ABSTRACT
The mammalian ubiquitin ligase Hrd1 is the central component of a complex facilitating degradation of misfolded proteins during the ubiquitin–proteasome-dependent process of ER-associated degradation (ERAD). Hrd1 associates with cofactors to execute ERAD, but their roles and how they assemble with Hrd1 are not well understood. Here, we identify crucial cofactor interaction domains within Hrd1 and report a previously unrecognised evolutionarily conserved segment within the intrinsically disordered cytoplasmic domain of Hrd1 (termed the HAF-H domain), which engages complementary segments in the cofactors FAM8A1 and Herp (also known as HERPUD1). This domain is required by Hrd1 to interact with both FAM8A1 and Herp, as well as to assemble higher-order Hrd1 complexes. FAM8A1 enhances binding of Herp to Hrd1, an interaction that is required for ERAD. Our findings support a model of Hrd1 complex formation, where the Hrd1 cytoplasmic domain and FAM8A1 have a central role in the assembly and activity of this ERAD machinery.
KEY WORDS: ER-associated degradation, Endoplasmic reticulum (ER), E3 ubiquitin ligase, Oligomerisation, Protein complex, FAM8A1, Herp, ER quality control
Highlighted Article: A cytoplasmic domain within the ubiquitin ligase Hrd1 recruits cofactors necessary for ligase oligomerisation and activity, which are important for ER-associated degradation (ERAD).
INTRODUCTION
Protein biosynthesis in the eukaryotic secretory pathway is an error-prone process that regularly produces irretrievably misfolded protein forms. Misfolded proteins arising spontaneously, constitutively, or due to stress in the endoplasmic reticulum (ER) are potentially toxic. To mitigate aggregation and ensure continuous flux through the pathway, non-native, aggregation-prone forms are promptly cleared from the organelle through ER-associated degradation (ERAD). The term ERAD collectively describes the coordinated process that identifies and guides aberrant protein conformers from the ER to their eventual degradation by the ubiquitin–proteasome system (UPS, reviewed by Christianson and Ye, 2014; Claessen et al., 2012). Multi-component complexes assembled around membrane-embedded E3 ubiquitin ligases organise the activities associated with ERAD. Accessory factors are responsible for: (1) identifying and escorting substrates to the ER membrane; (2) facilitating ‘retrotranslocation’ across and/or dislocation of substrates from the lipid bilayer; (3) enabling polyubiquitylation of substrates by proximal E3 ligases as they emerge into the cytoplasm; and (4) supporting delivery to and destruction of substrates by 26S proteasomes (reviewed by Olzmann et al., 2013a). The disparate range of structures, topologies, folding programs and post-translational modifications exhibited by secretory cargo implies that the ERAD mechanism needs to be adaptive and have a broad spectrum of removal capacities. The disproportionately high ratio of potential substrates (∼7000) to ER-resident E3 ligases (∼25; Neutzner et al., 2011) implies that the substrate range of each E3 ligase is likely to be extensive.
Hrd1 (also known as synoviolin) is an evolutionarily conserved ER-resident E3 ligase implicated in ERAD; it is present in all metazoans, fungi and plants (Amano et al., 2003; Chen et al., 2016; Hampton et al., 1996; Knop et al., 1996; Müller et al., 2005). Its polytopic architecture coordinates a diverse set of accessory factors and ubiquitylation machinery that collectively endow Hrd1 with extensive and broad ERAD capacity (Carvalho et al., 2006; Christianson et al., 2012). Mammalian Hrd1 putatively has at least six transmembrane domains (TMDs) spanning the ER lipid bilayer and possibly as many as eight, if structurally conserved with its yeast orthologue Hrd1p (Schoebel et al., 2017). The cytoplasmic C-terminus of Hrd1 contains a RING (really interesting new gene) domain that facilitates ubiquitin transfer from one (or more) E2 ubiquitin-conjugating enzymes to substrates. A short region capable of binding cytoplasmic p53 has also been reported (Yamasaki et al., 2006). In addition to its ubiquitylation role, photo-crosslinking and in vitro reconstitution studies of the yeast homologue Hrd1p suggest that the TMDs of Hrd1 may also form a retrotranslocation channel to mediate passage of lumenal substrates across the ER lipid bilayer during ERAD (Baldridge and Rapoport, 2016; Carvalho et al., 2010; Mehnert et al., 2014; Stein et al., 2014). Such a role is strongly supported by the recent cryo-EM structure of Hrd1p that shows a large aqueous cavity nearly spanning the ER membrane formed by five TMDs (Schoebel et al., 2017).
Together with its cofactor SEL1L (Hrd3p in yeast), Hrd1 forms the core of a functionally conserved ERAD complex (Gardner et al., 2000; Mueller et al., 2006). SEL1L scaffolds accessory factors that recognise, capture and guide misfolded lumenal and membrane proteins to Hrd1, including the ER lectins OS-9, XTP3-B/ERLEC1 and EDEM1 (Bernasconi et al., 2008; Christianson et al., 2008; Cormier et al., 2009; Hosokawa et al., 2008), the molecular chaperone BiP/Grp78 (Christianson et al., 2008; Hosokawa et al., 2008), the reductase ERdj5 (Ushioda et al., 2008), and the ER flavoprotein ERFAD (Riemer et al., 2009, 2011). Hrd1 and SEL1L associate with integral membrane proteins including: the rhomboid-like proteins Derlin-1 (Ye et al., 2005) and Derlin-2 (Lilley and Ploegh, 2005), factors related to lipid-droplet formation, including AUP1 (Christianson et al., 2012; Klemm et al., 2011; Mueller et al., 2008) and UbxD8 (Christianson et al., 2012; Mueller et al., 2008; Olzmann et al., 2013a), the tail-anchored E2 Ube2j1 (Mueller et al., 2008) and VIMP (Lilley and Ploegh, 2005), which is a cytoplasm-facing ER-resident protein able to reduce disulphide bonds (Christensen et al., 2012). The cytoplasmic AAA-ATPase VCP/p97 (and its cofactors Ufd1/Npl4), which is implicated in extraction of almost all substrates from the membrane during ERAD (Ye et al., 2005, 2001), can be recruited to the complex directly through a C-terminal VCP binding motif (VBM) in Hrd1 (Morreale et al., 2009) or indirectly through SHP domains in Derlin-1 and Derlin-2 (Greenblatt et al., 2011), a UBX domain in UbxD8 (Olzmann et al., 2013b) or a VIM domain in VIMP (Christensen et al., 2012).
Hrd1 also forms unique interactions with two integral membrane protein cofactors, Herp (also known as HERPUD1) and FAM8A1, to assemble higher-order ERAD complexes. Herp is a UBA-UBL domain containing protein implicated in ERAD of mainly lumenal Hrd1 substrates (Kny et al., 2011; Schulze et al., 2005) and is highly upregulated during ER stress (Kokame et al., 2000). Mammals also have a Herp homologue, Herp2 (also known as HERPUD2), which is constitutively expressed and not responsive to ER stress (Huang et al., 2014). Interactions with ubiquitylated substrates, ubiquilins and 26S proteasomes, have suggested that Herp has a role in delivering substrates from the ER membrane to cytoplasmic proteasomes (Kim et al., 2008; Okuda-Shimizu and Hendershot, 2007). In addition to Herp and Herp2, FAM8A1 has been shown to be a specific interactor of Hrd1 (Christianson et al., 2012). The majority of Hrd1 in the cell is in complex with FAM8A1 (Hwang et al., 2017). FAM8A1 is an ER-resident membrane protein conserved across metazoans, but with no orthologue in unicellular fungi. It contains a cytoplasmic N-terminus followed by a region containing three highly conserved TMDs that exhibit weak homology to the bacterial RDD domain. FAM8A1 binds exclusively to Hrd1 through interactions that are independent of SEL1L and other cofactors (Christianson et al., 2012). It has been proposed to contribute to Hrd1-dependent degradation of lumenal, but not cytoplasmic or membrane-bound, substrates. How interaction with FAM8A1 and Herp may influence Hrd1 activity or complex assembly remains undetermined.
As the archetypal ubiquitin ligase for ERAD, understanding how Hrd1 assembles cofactors to coordinate substrate delivery with ubiquitylation is fundamental to characterising the temporally and spatially regulated ERAD mechanism. The presence of metazoan-specific factors associated with Hrd1 suggests fundamental differences may have evolved between lower and higher eukaryotes, to promote distinct organisational strategies or substrate processing mechanisms. It is yet to be ascertained how this structurally diverse set of cofactors assemble with Hrd1 into a complex to facilitate (or augment) substrate processing. Here, we identify the specific domains of Hrd1 that interact with FAM8A1, Herp, SEL1L and other ERAD components. Based on these interactions, we propose a mechanism for how the interaction of Hrd1 with FAM8A1 promotes higher-order complex assembly, including recruitment of the cofactor Herp.
