Abstract
Animal cells use a conserved repertoire of intercellular signaling pathways to communicate with one another. These pathways are well-studied from a molecular point of view. However, we often lack an “operational” understanding that would allow us to use these pathways to rationally control cellular behaviors. This requires knowing what dynamic input features each pathway perceives and how it processes those inputs to control downstream processes. To address these questions, researchers have begun to reconstitute signaling pathways in living cells, analyzing their dynamic responses to stimuli, and developing new functional representations of their behavior. Here we review important insights obtained through these new approaches, and discuss challenges and opportunities in understanding signaling pathways from an operational point of view.
Introduction
Systems biology seeks to explain how molecular components function together in circuits to implement key cellular behaviors. An important test, and ultimate goal, of this endeavor is to be able to “operate” cells in a predictable manner, controlling their behaviors in rationally engineered cell-based genetic systems. The theme of this section – the future of systems biology – provides a timely opportunity to think about where we are in relation to this forward-looking goal, and how we might achieve it.
Here, we examine this larger goal in the context of the core intercellular signaling pathways found in animal cells, including Notch, Wnt, BMP/TGFβ, Hedgehog, growth factor signaling and others [1]. These pathways provide a central means of communication between cells in metazoan development. They also represent a set of “control knobs” that can induce or block differentiation into new cell fates [2], manipulate cellular behavior for biomedical applications [3,4] including regenerative medicine [5,6], and provide powerful drug targets [7,8]. Due to their prevalent and diverse roles, and their general conservation across species, these intercellular signaling pathways are now among the best studied systems in biology. At the molecular level, their ligands, receptors, intracellular effectors, transcription factors, and modulators have been identified and many of their interactions have been characterized. We now possess an astonishing amount of molecular information about these pathways, as well as the cellular and tissue-level processes they control.
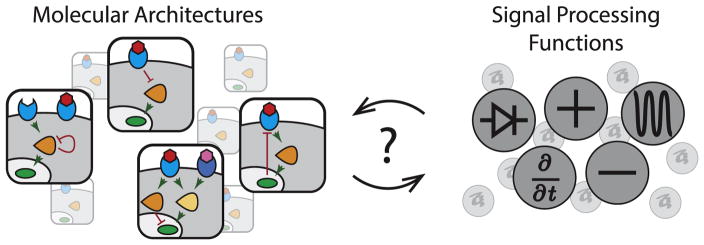
Oddly, however, despite the depth of molecular knowledge, some of the most basic operational questions about these pathways have remained obscure (Figure 1). Operational questions focus less on the description of specific molecular interactions, and more on how the pathway as a whole perceives, processes, and represents extracellular signals within the cell. For example, what quantitative features of its inputs, such as absolute concentration, rates of change in concentration, or relative concentrations of multiple ligands, does each of these pathways perceive? How are inputs processed and ultimately represented in the levels, states, and dynamics of intracellular molecules? Finally, from a comparative point of view, what are the functional differences among the pathways? If they all act to relay information from their ligands to various nuclear and cytoplasmic targets, why do they use such diverse molecular architectures? The answers to these questions are critical both for basic understanding, as well as for emerging applications that seek to use these pathways to direct cells into specific fates in a predictable manner. Without answers to these questions, our position is loosely analogous to knowing the parts of a car and how they are connected, but not knowing how to drive it.
Figure 1.
What signal processing capabilities do core signaling pathways provide? Animal cells utilize several core intercellular signaling pathways that share a similar overall structure (left), in which ligands (red) bind to receptors (blue) and activate transcription factors (green) through intermediate messengers (orange). Despite their similarity, each pathway uses a distinct molecular architecture of protein interactions (left). This representation highlights the pathway architecture but typically provides little information about its operational capabilities. A complementary representation could focus on the signal processing functions of each pathway, indicating how it processes and represents extracellular signals. More research is needed in order to reveal the map between each pathway architecture and the corresponding signal processing function, and to determine how specific interaction parameters quantitatively affect the signal processing functionality.
In this review, we discuss recent work that has begun to transform our understanding of signaling pathways by addressing these issues. We start with a motivating example from microbial signaling that illustrates the power of an integrated understanding of signal processing capabilities and molecular interactions. Next, moving to metazoan pathways, we describe new work that is beginning to provide an operational perspective on growth factor signaling through quantitative characterization of its input–output relationships. We then consider further examples showing how some pathways such as Wnt, TGF-β and EGF encode their inputs and outputs in distinct and unexpected ways, such as in fold-changes or component dynamics. We then turn to the Notch pathway, where recent work hints at a very different type of input–output capability that allows the pathway to effectively “address” signaling to specific cell types. These examples highlight the diverse types of signal processing that have been discovered thus far, but sometimes in a serendipitous manner. In the concluding section, we suggest possible approaches to more systematically map the unique repertoire of signal processing capabilities provided by each pathway. Finally, we discuss the general idea that knowledge of these capabilities could provide alternative conceptual representations of the pathways, complementary to the prevailing molecular representations we work with today, that could help us think about and understand how and why specific pathways are utilized in particular biological contexts. Due to space limitations, we don’t aim to be comprehensive, but rather to illustrate these issues with recent examples.
