ABSTRACT
Each mitochondrial compartment contains varying protein compositions that underlie a diversity of localized functions. Insights into the localization of mitochondrial intermembrane space-bridging (MIB) components will have an impact on our understanding of mitochondrial architecture, dynamics and function. By using the novel visualizable genetic tags miniSOG and APEX2 in cultured mouse cardiac and human astrocyte cell lines and performing electron tomography, we have mapped at nanoscale resolution three key MIB components, Mic19, Mic60 and Sam50 (also known as CHCHD3, IMMT and SAMM50, respectively), in the environment of structural landmarks such as cristae and crista junctions (CJs). Tagged Mic19 and Mic60 were located at CJs, distributed in a network pattern along the mitochondrial periphery and also enriched inside cristae. We discovered an association of Mic19 with cytochrome c oxidase subunit IV. It was also found that tagged Sam50 is not uniformly distributed in the outer mitochondrial membrane and appears to incompletely overlap with Mic19- or Mic60-positive domains, most notably at the CJs.
KEY WORDS: APEX, EFTEM, MICOS, Crista junction, Cristae, miniSOG
Highlighted Article: By using the novel genetic labels miniSOG and APEX2 combined with electron tomography, the sub-compartmental locations of Mic19, Mic60 and Sam50 were deciphered, which helps determine their physiological function and interaction partners.
INTRODUCTION
Mitochondria consist of two membranes of distinct architecture: the outer mitochondrial membrane (OMM) and the inner mitochondrial membrane (IMM). The IMM is heterogeneous and composed of distinct subdomains, including the inner boundary membrane (IBM), which faces the OMM, and the cristae membranes, which extend into the matrix space (Frey et al., 2002; Mannella, 2006; Perkins and Ellisman, 2007). The significance and protein constituents of the connection between the IBM and the cristae membrane, called the crista junction (CJ) (Perkins et al., 1997, 1998, 2001), are being systematically teased out (Anand et al., 2016; Barbot et al., 2015; Darshi et al., 2011; Dudkina et al., 2010; John et al., 2005; Koob et al., 2015; Körner et al., 2012; Mun et al., 2010; Rabl et al., 2009). The OMM, IBM and CJs are connected by a multi-subunit complex called the mitochondrial contact site and cristae organizing system (MICOS; Bohnert et al., 2015; Ding et al., 2015; Friedman et al., 2015; Hoppins et al., 2011; Huynen et al., 2016; Jans et al., 2013; Koob and Reichert, 2014; Ott et al., 2012, 2015; van der Laan et al., 2016; Xie et al., 2007; Zerbes et al., 2012b, 2016).
The study of mitochondrial intermembrane space bridging (MIB) composition and function has been a timely and prolific line of investigation, given the emerging data on its role in mitochondrial organization (Huynen et al., 2016; Ott et al., 2012, 2015; Xie et al., 2007). The centerpiece of the MIB is the MICOS, which has a crucial role in mitochondrial dynamics, the maintenance of cristae and in the formation of CJs and contact sites between the outer and inner membranes (Akabane et al., 2016; Alkhaja et al., 2012; Barbot et al., 2015; Bohnert et al., 2012; Genin et al., 2016; Harner et al., 2011; Hoppins et al., 2011; Jayashankar et al., 2016; John et al., 2005; Körner et al., 2012; Mun et al., 2010; Rabl et al., 2009; van der Laan et al., 2016; von der Malsburg et al., 2011; Zerbes et al., 2012a). Intriguingly, of the species studied, those without cristae never have MICOS proteins and all species with cristae possess at least one MICOS protein (Huynen et al., 2016). In mammals, which proteins are denoted as being part of the MICOS landscape has been changing and currently consists of nine proteins: Mic60 (also known as IMMT), Mic27 (APOOL), Mic25 (CHCHD6), Mic23 (Mic26 or APOO), Mic19 (CHCHD3), Mic14 (CHCHD5), Mic13, Mic10 (MINOS1) and DISC1 (Anand et al., 2016; Darshi et al., 2011; Genin et al., 2016; Guarani et al., 2015; Jans et al., 2013; Koob et al., 2015; Kozjak-Pavlovic, 2017; Li et al., 2016; Ott et al., 2015; Pfanner et al., 2014; Piñero-Martos et al., 2016; Weber et al., 2013).
It has been suggested that the most important components of the large 2200–2800 kDa MIB complex in mammalian mitochondria are Mic60 (mitofilin), which is tethered to the IMM, Mic19, which was first identified by our group as a cAMP-dependent protein kinase A substrate (Schauble et al., 2007) and resides in the intermembrane space (IMS), and Sam50, which resides in the OMM (Akabane et al., 2016; Darshi et al., 2011; Ott et al., 2012, 2015). Thus, key components of the MIB connect the IMM, IMS and OMM. Mic19 is a myristoylated peripheral membrane protein, whereas all other MICOS components contain at least one transmembrane segment (Alkhaja et al., 2012; Harner et al., 2011; Hoppins et al., 2011; van der Laan et al., 2012; von der Malsburg et al., 2011). The association of Mic19 with MICOS depends on Mic60 (Alkhaja et al., 2012; Bohnert et al., 2015; Darshi et al., 2011; Ding et al., 2015; Li et al., 2016; Ott et al., 2012; Xie et al., 2007), and Mic60 relies on Mic19 to regulate its distribution inside mitochondria (Ding et al., 2015; Friedman et al., 2015). The disulfide-bonded CHCH domain of Mic19 binds to Mic60, whereas the myristoylated N-terminus of Mic19 docks with Sam50, which extends into the IMS (Darshi et al., 2012). Stimulated emission depletion (STED) microscopy confirmed colocalization of Mic60 with Mic19 (Jans et al., 2013). Mic60 is in the IMM and has a large C-terminal domain facing the intracristal space or IMS, where it interacts with the sorting and assembly machinery (SAM) complex, also known as the translocase of outer membrane β-barrel proteins (TOB) complex (hereafter TOB/SAM complex) on the OMM (Harner et al., 2011; Hoppins et al., 2011; Körner et al., 2012; Odgren et al., 1996; von der Malsburg et al., 2011). The C-terminus of Mic60 is required for formation of CJs, which also requires Mic13 (Anand et al., 2016). CJs are lost and the cristae are detached from the IBM and re-arrange to large membrane stacks in mutant cells lacking Mic60 (John et al., 2005; Mun et al., 2010; Rabl et al., 2009; von der Malsburg et al., 2011). Knocking down Sam50 leads to the complete loss of cristae without modifying MIC19 and Mic60 protein levels (Ott et al., 2012). However, because Sam50 is part of the mitochondrial β-barrel protein import and assembly machinery, it is questionable whether the cristae depletion is caused directly by its knockdown or rather by a general impairment of protein import into mitochondria (Cogliati et al., 2016).
Deciphering the sub-compartmental location of a mitochondrial protein may help explain its physiological function and interaction partners. To localize Mic19, Mic60 and Sam50 at high spatial resolution in their mitochondrial milieu, we appended recently developed genetic tags, Mini Singlet Oxygen Generator (miniSOG, Shu et al., 2011) and engineered ascorbate peroxidase 2 (APEX2, Hung et al., 2014; Lam et al., 2015; Martell et al., 2012; Rhee et al., 2013), and fused them to these expressed proteins in cultured cells permitting high-quality ultrastructural preservation and three-dimensional protein localization via electron tomography (ET) in relation to structural landmarks. We explored the question of whether their high-resolution distributions in mitochondria are the same, indicating an obligatory association for proper functioning, or whether differences can be found in their compartmentation in relation to structural landmarks. We found that Mic19-positive domains are at CJs, distributed in a network pattern in the IMS and also line the intracristal space bounded by the cristae membranes. Based on the report that Mic19 associates with cristae (Harner et al., 2011), and our findings that this protein is inside the cristae and the fact that knockdown of Mic19 resulted in reduction of cytochrome c oxidase (Cox) subunit IV, we explored whether Mic19 associates with CoxIV and found that they indeed associate with each other when Mic19 is myristoylated. We also found that Sam50 is not uniformly distributed around the mitochondrial periphery and that its localization appears to incompletely overlap with the Mic19 and Mic60 domains, most notably at the CJs.
RESULTS
Engineering of Mic19–miniSOG for correlated light and electron microscopy
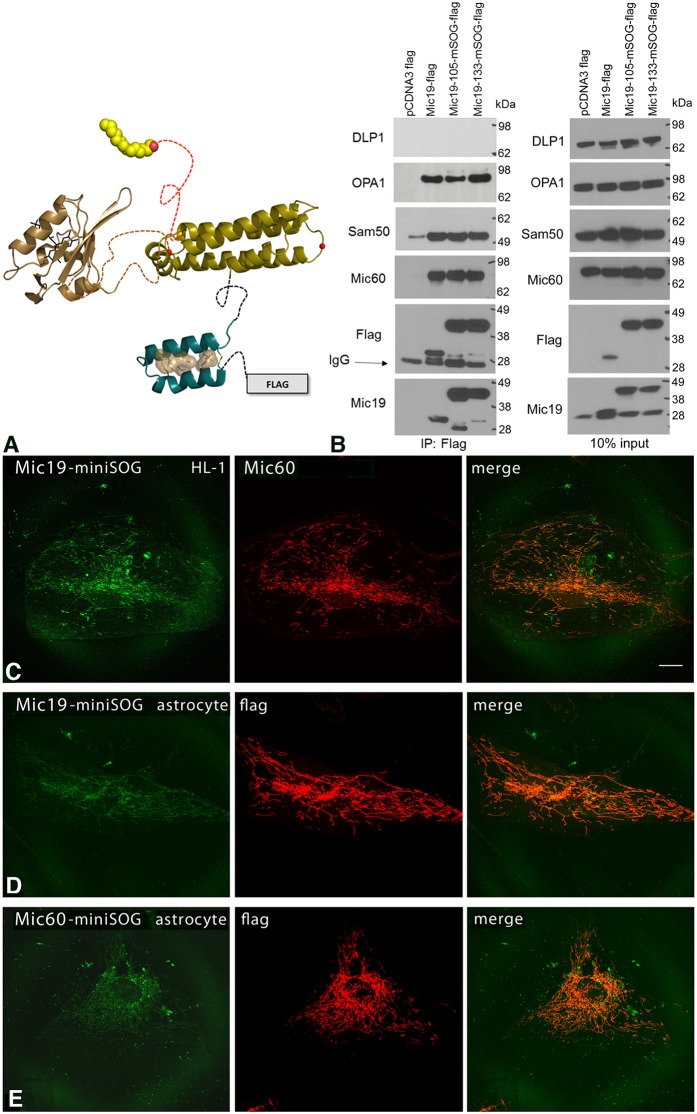
Mic19 has an N-terminal myristoylation motif preceded by a DUF737 domain (domain of unknown function), and a C-terminal coiled-coil-helix–coiled-coil-helix (CHCH) domain. Previously, we showed that both the N- and C-termini of the protein are critical for precise localization within mitochondrial compartments (Darshi et al., 2011, 2012). We found that attaching a fluorescent protein or other tags – except short peptide tags such as FLAG tags – to either terminus of Mic19 caused the protein to fail to localize to the mitochondria. Other studies have shown that although Mic19 with FLAG tags at the C-terminus localized to the mitochondria, 50% of the mitochondria lost their membrane potential (Ott et al., 2015). To overcome these hurdles, one possibility would have been to attach fluorescent tags in the loop regions of the protein; however, there is no known structure for Mic19 that would allow engineering such tags into the loop regions of the protein. Therefore, we used the threading program Phyre2 to search the Protein Data Bank (PDB) for a structural homolog of the DUF and CHCH domains of Mic19. Phyre2 identified 1Y4C, which is a designed IL-4 antagonist that was crystallized, as a candidate (Laporte et al., 2005). Based on this structure, we identified two loops that could serve as sites for introducing the genetic tag miniSOG, a fluorescent flavoprotein construct for correlated light and electron microscopy (Shu et al., 2011). The sites selected were in the loop regions present in the CHCH domain and were part of a model based on the conserved hairpin motif in Mia40 (also known as CHCHD4) and CHCHD5 (Banci et al., 2009, 2012). Both these sites, residues 105 and 133, are predicted to lie in loops between helices. Two constructs were engineered along with glycine serine linkers and the miniSOG was inserted in frame into each of these sites (Fig. 1A). HEK 293 cells expressing FLAG-tagged Mic19 did not interfere with binding to Mic60, Sam50 or OPA1 (Fig. 1B). Immunoisolated samples were analyzed by SDS-PAGE followed by immunoblotting (IB) and showed that Mic19 interacts with OPA1, Mic60 and Sam50. Mic19-133–miniSOG was subsequently used in all further studies.
