Abstract
Hyperoxaluria associated with renal deposition of calcium oxalate (CaOx) crystals causes renal injury and inflammation leading to number of diseases including chronic kidney disease (CKD). It is however, not been possible to separate the renal consequences of hyperoxaluria from that of CaOx crystal deposition. We decided to utilize ethylene glycol (EG) model where hyperoxaluria and CaOx crystal deposition can be separated in time. To test our hypothesis, male rats were made hyperoxaluric by administering EG, rats were euthanized and kidneys were extracted on day 14, when occasional crystal is seen in the kidneys and day 28, when all animals have developed renal CaOx crystal deposits. Total RNA was extracted for microarray analysis and genome wide analysis of differentially expressed genes was performed to investigate differences between hyperoxaluria and crystal induced alterations in the kidneys. Immunohistochemical and Hematoxylin and Eosin (H&E) staining was also done for macromolecules with significant role in stone formation. All EG fed rats became hyperoxaluric by day 7, showed a few crystal deposits on day 14, and had heavy crystal deposition by day 28. There were significant changes in the expression of genes encoding for NADPH Oxidases; macromolecular crystallization modulators; genes involved in inflammasome activation; and osteogenic marker genes. Results demonstrate major differences between hyperoxaluria and CaOx crystal induced changes in the kidneys. Injury and inflammation are mainly associated with crystal deposition indicating significant role played by crystal retention.
Introduction
Oxalate is an organic compound found naturally in many foods such as spinach, rhubarb, beets, wheat bran, strawberries, almonds and peanuts [1]. Higher concentration of oxalate in the human body can lead to a number of pathological conditions including hyperoxaluria which can further lead to a large spectrum of diseases such as nephrocalcinosis, cardiomyopathy, cardiac conductance disorders, systemic oxalosis, renal failure and specially CaOx kidney stones. Hyperoxaluria is excessive urinary excretion of oxalate and can be classified into different types such as primary, secondary, idiopathic and enteric hyperoxaluria caused by a variety of factors including genetic defects or mutation of specific genes (Primary hyperoxaluria), eating oxalate rich foods (Secondary hyperoxaluria), interplay of dietary, genetic and environmental factors with unknown causes (Idiopathic hyperoxaluria) and due to fat malabsorption, jejunal bypass surgery, and modern gastric bypass (Enteric hyperoxaluria) [2–6]. Kidney stone formation is a chronic disease which is becoming widespread nowadays, both in the United States and globally. Due to increased prevalence in the US, it is causing a significant economic burden on US health care [7]. According to National Health and Nutrition Examination Surveys (NHANES) of US adults, kidney stone prevalence increased from 5.2% (1988–1994) to 8.4% (2007–2010) [7] and a recent study showed that the number of kidney stone patients increased from 1 in 20 persons to 1 in 11 persons since 1994. Among men, the prevalence was 10.6% as compared to 7.1% among women and Caucasians were more likely to report a history of kidney stones as compared to African American and Hispanic individuals [8]. Also, recent studies show a link between stone formation and hypertension, chronic kidney disease and even end stage renal disease [9–12].
To understand the pathogenesis of hyperoxaluria and CaOx stone formation, many animal models have been developed [13–15]. Two rat models in which hyperoxaluria is induced by the administration of the ethylene glycol (EG) or hydroxy-L-proline (HLP) [16] have been studied in detail. Previous research has shown that hyperoxaluria and renal CaOx crystal deposition produced reactive oxygen species (ROS), upregulated mineralization modulators [17–20], caused lipid peroxidation, and renal cellular injury [16,21–23]. The free radicals or ROS appeared to play significant role in the expression of various factors and pathways [24,25], involved in the activation of inflammasomes [23], production of macromolecules implicated in crystal formation and retention [26,27], and osteogenesis [28]. These studies were however performed in the kidneys with extensive CaOx crystal deposition. As a result, nature of instigation, hyperoxaluria or CaOx crystals could not be established. Current study was performed using EG as hyperoxaluria inducing agent where, in our laboratory, administration of 0.75% EG in drinking water to male Sprague-Dawley rats lead to consistent hyperoxaluria by 2 weeks and CaOx crystal deposition by 4 weeks. We analyzed changes in global transcriptome of renal tissue, following development of hyperoxaluria at 2 weeks and CaOx crystal deposition or nephrolithiasis at 4 weeks of EG treatment. Emphasis was placed on the genes and pathways involved in production of reactive oxygen species, crystallization modulators, development of inflammation and osteogenesis. Results presented herein demonstrate that several pathways were commonly expressed at both day 14 and 28. Similarly, many significantly differentially expressed genes were common between day 14 and day 28. However significant differences were found between day 14 and 28, in the relative expression of genes involved in osteogenesis, activation of inflammasome and macromolecular production.
Materials and methods
Animal model
Eight weeks old, male Sprague- Dawley rats (n = 40), with average weight of about 110–120 grams, were bought from Harlan Laboratories (Harlan, Tampa, Florida). The rats were accustomed for 2 weeks within the Animal care facilities in their normal as well as metabolic cages for 2 days within the 2-week period at University of Florida before any experimental procedures and prior to start their dietary regimen. All the cages, food and water were sterilized by autoclaving before doing any experiments. Rats were divided into 2 groups (n = 20): Group 1 rats (control) were fed normal rat chow and sterile water and divided into 4 subgroups (day7, day14, day21 and day28) of 5 rats in each subgroup, Group 2 rats had a similar diet to Group 1 rats except were fed 1.25% Ethylene glycol (EG) in drinking water. Group 2 rats were also divided into 4 subgroups (day7, day14, day21, and day 28) of 5 rats in each subgroup. The rats were placed in metabolic cages for urine collection 1 day before sacrificing. On day 7, 5 rats from each group (total 10) belonging to day7 subgroup were euthanized and the kidneys freshly extracted. Rats were anesthetized with an injection of Pentobarbital IP. Once the rats were anesthetized and reached a surgical plane of anesthesia demonstrated by the lack of a pedal withdrawal reflex, their body cavity was opened to expose their internal organs. The rats were perfused with sterile saline and heparin was flushed throughout the body. Shortly after starting the perfusion process an over dose of Pentobarbital was added to ensure that the animal was dead. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institute of Health. All research was approved by the Institutional Animal Care and Use Committee (IACUC) at University of Florida (IACUC study # 201101850).
One Kidney was carefully divided into cortex and medulla and was stored in RNA later solution at -20 degrees for RNA isolation and at -80 degrees for protein isolation. The other kidney was cut into parts and was also kept in formalin to be embedded in paraffin for histological analysis. Similar procedure was followed for day14, 21, and day 28.
Urine collection and analysis
Every week, 24-hour urine was collected with 0.02% sodium azide to prevent bacterial growth. Rats were placed in metabolic cages for collection of urine. After determining volume and pH, urine was aliquoted for various assays. Light microscopy was used to determine crystalluria. Urinary lactate dehydrogenase (LDH) was measured using CytoTox® Non-Radioactive Cytotoxicity Assay (Cat. # G1780) as per the manufacturer’s instructions (Promega Corporation, Madison, WI). Urinary creatinine was measured using the Colorimetric Microplate Assay for Creatinine (Product No. CR 01) from Oxford Biomedical Research, Oxford, MI as per their protocol. Urinary Oxalate was measured using the Oxalate assay kit (Cat. # 591D) from Trinity Biotech, Bray CO as per the manufacturer’s instructions, similar to methods described in our previous studies [29]. The data was further analyzed using 2-way Analysis of Variance in Graph Pad Prism V 5.0 (La Jolla, CA).
RNA extraction and differential expression of genes by microarray analysis
The total RNA from each of the different specimens was isolated from the kidneys of rats within the two different treatment groups simultaneously using the RNeasy Mini-Kit (QIAGEN, Valencia, CA), as per the manufacturer’s instructions as described in our previous studies [23,30,31].
Microarray analysis and data mining
The microarray analysis was done in the Interdisciplinary Center for Biotechnology Research (ICBR), University of Florida, using the Agilent 8x60k single color arrays under the Agilent microarray incentive program. Under this program, we used 2 different time points as day 14 and day 28, with the control, and EG treated group. There were four replications for all the samples.
The data analysis of these samples was done at the Interdisciplinary Center for Biotechnology Research (ICBR) using the Bioconductor limma (Linear models for microarray analysis) package using R [32]. Before the analysis, the individual signal intensity values retrieved from the microarray probes were log transformed (using 2 as a base) and normalization was done for all the individual samples within all the groups in this study. After normalizing the signal intensity values for each of the arrays, the Student’s t-test was used to do a probe-by probe comparison between two groups concurrently. For each comparison, the fold change (FC) and p-value was calculated for each gene based on the n = 4 replicate samples within each experimental group and heat map, raw data box plot, relative log expression (RLE), and volcano plots were drawn for each comparison i.e. (Group2-1). The heat map was drawn using heat map package in R [32] and gene rows were clustered according to the fold change levels.
GO:TERM and KEGG pathway analyses based on differentially-expressed genes were carried out using DAVID (Database for Annotation, Visualization of Integrated Discovery) enrichment analysis tool from National Institute of Allergy and Infectious Diseases (NIAID), NIH [33]. The DAVID knowledgebase is built around DAVID gene concept and is intended to facilitate high throughput gene functional analysis. Cluster analyses of genes permitted identification of biological processes, cellular component, and molecular function ontology. All Microarray data have been deposited with Gene Expression Omnibus (GSE-89028).