Architecture determines signal processing in microbial two-component systems: a motivating example
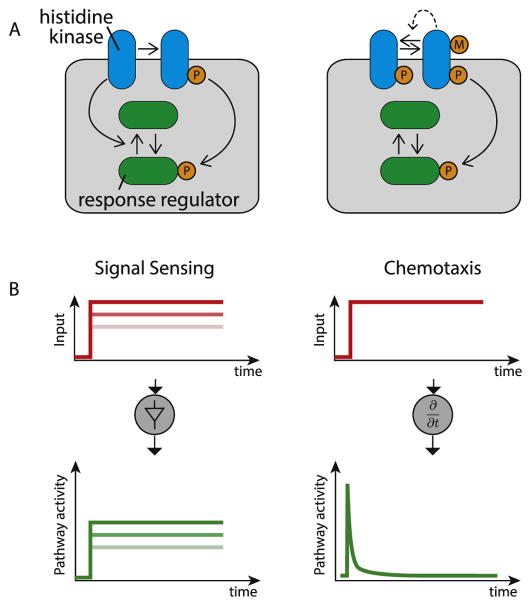
We start by considering the relatively tight integration of molecular and operational understanding that has been achieved in microbial two-component signaling pathways (Figure 2). In bacteria, two-component signaling systems enable cells to respond to diverse inputs and stresses. These systems respond to inputs using a sensor histidine kinase that can transfer phosphate groups to a ‘response regulator’, thereby modulating its activity [9,10]. Counterintuitively, in some two-component systems the sensor kinase is “bifunctional,” both phosphorylating and dephosphorylating the same response regulator (Figure 2A, left). What operational capability does such an apparently futile cycle provide? Computational and experimental work from multiple labs showed that it approximates an ideal linear amplifier, in which an output, the level of phosphorylated response regulator, remains linearly proportional to the rate of kinase activation (and inversely proportional to the phosphatase rate) over a broad range [11,12]. Thus, kinase bifunctionality can be understood to provide the specific signal processing capability of representing stimuli intracellularly with minimal distortion (Figure 2B, left). (Note that it may also provide other benefits such as robustness to component concentrations [13] and insulation between distinct pathways [12,14]).
Figure 2.
Microbial two component systems provide an ideal example in which the relationship between molecular architecture and signal processing functions has been mapped. (A) In two component systems a receptor histidine kinase (blue) phosphorylates a response regulator (green) inducing a response. In some cases (left), the kinase additionally dephosphorylates the response regulator giving rise to an apparent futile cycle. In contrast, the bacterial chemotaxis two component system has a distinct architecture (right). In this case, additional components methylate and demethylate the receptor to adjust its activity (M indicates methylation), and there is indirect negative feedback on kinase activity (dashed line). (B) The two different architectures produce distinct signal processing capabilities. Bifunctionality of the kinase can give rise to an approximately linear amplifier, in which outputs (green) are proportional to inputs (red intensities). By contrast, feedback (dashed line) within the bacterial chemotaxis architecture generate an adaptive response, allowing the system to sense temporal derivatives in its inputs.
By contrast, bacterial chemotaxis – also based on a two-component signaling system – uses a different molecular architecture (Figure 2A, right). In this case, dephosphorylation of the response regulator is catalyzed by a separate phosphatase, and the system uses receptor methylation for additional feedback loops. These molecular differences can be understood in terms of differences in signal processing. Rather than responding to the absolute concentration of the signal, such as an attractant or repellant in the environment, the chemo-taxis circuit tracks temporal changes in its concentration (Figure 2B, right) [15–17].
Thus, here we have two circuit architectures, each providing a distinct signal processing capability: linear amplification vs. temporal derivative. While these systems are not completely understood, the ability to relate molecular architecture directly to signal processing features, and vice versa, enables one to predict how changes in components impact the input–output behavior of the system as a whole, and could be used to forward engineer new signaling devices in bacteria [18]. To what extent is a similar depth of understanding possible in the more complex world of metazoan signaling pathways?
