SUMMARY
Interactions between signaling pathways help guide plant development. In this study, we found that brassinosteroid (BR) signaling converges with SUPPRESSOR OF PHYTOCHROME B4-#3 (SOB3) to influence both the transcription of genes involved in cell elongation and hypocotyl growth. Specifically, SOB3 mutant hypocotyl phenotypes, which are readily apparent when the seedlings are grown in dim white light, were attenuated by treatment with either brassinolide (BL) or the BR biosynthesis inhibitor brassinazole (BRZ). Hypocotyls of SOB3 mutant seedlings grown in white light with a higher fluence rate also exhibited altered sensitivities to BL, further suggesting a connection to BR signaling. However, the impact of BL treatment on SOB3 mutants grown in moderate-intensity white light was reduced when polar auxin transport was inhibited. BL treatment enhanced transcript accumulation for all six members of the SMALL AUXIN UP RNA19 (SAUR19) subfamily, which promote cell expansion, are repressed by SOB3 and light, and are induced by auxin. Conversely, BRZ inhibited the expression of SAUR19 and its homologs. Expression of these SAURs was also enhanced in lines expressing a constitutively active form of the BR signaling component BZR1, further indicating that the transcription of SAUR19 subfamily members are influenced by this hormone signaling pathway. Taken together, these results indicate that SOB3 and BR signaling converge to influence the transcription of hypocotyl growth-promoting SAUR19 subfamily members.
Keywords: SOB3, AHL, brassinosteroids, SAUR, BZR1, Arabidopsis thaliana, auxin
INTRODUCTION
Living organisms rely on complex signaling pathways to translate both internal and external signals into developmental changes. The rate of elongation in seedlings is one such process that is heavily influenced by external factors, including light quantity and quality, temperature, and day length (Downs, 1955; Gray et al., 1998; Nozue et al., 2007; Niwa et al., 2009), as well as internal factors, such as signals from various hormone pathways (Su and Howell, 1995; Neff et al., 1999; Alabadi et al., 2004; Bai et al., 2012; Hornitschek et al., 2012; Chen et al., 2013; Jia et al., 2014; reviewed in Vandenbussche et al., 2005). Much of our current understanding of how seedling elongation is modulated at the molecular level comes from studies on the hypocotyl of Arabidopsis thaliana (reviewed in Arsovski et al., 2012; Boron and Vissenberg, 2014; Braidwood et al., 2014).
SUPPRESSOR OF PHYTOCHROME B4-#3 (SOB3)/AHL29 and other members of the AT-HOOK MOTIF NUCLEAR LOCALIZED (AHL) family are important for proper regulation of hypocotyl elongation (Street et al., 2008; Xiao et al., 2009; Zhao et al., 2013, 2014; Favero et al., 2016). Specifically, SOB3 and its closest homolog, ESCAROLA (ESC), inhibit hypocotyl elongation in a fluence rate-dependent manner (Street et al., 2008; Zhao et al., 2013). The sob3–4 esc-8 double null mutant elongates normally when grown in darkness or in light of higher fluence rates but exhibits a tall-hypocotyl phenotype when grown in dim white, red, far-red, or blue light (Street et al., 2008). Furthermore, the sob3–4 esc-8 mutant exhibits enhanced elongation even in the absence of functional phytochrome B (phyB) and cryptochrome 1 (cry1) photoreceptors in red and blue light, respectively, although phyA is necessary for the tall-hypocotyl phenotype in far-red light (Street et al., 2008).
Removal of additional family members besides SOB3 and ESC generally results in even taller light-grown seedlings (Zhao et al., 2013). Interestingly, when mutant AHLs unable to bind DNA are expressed at high levels in seedlings, such as in the case of the sob3–6 mutant, they behave in a dominant-negative fashion, conferring extremely tall-hypocotyl phenotypes (Street et al., 2008; Zhao et al., 2013). This has been explained based on the fact that AHLs interact with other family members, as well as other non-AHL DNA-binding proteins (Zhao et al., 2013). As SOB3–6 is unaffected in its ability to engage in protein–protein interactions, the defective protein is thought to still associate with other transcription factors (TFs) in the plant, whereby it inhibits DNA binding and relieves the normally repressive effects of these TF complexes on hypocotyl growth.
At least in Arabidopsis, hypocotyl growth occurs mainly due to cell expansion, rather than cell division (Gendreau et al., 1997). Auxin, brassinosteroid (BR), and gibberellin (GA) hormone pathways all play major roles in regulating cell expansion (reviewed in Depuydt and Hardtke, 2011). However, of these three hormones, only one of them, auxin, has been directly connected to SOB3 function (Favero et al., 2016). SOB3 inhibits the expression of genes associated with the auxin pathway, including YUCCA8 (YUC8) and members of the SMALL AUXIN UP RNA19 (SAUR19) subfamily (Favero et al., 2016). YUC genes code for flavin monooxygenases which catalyze a rate-limiting step during auxin biosynthesis in plants (Zhao et al., 2001; Mashiguchi et al., 2011; Phillips et al., 2011; Stepanova et al., 2011; Won et al., 2011; Dai et al., 2013). In contrast with YUC8, the six members of the SAUR19 subfamily, SAUR19 to SAUR24, function at the end of the hormone signaling pathway, where they link auxin signals to a specific developmental output, cell expansion (Jain et al., 2006; Franklin et al., 2011; Spartz et al., 2012, 2014). YUC8 and SAUR19 subfamily members have primarily been associated with temperature-induced hypocotyl elongation mediated by auxin (Franklin et al., 2011; Sun et al., 2012; Delker et al., 2014; Johansson et al., 2014; Bours et al., 2015; Ma et al., 2016). However, many different signaling pathways likely converge to influence the expression of these genes. For example, both YUC8 and SAUR19 subfamily members are also influenced at the transcriptional level by light (Hornitschek et al., 2012; Li et al., 2012a; Spartz et al., 2012; Hersch et al., 2014; Sun et al., 2016). Further, RNA sequencing (RNA-seq) data also indicate that the SAUR19 subfamily member expression is influenced by GA and BR hormone signaling pathways (Bai et al., 2012; Oh et al., 2012).
Although one member of the AHL family, AT-HOOK PROTEIN OF GA FEEDBACK1 (AGF1)/AHL25, has been implicated in negative feedback regulation of a gene coding for a GA 3-oxidase, which is involved in biosynthesis of this hormone (Matsushita et al., 2007), connections between this family of TFs and BRs have not yet been studied. However, such a connection is likely to exist, as RNA-seq data indicate that SAUR19 subfamily member transcription is influenced by BR signaling (Oh et al., 2012), and these genes are also directly repressed by SOB3 (Favero et al., 2016). Additionally, a connection between BRs and AHLs is further suggested by the fact that ESC/AHL27 and AHL12 interact physically with ATAF2 (Zhao et al., 2013). ATAF2 is a NAC transcription factor which promotes hypocotyl elongation by repressing the expression of PHYTOCHROME B ACTIVATION-TAGGED SUPPRESSOR1 (BAS1) and SOB7, two genes which code for BR inactivating enzymes (Neff et al., 1999; Turk et al., 2003, 2005; Peng et al., 2015). In this study, we test the hypothesis that SOB3 interacts with the BR pathway in the modulation of Arabidopsis hypocotyl growth. Our results indicate that BR signaling converges with SOB3 in hypocotyls of light-grown seedlings at the level of transcriptional regulation of the SAUR19 subfamily.