RESULTS
Hrd1 scaffolds accessory factors through distinct domains
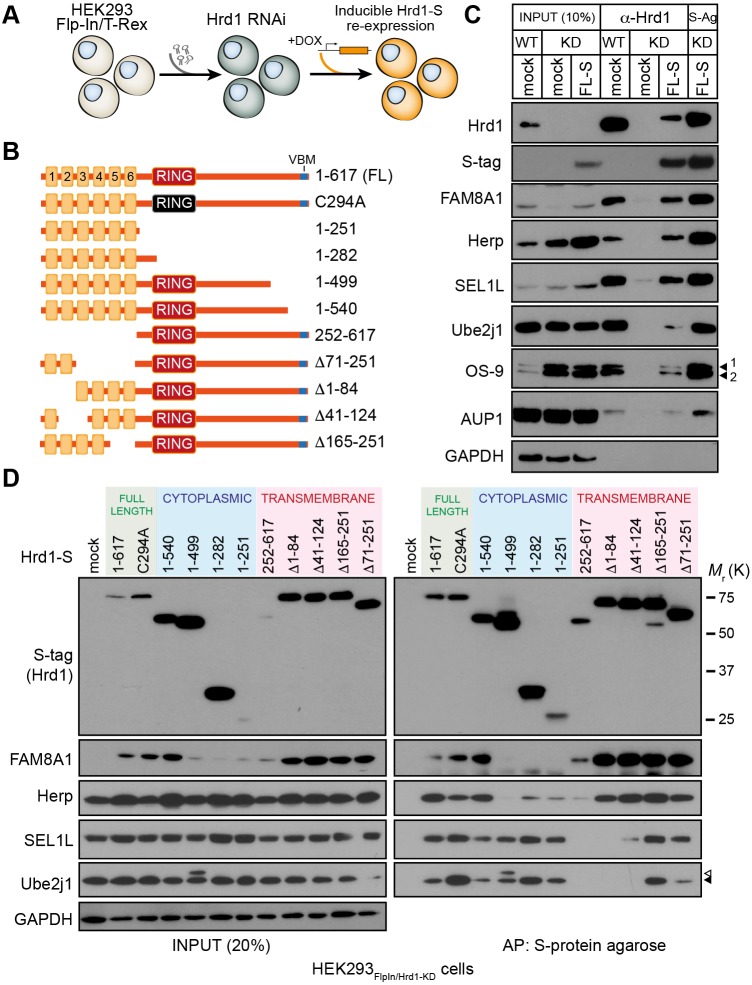
To define the Hrd1 domains through which functional complexes are assembled, we first generated a Flp-In T-REx 293 cell line stably expressing a validated shRNA targeted to the 3′ untranslated region (UTR) of Hrd1 (Fig. 1A, HEK293Flp-In/Hrd1-KD) (Christianson et al., 2012). Knockdown was estimated as >90% by qRT-PCR (Fig. S1A) and western blot (Fig. S1B). Epitope (S-peptide)-tagged Hrd1 truncations and deletions containing or excluding regions of interest or defined domains (e.g. TMDs, RING domain, VBM; Fig. 1B) were then re-introduced to HEK293Flp-In/Hrd1-KD cells through homologous recombination at the single doxycycline (DOX)-inducible locus. Affinity-purified full-length (FL) Hrd1FL-S (1-617) re-introduced into HEK293Flp-In/Hrd1-KD cells produced a co-precipitation profile comparable to endogenous Hrd1 from HEK293Flp-In/WT immunoprecipitated using a Hrd1 monoclonal antibody. The profile included associations with known cofactors SEL1L, FAM8A1, Herp, Ube2j1 and OS-9 (Fig. 1C). We preserved labile interactions that are well documented for the Hrd1 complex (Christianson et al., 2008; Hosokawa et al., 2008; Lilley and Ploegh, 2005) by solubilising in buffer containing 1% LMNG (lauryl maltose neopentyl glycol, Fig. S1C). Hrd1FL-S also co-sedimented with these cofactors in sucrose-gradient fractions, recapitulating the pattern observed for endogenous Hrd1 (Fig. S1D), indicating that DOX-induced Hrd1FL-S forms complexes in a similar manner to endogenous Hrd1.
Fig. 1.
Independent regions of Hrd1 scaffold accessory factors. (A) Schematic diagram for functional replacement of Hrd1 by stably expressing an shRNA targeting the 3′ UTR of Hrd1 in Flp-In T-REx 293 cells (HEK293Flp-In/Hrd1-KD). (B) Hrd1 deletions and truncations stably integrated into HEK293Flp-In/Hrd1-KD cells. Expression of Hrd1 variants induced with doxycycline (DOX) at empirically determined concentrations and durations to emulate Hrd1 levels observed in parental Flp-In T-REx 293 cells. Variants include 1-617 (FL), C294A, 1-540, 1-499, 1-282, 1-251 (TM), 252-617 (CY), Δ71-251, Δ1-84, Δ41-124 and Δ165-251. (C) Restoring Hrd1 complex formation upon DOX induction (1 µg/ml, 18 h) of full-length Hrd1-S in HEK293Flp-In/Hrd1-KD cells. Input (left), immunoprecipitation with anti-Hrd1 (H1) and affinity purification with S-protein agarose (S-Ag) of LMNG lysates of HEK293Flp-In/WT (parental) and HEK293Flp-In/Hrd1-KD cells. Resulting western blots were probed with antibodies against Hrd1, S-tag, SEL1L, OS-9 (isoforms 1 and 2 indicated), Herp, AUP1 and Ube2j1. GAPDH served as the loading control. (D) Co-precipitation profiles of Hrd1 variants induced by DOX (1 µg/ml, 18 h) in HEK293Flp-In/Hrd1-KD cells. LMNG-solubilised lysates from post-nuclear fractions were affinity purified (AP) by S-Ag (right) and separated by SDS-PAGE along with representative inputs (20% of AP, left). Resulting blots were probed with antibodies against Hrd1 (S-tag), FAM8A1, Herp, SEL1L, Ube2j1 and GAPDH. A phosphorylated form of Ube2j1 is indicated (white triangle).
We then compared interaction profiles for the series of Hrd1 truncations and deletions. The repertoire of interactors found with a RING domain mutant (C294A) or a C-terminal VBM truncation (1-540) closely resembled that of Hrd1FL (1-617) (Fig. 1D). Further truncation of the C-terminus (1-499, 1-282, 1-251), however, resulted in loss of co-precipitated FAM8A1 and Herp, whereas interactions between SEL1L and Ube2j1 were unaffected (Fig. 1D, right). A second Ube2j1 band appears with Hrd11-499 and corresponds to its phosphorylated form (Menon et al., 2013; Oh et al., 2006), as it was sensitive to treatment with phosphatase and not the deubiquitylase Usp2cc (Fig. S1E). Steady-state levels of endogenous Herp and most other Hrd1 interactors were comparable upon re-introduction of Hrd1 variants. In contrast, FAM8A1 was reduced (or absent) when the truncated cytoplasmic variants of Hrd1 were expressed, but stabilised by the Hrd1 cytoplasmic domain (252-617) alone (Fig. 1D, left). FAM8A1 was unstable and degraded rapidly in the absence of Hrd1 (Fig. S1F). This degradation was prevented by the proteasome inhibitor MG132 or by siRNA-mediated knockdown of the ER-resident E3 ligase MARCH6/TEB4 (Fig. S1G), indicating that FAM8A1 is an obligate component of Hrd1 complexes whose degradation, when unassembled, is enforced by a MARCH6-dependent process.
Co-precipitation profiles of Ube2j1 and OS-9 were consistent with their recruitment to the Hrd1 complex through SEL1L-dependent interactions (Fig. 1C,D) (Mueller et al., 2008). Hrd1 variants systematically lacking one or more of the six putative TMDs were re-introduced to determine which were important for SEL1L scaffolding. Consistent with the organisation observed in yeast (Carvalho et al., 2010; Schoebel et al., 2017), the Hrd1 N-terminus encompassing TM1 and TM2 (residues 1-84) was both necessary and sufficient for SEL1L interaction (Fig. 1D, right). Ube2j1 was absent from affinity-purified complexes that lacked SEL1L, whereas levels of FAM8A1 and Herp were unchanged in these complexes. Therefore, FAM8A1 and Herp interact with distinct regions of Hrd1 compared with Ube2j1 and SEL1L. Forms lacking any of the remaining TMDs (Δ71-253, Δ165-253) produced interaction profiles resembling Hrd1FL. Thus, Hrd1 scaffolds important cofactors through distinct TM and cytoplasmic domains to assemble functional complexes.