Fig. 1.
Engineering of Mic19–miniSOG. (A) Two sites were selected for insertion of miniSOG based on our modeling of the Mic19 DUF domain. Residues 105 and 133 are predicted to lie in loops between helices. Phyre2 was used for generating a 3D model of miniSOG (tan) and Mic19. (B) MiniSOG addition at 105 or 133 did not interfere with binding to Mic60, Sam50 or OPA1 in HEK 293 cells expressing Flag-tagged Mic19–miniSOG as determined by immunoprecipitation (IP). Immunoisolated samples were analyzed by SDS-PAGE and immunoblotting (IB) with the indicated antibodies. DLP1 is a negative control. 10% input control is shown on the right. Note that the bottom two Flag blots are also shown in Fig. 8A,B for comparison. (C) MiniSOG constructs of Mic19 and Mic60 correctly trafficked to mitochondria. Left, fluorescence confocal microscopy of Mic19–miniSOG labeling in a HL-1 cell, showing a mitochondrial labeling pattern. Middle, endogenous Mic60 labeling in the same HL-1 cell also showing mitochondrial labeling. Right, overlay of Mic19–miniSOG and Mic60 showing colocalization. Scale bar: 20 μm. (D) Fluorescence confocal microscopy of Mic19–miniSOG (left) and anti-Flag (middle) in a human primary astrocyte, showing mitochondrial labeling; an overlay is on the right. (E) Fluorescence confocal microscopy of Mic60–miniSOG (left) and anti-Flag antibody staining (middle) in a human primary astrocyte, showing mitochondrial labeling; an overlay is on the right. All results are representative of n=3 experiments.
MiniSOG constructs of Mic19, Mic60 and Sam50 were tested for localization in HL-1 cells and human primary astrocytes by using fluorescence confocal microscopy. We observed that only Mic19 and Mic60 showed proper fluorescence localization to mitochondria (Fig. 1C) whereas the Sam50–miniSOG failed to express in these cell lines (data not shown). Images of Mic19-133–miniSOG fluorescence labeling indicated that when the protein was overexpressed in HL-1 cells, it localized to the mitochondria and colocalized with the endogenous Mic60. Similar to HL-1 cells, Mic19–miniSOG showed mitochondrial localization in cultured primary human astrocytes (Fig. 1D). The miniSOG fluorescence was weak in the astrocytes. Therefore, to further verify the expression, immunostaining was performed with anti-Flag antibody. As with Mic19, overexpressed Mic60–miniSOG in human primary astrocytes localized to the mitochondria (Fig. 1E). Again, to further verify the expression, immunostaining was performed with anti-Flag antibody.
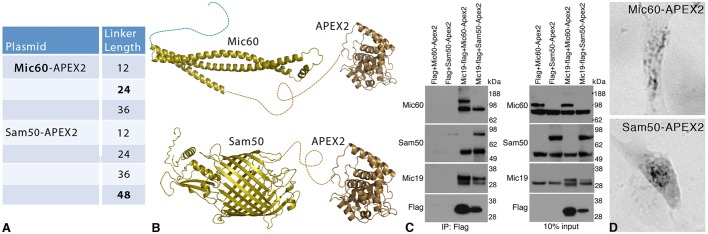
Because the Sam50–miniSOG failed to express and the Mic60–miniSOG showed only weak expression in astrocytes, an alternative strategy was adopted of using APEX2, which like miniSOG is used for correlative light and electron microscopy localization of proteins, for genetically tagging Sam50 Mic19 and Mic60. To obtain maximal expression, varying lengths of linker peptides between the C-terminal end of Mic60 or Sam50 and APEX2 were tested (Fig. 2A). We determined that only Mic60 with the 24-residue linker and Sam50 with the 48-residue linker showed any expression via light microscopy of human primary astrocytes. With these linker peptides, the partial structure prediction of the C-terminal region of Mic60 rendered with the structure of APEX2 is shown in Fig. 2B. Likewise, the predicted structure of Sam50 with APEX2 is shown. In HEK 293 cells, it was found that Mic19 interacted with both the endogenous and the APEX2-tagged Mic60 and Sam50 (Fig. 2C). The transfection efficiency and expression levels in astrocytes were extremely low and undetectable by western blotting. Bright-field light microscopy of the osmicated DAB labeling of transfected Mic60–APEX2 and Sam50–APEX2 showed proper mitochondrial localization in human primary astrocytes (Fig. 2D).
Fig. 2.
Engineering of APEX2 constructs of Mic60 and Sam50. (A) Clone and linker peptide length between the C-terminal end of Mic60 or Sam50 and APEX2. Only Mic60 with the 24-residue linker and Sam50 with the 48-residue linker showed expression. (B) Partial structure prediction of the C-terminal region of Mic60 and Sam50 rendered with the structure of APEX2 attached. (C) Flag-tagged Mic19 along with APEX2-tagged Mic60 and Sam50 were transiently expressed in HEK 293 cells and immunoprecipitated (IP) using FLAG resin. The eluted samples were analyzed by immunoblotting with antibodies against Flag, Mic19, Mic60 and Sam50. Mic19 interacted with both the endogenous and the APEX2-tagged MIc60 and Sam50. A 10% input control is shown on the right. The lower molecular mass band represents endogenous protein. (D) Bright-field light microscopy of Mic60–APEX2- and Sam50–APEX2-transfected human primary astrocytes showing mitochondrial labeling. Results are representative of n=10 experiments.
EFTEM confirmation of labeling
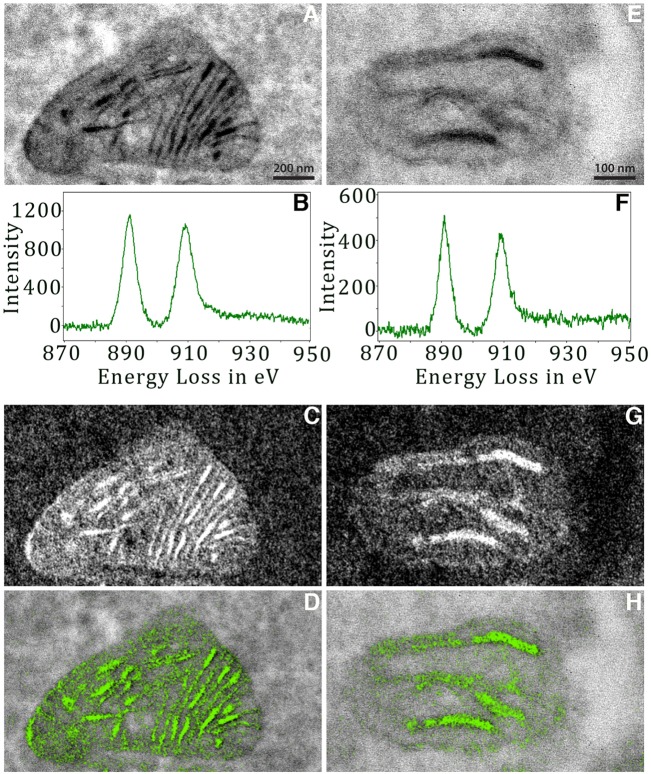
Because the genetically tagged Mic19, Mic60 or Sam50 that becomes negatively stained by the surrounding oxidized DAB precipitate may not be readily distinguishable from the deposition of the osmium, uranium, or lead heavy metals that stain the protein and lipid endogenous cellular structures, we used multi-color electron microscopy (EM) (Adams et al., 2016) to differentiate the DAB precipitate from the general staining of endogenous cellular material. Energy-filtered transmission electron microscopy (EFTEM) was used with cerium conjugated to DAB (Ce-DAB2) to generate electron energy-loss spectra (EELS) (Ahn and Krivanek, 1983) and elemental maps (Messaoudi et al., 2013; Ramachandra et al., 2014) (Fig. 3). We tested this new detection technology using Mic19-miniSOG in HL-1 cells and found that when the Ce map was overlaid as a color on the conventional grayscale electron micrograph (Fig. 3D,H), the strongest staining corresponded to the Ce-DAB2, and hence to the Mic19–miniSOG. This provided confidence moving forward that the darkest staining of the cellular ultrastructure seen in the conventional TEM image would correspond to the miniSOG- or APEX2-labeled protein and could be easily distinguished from the much lower contrast unlabeled cellular components (Fig. 3A,E).
Fig. 3.
EELS and EFTEM confirm Mic19–miniSOG labeling. (A) TEM image of a large Mic19–miniSOG Ce-DAB-labeled mitochondrion in a HL-1 cell (viewed at 15,000× magnification, 0.56 nm/pixel). (B) EELS showing a strong cerium (Ce) signal. Arbitrary intensity unit. (C) Ce EFTEM elemental map (viewed at 15,000× magnification). Pre-edge and post-edge were a sum of 12 drift-corrected images. (D) Overlay of the elemental map (green) on the TEM image. (E) TEM image, (F) EELS, (G) Ce-elemental map and (H) overlay of the elemental map (green) on the TEM image of a small miniSOG–Mic19 Ce-DAB-labeled mitochondrion in a HL-1 cell (viewed at 30,000× magnification, 0.28 nm/pixel). A Gaussian smoothing filter of radius 1 pixel, was applied to all images to reduce the noise. All results are representative of n=10 experiments.
