Abstract
Background
Left ventricular non-compaction (LVNC) is a genetically and phenotypically heterogeneous disease and, although increasingly recognized in clinical practice, there is a lack of widely accepted diagnostic criteria. We sought to identify novel genetic causes of LVNC and describe genotype-phenotype correlations.
Methods and Results
190 patients from 174 families with left ventricular hypertrabeculation (LVHT) or LVNC were referred for cardiac magnetic resonance and whole exome sequencing. 425 control individuals were included to identify variants of interest (VOIs). We found an excess of 138 VOIs in 102 (59%) unrelated patients in 54 previously identified LVNC or other known cardiomyopathy genes. VOIs were found in 68 of 90 probands with LVNC and 34 of 84 probands with LVHT (76% and 40%, respectively; p<0.001). We identified 0, 1, and ≥2 VOIs in 72, 74, and 28 probands, respectively. We found increasing number of VOIs in a patient strongly correlated with several markers of disease severity, including ratio of non-compacted to compacted myocardium (p<0.001) and left ventricular ejection fraction (p=0.01). The presence of sarcomeric gene mutations was associated with increased occurrence of late gadolinium enhancement (p=0.004).
Conclusions
LVHT and LVNC likely represent a continuum of genotypic disease with differences in severity and variable phenotype explained, in part, by the number of VOIs and whether mutations are present in sarcomeric or non-sarcomeric genes. Presence of VOIs is common in patients with LVHT. Our findings expand the current clinical and genetic diagnostic approaches for patients with LVHT and LVNC.
Keywords: genetics, human, noncompaction cardiomyopathy, left ventricular noncompaction, oligogenics
Introduction
Left ventricular non-compaction (LVNC) is a disorder of the heart characterized by a two-layered myocardium consisting of compacted and non-compacted segments, prominent ventricular trabeculations, and intertrabecular recesses.1 LVNC is increasingly recognized in clinical practice with a reported prevalence of 1:5000 individuals in the general population and 3-4% of adults with heart failure, although this is likely an underestimate.2 Patients with LVNC are at increased risk to develop heart failure, atrial and ventricular arrhythmias, and systemic embolic events, both in pediatric and adult populations. There is an ongoing debate about whether LVNC can be labeled as a distinct cardiomyopathy or simply as a trait shared by other primary cardiomyopathies.3-7 The American Heart Association classified LVNC as a distinct primary cardiomyopathy.8 However, the European Society of Cardiology has designated LVNC as an “unclassified cardiomyopathy” and does not take a firm stance on whether LVNC is a separate cardiomyopathy or a morphological trait.9 One of the key issues underpinning this debate is the paucity of data regarding specific genotype–phenotype correlations.10, 11 Numerous echocardiographic criteria have been proposed for diagnosing LVNC. However, poor correlation between these criteria has been reported.12 Cardiac magnetic resonance (CMR) depicts appearance of the myocardium with excellent quality and its role in diagnosing LVNC is increasing.13-15 LVNC is currently thought to be a genetically heterogeneous, monogenic disease with autosomal dominant, autosomal recessive, and X-linked forms reported.16-18 The mechanism responsible for the LVNC phenotype remains unclear but is thought to be related to abnormal cardiac embryogenesis with the majority of cases being familial or genetically triggered.19-22 The LVNC phenotype has been linked to mutations in sarcomeric genes in 30% of cases and Z-disc, cytoskeletal, and mitochondrial genes in the remainder.3,16,23-25
Herein, we describe the largest cohort of consecutive subjects with left ventricular hypertrabeculation (LVHT) or LVNC undergoing CMR and whole-exome sequencing (WES) in an attempt to better correlate genotype and phenotype for this patient population.
Methods
Patients
Between January 2013 and December 2013 we prospectively enrolled 190 patients from 174 families with suspected or known LVNC on the basis of clinical presentation (history, symptoms, electrocardiogram, presence of neuromuscular disease, and familial occurrence of LVNC) and transthoracic echocardiographic documentation of a distinct two-layered structure of non-compacted and compacted left ventricular (LV) myocardium. In an attempt to capture patients with borderline expression of the disease, no arbitrary echocardiographic threshold for LVNC diagnosis was applied. Only patients ≥7 years of age with the ability to hold their breath were included in the study. The study was approved by the local institutional ethics committees (IRB), and written informed consent was obtained from each subject and assent from pediatric subjects.
Clinical data collection
At initial presentation, the following clinical characteristics were collected: 1) presence and nature of cardiac symptoms, 2) reason for referral, 3) presence of concomitant neuromuscular disorders, and 4) family history. Previous medical history was reviewed on all subjects. The initial clinical evaluation was performed without knowledge of genotype status.
Cardiac magnetic resonance imaging
All CMR examinations were performed on a 1.5T clinical scanner (Signa HDxt, GE Healthcare, Milwaukee, WI) with standard acquisition methods to acquire cine and delayed enhancement images. Images were analyzed with commercial software (Qmass MR, Medis Medical Imaging Systems, Leiden, the Netherlands) by an experienced observer blinded to the patients' profile. The 17-segment model of LV segmentation was used26 and the presence of any degree of trabeculation/non-compaction was assessed in every LV segment. A segment was considered as non-compacted if a distinct two-layered appearance of trabeculated and compacted myocardium could be visually identified. For the segment with the most pronounced trabeculations, the maximal ratio of non-compacted to compacted myocardium (NC/C) was calculated. According to generally accepted CMR criteria proposed by Petersen et al.,13 NC/C ratio greater than 2.3:1 was considered diagnostic for LVNC. Subjects that failed to meet these criteria but were otherwise suspected of having LVNC were labeled as having LVHT. Details regarding imaging protocol and data analysis are provided in the Supplementary Appendix.
DNA Library preparation, Capture enrichment and Sequencing
Libraries were constructed into Illumina paired-end pre-capture libraries according to the manufacturer's protocol (Illumina Multiplexing_SamplePrep_Guide_1005361_D) with modifications as described in the BCM-HGSC protocol (https://www.hgsc.bcm.edu/content/protocols-sequencing-library-construction).
Primary Sequence Data Analysis and Variant Enrichment
Initial sequence analysis was performed using the HGSC Mercury analysis pipeline. Additionally, variants were filtered if they had a minor allele frequency >0.5% (averaged from Thousand Genomes, ExAc and ESP), >1% if they had been previously associated with disease (HGMD) or >2% if the variant was homozygous or there was a second variant in the gene. Variants were removed from consideration if they were annotated as benign or likely benign in ClinVar from multiple labs. Variants were prioritized based on function and disease association (for details, see the Supplementary Appendix). Analysis was conducted separately for each member in a family. We computationally examined a control cohort of 425 unselected European individuals to identify variants, obtained from the Atherosclerosis Risk in Communities study27, which met our criteria for a variants of interest (VOIs) in 2 different gene sets: a set of well characterized known LVNC genes28 (Table S1 in the Supplementary Appendix) and a larger set of known cardiomyopathy genes (Table S2 in the Supplementary Appendix).