Tunable input–output relationships in growth factor signaling pathways
Most progress towards this goal has been made with growth factor signaling, which is one of the best-studied metazoan signaling systems. For instance, recent experimental mapping of input–output relationships, in conjunction with mathematical modeling, has revealed the tunability of the input–output relationship as a central, functional, feature of these systems. Growth factor signaling is initiated at the cell-surface with the assembly of multimeric ligand-receptor complexes through sequential binding of ligands to receptors. By modeling such multi-step ligand-receptor interactions, Ha and Ferrell recently showed that varying the cooperativity, binding affinities, and ligand-receptor concentration ratio could essentially ‘tune’ the dose–response behavior [19]. Furthermore, the work showed that pathway behavior is not always easy to intuit even with such relatively simple systems, and requires quantitative modeling to develop an accurate understanding. For example, regimes of ‘negative’ cooperativity in ligand-receptor binding, where the rate at which a receptor binds to a first ligand is greater than the rate at which the resulting complex binds to a second ligand, might be expected to suppress sensitivity to ligand. Counterintuitively, however, it can in fact produce an ultrasensitive input-output response when ligands are in short supply relative to receptors.
Interestingly, O’Shaugnessy et al. also identified tunable sensitivity as a key feature of the Raf-MEK-ERK phosphorylation cascade, the step(s) subsequent to ligand-receptor binding in growth factor signaling [20]. By reconstituting the core mammalian kinase cascade in yeast, and thus bypassing the need for ligands and receptors, the authors effectively isolated this module from upstream inputs and downstream responses. They then systematically characterized its input–output features from an operational point of view, varying the levels of its components, and testing the effects of other accessory components. This work revealed that the phosphorylation cascade acts as a tunable amplifier, in which component concentrations can be used to modulate the ultrasensitivity, threshold and amplification of the pathway as a whole.
Thus, growth factor pathways can respond to the concentration of their ligand in a flexible manner, with a range of different sensitivities. These results highlight the need for quantitatively characterizing the input–output properties of each signaling pathway before constructing operational models of its behavior. However, this task is complicated by the fact that pathways may not represent the extracellular ligand concentration in the level of an intracellular protein. That is, the relevant inputs may not simply be instantaneous ligand concentrations, and the relevant ‘outputs’ may not be levels of an intracellular molecule. This is illustrated in the next few examples.
Fold-change signal encoding in the Wnt pathway
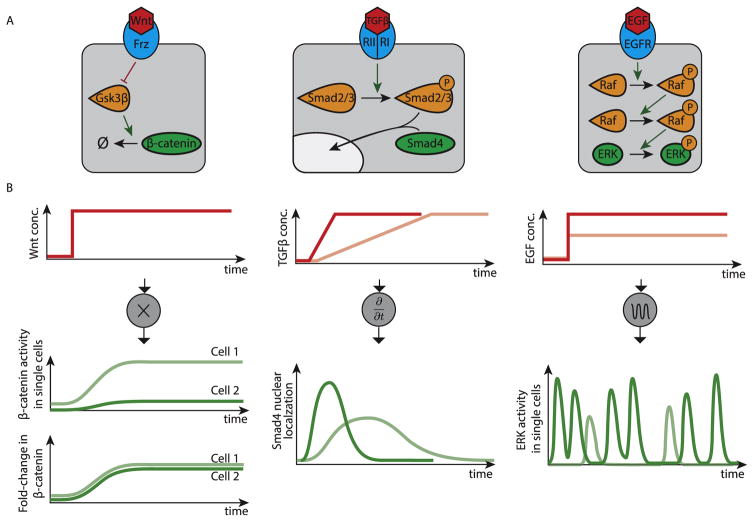
The Wnt pathway is critical for control of proliferation and cell fate, among many other functions. It employs a complex molecular interaction network to control the degradation rate of its second messenger, β-catenin, which acts as a transcriptional co-regulator (Figure 3A, left). When the pathway is inactive, β-catenin is rapidly degraded through a highly active destruction complex composed of multiple proteins. Upon stimulation, the ligand-bound receptors inhibit the activity of the destruction complex, resulting in accumulation of β-catenin and increased activation of downstream targets [21]. Based on these interactions, it was generally assumed that Wnt signaling involved the control of absolute β-catenin level by extracellular Wnt ligand concentration.
Figure 3.
Examples of input and output signal encoding in metazoan signaling pathways. (A) Signaling pathways possess different signal transduction architectures, which influence their signaling processing abilities, shown in B. (B) From left to right, signal processing in the Wnt, TGF-β, and EGF pathways, respectively. In the Wnt pathway, extracellular ligand concentration is encoded in the fold-change in β-catenin response. An increase in extracellular ligand concentration (red line, top) leads to increases in absolute β-catenin levels that vary between cells (green lines, middle), but the fold-change in β-catenin levels is uniform (green lines, bottom). The TGF-β pathway is rate-responsive. An increase in extracellular ligand concentration (red lines, top) leads to an adaptive response in Smad4 nuclear localization (green lines, bottom). The amplitude of the response depends on the rate of increase of ligand concentration (compare light and dark lines in top and bottom plots). The EGF signaling pathway encodes ligand concentration in the frequency of ERK activity pulses. Step increases in ligand concentration (red lines, top) result in sustained, stochastic, pulses in ERK activity (green lines, bottom). The concentration of ligand is reflected in the average frequency of ERK pulses (compare light and dark lines).
