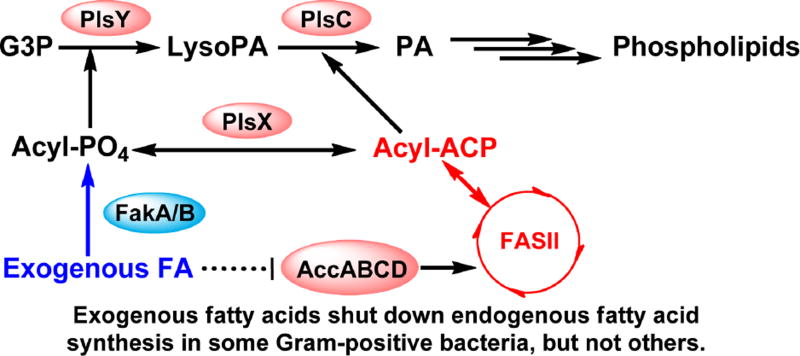
Abstract
Bacterial type II fatty acid synthesis (FASII) is a target for novel antibiotic development. All bacteria encode for mechanisms to incorporate exogenous fatty acids, and some bacteria can use exogenous fatty acids to bypass FASII inhibition. Bacteria encode three different mechanisms for activating exogenous fatty acids for incorporation into phospholipid synthesis. Exogenous fatty acids are converted into acyl-CoA in Gammaproteobacteria such as E. coli. Acyl-CoA molecules constitute a separate pool from endogenously synthesized acyl-ACP. Acyl-CoA can be used for phospholipid synthesis or broken down by β-oxidation, but cannot be used for lipopolysaccharide synthesis. Exogenous fatty acids are converted into acyl-ACP in some Gram-negative bacteria. The resulting acyl-ACP undergoes the same fates as endogenously synthesized acyl-ACP. Exogenous fatty acids are converted into acyl-phosphates in Gram-positive bacteria, and can be used for phospholipid synthesis or become acyl-ACP. Only the order Lactobacillales can use exogenous fatty acids to bypass FASII inhibition. FASII shuts down completely in presence of exogenous fatty acids in Lactobacillales, allowing Lactobacillales to synthesize phospholipids entirely from exogenous fatty acids. Inhibition of FASII cannot be bypassed in other bacteria because FASII is only partially down-regulated in presence of exogenous fatty acid or FASII is required to synthesize essential metabolites such as β-hydroxyacyl-ACP. Certain selective pressures such as FASII inhibition or growth in biofilms can select for naturally occurring one step mutations that attenuate endogenous fatty acid synthesis. Although attempts have been made to estimate the natural prevalence of these mutants, culture-independent metagenomic methods would provide a better estimate.
Graphical abstract
1. INTRODUCTION
Phospholipids are major components making the essential cellular membrane that forms the boundary between the organism and the environment [1]. Phosphatidic acid, the precursor to all phospholipid species, is synthesized through two successive acylation reactions of glycerol-3-phosphate (G3P) using acyl-acyl carrier protein (ACP) synthesized by the bacterial type II fatty acid synthesis system (FASII) (Fig. 1) [2]. Due to differences between FASII and the monofunctional mammalian type I fatty acid synthase [3], targeting FASII enzymes for novel antibiotic therapeutics is an active area of research [4–7]. However, all bacteria characterized to date can assimilate exogenous fatty acids [8–12], but only some species can bypass inhibition of FASII by incorporating exogenous fatty acids [13]. Several FASII inhibitors are currently in clinical development [14–20], making understanding the mechanism of exogenous fatty acid activation for phospholipid synthesis and whether exogenous fatty acids can bypass FASII inhibition in pathogenic bacterial species an important area of research. This review will summarize the pathways for the incorporation of exogenous fatty acids into phospholipids and whether exogenous fatty acids can bypass the inhibition of endogenous fatty acid synthesis in those species.
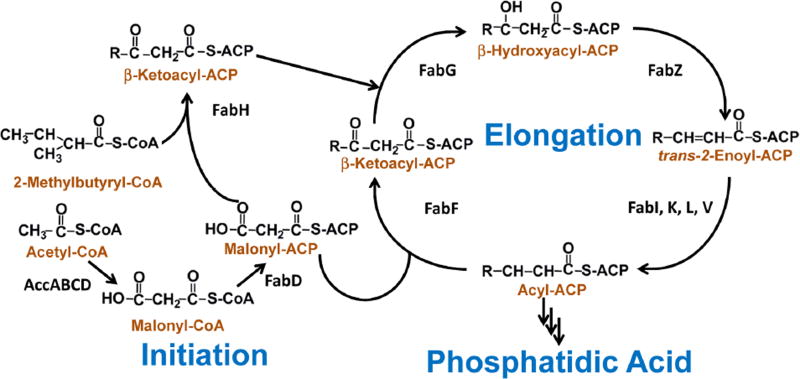
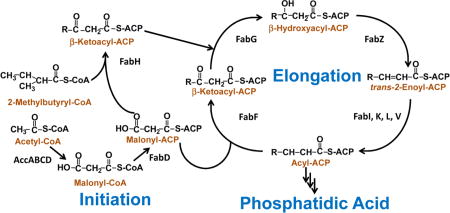
Fig. 1. The core enzymes in bacterial type II fatty acid synthesis.
The acetyl-CoA carboxylase enzyme complex (AccABCD) converts aceyl-CoA into malonyl-CoA, which is in turn converted into malonyl-ACP by the malonyl-CoA:ACP transacylase (FabD). The β-ketoacyl-ACP synthase III (FabH) catalyzes the condensation of malonyl-ACP with either acetyl-CoA or 2-methylbutyryl-CoA to form β-ketoacyl-ACP and initiate straight- or branched-chain anteiso fatty acid synthesis. The β-ketoacyl-ACP is reduced by β-ketoacyl-ACP reductase (FabG) to make β-hydroxyacyl-ACP. β-hydroxyacyl-ACP of the appropriate length is used for lipopolysaccharide synthesis in Gram-negative bacteria. β-hydroxyacyl-ACP is dehydrated by β-hydroxyacyl-ACP dehydratase (FabZ) to make trans-2-enoyl-ACP. The trans-2-enoyl-ACP is reduced by enoyl-ACP reductase (FabI) into acyl-ACP. Acyl-ACP of the appropriate length can be used for phospholipid synthesis. Acyl-ACP can also be lengthened by two carbons through condensation with malonyl-ACP by β-ketoacyl-ACP synthase II (FabF) to make β-ketoacyl-ACP, which can undergo another elongation cycle by FabG, FabZ, FabI, and FabF. The bacterial FASII enzymes are soluble, discrete, and monofunctional, in contrast to the multifunctional mammalian fatty acid synthase. There are multiple isoforms of the enoyl-ACP reductase enzyme (FabK, FabL, and FabV). Additional enzymes interact with the FASII intermediates to synthesize unsaturated fatty acid and lipoic acid. Please refer to reviews for more details in FASII operation [1,105].
2. PATHWAYS FOR EXOGENOUS FATTY ACID INCORPORATION
Fatty acid synthesis is an energy and material intensive process (Fig. 1) in phospholipid synthesis and the incorporation of useable exogenous fatty acids saves energy and metabolic building materials. The synthesis of the 16 carbon, saturated palmitoyl-CoA requires 8 acetyl-CoA molecules, 14 NADPH reducing equivalents, and the hydrolysis of 7 ATP molecules [1]. Two acyl-ACP molecules are used in successive acylation reactions to synthesize phosphatidic acid, the precursor to all bacterial phospholipid species [2]. There are two major types of acyltransferase systems found in bacteria. The PlsB pathway found in E. coli and other Gammaproteobacteria acylates the G3P using acyl-ACP or acyl-CoA [21–23]. The PlsX/PlsY pathway found in all other characterized bacterial taxa acylates G3P using an acyl-phosphate derived from acyl-ACP via PlsX [24,25]. PlsC acylates the resulting lysophosphatidic acid using acyl-ACP in both pathways [26,27]. Further details of these two pathways can be found in a recent review [2]. This review will focus on how the exogenous fatty acid activation systems present fatty acids to the acyltransferases.
The energy and material savings gained from being able to use exogenous fatty acids (when the correct acyl chain species are available) in lieu of endogenous synthesis suggests why all bacteria characterized to date encode for some mechanism to incorporate exogenous fatty acids [2]. However, the acyl chain composition of the phospholipid controls the membrane fluidity and function [28–32], so bacteria must balance the energy savings from incorporating exogenous fatty acids with synthesizing enough fatty acids endogenously to ensure the proper membrane biophysical properties. There are three characterized mechanisms for incorporating exogenous fatty acids: acyl-CoA synthetase, acyl-ACP synthetase, and fatty acid kinase. The acyl-CoA synthetase was first discovered in E. coli and found in Gammaproteobacteria bacteria [12]. Acyl-CoA synthetases are also found in mammalian lipid synthesis, and is an essential component because the free fatty acids generated by mammalian fatty acid synthase must be converted into acyl-CoA for phospholipid synthesis [33]. Acyl-ACP synthetase was the second mechanism discovered [34,35]. Acyl-ACP synthetases belong in the same structural family as acyl-CoA synthetases and are also found in Gram-negative bacteria [9]. Acyl-CoA synthetase and acyl-ACP synthetase are found in bacteria encoding either the PlsB or the PlsX/PlsY acyltranferase system [8,9]. Biochemical validation is needed to identify acyl-ACP synthetase versus acyl-CoA synthetase. Finally, the fatty acid kinase system is found in Gram-positive bacteria that exlusively encode the PlsX/PlsY acyltransferase system [36,37]. The fatty acid kinase generates an acyl-phosphate molecule which can enter into phospholipid synthesis in bacteria encoding the PlsX/PlsY acyltransferase pathway.