Histological and immunohistological examinations
Kidneys from all the rats were extracted after euthanasia, one of which was used for RNA preparation, while the second was placed in 10% phosphate buffered formalin for 24 hours for histological analysis as previously described [31]. The tissue already fixed with formalin were embedded in paraffin and cut into sections of 5μm thickness. Deparaffinization of paraffin-embedded slides was performed by xylene immersion and subsequent dehydration in ethanol. Kidney sections were processed for immunohistochemistry using specific primary antibodies reactive to Osteopontin (OPN- Polyclonal Rb-Anti-OPN, 1:100 dilute, Abcam Cat. # ab8448), Matrix Gla-Protein (MGP- Polyclonal Rb-Anti-MGP, 1:50 dilute, Santa Cruz Cat. # 66965), Kidney Injury Molecule (KIM-1 Polyclonal Rb-Anti-KIM1, 1:100 dilute, Abcam Cat. # ab47635), Collagen (Col1a1 Polyclonal Rb-Anti-Col1a1, 1:00 dilute, Novus Biologicals Cat. # NB600-408), Fibronectin 1 (Fn 1 Monoclonal Ms-Anti-FN1, 1:50 dilute, Santa Cruz Cat. # sc59826), Tamm-Horsfall Protein (THP Polyclonal Rb-Anti-THP, 1:50 dilute, Santa Cruz Cat. # sc20631), Fetuin B (Fetub Monoclonal Rb-Anti-Fetuin B, 1:50 dilute, Abcam Cat. # ab191569) NADPH oxidase 2 (NOX2 Polyclonal Rb-Anti-Nox2, 1:100 dilute, (Abcam Cat. # ab31092) and NADPH oxidase 4 (NOX4 Polyclonal Rb-Anti-Nox4, Abcam Cat. # ab61248). Isotype controls were performed using rabbit IgG. Slides were incubated for 30 min in biotinylated goat anti-rabbit IgG followed by incubation with biotinylated horseradish peroxidase using the Vectastain® ABC kit. Staining was developed by addition of diaminobenzidine (DAB) substrate (Vector Labs, Burlingame, CA) and counterstained with hematoxylin. To ensure that positive infiltrate staining for these antibodies was not due to high background staining, an additional run was performed using 10 mM citrate buffer for antigen-retrieval with all other procedures unchanged. Antigen-retrieval was carried out in 25 mMTris/EDTA buffers, pH 9.1 at 60°C for 20 min under 18 psi pressure. Images were taken using the Zeiss Axiovert 200M microscope (Carl Zeiss Microimaging, Inc., Thornwood, NY).
Results
Histology and urinary assays
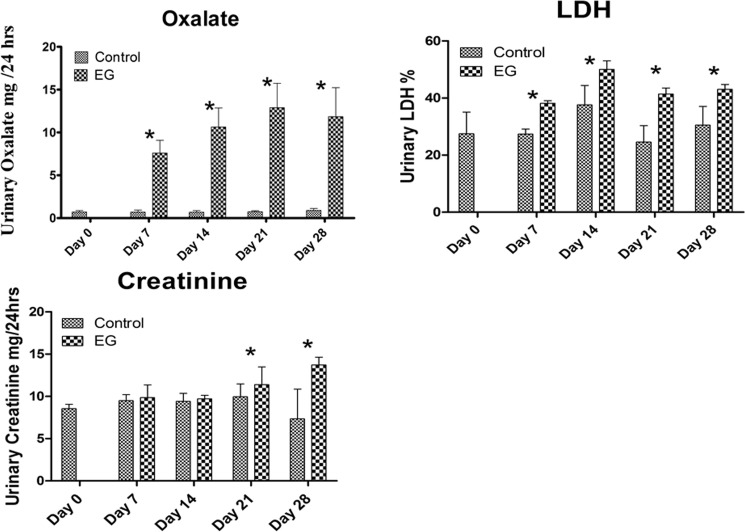
As anticipated, EG administration to the rats produced hyperoxaluria and CaOx nephrolithiasis i.e. crystal deposition in the kidneys. There was a significant increase in urinary excretion of oxalate by the rats consuming EG as compared to the control group peaking at day 14 and remaining significantly high till day 28. Urinary LDH increased significantly for Days 7, 14, 21and 28 for the EG consuming rats as compared to the control rats (Fig 1). Urinary creatinine increased significantly on day 21 and day 28 in the EG fed rats as compared to the control group (Fig 1).
Fig 1. Urinary excretion of control and ethylene glycol (EG) treated rats.
A) EG fed rats showed significant increase in urinary oxalate (mg/24hrs) excretion on day 7, 14, 21 and 28 respectively B) Urinary Lactate Dehydrogenase (LDH) was elevated in the EG fed rats on Day 7, 14, 21 and 28 respectively as compared to the control rats, Cytotoxicity level of LDH was measured as expressed as percentage (%) and C) Urinary Creatinine (mg/24hrs) was significantly higher on day 21 and 28 in the EG fed rats as compared to the control group. The data was analyzed using Graph Pad Prism with 2-way Analysis of Variance. *P < 0.05 Control versus EG within the same time period.
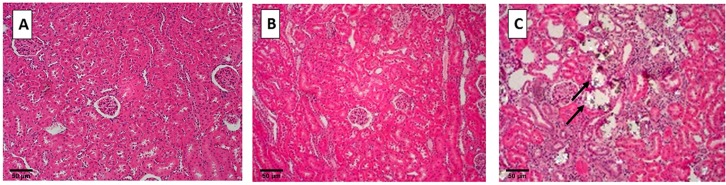
When sacrificed on day 28, kidneys of control rats appeared normal without any crystal deposits. Kidneys of rats sacrificed on day 14 of EG treatment also did not contain easily detectable CaOx crystal deposits. However, kidneys of rats sacrificed on day 28 of the EG administration showed copious amounts of birefringent CaOx crystals in the kidneys (Fig 2). Most of the crystals were found in the cortex and outer medulla, with few on the inner medulla. Crystals were generally located in the tubular lumens of the distal tubules and collecting ducts. Most of the renal tubules where crystals were present were distended. Crystal deposition also caused tissue disruption.
Fig 2. Hematoxylin and eosin staining of paraffin embedded kidneys examined under polarized light.
A) Control rats, no crystal deposition and B) rats fed 1.25% Ethylene Glycol (EG), observed on Day 14 (Hyperoxaluric stage) and C) rats fed 1.25% EG with lots of birefringent crystals observed on Day 28 (Nephrolithiasis stage), shown by black arrows Magnification 10x, Scale bar 50μm.
Microarray analysis and data mining
Of more than 30,000 rat genes represented on the arrays, 938 and 2400 genes were found to be significantly differentially expressed and also up regulated in the renal tissue, respectively for Day 14 and Day 28 (Log FC ≥ 0.3), in comparison between the control and the EG-fed rats. There were 440 (Day 14) and 1902 (Day28) genes that were significantly differentially expressed and highly up regulated.
Using Kyoto Encyclopedia of Genes and Genome Pathway analysis (KEGG) to curate the differentially-expressed genes, several gene clusters were identified that defined specific biological processes for the EG-fed rats, on Day 14 and Day 28 respectively as presented in Tables 1 and 2.
Table 1. Functional annotation charts showing differentially expressed pathways on Day 14 of highly expressed genes in the EG group.
| Category | Signaling pathway | Gene count | Percentage | p-value | Benjamini |
|---|---|---|---|---|---|
| KEGG-PATHWAY | Complement and coagulation cascades | 20 | 2.6 | 5.3E-10 | 7.8E-8 |
| KEGG-PATHWAY | ECM-receptor interaction | 18 | 2.3 | 2.8E-7 | 2.0E-5 |
| KEGG-PATHWAY | Focal adhesion | 25 | 3.2 | 2.6E-5 | 1.3E-3 |
| KEGG-PATHWAY | Leucocyte transendothelial migration | 18 | 2.3 | 4.1E-5 | 1.5E-3 |
| KEGG-PATHWAY | Cell adhesion molecules (CAMS) | 20 | 2.6 | 1.1E-4 | 3.1E-3 |
| KEGG-PATHWAY | Systemic lupus erythematosus | 13 | 1.7 | 1.4E-3 | 3.5E-2 |
| KEGG-PATHWAY | Chemokine signaling pathway | 18 | 2.3 | 4.3E-3 | 8.8E-2 |
| KEGG-PATHWAY | Natural killer cell mediated cytotoxicity | 12 | 1.6 | 9.8E-3 | 1.7E-1 |
| KEGG-PATHWAY | Prion diseases | 6 | 0.8 | 2.7E-2 | 3.7E-1 |
| KEGG-PATHWAY | Hematopoietic cell lineage | 9 | 1.2 | 3.7E-2 | 4.3E-1 |
| KEGG-PATHWAY | Pathways in cancer | 24 | 3.1 | 3.9E-2 | 4.1E-1 |
| KEGG-PATHWAY | Tight junction | 12 | 1.6 | 5.5E-2 | 5.0E-1 |
| KEGG-PATHWAY | Cytokine-cytokine receptor interaction | 16 | 2.1 | 6.2E-2 | 5.1E-1 |
| KEGG-PATHWAY | Viral myocarditis | 9 | 1.2 | 6.4E-2 | 5.0E-1 |
| KEGG-PATHWAY | Fc gamma R-mediated phagocytosis | 9 | 1.2 | 6.8E-2 | 5.0E-1 |
| KEGG-PATHWAY | Cell cycle | 11 | 1.4 | 9.2E-2 | 5.9E-1 |
| KEGG-PATHWAY | Glutathione metabolism | 6 | 0.8 | 9.9E-2 | 6.0E-1 |
The gene heading indicates number of genes mapped to an ontology category. P-values derived from Fisher’s exact test and Benjamini multiple test correlation. Gene ontology (GO) analysis and pathway analysis of genes also done using DAVID (Database for Annotation, Visualization of Integrated Discovery) enrichment analysis tool from National Institute of Allergy and Infectious diseases (NIAID), NIH.
