(201-242) E225A | This paper | N/A |
| pSmt3-PGRP-LC(201-242) G226A | This paper | N/A |
| pSmt3-PGRP-LC(201-242) V228A | This paper | N/A |
| pSmt3-PGRP-LC(201-242) ΔcRHIM | This paper | N/A |
| pSmt3-PGRP-LE(80-127) WT | This paper | N/A |
| pSmt3-PGRP-LE(80-127) I96A | This paper | N/A |
| pSmt3-PGRP-LE(80-127) S99A | This paper | N/A |
| pSmt3-PGRP-LE(80-127) V102A | This paper | N/A |
| pSmt3-PGRP-LE(80-127) H103A | This paper | N/A |
| pSmt3-PGRP-LE(80-127) I104A | This paper | N/A |
| pSmt3-PGRP-LE(80-127) G105A | This paper | N/A |
| pSmt3-PGRP-LE(80-127) G112A | This paper | N/A |
| pSmt3-PGRP-LE(80-127) I116A | This paper | N/A |
| pSmt3-PGRP-LE(80-127) ΔcRHIM | This paper | N/A |
| pSmt3-Imd(3-273) | This paper | N/A |
| pSmt3-Imd(60-273) | This paper | N/A |
| pOPINS-Imd(31-273) | This paper | N/A |
| pOPINS-Imd(31-167) | Paquette et al., 2010 | N/A |
| pOPINS-Imd(168-273) | This paper | N/A |
| pOPINS-ImdΔ(76-109) | This paper | N/A |
| pOPINS-ImdΔ(110-143) | This paper | N/A |
| pOPINS-ImdΔ(144-177) | This paper | N/A |
| pOPINS-ImdΔ(222-266) | This paper | N/A |
| pOPINS-ImdΔ(114-121) | This paper | N/A |
| pRmHa3-Imd-FLAG | Paquette et al., 2010 | N/A |
| pRmHa3-ImdΔ(114–121)-FLAG | This paper | N/A |
| pRmHa3-M45(1-277)-FLAG WT | This paper | N/A |
| pRmHa3-M45(1-277)-FLAG AAAA | This paper | N/A |
| pRmHa3-FLAG-PGRP-LE | Aggarwal et al., 2008 | N/A |
| pRmHa3-FLAG-PGRP-LEΔ(83–122) | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCxΔ(209–225) | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCxΔ(226-242) | This paper | N/A |
| pRmHa3-mCherry-PGRP-LE | This paper | N/A |
| pRmHa3-mCherry-PGRP-LEΔ(83–122) | This paper | N/A |
| pRmHa3-Imd-mCherry | This paper | N/A |
| pRmHa3-Imd Δ(114–121)-mCherry | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx Λ210A | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx N212A | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx S213A | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx D215A | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx V216A | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx T217A | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx F218A | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx G219A | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx D220A | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx E225A | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx G226A | This paper | N/A |
| pRmHa3-mCherry-PGRP-LCx V228A | This paper | N/A |
| pRmHa3-Flag-Pirk | Aggarwal et al., 2008 | N/A |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Neal Silverman (Neal.Silverman@umassmed.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Drosophila S2* cells were cultured in Schneider’s Drosophila medium (GIBCO) supplemented with 10% FBS (not heat-inactivated), 1% Glutamax (GIBCO), and 0.2% PenStrep (GIBCO) at 27°C. This cell line originated in the Hultmark laboratory (Samakovlis et al., 1992), and looks and behaves like S2 cells, being small (approximately 10 μm of diameter), round, semi-adherent, phagocytic, responsive to 20-hydroxyecdysone, and immune-inducible. S2 cells are male, by criterion of MSL complex assembly.
HeLa cells (female) were cultured in DMEM high sucrose medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Fly maintenance
w1118 flies were maintained at 22°C on food containing 6.5 g/l agar, 23.5 g/l dry yeast, 60 g/l cornmeal, 60 ml/l molasses, 4 ml/l acid mix, and 0.13% tegosept. The flies used in experiments were 10-days-old, healthy flies, which were first collected and mixed from different vials, and then randomly divided into treatment groups.