A conserved Hrd1 cytoplasmic domain interacts with FAM8A1 and Herp
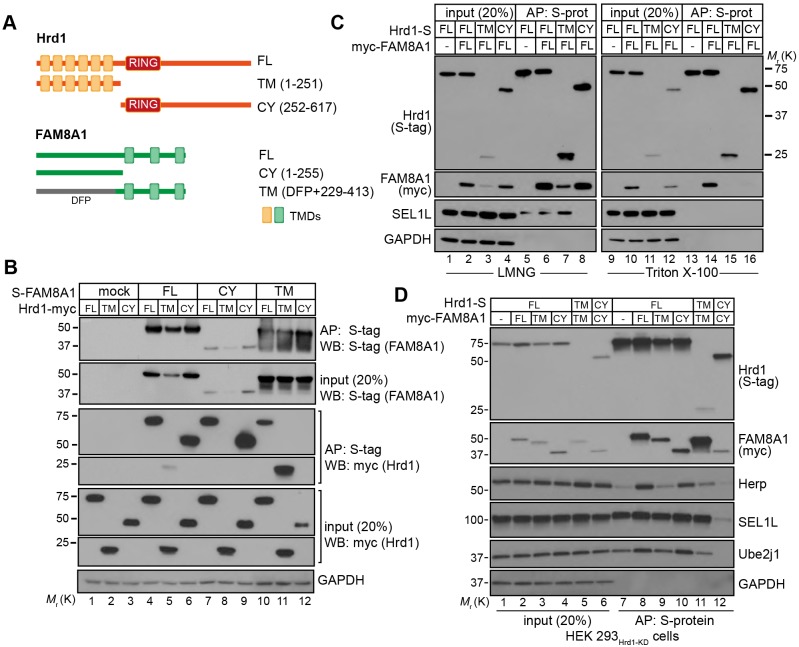
FAM8A1 was nearly undetectable by western blot in HEK293Flp-In/Hrd1-KD cells but could be restored with expression of Hrd1FL-S or variants containing the region between residues 251-540 (Fig. 1D, left). To determine how interaction with Hrd1 stabilises FAM8A1, we ectopically expressed transmembrane (TM) and cytoplasmic (CY) domains from Hrd1 and FAM8A1 in HEK293Hrd1-KD cells (TM: Hrd11-251, FAM8A1256-413) (CY: Hrd1252-617, FAM8A11-255) (Fig. 2A) and evaluated their co-precipitation profiles. FAM8A1CY and FAM8A1TM each co-precipitated Hrd1FL (Fig. 2B, lanes 4, 7 and 10) as well as their spatially corresponding Hrd1 domains (i.e. FAM8A1TM-Hrd1TM and FAM8A1CY-Hrd1CY) (Fig. 2B, lanes 9 and 11; D, lanes 11 and 12). Hrd1TM was sufficient to bring down both SEL1L and FAM8A1, but only under mild solubilisation conditions using LMNG (Fig. 2C, compare lane 7 to 15), consistent with the labile TMD interactions documented for this complex. Both FAM8A1FL and FAM8A1CY were less stable when co-expressed with Hrd1TM (Fig. 2B, lanes 5 and 8; C, lane 3), indicating that a stabilising motif resides within the Hrd1 cytoplasmic domain. Consistent with this result, the Hrd1FL-FAM8A1FL interaction persisted even when solubilised in the presence of the more stringent, non-ionic detergent Triton X-100 that disrupts TMD contact (Fig. 2C, lane 14). Additionally, co-precipitation of Herp by Hrd1FL required FAM8A1CY to maximise its interaction (Fig. 2D, compare lanes 9 and 10). In contrast, the TMDs of Hrd1 were necessary to interact with SEL1 L (Fig. 2C, lane 8; D, compare lanes 11 and 12). Thus, Hrd1 assembles with SEL1L and FAM8A1 through its TMDs, but interaction with its cytoplasmic domain is required to confer stability to FAM8A1 and enhance recruitment of Herp.
Fig. 2.
Hrd1 forms a complex with FAM8A1 through cytoplasmic and membrane interactions. (A) Illustration of Hrd1 and FAM8A1 full-length (FL), transmembrane (TM) and cytoplasmic (CY) domain expression constructs. DFP, a non-fluorescent GFP variant appended to the FAM8A1 TM region (aa 229-413) was used to stabilise expression. (B) S-tagged FAM8A1 (FL, TM and CY) and Hrd1–Myc (FL, TM and CY) co-expressed in HEK293Flp-In/Hrd1-KD cells were solubilised in 1% LMNG and complexes affinity purified (AP) by S-Ag. Input (20%) and AP are shown in resulting blots probed with anti-S-tag (FAM8A1) and anti-Myc (Hrd1) with GAPDH serving as a loading control. (C) Hrd1-S (FL, TM and CY) co-expressed with Myc–FAM8A1 (FL) in HEK293Flp-In/Hrd1-KD cells lysed in either 1% LMNG (left) or 1% Triton X-100 (right) and isolated using S-Ag. Input and AP are shown in resulting blots probed for S-tag (Hrd1), Myc (FAM8A1) and SEL1L. (D) Myc–FAM8A1 (FL, TM and CY) co-expressed with Hrd1-S (FL, TM and CY). Western blots were also probed for SEL1L, Herp and Ube2j1; GAPDH is presented as a loading control.
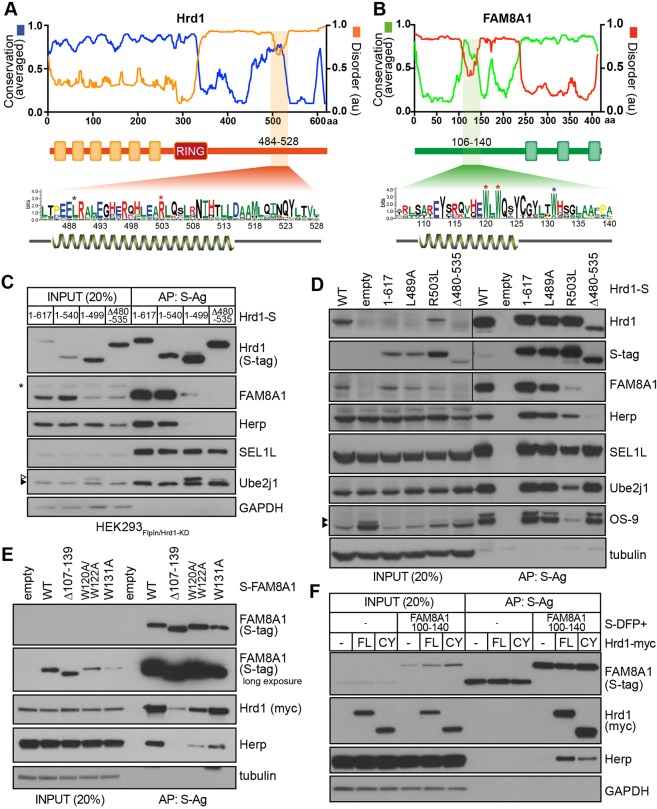
To determine the binding motifs underlying cytoplasmic interactions, we analysed predicted secondary structures and evolutionary conservation of Hrd1, FAM8A1 and Herp. Meta-analysis integrating multiple secondary structure algorithms (MetaDisorder; Kozlowski and Bujnicki, 2012) predicts intrinsic disorder within the cytoplasmic domains of Hrd1 (aa 330-617), FAM8A1 (aa 1-255) and Herp (aa 87-258). These disordered regions are interrupted by putative α-helices (predicted using Phyre2; Hrd1486-528, FAM8A1107-140, Herp1172-191) that are evolutionarily conserved (Consurf) among metazoans (Fig. 3A,B; Fig. S2B). Hrd1 variants deleted or truncated in this segment (Hrd1Δ480-535 and Hrd11-499, respectively) retained interactions with SEL1L and Ube2j1, whereas FAM8A1 and Herp were partially or completely lost (Fig. 3C and Fig. 1D). Mutating one of the conserved, positively charged residues within this region (R503L) was sufficient to disrupt interactions with FAM8A1 and Herp, as well as OS-9 and Ube2j1, which might signify more extensive structural changes (Fig. 3D). Based on this requirement, we have designated residues 480-535 of Hrd1 as the HAF-H domain (Hrd1 assembly with FAM8A1 and Herp).
Fig. 3.