Table 2. Functional annotation charts showing differentially expressed pathways of highly expressed genes in the EG group on Day 28.
| Category | Signaling pathway | Gene count | Percentage | p-value | Benjamini | ||
|---|---|---|---|---|---|---|---|
| KEGG-PATHWAY | Hematopoietic cell lineage | 24 | 1.7 | 1.0E-7 | 1.7E-5 | ||
| KEGG-PATHWAY | ECM-receptor interaction | 23 | 1.6 | 9.5E-7 | 7.9E-5 | ||
| KEGG-PATHWAY | Cell cycle | 29 | 2.0 | 2.7E-6 | 1.5E-4 | ||
| KEGG-PATHWAY | DNA replication | 14 | 1.0 | 3.7E-6 | 1.6E-4 | ||
| KEGG-PATHWAY | Complement and coagulation cascades | 19 | 1.3 | 2.1E-5 | 7.1E-4 | ||
| KEGG-PATHWAY | Cell adhesion molecules (CAMS) | 30 | 2.1 | 2.5E-5 | 6.9E-4 | ||
| KEGG-PATHWAY | Chemokine signaling pathway | 31 | 2.2 | 1.5E-4 | 3.7E-3 | ||
| KEGG-PATHWAY | Cytokine-cytokine receptor interaction | 34 | 2.4 | 1.8E-4 | 3.8E-3 | ||
| KEGG-PATHWAY | p53 signaling pathway | 16 | 1.1 | 4.5E-4 | 8.3E-3 | ||
| KEGG-PATHWAY | Ribosome | 18 | 1.2 | 7.0E-4 | 1.2E-2 | ||
| KEGG-PATHWAY | Focal adhesion | 32 | 2.2 | 7.2E-4 | 1.1E-2 | ||
| KEGG-PATHWAY | NOD-like receptor signaling pathway | 15 | 1.0 | 7.4E-2 | 1.0E-1 | ||
| KEGG-PATHWAY | Primary immunodeficiency | 10 | 0.7 | 2.0E-3 | 2.6E-2 | ||
| KEGG-PATHWAY | Leucocyte transendothelial migration | 20 | 1.4 | 4.9E-3 | 5.7E-2 | ||
| KEGG-PATHWAY | Natural killer cell mediated cytotoxicity | 18 | 1.2 | 5.6E-3 | 6.1E-2 | ||
| KEGG-PATHWAY | Systemic lupus erythematosus | 16 | 1.1 | 1.1E-2 | 1.1E-1 | ||
| KEGG-PATHWAY | Hypertrophic cardiomyopathy (HCM) | 15 | 1.0 | 1.4E-2 | 1.3E-1 | ||
| KEGG-PATHWAY | Pathways in cancer | 40 | 2.8 | 1.7E-2 | 1.5E-1 | ||
| KEGG-PATHWAY | Gap junction | 14 | 1.0 | 2.3E-2 | 1.8E-1 | ||
| KEGG-PATHWAY | Dilated cardiomyopathy | 15 | 1.0 | 2.4E-2 | 1.8E-1 | ||
| KEGG-PATHWAY | Glutathione metabolism | 10 | 0.7 | 2.8E-2 | 2.0E-1 | ||
| KEGG-PATHWAY | Terpenoid backbone biosynthesis | 5 | 0.3 | 2.9E-2 | 2.0E-1 | ||
| KEGG-PATHWAY | Graft-versus-host disease | 10 | 0.7 | 3.1E-2 | 2.1E-1 | ||
| KEGG-PATHWAY | Toll-like receptor signaling pathway | 14 | 1.0 | 4.9E-2 | 3.0E-1 | ||
| KEGG-PATHWAY | Small cell lung cancer | 13 | 0.9 | 5.7E-2 | 3.2E-1 | ||
| KEGG-PATHWAY | Apoptosis | 13 | 0.9 | 6.6E-2 | 3.6E-1 | ||
| KEGG-PATHWAY | Viral myocarditis | 13 | 0.9 | 7.6E-2 | 3.9E-1 | ||
| KEGG-PATHWAY | Type I diabetes mellitus | 10 | 0.7 | 7.6E-2 | 3.8E-1 | ||
| KEGG-PATHWAY | Steroid hormone biosynthesis | 8 | 0.6 | 7.8E-2 | 3.7E-1 | ||
| KEGG-PATHWAY | Fc gamma R-mediated phagocytosis | 13 | 0.9 | 8.1E-2 | 3.8E-1 | ||
| KEGG-PATHWAY | Drug metabolism | 8 | 0.6 | 8.6E-2 | 3.9E-1 | ||
| KEGG-PATHWAY | Renin-angiotensin system | 5 | 0.3 | 9.2E-2 | 4.0E-1 | ||
| KEGG-PATHWAY | T cell receptor signaling pathway | 15 | 1.0 | 9.3E-2 | 3.9E-1 | ||
The gene heading indicates number of genes mapped to an ontology category. P-values derived from Fisher’s exact test and Benjamini multiple test correlation.
Interestingly, the differentially-expressed genes identifying the various KEGG signaling pathways revealed that, there were 17 and 33 pathways that were significant on Day 14 and Day 28 respectively. Of these, 15 pathways were common between Day 14 and Day 28 (Table 3).
Table 3. Common pathways between Day 14 and Day 28 of highly expressed genes in the EG group.
| KEGG # | Signaling Pathway | Day 14 | Day 28 | ||
|---|---|---|---|---|---|
| Gene Count | Percentage | Gene Count | Percentage | ||
| 1 | Hematopoietic cell lineage | 9 | 1.2 | 24 | 1.7 |
| 2 | ECM-receptor interaction | 18 | 2.3 | 23 | 1.6 |
| 3 | Cell cycle | 11 | 1.4 | 29 | 2 |
| 4 | Complement and coagulation cascades | 20 | 2.6 | 19 | 1.3 |
| 5 | Cell adhesion molecules (CAMs) | 20 | 2.6 | 30 | 2.1 |
| 6 | Chemokine signaling pathway | 18 | 2.3 | 31 | 2.2 |
| 7 | Cytokine-cytokine receptor interaction | 16 | 2.1 | 34 | 2.4 |
| 8 | Focal adhesion | 25 | 3.2 | 32 | 2.2 |
| 9 | Leukocyte transendothelial migration | 18 | 2.3 | 20 | 1.4 |
| 10 | Natural killer cell mediated cytotoxicity | 12 | 1.6 | 18 | 1.2 |
| 11 | Systemic lupus erythematosus | 13 | 1.7 | 16 | 1.1 |
| 12 | Pathways in cancer | 24 | 3.1 | 40 | 2.8 |
| 13 | Glutathione metabolism | 6 | 0.8 | 10 | 0.7 |
| 14 | Viral myocarditis | 9 | 1.2 | 13 | 0.9 |
| 15 | Fc gamma R-mediated phagocytosis | 9 | 1.2 | 13 | 0.9 |
Gene ontology (GO) analysis and pathway analysis of genes also done using DAVID (Database for Annotation, Visualization of Integrated Discovery) enrichment analysis tool from National Institute of Allergy and Infectious diseases (NIAID), NIH
We also found that there were 18 unique pathways for the Day 28 (Nephrolithiasis) group (Table 4) and 2 pathways that were unique for Day 14 (Hyperoxaluria) group (Table 5) when we compared the pathways.
Table 4. Signaling pathways that are unique to day 28 with heavy CaOx crystal deposition (Nephrolithiasis) when compared with day 14 (Hyperoxaluric condition).
| KEGG Pathway | Signaling pathway | Gene Count | Percentage |
|---|---|---|---|
| 1 | DNA replication | 14 | 1.0 |
| 2 | P53 signaling pathway | 16 | 1.1 |
| 3 | Ribosome | 18 | 1.2 |
| 4 | Nod–like receptor signaling pathway | 15 | 1.0 |
| 5 | Primary immunodeficiency | 10 | 0.7 |
| 6 | Hypertrophic cardiomyopathy | 15 | 1.0 |
| 7 | Gap junction | 14 | 1.0 |
| 8 | Dilated cardiomyopathy | 15 | 1.0 |
| 9 | Terpenoid backbone biosynthesis | 5 | 0.3 |
| 10 | Graft-versus-host disease | 10 | 0.7 |
| 11 | Toll-like receptor signaling pathway | 14 | 1.0 |
| 12 | Small cell lung cancer | 13 | 0.9 |
| 13 | Apoptosis | 13 | 0.9 |
| 14 | Type-1 diabetes mellitus | 10 | 0.7 |
| 15 | Steroid hormone biosynthesis | 8 | 0.6 |
| 16 | Drug metabolism | 8 | 0.6 |
| 17 | Renin-angiotensin system | 5 | 0.3 |
| 18 | T-cell receptor signaling pathway | 15 | 1.0 |
Table 5. Signaling pathways that were unique to day 14 (Hyperoxaluria) only when compared with day 28.
| KEGG Pathway | Signaling pathway | Gene Count | Percentage |
|---|---|---|---|
| 1 | Prion disease | 6 | 0.8 |
| 2 | Tight junction | 12 | 1.6 |
Major pathways that were unique to nephrolithiasis group and were highly significantly expressed included P53 signaling, Nod-like receptor signaling, hypertrophic cardiomyopathy, toll like receptor signaling, and T cell receptor signaling. The two unique pathways for hyperoxaluric group were the prion disease and tight junction signaling.
Fig 3 shows microarray data as heat maps. It shows up and down regulated genes on day 14 and 28 after EG treatment compared to their controls. Significant differences are apparent between controls and EG treated rats as well as between 14 day and 28 days of treatments.
Fig 3. Heat map showing up as well as down regulated genes.