RESULTS
Brassinosteroid responses are altered in SOB3 mutants
In order to investigate if there is a connection between SOB3 and BRs, we generated dose–response curves for WT Col-0 and SOB3 mutants grown in the presence of exogenous brassinolide (BL), which generally promotes hypocotyl elongation in light-grown seedlings (Neff et al., 1999). Specifically, for this experiment, we chose two SOB3 mutants with opposite hypocotyl phenotypes, the short SOB3-D gain-of-function mutant and the tall sob3–6 mutant. Because SOB3 functions under red, far-red, and blue light (Street et al., 2008), we chose to use white light containing all three colors of light (see Experimental procedures) for this experiment and the rest of the study. The mutants both exhibited different responses to BL as compared with the WT (Figure 1a,b). SOB3-D was more sensitive to promotion of hypocotyl elongation by BL, while sob3–6 was less sensitive. This suggests that BRs are important for modulation of hypocotyl growth by SOB3. To further test this hypothesis, we also generated dose–response curves for all three genotypes grown in the presence of the BR biosynthesis inhibitor brassinazole (BRZ) (Asami et al., 2000). BRZ treatment had the opposite effect as compared with BL (Figure 1c,d). Hypocotyl elongation was inhibited by BRZ, and the inhibitory effect was less severe in SOB3-D as compared with the WT, but more severe in sob3–6. These results further suggest that there is a connection between SOB3 function and BRs in hypocotyls.
Figure 1. SOB3-D and sob3–6 exhibit opposite responses to exogenous BL and BRZ.
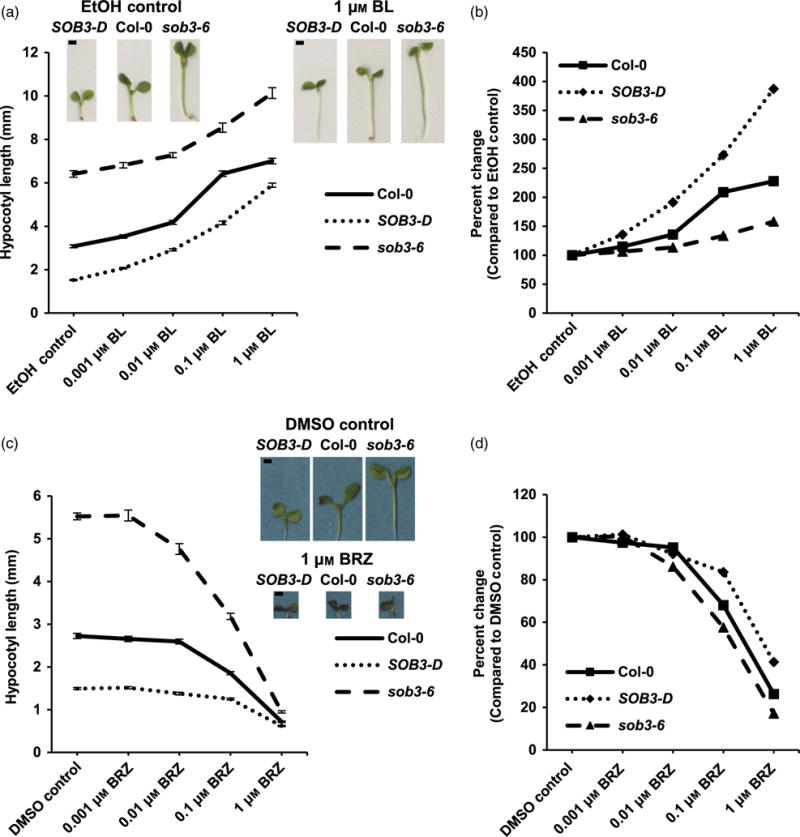
(a–d) Hypocotyl growth of Col-0, SOB3-D, and sob3–6 grown for 6 days in 23 μmol m−2 sec−1 white light on media containing the specified concentrations of brassinolide (BL). Values represent the mean of either the actual measured hypocotyl length (a) or the sensitivity to BL treatment (b) calculated as percent change in length compared with the same genotype on EtOH control plates. EtOH-Col-0, n = 39; SOB3-D, n = 41; sob3–6, n = 34. 0.001 μM BL-Col-0, n = 51; SOB3-D, n = 60; sob3–6, n = 51. 0.01 μM BL-Col-0, n = 54; SOB3-D, n = 53; sob3–6, n = 57. 0.1 μM BL-Col-0, n = 48; SOB3-D, n = 49; sob3–6, n = 45. 1 μM BL-Col-0, n = 41; SOB3-D, n = 49; sob3–6, n = 47.
(c, d) Hypocotyl growth of WT Col-0, SOB3-D, and sob3–6 grown for 6 days in 23 μmol m−2 sec−1 white light on media containing the specified concentrations of brassinazole (BRZ). DMSO-Col-0, n = 35; SOB3-D, n = 32; sob3–6, n = 43. 0.001 μM BRZ-Col-0, n = 37; SOB3-D, n = 44; sob3–6, n = 39. 0.01 μM BRZ-Col-0, n = 38; SOB3-D, n = 37; sob3–6, n = 36. 0.1 μM BRZ-Col-0, n = 39; SOB3-D, n = 41; sob3–6, n = 44. 1 μM BRZ-Col-0, n = 47; SOB3-D, n = 41; sob3–6, n = 42. Values represent the mean of either the actual measured hypocotyl length (c) or the sensitivity to BRZ treatment (d) calculated as percent change in length compared with the same genotype on dimethyl sulphoxide (DMSO) control plates. Error bars represent standard error of the mean. Photographs show seedlings of average length given the indicated genotype and plate type. Scale bars depict 1 mm. [Colour figure can be viewed at wileyonlinelibrary.com].
Although the dose–response curves generated for Col-0, SOB3-D, and sob3–6 grown in 23 μmol m−2 sec−1 on plates supplemented with BL or BRZ indicated that SOB3 likely influences BR metabolism and/or signaling, one could argue that these results are difficult to interpret accurately, because the hypocotyl lengths for all three genotypes are quite different on the control plates. Therefore, we investigated the effect of exogenous BL in conditions where hypocotyl lengths are much more similar between all three genotypes. First, we tested the effect of BL on seedlings grown in a higher fluence rate of white light, 140 μmol m−2 sec−1. Clear differences emerged when we compared the dose–response curves between the three genotypes (Figure 2). SOB3-D was less sensitive to BL-induced hypocotyl elongation in this light condition as compared with the WT, while sob3–6 was more sensitive. Therefore, as compared with the wild-type, BR signaling was reduced in SOB3-D but enhanced in sob3–6, further demonstrating that there is a connection between BRs and SOB3-mediated repression of hypocotyl growth. In addition, we generated BL dose–response curves for the three genotypes grown in the dark, as darkness also mostly abolishes defects in hypocotyl elongation for SOB3-D and sob3–6. In contrast with the situation observed in 140 μmol m−2 sec−1 white light, no clear pattern emerged for the three genotypes grown in the presence of BL in the dark (Figure S1).
Figure 2. SOB3-D and sob3–6 display different sensitivities to BL when grown under moderate-intensity white light.
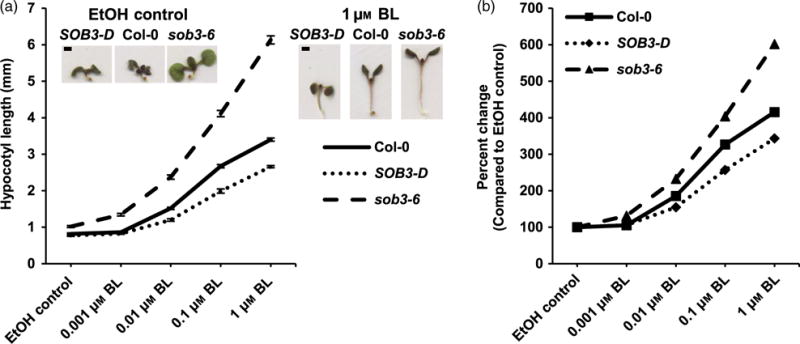
Hypocotyl growth of Col-0, SOB3-D, and sob3–6 grown for 6 days in 140 μmol m−2 sec−1 white light on media containing the specified concentrations of brassinolide (BL). EtOH-Col-0, n = 29; SOB3-D, n = 26; sob3–6, n = 34. 0.001 μM BL-Col-0, n = 31; SOB3-D, n = 33; sob3–6, n = 39. 0.01 μM BL Col-0, n = 40; SOB3-D, n = 27; sob3–6, n = 41. 0.1 μM BL-Col-0, n = 37; SOB3-D, n = 31; sob3–6, n = 39. 1 μM BL-Col-0, n = 38; SOB3-D, n = 41; sob3–6, n = 32.