METHOD DETAILS
Necroptosis cell death assay
HeLa cells, which do not express endogenous RIPK3, were transfected with wild-type RIPK3 or RIPK3 RHIM chimeras fused to GFP at the C-termini. Transfection was performed with Lipofectamine 2000 as per manufacturer’s protocol. Sixteen hours after transfection, cells were treated with 10 μM zVAD-fmk and 1 μM BV6 for 1 hour prior to stimulation with 50 ng/ml (Figure 1C) or 10 ng/ml (Figure 6H) human TNF. Cell death of the transfected populations were enumerated 16 hours later by FACS using propidium iodide. Cell death was calculated with the formula: % Cell death = (1-(# live GFP+ cells in sample/# live GFP cells in untreated group)) × 100%.
Cloning, protein expression, and purification of recombinant proteins
The fragments of PGRP-LC (residues 201-242), PGRP-LE (residues 80-127), Imd (residues 100-125), and Imd (residues 60-273, and 3-273) were amplified from plasmids described previously (Aggarwal et al., 2008; Kaneko et al., 2006) and subcloned into the pSMT3 vector with N-terminal 6xHis-Sumo tag. The proteins were expressed in E. coli BL21 (DE3) RIPL cells (Novagen). Bacterial cells were transformed with expression plasmids and grown in Luria broth supplemented with 40 μg/ml kanamycin and 25 μg/ml chloramphenicol. At an OD600 of 0.8, protein expression was induced with 0.5 mM isopropyl β-D-thiogalactoside (Fisher Scientific). After 16 hours of induction at 20°C, cells were harvested by centrifugation at 5,000 rpm for 10 min and lysed by sonication in a buffer containing 50 mM Tris-HCl (pH8.0), 300 mM NaCl, 1 mM phenylmethyl-sulfonyl fluoride, 1 mM β-mercaptoethanol, 5% Glycerol (v/v) and a pro-tease inhibitor cocktail (Roche). After spinning at 16,000 rpm for 45 min, the supernatant was incubated with Ni-NTA agarose (QIAGEN) at 4°C for 60 min. The subsequent purification procedure followed the standard manual from QIAGEN. The His-Sumo tag was removed by adding enzyme Ulp1 (1:1000) at 4°C overnight. The protein was further purified by gel filtration chromatography using the Superdex 200 10/300 GL or 16/600 column (GE Healthcare) and an ÄKTA FPLC (GE Healthcare) at either 4°C or room temperature with a running buffer of 20 mM Tris (pH 7.4), 150 mM NaCl, and 2 mM dithiothreitol. The purified protein was concentrated to 5 mg/ml, flash-frozen in liquid nitrogen and stored at −80°C. Protein concentrations were determined by the Bradford assay using BSA as a standard.
Active cleaved Imd (Imd31-273) (Paquette et al., 2010) and its mutant versions listed in the Key Resource Table were cloned into pOPIN S vector (from Oxford Protein Production Facility, UK) and produced as 6xHis-Sumo fusion proteins in E. coli BL21 (DE3) (pLysS). Protein production was induced by 0.8 mM IPTG when cultures reached OD600 of 0.6, and the cultures were grown at 26°C shaking for 4h. Bacterial pellets were lysed by sonication in buffer containing 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10 mM imidazole, and protease inhibitor cocktail. Lysates were cleared by centrifugation at 10,000 × g for 30 min. at 4°C, and the proteins were purified from cleared lysates using Millipore PureProteome Nickel Magnetic Beads according to the manufacturer’s protocol.
Thioflavin T (ThT) fluorimetry
Fluorescence measurements were performed as described previously (Meng et al., 2010) at 25°C in 96-well plates on a SpectraMax M5e plate reader (Molecular Devices) or an Enspire multimode plate reader (PerkinElmer) with an excitation wavelength of 430 nm. The final ThT concentrations ranged from 15 μM to 25 μM in 20 mM Tris-HCl, 100 mM NaCl (pH 7.4).
Congo red binding
Congo red (CR) binding was detected using a previously described method (Klunk et al., 1989) at 25°C in 96-well plates using a SpectraMax M5e plate reader (Molecular Devices). The incubation time with CR was 1 hour at room temperature and the concentration of CR was 15 μM.