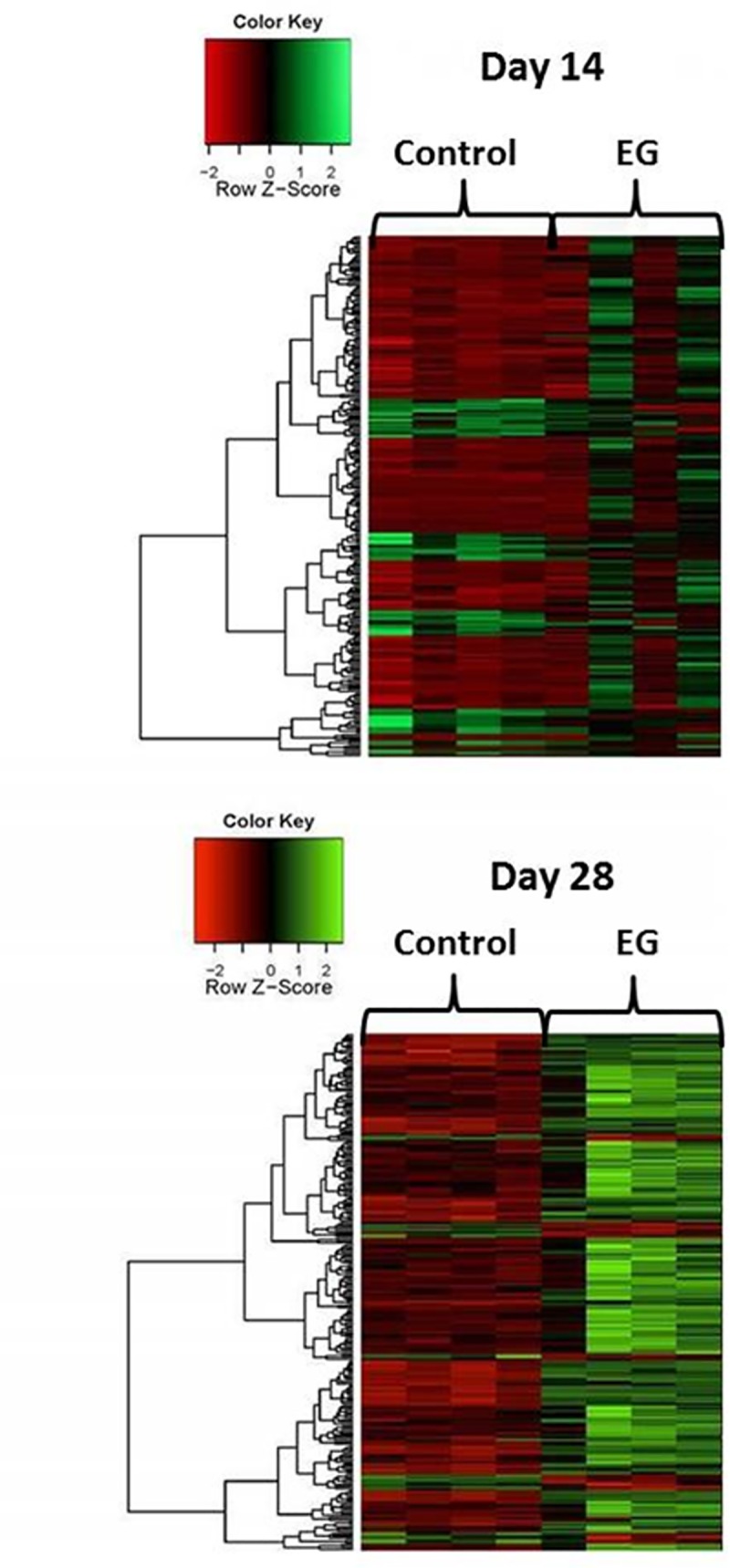
Top map compares 4 control rats to 4 rats after EG treatment on day 14 and bottom map on Day 28. Significant differences are apparent between control and EG treated rats on both days as well as between the treated rats on day 4 when rats were mostly hyperoxaluric and crystals free vs day 28 with heavy CaOx crystals deposits in the kidneys.
Based upon previous results and our earlier investigations with HLP fed rats [31], we looked for genes encoding for NOX 4, NOX 2, p22phox, GPX2 as well as KIM-1 (Table 6). They were all significantly upregulated in rats given EG. Expression of NOX 4 and NOX 2 was however higher on day 14 than on day 28. Expression of p22Pphox and Kim-1 was on the other hand higher on day 28 than on day 14.
Table 6. Relative expression of important genes in ethylene glycol (EG) fed rat vs control group, along with their p-values derived from microarray analysis and data mining.
| GENE ID | DESCRIPTION | DAY 14 | DAY 28 | ||
|---|---|---|---|---|---|
| Log Fold Change | p-value | Log Fold Change | p-value | ||
| NOX4 | NADPH Oxidase 4 / Renal NADPH Oxidase | 0.578 | 0.037 | 0.387 | 0.035 |
| NOX2/CYBB | NADPH Oxidase 2/ Cytochrome B-245 Beta Chain | 0.952 | 0.626 | 0.518 | 0.840 |
| P22/CYBA | P22Phox / Cytochrome B-245 Alpha Chain | 0.202 | 0.268 | 0.290 | 0.080 |
| GPX2 | Glutathione Peroxidase 2 | 1.583 | 0.028 | 2.742 | 0.0001 |
| KIM1 | Kidney Injury Molecule 1 | 1.870 | 0.065 | 4.390 | 0.0001 |
| OPN/SPP1 | Osteopontin / Secreted Phosphoprotein 1 | 2.400 | 0.022 | 4.160 | 0.00002 |
| MCP1 | Monocyte Chemoattractant Protein 1 | 0.175 | 0.638 | 1.740 | 0.201 |
| MGP | Matrix Gla Protein | 0.705 | 0.108 | 0.807 | 0.0019 |
| FETUB | Fetuin B | 0.285 | 0.643 | 0.420 | 0.053 |
| THP/UMOD | Tamm-Horsfall Protein / Uromodulin | -0.246 | 0.442 | -0.426 | 0.063 |
| LCN2 | Lipocalin 2 | 1.807 | 0.058 | 2.178 | 0.004 |
| FN1 | Fibronectin 1 | 0.078 | 0.393 | 1.095 | 0.047 |
| CLU | Clusterin | 1.773 | 0.041 | 2.086 | 0.0003 |
| CD44 | Cell Surface glycoprotein CD44 | 0.626 | 0.137 | 1.586 | 0.00001 |
| AMBP | Alpha-1- Microglobulin / Bikunin Precursor | 0.210 | 0.020 | 0.258 | 0.020 |
| PYCARD | PYD And CARD Domain-Containing Protein | 0.422 | 0.096 | 0.393 | 0.097 |
| NLRP3 | NLR Family Pyrin Domain Containing 3 | 0.024 | 0.807 | 0.090 | 0.903 |
| IL-18 | Interleukin 18 | 0.076 | 0.562 | 0.092 | 0.406 |
| TXNIP | Thioredoxin Interacting Protein | 0.183 | 0.448 | 0.130 | 0.498 |
| IL-1β | Interleukin 1 Beta | 0.194 | 0.345 | 0.799 | 0.007 |
| CASP1 | Caspase 1 | -0.0041 | 0.979 | 0.371 | 0.010 |
| RUNX1 | Runt Related Transcription Factor 1 | 0.557 | 0.136 | 1.063 | 0.00007 |
| RUNX2 | Runt Related Transcription Factor 2 | 0.042 | 0.794 | 0.099 | 0.372 |
| KRT18 | Keratin 18 / Cytokeratin 18 | 0.447 | 0.077 | 0.706 | 0.009 |
| KRT8 | Keratin 8 / Cytokeratin 8 | 0.213 | 0.170 | 0.238 | 0.104 |
| VIM | Vimentin | 0.237 | 0.405 | 0.412 | 0.008 |
| COL1A1 | Collagen Type 1 Alpha 1 Chain | 0.474 | 0.235 | 0.591 | 0.009 |
| COL1A2 | Collagen Type 1 Alpha 2 Chain | 0.587 | 0.100 | 0.761 | 0.005 |
| ALPL | Alkaline Phosphatase, Liver/Bone/Kidney | -0.228 | 0.147 | -0.200 | 0.227 |
| BMP7 | Bone Morphogenetic Protein 7 | -0.066 | 0.627 | -0.014 | 0.852 |
| BMPR2 | Bone Morphogenetic Protein Receptor Type 2 | -0.118 | 0.183 | -0.428 | 0.047 |
| BMP2 | Bone Morphogenetic Protein 2 | -0.031 | 0.824 | 0.339 | 0.039 |
Most of the genes had log Fold Change values >0.3 (> 2-fold ratio differential expression). The genes include those encoding for macromolecules, various components of NLRP3 inflammasome, and osteogenic proteins. All Microarray data have been deposited with Gene Expression Omnibus (GSE-89028).
Further we looked at the genes encoding for the macromolecules that have specific roles in modulating CaOx crystal formation and deposition in the hyperoxaluric rats. On comparing the relative gene expression of the control and the EG fed group, we found the results as anticipated for Osteopontin (OPN), Monocyte Chemoattractant Protein (MCP-1), Matrix Gla Protein (MGP), and Fetuin-B. For both Day 14 and Day 28, the genes encoding for the above said proteins were up-regulated and the relative level of up regulation was more for Day 28 as compared to Day 14. Similar results were found for the gene encoding for Lipocalin (Lcn2), Fibronectin (FN-1), Clusterin (CLU), CD44 and Alpha Microglobulin Bikunin precursor (AMBP) (Table 6). Genes encoding for THP were however significantly downregulated on days, 14 as well as 28.
Major genes involved in the activation of the NLRP3 inflammasome were significantly up regulated on both day 14 and day 28 (Table 6). These included PYCARD, NLRP3, IL-18, TXNIP, and IL-1β. Caspase-1 was non-significant on day 14 but was highly significant on day 28. Expression of the genes encoding for molecules involved in osteogenesis were also affected. Genes for Runt Related Transcription factor 1 (RUNX 1), Runt related Transcription factor 2 (RUNX 2), Cytokeratin -18 (KRT-18), Cytokeratin-8 (KRT-8), Vimentin (VIM), Collagen 1A1 (COL1A1), and Collagen 1A2 (COL 1A2) were up regulated on day 14 as well as day 28. Genes encoding for Alkaline Phosphatase (ALPL), Bone Morphogenetic Protein 7 (BMP 7), and Bone Morphogenetic Protein receptor 2 (BMPr2) were down regulated on both day 14 and day 28. Bone Morphogenetic Protein 2 (BMP 2) was down regulated on day 14 but was up regulated on Day 28 (Table 6).