Statistical analysis
Data are presented as mean±SD for continuous variables, and as counts (percentages) for categorical variables. Normality assumption of the continuous data was assessed on histograms and using Shapiro-Wilk test. Comparisons between two independent groups were assessed with the use of the Student t-test or Mann-Whitney test for continuous variables. Proportions comparison between groups were analyzed with Fisher's exact test or Pearson's chi-square test. Analysis of variance (ANOVA) was used to compare more than two groups with respect to continuous outcomes. For continuous variables, that exhibit a strong statistical significance (p≤0.01), Tukey's post-hoc test was used to conduct a pairwise comparison between the groups. Where appropriate, 95% confidence intervals are provided. All statistical tests were two-sided. A p-value was considered to indicate statistical significance at the nominal 0.05 level. Statistical analyses were performed using SPSS version 20.0 (SPSS Inc., Chicago, IL). To eliminate bias from multiple members in a single family, only the proband was included in the statistical analysis.
Results
Study participants
A total of 190 patients from 174 families were enrolled. After non-proband family members were removed, genotype-phenotype correlations were analyzed in the cohort of 174 probands (123 males). The mean age of probands was 26±16 years (range, 7 to 72). Seventy-nine subjects (45%) were under 18 years of age. All patients successfully underwent CMR imaging and whole exome sequencing of their genomic DNA. Baseline characteristics are presented as Table 1.
Table 1.
Baseline Characteristics of the Study Population.*
| Characteristic | VOI absent (N=72) | VOI present (N=102) | P Value |
|---|---|---|---|
| Age – yr | 26±15 | 27±17 | 0.68 |
| Male sex — no. (%) | 50 (69) | 73 (72) | 0.76 |
| Time delay from first symptom to diagnosis – yr | 2.5±4.0 | 3.3±4.4 | 0.27 |
| Familial occurrence of LVNC — no. (%) | 8 (11) | 16 (16) | 0.39 |
| Body-mass index – kg/m2 | 23±5 | 23±4 | 0.98 |
|
| |||
| NYHA functional class – no. (%) | 0.28 | ||
|
| |||
| I | 50 (69) | 61 (60) | |
| II | 13 (18) | 21 (21) | |
| III | 9 (13) | 16 (16) | |
| IV | 0 (0) | 4 (4) | |
| Atrial fibrillation – no. (%) | 4 (6) | 11 (11) | 0.28 |
| Neuromuscular disease – no. (%) | 0 (0) | 7 (7) | 0.04 |
|
| |||
| Medications – no. (%) | |||
|
| |||
| ACE inhibitor or ARB | 20 (28) | 53 (52) | 0.001 |
| Beta-blocker | 32 (44) | 60 (59) | 0.06 |
| Diuretic | 11 (15) | 25 (25) | 0.14 |
| Mineralocorticoid receptor antagonist | 16 (22) | 37 (36) | 0.05 |
| Aspirin | 21 (29) | 48 (47) | 0.02 |
| Vitamin K antagonists | 6 (8) | 22 (22) | 0.02 |
|
| |||
| LV volume parameters | |||
|
| |||
| LVEF <50% – no. (%) | 15 (21) | 35 (34) | 0.05 |
| LVEF – % | 53±12 | 48±16 | 0.04 |
| LVEDVI – ml/m2 | 106±46 | 116±49 | 0.22 |
| LVESVI – ml/m2 | 54±42 | 66±51 | 0.09 |
|
| |||
| Distribution of non-compaction | |||
|
| |||
| Number of affected segments | 7.3±2.8 | 8.3±3.0 | 0.02 |
| Inter-ventricular septum affected – no. (%) | 15 (21) | 25 (25) | 0.57 |
| Basal segments affected – no. (%) | 12 (17) | 31 (30) | 0.04 |
| NC/C ratio | 2.2±0.7 | 2.6±0.8 | <0.001 |
| Fulfilled CMR criteria for LVNC – no. (%) | 22 (31) | 68 (67) | <0.001 |
| Phenotype: LVNC+ DCM+ | 5 (7) | 26 (25) | 0.002 |
| Phenotype: LVNC+ DCM- | 17 (24) | 42 (41) | 0.02 |
| Presence of LGE – no. (%) | 3 (4) | 17 (17) | 0.01 |
Values are presented as number of patients (%), or the mean±SD. Multiple family members excluded.
ACE denotes angiotensin converting enzyme, ARB angiotensin receptor blocker, CMR cardiac magnetic resonance, DCM dilated cardiomyopathy, LGE late gadolinium enhancement, LV left ventricle, LVEDVI left ventricular end-diastolic volume index, LVEF left ventricular ejection fraction, LVESVI left ventricular end-systolic volume index, LVNC left ventricular non-compaction, NC/C ratio of non-compacted to compacted myocardium, NYHA New York Heart Association, and VOI variants of interest.
Genetic variants
We found 138 rare, protein altering variants in 54 previously identified LVNC or other known cardiomyopathy genes in 102 probands (59%) which were likely contributory to the patient phenotype (Table 2 and Table S3 in the Supplementary Appendix).
Table 2.