(a, b) Values represent the mean of either the actual measured hypocotyl length (a) or the sensitivity to BL treatment (b) calculated as percent change in length compared with the same genotype on EtOH control plates. Error bars represent standard error of the mean. Photographs show seedlings of average length given the indicated genotype and plate type. Scale bars depict 1 mm. [Colour figure can be viewed at wileyonlinelibrary.com].
Auxin converges with brassinosteroid signaling to impact hypocotyl phenotypes in SOB3 mutants
SOB3 is known to repress hypocotyl elongation at least in part by inhibiting the auxin pathway (Favero et al., 2016). Multiple auxin and BR signaling components physically interact with each other, and TFs activated by these two different signaling pathways bind to the promoters of many shared target genes, synergistically inducing their transcription (Nemhauser et al., 2004; Vert et al., 2008; Oh et al., 2014; reviewed in Hardtke, 2007). Therefore, we hypothesized that the altered sensitivities of SOB3-D and sob3–6 to BL and BRZ might be related to SOB3′s impact on auxin signaling. In order to test this hypothesis, we investigated hypocotyl phenotypes for SOB3 mutant seedlings grown in 150 μmol m−2 sec−1 white light on plates containing 1 μM BL and various concentrations of the polar auxin transport inhibitor N-1-naphthylphthalamic acid (NPA). Previously, we found that defects in hypocotyl elongation observed for SOB3 mutants could be attenuated by growth in the presence of NPA (Favero et al., 2016). Therefore, we thought that NPA might also attenuate the BL response phenotypes observed for SOB3-D and sob3–6 seedlings grown under a moderate intensity of white light. Interestingly, we found that NPA effectively blocked the enhanced sensitivity of sob3–6 to BL (Figure 3). Most notably, at 200 μM NPA, there was almost no difference in hypocotyl length between Col-0 and sob3–6 seedlings grown in the presence of 1 μM BL under 150 μmol m−2 sec−1 white light. In addition, NPA attenuated the SOB3-D short-hypocotyl phenotype observed in the presence of BL under 150 μmol m−2 sec−1 white light. These results suggest that auxin signaling is important for the BR-associated phenotypes observed in the SOB3 mutants.
Figure 3. NPA reduces BL sensitivity differences in SOB3 mutant hypocotyls.
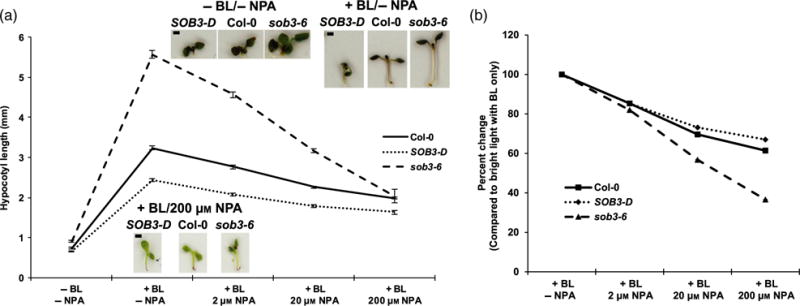
Hypocotyl growth of Col-0, SOB3-D, and sob3–6 grown for 6 days in 150 μmol m−2 sec−1 white light. ‘+ BL’ indicates that 1 μM brassinolide (BL) was included in the medium. EtOH/DMSO-Col-0, n = 41; SOB3-D, n = 44; sob3–6, n = 41. +BL/DMSO-Col-0, n = 59; SOB3-D, n = 53; sob3–6, n = 56. +BL/2 μM NPA-Col-0, n = 62; SOB3-D, n = 48; sob3–6, n = 63. +BL/20 μM NPA-Col-0, n = 56; SOB3-D, n = 47; sob3–6, n = 53. +BL/200 μM NPA-Col-0, n = 49; SOB3-D, n = 43; sob3–6, n = 37.
(a, b) Values represent the mean of either the actual measured hypocotyl length (a) or the sensitivity to NPA treatment (b) calculated as percent change in length compared with seedlings of the same genotype grown on +BL/DMSO plates. Error bars represent standard error of the mean. Photographs show seedlings of average length given the indicated genotype and plate type. Scale bars depict 1 mm. [Colour figure can be viewed at wileyonlinelibrary.com].
Brassinosteroid signaling promotes transcription of SOB3 target genes
One possible explanation for the results observed in our hypocotyl elongation assays performed in the presence of BL and NPA may be that BR signaling converges with auxin signaling and SOB3 to affect common downstream components. Specifically, we hypothesized that BR signaling could converge with auxin and SOB3 at the level of transcriptional control of members of the SAUR19 subfamily. The expression of SAUR19 subfamily members are influenced by both auxin (Spartz et al., 2012; Oh et al., 2014) and SOB3 (Favero et al., 2016), and there are numerous reports of SAUR gene transcription, particularly that of SAUR15, being influenced by brassinosteroids (Clouse et al., 1992; Zurek et al., 1994; Goda et al., 2002, 2004; Yin et al., 2002, 2005; Nakamura et al., 2003b; Nemhauser et al., 2004; Vert et al., 2008; Yan et al., 2009; Chung et al., 2012; Oh et al., 2012, 2014; Bittner et al., 2015). In order to test the hypothesis that BRs influence the expression of members of the SAUR19 subfamily, gene expression was examined in seedlings grown in the presence or absence of BL using real-time quantitative reverse transcription PCR (qRT-PCR) (Figures 4, S2 and S3). SAUR22 was chosen for initial qRT-PCR analysis because previous RNA-seq data indicate that this gene is one of the most misregulated members of the SAUR family in SOB3 mutants (Favero et al., 2016). In a preliminary experiment, we first checked SAUR22 expression in 4-day-old WT, SOB3-D, and sob3–6 seedlings grown in the presence or absence of BL under either a dim or moderate intensity of white light (Figure S2). Consistent with the tall-hypocotyl phenotypes caused by exogenous application of this hormone (Figure 1a,b), the expression of SAUR22 was higher in both WT and SOB3-D seedlings grown under dim white light in the presence of BL as compared with the EtOH control, although for Col-0, this increase was not quite statistically significant (P = 0.0553) (Figure S2a). In contrast, no increase in SAUR22 expression was observed for sob3–6 grown in the presence of BL in dim white light. This finding is not surprising considering that SAUR22 expression is already induced in sob3–6 compared with Col-0 (Figure S2a) (Favero et al., 2016), and this mutant exhibited reduced sensitivity to BL-induced hypocotyl elongation in dim light as compared with the WT (Figure 1a,b). SAUR22 expression was also enhanced in the presence of BL for all three genotypes grown in moderate-intensity white light (Figure S2b). These results indicate that BR signaling promotes SAUR22 transcription. Interestingly, in this experiment, we also observed that SAUR22 expression was reduced in SOB3-D in all four conditions tested and increased in sob3–6 in three of the conditions, lending further support to a previous study indicating that SOB3 influences the transcription of SAUR19 subfamily members (Figure S2) (Favero et al., 2016).
Figure 4. Expression levels of SOB3 target genes are affected by BL.