Immunohistochemical staining
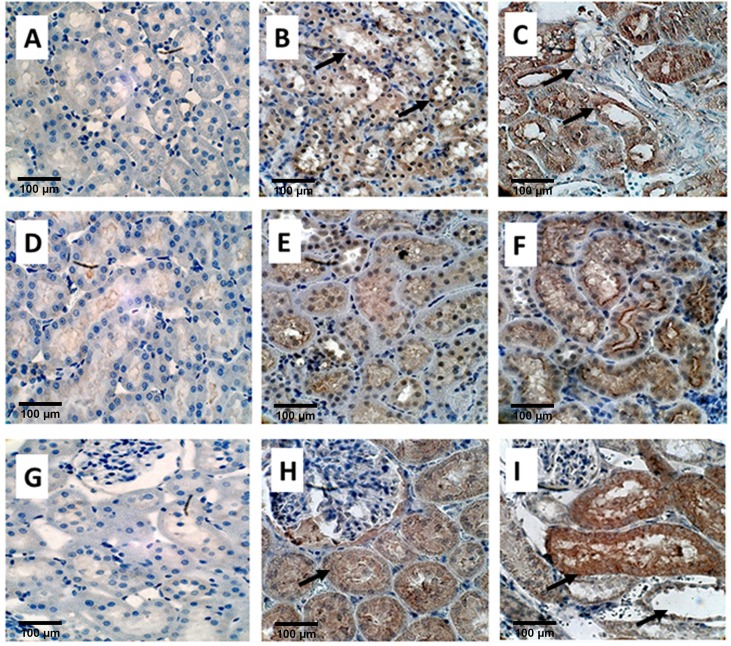
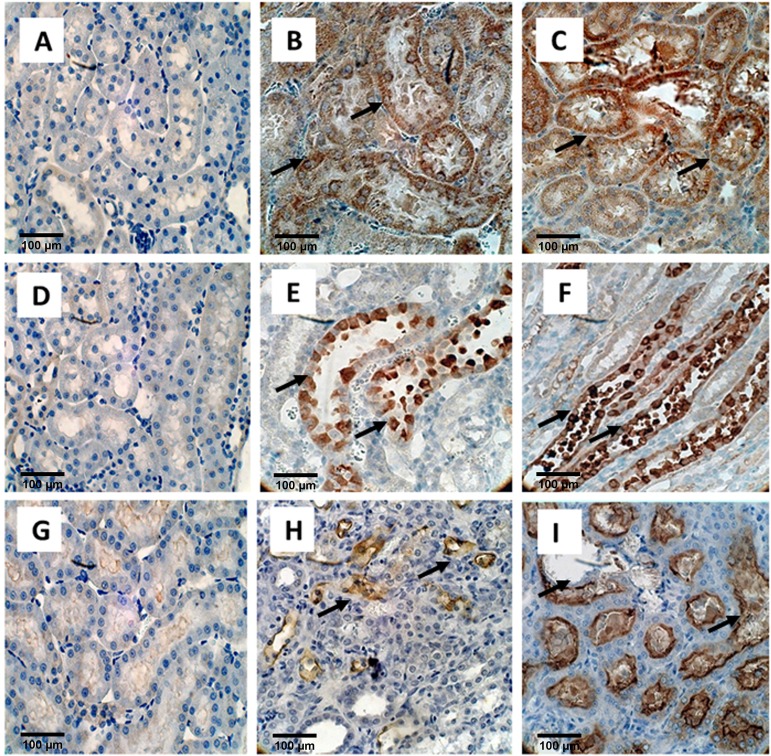
Immunostaining was used to determine the expression of OPN, KIM-1, MGP, Collagen, FN1, THP, FETUB, NOX2, and NOX4 in the kidneys (Figs 4, 5 and 6). To some extant these molecules were expressed in the kidneys on both day 14 and day 28. However, the expression of most of these proteins was more pronounced on day 28 than on day 14 as compared to the control. Expression of OPN (Fig 4A, 4B and 4C), MGP (Fig 4D, 4E and 4F), and Fetuin B (Fig 4G, 4H and 4I), was much higher on day 28 than on day 14 as compared with control. As has previously been shown, heavy staining for OPN, MGP, and Fetuin on day 28 was seen, in association with the crystals, in the tubular lumens of distal tubules and collecting ducts). Light and nonspecific collagen staining in tubular epithelial cells was seen on day 14, whereas no staining was there in the control. On the other hand, heavy staining of tubular basement membrane and interstitium was observed after 28 days for Collagen as compared to the control (Fig 5A, 5B and 5C). Staining for Fibronectin appeared limited to the tubular epithelial cells. (Fig 5D, 5E and 5F) THP expression was obvious in the tubular epithelial cells as well as tubular lumens on both day 14 and 28. THP staining was however seen in fewer tubules on day 28 (Fig 5G, 5H and 5I).
Fig 4. Immuno-histological staining of EG treated rat kidneys.
OPN, A) Control B) on Day 14 showing light staining of the tubular epithelial cells, shown by black arrows and C) on Day 28 showing strong staining of renal tubular epithelial cells as well as tubular contents (Black arrows), Magnification, X 40, Scale bar 100μm, MGP, D) Control E) light staining on Day 14 and F) heavy staining on D28 Magnification, X 40, Scale bar 100μm. Fetuin B, G) Control, H) Non-specific diffuse staining of epithelial cells on Day 14, shown by a single black arrow, and I) Heavy staining of cells as well as luminal contents (top black arrow), particularly those with crystals which have dropped out during processing shown by bottom right black arrow.
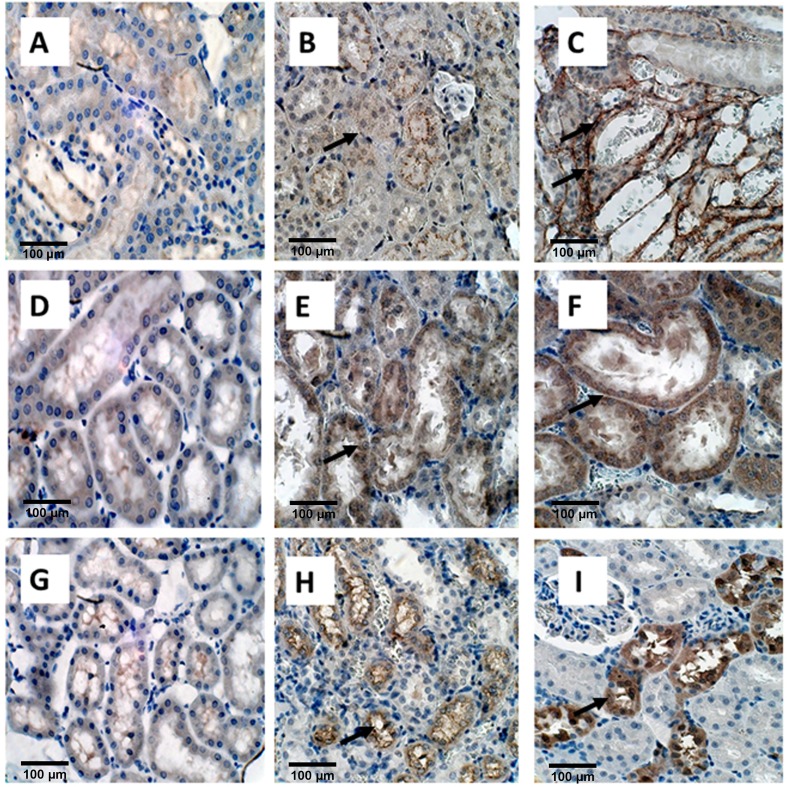
Fig 5. Immuno-histological staining of EG treated rat kidneys.
Collagen, A) Control B) Light staining nonspecific staining on Day 14 shown by black arrow. C) Staining of the tubular basement membrane and interstitium on Day 28 shown by 2 black arrows. Magnification X40. Fibronectin, D) Control, E) strong staining of tubular epithelial cells on Day 14 (black arrow) and F) day 28 shown by black arrow. THP, G) Control H) Tubular epithelial cells stained positive on both Day 14 (black arrow) and I) Day 28, but number of positive cells appeared less on day 28 then on Day 14. Magnification X40, Scale bar 100 μm.
Fig 6. Immuno-histological staining of EG treated rat kidneys.
NOX2, A) Control B) Day 14 C) Day 28. Staining is similar on both days and seen in both the renal tubular epithelial cells as well as interstitium shown by black arrows. NADPH oxidase 4 or NOX4, D) Control E) Day 14 and F) Day 28. Staining appears similar on both days and limited to the renal epithelial cells of the ascending limbs of the Loop of Henle (black arrows showing staining of renal epithelial cells). KIM1, G) Control H) Day 14 showing light localized staining (black arrows) and I) Day 28, showing large distended tubule from which crystals have fallen during processing (Black arrow) and there is heavy expression both in the tubular cells and lumens Magnification, X 40, Scale bar 100 μm.
Staining for various subunits of the NADPH oxidase was also affected by hyperoxaluria and the deposition of CaOx crystals in the kidneys. Nox 2 (Fig 6A, 6B and 6C) and Nox 4 (Fig 6D, 6E and 6F) appeared to increase in the cortical tubules, mostly in the cytoplasm of the collecting ducts, and distal tubules. Staining appeared more intense on day 28 than day 14 as compared to the control. Non- specific staining was also evident in the renal interstitium. Nox 4 staining was also seen in the ascending limbs of the loop of Henle. There was light localized staining on day 14 but strong staining with large distended tubules on day 28 for KIM1 and no staining for the control (Fig 6E, 6F and 6G).
Discussion
CaOx kidney stone formation depends not only on the urinary environment but also how it shapes the renal response. Many animal model and tissue culture studies have been performed to understand this relationship between various urinary factors and renal epithelial cells, particularly the interaction between renal epithelium, oxalate and CaOx crystals [34–41]. Most studies looked for signs of injury, such as membrane permeability, cell viability, DNA synthesis, production of ROS etc. All of them agreed that CaOx crystals are injurious to renal epithelial cells. Studies of oxalate exposure however led to a range of conclusions. Oxalate toxicity was determined to be cell specific and time and concentration dependent. Collecting duct cells were found to be less susceptible to injury than cells of the proximal tubular origin. Genome wide analysis of differentially expressed genes in a line of proximal tubular epithelial cells, the HK-2 cells, in response to oxalate showed 750 upregulated and 2276 downregulated genes and 20 genes were differentially regulated irrespective of the duration of exposure [34].
Genome wide analysis of mice kidneys with hyperoxaluria induced by the administration of glyoxalate showed at least 2-fold increase in the expression of 25 genes during deposition of CaOx crystals. Gene ontology analysis showed increased expression of genes related to inflammation, immune reactions and complement activation pathways [42]. We analyzed changes in global transcriptome in the kidneys of rats given hydroxyl-L-proline for 4 weeks. Rats were hyperoxaluric and their kidneys showed heavy deposition of CaOx crystals. Analysis of 22,226 genes displayed 20 and 24 significant pathways in the cortex and medulla respectively. Genes encoding for PYCARD (ASC), TXNIP, NLRP3, caspase-1, and IL-1β and IL-18 were significantly upregulated [23]. Genes involved in the activation of NADPH oxidase were highly expressed with concurrent decrease in the expression of genes encoding for ROS scavenger proteins [31]. Expression of genes involved in the epithelial transformation and bone morphogenesis was also altered [28]. In addition, CaOx nephrolithiasis affected the expression of genes encoding for various crystallization modulators [27].