Genotype-Phenotype Correlations.*
| Gene symbol | Number of patients | Gene | Location | LVEF <50% | Met CMR criteria for LVNC | NC/C ratio | Number of affected segments | LGE | NMD |
|---|---|---|---|---|---|---|---|---|---|
| TTN† | 14 | Titin | Sarcomere | 8/14 | 10/14 | 2.9 | 8.9 | 4/14 | 0/14 |
| MYH7 | 8 | Cardiac-myosin heavy chain | Sarcomere | 4/8 | 8/8 | 3.5 | 11.0 | 4/8 | 1/8 |
| MYPN | 6 | Myopalladin | Sarcomere | 2/6 | 4/6 | 2.5 | 8.3 | 2/6 | 0/6 |
| LDB3 | 6 | ZASP/LIM domain binding 3 | Sarcomere | 2/6 | 6/6 | 2.8 | 8.7 | 3/6 | 0/6 |
| DSP | 5 | Desmoplakin | Intercellular | 1/5 | 1/5 | 2.1 | 5.8 | 0/5 | 1/5 |
| LAMA4 | 5 | Laminin, alpha 4 | Extracellular | 0/5 | 5/5 | 2.6 | 9.0 | 0/5 | 0/5 |
| MYBPC3 | 5 | Myosin-binding protein C | Sarcomere | 2/5 | 3/5 | 2.5 | 6.6 | 0/5 | 0/5 |
| MYH6 | 5 | Myosin, Heavy Chain 6 | Sarcomere | 1/5 | 3/5 | 3.0 | 8.0 | 0/5 | 0/5 |
| RBM20 | 5 | RNA-binding protein 20 | Nucleus | 2/5 | 5/5 | 3.3 | 9.4 | 0/5 | 0/5 |
| SYNE2 | 5 | Spectrin Repeat Containing, Nuclear Envelope 2 | Nuclear envelope | 2/5 | 2/5 | 2.2 | 8.6 | 1/5 | 1/5 |
| TPM1 | 5 | Alpha -Tropomyosin | Sarcomere | 3/5 | 5/5 | 3.1 | 10.2 | 1/5 | 0/5 |
| DSC2 | 4 | Desmocollin 2 | Intercellular | 4/4 | 4/4 | 2.7 | 7.5 | 0/4 | 0/4 |
| NEXN | 4 | Nexilin | Sarcomere | 0/4 | 2/4 | 2.1 | 8.3 | 0/4 | 0/4 |
| RYR2 | 4 | Ryanodine receptor 2 | Sarcoplasmic reticulum | 2/4 | 2/4 | 2.4 | 8.0 | 0/4 | 0/4 |
| DES | 3 | Desmin | Cytoskeleton | 1/3 | 3/3 | 2.9 | 8.7 | 1/3 | 0/3 |
| FLNA | 3 | Filamin A, Alpha | Cytoskeleton | 0/3 | 1/3 | 1.9 | 5.0 | 0/3 | 1/3 |
| GATA4 | 3 | GATA Binding Protein 4 | Nucleus | 0/3 | 0/3 | 2.0 | 5.3 | 0/3 | 0/3 |
| SDHA | 3 | Succinate dehydrogenase complex, subunit A, flavoprotein | Mitochondrial membrane | 1/3 | 1/3 | 2.8 | 8.7 | 1/3 | 0/3 |
| BAG3 | 3 | BCL2-Associated Athanogene 3 | Cytosol | 1/3 | 2/3 | 2.8 | 7.3 | 1/3 | 0/3 |
| ABCC6 | 2 | ATP-binding cassette, sub-family C (CFTR/MRP), member 6 | Membrane | 1/2 | 2/2 | 2.5 | 10.5 | 0/2 | 0/2 |
| ACTC1 | 2 | Cardiac actin | Sarcomere | 2/2 | 2/2 | 3.2 | 8.0 | 1/2 | 0/2 |
| DMD | 2 | Dystrophin | Cytoskeleton | 0/2 | 1/2 | 2.3 | 6.5 | 0/2 | 1/2 |
| EYA4 | 2 | EYA transcriptional coactivator and phosphatase 4 | Nucleus | 1/2 | 1/2 | 2.2 | 6.5 | 0/2 | 0/2 |
| MYOZ2 | 2 | Myozenin 2 | Sarcomere | 1/2 | 1/2 | 2.7 | 9.5 | 1/2 | 1/2 |
| SYNE1 | 2 | Spectrin repeat containing, nuclear envelope 1 | Nuclear envelope | 0/2 | 2/2 | 2.8 | 6.0 | 0/2 | 0/2 |
| TNNT2 | 2 | Cardiac troponin T | Sarcomere | 0/2 | 2/2 | 2.4 | 6.0 | 1/2 | 0/2 |
| ANG | 1 | Angiogenin, ribonuclease, RNase A family, 5 | Nucleus | 1/1 | 1/1 | 4.1 | 11.0 | 0/1 | 0/1 |
| CITED2 | 1 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | Nucleus | 0/1 | 0/1 | 2.2 | 9.0 | 0/1 | 0/1 |
| CRELD1 | 1 | Cysteine-rich with EGF-like domains 1 | Membrane | 0/1 | 0/1 | 1.5 | 4.0 | 0/1 | 0/1 |
| CRYAB | 1 | Crystallin, alpha B | Cytosol | 1/1 | 1/1 | 2.4 | 9.0 | 0/1 | 0/1 |
| FKTN | 1 | Fukutin | Golgi | 1/1 | 1/1 | 4.4 | 11.0 | 0/1 | 0/1 |
| FLNC | 1 | Filamin C, gamma | Sarcomere | 1/1 | 1/1 | 2.6 | 8.0 | 0/1 | 0/1 |
| FLT1 | 1 | Fms-related tyrosine kinase 1 | Membrane | 0/1 | 1/1 | 2.7 | 11.0 | 0/1 | 1/1 |
| KCNE1 | 1 | Potassium channel, voltage gated subfamily E regulatory beta subunit 1 | Ion Channel | 0/1 | 1/1 | 3.4 | 10.0 | 0/1 | 0/1 |
| LMNA | 1 | Lamin A/C | Nucleoskeleton | 1/1 | 1/1 | 2.4 | 9.0 | 0/1 | 0/1 |
| MEF2A | 1 | Myocyte enhancer factor 2A | Nucleus | 0/1 | 0/1 | 2.1 | 7.0 | 0/1 | 0/1 |
| MTMR14 | 1 | Myotubularin related protein 14 | Cytosol | 0/1 | 1/1 | 3.6 | 12.0 | 1/1 | 0/1 |
| MYL2 | 1 | Myosin, light chain 2, regulatory, cardiac, slow | Sarcomere | 0/1 | 0/1 | 2.2 | 8.0 | 0/1 | 0/1 |
| MYLK2 | 1 | Myosin light chain kinase 2 | Sarcomere | 1/1 | 1/1 | 2.4 | 9.0 | 0/1 | 0/1 |
| PLEC | 1 | Plectin | Cytoskeleton | 0/1 | 0/1 | 2.1 | 7.0 | 0/1 | 0/1 |
| PRDM16 | 1 | PR domain containing 16 | Nucleus | 0/1 | 0/1 | 2.1 | 7.0 | 0/1 | 0/1 |
| PRKAG2 | 1 | Protein kinase, AMP-activated, gamma 2 non-catalytic subunit | Cytosol | 0/1 | 1/1 | 2.9 | 15.0 | 0/1 | 0/1 |
| PRKAR1A | 1 | Protein kinase, cAMP-dependent, regulatory, type I, alpha | Membrane | 0/1 | 1/1 | 2.8 | 8.0 | 0/1 | 0/1 |
| PSEN1 | 1 | Presenilin 1 | Nuclear membrane | 0/1 | 0/1 | 1.8 | 4.0 | 0/1 | 0/1 |
| RAF1 | 1 | Raf-1 Proto-Oncogene | Membrane | 0/1 | 0/1 | 1.7 | 4.0 | 0/1 | 0/1 |
| SCN5A | 1 | Sodium channel, voltage-gated, type V | Ion channel | 1/1 | 1/1 | 2.9 | 9.0 | 0/1 | 0/1 |
| TBX1 | 1 | T-box 1 | Nucleus | 0/1 | 1/1 | 2.5 | 8.0 | 0/1 | 0/1 |
| TBX20 | 1 | T-box 20 | Nucleus | 1/1 | 1/1 | 2.9 | 7.0 | 0/1 | 0/1 |
| TGFB3 | 1 | Transforming Growth Factor, Beta 3 | Extracellular | 1/1 | 1/1 | 3.9 | 7.0 | 1/1 | 0/1 |
| TMPO | 1 | Thymopoietin | Nuclear envelope | 1/1 | 1/1 | 3.3 | 12.0 | 0/1 | 0/1 |
| TNNC1 | 1 | Cardiac troponin C | Sarcomere | 1/1 | 1/1 | 2.8 | 13.0 | 0/1 | 0/1 |
| TNNI3 | 1 | Cardiac troponin I | Sarcomere | 1/1 | 1/1 | 2.8 | 13.0 | 1/1 | 1/1 |
| TTR | 1 | Transthyretin | Extracellular | 1/1 | 1/1 | 6.5 | 13.0 | 0/1 | 0/1 |
| VCL | 1 | Metavinculin | Cytoskeleton | 0/1 | 0/1 | 1.9 | 4.0 | 0/1 | 0/1 |
Multiple family members excluded,
Truncating mutations only.