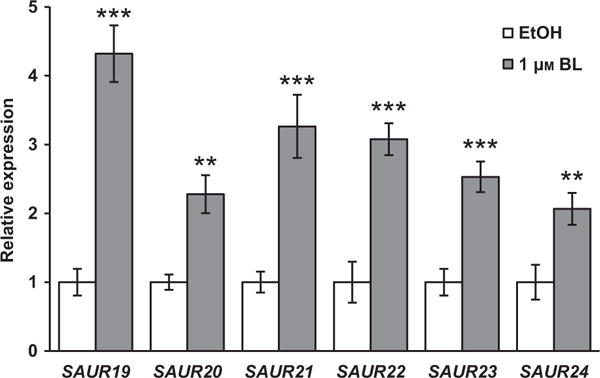
Relative expression of SAUR19 subfamily members, which are known targets of SOB3, in Col-0 4-day-old seedlings grown in 23 μmol m−2 sec−1 white light, as determined by qRT-PCR. Transcript levels are normalized based on the expression of the MDAR4 housekeeping gene. PCR was performed in duplicate and average expression values calculated and used for analysis. All values are shown as fold change compared with the EtOH control for the same gene. Error bars represent standard error of the mean from 11 (EtOH) or 10 (1 μM BL) biological replicates. In a Welch’s t-test (unpaired two-tailed t-test with unequal variance) compared with the EtOH control for the same gene **P < 0.01. ***P < 0.001. Note that a subset of the data for SAUR22 are also presented in Figure S2a.
In order to further evaluate if the expression of SAUR19 subfamily members are influenced by BRs, we generated a second set of cDNA samples from wild-type seedlings grown in the presence and absence of BL. Combining these cDNA samples with the previously generated set of samples for the wild-type, we performed qPCR for SAUR19 to 24, the six members of the SAUR19 subfamily (Jain et al., 2006; Spartz et al., 2012). Based on these results, we found that all six members of the SAUR19 subfamily were significantly induced in response to exogenous BL in dim white light (Figure 4). Similar results were achieved with seedlings grown under a moderate intensity of white light (Figure S3). These results indicate that BR promotes the expression of SAUR19 subfamily members.
We also tested if application of the BR biosynthesis inhibitor BRZ (Asami et al., 2000) influences the expression of SAUR19 to 24 (Figure 5). We found that the expression of all six SAURs was significantly lower in the seedlings grown on medium containing BRZ. These results indicate that inhibiting BR signaling represses the expression of SAUR19 to SAUR24, further indicating that the BR pathway promotes transcription of these genes. In contrast, expression of another known SOB3 target, YUC8 (Favero et al., 2016), was not significantly altered by BRZ application (Figure S4).
Figure 5. Effect of BRZ on SAUR expression.
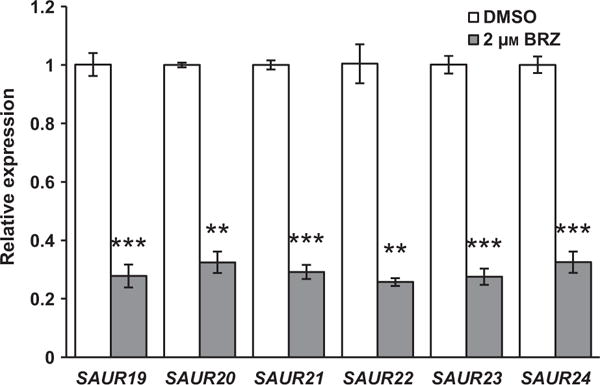
Relative expression of SAUR genes in the WT-like line (see Experimental procedures) grown in the presence or absence of the brassinosteroid biosynthesis inhibitor BRZ, as determined by qRT-PCR. Seedlings were grown in 23 μmol m−2 sec−1 white light. Transcript levels are normalized based on the expression of the MDAR4 housekeeping gene. PCR was performed in duplicate and average expression values calculated and used for analysis. Values are shown as fold change compared with the DMSO control for the same gene. Error bars represent standard error of the mean from three biological replicates. In a Welch’s t-test (unpaired two-tailed t-test with unequal variance) compared with the DMSO control for the same gene **P < 0.01. ***P < 0.001.
Data from a previous RNA-seq study indicate that the expression of SAUR19 subfamily members may be promoted by the transcription factor BRASSINAZOLE-RESISTANT1 (BZR1) (Oh et al., 2012), which is a major component of the BR signaling pathway (Wang et al., 2002). In order to test the hypothesis that BZR1 promotes the transcription of SAUR19 subfamily members in response to BR signals, we performed qRT-PCR using transgenic lines expressing a mutant, dominant allele of BZR1, denoted mBZR1 (He et al., 2002; Wang et al., 2002). BZR1 is normally phosphorylated and degraded in the absence of signals from the BR pathway, but a missense mutation in the mBZR1 protein confers reduced phosphorylation and enhanced stability. In turn, this leads to increased action of the mBZR1 transcription factor (He et al., 2005). We generated multiple T2 lines expressing mBZR1 and measured gene expression in these seedlings. First, we identified four lines that had significantly elevated levels of BZR1 transcripts, indicating that the mBZR1 construct was expressed in these seedlings (Figure 6a). We next examined the expression of SAUR19 to 24 in these four mBZR1-expressing lines (Figure 6b–g). SAUR19, SAUR21, SAUR22, and SAUR24 were significantly upregulated in all four mBZR1 lines (Figure 6b,d,e,g). SAUR20 and SAUR23 were significantly upregulated in three of the four lines, and nearly also significantly upregulated in the fourth line (SAUR20, P = 0.0628; SAUR23, P = 0.0524) (Figure 6c,f). Further, the expression levels of all six SAUR genes correlated well with that of BZR1 in the lines examined, as indicated by Pearson correlation coefficient (r) values ranging from 0.949 to 0.982 for the different members of the subfamily (Figure 6a–g). This likely indicates that BZR1 is at least partially responsible for inducing SAUR19 subfamily members in response to signals from the BR pathway. In contrast, YUC8 expression correlated less well with that of BZR1 (r = 0.807), and was not significantly altered in three of the four lines tested (Figure 6h).
Figure 6. BZR1 promotes SAUR transcription.
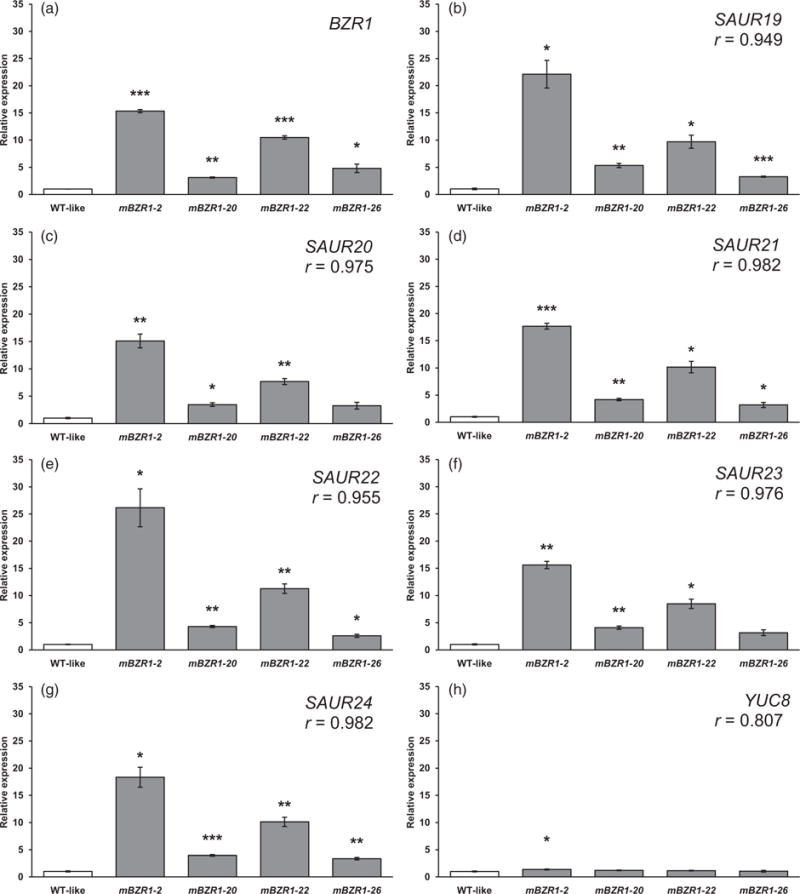
(a–h) Relative expression of (a) BZR1, (b) SAUR19, (c) SAUR20, (d) SAUR21, (e) SAUR22, (f) SAUR23, (g) SAUR24, and (h) YUC8 as determined by qRT-PCR, in T2 lines segregating for the mBZR1-GFP (mBZR1–2, etc.) construct or seedlings of the same genetic background lacking the construct (WT-like, see Experimental procedures). Previous work indicates that a C-terminal fluorescent protein tag does not interfere with BZR1 function (Wang et al., 2002). All seedlings were grown under 23 μmol m−2 sec−1 white light on plates containing 2 μM BRZ in order to degrade endogenous BZR1. Transcript levels are normalized based on the expression of the MDAR4 housekeeping gene. PCR was performed in duplicate and average expression values calculated and used for analysis. All values are shown as fold change compared with the WT-like samples for the same gene. Error bars represent standard error of the mean from three biological replicates. In a Welch’s t-test (unpaired two-tailed t-test with unequal variance) compared with the WT-like control for the same gene *P < 0.05. **P < 0.01. ***P < 0.001. Pearson correlation coefficient (r) values were calculated based on the expression levels of BZR1 and the indicated gene.