In the present study, we investigated differences in renal response to continuous exposure to high oxalate with or without the deposition of CaOx crystals by analyzing changes in global transcriptome of renal tissue, following EG induced hyperoxaluria in male rats. Emphasis was placed on the genes and pathways involved in the production of reactive oxygen species, crystallization modulators, development of inflammation and osteogenesis. We looked at the relative gene expression in hyperoxaluric and nephrolithic kidneys compared to the control group to determine the signaling pathways activated during hyperoxaluria and nephrolithiasis to better understand the pathophysiology of oxalate induced diseases of the kidneys. 15 pathways were common between hyperoxaluric and nephrolithic kidneys. Interesting thing to observe was that 18 unique pathways became significant only in nephrolithic kidneys while only 2 unique pathways were significant in the hyperoxaluric kidneys. Unique pathways in the nephrolithic rats included Nod-like receptor pathway which involves NFĸB signaling, MAPK signaling, apoptosis and activation of inflammasomes. Our previous study has already shown the involvement and activation of the Renin-angiotensin pathway along with DAG/PKC pathway of neutrophil activation [31].
Our study also considered important set of genes involved in the activation of NLRP3 (NACHT, LLR and PYD domains containing protein 3). We saw significant up regulation of genes encoding for PYCARD (PYD and CARD domain containing) also known as ASC (Apoptosis Associated Spec like protein), Caspase 1, IL-18 (Interleukin-18), TXNIP (Thioredoxin Interacting Protein), and IL-1β. Relative gene expression for Caspase-1 and IL-1β was, however, not significant at day 14 but when crystals were formed around day 28 we saw highly significant gene expression. Caspase-1 and IL-1β are important end products of NLRP3 inflammasome activation [43]. This indicates that inflammation pathways are activated during crystal formation as compared to hyperoxaluric conditions as shown in this study.
We further looked at genes encoding for proteins responsible for epithelial to osteogenic transformation including Runx-1 (Runt-related transcription factor-1), Runx-2, ALPL (Alkaline phosphatase), Bone morphogenetic proteins (BMP) genes such as BMP-2, BMP-7, BMPr2 as well as Cytokeratin’s such as KRT 8, KRT 18, Vimentin (VIM), Fibronectin (Fn-1), Collagen 1a (COL1a1) and COL1a2. Genes encoding for Runx1, Runx2, KRT 18, KRT 8, VIM, Fn-1, Col1a1 and Col1a2 were up regulated during hyperoxaluric conditions and further increased after crystal deposition or nephrolithiasis. Gene encoding for ALPL (Alkaline Phosphatase) was down regulated as shown in our previous study [28].
As mentioned earlier, CaOx nephrolithiasis in a rat model is associated with the activation of NADPH oxidase, production of ROS, development of oxidative stress and renal injury [22,44,45]. In our current study, Nox 2, Nox 4 as well as P22 were upregulated during hyperoxaluria as well as during nephrolithiasis suggesting early activation of NADPH oxidase. KIM-1, a recognized marker of renal injury [46], was highly expressed at both the gene and protein levels during nephrolithiasis. Glutathione Peroxidase (Gpx) is an antioxidant enzyme that is highly expressed in the kidney and removes peroxides and peroxynitrite that can cause renal damage. During oxidative stress, there is an increased expression of Gpx as shown previously. In our data, Gpx 2 was highly upregulated in the kidneys particularly during nephrolithiasis. Results indicate that CaOx crystals are more injurious to the kidneys.
Results also showed increased urinary excretion of LDH, a marker of membrane permeability and damage. The increase was evident early during hyperoxaluria indicating that oxalate alone can induce changes in the renal epithelial cells. Urinary creatinine levels were not affected during hyperoxaluria but increased after crystal deposition, highly significantly so on day 28 indicating renal damage.
As shown in previous studies [27,31] many macromolecular crystallization modulators are upregulated when renal epithelial cells are exposed to oxalate and CaOx crystals [24–27,47–49]. Results of our present study show that OPN, MGP, Fetuin B, Fn1, CD 44, Clusterin, Bikunin/AMBP genes are upregulated during hyperoxaluria and further increased with crystal deposition. Osteopontin is one of the most studied macromolecular modulator of CaOx and Calcium Phosphate (CaP) stone formation and plays a vital role in bio mineralization and crystallization of CaOx and CaP kidney stones [50–55]. Matrix Gla Protein (MGP) also known as the cell growth-inhibiting gene 36 protein is a vitamin K dependent protein whose main role is inhibitor of bone formation/vascular calcification [56]. Previous studies have shown increased expression of MGP in renal tubules of hyperoxaluric rats [57,58], as well as renal epithelial cell lines NRK-52E and MDCK on exposure to oxalate and CaOx crystals [58,59]. Fetuin-A and B are expressed in humans, rats and mice at both the mRNA and protein levels [60]. Both actively inhibit precipitation of basic calcium phosphate in vitro. Fetuin A is considered an important inhibitor of pathological calcification in the humans [61]. Fetuin A deficient mice develop soft tissue calcification, including nephrocalcinosis [62]. The role of fetuin in kidney stone formation has not been analytically examined; however, it has been reported that kidney stone patients have lower urinary Fetuin A levels than normal controls [63].
Clusterin, like Kim-1, is expressed on the tubular cells after kidney injury and is also induced in polycystic kidney disease [64], and renal cell carcinoma [65]. CD44 is a cell surface receptor of OPN and has been shown to be highly expressed during injury, inflammation and wound healing and plays a role in formation of peri cellular matrix on surface of proliferating and migrating cells. CD44 has been shown to be increased in the kidneys of hyperoxaluric rats [16]. The AMBP gene encodes for Bikunin and α1-microglobulin, both inhibitors of CaOx crystallization [66,67]. Previous studies have shown that expression of AMBP was increased in the renal tubular cells of hyperoxaluric rats [18,68]. Bikunin mRNA expression was increased when renal epithelial cells were exposed to oxalate and CaOx crystals [69]. Our microarray data also showed up regulation of AMBP gene in the hyperoxaluric rats. Fibronectin is a known inhibitor of CaOx kidney stones [70]. Previous studies based on microarray analysis of genes in renal epithelial cells exposed to CaOx crystals have shown high expression of Fibronectin [42,71,72]. Our study based on microarray analysis of genes obtained from the kidney of hyperoxaluric rat showed comparable results.
Studies have shown that renal epithelial cells on exposure to oxalate/CaOx crystals produce inflammatory molecules such as Monocyte Chemoattractant Protein (MCP-1) [73]. MCP-1 or Ccl2 (Chemokine -CC motif Ligand 2) recruits monocytes, T-cells and dendritic cells at site of renal injury/inflammation and plays a significant role in the progression of renal failure. Results of this study show that MCP-1 was only slightly increased during hyperoxaluria, but highly expressed in the kidneys with CaOx crystals indicating that crystal exposure plays a key role in its expression.
Lipocalin 2 (Lcn 2) also known as Neutrophil Gelatinase-Associated Lipocalin or NGAL is a critical inflammatory mediator, contributing to tubular damage and kidney failure [74]. In our data, Lcn2 was highly up regulated, indicating tubular damage due to CaOx crystals.
Interestingly THP gene was downregulated during hyperoxaluria as well as CaOx nephrolithiasis. Similar results were obtained in our previous study using hydroxyl-L-proline as a hyperoxaluria inducing agent [27], as well as another study where EG was used to induce hyperoxaluria [75]. Inactivation of THP gene increases the frequency and severity of CaOx crystal deposition during EG induced hyperoxaluria, compared to the wild type control mice. In addition absence of THP leads to OPN induction suggesting that THP serves as a constitutive inhibitor of crystallization while OPN may act as an inducible inhibitor [76].
In this study, we identified 15 pathways that were common between the hyperoxaluria (day 14) and nephrolithiasis (day 28) (Table 3). These common pathways do provide an up or down stream response to development of hyperoxaluria and subsequently CaOx crystal formation. Looking at the common pathways we see a number of immunological and pathological changes taking place in various cells of the renal tissue in response to an oxalate load. After the onset of hyperoxaluria, we see up regulation of different signaling pathways including the extracellular matrix, complement and coagulation cascades, cell adhesion molecules, focal adhesion and pathways in cancer, to name a few. We observe that the immune response of the animal is triggered as evident by the activation of signaling pathways such as chemokine signaling pathway, cytokine-cytokine receptor interaction, leukocyte transendothelial migration, natural killer cell mediated cytotoxicity, and Fc Gamma R-mediated phagocytosis. These pathways remain active and significantly expressed till crystal formation stage.
There were 18 unique pathways that were significantly expressed after or during the onset of crystals as shown in Table 4. These pathways include but are not limited to P53 signaling which can be triggered due to different stress signal and may result in cell cycle arrest and apoptosis, Nod-like receptor signaling which leads to the activation of various proinflammatory cytokines such as IL-18 and IL1-β, after the activation of various inflammasomes, Hypertrophic cardiomyopathy, dilated cardiomyopathy, Toll-like receptor signaling pathway, T-cell receptor signaling pathway and Renin-angiotensin system pathway. These unique pathways give us an insight into how to differentiate between the renal consequences of hyperoxaluria and nephrolithiasis.
Conclusions
This transcriptional study of the ethylene glycol fed rat hyperoxaluric model at two different time intervals provides a better insight on the different pathways involved in hyperoxaluria and CaOx crystal deposition in the EG fed rat kidneys. Hyperoxaluria alone upregulates several genes, but crystal deposition further increases their expression. Hyperoxaluria is also associated with the activation of NADPH oxidase which generally leads to the production of ROS [44,77]. In addition crystal deposition is associated with substantially increased renal expression of KIM-1, and its urinary excretion [29], which is a well-recognized sign of renal injury [44]. Hyperoxaluria alone did not cause a change in urinary excretion of creatinine which was significantly increased after crystal deposition indicating renal damage. Apparently changes brought about by the exposure to oxalate without crystal deposition are not sufficient for noticeable renal injury. ROS produced during hyperoxaluria are most likely taken care of by antioxidant defense of the kidneys such as the upregulation of glutathione peroxidase as seen here.
Acknowledgments
The authors thank Dr. Jin Yao and Dr. Yijun Sun from the University of Florida’s Interdisciplinary Center for Biotechnology Research (ICBR) for running the microarrays and data analyses, Dr. Ammon B. Peck for his advice and suggestions during studies, and Dr. Vineet Sangar from Institute of Systems Biology (ISB), Seattle, WA for providing expert assistance in data analysis and Bioinformatics. Supported by National Institutes of Health grant #RO1-DK078602, DK092311 and University of Florida Center for the Study of Lithiasis. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
All microarray data have been deposited with Gene Expression Omnibus (GSE-89028).