By starting with a mathematical model of known molecular components and interactions, and then simplifying it to identify key component combinations, Goentoro and coworkers showed that, across a broad set of parameters, the system effectively encoded the level of extracellular Wnt into a fold-change, rather than a linear increase, in β-catenin levels [22]. Importantly, while the levels of β-catenin showed a high degree of variability from cell to cell, likely due to their sensitivity to small variations in biochemical parameters, their fold-change (the ratio of post- to pre-stimulus levels) was found to be more uniform across cells for a given level of Wnt ligand (Figure 3B, left). This fold-change encoding functionality allows cells to sense ligand levels while being robust to most synthesis or degradation parameters, whose effects on basal and activated level cancel out [22]. It requires that at least some Wnt target genes sense fold-changes, rather than absolute levels, of β-catenin [22] giving rise to adaptive responses. Thus, Wnt may be optimized for controlling transient events, such as cell fate decisions, rather than for continuously transmitting information about extracellular ligand levels.
Rate-responsive signal encoding in the TGF-β pathway
While the Wnt pathway encodes input levels in fold-changes of β-catenin, TGF-β signaling appears to do the opposite, encoding changes in its input in levels of the Smad4 transcription factor. It was recently shown that the TGF-β pathway exhibits adaptive Smad4 dynamics in response to step increases in ligand [23]. Such dynamics allow cells to sense the rate of change of a signal, rather than its absolute level. To demonstrate rate-responsiveness, Sorre et al. used new microfluidic techniques to show that the rate of increase of TGF-β in the media controlled the magnitude of the response (Figure 3, middle) [24]. In this case, the molecular mechanism underlying rate-responsiveness remains incompletely understood, but the authors suggest a functional rationale in terms of accelerating cell fate decisions in response to morphogenetic gradients. This work parallels recent work in bacteria, which showed how cells use rate-responsiveness to control the specificity or generality of a stress response, in terms of target genes activation, depending on the rate of increase of stress [25]. Utilizing rate-responsiveness could open up new strategies for manipulating cells as we improve our ability to exert quantitative control over ligand dynamics, particularly in a pharmacological context [26].
Dynamic signal encoding in the EGF pathway
Other systems appear to encode inputs by continuously generating intracellular dynamics even when the cell is in a constant environment. For example, in the EGF (epidermal growth factor) pathway, activation of the EGF receptor (EGFR) triggers a MAPK (mitogen-activated protein kinase) phosphorylation cascade within the cell, ultimately activating the terminal kinase, ERK (Figure 3A, right). In order to understand how EGF levels modulated ERK activity, Albeck and coworkers monitored the dynamics of an ERK phosphorylation sensor at the single-cell level using timelapse microscopy. Unexpectedly, they discovered that ERK activity in individual cells occurred in discrete, stochastic, and repetitive ‘pulses’, even at constant EGF concentrations [27]. Furthermore, varying EGF concentration modulated the average frequency of these pulses (Figure 3B, right). In fact, this type of frequency-modulated pulsatile dynamics have now been observed across a remarkably diverse set of pathways in bacteria, yeast, and animal cells [28–34], suggesting that the encoding of a constant signal into a dynamic intracellular representation is a pervasive theme.
What functions could dynamic encoding provide? Recent work shows how different types of transcription factor dynamics can generate diverse types of input–output functionality [29,35–37]. For example, dynamic signal encoding allows cells to control intracellular transcription factor activities in time rather than in concentration. In frequency-modulated systems like Erk (as well as Crz1 in yeast), inputs effectively control the fraction of time a transcription factor is active [29,30]. As a result, the average expression of diverse target genes can be maintained in fixed proportions. Even more interestingly, dynamic signal encoding appears to provide powerful ways of integrating and processing signals in time. For example, in yeast, the response to glucose limitation is controlled by the temporal overlap, or relative timing, between two transcription factors that both pulse repetitively in and out of the nucleus and co-regulate some target genes when they are both in the nucleus simultaneously [32]. More recently, Hao and colleagues discovered logical signal processing functions enabled by two paralogous transcription factors exhibiting specific timing differences in their nuclear localization dynamics [38]. It remains to be seen how such distributed time-based strategies play out in core mammalian signaling pathways.