The following sections describe four characterized pathways to activate exogenous fatty acids for incorporation into phospholipid synthesis. Exogenous fatty acids are converted into acyl-CoA, which can be used by the PlsB/PlsC acyltransferase system in E. coli and other Gammaproteobacteria. Exogenous fatty acids are converted into acyl-ACP, which can enter into FASII and become indistinguishable from endogenously synthesized fatty acids in Chlamydia trachomatis and Neisseria gonorrhoeae. In C. trachomatis, the acyl-ACP is used directly by the PlsE/PlsC pathway for phosphatidic acid synthesis. In N. gonorrhoeae, the acyl-ACP is converted into acyl-phosphate by PlsX to initiate phosphatidic acid synthesis by PlsY. Finally, exogenous fatty acids are converted into acyl-phosphate by the fatty acid kinase system in Gram-positive bacteria. The resulting acyl-phosphate can by used by PlsY in the first step of phosphatidic acid synthesis, or converted back into acyl-ACP by PlsX. The resulting acyl-ACP can be elongated by FASII or used by PlsC in the second step of phosphatidic acid synthesis. The following section also notes other instances where fatty acid activation is needed such as β-oxidation and recycling free fatty acids generated in other processes.
2.1. Acyl-CoA Synthetase and Acyl-ACP Synthetase in E. coli
E. coli utilizes the acyl-CoA synthetase (FadD) to generate acyl-CoA from exogenous fatty acids (Fig. 2) [12]. Acyl-CoA synthetase catalyzes the formation of acyl-CoA from a fatty acid and a CoA using the energy generated from the hydrolysis of an ATP into AMP and pyrophosphate [38]. Members of this superfamily of proteins contain the AFD_class_I domain, which corresponds to the binding site for the acyl-adenylate intermediate. Acyl-CoA molecules cannot enter FASII and become elongated in E. coli [39], but undergoes two other fates. The E. coli acyltransferases PlsB and PlsC both use acyl-CoA molecules of the appropriate acyl chain length to acylate G3P and synthesize phosphatidic acid [26,40]. Acyl-CoA molecules are also broken down via β-oxidation to generate acetyl-CoA molecules and ATP [41,42]. E. coli also encodes for a short chain acyl-CoA synthetase (FadK) that is expressed during anaerobic conditions and in presence of the terminal electron acceptor fumarate [43]. Acyl-CoA generated through this pathway cannot enter phosphatidic acid synthesis due to the acyl chain length, and can only be broken down via β-oxidation. It is important to note that the E. coli FASII generates β-hydroxyacyl-ACP for lipopolysaccharide synthesis in addition to acyl-ACP [44]. There is no known mechanism for converting exogenous fatty acids into β-hydroxyacyl-ACP in E. coli. Because lipopolysaccharide synthesis is essential in E. coli [45], exogenous fatty acids cannot bypass genetic or biochemical inhibition of FASII in E. coli. The E. coli model is predicted to be representative of other Gammaproteobacteria.
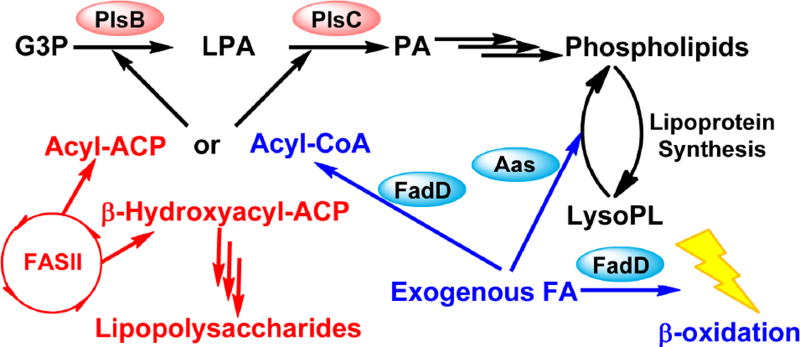
Fig. 2. Phosphatidic acid synthesis and exogenous fatty acid activation in E. coli.
E. coli and other Gammaproteobacteria utilize the PlsB pathway for the synthesis of phosphatidic acid. Phosphatidic acid is synthesized from the successive acylation of G3P by the sn-glycerol-3-phosphate acyltransferase PlsB and 1-acyl-sn-glycerol-3-phosphate acyltransferase PlsC in E. coli. Both PlsB and PlsC are integral membrane enzymes. Exogenous fatty acids are converted into acyl-CoA by the acyl-CoA synthetase (FadD) and metabolically compartmentalized from endogenously synthesized acyl-ACP. Exogenous fatty acid is used for phospholipid synthesis because both acyltransferases in E. coli use either acyl-ACP or acyl-CoA of the appropriate length as acyl donors. Acyl-CoA can also be broken down via β-oxidation to generate acetyl-CoA and energy. Exogenous fatty acid is also used by the 2-acylglycerophosphoethanolamine acyltransferase (Aas) to reacylate lysophospholipids generated by lipoprotein synthesis. This reacylation reaction proceeds through an acyl-ACP intermediate, but the acyl-ACP cannot dissociate from the Aas complex and is used only for the reacylation reaction. Exogenous fatty acids cannot rescue inhibition of endogenous fatty acid synthesis in E. coli because β-hydroxyacyl-ACP generated by FASII is required for the synthesis of essential lipopolysaccharides. A pathway to generate β-hydroxyacyl-ACP from exogenous fatty acids does not exist.
E. coli also encodes for a bifunctional 2-acylglycerophosphoethanolamine acyltransferase [23]. The carboxy-terminus of this bifunctional protein contains the acyl-ACP synthetase (Aas) domain that generates the acyl-ACP from ACP and exogenous fatty acids, while the amino-terminus of this protein catalyzes the acylation of lysophospholipids using acyl-ACP [46]. Lysophospholipids are generated from the synthesis of lipoproteins, where the 1-acyl chain of phospholipids is used to acylate the lipoprotein [47]. The lysophospholipid is reacylated into a phospholipid by the 2-acylglycerophosphoethanolamine acyltransferase [48]. The acyl-ACP generated by the carboxy-terminal Aas cannot dissociate from the enzyme complex, and therefore the acyl-ACP can only be used for acylating lysophospholipids [49,50]. The FadD is responsible for the bulk of exogenous fatty acid incorporation in E. coli, while the bifunctional 2-acylglycerophosphoethanolamine acyltransferase pathway is only involved in recycling lysophospholipids.
2.2. Acyl-ACP Synthetase in Vibrio harveyi
The first described soluble acyl-ACP synthetase is from Vibrio harveyi (VhAas) [34]. The expression of VhAas in E. coli allows the elongation of medium chain exogenous fatty acids and their subsequent incorporation into the membrane phospholipids [35,51]. VhAas accepts a broad array of acyl chains including unnatural fatty acid analogues, and has been extensively used as a chemical biology tool to generate unnatural acyl-ACP molecules [52]. However, the function of VhAas in the parent organism has been poorly characterized. The low acyl chain selectivity of the VhAas suggests that it plays a role other than incorporating exogenous fatty acids into the phospholipids because bacteria have high acyl chain selectivity for their phospholipids [53]. Bacterial bioluminescence is generated from the conversion of acyl aldehyde, O2, and reduced flavin mononucleotide into free fatty acids, H2O and oxidized flavin mononucleotide. This acyl-ACP synthetase could be involved in recycling the free fatty acid generated by the bacterial luciferase.
2.3. Acyl-ACP Synthetase in Chlamydia trachomatis
The obligate intracellular parasite C. trachomatis utilizes an acyl-ACP synthetase to incorporate exogenous fatty acids from the host into its phospholipid (Fig. 3) [8,54]. Isotopic labeling experiments found that deuterium labeled, exogenous palmitic acids were incorporated into both host and chlamydial synthesized phospholipids in a C. trachomatis infected HeLa cell culture. In contrast, deuterium labeled, exogenous lauric acid was incorporated only into chlamydial synthesized phospholipids as elongated palmitic acid. These labeling experiments show that C. trachomatis is able to elongate exogenous fatty acids before using them for phospholipids synthesis. Bioinformatic analysis found that C. trachomatis encodes for 2 separate genes homologous to the two separate domains of the bifunctional E. coli acyl-ACP synthetase/2-acylglycerophosphoethanolamine acyltransferase [8]. The independent encoding of the acyl-ACP synthetase domain (AasC) in C. trachomatis suggests that acyl-ACP generated from exogenous fatty acids can dissociate from the AasC and offers a mechanism by which exogenous fatty acids are elongated in C. trachomatis. Biochemical analysis confirmed that the AasC can catalyze the formation of acyl-ACP with preference for the saturated 16:0 fatty acid over the unsaturated 18:1 fatty acid. Expression of AasC in E. coli allowed elongation of exogenous fatty acids prior to incorporation into phospholipids. The function of the 2-acylglycerophosphoethanolamine acyltransferase was also confirmed using biochemical and genetic complementation experiments. Together, these experiments show that exogenous fatty acids are converted into acyl-ACP by the C. trachomatis AasC. The acyl-ACP synthesized from AasC undergoes the same fate as endogenously synthesized acyl-ACP: elongation by FASII or acylation by the acyltransferase system [54]. The most abundant fatty acids synthesized by the mammalian host are 16:0 and 18:1 fatty acids. AasC has high preference for 16:0 over 18:1 and 16:0 is the major constituent of the 1-position of C. trachomatis phospholipid. Therefore, the purpose of AasC is to selectively scavenge saturated fatty acids from the host.