NMD denotes neuro-muscular disease; other abbreviations as in Table 1.
Twenty-six of the 54 identified cardiomyopathy genes were found to be mutated in two or more subjects, with TTN, MYH7, MYPN and LDB3 mutated in 6 or more patients each (Figure S1A in the Supplementary Appendix), although when controlling for transcript length the most enriched genes were TPM1, LDB3 and MYOZ2 (Figure S1B in the Supplementary Appendix). We identified a strong enrichment for truncating mutations in TTN, RBM20 and MYH7 when compared to control and other disease cohorts. Although truncating mutations in MYH7 are generally considered not to be disease causing.29
Forty-six of 54 identified cardiomyopathy genes did not have a previously established link with LVNC (Table S1 in the Supplementary Appendix), although 16 of these genes have been associated with skeletal myopathy or cardiac dysfunction. Although it is difficult to assess the functional effects and potential pathogenicity of nonsynonymous variants, in a number of these genes (FLNC, KCNQ1, FKTN, MTMR14, MYLK2, RBM20, TBX20, PRKAR1A, MEF2A) the identified variants were rare, previously identified as disease causing and predicted deleterious. Thus, these mutations have high potential of proving pathogenic and it is likely that the subject's cardiac phenotype was associated with the genotype.
When considering only the well characterized set of LVNC genes, we observed a modest increase in the number of individuals in our cohort with VOIs when compared to our control group (20.35% to 21.5%, p=0.74, Fisher's exact test). A greater enrichment was found using the cardiomyopathy gene set, with approximately double the number of individuals having at least one VOI. This effect was enhanced to ∼5-fold difference when considering only loss of function mutations (Figure S2A in the Supplementary Appendix). The distribution of VOIs in cardiomyopathy genes was significantly different between the cohort and the control (p=0.01). The disease cohort was also significantly enriched for multiple VOIs in individual patients; approximately 1.7 as many patients had at least one VOI in our disease cohort, but 4.2 times as many individuals had 2 or more VOIs (Figure S2B in the Supplementary Appendix). Copy number analysis was performed from the WES data using XHMM30 and no high confidence copy number variants were identified in known cardiomyopathy genes.
Genotype-phenotype correlations
There were no significant differences between subjects with and those without VOIs with respect to age, sex, familial occurrence of LVNC, or New York Heart Association functional class (Table 1).
When compared to patients with zero VOIs, patients with one or more VOIs had lower LV ejection fraction (LVEF), higher number of non-compacted LV segments, higher NC/C ratio, were more likely to have neuromuscular disease, affected LV basal segments, late gadolinium enhancement (LGE) and CMR confirmatory diagnosis of LVNC (67% vs. 31%, p<0.001).
In total, 90 of 174 probands (52%) met current CMR criteria for LVNC.13 The remaining 84 probands were classified as left ventricular hypertrabeculation (LVHT). VOIs were identified significantly more frequently in the LVNC group, as compared with the LVHT group (76% vs. 40%, p<0.001).
Twenty-seven subjects were found to have VOIs in the very conservative set of LVNC genes reported in the Online Mendelian Inheritance in Man (OMIM) database (Table S4 in the Supplementary Appendix). Presence of VOIs in these genes was associated with several markers of disease severity including higher NC/C ratio (p=0.01) and more frequent occurrence of LVNC (p=0.004) and LGE (p=0.001) as compared to 75 subjects with VOIs in non-OMIM LVNC genes and other known cardiomyopathy genes.
Multiple variants of interest
In 28 probands, two or more variants met our criteria for being interesting (2, 3, and 4 VOIs found in 21, 6, and 1 patients, respectively). For the patient with 4 VOIs, all 4 variants were shared with an affected sibling. For further analyses, the entire cohort was divided into three subgroups on the basis of the number of VOIs (0, 1, and ≥2 VOIs, respectively) (Table 3).
Table 3. Baseline Characteristics of the Study Population, According to Number of Variants of Interest.
| Characteristic | 0 VOIs (N=72) | 1 VOIs (N=74) | ≥2 VOIs (N=28) | P Value |
|---|---|---|---|---|
| Age – yr | 26±15 | 26±16 | 28±18 | 0.81 |
| Male sex — no. (%) | 50 (69) | 54 (73) | 19 (68) | 0.84 |
| Atrial fibrillation – no. (%) | 4 (6) | 6 (8) | 5 (18) | 0.14 |
|
| ||||
| LV volume parameters | ||||
|
| ||||
| LVEF <50% – no. (%) | 15 (21) | 21 (28) | 14 (50) | 0.02 |
| LVEF – % | 53±12 | 50±15 | 43±19 | 0.01 |
| LVEDVI – ml/m2 | 106±46 | 112±45 | 124±59 | 0.27 |
| LVESVI – ml/m2 | 54±42 | 61±46 | 80±61 | 0.05 |
|
| ||||
| Distribution of non-compaction | ||||
|
| ||||
| Number of affected segments | 7.3±2.8 | 8.0±3.0 | 8.9±2.8 | 0.03 |
| Inter-ventricular septum affected – no. (%) | 15 (21) | 15 (20) | 10 (36) | 0.22 |
| Basal segments affected – no. (%) | 12 (17) | 20 (27) | 11 (39) | 0.05 |
| NC/C ratio | 2.2±0.7 | 2.5±0.8 | 2.9±1.0 | <0.001 |
| Fulfilled CMR criteria for LVNC – no. (%) | 22 (31) | 46 (62) | 22 (79) | <0.001 |
| Phenotype: LVNC+ DCM+ | 5 (7) | 14 (19) | 12 (43) | <0.001 |
| Phenotype: LVNC+ DCM- | 17 (24) | 32 (43) | 10 (36) | 0.04 |
| Presence of LGE – no. (%) | 3 (4) | 13 (18) | 4 (14) | 0.04 |
P values are for the overall comparison among the groups. Multiple family members excluded.
Abbreviations as in Table 1.