DISCUSSION
SOB3, BRs, and auxin influence SAUR transcription
Our findings indicate that SOB3 interacts with BRs in the modulation of hypocotyl growth (Figures 1, 2 and 3). Further, this is at least partially attributable to direct downstream targets of SOB3, members of the SAUR19 subfamily (Favero et al., 2016), being influenced at the transcriptional level by BR signaling (Figures 4, 5, 6b–g, S2 and S3). Considering these results in light of previous work, we propose a model for how SOB3, auxin, and BRs converge to influence hypocotyl growth (Figure 7). Hypocotyl growth-promoting SAUR19 subfamily members (Franklin et al., 2011; Spartz et al., 2012, 2014) are directly repressed by SOB3 (Favero et al., 2016). However, SOB3 also indirectly represses the expression of SAUR19 subfamily members, which are induced by auxin (Spartz et al., 2012; Oh et al., 2014), as it inhibits the transcription of YUC8 (Favero et al., 2016). YUC8 and other YUC genes code for a class of enzymes which play critical roles in auxin biosynthesis (Zhao et al., 2001; Mashiguchi et al., 2011; Phillips et al., 2011; Stepanova et al., 2011; Won et al., 2011; Dai et al., 2013). Results from the current study suggest that BRs converge with SOB3-mediated modulation of hypocotyl growth by promoting expression of SAUR19 subfamily members, via the transcription factor BZR1 (Figure 7).
Figure 7. Model for the convergence of SOB3 with brassinosteroids, auxin, and light in the modulation of hypocotyl elongation.
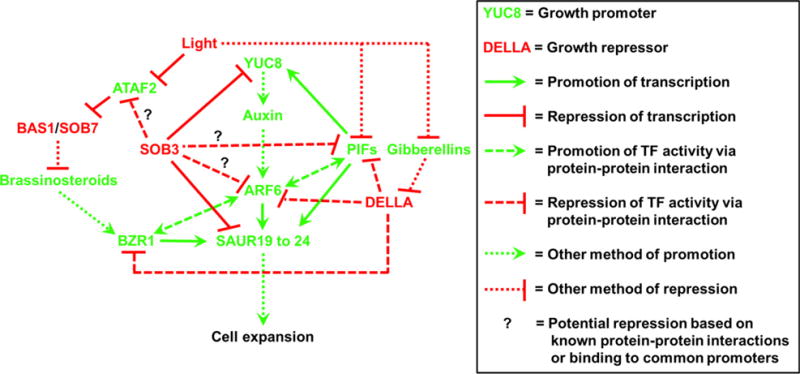
SOB3 represses hypocotyl elongation at least in part by directly inhibiting the transcription of the auxin biosynthetic gene YUC8 and the growth-promoting SAUR19 subfamily members. Based on our results here, we propose that BRs converge with SOB3 and auxin at the level of transcriptional control of SAUR19 to 24. BRs promote the expression of SAUR19 subfamily members via BZR1. BRs are deactivated by the photomorphogenesis-promoting BAS1 and SOB7 cytochrome P450s, and the genes encoding these enzymes are repressed at the transcriptional level by ATAF2. Light antagonizes the transcription of ATAF2 and the activity of PIFs, the latter which activate YUC8 expression. PIF4, ARF6, and BZR1 interact with each other and synergistically induce the expression of SAURs, while DELLAs repress the activating functions of these TFs. DELLAs are destabilized by GA signaling, while GA biosynthesis is inhibited by light.
In contrast with the SAUR19 subfamily, we found little evidence that YUC8 is regulated by BR signaling. Its expression was not significantly altered by BRZ application (Figure S4) or in the majority of mBZR1 lines evaluated (Figure 6h). This is not surprising when considering previous reports indicating that auxin biosynthesis is not influenced by BRs (Nakamura et al., 2003a; Nemhauser et al., 2004). Rather, it has been demonstrated that a BR signaling component, BES1, which is BZR1′s closest homolog, binds directly to the promoters of SAUR15 and SAUR68, suggesting that these genes can be directly regulated by this transcription factor in response to signals from the pathway (Nemhauser et al., 2004; Yin et al., 2005; Vert and Chory, 2006; Vert et al., 2008). Future work should test the hypothesis that BZR1 activates the expression of SAUR19 subfamily members directly, by binding to their promoters.
SOB3 and photomorphogenesis
Another interesting observation from this study is that SOB3 mutant phenotypes are largely dependent on light fluence rate (Figures 1a,c, 2a and 3a), in agreement with a previous study (Street et al., 2008). However, SOB3 function is not dependent on growth in the presence of a particular wavelength of light, and far-red light alone is sufficient to induce a tall-hypocotyl phenotype in the sob3–4 esc-8 double null mutant (Street et al., 2008). This indicates that the low fluence rate (LFR) response is not essential for SOB3 activity, because far-red light fails to activate this photomorphogenic response (reviewed in Casal et al., 1998). Conversely, since far-red light alone is sufficient to induce a phenotype in sob3–4 esc-8 (Street et al., 2008), it is possible that these genes act as downstream components in the very low fluence rate (VLFR) response (reviewed in Casal et al., 2014). However, this possibility seems unlikely, given that sob3–4 esc-8, which is in the Columbia-0 (Col-0) ecotype, exhibits a phenotype in red light. VLFR is mediated by phyA (reviewed in Casal et al., 2014), and the removal of this photoreceptor alone in Col-0 has no effect on hypocotyl growth in red light (Su et al., 2015). Therefore, in Col-0, inhibition of hypocotyl growth in red light is entirely attributable to LFR, rather than VLFR effects. Furthermore, although no hypocotyl phenotype was observed for sob3–4 esc-8 in a phyA mutant background in far-red light (Street et al., 2008), this is explainable by the fact that this photoreceptor alone is predominantly responsible for mediating de-etiolation in response to far-red light (Neff and Chory, 1998). Therefore, phyA mutants grown in far-red light lack an active photomorphogenic signaling pathway necessary for manifestation of the sob3–4 esc-8 tall-hypocotyl phenotype, similar to the situation in dark-grown seedlings (Street et al., 2008). Taken together, these observations indicate that SOB3 likely functions as a growth repressor downstream of multiple photoreceptors, and its function does not depend specifically on either VLFR or LFR.