Directionality in Notch signaling
In contrast to the systems above, which use diffusible ligands to transmit signals, the Notch signaling system uses cell-bound ligands for direct communication between adjacent cells, which endows this pathway with some unique properties. At first glance, Notch appears to operate in a relatively straightforward way: membrane-bound ligands expressed in one cell activate receptors in neighboring cells (i.e., in trans), causing cleavage and release of their intracellular domains, which then translocate to the nucleus to activate target genes. However, in addition to this productive interaction, the pathway also incorporates a parallel non-productive interaction, in which ligands and receptors within the same cell (in cis) mutually inactivate each other (Figure 3A). Depending on relative ligand and receptor levels, and the strength of this interaction, “cancellation” of ligand-receptor pairs can lead to preferential ‘sending’ or ‘receiving’ states (Figure 3B) [39]. This in turn could make signaling more unidirectional, i.e. occurring predominantly from cell A to cell B but not vice-versa. Thus, the molecular feature of cis-inhibitory receptor–ligand interactions can provide the signaling capability of transforming continuous variation in ligand (or receptor) levels into sharp differences in signaling ability and directionality.
Notch signaling often occurs in the context of more complex regulation. For example, in lateral inhibition patterning systems, activation of Notch leads to down-regulation of ligand in the same cell, reducing Notch signaling in the neighboring cell, allowing its ligand concentration to increase, and thereby further increasing Notch signaling in the first cell [40]. This intercellular positive feedback loop can generate steady-states that are anti-correlated between neighboring cells [41]. In the context of such a system, cis-inhibition could help to accelerate the dynamics of patterning [42], a capability that could make Notch ideally suited for the types of developmental processes that often deploy it, such as generation of spatially organized distributions of opposite cell fates [42,43].
cis-interactions can allow counter-intuitive response to perturbations. For example, a decrease in the level of a ligand could either increase signaling (due to cis interactions) or decrease signaling (due to trans interactions), or both, depending on the levels of other Notch pathway components expressed in the targeted cell and its neighbors. This has implications in cancer contexts where Notch signaling is often misregulated [7], and in regenerative medicine applications where Notch signaling is used to control cell fate decision-making [3].
From expression levels to communication channels: alternative representations for Notch signaling systems
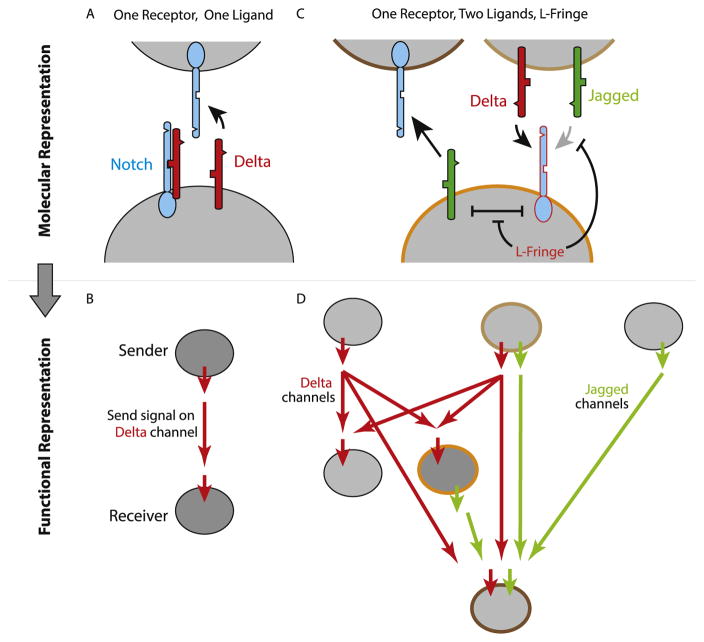
The send/receive property of Notch signaling suggests an alternative representation for intercellular signaling in terms of ‘signaling states’, rather than the molecular interactions of Notch components. A signaling state is defined by the ability of cells in that state to receive signal from, or send signal to, cells in other signaling states. Considering only a single type of Notch ligand and a single type of receptor, the signaling state representation consists only of a single ‘send’ state and a single ‘receive’ state, signaling through a single communication “channel” (Figure 4A and B).
Figure 4.
Expression of different combinations of Notch pathway components controls the specificity of signaling. (A) With one ligand and one receptor, when cis receptor ligand interactions (symmetrical inhibitory arrow) are strong, a cell of interest (lower cell) can predominantly send to Notch in a neighboring cell (as pictured) when Delta ligand (red) exceeds Notch receptor levels (blue), or receive when Delta levels are low (not shown). (B) This behavior can be represented more abstractly as a sending state (upper cell) and a receiving state (lower cell), with a connecting red arrow indicating the capability of sending from a cell in one state to a cell in the other. (C) More complex configurations of Notch components are possible and occur frequently. In this example one possibility is illustrated involving one type of Notch receptor, two ligands (Delta and Jagged in red and green, respectively), and Lunatic Fringe. In this configuration, Fringe suppresses cis and trans interactions between Notch and Jagged, but not between Notch and Delta. As a result, there is no inhibition between Jagged and Notch. This allows the cell to send signals using Jagged while receiving signals from trans Delta ligands using Notch. However, Fringe blocks the ability to receive signals from Jagged (inhibitory arrow, right). (D) A diagram of multiple signaling states possible from other component configurations. The cell states shown in C are highlighted in corresponding outline colors. Red and green arrows indicate communication channels, showing which states are capable of sending and receiving to and from other states. Note that this diagram forms an acyclic directed graph.