Funding Statement
This study was supported by National Institutes of Health grant #RO1-DK078602, DK092311 and University of Florida Center for the Study of Lithiasis. Publication of this article was funded in part by the University of Florida Open Access Publishing Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Holmes RP, Ambrosius WT, Assimos DG (2005) Dietary oxalate loads and renal oxalate handling. J Urol 174: 943–947; discussion 947. doi: 10.1097/01.ju.0000169476.85935.e2 [DOI] [PubMed] [Google Scholar]
- 2.Asplin JR (2002) Hyperoxaluric calcium nephrolithiasis. Endocrinol Metab Clin North Am 31: 927–949. [DOI] [PubMed] [Google Scholar]
- 3.Joshi S, Peck AB, Khan SR (2013) NADPH oxidase as a therapeutic target for oxalate induced injury in kidneys. Oxid Med Cell Longev 2013: 462361 doi: 10.1155/2013/462361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes RP, Kennedy M (2000) Estimation of the oxalate content of foods and daily oxalate intake. Kidney Int 57: 1662–1667. doi: 10.1046/j.1523-1755.2000.00010.x [DOI] [PubMed] [Google Scholar]
- 5.Monico CG, Persson M, Ford GC, Rumsby G, Milliner DS (2002) Potential mechanisms of marked hyperoxaluria not due to primary hyperoxaluria I or II. Kidney Int 62: 392–400. doi: 10.1046/j.1523-1755.2002.00468.x [DOI] [PubMed] [Google Scholar]
- 6.Canales BK, Reyes L, Reinhard MK, Khan SR, Goncalves CG, et al. (2012) Renal glomerular and tubular injury after gastric bypass in obese rats. Nutrition 28: 76–80. doi: 10.1016/j.nut.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stamatelou KK, Francis ME, Jones CA, Nyberg LM, Curhan GC (2003) Time trends in reported prevalence of kidney stones in the United States: 1976–1994. Kidney Int 63: 1817–1823. doi: 10.1046/j.1523-1755.2003.00917.x [DOI] [PubMed] [Google Scholar]
- 8.Scales CD Jr., Smith AC, Hanley JM, Saigal CS (2012) Prevalence of kidney stones in the United States. Eur Urol 62: 160–165. doi: 10.1016/j.eururo.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rule AD, Roger VL, Melton LJ 3rd, Bergstralh EJ, Li X, et al. (2010) Kidney stones associate with increased risk for myocardial infarction. J Am Soc Nephrol 21: 1641–1644. doi: 10.1681/ASN.2010030253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan SR (2012) Is oxidative stress, a link between nephrolithiasis and obesity, hypertension, diabetes, chronic kidney disease, metabolic syndrome? Urol Res 40: 95–112. doi: 10.1007/s00240-011-0448-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jungers P, Joly D, Barbey F, Choukroun G, Daudon M (2004) ESRD caused by nephrolithiasis: prevalence, mechanisms, and prevention. Am J Kidney Dis 44: 799–805. [PubMed] [Google Scholar]
- 12.Rule AD, Bergstralh EJ, Melton LJ 3rd, Li X, Weaver AL, et al. (2009) Kidney stones and the risk for chronic kidney disease. Clin J Am Soc Nephrol 4: 804–811. doi: 10.2215/CJN.05811108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan SR, Glenton PA (2010) Experimental induction of calcium oxalate nephrolithiasis in mice. J Urol 184: 1189–1196. doi: 10.1016/j.juro.2010.04.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan SR (1997) Animal models of kidney stone formation: an analysis. World J Urol 15: 236–243. [DOI] [PubMed] [Google Scholar]
- 15.Mandel NS, Henderson JD Jr., Hung LY, Wille DF, Wiessner JH (2004) A porcine model of calcium oxalate kidney stone disease. J Urol 171: 1301–1303. doi: 10.1097/01.ju.0000110101.41653.bb [DOI] [PubMed] [Google Scholar]
- 16.Khan SR, Glenton PA, Byer KJ (2006) Modeling of hyperoxaluric calcium oxalate nephrolithiasis: experimental induction of hyperoxaluria by hydroxy-L-proline. Kidney Int 70: 914–923. doi: 10.1038/sj.ki.5001699 [DOI] [PubMed] [Google Scholar]
- 17.Umekawa T, Hatanaka Y, Kurita T, Khan SR (2004) Effect of angiotensin II receptor blockage on osteopontin expression and calcium oxalate crystal deposition in rat kidneys. J Am Soc Nephrol 15: 635–644. [DOI] [PubMed] [Google Scholar]
- 18.Iida S, Peck AB, Johnson-Tardieu J, Moriyama M, Glenton PA, et al. (1999) Temporal changes in mRNA expression for bikunin in the kidneys of rats during calcium oxalate nephrolithiasis. J Am Soc Nephrol 10: 986–996. [DOI] [PubMed] [Google Scholar]
- 19.Moriyama MT, Glenton PA, Khan SR (2001) Expression of inter-alpha inhibitor related proteins in kidneys and urine of hyperoxaluric rats. J Urol 165: 1687–1692. [PubMed] [Google Scholar]
- 20.Gokhale JA, Glenton PA, Khan SR (2001) Characterization of Tamm-Horsfall protein in a rat nephrolithiasis model. J Urol 166: 1492–1497. [PubMed] [Google Scholar]
- 21.Huang HS, Ma MC, Chen J, Chen CF (2002) Changes in the oxidant-antioxidant balance in the kidney of rats with nephrolithiasis induced by ethylene glycol. J Urol 167: 2584–2593. [PubMed] [Google Scholar]
- 22.Thamilselvan S, Hackett RL, Khan SR (1997) Lipid peroxidation in ethylene glycol induced hyperoxaluria and calcium oxalate nephrolithiasis. J Urol 157: 1059–1063. [PubMed] [Google Scholar]
- 23.Joshi S, Wang W, Peck AB, Khan SR (2015) Activation of the NLRP3 Inflammasome in Association with Calcium Oxalate Crystal Induced Reactive Oxygen Species in Kidneys. J Urol 193: 1684–1691. doi: 10.1016/j.juro.2014.11.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan SR (2004) Crystal-induced inflammation of the kidneys: results from human studies, animal models, and tissue-culture studies. Clin Exp Nephrol 8: 75–88. doi: 10.1007/s10157-004-0292-0 [DOI] [PubMed] [Google Scholar]
- 25.Khan SR (2013) Reactive oxygen species as the molecular modulators of calcium oxalate kidney stone formation: evidence from clinical and experimental investigations. J Urol 189: 803–811. doi: 10.1016/j.juro.2012.05.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan SR, Kok DJ (2004) Modulators of urinary stone formation. Front Biosci 9: 1450–1482. [DOI] [PubMed] [Google Scholar]
- 27.Khan SR, Joshi S, Wang W, Peck AB (2014) Regulation of macromolecular modulators of urinary stone formation by reactive oxygen species: transcriptional study in an animal model of hyperoxaluria. Am J Physiol Renal Physiol 306: F1285–1295. doi: 10.1152/ajprenal.00057.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joshi S, Clapp WL, Wang W, Khan SR (2015) Osteogenic changes in kidneys of hyperoxaluric rats. Biochim Biophys Acta 1852: 2000–2012. doi: 10.1016/j.bbadis.2015.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuo J, Khan A, Glenton PA, Khan SR (2011) Effect of NADPH oxidase inhibition on the expression of kidney injury molecule and calcium oxalate crystal deposition in hydroxy-L-proline-induced hyperoxaluria in the male Sprague-Dawley rats. Nephrol Dial Transplant 26: 1785–1796. doi: 10.1093/ndt/gfr035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan SR, Khan A, Byer KJ (2011) Temporal changes in the expression of mRNA of NADPH oxidase subunits in renal epithelial cells exposed to oxalate or calcium oxalate crystals. Nephrol Dial Transplant 26: 1778–1785. doi: 10.1093/ndt/gfq692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joshi S, Saylor BT, Wang W, Peck AB, Khan SR (2012) Apocynin-treatment reverses hyperoxaluria induced changes in NADPH oxidase system expression in rat kidneys: a transcriptional study. PLoS One 7: e47738 doi: 10.1371/journal.pone.0047738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, et al. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47 doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 34.Koul S, Khandrika L, Meacham RB, Koul HK (2012) Genome wide analysis of differentially expressed genes in HK-2 cells, a line of human kidney epithelial cells in response to oxalate. PLoS One 7: e43886 doi: 10.1371/journal.pone.0043886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koul S, Chaturvedi LS, Sekhon A, Bhandari A, Menon M, et al. (2002) Effects of oxalate on the re-initiation of DNA synthesis in LLC-PK1 cells do not involve p42/44 MAP kinase activation. Kidney Int 61: 525–533. doi: 10.1046/j.1523-1755.2002.00163.x [DOI] [PubMed] [Google Scholar]
- 36.Koul S, Khandrika L, Pshak TJ, Iguchi N, Pal M, et al. (2014) Oxalate upregulates expression of IL-2Rbeta and activates IL-2R signaling in HK-2 cells, a line of human renal epithelial cells. Am J Physiol Renal Physiol 306: F1039–1046. doi: 10.1152/ajprenal.00462.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schepers MS, van Ballegooijen ES, Bangma CH, Verkoelen CF (2005) Crystals cause acute necrotic cell death in renal proximal tubule cells, but not in collecting tubule cells. Kidney Int 68: 1543–1553. doi: 10.1111/j.1523-1755.2005.00566.x [DOI] [PubMed] [Google Scholar]
- 38.Thamilselvan S, Hackett RL, Khan SR (1999) Cells of proximal and distal tubular origin respond differently to challenges of oxalate and calcium oxalate crystals. J Am Soc Nephrol 10 Suppl 14: S452–456. [PubMed] [Google Scholar]
- 39.Khan SR, Thamilselvan S (2000) Nephrolithiasis: a consequence of renal epithelial cell exposure to oxalate and calcium oxalate crystals. Mol Urol 4: 305–312. [PubMed] [Google Scholar]