High-resolution distribution of Mic19, Mic60 and Sam50
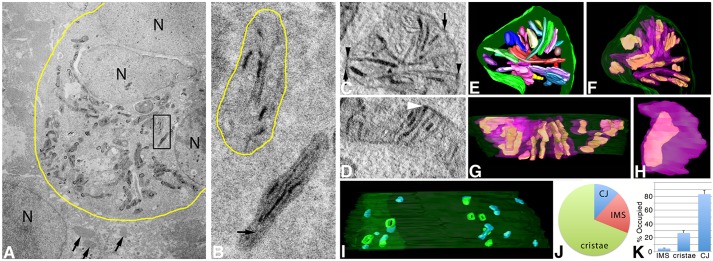
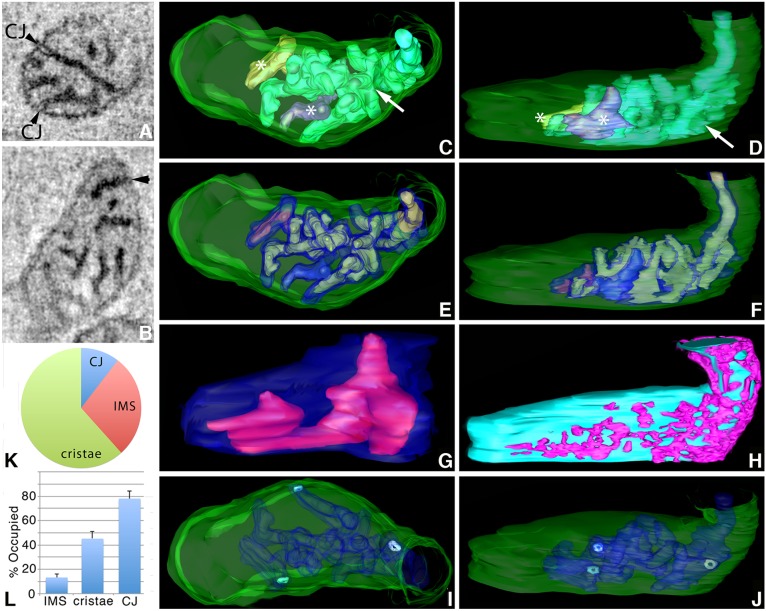
ET was used to determine the high-resolution 3D distribution of labeled Mic19, Mic60 and Sam50 molecules inside cultured cells. Cells expressing a miniSOG construct that were also photooxidized were easily distinguishable from their un-photooxidized neighbors by using conventional electron microscopy because of their highly contrasted mitochondria. An example is shown in Fig. 4A of a cluster of HL-1 cells expressing Mic19–miniSOG. Inside the photooxidized region (yellow boundary curve), the mitochondria are darker and highly localized regions of labeling are seen at places around the mitochondrial periphery and in the cristae. The matrix is sometimes darker than the cytoplasm, which provides information about the mitochondrial shape. When the matrix is the same contrast as the cytoplasm, the shape of the mitochondrion can still be roughly deduced from the extent of the labeled cristae (Fig. 4B). ET maps showed that, in HL-1 cells, Mic19-positive domains are at CJs, distributed in the IMS and also line the intracristal space bounded by the cristae membranes (Fig. 4C,D). The segmented and surface-rendered volumes confirmed that the Mic19-positive domains found in the cristae extended throughout the volume (Fig. 4E–G). However, the labeled regions never filled the cristae volumes and were in one discrete shape rather than speckled throughout (Fig. 4H). No pattern could be detected with the Mic19-positive shapes, nor in how they were distributed inside the cristae. It appeared that all the CJs were positive for Mic19 and some of the CJ openings were filled with label, whereas others were not (Fig. 4I). Mic19–miniSOG expression did not alter the CJ architecture. The CJ opening size of ∼15 nm and roundish shape were similar to what was reported previously (Darshi et al., 2011). Localized regions in the IMS not associated with CJs were also positive for Mic19 (Fig. 4I). Measurements on how the label was distributed between CJ, IMS and cristae using 10 mitochondrial tomographic volumes indicated that the majority of the Mic19-positive domains were in the cristae (69±5%, mean±s.e.m.) with lesser amounts in the IMS (19±4%) and CJs (12±2%) (Fig. 4J). It should be understood that the smaller value for the CJs is due to this structure being much smaller than a crista and thus having less volume. 82±6% (mean±s.e.m.) of the CJs were labeled, whereas only part of the IMS (4±1%) and cristae (27±4%) volumes were filled with Mic19-positive domains (Fig. 4K). The IMS in particular was only sparsely occupied by Mic19-positive domains.
Fig. 4.
ET volumes of Mic19–miniSOG show that it localizes to discrete domains in CJ, IMS and cristae in HL-1 cells. (A) TEM of a cluster of HL-1 cells expressing Mic19–miniSOG. The yellow boundary curve shows the photooxidized region. The mitochondria display dark labeling around their periphery and in the cristae. Outside the photooxidized region, the mitochondria show no dark labeling (arrows) and serve as negative controls. N, nucleus. Results are representative of n=6 cells. (B) Enlarged view of the boxed region in A showing a mitochondrion (bottom) with a relatively dark matrix (arrow points to a portion of the matrix) in contrast to the mitochondrion directly above it with a matrix as light as the surrounding cytoplasm. Here, the OMM (yellow boundary curve) can be deduced by using the labeled cristae and IMS patches as guideposts. In both mitochondria, the cristae labeling is dark. (C) A 1-nm slice through the center of a tomographic volume from a HL-1 cell showing that Mic19-positive domains are at CJs (arrowheads), distributed in the IMS (arrows) and also partially line the intracristal space. Results are representative of n=38 mitochondria. (D) Another example of a slice through the center of a tomographic volume from a different HL-1 cell that has a darkly labeled Mic19-positive domain at a CJ (arrowhead). (E) The segmented and surface-rendered volume of the mitochondrion shown in C. The OMM is green, the cristae are in various colors. HL-1 cells are densely packed with cristae. (F) As in E in the same orientation but showing the Mic19-positive domains (green) in the cristae (translucent magenta) typical of the 38 mitochondrial volumes examined. (G) As in E but in a side orientation showing the Mic19-positive domains extending throughout the cristae. However, the labeled regions never filled the cristae volumes and were in contiguous shapes rather than speckled throughout. (H) A typical Mic19-positive domain inside a crista (translucent magenta) showing that the label filled less than half the crista volume. (I) Side view of the entire complement of CJ (green) and IMS (magenta) Mic19-positive domains. The OMM is translucent. This volume had eight CJs, four of which had openings filled with label and four others did not. The IMS domains not associated with CJs were similar in size to the CJs with no apparent pattern to their distribution. (J) Pie chart showing the distribution of Mic19-positive domains between CJ, IMS and cristae. (K) Histogram showing the percentage of the IMS, cristae or CJ volumes occupied by Mic19-positive domains (mean±s.e.m.). Results are from three biological replicates and 10 total technical replicates (J,K).
To test the consistency of the 3D labeling pattern of Mic19–miniSOG observed in HL-1 cells with another cell type and to correlate with a cell type where genetically tagged Mic60 and Sam50 had strong expression, human primary astrocytes labeled with Mic19–miniSOG were examined via ET. CJs in astrocyte mitochondria often show a light, open region near their connection to the IBM where the two sides of the cristae membrane show Mic19 labeling, but the intracristal space in between the two membrane sides at the junction is devoid of the label indicating low Mic19 occupancy in this small, localized region and suggesting another protein occupying this Mic-19 excluded space (Fig. 5A). As seen, the other portions of these two cristae are filled with label. Again, the finding that Mic19 lines the cristae membranes facing the intracristal space was observed (Fig. 5A,B). On occasion, the cristae in astrocytes were highly networked, which was discovered only after cristae segmentation. For example, in the particular mitochondrion shown (Fig. 5C,D), there was one highly networked crista that had nine branched segments (arrow) and only two other much smaller, un-networked cristae (Fig. 5C,D, asterisks). Nevertheless, as with HL-1 cells, the segmented and surface-rendered astrocyte volume confirmed that the Mic19-positive domains found in the cristae extended throughout the volume, but did not completely fill the intracristal space and were in one discrete shape rather than speckled throughout (Fig. 5E–G). As with HL-1 cells, no pattern could be detected with the Mic19-positive shapes nor in how they were distributed inside the cristae. In contrast to HL-1 cells, Mic19-positive domains did not have a patchy pattern in the IMS, but rather were distributed in a network pattern (Fig. 5H; Movie 1). This network occupied significantly more of the IMS volume than in HL-1 cells (13±3% astrocytes versus 4±1% HL-1 cells, two-tailed t-test, P=0.013). As before, all the CJs were positive for Mic19, and the CJ architecture was not altered upon tagging Mic19 with APEX2 (Fig. 5I,J). As with HL-1 cells, measurements on how the label was distributed between CJ, IMS and cristae using ten human primary astrocyte mitochondrial tomographic volumes indicated that the majority of the Mic19-positive domains were in the cristae (62±4%, two-tailed t-test compared with HL-1 cells, P=0.26) with lesser amounts in the IMS (28±4%, two-tailed t-test compared with HL-1 cells, P=0.12) and CJs (10±3%, two-tailed t-test compared with HL-1 cells, P=0.72) (Fig. 5K). As with HL-1 cells, as far as could be determined, all the CJs were labeled in human primary astrocytes. As before, most of the CJs were labeled (78±6%, mean±s.e.m.), but only part of the IMS (13±3%, two-tailed t-test compared with HL-1 cells, P=0.013) and cristae (45±6%, two-tailed t-test compared with HL-1 cells, P=0.019) volumes were filled with Mic19-positive domains (Fig. 5L), yet both IMS and intracristal space were more substantially filled with these domains in the astrocytes compared with the HL-1 cells. In summary, the 3D labeling patterns of Mic19 were similar in HL-1 and human primary astrocytes, including the cristae distribution. The only two differences were: (1) the networked Mic19-positive domain in astrocytes in contrast to the patchy pattern in HL-1 cells and (2) the greater filling of IMS and cristae with Mic19 in the astrocytes.
Fig. 5.