Hrd1 and FAM8A1 interact through conserved cytoplasmic domains to enhance recruitment of Herp. Predicted intrinsic protein disorder (MetaDisorder, red/orange) and evolutionary conservation (ConSurf, blue/green) for Hrd1 (A) and FAM8A1 (B). Consensus sequences for Hrd1 (aa 484-528) and FAM8A1 (aa 106-140) generated by WebLogo 3.0 are shown below. Letter sizes correspond to their conservation. Asterisks indicate conserved residues selected for mutation. (C) Co-precipitation profiles of Hrd1-S variants lacking conserved cytoplasmic domain from A (1-499 and Δ480-535) expressed in HEK293Flp-In/Hrd1-KD cells. Hrd1 1-617 (FL), 1-540 are included for comparison. Samples affinity purified by S-Ag from 1% LMNG lysates are shown along with inputs (20% of AP) with resulting blots probed for Hrd1 (S-tag), FAM8A1, SEL1L, Herp, Ube2j1 and GAPDH. Asterisk indicates a non-specific band. (D) DOX-induced expression of Hrd1 variants (1-617, L489A, R503R and Δ480-535) in HEK293Flp-In/Hrd1-KD cells. Samples affinity purified by S-Ag from 1% LMNG lysates are shown along with inputs (20% of AP) with resulting blots probed for Hrd1 (S-tag), FAM8A1, SEL1L, Herp, Ube2j1, OS-9 and tubulin. (E) S-tagged FAM8A1 variants (FL, Δ107-139, W120A/W122A and W131A were transiently co-expressed along with Hrd1–Myc (FL) in HEK293Flp-In/Hrd1-KD cells. Following isolation by S-Ag from cells 1% LMNG lysate, resulting blots of input (20% of AP) and AP samples were probed for Hrd1 (Myc), FAM8A1 (S-tag), Herp and tubulin. (F) Co-expression of S-DFP or S-DFP-FAM8A1100-140 fusion protein in HEK293Hrd1-KD cells together with Hrd1–Myc FL, TM and CYTO. Following isolation by S-Ag, the resulting blots of LMNG lysates (20% of AP) and AP samples were probed for Hrd1 (anti-Myc), FAM8A1 (anti-S-tag), Herp and GAPDH.
FAM8A1 lacking or mutated in its corresponding conserved cytoplasmic domain (FAM8A1Δ107-139 or FAM8A1W120A/W122A, respectively) interacted poorly with both Hrd1 and Herp. Mutation of a conserved Trp residue beyond the predicted α-helix (W131A) brought down Hrd1 at almost wild-type (WT) levels, but still co-precipitated less Herp. This interaction was only detectable upon solubilisation with LMNG (Fig. 3E) and not with Triton X-100 (Fig. S2A). A fusion protein of FAM8A1100-140 with DFP (non-fluorescent GFP) was sufficient to bring down Herp, but only when the Hrd1 cytoplasmic domain was present (Hrd1FL or Hrd1CY, Fig. 3F). More Herp co-precipitated with Hrd1FL and Hrd1CY, which is consistent with Herp interactions (direct or indirect) through Hrd1 TMDs (Fig. 2D). These results indicated that this conserved segment of FAM8A1 is both necessary and sufficient to interact with Hrd1 and suggest that FAM8A1 binding promotes recruitment of Herp to the Hrd1 complex.
Like Hrd1 and FAM8A1, Herp contains a highly conserved cytoplasmic region distal to the N-terminal UBL domain, with residues 170-195 exhibiting more than 70% identity across mammals (Fig. S2B) and with its homologue Herp2 (Fig. S2C). Using a nested deletion series, we determined that residues 170-190 of Herp are required for interaction with Hrd1 (Fig. S2D, S2E), which is within a region that has been reported previously (Huang et al., 2014; Schulze et al., 2005). Collectively, these findings illustrate that the Hrd1 HAF-H is a domain that binds a complementary region within FAM8A1 (aa 107-139) to enhance assembly with Herp via its conserved cytoplasmic segment (aa 170-190).
The HAF-H domain and FAM8A1 are necessary to form higher-order Hrd1 complexes
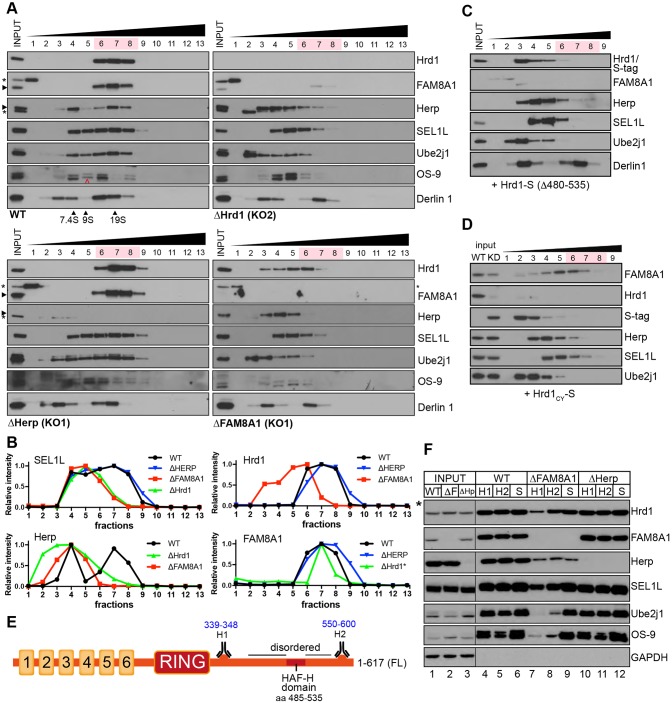
Oligomerisation of E3 ligases with accessory factors forms membrane-bound complexes associated with substrate retrotranslocation and ubiquitylation during ERAD (Horn et al., 2009), but both their composition and stoichiometry have remained elusive. To assess the contributions made by FAM8A1 and Herp to the formation of mature Hrd1 complexes, we compared velocity sedimentation profiles of Flp-In T-REx 293 cells modified by CRISPR/Cas-9-mediated genomic editing to preclude Hrd1, FAM8A1 or Herp expression (Fig. 4A; Fig. S3A). In parental WT cells, Hrd1 co-sediments with known interactors (FAM8A1, SEL1L, Herp, Ube2j1, Derlin-1) primarily in fractions 6-8, with distribution outside these fractions exhibited by all factors except FAM8A1 (Fig. 4A, top left). Hrd1 deletion (ΔHrd1) resulted in loss of FAM8A1 and sedimentation of SEL1L, Ube2j1 and Herp (but not Derlin-1) in lighter fractions (3-6), which corresponds to smaller complexes (Fig. 4A, top right). This is consistent with the pattern observed for HEK293Flp-In/Hrd1-KD cells (Fig. S1D). Loss of Herp (ΔHerp) did not grossly alter sedimentation of Hrd1 or its cofactors (Fig. 4A, bottom left). Strikingly, we observed a marked shift of Hrd1, SEL1L, Ube2j1 and Herp to lighter fractions (4-6) in cells lacking FAM8A1 (ΔFAM8A1) (Fig. 4A, bottom right). This shift reflects significant disruption to the components of higher-order assemblies of Hrd1 complexes (Fig. 4B). HEK293FlpIn/Hrd1-KD cells restored by Hrd1Δ480-535 (i.e. lacking the HAF-H domain) produced a sedimentation pattern comparable to that of ΔFAM8A1 (Fig. 4C), indicating that FAM8A1 interaction with the HAF-H domain is crucial to enable formation of native, higher-order Hrd1 complexes.
Fig. 4.
The HAF-H domain and FAM8A1 are required for Hrd1 to form mature complexes. (A) Velocity sedimentation of 1% LMNG lysates from WT and CRISPR/Cas-9 edited Flp-In T-REx 293 cells (ΔHrd1, ΔFAM8A1 and ΔHerp) on 10-40% sucrose gradients. Individual TCA-precipitated fractions (1-13) were separated by SDS-PAGE with the resulting blots probed for Hrd1, FAM8A1, Herp, SEL1L, OS-9, Ube2j1 and Derlin-1. Asterisks indicate non-specific bands and red arrowhead, an underloaded OS-9 lane. Gel filtration standards solubilised and sedimented in equivalent buffer conditions are shown for comparison. 7.4S, yeast alcohol dehydrogenase (150 kDa); 9S, bovine serum albumin (200 kDa); 19S, bovine thyroglobulin (669 kDa). (B) Relative band intensities (normalised to strongest band) indicating shifts of SEL1L, Hrd1, FAM8A1 and Herp from gradients in A. Asterisk indicates a weak FAM8A1 signal for the ΔHrd1 sample. (C,D) Velocity sedimentation of LMNG lysates from HEK293Flp-In/Hrd-KD cells expressing S-tagged Hrd1Δ485-528 (C) and Hrd1CY (aa 252-617) (D). Samples and blots were processed as in A. Only fractions 1-9 (of 13) are shown. (E) Illustration of Hrd1 topology indicating cytoplasmic epitopes recognised by the anti-Hrd1 antibodies H1 (aa 339-348) and H2 (aa 550-600). (F) Comparative immunoprecipitation of Hrd1 complexes by antibodies against Hrd1 (H1, H2) and SEL1L from WT, ΔFAM8A1 and ΔHerp cells. Resulting blots were probed for Hrd1, FAM8A1, Herp, SEL1L, Ube2j1, OS-9 and GAPDH. Input lysates are also shown; note that asterisk indicates a longer exposure of Hrd1 lysate necessary for visualisation.