X-ray diffraction from fibrils
A 5 mg/ml solution of the 6xHis-Sumo-tagged PGRP-LC or PGRP-LE was dried using a SpeedVac System (Savant). Dried and partially aligned fibrils were mounted in a cryo-loop and exposed to Cu Kα radiation from a Bruker X-ray generator operating at 40 kV and 60 mA. Data were collected at room temperature for 5 min with a 5° oscillation on a Mar345 imaging plate detector.
Electron microscopy and image processing
The purified samples (6xHis-Sumo-tagged PGRP-LC201-242, PGRP-LE80-127, or IMD100-125) were loaded onto copper grids. After 30 s of incubation on the grid at room temperature, the samples were negatively stained with 1.5% uranyl acetate, and imaged with a 1K × 1K CCD camera on a Tecnai G2 Spirit BioTWIN electron microscope equipped with a LaB6 filament and operated at 80 kV. The nominal magnification is around 50,000.
Seeding experiments using ThT fluorescence
Imd3-273 was produced as a 6xHis-Sumo fusion protein, and denatured in 8 M urea. The denatured Imd3-273 was diluted 50-fold into a native buffer (20 mM Tris pH 7.4, 100 mM NaCl, 25 μM ThT), to a final concentration of 6 μM. ThT fluorescence was measured on 96-well plates at 25°C for 15,000 s on an Envision Multi-mode plate reader (PerkinElmer) with excitation and emission wavelengths of 430 nm and 485 nm, respectively. Seeding experiments were performed as described above with the exception that 6xHis-Sumo-PGRP-LE80-127 preformed amyloidal seeds were added to the reaction to the final concentration of 0.6 μM (10%). To analyze Pirk’s ability to inhibit amyloid polymerization, Imd60-273 and full-length Pirk were denatured in 8M urea, 100 mM NaCl, 20 mM Tris-HCl (pH7.5), followed by heating at 95°C for 20 min, and immediate sonication for 2 min. Precipitated proteins were removed by centrifugation at 13,000 rpm for 1 min, and the supernatants containing the denatured proteins were diluted into 1mg/ml with the ThT buffer (20 mM Tris-HCl pH7.4, 100 mM NaCl). The final concentration for Imd and Pirk was 2.5 uM. All reactions were measured kinetically at 37°C in 96-well plates with excitation and emission wavelengths of 430 nm and 485 nm, respectively.
Semi-denaturing detergent agarose gel electrophoresis (SDD-AGE)
The formation of amyloidal aggregates of 6xHis-Sumo-tagged PGRP-LC201-242, PGRP-LE80-127, and their mutants was analyzed by SDD-AGE according to a published protocol (Halfmann and Lindquist, 2008) with minor modifications. For Figure 3A,B, purified proteins were quantified and mixed with 4× protein loading buffer with 8% SDS at room temperature for 5 min. After the samples were loaded onto a vertical 1.5% agarose gel with 0.1% SDS (Bio-Rad), and separated by electrophoresis in the running buffer (1xTBE and 0.1% SDS) for 3 hours with a constant voltage of 25 V at 4°C, the proteins were transferred to PVDF membrane (Millipore) for immunoblotting with anti-His monoclonal antibody (Sigma) at 1:10000 ratio.
6xHis-Sumo-Imd31-273 and Imd mutant forms used in Figure 3C were analyzed according to the published protocol (Halfmann and Lindquist, 2008). 0.5 mg of purified protein was loaded in each lane. Equal loading was confirmed by running replicate samples on SDS-PAGE, and staining the gel with InstantBlue Protein Stain (Expedeon). SDD-AGE samples were separated by running a horizontal 1.5% agarose gel with 0.1% SDS in a buffer containing 1xTAE with 0.1% SDS for 5h at constant voltage of 35V at 4°C. Proteins were then transferred to PVDF by capillary transfer and immunoblotted with mouse anti-His IgG (Abgent #AM1010a) at 1:5000.
To analyze detergent resistant protein aggregates in cell lysates, S2* cells expressing FLAG-tagged proteins were lysed in buffer containing 20 mM Tris-HCl (pH 7.6), 150 mM NaCl, 25 mM β-glycerophosphate, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM DTT, 1 mM NaVO4, and 1× protease inhibitor cocktail. Lysates were mixed with 4× loading buffer and run as described above and in (Halfmann and Lindquist, 2008). Proteins were transferred to PVDF by capillary transfer, and immunoblotted with monoclonal mouse anti-FLAG M2 antibody (Sigma #F3165) at 1:5000.