ET volumes of Mic19-miniSOG label similar domains in human primary astrocytes, including networks in the IMS. (A) A 1-nm slice through the center of a tomographic volume from an astrocyte showing Mic19-positive domains at CJs, IMS and also substantially in the intracristal space. CJs often showed a light open region (arrowheads) indicating low Mic19 occupancy in this small localized region. Results are representative of n=9 cells and 60 mitochondria. (B) Another example of a 1-nm slice through the center of a tomographic volume from a different astrocyte that has a darkly labeled Mic19-positive CJ (arrowhead). (C) The segmented and surface-rendered volume of the mitochondrion in B (OMM green, three cristae in various colors). There was one highly networked crista (arrow), and two much smaller and non-networked cristae (*). (D) As in C, but with a side orientation showing that the networked crista was far larger than the only two other cristae present. (E) A substantial portion of the cristae (translucent blue), typical of the 60 mitochondrial volumes examined, was occupied by the Mic19-positive domains (green). (F) Side orientation showing the Mic19-positive domains extending throughout the cristae and some of the cristae segments were filled with label. (G) One of the two smaller cristae showing a typical Mic19-positive domain inside that occupied about half the crista volume. The shapes of these domains were contiguous rather than speckled throughout. (H) IMS (magenta) Mic19-positive network against the IBM background (cyan). (I) Only three CJs (cyan) were present in this volume and are shown in relation to the OMM (translucent green) and cristae (translucent blue). (J) Side view showing the CJ openings. (K) Pie chart showing the distribution of Mic19-positive domains between CJ, IMS and cristae. (L) Histogram showing the percentage of the IMS, cristae or CJ volumes occupied by Mic19-positive domains (mean±s.e.m.). Results are from three biological replicates and 10 total technical replicates (K,L).
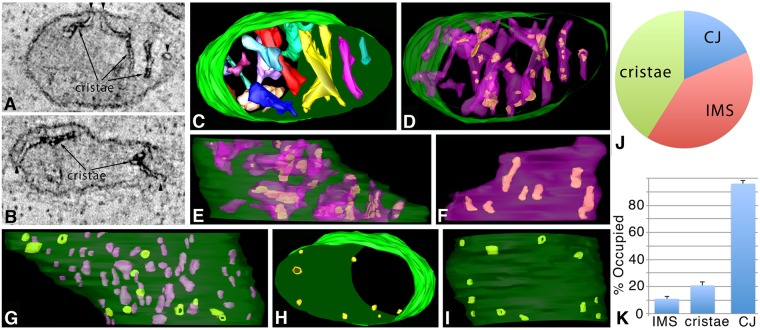
We investigated the 3D distribution of Mic60–APEX2 in relation to Mic19–miniSOG in human primary astrocytes. ET showed that Mic60-positive domains are at CJs (arrowheads), which was to be expected based on the literature (Fig. 6A,B), and also line the intracristal space, but in a variegated pattern distinct from the mostly continuous pattern found with Mic19. Mic60–APEX2 was also found in discrete stretches along the IMM. Note, that the label appears in the IMS and intracristal space because the tagged portion of the amino acid sequence extends into these spaces; however, we will continue to use ‘IBM’ or ‘IMM’ because Mic60 is inserted into the membrane. The cristae architecture was not altered by expressing Mic60–APEX2 in human primary astrocytes (Fig. 6C). As with Mic19, the Mic60-positive domains in the cristae were distributed throughout the volume (Fig. 6D,E). The novel finding here is that Mic60 has substantial presence in the cristae. However, instead of one discrete shape per crista, the Mic60-positive domains had several smaller shapes distributed throughout that did not fill the intracristal space to the same extent as Mic19–miniSOG (Fig. 6F). As before, no pattern could be detected for the Mic60-positive shapes nor in how they were distributed inside the cristae. Localized regions distributed throughout the IBM and not associated with CJs were also Mic60-positive (Fig. 6G). These regions had sizes ranging from about the size of a CJ to several times larger. The Mic60-labeled CJ architecture was found to be similar to that of the Mic19-positive CJs (Fig. 6H,I). Of the 12 CJs found in this volume, five had a clear opening and seven had their openings filled with Mic60 label. As with Mic19 cells, measurements were made on how the Mic60-positive domains were distributed between CJ, IMS and cristae using 10 human primary astrocyte mitochondrial tomographic volumes. In contrast to Mic19-positive domains in both HL-1 and astrocytes, the percent of the total labeled volume of Mic60-positive domains in the cristae was not the majority (41±3%, compare with Figs 4J and 5K; two-tailed t-test compared with Mic19 astrocytes, P=0.00046) (Fig. 6J). Although the Mic60-APEX2 labeling of the CJs was high (96±3%) in the astrocytes, there was no statistical difference in the percentage of Mic19- and Mic60-filled IMS or CJ labeling (Fig. 6K) (compare with Fig. 5L). Taken together, the cristae and IMS results suggest that there may be a closer association between Mic19 and Mic60 in the IMS than in the cristae, where Mic19 associates with Cox, as described below.
Fig. 6.
ET volumes of Mic60–APEX2 in human primary astrocytes show that it has a distinct localization pattern. (A) A 1-nm slice through the center of a tomographic volume from an astrocyte showing Mic60-positive domains at CJs (arrowheads), discrete patterns in the IMS and a patchy pattern in the intracristal space (arrows). Results are representative of n=6 cells and 39 mitochondria. (B) Another example of a 1-nm slice through the center of a tomographic volume from a different astrocyte having similar Mic60–APEX2 positive domains in the CJ, IMS and cristae. (C) Segmented and surface-rendered volume of the mitochondrion shown in A (OMM green, cristae various colors). (D) The 3D patchy pattern of Mic60-positive domains (green) inside cristae (translucent blue), typical of the 39 mitochondrial volumes examined. (E) Side orientation showing Mic60-positive domains extended throughout the cristae. (F) A crista in this volume showing typical Mic60-positive patches, with seven seen here. (G) IMS (magenta) and CJ (green) Mic60-positive domains. A total of 12 Mic60-positive CJs shown in top (H) and side (I) views in relation to the OMM (translucent green). Five had a clear opening and seven had their openings filled with Mic60 label. (J) Pie chart showing the distribution of Mic60-positive domains between CJ, IMS and cristae. (K) Histogram showing the percentage of the IMS, cristae or CJ volumes occupied by Mic60-positive domains (mean±s.e.m.). Results are from three biological replicates and 10 total technical replicates (J,K).
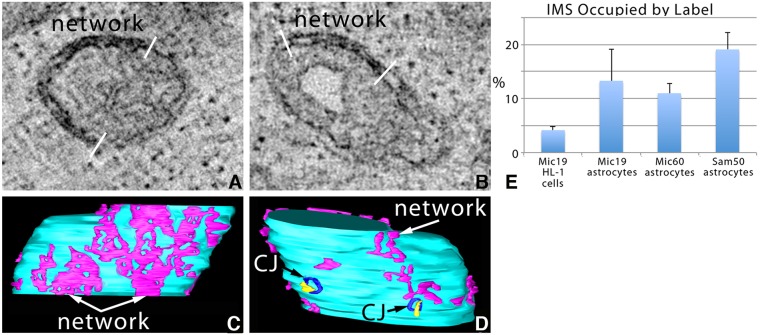
We investigated the 3D distribution of Sam50-APEX2 in relation to Mic19 and Mic60 in human primary astrocytes. The secondary structure of Sam50 indicates that it is an integral membrane protein with the APEX2-tagged c-terminus extending into the IMS. Discrete labeling was observed in the IMS (Fig. 7A,B). This labeling pattern did not extend uniformly around the IMS, though (bounded by white bars). Upon segmentation and surface rendering of the Sam50-positive domains, it was found that they formed an extensive network in the mitochondrial IMS (Fig. 7C,D). Occasionally, the Sam50-positive domain in the IMS overlapped CJs in the inner membrane that were not labeled by Sam50, yet were visible faintly due to general membrane contrast provided by osmium tetroxide (Fig. 7D). However, the shape of the Sam50-positive domains did not match the CJ shape indicating that Sam50 is not part of the protein scaffold of the CJ. Indeed, only 16% (mean, s.e.m.=7%, n=10 mitochondria) of the CJs overlapped with the Sam50 network, implying that Sam50 and Mic19 associate in the IMS at places other than CJs. Interestingly, the network-like IMS labeling of Sam50–APEX2 appeared to be similar to the Mic19–miniSOG labeling pattern. A side-by-side comparison of labeling in the IMS showed that, not surprisingly, the two proteins, Mic19 and Sam50, that formed a network in human astrocytes, had the greatest percentage of IMS volume occupied by the label (Fig. 7E).
Fig. 7.
ET volume analysis shows that Sam50–APEX2 is only found in the OMM in human primary astrocytes. (A,B) 1-nm slices through the center of tomographic volumes from two separated astrocytes showing that Sam50-positive domains did not extend uniformly around the IMS (bounded by white bars). Results are representative of n=12 cells and 31 mitochondria. (C,D) Opposite sides of the segmented and surface-rendered volume of the mitochondrion shown in A with the IMS (magenta) Sam50-positive network against the background of the IBM (cyan). In D, 2 CJs (blue) overlap with Sam50-positive domains (yellow) in the IMS, but do not have the same shape. (E) Side-by-side comparison of the percent of the IMS volume occupied by the label for all samples examined (mean±s.e.m., n=10 mitochondria from three biological replicates for each population).
Mic19 associates with CoxIV
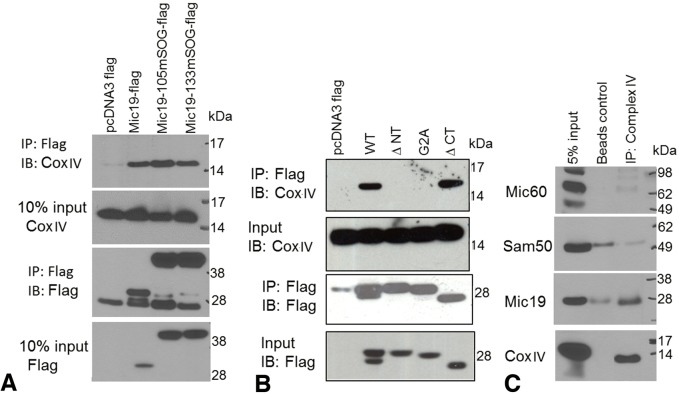
Guided by the 3D localization results in two distinct cell lines showing a strong presence of Mic19 in cristae, and based on our previous findings that knockdown of Mic19 results in a significant reduction of oxidative phosphorylation and protein levels of CoxIV, we investigated whether Mic19 associates with Cox. It is known that Cox is heavily enriched in the cristae membranes and that little is present in the IBM (Gilkerson et al., 2003; Vogel et al., 2006; Wurm and Jakobs, 2006). We used transiently expressed C-terminal Flag-tagged mutants of Mic19 that lack the N-terminal 14 amino acids (ΔNT) or the CHCH domain (ΔCT) in HEK 293 cells and immunoprecipitated using anti-Flag resin. We analyzed the immunoprecipitated samples for CoxIV and observed that, while the full-length Mic19 and the ΔCT can bind efficiently to CoxIV, deletion of the N-terminal 14 amino acids (ΔNT) or blocking the N-terminal myristoylation motif (G2A) disrupts this binding (Fig. 8A,B), suggesting that Mic19 binds to CoxIV and that this interaction may be through the N-terminus. Previously, we had shown that this same region was essential in binding Sam50, but not Mic60, which instead binds through the CHCH domain of Mic19 (Darshi et al., 2012). Moreover, the deletion mutants ΔNT and ΔCT do not localize to mitochondria and thus are used as negative controls. To further confirm this interaction, we immunoprecipitated CoxIV from isolated mouse liver mitochondria and analyzed for the presence of Mic19. As shown in Fig. 8C, the CoxIV-immunoprecipitated sample showed CoxIV associated with Mic19, but no association with Mic60 or Sam50. This finding not only provides functional insight into the 3D localization pattern of Mic19 but also supports and extends the ET finding by suggesting a direct regulatory role of Mic19 on Cox function.