In ΔHrd1 cells, the residual FAM8A1 sedimented on sucrose gradients around fraction 7 (Fig. 4A, top right), suggesting that FAM8A1 could be the principal organiser (or enforcer) of higher-order assemblies, independent of Hrd1. To test this, we expressed HAF-H domain-containing Hrd1CY (aa 252-617) in HEK293Hrd1-KD cells, which stabilises endogenous FAM8A1 but does not assemble with other cofactors (Fig. 1D and Fig. 2B). When FAM8A1 partially stabilised by Hrd1CY was separated on sucrose gradients (Fig. 4D), it sedimented in fractions (5-7) that were closer in size to endogenous Hrd1 complexes from WT cells (Fig. 4A, top left) and HEK293Hrd1-KD cells, restored with Hrd1FL-S (Fig. S1D, bottom). Sedimentation profiles of SEL1L and Ube2j1 mirrored those seen for ΔHrd1 (Fig. 4A, top right) and HEK293Flp-In/Hrd1-KD (Fig. S1D, middle), confirming that neither contributed to stable complexes with FAM8A1 or Hrd1CY (Fig. 1D). These patterns imply that once stabilised, FAM8A1 may be intrinsically able to oligomerise. Assembly is more likely to occur through the highly conserved TMDs in FAM8A1; however, as stabilisation by the Hrd1 HAF-H domain is a prerequisite, Hrd1 would likely be at least in a 1:1 stoichiometry with FAM8A1 in mature complexes.
FAM8A1 enforces the native Hrd1 conformation through the HAF-H domain
The dramatic change to Hrd1 complex sedimentation assembled without FAM8A1 (Fig. 4A,B) or HAF-H domain (Fig. 4C) reflects significant disruption to Hrd1 conformation and/or organisation. To further probe this disturbance, we used a monoclonal antibody raised against a region neighbouring the RING domain (aa 339-348, H1, Fig. 4E). The ability of the H1 antibody to immunoprecipitate native Hrd1 complexes from LMNG-solubilised WT lysates was markedly diminished in ΔFAM8A1 cells (Fig. 4F, compare lanes 4 and 7), even though the total amount of cellular Hrd1 present and available for immunoprecipitation did not differ (Fig. 4F, lanes 1-3; Fig. S3B-D). Immunoprecipitation with antibodies recognising the Hrd1 C-terminus (H2, aa 550-617) or TMD region (H3, aa 198-207) also produced patterns that were similar to those obtained with the H1 antibody (Fig. 4F, lane 8; Fig. S3D). Inaccessibility of these epitopes would seem to indicate that Hrd1 secondary structure was altered without FAM8A1 interaction. Although we cannot rule out a new interaction partner that blocks epitope access, the shift of Hrd1 to lower sucrose gradient fractions in ΔFAM8A1 cells argues against this. Hrd1 complexes immunoprecipitated through SEL1L were comparable between cell lines except for Herp (Fig. 4F, lanes 6, 9, 12). This confirms that the cytoplasmic Hrd1–FAM8A1 interaction enhances, but is not absolutely required for Herp recruitment (Fig. 4F, lanes 7-9), whereas the luminal Hrd1–SEL1L interaction is independent of FAM8A1. Some Herp residually binds to Hrd1–SEL1L in ΔFAM8A1 cells, probably through its TMDs, indicating the presence of secondary assembly surfaces. These findings indicate that interaction with FAM8A1 promotes the native conformation of the Hrd1 cytoplasmic domain, which is then able to maximally recruit Herp to the complex.
The HAF-H domain contributes to ERAD of Hrd1-dependent substrates
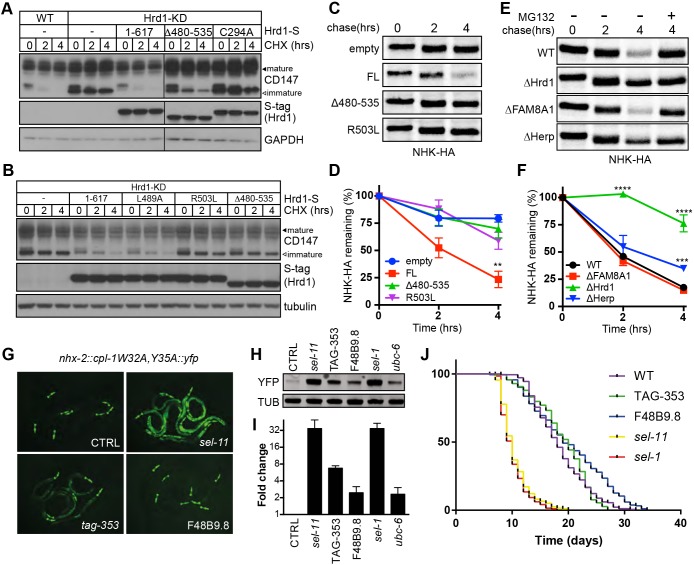
To determine whether the cytoplasmic interactions of Hrd1 with FAM8A1 and Herp are required for ERAD substrate processing, we monitored degradation of the endogenous Hrd1 substrate CD147 (Tyler et al., 2012) in HEK293FlpIn/Hrd1-KD cells reconstituted with Hrd1 variants. Translational shut-off assays showed that expressing Hrd1Δ480-535, lacking the HAF-H domain impaired degradation of core-glycosylated CD147, albeit to a lesser degree than disrupting ubiquitylation with a mutant RING domain (C294A, Fig. 5A). The Hrd1 HAF-H domain mutant Hrd1R503L, which has attenuated interactions with FAM8A1, Herp and SEL1L (Fig. 3D), recapitulated this observation (Fig. 5B,C). Next, we performed 35S-Met/Cys pulse-chase assays of model ERAD substrate α1-antitrypsin null Hong Kong (NHK) transiently expressed in HEK293FlpIn/Hrd1-KD cells reconstituted with Hrd1Δ480-535, Hrd1R503L or Hrd1FL (Fig. 5D,E). As anticipated, NHK degradation was attenuated in Hrd1-knockdown cells (t1/2=11.2 h) and restored with Hrd1FL expression (t1/2=2.0 h) to a rate comparable to that observed in WT parental cells (Fig. 5G, t1/2=1.7 h). Neither Hrd1Δ480-535 nor Hrd1R503L were able to restore NHK degradation (Fig. 5D,E, t1/2=7.5 h and 5.8 h, respectively). These data confirm that the Hrd1 HAF-H domain is contributing to the degradation of both misfolded lumenal and membrane-bound substrates during ERAD.
Fig. 5.
The cytoplasmic interaction domain underlies Hrd1-dependent ERAD. (A) Degradation of CD147 monitored by CHX chase (0, 2, 4 h; 10 μg/ml) and western blotting in Flp-In T-REx 293 cells, and HEK293Flp-In/Hrd1-KD cells stably reintroduced with S-tagged Hrd1 variants. Lysates of LMNG-solubilised post-nuclear fractions were separated by SDS-PAGE and the resulting blots probed with antibodies against CD147 and Hrd1 (S-tag). Black arrowheads indicate the immature, ER-localised form of CD147 while asterisks designate the post-ER, mature forms. (B) Degradation of CD147 monitored in HEK293Flp-In/Hrd1-KD cells expressing Hrd1 mutants (L489A, R503L). (C) Representative 35S-Met/Cys pulse-chase assays (0, 2 and 4 h) of the HA-tagged ERAD substrate NHK-HA expressed in HEK293FlpIn/Hrd1-KD cells with or without DOX-induced expression of Hrd1-S (empty, FL, Δ480-535 or R503L). Radiolabelled substrates immunoprecipitated from 1% Triton X-100 lysates by anti-HA were separated by SDS-PAGE. (D) Quantification of biological replicates in C. Band intensities quantified by phosphorimaging (BioRad) were normalised to values at t=0 h. Results are presented as mean±s.e.m. for each (n=3). Significance as determined by Students t-test is shown. **P≤0.01. (E) Representative 35S-Met/Cys pulse-chase assays of NHK-HA transiently expressed in Flp-In T-REx 293 cells WT, ΔHrd1, ΔFAM8A1 and ΔHerp cells. Samples were collected and processed as in C. (F) Quantification of biological replicates in E. Band intensities quantified by phosphorimaging were normalised to values at t=0 h. Mean±s.e.m. are plotted for each (n=3). Significance as determined by Students t-test is shown. ***P≤0.001, ****P≤0.0001. (G) Fluorescence images of the ERAD-substrate CPL-1W32A,Y35A-YFP in C. elegans disrupted for Hrd1 (sel-11), Herp (tag-353) or FAM8A1 (F48B9.8). (H) Western blot of CPL-1W32A,Y35A-YFP and tubulin from the mutants of Hrd1 complex. (I) Quantification of band intensities from I. Displayed as fold-change in YFP or tubulin with mean±s.e.m. are shown for each (n=3). (J) Kaplan–Meyer survival curves of Hrd1 complex mutant worms. Calculated mean lifespans are: WT, 18.5 days; cup-2 and tag-353 (gk443)l, 18.9 days; F48B9.8 (gk272969), 20.2 days; sel-11 (nDf59) V, 10.5 days; sel-1 (e1948)V, 10.1 days. Raw data from biological replicates (n=4) is presented in Table S4.