One likely reason SOB3 functions in a fluence rate-dependent manner is because the expression of YUC8 and SAUR19 subfamily members are influenced by light (Hornitschek et al., 2012; Li et al., 2012a; Spartz et al., 2012; Hersch et al., 2014; Sun et al., 2016). PHYTOCHROME INTERACTING FACTOR (PIF) TFs, which are generally destabilized by light-activated phytochromes (Ni et al., 1998; Huq and Quail, 2002; Lorrain et al., 2008; reviewed in de Lucas and Prat, 2014), regulate YUC8 expression in response to light (Figure 7) (Hornitschek et al., 2012; Li et al., 2012a; Hersch et al., 2014). YUC8 expression is substantially elevated during shade avoidance, i.e. in light with a low red to far-red (R:FR) ratio, an effect which is mediated largely by PIF7 (Li et al., 2012a; Hersch et al., 2014). In contrast, there is evidence that PIF4 and PIF5 activate YUC8 in response to low fluence-rate light when the R:FR ratio is higher (Hornitschek et al., 2012). Similar to YUC8, the expression of SAUR19, 21, 23, and 24 is induced by light with a low ratio of R:FR (Spartz et al., 2012), and it is likely that these changes in expression are caused at least partially by increased auxin levels in shade-avoiding seedlings (Li et al., 2012a; Hersch et al., 2014). However, chromatin immunoprecipitation sequencing (ChIP-seq) data indicate that both PIF4 and PIF5 bind in the vicinity of all six members of the SAUR19 subfamily (Hornitschek et al., 2012; Oh et al., 2012), and, therefore, that these SAURs are also direct downstream targets of PIFs (Figure 7). Although PIFs were named based on their ability to interact with phytochromes (Ni et al., 1998), cryptochromes also interact with these TFs, altering their ability to influence target gene transcription (Ma et al., 2016; Pedmale et al., 2016). Therefore, it seems likely that SAUR transcription is influenced by both red and far-red light, through the phytochromes, as well as by blue light, via the cryptochromes. Future studies directly testing how the expression of SAUR19 subfamily members is influenced by monochromatic red, far-red, and blue light, as well as if it is altered in phytochrome and cryptochrome mutants, will be useful for confirming this hypothesis.
The connection established in this study between the BR signaling pathway and the expression of SAUR19 subfamily members (Figures 4, 5 and 6b–g) provides another indirect route by which light may affect SAUR transcription (Figure 7). ATAF2 transcription correlates inversely with light intensity, and the TF encoded by this gene promotes hypocotyl elongation in a fluence rate-dependent manner by repressing BAS1 and SOB7 expression (Peng et al., 2015). This indicates that ATAF2 inhibits BR inactivation, while light antagonizes this effect. Therefore, BZR1-mediated SAUR induction through the BR signaling pathway should also be antagonized by light, particularly that of higher fluence rates (Figure 7).
Interestingly, BZR1 interacts physically with PIF4 as well as a transcription factor which is activated by the auxin signaling pathway, AUXIN RESPONSE FACTOR6 (ARF6) (Nagpal et al., 2005). Together, the three TFs synergistically activate the expression of SAUR genes, including SAUR19 (Figure 7) (Oh et al., 2014). SAUR19 subfamily members contain a number of putative binding sites for all three TFs in their promoters, with many of these located near AHL-binding motifs (Favero et al., 2016). Therefore, activation of SAUR19 subfamily members via the BZR1/ARF6/PIF4 (BAP) complex (Oh et al., 2014) may be directly inhibited by SOB3 (Figure 7). Future studies should investigate if SOB3 antagonizes BAP function by physically disrupting the assembly of the complex on the promoters of target genes. This mode of regulation would not be without precedent, as DELLA repressor proteins physically interfere with the formation of BAP-DNA complexes by disrupting PIF4-ARF6 interactions as well as by blocking BZR1, ARF6 and PIF4 from binding to target promoters (de Lucas et al., 2008; Bai et al., 2012; Li et al., 2012b; Oh et al., 2014). DELLAs are themselves destabilized by activation of the GA signaling pathway (Dill et al., 2001). GA levels are reduced in light, where DELLAs promote photomorphogenesis in a fluence rate-dependent manner (Achard et al., 2007). This presents another route by which light should impact the expression of SAUR19 subfamily members (Figure 7), a hypothesis which is supported by RNA-seq data indicating that SAUR19, SAUR20, SAUR21, SAUR23, and SAUR24 are all induced by a 12-h GA treatment (Bai et al., 2012). Interestingly, another AHL known to influence hypocotyl growth, AHL6 (Zhao et al., 2013), interacts with the DELLA protein RGA1 based on results from a blind, high-throughput yeast two-hybrid (Y2H) assay (Stark et al., 2006; Arabidopsis Interactome Mapping Consortium, 2011). This suggests that DELLAs may physically interact with AHL complexes containing SOB3, potentially leading to the formation of large repressor complexes which antagonize the activation of genes induced by BAP. This possibility should be investigated in future studies.
Finally, TCP TFs may be important for the effects of both SOB3 and light on SAUR19 subfamily member expression. SOB3 physically interacts with TCPs, and the sob3–6 tall-hypocotyl phenotype is completely abolished in a mutant exhibiting reduced levels of several TCPs (Zhao et al., 2013). Additionally, several SAUR genes are misregulated in TCP mutants (Koyama et al., 2010; Sarvepalli and Nath, 2011; Danisman et al., 2013), and TCP2 protein levels are influenced by light (He et al., 2016). Future experiments should elucidate precisely how SOB3–TCP interactions influence the expression of SAUR19 to SAUR24.
Alternative explanations for the altered sensitivities of SOB3-D and sob3–6 to BL and BRZ
Although our gene expression analyses suggest that the physiological results from this study are at least partially explained by the convergence of SOB3, auxin, and BRs at the level of transcriptional regulation of SAUR19 subfamily members, there are other potential explanations for the altered sensitivities of the SOB3 mutant hypocotyls to BL and BRZ. For example, SOB3 could reduce BR levels by associating with ATAF2-containing complexes and relieving repression of BAS1 and SOB7 by this TF (Figure 7) (Peng et al., 2015). This idea is consistent with the current model in which AHLs are believed to modulate hypocotyl growth by functioning as components of large complexes composed of multiple family members and other non-AHL TFs (Zhao et al., 2013). Furthermore, ESC and AHL12 physically interact with ATAF2 based on Y2H assays (Zhao et al., 2013). Future studies should investigate if interactions between AHLs and ATAF2 are important for photomorphogenesis, as well as if they contribute to the BL- and BRZ-response phenotypes observed in this study. Additionally, another potential explanation for the physiological results obtained in this study, which we cannot rule out entirely, is that SOB3 directly represses the expression of genes encoding enzymes which are involved more directly in promoting hypocotyl elongation, e.g. expansins and pectin methylesterases (reviewed in Braidwood et al., 2014). An in-depth transcriptomics study using RNA extracted from multiple AHL mutants at different stages of seedling development would be informative for testing this hypothesis.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
All plants used in this study are in the Col-0 ecotype. The SOB3-D and sob3–6 mutants were described previously (Street et al., 2008; Zhao et al., 2013). Seeds were sown on half-strength LS medium with 1.5% sucrose and 1% Phytagel (Sigma, St. Louis, MO, USA). For assays with brassinolide (BL), brassinazole (BRZ), or N-1-naphthylphthalamic acid (NPA), the compound was added in solution to the medium to achieve the specified concentration, with the amount of solvent held constant among all plates used for the same experiment. In order to synchronize germination, seeds were incubated in the dark at 4°C for about 4 days, then transferred to red light and 25°C for 12 h, prior to being grown in a chamber. Seedlings were grown in an E-30B (Percival Scientific, Perry, IA, USA) growth chamber, where both fluorescent and incandescent bulbs were used to supply continuous white light at the specified fluence rate. All seedlings were grown at 25°C.