Fig. 8.
Mic19, but not Mic60 or Sam50, interacts with CoxIV, perhaps through its N-terminus. (A,B) Flag-tagged Mic19 constructs were transiently expressed in HEK 293 cells and immunoprecipitated (IP) with Flag resin. Note that the bottom two Flag blots are reproduced from panels shown in Fig. 1B for comparison. (C) CoxIV was immunoprecipitated from Histodenz-gradient-purified mouse liver mitochondria with a CoxIV immunoprecipitation (IP) kit. Protein G–agarose beads were used as a negative control. Immunoisolated samples were analyzed by SDS-PAGE followed by immunoblotting with the indicated antibodies. WT, wild-type. 5% of the input is shown. Results are representative of n=3 for all.
DISCUSSION
As proteomics is identifying mitochondrial proteins and new genetic tag technology is starting to assign these proteins to the various mitochondrial compartments (Rhee et al., 2013; Hung et al., 2014; Lee et al., 2016), there now a need to accurately map the 3D distribution of these proteins at nanoscale resolution in diverse cell types to gain insight into their functional roles and to tease out possible interacting partners. Even as we chose to label the same MICOS proteins examined previously by other groups employing immunogold cryo-electron microscopy (Harner, et al., 2011; Jans et al., 2013; Rabl et al., 2009), we chose distinct cells, cardiomyocyte and astrocyte cell lines in which to apply our new genetic tags designed for electron microscopy. HL-1 cells, derived from adult mouse atria, are an established model system that mimic cardiomyocytes and have been used to study energy metabolism processes, such as the cellular ATP production, bioenergetics, function and morphology of mitochondria (Claycomb et al., 1998; Kuznetsov et al., 2015; Ong and Hausenloy, 2010). The study of mitochondria in astrocytes is a growing endeavor to investigate the complex bidirectional relationship between astrocytes and neurons in health and disease, including the concept of transmitophagy (Davis et al., 2014; Stephen et al., 2014).
Mic19–miniSOG, Mic19–APEX2 and Mic60–APEX2 distribution in the IMS and cristae
It should be no surprise that tagged Mic19 and Mic60 show distinct patterns of distribution between the IMS and cristae because prior studies have reported an uneven distribution of several classes of integral membrane proteins between the IBM and cristae membranes (Gilkerson et al., 2003; Vogel et al., 2006; Wurm and Jakobs, 2006; Jakobs and Wurm, 2014), suggesting compartmentation of function in the IMM. It was proposed that Mic19 controls the copy number and position of CJs within mitochondria as well as directing the IMM distribution of the two MICOS subcomplexes: the Mic60–Mic19 subcomplex and the Mic27–Mic10–Mic12 subcomplex (Friedman et al., 2015). Interestingly, of all the MIB proteins, only the knockdown of Mic19, Mic60 or Sam50 affects cristae abundance and structure (Darshi et al., 2011; Ding et al., 2015; John et al., 2005; Ott et al., 2012, 2015). In addition to the predicted localization at CJs and IMS (Darshi et al., 2011; Jans et al., 2013), we found that Mic19-positive domains in the intracristal space, consistent with previous findings by Harner and co-workers (2011). Interestingly, both the IMS and intracristal space were more substantially filled with these domains in the astrocytes than in the HL-1 cells. Whereas we report that there was no difference in the percentage of the total labeled volume of tagged Mic19 or Mic60 in the IMS, there was a significant difference in the total labeled volume of Mic19 and Mic60 inside the cristae (compare Figs 4J, 5K and 6J). This finding suggests that there may be a closer association between Mic19 and Mic60 in the IMS than in the cristae, where Mic19 associates with Cox. Interestingly, it was reported that another protein with a CHCH domain like that of Mic19, CHCHD2 binds to Cox and is required for full Cox activity (Aras et al., 2015). However, unlike Mic19, CHCHD2 does not appear to be part of MICOS.
Immunogold-labeled Mic60 was reported to be associated with the IMM, being heavily enriched at the CJ (Harner et al., 2011), and additionally to be present inside the cristae (Rabl et al., 2009), consistent with our finding that Mic60–APEX2 is in the IBM, at the CJ and in the cristae membranes. However, we found more label inside the cristae than the previous reports. Recently, it was suggested that Mic60 was present inside cristae because of a physical interaction between Cox17, a protein involved in the assembly of Cox but not actually a subunit of Cox, and Mic60 (Chojnacka et al., 2015). Furthermore, Mic60 modulates the levels of ATP synthase dimers (Rabl et al., 2009), which help to shape mitochondria (Jayashankar et al., 2016). However, a direct interaction between the two was placed in question by the report that mitochondria isolated from a MIC60-null yeast strain had intact respiratory complexes and no change in ATP synthase activity (Friedman et al., 2015). Our finding of a patchy distribution of Mic19–miniSOG and Mic60–APEX2 inside cristae may be due to a likewise patchy or mosaic distribution of respiratory chain complexes after mitochondrial fusion (Muster et al., 2010; Wilkens et al., 2013) since we have shown a physical interaction of Mic19 with CoxIV, and others have shown a physical interaction of Mic60 with Cox17.
Sam50–APEX2 is in a network in the OMM
In fibroblasts, HeLa and kidney epithelial cells, Sam50 appeared to be evenly distributed around the periphery of mitochondria (Jans et al., 2013), whereas we found that in astrocytes, Sam50 was not uniformly distributed around the mitochondrial periphery and appeared to incompletely overlap with Mic19 or Mic60 domains; instead, it formed a network pattern. These findings suggest that it would be fruitful to examine more cell types to determine whether differences in Mic19, Mic60 or Sam50 labeling patterns might be consistently specific to the cell type. Even though the network pattern appeared similar between Mic19- and Sam50-positive domains in the IMS, it was found that only 16% of the CJs overlap with the Sam50-positive IMS network. In contrast, Mic19 and Mic60 strongly labeled the CJs (Mic60 more so than Mic19) (Figs 4K, 5L and 6K). Taken together, it is likely that Mic19 molecules interact closely with the portion of Sam50 extending into the IMS, yet not exclusively at the CJs.
Interestingly, Sam50, which is necessary for cristae organization and facilitates the interaction between OMM and IMM proteins, has been implicated in respiratory regulation (Ott et al., 2012). In yeast, moderate Sam50 reduction generated acutely comprised cristae morphology though with initial preservation of mitochondrial function; in contrast, long-term depletion caused a significant reduction in the amounts of complex I and Cox resulting in abrogation of function (Ott et al., 2012). Furthermore, depletion of Mic60 not only severely repressed Sam50 and Mic19 expression but also the activity of Cox (Ott et al., 2012). Even though there is strong evidence in the literature that Mic19, Mic60 and Sam50 are interacting partners, the distinct labeling pattern of each suggests that they can exist naturally outside of a stable MICOS scaffold. Further high-resolution EM with double-labeling of pairs of Mic19, Mic60 and Sam50 is warranted to determine the extent of colocalization.
Mic19 associates with Cox IV
We found that Mic19 associates with CoxIV and that this interaction may be mediated by the N-terminus and myristoylation motif of Mic19. Previously, we showed that Mic19 is a substrate for PKA (Schauble et al., 2007). With external stimuli, mitochondrial function is modified by reversible phosphorylation/dephosphorylation of the supercomplexes (Acin-Perez and Enriquez, 2014; Chaban et al., 2014; Cruciat et al., 2000; Hüttemann et al., 2007; Kadenbach and Hüttemann, 2015). CoxIV has been shown to be phosphorylated by PKA and this phosphorylation is essential for yeast mitochondria to toggle between the ‘low ATP consumption mode’ and the ‘high ATP consumption mode’ (Acin-Perez et al., 2011). Furthermore, in heart mitochondria, it has been found that unphosphorylated Cox associates with the ‘respirasome’, while upon phosphorylation by PKA, it was present as a free complex (Rosca et al., 2011) indicating that phosphorylation prevents respirasome association. Mic19 also interacts with sphingosine kinase interacting protein (SKIP; also known as SNW1) and this interaction was essential for phosphorylation of Mic19 by PKA (Means et al., 2011). SKIP was initially identified as a negative regulator of sphingosine kinase 1 and compelling evidence suggests that sphingosine kinases are important in the assembly and function of Cox (Strub et al., 2011). These reports suggest that Cox is in close proximity to Mic19 and its interacting proteins.
Previously, we observed a significant reduction in CoxIV and no changes in complexes III and V upon Mic19 knockdown (Darshi et al., 2011). Mutations in Cox have been linked to a number of neurological diseases including Alzheimer's disease (AD) and Parkinson's disease (PD). In AD, the activity of Cox is attenuated and is an early indicator of the disease (Kish et al., 1992; Arnold, 2012). In PD, Cox causes neurotoxicity of the substantia nigra (Arnold, 2012; Itoh et al., 1996). Although Mic19 has not been studied particularly in PD, a genome-wide study of copy number variants in the PD genome and ingenuity pathway analysis has shown that Mic19 was present in the same network with other PD genes (Liu et al., 2013). However, it was recently found that Mic19 and Mic60 are involved with Parkin recruitment to mitochondria and this Parkin translocation is controlled by PKA-mediated phosphorylation (Akabane et al., 2016), suggesting that these proteins have a role in the protection of mitochondria with low membrane potential from selective autophagy in neurons. Finally, it was recently reported that Mic19 is part of the Hippo pathway in Drosophila development (Deng et al., 2016) promoting mitochondrial fusion and biogenesis by activating mitochondrial genes. In the bigger picture, this pathway is a signaling cascade that controls tissue growth and apoptosis.
Conclusion and perspectives
By coupling miniSOG and APEX2 with ET and EFTEM, expanding tools for dissecting the protein landscape useful for providing clues on interacting partners and their functions at high spatial resolution, we discovered two unforeseen aspects of Mic19, Mic60 and Sam50. One, even though Mic19, Mic60 and Sam50 are interacting partners, and MICOS has a stable stoichiometry (Bohnert et al., 2015), they also exist separately displaying distinct patterns inside mitochondria. Two, Mic19 has a physical association with Cox subunit IV.
From a larger perspective, the study of the interaction of Mic19, Mic60 and Sam50 is important because disruption of MIB function produces distinct disease phenotypes (Genin et al., 2016; Guarani et al., 2016; Zeharia et al., 2016). For example, defective mitochondrial structure–function relationships are implicated in several human diseases including neurodegenerative diseases, aging, metabolic disorders and cancer (Jayashankar et al., 2016). The mechanisms by which the MIB couples with mitochondrial architecture to contribute to the pathology of these diseases are of high interest and constitute an active area of mitochondrial research. The way in which the MIB components interact, which we now know occurs in every mitochondrial compartment, and leads to proper CJ and cristae organization, remains to be discovered. Exploring the nature of their regulation will be crucial for a molecular understanding of how IMM architecture is generated and remodeled in space and time in response to the functional needs of the cell.