Next, we examined whether FAM8A1 and Herp differentially contributed to degradation of specific ERAD substrate classes by Hrd1. We found that degradation of immature CD147 was impaired without Hrd1 (ΔHrd1), but not FAM8A1 or Herp (Fig. S4A). 35S-Met/Cys pulse-chase assays of NHK expressed in ΔHrd1, ΔFAM8A1 and ΔHerp Flp-In T-REx 293 cells, showed that while deletion of Hrd1, and to a lesser extent Herp, stabilised NHK (ΔHrd1: t1/2=11.5 h; ΔHerp: t1/2=2.5 h), degradation rates in ΔFAM8A1 and WT cells were similar (ΔFAM8A1: t1/2=1.5 h; WT: t1/2=1.7 h, Fig. 5F,G,). In ΔFAM8A1 cells, NHK degradation remained dependent upon Hrd1 and p97/VCP (Fig. S4B). These findings were confirmed using HeLa cell lines stably expressing the substrate GFP-tagged MHC Class I (GFP-HLA-A2), either as a full length, integral membrane protein (WT) or as a truncated ER lumenal form (SOL) (Fig. S4C, S4D). Degradation of GFP-HLA-A2 forms [following β2-microglobulin (β2 m) depletion] has previously been shown to depend on Hrd1, SEL1L and Ube2j1 (Burr et al., 2011), and we found that this degradation also requires Herp, but not FAM8A1 (Fig. S4C, S4D). These results indicate that the HAF-H domain is necessary to maximise processing of canonical Hrd1 substrates during ERAD, but this may be, in part, due to the loss of Herp recruitment.
With very little effect on ERAD activity of the Hrd1 complex in vitro, FAM8A1 may play a more prominent cellular role during development. Therefore, we asked which components of the Hrd1 complex contributed to organismal viability or longevity by disrupting orthologues of ERAD components in Caenorhabditis elegans. As observed in mammalian cells, impaired degradation of a fluorescent model ERAD substrate CPL-1* (nhx-2::cpl-1W32A,Y35A::yfp) occurred after RNAi knockdown of Hrd1 complex orthologues SEL-11 (Hrd1), SEL-1 (SEL1L) and, to a lesser degree, TAG-353 (Herp) and Ubc-6 (Ube2j1) (Fig. 5G-I). Knockdown of F48B9.8 (FAM8A1) partially stabilised steady-state levels of CPL-1*, but to a lesser extent than observed upon silencing either SEL-1 or SEL-11. As anticipated, disrupting either SEL-11 (Hrd1) or SEL-1 (SEL1L) severely attenuated mean lifespans when compared with the WT (10.5 and 10.1 days, respectively versus 18.5 days), whereas the mean lifespans of the F48B9.8 (FAM8A1) and TAG-353 (Herp) mutants (20.2 and 18.9 days, respectively) were slightly longer, but not statistically different from WT worms (Fig. 5J; Table S4). These data highlight the key physiological roles Hrd1 and SEL1L play in both ERAD and organismal longevity, while suggesting that contributions from FAM8A1 and Herp (and Hrd1 complex oligomerisation) also help to shape these activities.
DISCUSSION
ERAD via the Hrd1 ubiquitin ligase complex has a prominent role in a eukaryotic cell's arsenal of mechanisms to maintain homeostasis in the ER; a constitutively active process that is reinforced to ameliorate proteotoxic stress (Kaneko et al., 2002). As the nexus of a hetero-oligomeric complex serving as the primary route for ERAD, Hrd1 assembles accessory components principally through its cofactor SEL1L but also through direct recruitment in the case of FAM8A1. This assembly affords proteins misfolded in the ER lumen, membrane or cytoplasmic environments access to the ubiquitin ligase activity of Hrd1, but how each cofactor contributes to diversifying the roles of Hrd1 has remained elusive. Here, we identify the HAF-H domain within the Hrd1 cytoplasmic tail as an organisational hub for both FAM8A1 and Herp that participates in ERAD of lumenal and membrane substrates, and where FAM8A1 is the component necessary for oligomerisation of the Hrd1 complex.
Serial truncations and deletions of the Hrd1 C-terminus revealed a domain that binds and stabilises FAM8A1 and enhances recruitment of the functionally important ERAD cofactor Herp. The HAF-H domain is evolutionarily conserved in higher eukaryotes, but only partially represented in yeast, consistent with absence of a FAM8A1 orthologue in fungi. It lies within an intrinsically disordered region between the RING and VBM domains, but distal to the so-called ‘proline-rich’ region (aa 339-478; Omura et al., 2006). Its position within a region anticipated to be highly mobile may reflect a requirement by the Hrd1 complex to have the flexibility to organise cytoplasmic interactions for stability, coordinate ubiquitin transfer and/or bind directly to misfolded proteins (Rosenbaum et al., 2011). Hrd1p reconstructions did not show any density for the cytoplasmic RING-containing domain, consistent with this region being one of high mobility (Schoebel et al., 2017). The corresponding segments of FAM8A1 (aa 100-140) and Herp (aa 170-190) that interact with the HAF-H domain are also short, conserved α-helical stretches flanked by disordered regions. Isolated binding domains flanked by extended, intrinsically disordered regions are commonly found in E3 ligases and may provide the plasticity required for ubiquitin transfer when the range of substrates is diverse (Boomsma et al., 2016). Efficient ubiquitin conjugation requires RING-type E3 ligases to orient target substrates, coordinating and positioning an E2 to facilitate ubiquitin transfer (Deshaies and Joazeiro, 2009). Untethered cytoplasmic ubiquitylation machinery (e.g. SCF complexes; Lydeard et al., 2013) freely accesses substrates with limited spatial constraint. In contrast, movements of lumenal substrates emerging from the ER through a putative retrotranslocation or membrane-bound substrates accessing laterally through the lipid bilayer are inherently restricted, possibly limiting access to the Hrd1 membrane-tethered RING domain. Flexibility within the disordered cytoplasmic domains of Hrd1 and FAM8A1 (and to a lesser extent Herp) may provide greater opportunity to engage emerging, partially unfolded, lumenal substrates or orient the RING–E2 interaction (or both). Correct positioning of the coordinating Hrd1 RING domain and the E2 Ube2j1 could be expected to maximise ubiquitin transfer efficiency to substrates.
We found that FAM8A1 plays the key role in organising higher-order Hrd1 assembly. Herp shares a common topology (two TMDs) and a UBL domain (aa 10-72) with Usa1p, which mediates oligomerisation in yeast by linking Hrd1p to Der1p through its N-terminus (Horn et al., 2009). Usa1p is required to degrade misfolded lumenal substrates (ERAD-L) (Carroll and Hampton, 2010; Carvalho et al., 2006; Horn et al., 2009; Kanehara et al., 2010) and the ability of Herp to partially reconstitute Usa1p function prompted speculation of an analogous role in metazoans (Carvalho et al., 2006; Yan et al., 2014). While we also found that Herp was required for ERAD of lumenal substrates (Huang et al., 2014; Kny et al., 2011; Okuda-Shimizu and Hendershot, 2007), it played no part in Hrd1 complex oligomerisation (Fig. 4A) (Huang et al., 2014). The contribution made by FAM8A1 instead of Herp to enable higher-order assembly reflects an organisational departure from the model proposed from analyses of yeast Hrd1p (Carvalho et al., 2010; Horn et al., 2009). Since the functions associated with Usa1p are segregated in mammals, this permits dissection of their relative contributions to the Hrd1 complex. The FAM8A1 sequence (aa 100-140) binding the HAF-H domain bears no homology to corresponding domain of Usa1p (aa 437-490) underlying both Usa1p–Hrd1p binding and Hrd1p oligomerisation (Carvalho et al., 2010), even though interaction is likely through the 34 amino acids of Hrd1p (Horn et al., 2009), a region most closely corresponding to the HAF-H domain. Thus, metazoans have evolved additional factors and an alternative strategy to organise Hrd1 into higher-order, functional assemblies for ERAD. A recent study of endogenous Hrd1 complex composition and stoichiometry found three high-molecular weight complexes, where the most abundant one notably contained both FAM8A1 and Herp (Hwang et al., 2017). This Hrd1-containing subcomplex was largely distinct from those containing SEL1L, OS-9 and XTP3-B, highlighting some heterogeneity of Hrd1 complexes that may be linked to its functionality.