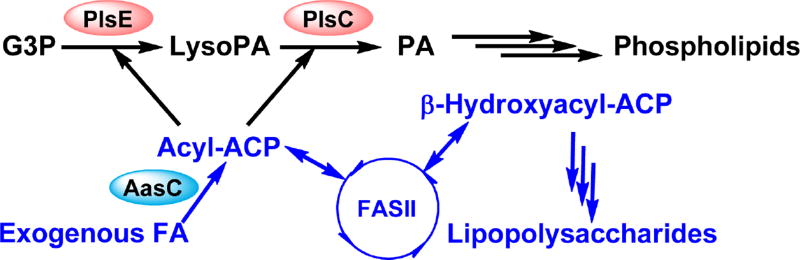
Fig. 3. Phosphatidic acid synthesis and exogenous fatty acid incorporation in C. trachomatis.
Phosphatidic acid is synthesized from the successive acylation of G3P by the G3P acyltransferase PlsE and 1-acyl-sn-glycerol-3-phosphate acyltransferase PlsC in C. trachomatis. The C. trachomatis PlsE is a soluble G3P acyltransferase. Host fatty acids are converted into acyl-ACP by the C. trachomatis acyl-ACP synthetase (AasC). The C. trachomatis acyl-ACP synthetase and lysophospholipid acyltransferase are encoded as separate enzymes rather than a bifunctional enzyme as found in E. coli. This organizational change allows the resulting acyl-ACP to undergo similar fates as endogenously synthesized acyl-ACP: elongated by the FASII or used as an acyl-donor by the acyltransferases. Despite actively incorporating host fatty acids for phospholipid synthesis, C. trachomatis cannot bypass FabI inhibition. The 2-position of C. trachomatis phospholipids has high preference for anteiso15:0 fatty acid, which cannot be obtained from the host.
An alternative model proposed that C. trachomatis also synthesized phospholipids by deacylating the 2-position of host phospholipids using the host cytosolic phospholipase A2 and reacylating it with endogenously synthesized branched-chain fatty acids [55,56]. Additional reports propose that endogenously synthesize fatty acids are exported out of the C. trachomatis cell and into the chlamydial inclusion, converted to acyl-CoA by the host acyl-CoA synthetase, and used to acylate the deacylated host phospholipids in the inclusion [57,58]. These models for phospholipid synthesis are unprecedented in literature and are not consistent with other experimental results. Molecular species analysis by mass spectrometry found no evidence for the products of the proposed deacylation/reacylation in C. trachomatis infected HeLa cell cultures [54]. The deacylation/reacylation model also proposes the extracellular synthesis of phospholipids, which has never been described and would seem to violate the principle of thermodynamics. It is unclear why C. trachomatis would encode for such a convoluted mechanism for synthesizing phospholipids when it has a vastly reduced genome [59] and already encodes for all the normal bacterial enzymes required for phospholipid synthesis and exogenous fatty acid incorporation [8,54].
2.4. Acyl-ACP and Acyl-CoA Synthetase in Neisseria gonorrhoeae
Isotopic labeling experiments found that exogenous lauric acids are also elongated before being incorporated into the phospholipids of N. gonorrhoeae (Fig. 4) [9]. Bioinformatic analysis found 2 predicted acyl-CoA/ACP synthetase genes in the N. gonorrhoeae genome. Genetic complementation studies identified NGO0530 as an acyl-ACP synthetase and NGO1213 as an acyl-CoA synthetase [9]. The elongation of exogenous fatty acids by N. gonorrhoeae suggests that exogenous fatty acids are converted into acyl-ACP by NGO0530. The acyl-ACP is subsequently used by the PlsX/PlsY acyltransferase system to make phosphatidic acid. How the acyl-CoA synthetase, NGO1213, functions in N. gonorrhoeae biology has yet to be determined. The synthesis of acyl-CoA molecules in N. gonorrhoeae is unexpected because N. gonorrhoeae lacks a β-oxidation system to breakdown acyl-CoA [60]. It is unknown if the N. gonorrhoeae acyltransferases can use acyl-CoA as substrates. PlsB and PlsC acyltransferases from species such as E. coli can use acyl-CoA as the acyl donor. However, PlsX cannot function as an acyl-CoA/phosphate transacylase meaning that acyl-CoA cannot be converted to form acyl-phosphate or acyl-ACP in the cases where the acyltransferases are specific for this acyl donors.
Fig. 4. Phosphatidic acid synthesis and exogenous fatty acid incorporation in Neisseria.
Neisseria utilize the PlsX/PlsY pathway for the synthesis of phosphatidic acid. Acyl-ACP is converted into acyl-phosphate by the acyl-ACP:phosphate transacylase PlsX. The G3P acyltransferase PlsY utilizes the acyl-phosphate to acylate G3P in the first acylation step of phosphatidic acid synthesis. The acyltransferase PlsC acylates the resulting lysophosphatidic acid using acyl-ACP in the second step to generate phosphatidic acid. Exogenous fatty acids are introduced into phosphatidic acid synthesis via conversion into acyl-ACP by the acyl-ACP synthetase in Neisseria. The resulting acyl-ACP undergoes the same fate as endogenously synthesized acyl-ACP. Lipopolysaccharides are not essential in Neisseria, and the major acyl chains found in Neisseria are 16:0 and 16:1 fatty acids. However, 16:0 and 16:1 fatty acid complementation cannot bypass the inhibition of endogenous fatty acid synthesis. It is unclear why this is the case.
2.5. Exogenous Fatty Acids Cannot Bypass the Inhibition of Endogenous Fatty Acid Synthesis in C. trachomatis and N. gonorrhoeae
Experiments show that exogenous fatty acids cannot bypass inhibition of endogenous fatty acid synthesis in either C. trachomatis or N. gonorrhoeae. Despite actively incorporating exogenous fatty acids synthesized from the host and available from the growth media, inhibition of FabI blocks the replication of the metabolically active C. trachomatis reticulate body over the entire growth phase [61]. The essentiality of endogenous fatty acid synthesis is likely due to a combination of the selectivity of the chlamydial PlsC for endogenously synthesized anteiso15:0 fatty acid and the essentiality of β-hydroxyacyl-ACP for lipopolysaccharide synthesis.
Similarly, addition of the fatty acid species found in N. gonorrhoeae phospholipids cannot rescue FabI inhibition [9]. Why exogenous fatty acids cannot rescue the inhibition of FASII in N. gonorrhoeae is unclear. Lipopolysaccharides are not essential for N. gonorrhoeae so FASII is not required to generate β-hydroxyacyl-ACP [62,63]. The same fatty acid species found in the phospholipids were supplemented so the lack of bypass is not due to the incorrect acyl chain species. These results highlight the need to experimentally verify the efficacy of FASII inhibition against distinct bacterial groups because bypass cannot be reliably predicted by the existence of an acyl-ACP synthetase in the genome.
2.6. Fatty Acid Kinase in Gram-Positive Bacteria
Gram-positive bacteria use a fatty acid kinase system for the incorporation of exogenous fatty acids (Fig. 5) [36,37,64]. The exogenous fatty acids are bound by the fatty acid binding protein, FakB, and then phosphorylated by the fatty acid kinase, FakA. FakB can deliver the resulting acyl-phosphate to the membrane where it is used by the acyltransferase PlsY to acylate G3P in the first step of phosphatidic acid synthesis. The acyl-phosphate can also be converted into acyl-ACP by the acyl-ACP/phosphate transacylase PlsX [36]. The resulting acyl-ACP undergoes the same fate as the endogenously synthesized acyl-ACP –it can be elongated by FASII and converted back into acyl-phosphate or used for phosphatidic acid synthesis by PlsC. Gram-positive bacteria do not have an outer membrane and fatty acids derived from FASII are used for phospholipid and glycolipid synthesis. One might think that providing the correct species of exogenous fatty acids should be able to bypass the need for endogenous fatty acid synthesis. However, whether exogenous fatty acids can actually bypass endogenous synthesis varies by bacterial taxa and how exogenous fatty acids regulate endogenous synthesis [11,65,66].
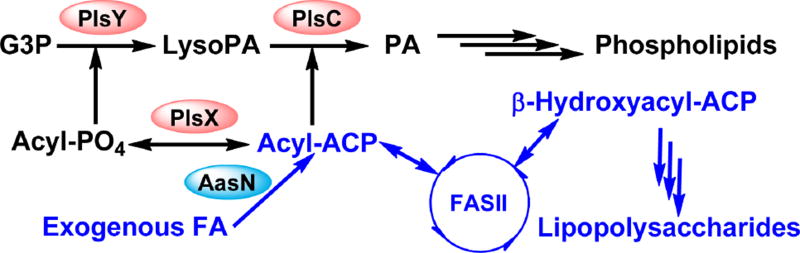
Fig. 5. Phosphatidic acid synthesis and exogenous fatty acid incorporation in Gram-positive bacteria.