Cell culture and transfections
For confocal microscopy, S2* cells were plated 0.5×106 cells/ml on 6-well plates, and transiently transfected one hour later with 0.5 μg of Cu2+-inducible constructs in pRmHa3 backbone listed in the Key Resource Table using calcium phosphate precipitation method. Half of the old medium was carefully replaced with fresh medium 24 hours after transfections. Expression of constructs was induced by adding CuSO4 to the culture medium into 250 μM final concentration. For SDD-AGE analysis, S2* cells were plated 1×106 cells/ml on 6-well plates and transfected on hour later with 0.5 μg of FLAG-tagged Cu2+-inducible constructs using calcium phosphate. 24 hours post-transfection the cells were split into two wells (+/− Cu2+) and supplemented with fresh medium. Protein expression was induced with 250 μM Cu2+ 24 hours later, and cells harvested 18h post-induction.
qRT-PCR
Total RNAs from S2* cells were extracted using TRIzol® reagent (Invitrogen) according to the manufacturer’s protocol, and dissolved in nuclease-free water. 1 μg of total RNA was treated with amplification grade DNase I (Invitrogen) according to the manufacturer’s protocol, and the enzyme was heat-inactivated at +65°C for 10 min. at the presence of 2.5mM EDTA. Efficiency of the DNase I treatment was controlled by running a regular PCR with Rp49 primers using diluted RNA samples as templates. cDNA was synthesized from 0.5 μg of each DNase I-treated RNA using the BioRad iScript cDNA synthesis kit, and real-time PCR was performed using Bio-Rad SYBR Green Supermix according to the manufacturer’s protocols. Primers for Diptericin (Dpt), Attacin A (AttA), Cecropin A1 (CecA1), Drosomycin (Drs), and Rp49 (also known as RpL32) are listed in the Key Resources Table.
Confocal microscopy
Drosophila S2* cells transfected with mCherry-tagged constructs were resuspended 18h after Cu2+ induction, and transferred on Alcian Blue-treated coverslips. The cells were allowed to attach for 40 min before the medium was removed and the cells fixed with 3% PFA in PBS for 20 min. The cells were gently rinsed with PBS and stained with 3 mM Thioflavin T (Santa Cruz Biotechnologies) in 30:70 ethanol:PBS for 20 min, washed twice in 70% ethanol and thrice in 1xPBS, mounted with Mowiol, and dried overnight at 37°C in darkness. The cells were imaged using a Leica TCS SP8 confocal microscope with HyD detectors and a 63× oil immersion objective using sequential scanning. ThT was excited at 458 nm and emission detected at 485–530 nm, and mCherry was excited at 561 nm and emission detected at 610–620 nm.
Analysis of confocal microscopy images
Confocal microscopy images of ThT-stained S2* cells were taken so that there would always be both mCherry-expressing (transfected) and untransfected cells in the same image to control the background of ThT staining. Image analysis was performed using ImageJ/Fiji. The images were thresholded so that only the signal exceeding the background signal of untransfected cells was measured. Each cell expressing the mCherry-tagged protein was measured separately for the mCherry signal and the ThT signal, and the ThT binding (relative integrated density) represents the sum of the pixel intensity values of ThT divided by the sum of pixel intensity values of the mCherry signal. Hence, ThT signal is normalized to mCherry signal strength to avoid any bias caused by variable protein expression levels. Linear adjustment of brightness and contrast has been performed by ImageJ/Fiji to representative images at Figure 4B, and Figure 6A,C,E. Note that all quantifications in Figure 4C,D and in Figure 6B,D,F were performed using the raw, unenhanced images.
ThT inhibition assay in S2* cells
S2* cells were plated on 12-well plates, 1×106 cells/ml, and 20-hydroxyecdysone was added to 1 μM concentration on the following morning. 24 hours later, ThT or equal volume of vehicle (isopropanol) was added in the cell culture medium. The Imd pathway activation was triggered 30 min. later by adding E. coli PGN to the cells. The Toll pathway was triggered by adding recombinant cleaved Spätzle-C106 to 10 nM concentration to the cells. Cells were harvested 6h after PGN addition, and 18h after Spätzle-C106 addition.