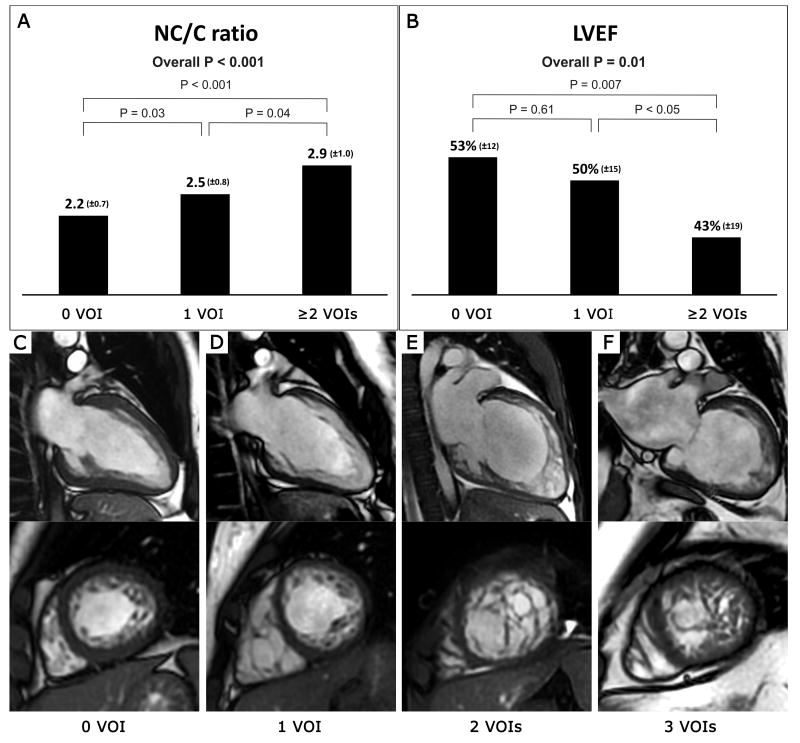
A higher number of VOIs was correlated with higher NC/C ratio and lower LVEF (Figure 1). A number of VOIs was also correlated with left ventricular end-systolic volume index, number of non-compacted LV segments, affected LV basal segments, rates of CMR confirmatory diagnosis of LVNC and presence of LGE.
Figure 1.
Impact of the number of variants of interest on NC/C ratio (Panel A) and LVEF (Panel B). Values are presented as mean (±SD). CMR imaging in four patients shows increasing degree of non-compaction with increasing number of VOIs (Panel C to F). Note the preservation of compacted myocardial thickness in a subject without a VOI with LVHT (Panel C). WES revealed one VOI in a patient with moderate LVNC (Panel D). Two subjects with multiple VOIs and extensive LVNC presented with severe LV systolic dysfunction (Panel E and F).
Sarcomere vs non-sarcomere genes
VOIs in either sarcomeric or non-sarcomeric genes were found in 39 and 47 unrelated patients, respectively, with 16 subjects having mutations in both sarcomeric and non-sarcomeric genes. The presence of sarcomeric gene mutations was associated with more frequent occurrence of trabeculations in the inter-ventricular septum (p=0.01) and LGE, likely indicating fibrosis of the myocardium (p=0.004) (Table 4). There was a nonsignificant trend toward lower LVEF and higher number of non-compacted LV segments among patients with sarcomeric gene mutations as compared to patients with non-sarcomeric gene mutations (both p=0.07).
Table 4.
Presence of Non-sarcomere or Sarcomere Gene Mutations.
| Characteristic | Sarcomere gene mutation (N=39) | Non-sarcomere gene mutation (N=47) | P Value |
|---|---|---|---|
| Age – yr | 26±14 | 26±18 | 0.90 |
| Male sex — no. (%) | 31 (79) | 33 (70) | 0.33 |
|
| |||
| LV volume parameters | |||
|
| |||
| LVEF <50% – no. (%) | 15 (38) | 11 (23) | 0.13 |
| LVEF – % | 46±18 | 52±13 | 0.07 |
| LVEDVI – ml/m2 | 120±54 | 107±38 | 0.21 |
| LVESVI – ml/m2 | 72±56 | 54±37 | 0.09 |
|
| |||
| Distribution of non-compaction | |||
|
| |||
| Number of affected segments | 8.8±3.1 | 7.6±2.9 | 0.07 |
| Inter-ventricular septum affected – no. (%) | 14 (36) | 6 (13) | 0.01 |
| Basal segments affected – no. (%) | 14 (36) | 10 (21) | 0.13 |
| NC/C ratio | 2.7±0.9 | 2.5±0.8 | 0.30 |
| Fulfilled CMR criteria for LVNC – no. (%) | 27 (69) | 27 (57) | 0.26 |
| Phenotype: LVNC+ DCM+ | 11 (28) | 8 (17) | 0.21 |
| Phenotype: LVNC+ DCM- | 16 (41) | 19 (40) | 0.96 |
| Presence of LGE – no. (%) | 12 (31) | 3 (6) | 0.004 |
Multiple family members and subjects with simultaneous presence of mutations in both non-sarcomere and sarcomere genes were excluded. Abbreviations as in Table 1
Long-QT syndrome genes
Our initial screening identified a homozygous variant in KCNE1, a heterozygous frameshift mutation in KCNQ1, and a heterozygous variant in SCN5A, a known LVNC gene.31 All these genes are known causes of long-QT syndrome (LQTS). We investigated whether other known disease-causing or highly likely deleterious variants occured in our cohort in other long-QT syndrome genes (Table S5 in the Supplementary Appendix) but utilized a less stringent minor allele frequency cut-off of 1%. We found a significant number of subjects with variants in KCNH2 (N=4, p=0.03 compared to control cohort). All variants had a MAF of 0, and of these, 3 were previously described as disease causing and the remaining variant was proximal (3 amino acids distant) from a previously described disease mutation. Although the mechanism by which mutations in KCNH2 might cause LVNC is unclear, the data shows clear enrichment of rare variants in this gene when compared to a large population database as well as an arrhymia cohort (2.2% vs 0.6% and 0.8%, respectively).32 Nine additional variants (10 total) were found in SCN5A, of these 7 were identical (p.P1973A) and were significantly enriched over the expected population frequency (1.84% vs. 0.2%, p<0.001). One subject with the clinical phenotype of LVNC and LQTS type 3 was found to have two variants in SCN5A and an additional variant in TPM1. The clinical course has been significant for cardiac arrest secondary to ventricular tachycardia with subsequent implantable cardioverter defibrillator placement and the patient was awaiting heart transplant secondary to end-stage heart failure.