Gram-positive bacteria utilize the PlsX/PlsY pathway for the synthesis of phosphatidic acid. PlsX converts acyl-ACP into acyl-phosphate, which is used as the acyl donor by the acyltransferase PlsY in the synthesis of lysophosphatidic acid. PlsC utilizes acyl-ACP as the acyl donor to generate phosphatidic acid. Exogenous fatty acids are converted into acyl-phosphates by the fatty acid kinase system FakA/B. The resulting acyl-phosphate can be converted into acyl-ACP by the acyl-ACP:phosphate transacylase PlsX. The resulting acyl-ACP can be elongated by the FASII, used by PlsC, or converted back into acyl-phosphate. Exogenous fatty acids can bypass the inhibition of FASII in Lactobacillales but not Bacillales. This difference is due to how exogenous fatty acids regulate endogenous fatty acid synthesis in these two bacterial orders. In Lactobacillales, exogenous fatty acids cause a dose dependent and complete inhibition of the endogenous synthesis of fatty acids, effectively rendering FASII inactive and bypassed. In Bacillales, exogenous fatty acids cause a dose dependent, but only partial inhibition of the endogenous synthesis of fatty acids. Inhibition of FASII in Bacillales causes depletion of the free ACP pool by tying them up as short chain acyl-ACP. Exogenous fatty acids cannot be converted into acyl-ACP, and phospholipid synthesis is halted.
2.7. Fatty Acid Kinase in Lactobacillales Allows Bypass of Endogenous Synthesis
Members of the order Lactobacillales, which includes Streptococcus, Lactococcus, Lactobacillus, and Clostridia, can bypass endogenous fatty acid synthesis using exogenous fatty acids [65]. FASII can be completely shut down in presence of exogenous fatty acids, allowing the synthesis of phospholipids entirely from exogenous fatty acids. In the most extreme example, certain Lactobacillus species don’t encode the genes for FASII and synthesize phospholipids entirely from exogenous fatty acids [67]. Clearly, FASII inhibition would be ineffective in such bacteria. Exogenous fatty acids regulate FASII activity via both transcriptional and biochemical mechanisms. First, FASII genes are encoded in a single locus in these bacteria, and are coordinately regulated by the transcriptional repressor FabT [68]. FabT binding to long-chain acyl-ACP increases the FabT affinity for the repressor binding sites and can completely down-regulate the expression of the FASII genes [69]. Second, exogenous fatty acids cause a dose dependent repression of endogenous fatty acid synthesis in these bacteria, including production of malonyl-CoA, the rate-determining intermediate required for endogenous fatty acid synthesis [11]. The metabolite(s) and enzyme(s) responsible for this biochemical feedback is unknown, but the complete inhibition of fatty acid synthesis and the reduction of malonyl-CoA points to acyl-phosphate or acyl-ACP inhibition of the acetyl-CoA carboxylase complex, the initiator of FASII [70].
2.8. Fatty Acid Kinase in Bacillales Cannot Bypass Endogenous Synthesis
In contrast, members of the order Bacillales, which includes Staphylococcus and Listeria, cannot bypass FASII inhibition using exogenous fatty acids. No combination of exogenous fatty acids can rescue FASII inhibition in S. aureus without genetic modification to the reference organism [11,16]. Host fatty acids cannot rescue the FabI inhibition of intracellular Listeria monocytogenes, and exogenous fatty acid combinations cannot replace endogenous synthesis in extracellular cultures [10]. Exogenous fatty acids cause only a dose dependent, partial inhibition of fatty acid synthesis in these bacteria [11]. Malonyl-CoA levels remain elevated and endogenous fatty acids made up half or more of the acyl chains in phospholipid synthesis even in presence of exogenous fatty acids [10,11]. The inhibition of the remaining FASII activity causes accumulation of short chain acyl-ACP and depletion of free ACP [16]. The lack of free ACP prevents the synthesis of acyl-ACP from exogenous fatty acid. Because the PlsC acyltransferase requires acyl-ACP of the appropriate chain length in the second step of phosphatidic acid synthesis, phospholipid synthesis is halted by FASII inhibitors even in presence of exogenous fatty acids. Exogenous fatty acids also do not down-regulate the expression of FASII genes. The FASII genes are spread throughout the genome, and regulated by the FapR repressor [71]. The FapR repressor is released from the DNA binding site after binding to malonyl-CoA to allow for the expression of the FASII genes [72]. Because malonyl-CoA levels remain high in presence of exogenous fatty acids, FASII genes continue to be transcribed.
One major difference between Bacillales and Lactobacillales that might explain the difference in the regulation of FASII in presence of exogenous fatty acids is their respective acyl-chain composition. Membrane fluidity is controlled by the ratio of saturated to unsaturated fatty acids in the phospholipids of mammals and bacteria such as order Lactobacillales [73]. The fatty acid species produced by Lactobacillales FASII can be salvaged from the mammalian host, meaning that exogenous fatty acids from the host could fully replace endogenous synthesis. In contrast, anteiso15:0 fatty acid is found at the 2-position of Staphylococcus and Listeria phospholipids [11,65]. Because branched-chain anteiso15:0 fatty acids are not made by the host, endogenous synthesis of branched-chain fatty acids must continue even in presence of exogenous host fatty acids to ensure proper membrane function. Decreasing the ratio of branched-chain fatty acids severely decreased the extracellular growth rate of Staphylococcus and Listeria, while restoring the ratio of branched-chain fatty acids restored the growth rate [31,32]. Decreasing the content of branched-chain fatty acids also inhibited intracellular pathogenesis and proliferation in a mouse peritonitis model [10,74,75]. Branched-chain amino acids are precursors to branched-chain fatty acids, and the increased import of branched-chain amino acids correlated with increased virulence while the decreased import of branched-chain amino acids correlated with decreased virulence [76]. Existing data in literature strongly support the importance of anteiso15:0 fatty acids in Bacillale phospholipids.
There are disagreements about what fraction of the 2-position of S. aureus phospholipids can be occupied by mammalian fatty acids such as 16:0 or 18:1 when endogenous fatty acid synthesis is functional. A paper in 2011 [11] reported that the 2-position is exclusively occupied by 15:0 fatty acids even in presence of millimolar concentrations of 18:1, while a paper in 2016 [77] reported that branched-chain fatty acids were reduced to less than 40% of the total cellular acyl chain composition so 16:0 and 18:1 is assumed to make up at least 10% of the 2-position composition. The difference between these two papers is what was measured and how it was measured. Parsons and coworkers [11] used molecular species analysis via mass spectrometry to look specifically at the acyl chain position of phosphatidylglycerol, the major phospholipid class in S. aureus. It was found that all phosphatidylglycerol species contained 15:0 fatty acids. In contrast, Sen and coworkers [77] performed gas chromatography analysis on fatty acid methyl esters generated from hydrolyzing the total lipid extracts. Because lipid extracts contain non-phospholipid components like staphyloxanthin and media components such as free fatty acids and lipoproteins adsorbed to the bacterial cell surface, this form of analysis is less accurate at measuring phospholipid acyl chain composition. In our experience, free fatty acids from the media associate with the bacterial cell wall and are difficult to remove by washing with buffers.
3. EVOLUTIONARY CHANGES IN BACTERIAL PHOSPHOLIPID SYNTHESIS AND EXOGENOUS FATTY ACID INCORPORATION
Bacteria evolve rapidly through genetic alterations in response to changes in their environment. The models of exogenous fatty acid incorporation described in the previous section hold true to the particular reference strain in the specific experimental context. However, evolution is an ongoing processing, and this section discusses some of the recently characterized changes that can occur to the bacterial exogenous fatty acid incorporation and phospholipid synthesis systems, as well as their biological consequences. Potential alterations to how bacteria use exogenous fatty acids have profound consequences on biology and medicine.
3.1. Bypassing FASII Inhibition through Deactivating Endogenous Fatty Acid Synthesis in S. aureus
A current topic is whether S. aureus can become functional, obligate fatty acid auxotrophs through genetic mutations in FASII to bypass the inhibition of bacterial fatty acid synthesis inhibitors [11,65,66]. With the anti-staphylococcal FabI inhibitor AFN-1252 in late phase clinical trials [14,78–80], this debate takes on a greater significance. AFN-1252 targets the enoyl-ACP reductase (FabI) of S. aureus. FabI, along with another FASII enzyme FabH, was validated as an antibiotic target by the combined genetic and high-throughput screens conducted by GlaxoSmithKline [81,82]. Lead compounds against FabI and FabH generated by the GlaxoSmithKline efforts was demonstrated to work in the animal model against S. aureus [83,84]. AFN-1252 was derived from the scaffold generated by the GlaxoSmithKline lead through rounds of structure based optimization [15].
In 2009, Brinster et al argued that fatty acid synthesis enzymes are not valid antibiotic targets in Gram-positive bacteria because Gram-positive bacteria are able to bypass FASII inhibition by incorporating exogenous fatty acids when endogenous synthesis is inhibited [65]. They demonstrated that Streptococcus agalactiae shuts down endogenous fatty acid synthesis in presence of human serum fatty acids, and can grow with deletions in the endogenous fatty acid synthesis genes in presence of human serum fatty acids. This result was generalized to all Gram-positive bacteria and used to argue against fatty acid synthesis as a drug target in Gram-positive bacteria including S. aureus because exogenous fatty acids are abundant in animal models. This study was met with intense skepticism by several research groups because a variety of FASII inhibitors have been demonstrated to be effective against S. aureus in mouse models and in human clinical trials [14,79,84]. Two papers comparing fatty acid metabolism between Streptococcus and Staphylococcus found that while exogenous fatty acids can bypass FASII inhibition in Streptococcus, exogenous fatty acids cannot bypass FASII inhibition in Staphylococcus [11,66]. This differential susceptibility is due to differences in the regulation of endogenous fatty acid synthesis by exogenous fatty acids as described in sections 1.2.6–1.2.8. S. aureus could bypass FASII inhibition using exogenous fatty acids if the acetyl-CoA carboxylase (acc) or malonyl CoA-acyl carrier protein transacylase (fabD) genes were inactivated, and such a strain has been created [11]. However, this strain experienced significant growth defects in planktonic culture even when supplemented with human serum, and failed to proliferate in a mouse peritonitis model [74].