Injections in flies
In Figure 5C, and in related Figure S2, male and female flies were injected with 64 nL of the following solutions using Nanoject II injector (Drummond Scientific): untreated: no injection; DMSO: 5% DMSO in sterile Dulbecco’s modified PBS (Corning); ThT: 1 mM ThT, 5% DMSO in PBS; DMSO+PGN: 5% DMSO, 0.5 mg/ml E. coli strain 1106 PGN in PBS; ThT+PGN: 1 mM ThT, 0.5 mg/ml PGN in PBS. In Figure 5D, male flies were injected with 64 nL of indicated concentrations of ThT diluted in 5% DMSO in PBS, +/− 0.5 mg/ml PGN. Flies were harvested 1 h post-injection, snap-frozen on dry-ice, homogenized in TRIzol® reagent (Invitrogen), and total RNAs were extracted according to the manufacturer’s protocol. In Figure 5E, and in related Figure S3, flies were injected and harvested as described above, but 40 μM TCT or 400 μM TCT was used instead of PGN.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis for all the numerical data was performed using GraphPad Prism 7.0a software. In cell-based assays, N was ≥3, and in assays using live animals, where bigger variation between samples was expected, N was ≥5. In Figures 1C and 6H, the necroptosis cell death assay was repeated three times, and a representative experiment with three biological replicates is shown. Data was analyzed using two-way ANOVA with Bonferroni’s post test comparing all other samples to non-functional AAAA mutant (Figure 1C), or to WT (Figure 6H) treated with zVAD-fmk, BV6, and TNF. Figure 4C and 4D, and Figure 6B, 6D, and 6F present data pooled from 3 independent experiments. In Figure 4C, N for each individual sample is the following: PGRP-LCx: 36; PGRP-LCxΔ(209-225): 27; PGRP-LCxΔ(226-242): 29; PGRP-LE:28; PGRP-LEΔ(83-122): 15; Imd: 26; ImdΔ(114-121): 31. Values of wild-type versus RHIM mutant PGRP-LCx were analyzed by one-way ANOVA with Bonferroni’s post test. PGRP-LE and Imd versus corresponding RHIM mutants were analyzed using unpaired t test. In Figure 4D, N for each individual sample is the following: WT: 28; Δ(209-225): 32; L210A: 30; N212A: 23; S213A: 28; D215A: 25; V216A: 27; T217A: 35; F218A: 24; G219A: 23; D220A: 22; E225A: 31; G226A: 20; V228A: 31. Data was analyzed using one-way ANOVA with Bonferroni post test, and all mutants were compared to the WT. Columns represent mean ± SEM. In Figure 6B,D,F N for each individual sample is the following: LCx + mock: 147, LCx + Pirk: 78; LE + mock: 71, LE + Pirk: 81; Imd + mock: 48, Imd + Pirk: 39. Statistical significance was analyzed by unpaired t test. Figure 5A,B, and related Figure S2 show data from 3 or 4 biological replicates performed on different days. Data was analyzed using two-way ANOVA with Bonferroni’s post test, and the effect of different ThT concentrations was compared to vehicle control stimulated with PGN (Figure 5A, Figure S2) or Spz-C106 (Figure 5B). Individual data points are shown with a line indicating the mean. Figure 5C–E, and related Figure S3, show individual data points of 6 (Figure 5C) or 5 (Figure 5D,E, and Figure S3) biological replicates, each replicate representing the RNA extracted from 6 flies. Data was analyzed using two-way ANOVA with Bonferroni’s post test, and comparing vehicle + PGN/TCT-treated flies to ThT+PGN/TCT -treated flies.
Supplementary Material
Highlights.