Discussion
We identified at least one VOI in 59% of our cohort subjects in 54 different cardiomyopathy genes. Multiple variants were found in genes not previously reported in association with LVNC including LQTS genes (KCNH2, KCNE1, KCNQ1, and KCNJ2), the cardio- and muscular myopathy gene FLNC, and MEF2A, which is associated with premature coronary artery disease. In many cases these novel associations were made to genes harboring rare, highly deleterious, often previously described as disease causing, mutations in autosomal dominant genes; molecular diagnoses consistent with the contention that the subject either has or is likely to develop the disease. These findings underscore the diversity of genes already associated with LVNC and offer insight into varying clinical phenotypes associated with LVNC. Mutations in SCN5A have been previously reported to result in dilated cardiomyopathy and LQTS33 and our results indicate that other LQTS genes may also result in cardiomyopathy phenotypes. Although the mechanism for this is unclear, several hypothesis have been proposed including direct protein-protein interaction of the LQTS gene with the sarcomere or that LVNC is an acquired adaptive remodeling feature in response to impaired conduction. It is also possible that there is an underlying mutation driving the LVNC phenotype and mutations in LQTS genes act epigenetically to modify the penetrance or expressivity of the phenotype. Whatever the mechanism our data shows a very strong enrichment for mutations in these genes when compared to control cohorts.32
We identified 14 probands with truncating mutations in TTN in our cohort, firmly establishing the importance of this gene to LVNC.34 Truncating mutations in TTN have been reported in association with dilated cardiomyopathy, however, there was no evidence of LVNC in their cohort.35 We identified 14 probands with truncating TTN mutations and 8 had reduced LVEF. Mutations were identified in both the A disc and I band regions of titin but there were no differences in NC/C ratio or LVEF between the two subgroups. Although truncating mutations in titin can be abrogated by alternate splicing, 11 of the 14 unique truncating mutations identified in this study occurred in constitutive exons (Table S6 in the Supplementary Appendix). Interestingly, across our cohort, titin showed significant (p=0.01) enrichment for co-occurrence with other VOIs suggesting that additional mutational burden is required to develop a phenotype.
Twenty-eight unrelated patients had two or more identified VOIs. We found increased mutational burden in these genes between our LVNC cohort and an unselected population. Evidence for a mutational burden has also been reported for neuropathy and oligogenic inheritance in a family with HCM.36,37 Strong correlations were found between mutational burden in individual patients and cardiac phenotypes, including NC/C ratio, number and distribution of non-compacted LV segments, LVEF, and presence of LGE. Interestingly, the probability of meeting current CMR criteria for LVNC was strongly associated with the number of VOIs. Multiple mutations, which alone are benign, have been shown to elicit a severe phenotype when present in combination,38 in a similar way our findings indicate that LVNC may be caused by, or the phenotype exacerbated by, multiple, uncommon variants that act in a synergistic manner. This, in part, could explain the tremendous clinical heterogeneity of this disease specifically related to the presence or absence of ventricular dysfunction and remodeling as well as the presence or absence of arrhythmia burden. This finding also potentially bridges the gap between the clinical diagnosis of LVNC and LVHT.
In 41% of subjects in our cohort, no VOI was found. This suggests that there are other causes of LVNC that are not captured by a WES approach. Such causes may include hitherto unrecognized LVNC/cardiomyopathy genes and variants which our analysis incorrectly determined were not pathogenic. Such variants may appear benign (e.g. synonymous variants) but impact transcription, translation or the function of the protein alone or in conjunction with other variants, or environmental factors, although the relatively young age of onset for the cohort implicate a genetic cause of LVNC in these cases. WES, typically, does not interrogate the intronic or untranslated regions of genes, nor intergenic regions, and is limited in its ability to detect copy number variants, all of which may contribute to the genetic landscape of LVNC. These latter challenges can be better addressed by whole genome sequencing, however, interpretation of non-coding variants will remain challenging for the foreseeable future.
The term LVHT has traditionally been used to describe an increased number of trabeculations with normal histological appearance and was considered as a normal anatomic variant.4 However, a distinction between LVNC and LVHT is often extremely difficult because there is a continuum from normal, hypertrabeculated myocardium to pathological appearance of the LV myocardium. In our study, we found presence of VOIs to be very common (40%) in patients with LVHT. We also found that the NC/C ratio was directly correlated with the number of VOIs. Thus, there is a continuum of clinical expression and the distinction between LVHT and LVNC may not be clinically relevant. The presence of LVHT warrants careful, longitudinal surveillance for change in clinical status.
The potential prognostic implications of LVHT and LVNC in patients with nonischemic dilated cardiomyopathy has been recently studied.39 The authors reported that the presence of trabeculations had no influence on a variety of clinical outcomes. Although we did not assess these endpoints, our study suggests that the presence of LVHT/LVNC strongly impacts LV systolic function and fibrosis burden, both of which have been previously reported to result in increased mortality in the setting of LVNC.40-42
Our data argue in favor of LVNC being a phenotype shared by different cardiac diseases and suggest that the presence of VOIs directly influences not only the morphologic findings used currently for diagnosis but also LV systolic dysfunction and fibrosis burden. Based on our data, the use of currently promoted CMR diagnostic criteria may result in many patients failing to be appropriately diagnosed even though they harbor pathogenic mutations and may benefit from genetic testing. This would include interrogating all known cardiomyopathy genes, or all genes using WES, to give a clearer picture of the genetic burden than achieved testing the smaller number of known LVNC genes.
Limitations
A challenge to our study is the lack of consensus regarding criteria for LVNC and by including patients with LVHT we may confound our findings. Further, a proportion of normal adults may show significant NC/C ratios in at least one region.43 Nevertheless, to increase pre-test probability of having LVNC both imaging and clinical data were used.44 Our approach of including patients with LVHT was intentional to assist in deconstructing the current diagnostic approach and our data support the continuum concept of disease. We also recognize that the LVNC phenotype may be secondary to causes other than genetic triggers, for example unfavorable LV remodeling.7,45 With this in mind, we recognize that our study does not have longitudinal data and that we cannot document presence or absence of LV trabeculations early in life in our cohort. Although numerous potentially contributory mutations were identified using strict criteria (population conservation, evolutionary conservation and known deleteriousness of the variant, gene function), these variants may still not be disease causing.32 Functional methods of validation can be laborious and expensive and cannot be conducted on every novel variant. In the future, as sequencing becomes more ubiquitous, discovery of identical variants in multiple unrelated subjects with LVNC or LVHT may become the best method for determining pathogenicity.
Conclusions
Our study identifies potentially causative variants in multiple genes not previously implicated in LVNC including truncating mutations in titin, an important cardiomyopathy and musclular dystrophy gene. The phenotype of LVNC may be influenced by the co-occurrence of multiple genetic mutations interacting epistatically. The severity of cardiac involvement, including degree of non-compaction and LV systolic dysfunction, is more pronounced in the presence of increasing number of variants of interest, consistent with a mutational burden hypothesis. Although sarcomeric genes may be responsible for LVNC, other genetic triggers, such as non-sarcomeric genes, may exist that result in the cardiac phenotype. Presence of VOIs is common in patients with LVHT and may represent partial expression of LVNC. LVHT and LVNC may represent a continuum of genotypic disease, with LVHT possibly representing a less advanced form that also warrants careful, longitudinal surveillance.