A recent paper proposed that exogenous fatty acids can enable the emergence of infectious, fatty acid auxotrophic S. aureus resistant to FASII inhibitors [85]. S. aureus was selected on triclosan and a mixture of mammalian fatty acids. Mutants with defects in the FabD gene were found in the selection. Two classes of FabD mutations were found - point mutations in the 196 position (G196V, G196R) of FabD and truncations of the C-terminal of the FabD protein. The FabD truncation mutants were fatty acid auxotrophs, while the point mutants could grow without exogenous fatty acids. These mutants synthesized their phospholipids using exogenous fatty acids in lieu of endogenously synthesized branched-chain fatty acids when grown under exogenous fatty acid supplementation, and are resistant to FASII inhibition when supplemented with exogenous fatty acids. The fitness of these mutants was evaluated in a mouse bacteremia model by injecting the bacteria (106 CFU) into the tail vein of mice. The FabD point mutant had minimal fitness cost in this model, while the FabD truncation mutant had up to a 104 decrease in growth. Bioinformatic analysis found that 3 strains of the ~ 4000 strains of sequenced S. aureus in the NCBI database harbored mutations in the 196 position of FabD. Two strains encoded the FabD[G196R] mutation, while one strain harbored the FabD[G196S] mutation. From these results, the authors suggested that FASII inhibitors will make poor therapy in the clinic due to the quick emergence of mutants refractory to FASII inhibition by incorporating exogenous fatty acids. It is unclear if this conclusion is warranted based on the experimental results. The FabD truncation mutant had a significant growth defect (~104 decrease in the liver), demonstrating that endogenous fatty acid synthesis in S. aureus is important for fitness within the mouse model. The FabD point mutant does not have an apparent fitness cost, but would not be expected to because it can synthesize fatty acids endogenously. The clearest experiment to test the conclusion of the authors would be to directly test the susceptibility of the two classes of FabD mutants against a FASII inhibitor using a previously described mouse model [14,74]. This experiment would clearly show whether the two classes of mutants could bypass the effects of a FASII inhibitor.
The key point of disagreement between this paper and previous literature is the fitness of S. aureus with altered acyl chain composition. Previous work showed that S. aureus with decreased proportion of branched-chain fatty acids have significant growth defects in planktonic culture even when supplemented with human serum and in mouse models [32,74]. This branched-chain fatty acid dependency is also observed in the related Listeria bacteria family [31,86]. One possible explanation of the discrepancy is the mouse model used for the experiment [87]. It is plausible that endogenous fatty acid synthesis is not required in the bacteremia model, but is required for the peritonitis model. It is also important to note that S. aureus lineages are largely host species specific, and mice are not natural hosts for many of the S. aureus strains derived from human infections [88]. Because there is no gold standard mouse model for mimicking Staphylococcus infection in humans, the FabD mutant should be tested in other models simulating other phases of infection beyond blood growth to determine if FASII inhibitors are effective therapeutics. In particular, S. aureus predominantly causes skin and soft tissue infections in humans rather than sepsis and bacteremia, so it is important to evaluate the fitness of the fatty acid auxotroph within a skin and soft tissue infection model [89]. Furthermore, selection screens can introduce other mutations in the genome, making it difficult to assign a specific mutated gene to an altered phenotype without extensively characterizing each mutation. Recapitulating the FASII bypass phenotype by knocking out the fabD in a reference Staphylococcus strain is a classic microbiology experiment that would give greater confidence that FASII inhibitor resistance is due to fabD mutations alone.
3.2. Prevalence of S. aureus fatty acid auxotrophs in nature
The question of whether fatty acid auxotrophs thrive in infected tissue is important to evaluating the potential of FASII therapeutics. Recently, Gloux et al. [90] addressed this question by collecting S. aureus samples for clinical and veterinary settings and plating them on selective media containing exogenous fatty acids and/or triclosan. They report 12% of 695 the isolates had modification in FASII genes. A major challenge with interpreting these results is determining whether the S. aureus FASII mutations existed prior to the selection. Missense mutations at a particular base pair occur at frequencies approximating the error rate in DNA replication [91], and mutations causing inactivation of a protein function occur at even higher frequencies (~1 in 106). Given the number of bacteria spotted on selective media, mutants will arise from a population of wild-type organisms [11,85]. Thus, it is not possible to determine for this approach whether the mutants were selected or whether they existed prior to the experiment. What is required to answer this important question is to use deep sequencing of infected samples to determine the frequency of FASII mutants without amplification or selection of the cells.
Determining the rate and mechanism of resistance mutations against an antibiotic candidate by plating the susceptible bacteria against the antibiotic candidate is a well described and routine step in antibiotic discovery [4,92,93]. Resistant mutants against single-target antibiotics are practically guaranteed to occur and are often used to determine/validate the mechanism of the antibiotic. Extensive literature exists showing the rise of Staphylococcal resistance to the FASII/FabI inhibitor triclosan [5,94,95]. Amino acid mutations to the Staphylococcal FabI can cause significant increase in resistance against triclosan (or AFN-1252) with minimal reduction in fitness [5,16]. FabI mutants pose the greatest threat to FabI therapeutics because these mutants are resistant and grow without exogenous fatty acid supplementation. Significant reservoirs of such mutants can accumulate in environmental S. aureus populations, unlike the fatty acid auxotrophs which can only grow in environments with sufficient exogenous fatty acids. Whether single step mutations leading to high level resistance affect the efficacy of single-target antibiotics is under active discussion and play into the strategy of developing novel antibiotics [4,92,96]. Well-designed and comprehensive experiments characterizing the fitness consequences of these mutations will be essential to interpret their importance to successful antibiotic therapy.
3.3. Fatty Acid Metabolism Alterations in Biofilm Synthesis
Biofilms are bacteria produced matrices of extracellular polymers that allow the surface adherence of normally planktonic bacteria [97,98]. Biofilms are made of primarily polysaccharides, DNA, and peptides. Biofilm formation promotes bacterial persistence and antibiotic resistance by forming dense matrices that allow the cells in the outer layer to protect the cells in the matrix [99]. Biofilm formation also promotes lateral gene transfer and cooperation between different bacterial species within the matrix [100]. Fatty acid derived quorum sensing molecules are known as communication molecules within the biofilm [101,102], but recent research demonstrates that alterations of fatty acid and phospholipid metabolism are also associated with the formation of biofilms.
A transposon insertion library of S. aureus strain USA300 was screened for change in biofilm production [103]. A mutant bearing a transposon insertion in the fakA gene had a 1.9–4.6 fold increase in biofilm formation. Transduction of the transposon and complementation with the wild type fakA gene confirmed that FakA alone is responsible for the change in biofilm production. This experiment is the first to demonstrate that biofilm production is linked to the molecular machinery for incorporating exogenous fatty acids in S. aureus, and offers a molecular link for biofilm formation and fatty acid metabolism. However, further research is necessary to elucidate how these two processes are connected.
Mutations to fatty acid and phospholipid synthesis genes were observed in the evolution of Burkholderia multivorans during a chronic cystic fibrosis infection [104]. B. multivorans is an opportunistic pathogen responsible for chronic lung infections in cystic fibrosis patients. The evolution of a B. multivorans infection in a cystic fibrosis patient was documented over 20 years. Ten gene loci acquired three or more independent mutations over the infection, and three of these gene loci map to key steps in fatty acid and phospholipid metabolism (fabD, accD, and plsX). The other seven loci mapped to gene regulation proteins or membrane transporters. Nonsynonymous substitutions in the fabD gene and ABC transport permease were the first to arise, and these mutants had decreased growth rate, decreased biomass, but no change in the production of biofilms. Additional and subsequent nonsynonymous substitutions in accD, plsX, and OmpR-like response regulator caused increase in biofilm production. Bioinformatic analysis performed for this review found that the nonsynonymous amino acid mutations in the fabD and accD genes do not map to essential catalytic residues. However, some of the mutations such as H89Y in FabD do occur in active site residues, suggesting that these mutations decrease the activity of the enzymes rather than completely abolish the activity. One blind spot in this experiment is that the selection media was not supplemented with exogenous fatty acids, and therefore mutants with severely decreased fatty acid synthesis could never be isolated. Again, this work shows a clear link between fatty acid metabolism and biofilm production, although the underlying mechanism is unclear.
4. Perspectives
Fatty acid synthesis is the most energy and material intensive part of phospholipid synthesis [1], and bacteria have long been recognized to incorporate exogenous fatty acids from the environment [13]. With the discovery of the fatty acid kinase system [37] and the characterization of acyl-ACP synthetase in pathogenic Gram-negative bacteria species [8,9], the molecular mechanisms for the incorporation of exogenous fatty acids in bacteria are largely characterized. However, more research is required to understand how exogenous fatty acid incorporation effects bacterial physiology. One current area of research focuses on understanding whether exogenous fatty acids can bypass the inhibition of endogenous fatty acid synthesis [11,65,66]. The result of this research will determine whether FASII inhibitors will make therapeutically successful antibiotics for the targeted bacterial species. Only Lactobacillales encode for the ability to bypass the inhibition of endogenous fatty acid synthesis using exogenous fatty acids, but can single genetic mutations allow other pathogenic species to bypass FASII inhibition? Another important question is what are the fitness implications of these mutations and can the mutants persist in the environment? More work is required to characterize the fitness of the S. aureus fabD mutants generated from the selection experiments and the putative biochemical defects in the mutant proteins [85]. These questions are also relevant for pathogens like N. gonorrhoeae that could scavenge all the essential FASII products from the host [9].