Formation of functional Imd amyloid fibril is required for Drosophila NF-κB signaling
Cryptic RHIM motifs in peptidoglycan receptors PGRP-LC and PGRP-LE form amyloids
Protofibrils of PGRP-LC and PGRP-LE seed Imd amyloid polymerization via Imd cRHIMs
Pirk, an NF-κB target, acts as a feedback inhibitor of Imd amyloid formation
Acknowledgments
We thank M. Ericsson (Harvard Medical School, Boston) for EM technical assistance and T. McQuade for protocols and advice. This work was supported by the NIH AI045937 to H.W., AI119030 to F.K.-M.C., and AI060025 to N.S., the EMBO Long-Term Fellowship (ALTF 357-2011), Sigrid Juselius Foundation, K. Albin Johansson Foundation, and Maud Kuistila Memorial Foundation to A.K., and the National Key Research and Development Program (2016YFA0500600, 2015CB943300), the National Natural Science Foundation of China (31670878, 31470724), and the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (TP2014010) to J.L.
Footnotes
Supplemental Information includes four figures and can be found with this article online at https://doi.org/10.1016/j.immuni.2017.09.011.
AUTHOR CONTRIBUTIONS
A.K., G.B., and J.L. designed and performed experiments and wrote the manuscript. N.F.R., J.H., J.N., Y.S., H.N., and M.G. provided reagents and performed experiments. F.K.-M.C., H.W., and N.S. designed experiments, provided funding, and wrote the manuscript.
References
- Aggarwal K, Rus F, Vriesema-Magnuson C, Ertürk-Hasdemir D, Paquette N, Silverman N. Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog. 2008;4:e1000120. doi: 10.1371/journal.ppat.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar MA, Mandraju R, Tracy C, Hu W, Pasare C, Krämer H. ARC syndrome-linked Vps33B protein is required for inflammatory endosomal maturation and signal termination. Immunity. 2016;45:267–279. doi: 10.1016/j.immuni.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavez S, Vantipalli MC, Zucker DJ, Klang IM, Lithgow GJ. Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature. 2011;472:226–229. doi: 10.1038/nature09873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L, Magoc L, Dejardin S, Cappillino M, Paquette N, Hinault C, Charriere GM, Ip WK, Fracchia S, Hennessy E, et al. Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity. 2011;35:536–549. doi: 10.1016/j.immuni.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Chtarbanova S, Petersen AJ, Ganetzky B. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc. Natl. Acad. Sci. USA. 2013;110:E1752–E1760. doi: 10.1073/pnas.1306220110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Paquette N, Mamoor S, Rus F, Nandy A, Leszyk J, Shaffer SA, Silverman N. Innate immune signaling in Drosophila is regulated by transforming growth factor β (TGFβ)-activated kinase (Tak1)-triggered ubiquitin editing. J Biol Chem. 2017;292:8738–8749. doi: 10.1074/jbc.M117.788158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. Phosphorylation-driven assembly of the RIP1–RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci USA. 2005;102:1122–1126. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson BT, Cho HL, Herwaldt LA, Goldman WE. Biological activities and chemical composition of purified tracheal cytotoxin of Bordetella pertussis. Infect Immun. 1989;57:2223–2229. doi: 10.1128/iai.57.7.2223-2229.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R, Gupta D. The peptidoglycan recognition proteins (PGRPs) Genome Biol. 2006;7:232. doi: 10.1186/gb-2006-7-8-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, Kim CH, Stöven S, Meier P, Silverman N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci USA. 2009;106:9779–9784. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid–from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Franklin BS, Bossaller L, De Nardo D, Ratter JM, Stutz A, Engels G, Brenker C, Nordhoff M, Mirandola SR, Al-Amoudi A, et al. The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nat Immunol. 2014;15:727–737. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrin M, Zaidman-Rémy A, Broderick NA, Paredes J, Poidevin M, Roussel A, Lemaitre B. Functional analysis of PGRP-LA in Drosophila immunity. PLoS ONE. 2013;8:e69742. doi: 10.1371/journal.pone.0069742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle IE, McHenry KT, Weber A, Metz A, Kretz O, Porter D, Häcker G. TIR-domain-containing adapter-inducing interferon-β (TRIF) forms filamentous structures, whose pro-apoptotic signalling is terminated by autophagy. FEBS J. 2017;284:1987–2003. doi: 10.1111/febs.14091. [DOI] [PubMed] [Google Scholar]
- Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- Guo H, Omoto S, Harris PA, Finger JN, Bertin J, Gough PJ, Kaiser WJ, Mocarski ES. Herpes simplex virus suppresses necroptosis in human cells. Cell Host Microbe. 2015;17:243–251. doi: 10.1016/j.chom.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J Vis Exp. 2008;(17):838. doi: 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Yang X. dFADD, a novel death domain-containing adapter protein for the Drosophila caspase DREDD. J Biol Chem. 2000;275:30761–30764. doi: 10.1074/jbc.C000341200. [DOI] [PubMed] [Google Scholar]
- Huang Z, Wu SQ, Liang Y, Zhou X, Chen W, Li L, Wu J, Zhuang Q, Chen C, Li J, et al. RIP1/RIP3 binding to HSV-1 ICP6 initiates necroptosis to restrict virus propagation in mice. Cell Host Microbe. 2015;17:229–242. doi: 10.1016/j.chom.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Magupalli VG, Wu H. SMOCs: supramolecular organizing centres that control innate immunity. Nat Rev Immunol. 2014;14:821–826. doi: 10.1038/nri3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WJ, Offermann MK. Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J Immunol. 2005;174:4942–4952. doi: 10.4049/jimmunol.174.8.4942. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Mocarski ES. Receptor-interacting protein homotypic interaction motif-dependent control of NF-kappa B activation via the DNA-dependent activator of IFN regulatory factors. J Immunol. 2008;181:6427–6434. doi: 10.4049/jimmunol.181.9.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV, Klopffleisch K, Chen S, Hofmann K. Evolutionary link between metazoan RHIM motif and prion-forming domain of fungal heterokaryon incompatibility factor HET-s/HET-s. Sci Rep. 2014;4:7436. doi: 10.1038/srep07436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, Silverman N. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Golenbock D, Silverman N. Peptidoglycan recognition by the Drosophila Imd pathway. J Endotoxin Res. 2005;11:383–389. doi: 10.1179/096805105X76823. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh BH, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- Kim CH, Paik D, Rus F, Silverman N. The caspase-8 homolog Dredd cleaves Imd and Relish but is not inhibited by p35. J Biol Chem. 2014;289:20092–20101. doi: 10.1074/jbc.M113.544841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleino A, Silverman N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol. 2014;42:25–35. doi: 10.1016/j.dci.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleino A, Myllymäki H, Kallio J, Vanhaaho LM, Oksanen K, Ulvila J, Hultmark D, Valanne S, Rämet M. Pirk is a negative regulator of the Drosophila Imd pathway. J Immunol. 2008;180:5413–5422. doi: 10.4049/jimmunol.180.8.5413. [DOI] [PubMed] [Google Scholar]
- Klunk WE, Pettegrew JW, Abraham DJ. Quantitative evaluation of congo red binding to amyloid-like proteins with a beta-pleated sheet conformation. J Histochem Cytochem. 1989;37:1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci USA. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F, Vidal S, Saigo K, Ueda R, Lemaitre B. Inducible expression of double-stranded RNA reveals a role for dFADD in the regulation of the antibacterial response in Drosophila adults. Curr Biol. 2002;12:996–1000. doi: 10.1016/s0960-9822(02)00873-4. [DOI] [PubMed] [Google Scholar]
- LeVine H., 3rd Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- Lhocine N, Ribeiro PS, Buchon N, Wepf A, Wilson R, Tenev T, Lemaitre B, Gstaiger M, Meier P, Leulier F. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. 2008;4:147–158. doi: 10.1016/j.chom.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Wu H. Structural mechanisms of inflammasome assembly. FEBS J. 2015;282:435–444. doi: 10.1111/febs.13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Li Y, Schmidt FI, Yin Q, Chen S, Fu TM, Tong AB, Ploegh HL, Mao Y, Wu H. Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism. Nat Struct Mol Biol. 2016;23:416–425. doi: 10.1038/nsmb.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinander A, Runchel C, Tenev T, Chen L, Kim CH, Ribeiro PS, Broemer M, Leulier F, Zvelebil M, Silverman N, Meier P. Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 2012;31:2770–2783. doi: 10.1038/emboj.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Abedini A, Plesner A, Verchere CB, Raleigh DP. The flavanol (−)-epigallocatechin 3-gallate inhibits amyloid formation by islet amyloid polypeptide, disaggregates amyloid fibrils, and protects cultured cells against IAPP-induced toxicity. Biochemistry. 2010;49:8127–8133. doi: 10.1021/bi100939a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mompeán M, Nogales A, Ezquerra TA, Laurents DV. Complex system assembly underlies a two-tiered model of highly delocalized electrons. J Phys Chem Lett. 2016;7:1859–1864. doi: 10.1021/acs.jpclett.6b00699. [DOI] [PubMed] [Google Scholar]