However, the Notch pathway comprises multiple ligands, receptors, and other modulators such as the Fringe glycosyltransferases, which alter ligand-receptor interaction strengths in both cis and trans [44]. This molecular diversity could generate a larger set of signaling states and communication channels. Depending on the combination of components expressed in any given cell type, one could expect a variety of signaling states with differing abilities to send to or receive from one another using different ligands. For example, in mammalian cells, Lunatic Fringe strengthens cis and trans interactions between the Notch1 receptor and the Dll1 ligand, while weakening cis and trans interactions between Notch1 and the Jag1 ligand. As a result, cells expressing Lunatic (L-) Fringe, Notch1, and Jag1 at appropriate levels could send signals using Jag1 ligands, receive signals from Dll1 ligands, but not be able to receive signals from Jag1 ligands (Figure 4C) [44].
A representation of Notch in terms of such signaling states could offer insights that would be difficult to obtain at the molecular level. For example, initial work suggests that communication channels appear to form an acyclic directed graph, in which signaling is hierarchical, and homotypic signaling, even when indirect, is suppressed (Figure 4D) [44]. Looking ahead, it will be interesting to see whether a more complete map of signaling states could enable researchers to predict which cell types are capable of signaling to which others in natural contexts.
Conclusions: towards an ‘operating manual’ for intercellular signaling pathways
Together, the results above show that when examined from an operational, signal processing-centric point of view, many pathways offer unexpected, and often counterintuitive, capabilities that could not have been inferred in a straightforward, or qualitative, fashion from known molecular interactions. Most pathways are yet to be analyzed from this operational point of view. However, a few key strategies should help to extend this paradigm in a more systematic and inclusive manner: First, dynamic single cell analysis is critical. Many pathways are already known to be highly dynamic, and most signaling pathways are likely to include at least some dynamic features. However, these dynamics are generally unsynchronized across cells, severely limiting what can be learned even from static, single-cell measurements (let alone population averages). Second, dynamic and quantitative control of inputs is essential. Pathways are as likely to perceive rates of change or, potentially, frequencies of pulsing or oscillation, as they are static ligand concentrations. Techniques for accurate dynamic manipulation of pathway inputs are therefore essential. Third, isolation is powerful: by reconstituting minimal versions of these pathways in cells, isolated as much as possible from natural inputs and outputs, one can study signal processing capabilities more systematically, minimizing confounding downstream effects and other interactions. Different methods must be used to isolate different pathways, given their tight integration with other cellular components, and their diversity of molecular mechanisms. Isolation relies on genetic manipulation of cells, something that is becoming faster and easier, thanks in part to CRISPR technologies, although larger scale genetic circuit engineering remains challenging. Fourth, mathematical modeling plays a powerful, and often essential, role in exploring the potential and actual behavior of core pathways across parameter regimes, and understanding the extent to which operational behaviors can be explained (or not) in terms of known interactions. In most examples above, models were essential for synthesizing the results of experiments and formulating predictions.
These approaches, pursued more systematically and perhaps in a more coordinated manner across laboratories, could begin to yield a kind of ‘operating manual’ representation for the canonical signaling pathways. This representation would provide insights into what kinds of modes each pathway can operate in and how they effectively perceive and process their inputs. It would provide the instructions needed to program cells to interact in predictable ways with endogenous cellular systems for diverse applications including emerging cell-based therapies. At the same time, it would also suggest strategies, inspired by natural pathways, that could help enable engineering of new or modified pathways for synthetic biology applications [45,46].
This operational view, which emphasizes manipulation, engineering, and control, is not a replacement for the ubiquitous molecular circuit diagrams that we rely on today, but rather a complement to them. We anticipate that in the future we will be able to map between molecular and signal processing representations for specific systems. For example, given the gene expression profiles of tumor cells, one should be able to predict how it will interpret particular signals or respond to inhibitors. Similarly, the cell type specific expression of signaling pathway components should help predict which cells are communicating with which others at each stage of development. Throughout the history of science, changes in representation have often led directly to major transformations in understanding. It will be exciting to see whether and how emerging operational representations lead to new conceptual understanding of cellular communication and other systems.