Recent research shows that the fatty acid and phospholipid biosynthetic genes in B. multivorans are undergoing long term evolutionary changes in cystic fibrosis patients [104]. The mutations that are predicted to decrease FASII activity are associated with increased biofilm formation. FakA inactivation mutants in S. aureus also have increased biofilm production and decreased virulence [37,103], providing another link between phospholipid metabolism and virulence in bacteria. Further work is required to characterize the molecular mechanism linking phospholipid metabolism, biofilm formation, and virulence. Another key question is whether these mutants are able to persist in the environment or if they are dead end mutations that evolve to optimize growth in a specific host niche. Applying culture-independent metagenomic sequencing methods would give a better understanding of the extent of FASII mutations that persist in nature.
Highlights.
Bacterial fatty acid synthesis is a target for antibiotic development
Bacteria use one of three enzymes for the activation of exogenous fatty acid for incorporation into phospholipid
Lactobacillales are the only bacterial order where exogenous fatty acid lead to a shutdown of endogenous fatty acid synthesis
Fatty acid synthesis cannot be bypassed by exogenous fatty acid in other bacteria
Culture-independent metagenomic methods are needed to assess the prevalence of fatty acid synthesis mutations in bacterial infections.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rock CO, Jackowski S. Forty years of fatty acid biosynthesis. Biochem. Biophys. Res. Commun. 2002;292:1155–1166. doi: 10.1006/bbrc.2001.2022. [DOI] [PubMed] [Google Scholar]
- 2.Yao J, Rock CO. Phosphatidic acid synthesis in bacteria. Biochim. Biophys. Acta. 2013;1831:495–502. doi: 10.1016/j.bbalip.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amy CM, Williams-Ahlf B, Naggert J, Smith S. Molecular cloning of the mammalian fatty acid synthase gene and identification of the promoter region. Biochem. J. 1990;271:675–679. doi: 10.1042/bj2710675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao J, Rock CO. Bacterial fatty acid metabolism in modern antibiotic discovery. Biochim. Biophys. Acta. 2016 doi: 10.1016/j.bbalip.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao J, Rock CO. Resistance mechanisms and the future of bacterial enoyl-acyl carrier protein reductase (FabI) antibiotics. Cold Spring Harb. Perspect. Med. 2016;6:a027045. doi: 10.1101/cshperspect.a027045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace J, Bowlin NO, Mills DM, Saenkham P, Kwasny SM, Opperman TJ, Williams JD, Rock CO, Bowlin TL, Moir DT. The discovery of bacterial FASII inhibitors using a novel cellular bioluminescent reporter assay. Antimicrob. Agents Chemother. 2015;59:5775–5787. doi: 10.1128/AAC.00686-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Ma S. Recent advances in inhibitors of bacterial fatty acid synthesis type II (FASII) system enzymes as potential antibacterial agents. ChemMedChem. 2013;8:1589–1608. doi: 10.1002/cmdc.201300209. [DOI] [PubMed] [Google Scholar]
- 8.Yao J, Dodson VJ, Frank MW, Rock CO. Chlamydia trachomatis scavenges host fatty acids for phospholipid synthesis via an acyl-acyl carrier protein synthetase. J. Biol. Chem. 2015;290:22163–22173. doi: 10.1074/jbc.M115.671008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao J, Bruhn DF, Frank MW, Lee RE, Rock CO. Activation of exogenous fatty acids to acyl-acyl carrier protein cannot bypass FabI inhibition in Neisseria. J. Biol. Chem. 2016;291:171–181. doi: 10.1074/jbc.M115.699462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao J, Ericson ME, Frank MW, Rock CO. Enoyl-acyl carrier protein reductase I (FabI) is essential for the intracellular growth of Listeria monocytogenes. Infect. Immun. 2016;84:3597–3607. doi: 10.1128/IAI.00647-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15378–15383. doi: 10.1073/pnas.1109208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black PN, DiRusso CC, Metzger AK, Heimert TL. Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J. Biol. Chem. 1992;267:25513–25520. [PubMed] [Google Scholar]
- 13.Yao J, Rock CO. How bacterial pathogens eat host lipids: Implications for the development of fatty acid synthesis therapeutics. J. Biol. Chem. 2015;290:5940–5946. doi: 10.1074/jbc.R114.636241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banevicius MA, Kaplan N, Hafkin B, Nicolau DP. Pharmacokinetics, pharmacodynamics and efficacy of novel FabI inhibitor AFN-1252 against MSSA and MRSA in the murine thigh infection model. J. Chemother. 2013;25:26–31. doi: 10.1179/1973947812Y.0000000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan N, Albert M, Awrey D, Bardouniotis E, Berman J, Clarke T, Dorsey M, Hafkin B, Ramnauth J, Romanov V, Schmid MB, Thalakada R, Yethon J, Pauls HW. Mode of action, in vitro activity, and in vivo efficacy of AFN-1252, a selective antistaphylococcal FabI inhibitor. Antimicrob. Agents Chemother. 2012;56:5865–5874. doi: 10.1128/AAC.01411-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao J, Maxwell JB, Rock CO. Resistance to AFN-1252 arises from missense mutations in Staphylococcus aureus enoyl-acyl carrier protein reductase (FabI) J. Biol. Chem. 2013;288:36261–36271. doi: 10.1074/jbc.M113.512905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao J, Carter RA, Vuagniaux G, Barbier M, Rosch JW, Rock CO. A pathogen-selective antibiotic minimizes disturbance to the microbiome. Antimicrob. Agents Chemother. 2016;60:4264–4273. doi: 10.1128/AAC.00535-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park HS, Yoon YM, Jung SJ, Yun IN, Kim CM, Kim JM, Kwak JH. CG400462, a new bacterial enoyl-acyl carrier protein reductase (FabI) inhibitor. Int. J Antimicrob. Agents. 2007;30:446–451. doi: 10.1016/j.ijantimicag.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Park HS, Yoon YM, Jung SJ, Kim CM, Kim JM, Kwak JH. Antistaphylococcal activities of CG400549, a new bacterial enoyl-acyl carrier protein reductase (FabI) inhibitor. J. Antimicrob. Chemother. 2007;60:568–574. doi: 10.1093/jac/dkm236. [DOI] [PubMed] [Google Scholar]
- 20.Escaich S, Prouvensier L, Saccomani M, Durant L, Oxoby M, Gerusz V, Moreau F, Vongsouthi V, Maher K, Morrissey I, Soulama-Mouze C. The MUT056399 inhibitor of FabI is a new antistaphylococcal compound. Antimicrob. Agents Chemother. 2011;55:4692–4697. doi: 10.1128/AAC.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lightner VA, Larson TJ, Tailleur P, Kantor GD, Raetz CRH, Bell RM, Modrich P. Membrane phospholipid synthesis in Escherichia coli: Cloning of a structural gene (plsB) of the sn-glycerol-3-phosphate acyltransferase. J. Biol. Chem. 1980;255:9413–9420. [PubMed] [Google Scholar]
- 22.Larson TJ, Lightner VA, Green PR, Modrich P, Bell RM. Membrane phospholipid synthesis in Escherichia coli: Indentification of the sn-glycerol-3-phosphate acyltransferase polypeptide as the plsB gene product. J. Biol. Chem. 1980;255:9421–9426. [PubMed] [Google Scholar]
- 23.Ray TK, Cronan JE., Jr Activation of long chain fatty acids with acyl carrier protein: demonstration of a new enzyme, acyl-acyl carrier protein synthetase, in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1976;73:4374–4378. doi: 10.1073/pnas.73.12.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Y-J, Zhang Y-M, Grimes KD, Qi J, Lee RE, Rock CO. Acyl-phosphates initiate membrane phospholipid synthesis in gram-positive pathogens. Molec. Cell. 2006;23:765–772. doi: 10.1016/j.molcel.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 25.Hara Y, Seki M, Matsuoka S, Hara H, Yamashita A, Matsumoto K. Involvement of PlsX and the acyl-phosphate dependent sn-glycerol-3-phosphate acyltransferase PlsY in the initial stage of glycerolipid synthesis in Bacillus subtilis. Genes Genet. Syst. 2008;83:433–442. doi: 10.1266/ggs.83.433. [DOI] [PubMed] [Google Scholar]
- 26.Coleman J. Characterization of the Escherichia coli gene for 1-acyl-sn-glycerol-3-phosphate acyltransferase (plsC) Mol. Gen. Genet. 1992;232:295–303. doi: 10.1007/BF00280009. [DOI] [PubMed] [Google Scholar]
- 27.Suwabe A, Mason RJ, Smith D, Firestone JA, Browning MD, Voelker DR. Pulmonary surfactant secretion is regulated by the physical state of extracellular phosphatidylcholine. J. Biol. Chem. 1992;267:19884–19890. [PubMed] [Google Scholar]
- 28.Kaneda T. Steroselectivity in the 2-methylbutyrate incorporation into anteiso fatty acids in Bacillus subtilis mutants. Biochim. Biophys. Acta. 1988;960:10–18. doi: 10.1016/0005-2760(88)90003-3. [DOI] [PubMed] [Google Scholar]