- Naitza S, Rossé C, Kappler C, Georgel P, Belvin M, Gubb D, Camonis J, Hoffmann JA, Reichhart JM. The Drosophila immune defense against gram-negative infection requires the death protein dFADD. Immunity. 2002;17:575–581. doi: 10.1016/s1074-7613(02)00454-5. [DOI] [PubMed] [Google Scholar]
- Neyen C, Poidevin M, Roussel A, Lemaitre B. Tissue- and ligand-specific sensing of gram-negative infection in Drosophila by PGRP-LC isoforms and PGRP-LE. J Immunol. 2012;189:1886–1897. doi: 10.4049/jimmunol.1201022. [DOI] [PubMed] [Google Scholar]
- Neyen C, Runchel C, Schüpfer F, Meier P, Lemaitre B. The regulatory isoform rPGRP-LC induces immune resolution via endosomal degradation of receptors. Nat Immunol. 2016;17:1150–1158. doi: 10.1038/ni.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik D, Monahan A, Caffrey DR, Elling R, Goldman WE, Silverman N. SLC46 family transporters facilitate cytosolic innate immune recognition of monomeric peptidoglycans. J Immunol. 2017;199:263–270. doi: 10.4049/jimmunol.1600409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette N, Broemer M, Aggarwal K, Chen L, Husson M, Ertürk-Hasdemir D, Reichhart JM, Meier P, Silverman N. Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-kappaB signaling. Mol. Cell. 2010;37:172–182. doi: 10.1016/j.molcel.2009.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Kim CH, Kim JH, Je BR, Roh KB, Kim SJ, Lee HH, Ryu JH, Lim JH, Oh BH, et al. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc Natl Acad Sci USA. 2007;104:6602–6607. doi: 10.1073/pnas.0610924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebsamen M, Heinz LX, Meylan E, Michallet MC, Schroder K, Hofmann K, Vazquez J, Benedict CA, Tschopp J. DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO Rep. 2009;10:916–922. doi: 10.1038/embor.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samakovlis C, Asling B, Boman HG, Gateff E, Hultmark D. In vitro induction of cecropin genes–an immune response in a Drosophila blood cell line. Biochem. Biophys Res Commun. 1992;188:1169–1175. doi: 10.1016/0006-291x(92)91354-s. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- Silverman N, Zhou R, Stöven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöven S, Ando I, Kadalayil L, Engström Y, Hultmark D. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, Maniatis T, Hultmark D. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc Natl Acad Sci USA. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Yin J, Starovasnik MA, Fairbrother WJ, Dixit VM. Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J Biol Chem. 2002;277:9505–9511. doi: 10.1074/jbc.M109488200. [DOI] [PubMed] [Google Scholar]
- Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CC. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, Kurata S. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci USA. 2002;99:13705–13710. doi: 10.1073/pnas.212301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li Y, Liu S, Yu X, Li L, Shi C, He W, Li J, Xu L, Hu Z, et al. Direct activation of RIP3/MLKL-dependent necrosis by herpes simplex virus 1 (HSV-1) protein ICP6 triggers host antiviral defense. Proc Natl Acad Sci USA. 2014;111:15438–15443. doi: 10.1073/pnas.1412767111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelièvre E, Gascan H, Ray KP, Morse MA, Imler JL, Gay NJ. Binding of the Drosophila cytokine Spätzle to Toll is direct and establishes signaling. Nat Immunol. 2003;4:794–800. doi: 10.1038/ni955. [DOI] [PubMed] [Google Scholar]
- Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, et al. Autophagic control of listeria through intracellular innate immune recognition in Drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Fu TM, Li J, Wu H. Structural biology of innate immunity. Annu Rev Immunol. 2015;33:393–416. doi: 10.1146/annurev-immunol-032414-112258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.