Acknowledgments
This work was supported by the Gordon and Betty Moore Foundation through Grant GBMF2809 to the Caltech Programmable Molecular Technology Initiative, by the Human Frontiers Science Program (EFRI-11137269), by NIH R01 HD075335, by grant W911NF-11-2-0055 from the U.S. Army Research Office and by the Institute for Collaborative Bio-technologies through grant W911NF-09-0001 from the U.S. Army Research Office. This work does not necessarily reflect the position or policy of the Government and no official endorsement should be inferred. N.N was supported by HHMI as an International Student Research fellow.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Housden BE, Perrimon N. Spatial and temporal organization of signaling pathways. Trends Biochem Sci. 2014;39:457–464. doi: 10.1016/j.tibs.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zúñiga-Pflücker JC. T-cell development made simple. Nat Rev Immunol. 2004;4:67–72. doi: 10.1038/nri1257. [DOI] [PubMed] [Google Scholar]
- 3.Delaney C, Heimfeld S, Brashem-Stein C, Voorhies H, Manger RL, Bernstein ID. Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat Med. 2010;16:232–236. doi: 10.1038/nm.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim M, Choe S. BMPs and their clinical potentials. BMB Rep. 2011;44:619–634. doi: 10.5483/BMBRep.2011.44.10.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Date S, Sato T. Mini-gut organoids: reconstitution of the stem cell niche. Annu Rev Cell Dev Biol. 2015;31:269–289. doi: 10.1146/annurev-cellbio-100814-125218. [DOI] [PubMed] [Google Scholar]
- 6.Fordham RP, Yui S, Hannan NRF, Soendergaard C, Madgwick A, Schweiger PJ, Nielsen OH, Vallier L, Pedersen RA, Nakamura T, et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–744. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling are we there yet? Nat Rev Drug Discov. 2014;13:357–378. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]
- 8.Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov. 2014;13:928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 9.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 10.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinar G, Milo R, Martínez MR, Alon U. Input output robustness in simple bacterial signaling systems. Proc Natl Acad Sci U S A. 2007;104:19931–19935. doi: 10.1073/pnas.0706792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulian M. Two-component signaling circuit structure and properties. Curr Opin Microbiol. 2010;13:184–189. doi: 10.1016/j.mib.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batchelor E, Goulian M. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc Natl Acad Sci U S A. 2003;100:691–696. doi: 10.1073/pnas.0234782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Block SM, Segall JE, Berg HC. Impulse responses in bacterial chemotaxis. Cell. 1982;31:215–226. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- 16.Segall JE, Block SM, Berg HC. Temporal comparisons in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1986;83:8987– 8991. doi: 10.1073/pnas.83.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 18.Whitaker WR, Davis SA, Arkin AP, Dueber JE. Engineering robust control of two-component system phosphotransfer using modular scaffolds. Proc Natl Acad Sci U S A. 2012;109:18090–18095. doi: 10.1073/pnas.1209230109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Ha SH, Ferrell JE., Jr Thresholds and ultrasensitivity from negative cooperativity. Science. 2016;352:990–993. doi: 10.1126/science.aad5937. The authors developed a mathematical framework to explore the range of signal processing possibilities at the level of assembly of multimeric ligand-receptor complexes, and discovered a novel, and surprising, role for negative cooperativity in enabling ultrasensitive dose-responses. This study highlights the need for quantitative modeling in understanding signal transduction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Shaughnessy EC, Palani S, Collins JJ, Sarkar CA. Tunable signal processing in synthetic MAP kinase cascades. Cell. 2011;144:119–131. doi: 10.1016/j.cell.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacDonald BT, Tamai K, He X. Wnt/β-Catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9– 26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Goentoro L, Kirschner MW. Evidence that fold-change, and not absolute level, of beta-catenin dictates Wnt signaling. Mol Cell. 2009;36:872–884. doi: 10.1016/j.molcel.2009.11.017. By starting with a complete mathematical model of the Wnt pathway and identifying meaningful combination of parameters the authors showed the existence of a regime in which Wnt induces fold change of beta-catenin. They further demonstrated this experimentally in Xenopus. This study also shows the advantages of a fold change response in generating a robust response, and demonstrates the power of strategic mathematical modeling in revealing new functional abilities of a molecularly well-characterized system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warmflash A, Zhang Q, Sorre B, Vonica A, Siggia ED, Brivanlou AH. Dynamics of TGF-β signaling reveal adaptive and pulsatile behaviors reflected in the nuclear localization of transcription factor Smad4. Proc Natl Acad Sci U S A. 2012;109:E1947–E1956. doi: 10.1073/pnas.1207607109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Sorre B, Warmflash A, Brivanlou AH, Siggia ED. Encoding of temporal signals by the TGF-β pathway and implications for embryonic patterning. Dev Cell. 2014;30:334–342. doi: 10.1016/j.devcel.2014.05.022. The authors previously analyzed TGF-β signaling at the single-cell level using time-lapse microscopy and discovered the surprising dynamic feature of adaptive signaling. Further mathematical modeling predicted a rate-responsive behavior of the pathway, which the authors tested and confirmed by precisely controlling the dynamics of ligand exposure using microfluidic technologies. This study highlights the potential need for such control over input dynamics in order to better understand the capabilities of signaling pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young JW, Locke JCW, Elowitz MB. Rate of environmental change determines stress response specificity. Proc Natl Acad Sci U S A. 2013;110:4140–4145. doi: 10.1073/pnas.1213060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behar M, Barken D, Werner SL, Hoffmann A. The dynamics of signaling as a pharmacological target. Cell. 2013;155:448–461. doi: 10.1016/j.cell.