Hypocotyl measurements
Six-day-old seedlings were harvested for hypocotyl measurements. A ScanJet3500 (Hewlett-Packard, Palo Alto, CA, USA) or a Perfection V600 Photo (Epson, Nagano, Japan) flat-bed scanner was used to generate TIFF or JPEG format images of the seedlings. National Institutes of Health (NIH) ImageJ 1.48 (Schneider et al., 2012) was used to measure hypocotyl lengths and data were analyzed and graphs generated using Excel (Microsoft, Redmond, WA, USA) software.
qRT-PCR
RNA was extracted from 4-day-old seedlings using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). During extraction, genomic DNA contamination was eliminated via the On-Column DNase I Digestion Set (Sigma). First strand cDNA synthesis was conducted using the iScript Reverse Transcription Supermix for RT-qPCR (BioRad, Hercules, CA, USA). Quantitative PCR was carried out using the SsoAdvanced Universal SYBR Green Supermix (BioRad) in a CFX96 Touch Real-Time PCR Detection System (BioRad). Primers used for amplification are as follows: BZR1, 5′TTATCGCAAGGGATGCAAGC-3′ and 5′-GGGCTCTGGTTCTGTGATGA-3′; MDAR4, 5′-GCGGTGGCTATATCGGTATGG-3 and 5′-AAAGAGACGTGCCATGCAGTG-3′; SAUR19, 5′-GCTCTCATACTTGAGCCAACCG-3′ and 5′-AGGGATCGTTAAGCCACCCATC-3′; SAUR20, 5′-CGATCATCCAATGGGTGGCT-3′ and 5′-GCTCATCATCGTTGGAACCG-3′; SAUR21, 5′-GCTCTCATACTTGAGCCAACCTTC-3′ and 5-GGGATCGTTAAGCCACCCATTG-3′; SAUR22, 5′-TGTGACTTCTCGGCTCCAAT-3′ and 5′-CAAAAATGGCATCCATTCTCTAAAC-3′; SAUR23, 5-AATCCGAAGAAGAGTTTGGGTTCG-3′ and 5′-AATCATCAATGGAGCCGAGAAGTC-3′; SAUR24, 5′-ACCAGCCTTCATTTCAAGCTCTTC-3′ and 5′-AGGGATCGTTAAGCCTCCCATC-3′; YUC8, 5′-GAGGAAAGGGCTCTCAGGTG-3′ and 5′-GAAGAGAACCCCTTGAGCGT-3′. Excel (Microsoft) software was used to analyze and compare data using the DDCT method.
Generation of mBZR1 Lines
The BZR1 coding sequence was amplified using primers (5′-CCTAGGCCGTCAAGGCCAATGACTTCGGATGGAGCTACGTC-3′ and 5′-CTATGGCCCATGAGGCCTCAACCACGAGCCTTCCCATTTC-3′) and cloned into the pENTR223.1-Sfi vector. The QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) was used to generate the previously described mBZR1 allele, which contains a proline to leucine mutation at residue 234 (Wang et al., 2002), using the primer pair (5′ATTCAGGTATAGTAGCCAGGGTATGAAACTGGTGG-3′ and 5′CCACCAGTTTCATACCCTGGCTACTATACCTGAAT-3′). Subsequently, the presence of the desired mutation and absence of other errors in the BZR1 coding sequence was confirmed via sequencing. PCR was then performed to amplify mBZR1 using primers (5′CAGTTCGCTAGCATGACTTCGGATGGAGCTACGTC-3′ and 5′-TGTGTCGGCGCGCCAACCACGAGCCTTCCCATTTCC-3′). The resulting PCR product was cleaved with AscI and NheI and ligated into pMDC83 cleaved with AscI and SpeI, 5′ of and in frame with GFP. The resulting plasmid was sequenced to confirm the absence of errors, then cloned into Agrobacterium, which was subsequently used to transform the ‘WT-like’ line, homozygous for a ProSOB3: SOB3-FLAG construct in the previously described sob3–4 esc-8 ahl6 triple null (Zhao et al., 2013), using the floral dip method (Clough and Bent, 1998). Successful transformants were identified by screening on LS plates containing 25 ng ml−1 Hygromycin B (PhytoTechnology Laboratories, Lenexa, KS, USA). Single-locus insertion lines were identified by growing T2 seedlings on selection plates, scoring the number of resistant and susceptible individuals, and selecting lines which segregated at a 3:1 ratio based on chi-square analysis.
Supplementary Material
Figure S1. Response of SOB3 mutants to BL in the dark.
Figure S2. Expression levels of SAUR22 are induced by brassinos-teroids in WT, SOB3-D, and sob3–6.
Figure S3. Expression levels of SOB3 target genes are affected by BL in seedlings grown under moderate-intensity white light.
Figure S4. Effect of BRZ on YUC8 expression.
Acknowledgments
We thank Caitlin Jacques for help with extracting RNA samples. We also thank Dr. Jianfei Zhao for help with generating the WT-like line. This project was supported by the Agriculture and Food Research Initiative competitive grant #2013-67013-21666 of the USDA National Institute of Food and Agriculture (to M.M.N.), the USDA National Institute of Food and Agriculture, HATCH project # 1007178 (to M.M.N.) and the Brubbaken and Reinbold Monocot Breeding Fund (to M.M.N.). This project was also supported by the Global Plant Sciences Initiative Research Fellowship (Washington State University, to D.S.F.).
Footnotes
ACCESSION NUMBERS
Arabidopsis Genome Initiative numbers for the sequences used in this study are as follows: BZR1 (AT1G75080), MDAR4 (AT3G27820), SAUR19 (AT5G18010), SAUR20 (AT5G18020), SAUR21 (AT5G18030), SAUR22 (AT5G18050), SAUR23 (AT5G18060), SAUR24 (AT5G18080), YUC8 (AT4G28720).
The authors declare no conflict of interest.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
References
- Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, Harberd NP. DELLAs contribute to plant photomorphogenesis. Plant Physiol. 2007;143:1163–1172. doi: 10.1104/pp.106.092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadi D, Gil J, Blazquez MA, Garcia-Martinez JL. Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 2004;134:1050–1057. doi: 10.1104/pp.103.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Galstyan A, Guseman JM, Nemhauser JL. Photomorphogenesis. Arabidopsis Book. 2012;10:e0147. doi: 10.1199/tab.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S. Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 2000;123:93–99. doi: 10.1104/pp.123.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol. 2012;14:810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner T, Nadler S, Schulze E, Fischer-Iglesias C. Two homolog wheat Glycogen Synthase Kinase 3/SHAGGY – like kinases are involved in brassinosteroid signaling. BMC Plant Biol. 2015;15:247. doi: 10.1186/s12870-015-0617-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boron AK, Vissenberg K. The Arabidopsis thaliana hypocotyl, a model to identify and study control mechanisms of cellular expansion. Plant Cell Rep. 2014;33:697–706. doi: 10.1007/s00299-014-1591-x. [DOI] [PubMed] [Google Scholar]
- Bours R, Kohlen W, Bouwmeester HJ, van der Krol A. Thermoperiodic control of hypocotyl elongation depends on auxin-induced ethylene signaling that controls downstream PHYTOCHROME INTERACTING FACTOR3 activity. Plant Physiol. 2015;167:517–530. doi: 10.1104/pp.114.254425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braidwood L, Breuer C, Sugimoto K. My body is a cage: mechanisms and modulation of plant cell growth. New Phytol. 2014;201:388–402. doi: 10.1111/nph.12473. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Sanchez RA, Botto JF. Modes of action of phytochromes. J Exp Bot. 1998;49:127–138. [Google Scholar]
- Casal JJ, Candia AN, Sellaro R. Light perception and signalling by phytochrome A. J Exp Bot. 2014;65:2835–2845. doi: 10.1093/jxb/ert379. [DOI] [PubMed] [Google Scholar]
- Chen J, Sonobe K, Ogawa N, Masuda S, Nagatani A, Kobayashi Y, Ohta H. Inhibition of Arabidopsis hypocotyl elongation by jasmonates is enhanced under red light in phytochrome B dependent manner. J Plant Res. 2013;126:161–168. doi: 10.1007/s10265-012-0509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Choe V, Fujioka S, Takatsuto S, Han M, Jeon JS, Park YI, Lee KO, Choe S. Constitutive activation of brassinosteroid signaling in the Arabidopsis elongated-D/bak1 mutant. Plant Mol Biol. 2012;80:489–501. doi: 10.1007/s11103-012-9963-5. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Zurek DM, McMorris TC, Baker ME. Effect of brassinolide on gene expression in elongating soybean epicotyls. Plant Physiol. 1992;100:1377–1383. doi: 10.1104/pp.100.3.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Mashiguchi K, Chen Q, Kasahara H, Kamiya Y, Ojha S, DuBois J, Ballou D, Zhao Y. The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J Biol Chem. 2013;288:1448–1457. doi: 10.1074/jbc.M112.424077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S, van Dijk AD, Bimbo A, van der Wal F, Hennig L, de Folter S, Angenent GC, Immink RG. Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J Exp Bot. 2013;64:5673–5685. doi: 10.1093/jxb/ert337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, Sonntag L, James GV, et al. The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 2014;9:1983–1989. doi: 10.1016/j.celrep.2014.11.043. [DOI] [PubMed] [Google Scholar]