Supplementary Material
Clinical Perspective.
Left ventricular non-compaction (LVNC) is a genetically and phenotypically heterogeneous disease. In the current study, we described the largest cohort of patients with LVNC or left ventricular hypertrabeculation (LVHT) undergoing cardiac magnetic resonance and whole-exome sequencing. We sought to identify novel genetic causes of LVNC and describe genotype-phenotype correlations. Potentially causative variants were found in multiple sarcomeric and non-sarcomeric genes not previously reported in association with LVNC including titin and long-QT syndrome genes. These findings underscore the diversity of genes already associated with LVNC and offer insight into varying clinical phenotypes associated with LVNC. We also demonstrated the presence of sarcomeric gene mutations was associated with increased occurrence of late gadolinium enhancement, likely indicating fibrosis of the myocardium. The phenotype of LVNC is influenced by the co-occurrence of multiple genetic mutations interacting epistatically. In our study, significant proportion of subjects had two or more identified variants of interest (VOIs). We found increasing number of VOIs in a patient strongly correlated with several markers of disease severity, including degree of non-compaction and left ventricular systolic dysfunction, consistent with a mutational burden hypothesis. Presence of VOIs was very common (40%) in patients with LVHT. LVHT and LVNC likely represent a continuum of genotypic disease. LVHT may represent partial expression of LVNC and warrants careful, longitudinal surveillance. Our data argue in favor of LVNC being a marker of underlying different cardiac diseases. Comprehensives panels or whole exome sequencing tests may offer better insight into genetic burden of LVNC in the future.
Acknowledgments
We would like to thank the families for participating and Drs. John Belmont and Thomas Ryan for helpful discussions.
Sources of Funding: This work was funded in part by 5U54 HG006542 to the Baylor Hopkins Center for Mendelian Genomics and 5U54 HG003273-12 (Gibbs), the Lindner Center for Research and Education and the The Christ Hospital Lindner Research Center.
Footnotes
Disclosures: MNB and EV are founders of Codified Genomics LLC. Remaining authors have no disclosures.
References
- 1.Towbin JA, Lorts A, Jefferies JL. Left ventricular non-compaction cardiomyopathy. Lancet. 2015;386:813–825. doi: 10.1016/S0140-6736(14)61282-4. [DOI] [PubMed] [Google Scholar]
- 2.Kovacevic-Preradovic T, Jenni R, Oechslin EN, Noll G, Seifert B, Attenhofer Jost CH. Isolated left ventricular noncompaction as a cause for heart failure and heart transplantation: a single center experience. Cardiology. 2009;112:158–164. doi: 10.1159/000147899. [DOI] [PubMed] [Google Scholar]
- 3.Sen-Chowdhry S, McKenna WJ. Left ventricular noncompaction and cardiomyopathy: cause, contributor, or epiphenomenon? Curr Opin Cardiol. 2008;23:171–175. doi: 10.1097/HCO.0b013e3282fdc939. [DOI] [PubMed] [Google Scholar]
- 4.Oechslin E, Jenni R. Left ventricular non-compaction revisited: a distinct phenotype with genetic heterogeneity? Eur Heart J. 2011;32:1446–1456. doi: 10.1093/eurheartj/ehq508. [DOI] [PubMed] [Google Scholar]
- 5.Hussein A, Karimianpour A, Collier P, Krasuski RA. Isolated Noncompaction of the Left Ventricle in Adults. J Am Coll Cardiol. 2015;66:578–585. doi: 10.1016/j.jacc.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 6.Arbustini E, Weidemann F, Hall JL. Left ventricular noncompaction: a distinct cardiomyopathy or a trait shared by different cardiac diseases? J Am Coll Cardiol. 2014;64:1840–1850. doi: 10.1016/j.jacc.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weir-McCall JR, Yeap PM, Papagiorcopulo C, Fitzgerald K, Gandy SJ, Lambert M, et al. Left Ventricular Noncompaction: Anatomical Phenotype or Distinct Cardiomyopathy? J Am Coll Cardiol. 2016;68:2157–2165. doi: 10.1016/j.jacc.2016.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 9.Elliott P, Andersson B, Arbustini E, Bilinska Z, Cecchi F, Charron P, et al. Classification of the cardiomyopathies: a position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2008;29:270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 10.Charron P, Arad M, Arbustini E, Basso C, Bilinska Z, Elliott P, et al. Genetic counselling and testing in cardiomyopathies: a position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2010;31:2715–2726. doi: 10.1093/eurheartj/ehq271. [DOI] [PubMed] [Google Scholar]
- 11.Probst S, Oechslin E, Schuler P, Greutmann M, Boye P, Knirsch W, et al. Sarcomere gene mutations in isolated left ventricular noncompaction cardiomyopathy do not predict clinical phenotype. Circ Cardiovasc Genet. 2011;4:367–374. doi: 10.1161/CIRCGENETICS.110.959270. [DOI] [PubMed] [Google Scholar]
- 12.Kohli SK, Pantazis AA, Shah JS, Adeyemi B, Jackson G, McKenna WJ, et al. Diagnosis of left-ventricular non-compaction in patients with left-ventricular systolic dysfunction: time for a reappraisal of diagnostic criteria? Eur Heart J. 2008;29:89–95. doi: 10.1093/eurheartj/ehm481. [DOI] [PubMed] [Google Scholar]
- 13.Petersen SE, Selvanayagam JB, Wiesmann F, Robson MD, Francis JM, Anderson RH, et al. Left ventricular non-compaction: insights from cardiovascular magnetic resonance imaging. J Am Coll Cardiol. 2005;46:101–105. doi: 10.1016/j.jacc.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 14.Jacquier A, Thuny F, Jop B, Giorgi R, Cohen F, Gaubert JY, et al. Measurement of trabeculated left ventricular mass using cardiac magnetic resonance imaging in the diagnosis of left ventricular non-compaction. Eur Heart J. 2010;31:1098–1104. doi: 10.1093/eurheartj/ehp595. [DOI] [PubMed] [Google Scholar]
- 15.Andreini D, Pontone G, Bogaert J, Roghi A, Barison A, Schwitter J, et al. Long-Term Prognostic Value of Cardiac Magnetic Resonance in Left Ventricle Noncompaction: A Prospective Multicenter Study. J Am Coll Cardiol. 2016;68:2166–2181. doi: 10.1016/j.jacc.2016.08.053. [DOI] [PubMed] [Google Scholar]
- 16.Sasse-Klaassen S, Gerull B, Oechslin E, Jenni R, Thierfelder L. Isolated noncompaction of the left ventricular myocardium in the adult is an autosomal dominant disorder in the majority of patients. Am J Med Genet A. 2003;119A:162–167. doi: 10.1002/ajmg.a.20075. [DOI] [PubMed] [Google Scholar]