MATERIALS AND METHODS
Antibodies and plasmids
The cDNA clones for Mic19–Flag, Mic60–Flag and Sam50–Flag were described previously (Darshi et al., 2011). MiniSOG and APEX2 in PCDNA3 were generous gifts from the laboratory of Roger Tsien (Chemistry and Biochemistry Department, University of California, San Diego, USA). To generate Mic19-105–miniSOG and Mic19-133–miniSOG, EcoR1 and BamH1 restriction sites were introduced at these positions by site-directed mutagenesis. The circular polymerase extension cloning (CPEC) method (Quan and Tian, 2011) was used to sub-clone miniSOG with a (GlySer)4 linker at the 5′ and 3′ ends into Mic19–Flag at positions 105 or 133. Similarly, miniSOG was introduced between Mic60 and Flag, or Sam50 and Flag to generate the Mic60–miniSOG–Flag and Sam50–miniSOG–Flag constructs, respectively, using CPEC. Mic60–APEX2 and Sam50–APEX2 with linkers of different lengths described in the results section were generated by using the CPEC method by sub-cloning Mic60 and Sam50 into APEX2–Flag at the N-terminus of the PCDNA3 APEX2. The following antibodies were used in this study: the rabbit polyclonal antibody for Mic19, as described previously (Darshi et al., 2012), anti-mitofilin/Mic60 (Abcam ab137057, Cambridge, MA), anti-Sam50 (Santa Cruz Biotechnology, sc-100493, Dallas, TX), anti-OPA1 (BD Biosciences 612606, San Jose, CA), anti-Flag (Sigma-Aldrich F7425, St. Louis, MO), anti-CoxIV (Abcam 33985), Cy5-labeled donkey anti-rabbit IgG antibodies (Jackson ImmunoResearch Laboratories 711-175-152, West Grove, PA), and DyLight 649 (Jackson Laboratories 011-490-003). All antibodies were used at 1:1000 for western blot analysis and at 1:200 for immunoprecipitation and imaging experiments.
Growth and transfections of HL-1 and human primary astrocytes
HL-1 cells
The HL-1 cardiac immortalized cell line (Claycomb et al., 1998) was kindly provided by William Claycomb and confirmed free from contamination. Cells were grown in Claycomb medium (JRH Biosciences, Lenexa, KS). For the imaging studies, the cells were plated on 35 mm-glass bottom MatTek dishes (MatTek, Ashland, MA) at 20,000 cells/well and incubated at 37°C in 5% CO2 for 12–16 h. Transfections were performed using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA) and processed for light microscopy or for ET.
Human primary astrocytes
Normal human astrocytes were purchased from Sciencell Research Laboratories, Carlsbad, CA (Müller et al., 2012) and plated in MatTek EM dishes at 200,000 cells/well, incubated at 37°C in 5% CO2 for 1–2 days prior to plasmid transfection using Lipofectamine 3000 (Thermo Fisher) and confirmed to be contamination free and processed for light microscopy or ET.
Immunoprecipitation and immunofluorescence
Immunoprecipitation with Mic19–miniSOG–Flag in HEK 293 cells were performed as described in Darshi et al. (2011). Cox was immunoprecipitated from purified mouse liver mitochondria (5 mg) using a Cox rodent immunocapture kit (Abcam). For immunofluorescence, the miniSOG–Flag- or APEX-2–Flag-tagged DNA-transfected cells were imaged as described previously (Darshi et al., 2011).
MiniSOG, APEX2, ET and EFTEM
MiniSOG- or APEX2-transfected HL-1 or human primary astrocyte cells were prepared for ET (Perkins, 2014) or EFTEM (Adams et al., 2016) as described previously, including using mersalyl acid to reduce background nonspecific labeling of mitochondria by poisoning the electron transport chain to block its singlet oxygen production. For ET, sections of thickness 300–400 nm were coated with 15 nm colloidal gold particles for alignment of the tilt series. Tilt images were captured using a Tecnai Titan TEM (FEI) operated at 300 kV equipped with SerialEM software (University of Colorado, Boulder, CO) as described by Lawrence et al. (2006). To provide adequate sampling, a large number of mitochondrial tomographic volumes were analyzed from three plates for each sample type. Transfected cells were easily distinguished from untransfected cells because of the dark DAB-labeling inside mitochondria. Segmentations, surface-renderings and movies of miniSOG or APEX2-positive domains inside mitochondria were made using IMOD (University of Colorado). Imodinfo was used to calculate the percent of mitochondrial compartments occupied by labeled Mic19, Mic60 or Sam50. Paired Student's t-tests were calculated to determine statistical significance, P<0.05 used for significant difference.
Supplementary Material
Acknowledgements
In memory of our friend and colleague Roger Tsien, Nobel Prize winner and co-developer with Mark Ellisman of the miniSOG technology.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.E., S.T., G.P.; Methodology: M.D., M.M., G.P.; Software: S.P.; Validation: M.D., G.P.; Formal analysis: M.S., M.D., G.P.; Investigation: M.S., M.D., M.M., R.R., S.J., K.S., C.D., J.J.K., G.P.; Resources: S.A., M.E., S.T.; Data curation: M.D., G.P.; Writing - original draft: G.P.; Writing - review & editing: M.S., M.D., M.M., R.R., M.E., S.T.; Visualization: S.J., G.P.; Supervision: M.S., M.D., M.M., M.E., S.T., G.P.; Project administration: M.E., S.T., G.P.; Funding acquisition: M.E., S.T.
Funding
This work was supported by the National Institutes of Health (P41GM103412 and GM086197 to M.H.E., DK054441 to S.S.T.) and Larry L. Hillblom Foundation grant 2014-D-005-NET to S.S.T. Deposited in PMC for release after 12 months.
Data availability
All ET datasets have been deposited in the Cell-Centered Database (Martone et al., 2003). The accession numbers are: 20276 for Mic19-miniSOG, 20343 for Sam50-APEX2 and 20336 for Mic600-APEX2.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.201400.supplemental
References
- Acin-Perez R., Gatti D. L., Bai Y. and Manfredi G. (2011). Protein phosphorylation and prevention of cytochrome oxidase inhibition by ATP: coupled mechanisms of energy metabolism regulation. Cell Metab. 13, 712-719. 10.1016/j.cmet.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams S. R., Mackey M. R., Ramachandra R., Palida Lemieux S. F., Steinbach P., Bushong E. A., Butko M. T., Giepmans B. N. G., Ellisman M. H. and Tsien R. Y. (2016). Multicolor electron microscopy for simultaneous visualization of multiple molecular species. Cell Chem. Biol. 23, 1417-1427. 10.1016/j.chembiol.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn C. C. and Krivanek O. L. (1983). EELS Atlas: A Reference Collection of Electron Energy Loss Spectra Covering All Stable Elements. Gatan, Pleasanton, CA. [Google Scholar]
- Akabane S., Uno M., Tani N., Shimazaki S., Ebara N., Kato H., Kosako H. and Oka T. (2016). PKA regulates PINK1 stability and Parkin recruitment to damaged mitochondria through phosphorylation of MIC60. Mol. Cell 62, 371-384. 10.1016/j.molcel.2016.03.037 [DOI] [PubMed] [Google Scholar]
- Alkhaja A. K., Jans D. C., Nikolov M., Vukotic M., Lytovchenko O., Ludewig F., Schliebs W., Riedel D., Urlaub H., Jakobs S. et al. (2012). MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol. Biol. Cell 23, 247-257. 10.1091/mbc.E11-09-0774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R., Strecker V., Urbach J., Wittig I. and Reichert A. S. (2016). Mic13 is essential for formation of crista junctions in mammalian cells. PLoS ONE 11, e0160258 10.1371/journal.pone.0160258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aras S., Bai M., Lee I., Springett R., Hüttemann M. and Grossman L. I. (2015). MNRR1 (formerly CHCHD2) is a bi-organellar regulator of mitochondrial metabolism. Mitochondrion 20, 43-51. 10.1016/j.mito.2014.10.003 [DOI] [PubMed] [Google Scholar]
- Arnold S. (2012). Cytochrome c oxidase and its role in neurodegeneration and neuroprotection. Adv. Exp. Med. Biol. 748, 305-339. 10.1007/978-1-4614-3573-0_13 [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gallo A., Martinelli M., Sideris D. P., Katrakili N. and Tokatlidis K. (2009). MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 16, 198-206. 10.1038/nsmb.1553 [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Ciofi-Baffoni S., Jaiswal D., Neri S., Peruzzini R. and Winkelmann J. (2012). Structural characterization of CHCHD5 and CHCHD7: two atypical human twin CX9C proteins. J. Struct. Biol. 180, 190-200. 10.1016/j.jsb.2012.07.007 [DOI] [PubMed] [Google Scholar]
- Barbot M., Jans D. C., Schulz C., Denkert N., Kroppen B., Hoppert M., Jakobs S. and Meinecke M. (2015). Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab. 21, 756-763. 10.1016/j.cmet.2015.04.006 [DOI] [PubMed] [Google Scholar]
- Bohnert M., Wenz L.-S., Zerbes R. M., Horvath S. E., Stroud D. A., von der Malsburg K., Müller J. M., Oeljeklaus S., Perschil I., Warscheid B. et al. (2012). Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane. Mol. Biol. Cell 23, 3948-3956. 10.1091/mbc.E12-04-0295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert M., Zerbes R. M., Davies K. M., Mühleip A. W., Rampelt H., Horvath S. E., Boenke T., Kram A., Perschil I., Veenhuis M. et al. (2015). Central role of Mic10 in the mitochondrial contact site and cristae organizing system. Cell Metab. 21, 747-755. 10.1016/j.cmet.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Chaban Y., Boekema E. J. and Dudkina N. V. (2014). Structures of mitochondrial oxidative phosphorylation supercomplexes and mechanisms for their stabilisation. Biochim. Biophys. Acta 1837, 418-426. 10.1016/j.bbabio.2013.10.004 [DOI] [PubMed] [Google Scholar]
- Chojnacka M., Gornicka A., Oeljeklaus S., Warscheid B. and Chacinska A. (2015). Cox17 protein is an auxiliary factor involved in the control of the mitochondrial contact site and cristae organizing system. J. Biol. Chem. 290, 15304-15312. 10.1074/jbc.M115.645069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb W. C., Lanson N. A., Stallworth B. S., Egeland D. B., Delcarpio J. B., Bahinski A. and Izzo N. J. (1998). HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA 95, 2979-2984. 10.1073/pnas.95.6.2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S., Enriquez J. A. and Scorrano L. (2016). Mitochondrial cristae: where beauty meets functionality. Trends Biochem. Sci. 41, 261-273. 10.1016/j.tibs.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Cruciat C.-M., Brunner S., Baumann F., Neupert W. and Stuart R. A. (2000). The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 275, 18093-18098. 10.1074/jbc.M001901200 [DOI] [PubMed] [Google Scholar]