- 29.Kaneda T. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol. Rev. 1991;55:288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu K, Ding X, Julotok M, Wilkinson BJ. Exogenous isoleucine and fatty acid shortening ensure the high content of anteiso-C15:0 fatty acid required for low-temperature growth of Listeria monocytogenes. Appl. Environ. Microbiol. 2005;71:8002–8007. doi: 10.1128/AEM.71.12.8002-8007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giotis ES, McDowell DA, Blair IS, Wilkinson BJ. Role of branched-chain fatty acids in pH stress tolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 2007;73:997–1001. doi: 10.1128/AEM.00865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh VK, Hattangady DS, Giotis ES, Singh AK, Chamberlain NR, Stuart MK, Wilkinson BJ. Insertional inactivation of branched-chain a-keto acid dehydrogenase in Staphylococcus aureus leads to decreased branched-chain membrane fatty acid content and increased susceptibility to certain stresses. Appl. Environ. Microbiol. 2008;74:5882–5890. doi: 10.1128/AEM.00882-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Athenstaedt K, Daum G. Phosphatidic acid, a key intermediate in lipid metabolism. Eur. J. Biochem. 1999;266:1–16. doi: 10.1046/j.1432-1327.1999.00822.x. [DOI] [PubMed] [Google Scholar]
- 34.Fice D, Shen Z, Byers DM. Purification and characterization of fatty acyl-acyl carrier protein synthetase from Vibrio harveyi. J. Bacteriol. 1993;175:1865–1870. doi: 10.1128/jb.175.7.1865-1870.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Y, Morgan-Kiss RM, Campbell JW, Chan CH, Cronan JE. Expression of Vibrio harveyi acyl-ACP synthetase allows efficient entry of exogenous fatty acids into the Escherichia coli fatty acid and lipid A synthetic pathways. Biochemistry. 2010;49:718–726. doi: 10.1021/bi901890a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsons JB, Frank MW, Jackson P, Subramanian C, Rock CO. Incorporation of extracellular fatty acids by a fatty acid kinase-dependent pathway in Staphylococcus aureus. Mol. Microbiol. 2014;92:234–245. doi: 10.1111/mmi.12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsons JB, Broussard TC, Bose JL, Rosch JW, Jackson P, Subramanian C, Rock CO. Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 2014;111:10532–10537. doi: 10.1073/pnas.1408797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hisanaga Y, Ago H, Nakagawa N, Hamada K, Ida K, Yamamoto M, Hori T, Arii Y, Sugahara M, Kuramitsu S, Yokoyama S, Miyano M. Structural basis of the substrate-specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer. J. Biol. Chem. 2004;279:31717–31726. doi: 10.1074/jbc.M400100200. [DOI] [PubMed] [Google Scholar]
- 39.Cronan JE, Jr, Subrahmanyam S. FadR, transcriptional co-ordination of metabolic expediency. Mol. Microbiol. 1998;29:937–943. doi: 10.1046/j.1365-2958.1998.00917.x. [DOI] [PubMed] [Google Scholar]
- 40.Green PR, Merrill AH, Jr, Bell RM. Membrane phospholipid synthesis in Escherichia coli: Purification, reconstitution, and characterization of sn-glycerol-3-phosphate acyltransferase. J. Biol. Chem. 1981;256:11151–11159. [PubMed] [Google Scholar]
- 41.Weeks G, Shapiro M, Burns RO, Wakil SJ. Control of fatty acid metabolism. I. Induction of the enzymes of fatty acid oxidation in Escherichia coli. J. Bacteriol. 1969;97:827–836. doi: 10.1128/jb.97.2.827-836.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overath P, Raufuss EM. The induction of the enzymes of fatty acid degradation in Escherichia coli. Biochem. Biophys. Res. Commun. 1967;29:28–33. doi: 10.1016/0006-291x(67)90535-9. [DOI] [PubMed] [Google Scholar]
- 43.Morgan-Kiss RM, Cronan JE. The Escherichia coli fadK (ydiD) gene encodes an anerobically-regulated short-chain Acyl-CoA synthetase. J. Biol. Chem. 2004 doi: 10.1074/jbc.M405233200. [DOI] [PubMed] [Google Scholar]
- 44.Raetz CRH. Structure and Biosyntheis of Lipid A in Escherichia Coli. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger H, editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology; Washington, D.C.: 1987. pp. 498–503. [Google Scholar]
- 45.Zhang G, Meredith TC, Kahne D. On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr. Opin. Microbiol. 2013;16:779–785. doi: 10.1016/j.mib.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackowski S, Jackson PD, Rock CO. Sequence and function of the aas gene in Escherichia coli. J. Biol. Chem. 1994;269:2921–2928. [PubMed] [Google Scholar]
- 47.Jackowski S, Rock CO. Transfer of fatty acids from the 1-position of phosphatidylethanolamine to the major outer membrane lipoprotein of Escherichia coli. J. Biol. Chem. 1986;261:11328–11333. [PubMed] [Google Scholar]
- 48.Jackowski S, Hsu L, Rock CO. 2-Acylglycerophosphoethanolamine acyltransferase/acyl-[acyl-carrier-protein] synthetase from Escherichia coli. Methods Enzymol. 1992;209:111–117. doi: 10.1016/0076-6879(92)09015-u. [DOI] [PubMed] [Google Scholar]
- 49.Hsu L, Jackowski S, Rock CO. Isolation and characterization of Escherichia coli K-12 mutants lacking both 2-acyl-glycerophosphoethanolamine acyltransferase and acyl-acyl carrier protein synthetase activity. J. Biol. Chem. 1991;266:13783–13788. [PubMed] [Google Scholar]
- 50.Rock CO, Garwin JL. Preparative enzymatic synthesis and hydrophobic chromatography of acyl-acyl carrier protein. J. Biol. Chem. 1979;254:7123–7128. [PubMed] [Google Scholar]
- 51.Jiang Y, Chan CH, Cronan JE. The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain acyl-CoA synthetase family. Biochemistry. 2006;45:10008–10019. doi: 10.1021/bi060842w. [DOI] [PubMed] [Google Scholar]
- 52.Beld J, Finzel K, Burkart MD. Versatility of acyl-acyl carrier protein synthetases. Chem. Biol. 2014;21:1293–1299. doi: 10.1016/j.chembiol.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y-M, Rock CO. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 54.Yao J, Cherian PT, Frank MW, Rock CO. Chlamydia trachomatis relies on autonomous phospholipid synthesis for membrane biogenesis. J. Biol. Chem. 2015;290:18874–18888. doi: 10.1074/jbc.M115.657148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su H, McClarty G, Dong F, Hatch GM, Pan ZK, Zhong G. Activation of Raf/MEK/ERK/cPLA2 signaling pathway is essential for chlamydial acquisition of host glycerophospholipids. J. Biol. Chem. 2004;279:9409–9416. doi: 10.1074/jbc.M312008200. [DOI] [PubMed] [Google Scholar]
- 56.Hatch GM, McClarty G. Phospholipid composition of purified Chlamydia trachomatis mimics that of the eucaryotic host cell. Infect. Immun. 1998;66:3727–3735. doi: 10.1128/iai.66.8.3727-3735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Recuero-Checa MA, Sharma M, Lau C, Watkins PA, Gaydos CA, Dean D. Chlamydia trachomatis growth and development requires the activity of host long-chain acyl-CoA synthetases (ACSLs) Sci. Rep. 2016;6:23148. doi: 10.1038/srep23148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soupene E, Wang D, Kuypers FA. Remodeling of host phosphatidylcholine by Chlamydia acyltransferase is regulated by acyl-CoA binding protein AcBD6 associated with lipid droplets. MicrobiologyOpen. 2015;4:235–251. doi: 10.1002/mbo3.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 60.Isabella VM, Clark VL. Identification of a conserved protein involved in anaerobic unsaturated fatty acid synthesis in Neiserria gonorrhoeae: implications for facultative and obligate anaerobes that lack FabA. Mol. Microbiol. 2011;82:489–501. doi: 10.1111/j.1365-2958.2011.07826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao J, Abdelrahman YM, Robertson RM, Cox JV, Belland RJ, White SW, Rock CO. Type II fatty acid synthesis is essential for the replication of Chlamydia trachomatis. J. Biol. Chem. 2014;289:22365–22376. doi: 10.1074/jbc.M114.584185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bos MP, Tommassen J. Viability of a capsule- and lipopolysaccharide-deficient mutant of Neisseria meningitidis. Infect. Immun. 2005;73:6194–6197. doi: 10.1128/IAI.73.9.6194-6197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Piet JR, Zariri A, Fransen F, Schipper K, van der Ley P, van de Beek D, van der Ende A. Meningitis caused by a lipopolysaccharide deficient Neisseria meningitidis. J. Infect. 2014;69:352–357. doi: 10.1016/j.jinf.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 64.Broussard TC, Miller DJ, Jackson P, Nourse A, White SW, Rock CO. Biochemical roles for conserved residues in the bacterial fatty acid binding protein family. J. Biol. Chem. 2016;291:6292–6303. doi: 10.1074/jbc.M115.706820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature (London) 2009;458:83–86. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 66.Balemans W, Lounis N, Gilissen R, Guillemont J, Simmen K, Andries K, Koul A. Essentiality of FASII pathway for Staphylococcus aureus. Nature (London) 2010;463:E3. doi: 10.1038/nature08667. [DOI] [PubMed] [Google Scholar]
- 67.Muller JA, Ross RP, Sybesma WFH, Fitzgerald GF, Stanton C. Modification of the technical properties of Lactobacillus johnsonii NCC 533 by supplementing the growth medium with unsaturated fatty acids. Appl. Environ. Microbiol. 2011;77:6889–6898. doi: 10.1128/AEM.05213-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu Y-J, Rock CO. Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 2006;59:551–566. doi: 10.1111/j.1365-2958.2005.04951.x. [DOI] [PubMed] [Google Scholar]