Even though it is not a prerequisite for Hrd1-mediated ERAD, binding of FAM8A1 to the HAF-H domain promotes a conformation within the Hrd1 C-terminus that enhances Herp recruitment to the complex (Fig. 4F). Some natively unstructured proteins conditionally fold in the presence of cofactors to form stable, ordered domains (Bardwell and Jakob, 2012). Whether FAM8A1 binding stimulates structural rearrangement within the Hrd1 HAF-H domain to form a bona fide Herp binding site is not known. The altered conformation of Hrd1 in complexes without FAM8A1 suggests either a decrease in affinity or accessibility to the HAF-H domain for Herp, but currently we are not able to differentiate between the two. Herp only associates transiently with Hrd1 complexes, rather than as an obligate and stable interaction (Kny et al., 2011). How Herp modulates Hrd1 activity is not mechanistically understood, although it may enhance ubiquitylation efficiency through its UBL domain (Kny et al., 2011). Herp is required for efficient degradation of both glycosylated and non-glycosylated lumenal substrates (Huang et al., 2014; Kny et al., 2011; Okuda-Shimizu and Hendershot, 2007) and some (CD3δ; Schulze et al., 2005; MHC Class I, Fig. S4C), but not all (e.g. CD147, Fig. S4A), integral membrane targets. Intrinsic instability and a central role in Hrd1 activation might explain why Herp is the Hrd1 complex gene most strongly upregulated by the UPR during ER stress (Kokame et al., 2000).
In addition to the HAF-H organisational hub for FAM8A1 and Herp, we also confirmed the region containing TM1 and TM2 (aa 1-84) as both necessary and sufficient for assembly of SEL1L and Ube2j1 into the Hrd1 complex (Carvalho et al., 2010; Jeong et al., 2016). As the only TMD of SEL1L (and Hrd3p) is dispensable for interaction with Hrd1 (Christianson et al., 2008; Gauss et al., 2006), the lumen-exposed loop bridging TM1 and TM2 is likely to be the site of important contacts (Jeong et al., 2016), a prediction consistent with the recent cryo-EM structure of the Hrd1p–Hrd3p interface (Schoebel et al., 2017). SEL1L stabilises Hrd1 in the ER membrane (Sun et al., 2014) but is not obligatorily assembled with it, suggesting that SEL1L might be produced in excess to enable prompt assembly into the Hrd1 complex. Maintaining a functional Hrd1–SEL1L interaction is important for organismal viability as we found nematode longevity to be severely attenuated in animals lacking orthologues of either Hrd1 (SEL-11) or SEL1L (SEL-1) (Fig. 5J). This is consistent with the embryonic lethality found for heterozygous knockout mice of both Hrd1 (Yagishita et al., 2005) and SEL1L (Francisco et al., 2010, 2011) and illustrates the critical role played by these core components that is not always immediately observed with accessory factors of the complex such as Herp (Eura et al., 2012) and FAM8A1.
To date, no forward genetic screens for mediators of ERAD have identified FAM8A1 among their candidates (Grotzke et al., 2013; Krishnan et al., 2008; Ma et al., 2015). This would be consistent with a minor, non-essential role for FAM8A1 in degradation activities of the Hrd1 complex. Degradation of NHK, MHC class I and CD147 were predominantly reliant upon the Hrd1, its HAF-H domain and Herp, but in the case of CD147 and the C. elegans substrate, FAM8A1 loss did mildly disturb degradation. Since FAM8A1 binding to the HAF-H domain enhances the interaction of Herp with the Hrd1 complex, this suggests FAM8A1 could serve as an accessory factor that is necessary to assemble (or at least stabilise) higher-order structures of independently active Hrd1 complexes, or fine tune ubiquitin transfer by facilitating proximity of Hrd1 to the substrate, whereas Herp primes the reaction (Kny et al., 2011). If Hrd1 does not require higher order assembly for activity, then the evolutionary selection pressure conserving FAM8A1 TMDs and Hrd1-binding domain may arise from outside the realm of ERAD, but further studies will be required for this to be ascertained.
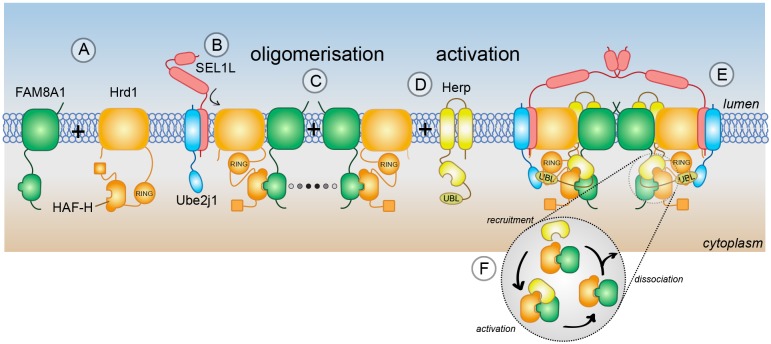
How the interactions through the HAF-H domain and the metazoan-specific component FAM8A1 might participate in the formation of ERAD-competent Hrd1 complexes is depicted in the model in Fig. 6. Interaction with FAM8A1 through the HAF-H domain and TMDs (Fig. 6A) confers stability and allows the recruitment of SEL1L and Ube2j1 to the complex through a region including TM1 and TM2 of Hrd1 (Fig. 6B), while at the same time enabling FAM8A1 to promote oligomerisation of Hrd1 either directly or by stabilising a Hrd1–Hrd1 interaction (Fig. 6C). The presence of SEL1L provides a scaffold for lumenal substrate recruitment factors such as OS-9, EDEM and XTP3-B (not shown). Other membrane-bound and cytoplasmic factors (e.g. VIMP, VCP/p97, Derlin proteins) may also be recruited at this time. After contact made initially through Hrd1–SEL1L (or other factor), Herp can fully engage the Hrd1 complex by binding to or near the HAF-H domain with the help of FAM8A1 as part of a bipartite assembly (Fig. 6D). As part of an oligomerised complex (Fig. 6E), Herp may activate Hrd1 through its UBL domain to trigger a round of substrate ubiquitylation, then dissociate to be degraded by the UPS (Yan et al., 2014) and be replaced by de novo synthesised Herp (Kny et al., 2011) (Fig. 6F). FAM8A1 remains within the Hrd1 complex, stabilising the disordered cytoplasmic regions and awaiting arrival of the next Herp molecule. Although speculative, the identification of an important cytoplasmic assembly platform for ERAD raises new mechanistic questions of processing by metazoan Hrd1 complexes. Further studies will be required to fully appreciate how the temporal assembly and spatial organisation of the Hrd1 complex underlie efficient processing of lumenal ERAD substrates.
Fig. 6.
Model for assembly and regulation of Hrd1 ligase complexes (details in Discussion).
MATERIALS AND METHODS
Plasmids and expression constructs
Mammalian expression plasmids encoding full-length and variants of human Hrd1, FAM8A1 and Herp were generated by cloning individual open reading frames into the pcDNA3.1 parental vector (Life Technologies) with an S (KETAAAKFERQHMDS), Myc (EQKLISEEDL) or HA (YPYDVPDYA) tag in-frame at either the C- or N-terminus. S-tagged Hrd1 and affiliated variants were subsequently subcloned into the pcDNA5/FRT/TO vector (Life Technologies) between the NheI and NotI sites to facilitate stable, single-site integration into Flp-In T-REx 293 cell lines, by co-expression with the Flp recombinase pOG44 (Life Technologies). Truncations and deletions of Hrd1, FAM8A1 and Herp were generated using either PCR or Gibson Assembly. Point mutations were introduced by QuikChange site-directed mutagenesis (Agilent) according to the manufacturer's protocol and are listed in Table S1. The dead fluorescent protein (DFP) used to tag FAM8A1 fragments as well as the ERAD substrates tagged with C-terminal HA tags (YPYDVPDYA) have been described previously (Christianson et al., 2008, 2012). Lentiviral expression vectors encoding GFP-HLA-A2 full length and GFP-HLA-A2 soluble were a gift from Paul Lehner (Cambridge University, UK) and have been reported previously (Burr et al., 2011, 2013).