- Darshi M., Mendiola V. L., Mackey M. R., Murphy A. N., Koller A., Perkins G. A., Ellisman M. H. and Taylor S. S. (2011). ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J. Biol. Chem. 286, 2918-2932. 10.1074/jbc.M110.171975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshi M., Trinh K. N., Murphy A. N. and Taylor S. S. (2012). Targeting and import mechanism of coiled-coil helix coiled-coil helix domain-containing protein 3 (ChChd3) into the mitochondrial intermembrane space. J. Biol. Chem. 287, 39480-39491. 10.1074/jbc.M112.387696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C.-O., Kim K.-Y., Bushong E. A., Mills E. A., Boassa D., Shih T., Kinebuchi M., Phan S., Zhou Y., Bihlmeyer N. A. et al. (2014). Transcellular degradation of axonal mitochondria. Proc. Natl. Acad. Sci. USA 111, 9633-9638. 10.1073/pnas.1404651111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q., Guo T., Zhou X., Xi Y., Yang X. and Ge W. (2016). Cross-talk between mitochondrial fusion and the hippo pathway in controlling cell proliferation during Drosophila development. Genetics 203, 1777-1788. 10.1534/genetics.115.186445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., Wu Z., Huang L., Wang Y., Xue J., Chen S., Deng Z., Wang L., Song Z. and Chen S. (2015). Mitofilin and CHCHD6 physically interact with Sam50 to sustain cristae structure. Sci. Rep. 5, 16064 10.1038/srep16064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina N. V., Kouřil R., Bultema J. B. and Boekema E. J. (2010). Imaging of organelles by electron microscopy reveals protein-protein interactions in mitochondria and chloroplasts. FEBS Lett. 584, 2510-2515. 10.1016/j.febslet.2010.03.027 [DOI] [PubMed] [Google Scholar]
- Frey T. G., Renken C. W. and Perkins G. A. (2002). Insight into mitochondrial structure and function from electron tomography. Biochim. Biophys. Acta 1555, 196-203. 10.1016/S0005-2728(02)00278-5 [DOI] [PubMed] [Google Scholar]
- Friedman J. R., Mourier A., Yamada J., McCaffery J. M. and Nunnari J. (2015). MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. Elife 4, e07739 10.7554/eLife.07739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin E. C., Plutino M., Bannwarth S., Villa E., Cisneros-Barroso E., Roy M., Ortega-Vila B., Fragaki K., Lespinasse F., Pinero-Martos E. et al. (2016). CHCHD10 mutations promote loss of mitochondrial cristae junctions with impaired mitochondrial genome maintenance and inhibition of apoptosis. EMBO Mol. Med. 8, 58-72. 10.15252/emmm.201505496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson R. W., Selker J. M. L. and Capaldi R. A. (2003). The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 546, 355-358. 10.1016/S0014-5793(03)00633-1 [DOI] [PubMed] [Google Scholar]
- Guarani V., McNeill E. M., Paulo J. A., Huttlin E. L., Fröhlich F., Gygi S. P., Van Vactor D. and Harper J. W. (2015). QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. Elife 4, e06265 10.7554/eLife.06265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarani V., Jardel C., Chrétien D., Lombès A., Bénit P., Labasse C., Lacène E., Bourillon A., Imbard A., Benoist J. F. et al. (2016). QIL1 mutation causes MICOS disassembly and early onset fatal mitochondrial encephalopathy with liver disease. Elife 5, e17163 10.7554/eLife.17163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M., Körner C., Walther D., Mokranjac D., Kaesmacher J., Welsch U., Griffith J., Mann M., Reggiori F. and Neupert W. (2011). The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30, 4356-4370. 10.1038/emboj.2011.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Collins S. R., Cassidy-Stone A., Hummel E., Devay R. M., Lackner L. L., Westermann B., Schuldiner M., Weissman J. S. and Nunnari J. (2011). A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195, 323-340. 10.1083/jcb.201107053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V., Zou P., Rhee H.-W., Udeshi N. D., Cracan V., Svinkina T., Carr S. A., Mootha V. K. and Ting A. Y. (2014). Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell 55, 332-341. 10.1016/j.molcel.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttemann M., Lee I., Samavati L., Yu H. and Doan J. W. (2007). Regulation of mitochondrial oxidative phosphorylation through cell signaling. Biochim. Biophys. Acta 1773, 1701-1720. 10.1016/j.bbamcr.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Huynen M. A., Mühlmeister M., Gotthardt K., Guerrero-Castillo S. and Brandt U. (2016). Evolution and structural organization of the mitochondrial contact site (MICOS) complex and the mitochondrial intermembrane space bridging (MIB) complex. Biochim. Biophys. Acta 1863, 91-101. 10.1016/j.bbamcr.2015.10.009 [DOI] [PubMed] [Google Scholar]
- Itoh K., Weis S., Mehraein P. and Müller-Höcker J. (1996). Cytochrome c oxidase defects of the human substantia nigra in normal aging. Neurobiol. Aging 17, 843-848. 10.1016/S0197-4580(96)00168-6 [DOI] [PubMed] [Google Scholar]
- Jakobs S. and Wurm C. A. (2014). Super-resolution microscopy of mitochondria. Curr. Opin. Chem. Biol. 20, 9-15. 10.1016/j.cbpa.2014.03.019 [DOI] [PubMed] [Google Scholar]
- Jans D. C., Wurm C. A., Riedel D., Wenzel D., Stagge F., Deckers M., Rehling P. and Jakobs S. (2013). STED super-resolution microscopy reveals an array of MINOS clusters along human mitochondria. Proc. Natl. Acad. Sci. USA 110, 8936-8941. 10.1073/pnas.1301820110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayashankar V., Mueller I. A. and Rafelski S. M. (2016). Shaping the multi-scale architecture of mitochondria. Curr. Opin. Cell Biol. 38, 45-51. 10.1016/j.ceb.2016.02.006 [DOI] [PubMed] [Google Scholar]
- John G. B., Shang Y., Li L., Renken C., Mannella C. A., Selker J. M. L., Rangell L., Bennett M. J. and Zha J. (2005). The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell 16, 1543-1554. 10.1091/mbc.E04-08-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadenbach B. and Hüttemann M. (2015). The subunit composition and function of mammalian cytochrome c oxidase. Mitochondrion 24, 64-76. 10.1016/j.mito.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Kish S. J., Bergeron C., Rajput A., Dozic S., Mastrogiacomo F., Chang L.-J., Wilson J. M., DiStefano L. M. and Nobrega J. N. (1992). Brain cytochrome oxidase in Alzheimer's disease. J. Neurochem. 59, 776-779. 10.1111/j.1471-4159.1992.tb09439.x [DOI] [PubMed] [Google Scholar]
- Koob S. and Reichert A. S. (2014). Novel intracellular functions of apolipoproteins: the ApoO protein family as constituents of the Mitofilin/MINOS complex determines cristae morphology in mitochondria. Biol. Chem. 395, 285-296. 10.1515/hsz-2013-0274 [DOI] [PubMed] [Google Scholar]
- Koob S., Barrera M., Anand R. and Reichert A. S. (2015). The non-glycosylated isoform of MIC26 is a constituent of the mammalian MICOS complex and promotes formation of crista junctions. Biochim. Biophys. Acta 1853, 1551-1563. 10.1016/j.bbamcr.2015.03.004 [DOI] [PubMed] [Google Scholar]
- Körner C., Barrera M., Dukanovic J., Eydt K., Harner M., Rabl R., Vogel F., Rapaport D., Neupert W. and Reichert A. S. (2012). The C-terminal domain of Fcj1 is required for formation of crista junctions and interacts with the TOB/SAM complex in mitochondria. Mol. Biol. Cell 23, 2143-2155. 10.1091/mbc.E11-10-0831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozjak-Pavlovic V. (2017). The MICOS complex of human mitochondria. Cell Tissue Res. 367, 83-93. 10.1007/s00441-016-2433-7 [DOI] [PubMed] [Google Scholar]
- Kuznetsov A. V., Javadov S., Sickinger S., Frotschnig S. and Grimm M. (2015). H9c2 and HL-1 cells demonstrate distinct features of energy metabolism, mitochondrial function and sensitivity to hypoxia-reoxygenation. Biochim. Biophys. Acta 1853, 276-284. 10.1016/j.bbamcr.2014.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S. S., Martell J. D., Kamer K. J., Deerinck T. J., Ellisman M. H., Mootha V. K. and Ting A. Y. (2015). Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51-54. 10.1038/nmeth.3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte S. L., Forsyth C. M., Cunningham B. C., Miercke L. J., Akhavan D. and Stroud R. M. (2005). De novo design of an IL-4 antagonist and its structure at 1.9 A. Proc. Natl. Acad. Sci. USA 102, 1889-1894. 10.1073/pnas.0408890102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence A., Bouwer J. C., Perkins G. and Ellisman M. H. (2006). Transform-based backprojection for volume reconstruction of large format electron microscope tilt series. J. Struct. Biol. 154, 144-167. 10.1016/j.jsb.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Lee S.-Y., Kang M.-G., Park J.-S., Lee G., Ting A. Y. and Rhee H.-W. (2016). APEX fingerprinting reveals the subcellular localization of proteins of interest. Cell Rep. 15, 1837-1847. 10.1016/j.celrep.2016.04.064 [DOI] [PubMed] [Google Scholar]
- Li H., Ruan Y., Zhang K., Jian F., Hu C., Miao L., Gong L., Sun L., Zhang X., Chen S. et al. (2016). Mic60/Mitofilin determines MICOS assembly essential for mitochondrial dynamics and mtDNA nucleoid organization. Cell Death Differ. 23, 380-392. 10.1038/cdd.2015.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Cheng R., Ye X., Verbitsky M., Kisselev S., Mejia-Santana H., Louis E. D., Cote L. J., Andrews H. F., Waters C. H. et al. (2013). Increased rate of sporadic and recurrent rare genic copy number variants in Parkinson's disease among Ashkenazi Jews. Mol. Genet. Genomic Med. 1, 142-154. 10.1002/mgg3.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannella C. A. (2006). Structure and dynamics of the mitochondrial inner membrane cristae. Biochim. Biophys. Acta 1763, 542-548. 10.1016/j.bbamcr.2006.04.006 [DOI] [PubMed] [Google Scholar]
- Martell J. D., Deerinck T. J., Sancak Y., Poulos T. L., Mootha V. K., Sosinsky G. E., Ellisman M. H. and Ting A. Y. (2012). Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 30, 1143-1148. 10.1038/nbt.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone M. E., Zhang S., Gupta A., Qian X., He H., Price D. L., Wong M., Santini S. and Ellisman M. H. (2003). The cell-centered database: a database for multiscale structural and protein localization data from light and electron microscopy. Neuroinformatics 1, 379-396. 10.1385/NI:1:4:379 [DOI] [PubMed] [Google Scholar]