- Depuydt S, Hardtke CS. Hormone signalling crosstalk in plant growth regulation. Curr Biol. 2011;21:R365–R373. doi: 10.1016/j.cub.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Dill A, Jung HS, Sun TP. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA. 2001;98:14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs RJ. Photoreversibility of leaf and hypocotyl elongation of dark grown red kidney bean seedlings. Plant Physiol. 1955;30:468–473. doi: 10.1104/pp.30.5.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favero DS, Jacques CN, Iwase A, Le KN, Zhao J, Sugimoto K, Neff MM. SUPPRESSOR OF PHYTOCHROME B4-#3 represses genes associated with auxin signaling to modulate hypocotyl growth. Plant Physiol. 2016;171:2701–2716. doi: 10.1104/pp.16.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Hofte H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997;114:295–305. doi: 10.1104/pp.114.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2002;130:1319–1334. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS. Transcriptional auxin-brassinosteroid crosstalk: who’s talking? BioEssays. 2007;29:1115–1123. doi: 10.1002/bies.20653. [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Zhao X, Kong F, Zuo Z, Liu X. TCP2 positively regulates HY5/HYH and photomorphogenesis in Arabidopsis. J Exp Bot. 2016;67:775–785. doi: 10.1093/jxb/erv495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch M, Lorrain S, de Wit M, Trevisan M, Ljung K, Bergmann S, Fankhauser C. Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:6515–6520. doi: 10.1073/pnas.1320355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71:699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa) Genomics. 2006;88:360–371. doi: 10.1016/j.ygeno.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Jia KP, Luo Q, He SB, Lu XD, Yang HQ. Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in Arabidopsis. Mol Plant. 2014;7:528–540. doi: 10.1093/mp/sst093. [DOI] [PubMed] [Google Scholar]
- Johansson H, Jones HJ, Foreman J, Hemsted JR, Stewart K, Grima R, Halliday KJ. Arabidopsis cell expansion is controlled by a photothermal switch. Nat Commun. 2014;5:4848. doi: 10.1038/ncomms5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell. 2010;22:3574–3588. doi: 10.1105/tpc.110.075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012a;26:785–790. doi: 10.1101/gad.187849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QF, Wang C, Jiang L, Li S, Sun SS, He JX. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci Signal. 2012b;5:ra72. doi: 10.1126/scisignal.2002908. [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Prat S. PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 2014;202:1126–1141. doi: 10.1111/nph.12725. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA. 2016;113:224–229. doi: 10.1073/pnas.1511437113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, et al. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:18512–18517. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita A, Furumoto T, Ishida S, Takahashi Y. AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol. 2007;143:1152–1162. doi: 10.1104/pp.106.093542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S. Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 2003a;133:1843–1853. doi: 10.1104/pp.103.030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Shimada Y, Goda H, Fujiwara MT, Asami T, Yoshida S. AXR1 is involved in BR-mediated elongation and SAUR-AC1 gene expression in Arabidopsis. FEBS Lett. 2003b;553:28–32. doi: 10.1016/s0014-5793(03)00945-1. [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 1998;118:27–36. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, et al. BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Niwa Y, Yamashino T, Mizuno T. The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 2009;50:838–854. doi: 10.1093/pcp/pcp028. [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife. 2014;3:e03031. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Huang SS, Zander M, et al. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell. 2016;164:233–245. doi: 10.1016/j.cell.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Zhao J, Neff MM. ATAF2 integrates Arabidopsis brassinosteroid inactivation and seedling photomorphogenesis. Development. 2015;142:4129–4138. doi: 10.1242/dev.124347. [DOI] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P. vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell. 2011;23:550–566. doi: 10.1105/tpc.110.075267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K, Nath U. Interaction of TCP4-mediated growth module with phytohormones. Plant Signal Behav. 2011;6:1440–1443. doi: 10.4161/psb.6.10.17097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inze D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012;70:978–990. doi: 10.1111/j.1365-313X.2012.04946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell. 2014;26:2129–2142. doi: 10.1105/tpc.114.126037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 2011;23:3961–3973. doi: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street IH, Shah PK, Smith AM, Avery N, Neff MM. The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J. 2008;54:1–14. doi: 10.1111/j.1365-313X.2007.03393.x. [DOI] [PubMed] [Google Scholar]
- Su W, Howell SH. The effects of cytokinin and light on hypocotyl elongation in Arabidopsis seedlings are independent and additive. Plant Physiol. 1995;108:1423–1430. doi: 10.1104/pp.108.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Hou P, Song M, Zheng X, Guo L, Xiao Y, Yan L, Li W, Yang J. Synergistic and antagonistic action of phytochrome (Phy) A and PhyB during seedling de-etiolation in Arabidopsis thaliana. Int J Mol Sci. 2015;16:12199–12212. doi: 10.3390/ijms160612199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Wang J, Gao Z, Dong J, He H, Terzaghi W, Wei N, Deng XW, Chen H. Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA. 2016;113:6071–6076. doi: 10.1073/pnas.1604782113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Denzel MA, Torres QI, Neff MM. CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogene-sis and plant steroid signal transduction. Plant Physiol. 2003;133:1643–1653. doi: 10.1104/pp.103.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, et al. BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J. 2005;42:23–34. doi: 10.1111/j.1365-313X.2005.02358.x. [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Verbelen JP, Van Der Straeten D. Of light and length: regulation of hypocotyl growth in Arabidopsis. BioEssays. 2005;27:275–284. doi: 10.1002/bies.20199. [DOI] [PubMed] [Google Scholar]
- Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105:9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:18518–18523. doi: 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Chen F, Yu X, Lin C, Fu YF. Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol. 2009;71:39–50. doi: 10.1007/s11103-009-9507-9. [DOI] [PubMed] [Google Scholar]
- Yan Z, Zhao J, Peng P, Chihara RK, Li J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 2009;150:710–721. doi: 10.1104/pp.109.138099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- Zhao J, Favero DS, Peng H, Neff MM. Arabidopsis thaliana AHL family modulates hypocotyl growth redundantly by interacting with each other via the PPC/DUF296 domain. Proc Natl Acad Sci USA. 2013;110:E4688–E4697. doi: 10.1073/pnas.1219277110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Favero DS, Qiu J, Roalson EH, Neff MM. Insights into the evolution and diversification of the AT-hook Motif Nuclear Localized gene family in land plants. BMC Plant Biol. 2014;14:266. doi: 10.1186/s12870-014-0266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek DM, Rayle DL, McMorris TC, Clouse SD. Investigation of gene expression, growth kinetics, and wall extensibility during brassinosteroid-regulated stem elongation. Plant Physiol. 1994;104:505–513. doi: 10.1104/pp.104.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Response of SOB3 mutants to BL in the dark.
Figure S2. Expression levels of SAUR22 are induced by brassinos-teroids in WT, SOB3-D, and sob3–6.
Figure S3. Expression levels of SOB3 target genes are affected by BL in seedlings grown under moderate-intensity white light.
Figure S4. Effect of BRZ on YUC8 expression.