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Albeck JG, Mills GB, Brugge JS. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol Cell. 2013;49:249–261. doi: 10.1016/j.molcel.2012.11.002. This study sought to analyze the signal processing behavior of growth factor signaling quantitatively and at the single-cell level. This effort revealed unexpected pulsatile dynamics in ERK activity, which could have important implications for understanding and perturbing the pathway in many contexts, especially in disease. Like many of the other highlighted work, this work again highlights the need for analyzing signaling dynamically, quantitatively, and at the single-cell level. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Locke JCW, Young JW, Fontes M, Hernández Jiménez MJ, Elowitz MB. Stochastic pulse regulation in bacterial stress response. Science. 2011;334:366–369. doi: 10.1126/science.1208144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao N, O’Shea EK. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat Struct Mol Biol. 2012;19:31– 39. doi: 10.1038/nsmb.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485–490. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yissachar N, Sharar Fischler T, Cohen AA, Reich-Zeliger S, Russ D, Shifrut E, Porat Z, Friedman N. Dynamic response diversity of NFAT isoforms in individual living cells. Mol Cell. 2013;49:322–330. doi: 10.1016/j.molcel.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Lin Y, Sohn CH, Dalal CK, Cai L, Elowitz MB. Combinatorial gene regulation by modulation of relative pulse timing. Nature. 2015;527:54–58. doi: 10.1038/nature15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine JH, Lin Y, Elowitz MB. Functional roles of pulsing in genetic circuits. Science. 2013;342:1193–1200. doi: 10.1126/science.1239999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni Q, Ganesan A, Aye-Han N-N, Gao X, Allen MD, Levchenko A, Zhang J. Signaling diversity of PKA achieved via a Ca2+-cAMP-PKA oscillatory circuit. Nat Chem Biol. 2011;7:34– 40. doi: 10.1038/nchembio.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen AS, O’Shea EK. Encoding four gene expression programs in the activation dynamics of a single transcription factor. Curr Biol. 2016;26:R269–R271. doi: 10.1016/j.cub.2016.02.058. [DOI] [PubMed] [Google Scholar]
- 36.Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao N, Budnik BA, Gunawardena J, O’Shea EK. Tunable signal processing through modular control of transcription factor translocation. Science. 2013;339:460– 464. doi: 10.1126/science.1227299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.AkhavanAghdam Z, Sinha J, Tabbaa OP, Hao N. Dynamic control of gene regulatory logic by seemingly redundant transcription factors. Elife. 2016:5. doi: 10.7554/eLife.18458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. This study used a reconstitution approach to systematically measure the input–output properties of the Notch signaling pathway. In combination with mathematical modeling and single-cell analysis of signaling using timelapse microscopy, this approach revealed an important capability of the pathway, its ability to generate mutually exclusive signaling states. This property of Notch signaling arises as a consequence of a simple ligand-receptor interaction, but could play an important role in many Notch-dependent processes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. 2016;17:722–735. doi: 10.1038/nrm.2016.94. [DOI] [PubMed] [Google Scholar]
- 41.Collier JR, Monk NA, Maini PK, Lewis JH. Pattern formation by lateral inhibition with feedback: a mathematical model of delta-notch intercellular signalling. J Theor Biol. 1996;183:429–446. doi: 10.1006/jtbi.1996.0233. [DOI] [PubMed] [Google Scholar]
- 42.Sprinzak D, Lakhanpal A, LeBon L, Garcia-Ojalvo J, Elowitz MB. Mutual inactivation of Notch receptors and ligands facilitates developmental patterning. PLoS Comput Biol. 2011;7:e1002069. doi: 10.1371/journal.pcbi.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barad O, Rosin D, Hornstein E, Barkai N. Error minimization in lateral inhibition circuits. Sci Signal. 2010;3:ra51. doi: 10.1126/scisignal.2000857. [DOI] [PubMed] [Google Scholar]
- 44.LeBon L, Lee TV, Sprinzak D, Jafar-Nejad H, Elowitz MB. Fringe proteins modulate Notch-ligand cis and trans interactions to specify signaling states. Elife. 2014;3:e02950. doi: 10.7554/eLife.02950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morsut L, Leonardo M, Roybal KT, Xin X, Gordley RM, Coyle SM, Matthew T, Lim WA. Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell. 2016;164:780–791. doi: 10.1016/j.cell.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gordley RM, Bugaj LJ, Lim WA. Modular engineering of cellular signaling proteins and networks. Curr Opin Struct Biol. 2016;39:106–114. doi: 10.1016/j.sbi.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]