- 69.Jerga A, Rock CO. Acyl-acyl carrier protein regulates transcription of fatty acid biosynthetic genes via the FabT repressor in Streptococcus pneumoniae. J. Biol. Chem. 2009;284:15364–15368. doi: 10.1074/jbc.C109.002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis MS, Cronan JE., Jr Inhibition of Escherichia coli acetyl coenzyme A carboxylase by acyl-acyl carrier protein. J. Bacteriol. 2001;183:1499–1503. doi: 10.1128/JB.183.4.1499-1503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schujman GE, Paoletti L, Grossman AD, de Mendoza D. FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev. Cell. 2003;4:663–672. doi: 10.1016/s1534-5807(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 72.Albanesi D, Reh G, Guerin ME, Schaeffer F, Debarbouille M, Buschiazzo A, Schujman GE, de Mendoza D, Alzari PM. Structural basis for feed-forward transcriptional regulation of membrane lipid homeostasis in Staphylococcus aureus. PLoS Pathog. 2013;9:e1003108. doi: 10.1371/journal.ppat.1003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parsons JB, Rock CO. Bacterial lipids: Metabolism and membrane homeostasis. Prog. Lipid Res. 2013;52:249–276. doi: 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parsons JB, Frank MW, Rosch JW, Rock CO. Staphylococcus aureus fatty acid auxotrophs do not proliferate in mice. Antimicrob. Agents Chemother. 2013;57:5729–5732. doi: 10.1128/AAC.01038-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun Y, Wilkinson BJ, Standiford TJ, Akinbi HT, O’Riordan MXD. Fatty acids regulate stress resistance and virulence factor production for Listeria monocytogenes. J. Bacteriol. 2012;194:5274–5284. doi: 10.1128/JB.00045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaiser JC, Omer S, Sheldon JR, Welch I, Heinrichs DE. Role of BrnQ1 and BrnQ2 in branched-chain amino acid transport and virulence in Staphylococcus aureus. Infect. Immun. 2015;83:1019–1029. doi: 10.1128/IAI.02542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sen S, Sirobhushanam S, Johnson SR, Song Y, Tefft R, Gatto C, Wilkinson BJ. Growth-environment dependent modulation of Staphylococcus aureus branched-chain to straight-chain fatty acid ratio and incorporation of unsaturated fatty acids. PLoS ONE. 2016;11:e0165300. doi: 10.1371/journal.pone.0165300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hafkin B, Kaplan N, Murphy B. Efficacy and Safety of AFN-1252, the First Staphylococcus-Specific Antibacterial Agent, in the Treatment of Acute Bacterial Skin and Skin Structure Infections, Including Those in Patients with Significant Comorbidities. Antimicrob. Agents Chemother. 2016;60:1695–1701. doi: 10.1128/AAC.01741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hunt T, Kaplan N, Hafkin B. Safety, tolerability and pharmacokinetics of multiple oral doses of AFN-1252 administered as immediate release (IR) tablets in healthy subjects. J. Chemother. 2015 doi: 10.1179/1973947815Y.0000000075. (PM:26431470) [DOI] [PubMed] [Google Scholar]
- 80.Kaplan N, Garner C, Hafkin B. AFN-1252 in vitro absorption studies and pharmacokinetics following microdosing in healthy subjects. Eur. J. Pharm. Sci. 2013;50:440–446. doi: 10.1016/j.ejps.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 81.McDevitt D, Payne DJ, Holmes DJ, Rosenberg M. Novel targets for the future development of antibacterial agents. J. Appl. Microbiol. 2002;92(Suppl):28S–34S. [PubMed] [Google Scholar]
- 82.Payne DJ, Warren PV, Holmes DJ, Ji Y, Lonsdale JT. Bacterial fatty-acid biosynthesis: a genomics-driven target for antibacterial drug discovery. Drug Discov. Today. 2001;6:537–544. doi: 10.1016/s1359-6446(01)01774-3. [DOI] [PubMed] [Google Scholar]
- 83.Miller WH, Seefeld MA, Newlander KA, Uzinskas IN, Burgess WJ, Heerding DA, Yuan CC, Head MS, Payne DJ, Rittenhouse SF, Moore TD, Pearson SC, Berry V, DeWolf WE, Jr, Keller PM, Polizzi BJ, Qiu X, Janson CA, Huffman WF. Discovery of aminopyridine-based inhibitors of bacterial enoyl-ACP reductase (FabI) J. Med. Chem. 2002;45:3246–3256. doi: 10.1021/jm020050+. [DOI] [PubMed] [Google Scholar]
- 84.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 85.Morvan C, Halpern D, Kénanian G, Hays C, Anba-Mondoloni J, Brinster S, Kennedy S, Trieu-Cuot P, Poyart C, Lamberet G, Gloux K, Gruss A. Environmental fatty acids enable emergence of infectious Staphylococcus aureus resistant to FASII-targeted antimicrobials. Nat. Commun. 2016;7:12944. doi: 10.1038/ncomms12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu K, Bayles DO, Xiong A, Jayaswal RK, Wilkinson BJ. Precursor and temperature modulation of fatty acid composition and growth of Listeria monocytogenes cold-sensitive mutants with transposon-interrupted branched-chain a-keto acid dehydrogenase. Microbiology. 2005;151:615–623. doi: 10.1099/mic.0.27634-0. [DOI] [PubMed] [Google Scholar]
- 87.Kim HK, Missiakas D, Schneewind O. Mouse models for infectious diseases caused by Staphylococcus aureus. J. Immunol. Methods. 2014;410:88–99. doi: 10.1016/j.jim.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Holtfreter S, Radcliff FJ, Grumann D, Read H, Johnson S, Monecke S, Ritchie S, Clow F, Goerke C, Bröker BM, Fraser JD, Wiles S. Characterization of a mouse-adapted Staphylococcus aureus strain. PLoS ONE. 2013;8:e71142. doi: 10.1371/journal.pone.0071142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bunce C, Wheeler L, Reed G, Musser J, Barg N. Murine model of cutaneous infection with gram-positive cocci. Infect. Immun. 1992;60:2636–2640. doi: 10.1128/iai.60.7.2636-2640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gloux K, Guillemet M, Soler C, Morvan C, Halpern D, Pourcel C, Vu Thien H, Lamberet G, Gruss A. Clinical relevance of FASII bypass in Staphylococcus aureus. Antimicrob. Agents. Chemother. 2017 doi: 10.1128/AAC.02515-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meyerovich M, Mamou G, Ben-Yehuda S. Visualizing high error levels during gene expression in living bacterial cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11543–11548. doi: 10.1073/pnas.0912989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Silver LL, Bostian KA. Discovery and development of new antibiotics: the problem of antibiotic resistance. Antimicrob. Agents Chemother. 1993;37:377–383. doi: 10.1128/aac.37.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Silver LL. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Russell AD. Whither triclosan? J Antimicrob. Chemother. 2004;53:693–695. doi: 10.1093/jac/dkh171. [DOI] [PubMed] [Google Scholar]
- 95.Fan F, Yan K, Wallis NG, Reed S, Moore TD, Rittenhouse SF, DeWolf WE, Jr, Huang J, McDevitt D, Miller WH, Seefeld MA, Newlander KA, Jakas DR, Head MS, Payne DJ. Defining and combating the mechanisms of triclosan resistance in clinical isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 2002;46:3343–3347. doi: 10.1128/AAC.46.11.3343-3347.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Silver LL. Multi-targeting by monotherapeutic antibacterials. Nat. Rev. Drug Discov. 2007;6:41–55. doi: 10.1038/nrd2202. [DOI] [PubMed] [Google Scholar]
- 97.Branda SS, Vik A, Friedman L, Kolter R. Biofilms: the matrix revisited. TIM. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 98.López D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harbor Perspectives in Biology. 2010;2 doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Van Acker H, Coenye T. The role of efflux and physiological adaptation in biofilm tolerance and resistance. J. Biol. Chem. 2016;291:12565–12572. doi: 10.1074/jbc.R115.707257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 2012;65:183. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 101.Davies DG, Marques CN. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shih PC, Huang CT. Effects of quorum-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. J. Antimicrob. Chemother. 2002;49:309–314. doi: 10.1093/jac/49.2.309. [DOI] [PubMed] [Google Scholar]
- 103.Sabirova JS, Hernalsteens JP, De Backer S, Xavier BB, Moons P, Turlej-Rogacka A, De Greve H, Goossens H, Malhotra-Kumar S. Fatty acid kinase A is an important determinant of biofilm formation in Staphylococcus aureus USA300. BMC Genomics. 2015;16:861. doi: 10.1186/s12864-015-1956-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Silva IN, Santos PM, Santos MR, Zlosnik JEA, Speert DP, Buskirk SW, Bruger EL, Waters CM, Cooper VS, Moreira LM. Long-term evolution of Burkholderia multivorans during a chronic cystic fibrosis infection reveals shifting forces of selection. MSystems. 2016;1 doi: 10.1128/mSystems.00029-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Parsons JB, Rock CO. Is bacterial fatty acid synthesis a valid target for antibacterial drug discovery? Curr. Opin. Microbiol. 2011;14:544–549. doi: 10.1016/j.mib.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]