- 40.Khan SR, Byer KJ, Thamilselvan S, Hackett RL, McCormack WT, et al. (1999) Crystal-cell interaction and apoptosis in oxalate-associated injury of renal epithelial cells. J Am Soc Nephrol 10 Suppl 14: 457–463. [PubMed] [Google Scholar]
- 41.Khan SR (2011) Crystal/cell interaction and nephrolithiasis. Arch Ital Urol Androl 83: 1–5. [PubMed] [Google Scholar]
- 42.Okada A, Yasui T, Hamamoto S, Hirose M, Kubota Y, et al. (2009) Genome-wide analysis of genes related to kidney stone formation and elimination in the calcium oxalate nephrolithiasis model mouse: detection of stone-preventive factors and involvement of macrophage activity. J Bone Miner Res 24: 908–924. doi: 10.1359/jbmr.081245 [DOI] [PubMed] [Google Scholar]
- 43.Mulay SR, Kulkarni OP, Rupanagudi KV, Migliorini A, Darisipudi MN, et al. (2013) Calcium oxalate crystals induce renal inflammation by NLRP3-mediated IL-1beta secretion. J Clin Invest 123: 236–246. doi: 10.1172/JCI63679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuo J, Khan A, Glenton PA, Khan SR (2011) Effect of NADPH oxidase inhibition on the expression of kidney injury molecule and calcium oxalate crystal deposition in hydroxy-L-proline-induced hyperoxaluria in the male Sprague-Dawley rats. Nephrol Dial Transplant 26: 1785–1796. doi: 10.1093/ndt/gfr035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khand FD, Gordge MP, Robertson WG, Noronha-Dutra AA, Hothersall JS (2002) Mitochondrial superoxide production during oxalate-mediated oxidative stress in renal epithelial cells. Free Radic Biol Med 32: 1339–1350. [DOI] [PubMed] [Google Scholar]
- 46.Han WK, Bailly V, Abichandani R, Thadhani R, Bonventre JV (2002) Kidney Injury Molecule-1 (KIM-1): a novel biomarker for human renal proximal tubule injury. Kidney Int 62: 237–244. doi: 10.1046/j.1523-1755.2002.00433.x [DOI] [PubMed] [Google Scholar]
- 47.Asselman M, Verhulst A, De Broe ME, Verkoelen CF (2003) Calcium oxalate crystal adherence to hyaluronan-, osteopontin-, and CD44-expressing injured/regenerating tubular epithelial cells in rat kidneys. J Am Soc Nephrol 14: 3155–3166. [DOI] [PubMed] [Google Scholar]
- 48.Ryall RL (2004) Macromolecules and urolithiasis: parallels and paradoxes. Nephron Physiol 98: p37–42. doi: 10.1159/000080262 [DOI] [PubMed] [Google Scholar]
- 49.Stapleton AM, Timme TL, Ryall RL (1998) Gene expression of prothrombin in the human kidney and its potential relevance to kidney stone disease. Br J Urol 81: 666–671. [DOI] [PubMed] [Google Scholar]
- 50.Kleinman JG, Wesson JA, Hughes J (2004) Osteopontin and calcium stone formation. Nephron Physiol 98: p43–47. doi: 10.1159/000080263 [DOI] [PubMed] [Google Scholar]
- 51.Khan SR, Kok DJ (2004) Modulators of urinary stone formation. Front Biosci 9: 1450–1482. [DOI] [PubMed] [Google Scholar]
- 52.Hunter GK (2013) Role of Osteopontin in Modulation of Hydroxyapatite Formation. Calcif Tissue Int. [DOI] [PubMed] [Google Scholar]
- 53.Hunter GK, Grohe B, Jeffrey S, O'Young J, Sorensen ES, et al. (2009) Role of phosphate groups in inhibition of calcium oxalate crystal growth by osteopontin. Cells Tissues Organs 189: 44–50. doi: 10.1159/000151430 [DOI] [PubMed] [Google Scholar]
- 54.Kohri K, Yasui T, Okada A, Hirose M, Hamamoto S, et al. (2012) Biomolecular mechanism of urinary stone formation involving osteopontin. Urol Res 40: 623–637. doi: 10.1007/s00240-012-0514-y [DOI] [PubMed] [Google Scholar]
- 55.Wesson JA, Johnson RJ, Mazzali M, Beshensky AM, Stietz S, et al. (2003) Osteopontin is a critical inhibitor of calcium oxalate crystal formation and retention in renal tubules. J Am Soc Nephrol 14: 139–147. [DOI] [PubMed] [Google Scholar]
- 56.Schurgers LJ, Cranenburg EC, Vermeer C (2008) Matrix Gla-protein: the calcification inhibitor in need of vitamin K. Thromb Haemost 100: 593–603. [PubMed] [Google Scholar]
- 57.Yasui T, Fujita K, Sasaki S, Sato M, Sugimoto M, et al. (1999) Expression of bone matrix proteins in urolithiasis model rats. Urol Res 27: 255–261. [DOI] [PubMed] [Google Scholar]
- 58.Khan A, Wang W, Khan SR (2014) Calcium oxalate nephrolithiasis and expression of matrix GLA protein in the kidneys. World J Urol 32: 123–130. doi: 10.1007/s00345-013-1050-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao B, Yasui T, Lu X, Zhou H, Liu J, et al. (2010) Matrix Gla protein expression in NRK-52E cells exposed to oxalate and calcium oxalate monohydrate crystals. Urol Int 85: 237–241. doi: 10.1159/000314947 [DOI] [PubMed] [Google Scholar]
- 60.Denecke B, Graber S, Schafer C, Heiss A, Woltje M, et al. (2003) Tissue distribution and activity testing suggest a similar but not identical function of fetuin-B and fetuin-A. Biochem J 376: 135–145. doi: 10.1042/BJ20030676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herrmann M, Kinkeldey A, Jahnen-Dechent W (2012) Fetuin-A function in systemic mineral metabolism. Trends Cardiovasc Med 22: 197–201. doi: 10.1016/j.tcm.2012.07.020 [DOI] [PubMed] [Google Scholar]
- 62.Schafer C, Heiss A, Schwarz A, Westenfeld R, Ketteler M, et al. (2003) The serum protein alpha 2-Heremans-Schmid glycoprotein/fetuin-A is a systemically acting inhibitor of ectopic calcification. J Clin Invest 112: 357–366. doi: 10.1172/JCI17202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stejskal D, Karpisek M, Vrtal R, Student V, Solichova P, et al. (2008) Urine fetuin-A values in relation to the presence of urolithiasis. BJU Int 101: 1151–1154. doi: 10.1111/j.1464-410X.2007.07432.x [DOI] [PubMed] [Google Scholar]
- 64.Harding MA, Chadwick LJ, Gattone VH 2nd, Calvet JP (1991) The SGP-2 gene is developmentally regulated in the mouse kidney and abnormally expressed in collecting duct cysts in polycystic kidney disease. Dev Biol 146: 483–490. [DOI] [PubMed] [Google Scholar]
- 65.Correa-Rotter R, Ibarra-Rubio ME, Schwochau G, Cruz C, Silkensen JR, et al. (1998) Induction of clusterin in tubules of nephrotic rats. J Am Soc Nephrol 9: 33–37. [DOI] [PubMed] [Google Scholar]
- 66.Atmani F, Khan SR (1999) Role of urinary bikunin in the inhibition of calcium oxalate crystallization. J Am Soc Nephrol 10 Suppl 14: S385–388. [PubMed] [Google Scholar]
- 67.Atmani F, Mizon J, Khan SR (1996) Identification of uronic-acid-rich protein as urinary bikunin, the light chain of inter-alpha-inhibitor. Eur J Biochem 236: 984–990. [DOI] [PubMed] [Google Scholar]
- 68.Grewal JS, Tsai JY, Khan SR (2005) Oxalate-inducible AMBP gene and its regulatory mechanism in renal tubular epithelial cells. Biochem J 387: 609–616. doi: 10.1042/BJ20041465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iida S, Peck AB, Byer KJ, Khan SR (1999) Expression of bikunin mRNA in renal epithelial cells after oxalate exposure. J Urol 162: 1480–1486. [PubMed] [Google Scholar]
- 70.Tsujihata M, Miyake O, Yoshimura K, Kakimoto KI, Takahara S, et al. (2000) Fibronectin as a potent inhibitor of calcium oxalate urolithiasis. J Urol 164: 1718–1723. [PubMed] [Google Scholar]
- 71.Miyazawa K, Aihara K, Ikeda R, Moriyama MT, Suzuki K (2009) cDNA macroarray analysis of genes in renal epithelial cells exposed to calcium oxalate crystals. Urol Res 37: 27–33. doi: 10.1007/s00240-008-0164-2 [DOI] [PubMed] [Google Scholar]
- 72.Okada A, Yasui T, Fujii Y, Niimi K, Hamamoto S, et al. (2010) Renal macrophage migration and crystal phagocytosis via inflammatory-related gene expression during kidney stone formation and elimination in mice: Detection by association analysis of stone-related gene expression and microstructural observation. J Bone Miner Res 25: 2701–2711. doi: 10.1002/jbmr.158 [DOI] [PubMed] [Google Scholar]
- 73.Umekawa T, Chegini N, Khan SR (2002) Oxalate ions and calcium oxalate crystals stimulate MCP-1 expression by renal epithelial cells. Kidney Int 61: 105–112. doi: 10.1046/j.1523-1755.2002.00106.x [DOI] [PubMed] [Google Scholar]
- 74.Glaros T, Fu Y, Xing J, Li L (2012) Molecular mechanism underlying persistent induction of LCN2 by lipopolysaccharide in kidney fibroblasts. PLoS One 7: e34633 doi: 10.1371/journal.pone.0034633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marengo SR, Chen DH, Kaung HL, Resnick MI, Yang L (2002) Decreased renal expression of the putative calcium oxalate inhibitor Tamm-Horsfall protein in the ethylene glycol rat model of calcium oxalate urolithiasis. J Urol 167: 2192–2197. [PubMed] [Google Scholar]
- 76.Mo L, Liaw L, Evan AP, Sommer AJ, Lieske JC, et al. (2007) Renal calcinosis and stone formation in mice lacking osteopontin, Tamm-Horsfall protein, or both. Am J Physiol Renal Physiol 293: F1935–1943. doi: 10.1152/ajprenal.00383.2007 [DOI] [PubMed] [Google Scholar]
- 77.Khan SR, Khan A, Byer KJ (2011) Temporal changes in the expression of mRNA of NADPH oxidase subunits in renal epithelial cells exposed to oxalate or calcium oxalate crystals. Nephrol Dial Transplant 26: 1778–1785. doi: 10.1093/ndt/gfq692 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All microarray data have been deposited with Gene Expression Omnibus (GSE-89028).