- 17.Xing Y, Ichida F, Matsuoka T, Isobe T, Ikemoto Y, Higaki T, et al. Genetic analysis in patients with left ventricular noncompaction and evidence for genetic heterogeneity. Mol Genet Metab. 2006;88:71–77. doi: 10.1016/j.ymgme.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Jefferies JL. Barth syndrome. Am J Med Genet C Semin Med Genet. 2013;163C:198–205. doi: 10.1002/ajmg.c.31372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Chen H, Qu X, Chang CP, Shou W. Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC) Am J Med Genet C Semin Med Genet. 2013;163C:144–156. doi: 10.1002/ajmg.c.31369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dusek J, Ostadal B, Duskova M. Postnatal persistence of spongy myocardium with embryonic blood supply. Arch Pathol. 1975;99:312–317. [PubMed] [Google Scholar]
- 21.Klaassen S, Probst S, Oechslin E, Gerull B, Krings G, Schuler P, et al. Mutations in sarcomere protein genes in left ventricular noncompaction. Circulation. 2008;117:2893–2901. doi: 10.1161/CIRCULATIONAHA.107.746164. [DOI] [PubMed] [Google Scholar]
- 22.Towbin JA. Left ventricular noncompaction: a new form of heart failure. Heart Fail Clin. 2010;6:453–469. doi: 10.1016/j.hfc.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Bagnall RD, Molloy LK, Kalman JM, Semsarian C. Exome sequencing identifies a mutation in the ACTN2 gene in a family with idiopathic ventricular fibrillation, left ventricular noncompaction, and sudden death. BMC Med Genet. 2014;15:99. doi: 10.1186/s12881-014-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang S, Batra A, Zhang Y, Ebenroth ES, Huang T. Left ventricular noncompaction is associated with mutations in the mitochondrial genome. Mitochondrion. 2010;10:350–357. doi: 10.1016/j.mito.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Bainbridge MN, Davis EE, Choi WY, Dickson A, Martinez HR, Wang M, et al. Loss of Function Mutations in NNT Are Associated With Left Ventricular Noncompaction. Circ Cardiovasc Genet. 2015;8:544–552. doi: 10.1161/CIRCGENETICS.115.001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerqueira MD, Weissman NJ, Dilsizian V, Jacobs AK, Kaul S, Laskey WK, et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart: a statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation. 2002;105:539–542. doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
- 27.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 28.Arbustini E, Favalli V, Narula N, Serio A, Grasso M. Left Ventricular Noncompaction: A Distinct Genetic Cardiomyopathy? J Am Coll Cardiol. 2016;68:949–966. doi: 10.1016/j.jacc.2016.05.096. [DOI] [PubMed] [Google Scholar]
- 29.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyatake S, Koshimizu E, Fujita A, Fukai R, Imagawa E, Ohba C, et al. Detecting copy-number variations in whole-exome sequencing data using the eXome Hidden Markov Model: an ‘exome-first’ approach. J Hum Genet. 2015;60:175–182. doi: 10.1038/jhg.2014.124. [DOI] [PubMed] [Google Scholar]
- 31.Shan L, Makita N, Xing Y, Watanabe S, Futatani T, Ye F, et al. SCN5A variants in Japanese patients with left ventricular noncompaction and arrhythmia. Mol Genet Metab. 2008;93:468–474. doi: 10.1016/j.ymgme.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2016;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36:1123–1135a. doi: 10.1093/eurheartj/ehu301. [DOI] [PubMed] [Google Scholar]
- 34.Bodian M, Jobe M, Leye M, Ndiaye MB, Kane A, Sarr SA, et al. Combination of left ventricular noncompaction and partial atrioventricular canal defect in a 21-year-old male: a case report. Clin Med Insights Case Rep. 2013;6:9–13. doi: 10.4137/CCRep.S10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts AM, Ware JS, Herman DS, Schafer S, Baksi J, Bick AG, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med. 2015;7:270–276. doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzaga-Jauregui C, Harel T, Gambin T, Kousi M, Griffin LB, Francescatto L, et al. Exome Sequence Analysis Suggests that Genetic Burden Contributes to Phenotypic Variability and Complex Neuropathy. Cell Rep. 2015;12:1169–1183. doi: 10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Bainbridge MN, Tan Y, Willerson JT, Marian AJ. A Potential Oligogenic Etiology of Hypertrophic Cardiomyopathy, A Classic Single Gene Disorder. Circ Res. 2017;120:1084–1090. doi: 10.1161/CIRCRESAHA.116.310559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blankenburg R, Hackert K, Wurster S, Deenen R, Seidman JG, Seidman CE, et al. Beta-Myosin heavy chain variant Val606Met causes very mild hypertrophic cardiomyopathy in mice, but exacerbates HCM phenotypes in mice carrying other HCM mutations. Circ Res. 2014;115:227–237. doi: 10.1161/CIRCRESAHA.115.303178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amzulescu MS, Rousseau MF, Ahn SA, Boileau L, de Meester de Ravenstein C, Vancraeynest D, et al. Prognostic Impact of Hypertrabeculation and Noncompaction Phenotype in Dilated Cardiomyopathy: A CMR Study. JACC Cardiovasc Imaging. 2015;8:934–946. doi: 10.1016/j.jcmg.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 40.Brescia ST, Rossano JW, Pignatelli R, Jefferies JL, Price JF, Decker JA, et al. Mortality and Sudden Death in Pediatric Left Ventricular Noncompaction in a Tertiary Referral Center. Circulation. 2013;127:2202–2208. doi: 10.1161/CIRCULATIONAHA.113.002511. [DOI] [PubMed] [Google Scholar]
- 41.Cheng H, Lu M, Hou C, Chen X, Li L, Wang J, et al. Comparison of cardiovascular magnetic resonance characteristics and clinical consequences in children and adolescents with isolated left ventricular non-compaction with and without late gadolinium enhancement. J Cardiovasc Magn Reson. 2015;17:44. doi: 10.1186/s12968-015-0148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stanton C, Bruce C, Connolly H, Brady P, Syed I, Hodge D, et al. Isolated left ventricular noncompaction syndrome. Am J Cardiol. 2009;104:1135–1138. doi: 10.1016/j.amjcard.2009.05.062. [DOI] [PubMed] [Google Scholar]
- 43.Kawel N, Nacif M, Arai AE, Gomes AS, Hundley WG, Johnson WC, et al. Trabeculated (noncompacted) and compact myocardium in adults: the multi-ethnic study of atherosclerosis. Circ Cardiovasc Imaging. 2012;5:357–366. doi: 10.1161/CIRCIMAGING.111.971713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petersen SE. CMR and LV noncompaction: does it matter how we measure trabeculations? JACC Cardiovasc Imaging. 2013;6:941–943. doi: 10.1016/j.jcmg.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 45.Statile CJ, Taylor MD, Mazur W, Cripe LH, King E, Pratt J, et al. Left ventricular noncompaction in Duchenne muscular dystrophy. J Cardiovasc Magn Reson. 2013;15:67. doi: 10.1186/1532-429X-15-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.