- Means C. K., Lygren B., Langeberg L. K., Jain A., Dixon R. E., Vega A. L., Gold M. G., Petrosyan S., Taylor S. S., Murphy A. N. et al. (2011). An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc. Natl. Acad. Sci. USA 108, E1227-E1235. 10.1073/pnas.1107182108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi C., Aschman N., Cunha M., Oikawa T., Sorzano C. O. S. and Marco S. (2013). Three-dimensional chemical mapping by EFTEM-TomoJ including improvement of SNR by PCA and ART reconstruction of volume by noise suppression. Microsc. Microanal. 19, 1669-1677. 10.1017/S1431927613013317 [DOI] [PubMed] [Google Scholar]
- Muller F. L., Colla S., Aquilanti E., Manzo V. E., Genovese G., Lee J., Eisenson D., Narurkar R., Deng P., Nezi L. et al. (2012). Passenger deletions generate therapeutic vulnerabilities in cancer. Nature 488, 337-342. 10.1038/nature11331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun J. Y., Lee T. H., Kim J. H., Yoo B. H., Bahk Y. Y., Koo H.-S. and Han S. S. (2010). Caenorhabditis elegans mitofilin homologs control the morphology of mitochondrial cristae and influence reproduction and physiology. J. Cell. Physiol. 224, 748-756. 10.1002/jcp.22177 [DOI] [PubMed] [Google Scholar]
- Muster B., Kohl W., Wittig I., Strecker V., Joos F., Haase W., Bereiter-Hahn J. and Busch K. (2010). Respiratory chain complexes in dynamic mitochondria display a patchy distribution in life cells. PLoS ONE 5, e11910 10.1371/journal.pone.0011910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odgren P. R., Toukatly G., Bangs P. L., Gilmore R. and Fey E. G. (1996). Molecular characterization of mitofilin (HMP), a mitochondria-associated protein with predicted coiled coil and intermembrane space targeting domains. J. Cell Sci. 109, 2253-2264. [DOI] [PubMed] [Google Scholar]
- Ong S.-B. and Hausenloy D. J. (2010). Mitochondrial morphology and cardiovascular disease. Cardiovasc. Res. 88, 16-29. 10.1093/cvr/cvq237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott C., Ross K., Straub S., Thiede B., Götz M., Goosmann C., Krischke M., Mueller M. J., Krohne G., Rudel T. et al. (2012). Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell. Biol. 32, 1173-1188. 10.1128/MCB.06388-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott C., Dorsch E., Fraunholz M., Straub S. and Kozjak-Pavlovic V. (2015). Detailed analysis of the human mitochondrial contact site complex indicate a hierarchy of subunits. PLoS ONE 10, e0120213 10.1371/journal.pone.0120213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G. A. (2014). The use of miniSOG in the localization of mitochondrial proteins. Meth. Enzymol. 547, 165-179. 10.1016/B978-0-12-801415-8.00010-2 [DOI] [PubMed] [Google Scholar]
- Perkins G. and Ellisman M. (2007). Mitochondrial architecture and heterogeneity. In Neural Energy Utilization (Eds, A. Lajtha, G. E. Gibson, G. A. Dienel) pp. 262-295. Springer. [Google Scholar]
- Perkins G., Renken C., Martone M. E., Young S. J., Ellisman M. and Frey T. (1997). Electron tomography of neuronal mitochondria: three-dimensional structure and organization of cristae and membrane contacts. J. Struct. Biol. 119, 260-272. 10.1006/jsbi.1997.3885 [DOI] [PubMed] [Google Scholar]
- Perkins G. A., Song J. Y., Tarsa L., Deerinck T. J., Ellisman M. H. and Frey T. G. (1998). Electron tomography of mitochondria from brown adipocytes reveals crista junctions. J. Bioenerg. Biomembr. 30, 431-442. 10.1023/A:1020586012561 [DOI] [PubMed] [Google Scholar]
- Perkins G. A., Renken C. W., Frey T. G. and Ellisman M. H. (2001). Membrane architecture of mitochondria in neurons of the central nervous system. J. Neurosci. Res. 66, 857-865. 10.1002/jnr.10050 [DOI] [PubMed] [Google Scholar]
- Pfanner N., van der Laan M., Amati P., Capaldi R. A., Caudy A. A., Chacinska A., Darshi M., Deckers M., Hoppins S., Icho T. et al. (2014). Uniform nomenclature for the mitochondrial contact site and cristae organizing system. J. Cell Biol. 204, 1083-1086. 10.1083/jcb.201401006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero-Martos E., Ortega-Vila B., Pol-Fuster J., Cisneros-Barroso E., Ruiz-Guerra L., Medina-Dols A., Heine-Suñer D., Lladó J., Olmos G. and Vives-Bauzà C. (2016). Disrupted in schizophrenia 1 (DISC1) is a constituent of the mammalian mitochondrial contact site and cristae organizing system (MICOS) complex, and is essential for oxidative phosphorylation. Hum. Mol. Genet. 25, 4157-4169. 10.1093/hmg/ddw250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J. and Tian J. (2011). Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 6, 242-251. 10.1038/nprot.2010.181 [DOI] [PubMed] [Google Scholar]
- Rabl R., Soubannier V., Scholz R., Vogel F., Mendl N., Vasiljev-Neumeyer A., Körner C., Jagasia R., Keil T., Baumeister W. et al. (2009). Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell Biol. 185, 1047-1063. 10.1083/jcb.200811099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandra R., Bouwer J. C., Mackey M. R., Bushong E., Peltier S. T., Xuong N.-H. and Ellisman M. H. (2014). Improving signal to noise in labeled biological specimens using energy-filtered TEM of sections with a drift correction strategy and a direct detection device. Microsc. Microanal. 20, 706-714. 10.1017/S1431927614000452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H.-W., Zou P., Udeshi N. D., Martell J. D., Mootha V. K., Carr S. A. and Ting A. Y. (2013). Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328-1331. 10.1126/science.1230593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosca M., Minkler P. and Hoppel C. L. (2011). Cardiac mitochondria in heart failure: normal cardiolipin profile and increased threonine phosphorylation of complex IV. Biochim. Biophys. Acta 1807, 1373-1382. 10.1016/j.bbabio.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Schauble S., King C. C., Darshi M., Koller A., Shah K. and Taylor S. S. (2007). Identification of ChChd3 as a novel substrate of the cAMP-dependent protein kinase (PKA) using an analog-sensitive catalytic subunit. J. Biol. Chem. 282, 14952-14959. 10.1074/jbc.M609221200 [DOI] [PubMed] [Google Scholar]
- Shu X., Lev-Ram V., Deerinck T. J., Qi Y., Ramko E. B., Davidson M. W., Jin Y., Ellisman M. H. and Tsien R. Y. (2011). A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 9, e1001041 10.1371/journal.pbio.1001041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen T.-L., Gupta-Agarwal S. and Kittler J. T. (2014). Mitochondrial dynamics in astrocytes. Biochem. Soc. Trans. 42, 1302-1310. 10.1042/BST20140195 [DOI] [PubMed] [Google Scholar]
- Strub G. M., Paillard M., Liang J., Gomez L., Allegood J. C., Hait N. C., Maceyka M., Price M. M., Chen Q., Simpson D. C. et al. (2011). Sphingosine-1-phosphate produced by sphingosine kinase 2 in mitochondria interacts with prohibitin 2 to regulate complex IV assembly and respiration. FASEB J. 25, 600-612. 10.1096/fj.10-167502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M., Horvath S. E. and Pfanner N. (2016). Mitochondrial contact site and cristae organizing system. Curr. Opin. Cell Biol. 41, 33-42. 10.1016/j.ceb.2016.03.013 [DOI] [PubMed] [Google Scholar]
- Vogel F., Bornhövd C., Neupert W. and Reichert A. S. (2006). Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 175, 237-247. 10.1083/jcb.200605138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Malsburg K., Müller J. M., Bohnert M., Oeljeklaus S., Kwiatkowska P., Becker T., Loniewska-Lwowska A., Wiese S., Rao S., Milenkovic D. et al. (2011). Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell 21, 694-707. 10.1016/j.devcel.2011.08.026 [DOI] [PubMed] [Google Scholar]
- Weber T. A., Koob S., Heide H., Wittig I., Head B., van der Bliek A., Brandt U., Mittelbronn M. and Reichert A. S. (2013). APOOL is a cardiolipin-binding constituent of the Mitofilin/MINOS protein complex determining cristae morphology in mammalian mitochondria. PLoS ONE 8, e63683 10.1371/journal.pone.0063683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens V., Kohl W. and Busch K. (2013). Restricted diffusion of OXPHOS complexes in dynamic mitochondria delays their exchange between cristae and engenders a transitory mosaic distribution. J. Cell Sci. 126, 103-116. 10.1242/jcs.108852 [DOI] [PubMed] [Google Scholar]
- Wurm C. A. and Jakobs S. (2006). Differential protein distributions define two sub-compartments of the mitochondrial inner membrane in yeast. FEBS Lett. 580, 5628-5634. 10.1016/j.febslet.2006.09.012 [DOI] [PubMed] [Google Scholar]
- Xie J., Marusich M. F., Souda P., Whitelegge J. and Capaldi R. A. (2007). The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 581, 3545-3549. 10.1016/j.febslet.2007.06.052 [DOI] [PubMed] [Google Scholar]
- Zeharia A., Friedman J. R., Tobar A., Saada A., Konen O., Fellig Y., Shaag A., Nunnari J. and Elpeleg O. (2016). Mitochondrial hepato-encephalopathy due to deficiency of QIL1/MIC13 (C19orf70), a MICOS complex subunit. Eur. J. Hum. Genet. 24, 1778-1782. 10.1038/ejhg.2016.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbes R. M., Bohnert M., Stroud D. A., von der Malsburg K., Kram A., Oeljeklaus S., Warscheid B., Becker T., Wiedemann N., Veenhuis M. et al. (2012a). Role of MINOS in mitochondrial membrane architecture: cristae morphology and outer membrane interactions differentially depend on mitofilin domains. J. Mol. Biol. 422, 183-191. 10.1016/j.jmb.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Zerbes R. M., van der Klei I. J., Veenhuis M., Pfanner N., van der Laan M. and Bohnert M. (2012b). Mitofilin complexes: conserved organizers of mitochondrial membrane architecture. Biol. Chem. 393, 1247-1261. 10.1515/hsz-2012-0239 [DOI] [PubMed] [Google Scholar]
- Zerbes R. M., Höß P., Pfanner N., van der Laan M. and Bohnert M. (2016). Distinct roles of Mic12 and Mic27 in the mitochondrial contact site and cristae organizing system. J. Mol. Biol. 428, 1485-1492. 10.1016/j.jmb.2016.02.031 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.