Chemicals and compounds
The following compounds were used in this study: MG132 (10 μM, Merck Chemicals Ltd), tunicamycin (10 μg/ml, Abcam), NMS-873 (10 µM, Sigma-Aldrich), puromycin (2 µg/ml), Invivogen), hygromycin (100 μg/ml, Invivogen), doxycycline (1-20 ng/ml, Sigma-Aldrich), cycloheximide (100 μg/ml, Abcam), N-ethylmaleamide (NEM, Fluka Analytical), bortezomib (5 nM, Merck Millipore), lambda protein phosphatase (400 units, NEB) and Usp2cc (Baker et al., 2005) (gift from Ron Kopito, Stanford University, CA, USA). Detergents including; n-decyl-β-maltopyanoside (DM, D332), n-dodecyl-β-maltopyanoside (DDM, D310), octyl glucose neopentyl glycol (OGNG, NG311), CYMAL-5 (C325) and octaethylene glycol monododecyl ether (C12E8, O338) were from Anatrace. Digitonin was from EMD Millipore (high purity, CAS 11034-24-1).
Antibodies
Experiments made use of the following antibodies: anti-S-tag (Thermo Scientific, mouse mAb, #MA1-981, 1:10.000), anti-Myc (9B11,Cell Signaling, mouse mAb, #2276, 1:5000), anti-Hrd1 (Bethyl Laboratories, rabbit pAb, #A302-946A, 1:2000, #H2), anti-SEL1L (Santa Cruz, goat pAb, #sc-48081, 1:2000), anti-CD147 (Santa Cruz, mouse mAb, #sc-25273, 1:2000), anti-Ube2j1 (Abcam, rabbit pAb, #ab39104, 1:10.000), anti-Herp (Abcam, rabbit pAb, #ab150424, 1:10.000), anti-AUP1 (Atlas Antibodies, rabbit pAb, #HPA007674, 1:5000), anti-HA (H7, Sigma-Aldrich, mouse mAb, #H9658, 1:2000), anti-GAPDH (Abcam, #ab9484, mouse pAb, 1:5000), anti-α-tubulin (Sigma-Aldrich, #T6074, mouse mAb, 1:10.000), anti-OS-9 (gift from Ron Kopito, Stanford University, CA, USA; rabbit pAb, 1:5000), anti-Derlin-1 (gift from Yihong Ye, NIH, USA; rabbit pAb, 1:1000). The polyclonal rabbit anti-FAM8A1 antibody has been reported previously (Christianson et al., 2012). Mouse monoclonal antibodies for immunoprecipitations were generated against synthetic peptides for human Hrd1 (H1, aa 339-348, PAQSPPPPEP; H3, aa 198-207, DLQSENPWDN) and SEL1L (aa 785-794, EGPPEQQPPQ) peptides (Abmart). Horseradish peroxidase (HRP)-conjugated secondary antibodies to mouse, goat and rabbit were obtained from Santa Cruz, and used in 1:10,000 dilution.
Cell culture, transfection and stable cell lines
HEK293 and Flp-In T-REx 293 cells (Invitrogen) were maintained in DMEM (Lonza) with 10% foetal bovine serum (Biosera) at 37°C and 5% CO2. All cell lines were confirmed to be negative for mycoplasma. Cells were transfected using the calcium-phosphate co-precipitation technique, with FuGENE6 (Roche) or with Lipofectamine 2000 (Invitrogen). HEK293 and Flp-In T-REx 293 cells stably expressing an shRNA against Hrd1 were generated by selecting for resistance to puromycin (2 μg/ml). Flp-In T-REx 293Hrd1-KD cells expressing S-tagged Hrd1 variants were selected by resistance to hygromycin (100 μg/ml).
RNAi and gene silencing
The plasmid encoding an shRNA directed at the Hrd1 3′UTR has been described previously (Christianson et al., 2012). For targeted mRNA depletion of selected ER-resident E3 ligases, ON-TARGETplus siRNA SmartPools (4×25 nM each, GE Dharmacon) were reverse-transfected into cells in six-well plates using Dharmafect I (GE Dharmacon) and expanded subsequently for 72 h in 6-cm dishes.
CRISPR/Cas-9-mediated genomic editing
CRISPR/Cas-9 gene editing was used according to methods that have been described previously (Ran et al., 2013). Flp-In T-REx 293 cells were transfected with pSpCas9(BB)-2A-Puro (PX459) plasmid (Addgene) containing target-specific sgRNA for Hrd1, FAM8A1 and Herp (sequences listed in Table S2). Cells edited by CRISPR/Cas-9 were selected by resistance to puromycin (2 μg/ml). Single-cell clones from the transfected cell populations were isolated using serial dilution. Validation of gene disruption in each cell line by CRISPR/Cas-9 was confirmed by western blotting of lysates and/or immunoprecipitations (Fig. S3) or by using a Surveyor Mutation Detection Kit (IDT), according to the manufacturer's protocol.
Immunopurification, affinity purification and immunoblotting
Both HEK293 and Flp-In T-REx 293 cells were rinsed in phosphate-buffered saline (PBS), mechanically lifted, harvested, and lysed in buffer [150 mM NaCl, 50 mM Tris-HCl, 5 mM EDTA, cOmplete protease inhibitor cocktail (Roche) and NEM (2 μM)] containing 1% v/v lauryl maltose neopentyl glycol (LMNG, Anatrace) or 1% Triton X-100 (Fischer Scientific), as indicated. In some instances, PhosSTOP (Roche) was also included in the lysis buffer. Detergent-soluble, post-nuclear supernatant fractions were collected by sequential centrifugation steps (750 g, 5 min, 20,000 g, 30 min), pre-cleared with Sepharose CL-4B (50 μl, 50:50 slurry), and subsequent affinity and immunopurifications carried out from the resulting lysate (1 mg). When total Hrd1 amounts were assessed, prior to immunoprecipitation, cell lysates were denatured by addition of SDS to 1% v/v to dissociate protein complexes and subsequently by diluting SDS to a final concentration of 0.1% with 1% Triton X-100. S-tagged proteins were affinity purified by S-protein agarose (EMD-Millipore) while endogenous Hrd1, SEL1L, and FAM8A1 and HA-tagged substrates were immunoprecipitated with specific antibodies (described above) followed by isolation with Protein-G–Sepharose (Roche). All bead-bound samples were washed three times in lysis buffer lacking detergent (20× bead volume). Where indicated, bead-bound material was treated with lambda protein phosphatase (according to the manufacturer's instructions) or Usp2cc (37°C, 30 min in lysis buffer+0.5% Triton X-100+1 mM DTT). Samples were resuspended in Laemmli buffer+20 mM DTT after the final wash, separated by SDS-PAGE and transferred to PVDF membrane for western blotting. Western blots were performed by incubating membranes in PBST blocking buffer (PBS+1% Tween-20 supplemented with 5% non-fat dry milk), with subsequent primary and secondary antibody incubations in PBST+1.5% non-fat dry milk. Secondary antibodies conjugated with horseradish peroxidase were used to detect proteins bound to primary antibodies for enhanced chemiluminescence.
qRT-PCR
Total RNA was isolated from cells using RNeasy columns (Qiagen) and transcribed into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen), according to the manufacturer's protocol. Transcript levels were measured in triplicate reaction by qRT-PCR using GoTaq qPCR Master Mix (Promega) and Rotor-Gene Q PCR cycler (Qiagen), according to the manufacturer's instructions. Amplification efficiency for each primer pair was confirmed to be between 90 and 105%. Fold change in gene expression was calculated by the ΔΔCt method using β-actin as the reference gene. Primer sequences are listed in Table S3. All statistical analyses were performed in Prism 6 (GraphPad).
Sucrose gradient fractionation
Flp-In T-REx 293 cells were lysed in buffer containing 1% LMNG, pre-cleared as described above and the resulting lysate (1 mg total at 4 mg/ml) loaded onto 10-40% continuous sucrose gradients prepared in lysis buffer containing 1% LMNG using a Gradient Master (BioComp). Samples were centrifuged in a SW41 rotor (Beckman) at 39,000 rpm for 16 h at 4°C. To approximate the hydrodynamic radii of individual protein complexes, a mixture of purified, native gel filtration standards (Gel Filtration Markers Kit, MWGF1000, Sigma) was subjected to the same sucrose gradient sedimentation conditions for comparison. Thirteen equal fractions were collected (930 μl each) manually and proteins precipitated by addition of 240 µl trichloroacetic acid (TCA) to yield a final concentration of ∼20% (v/v), incubation at −20°C, and centrifugation at 4°C for 20 min, rinsed twice with ice-cold acetone and dried. Precipitated proteins were resuspended in 1× Laemmli buffer+20 mM DTT as described above for separation by SDS-PAGE.
35S-methionine/cysteine pulse-chase assay
Radiolabel pulse-chase assays of Flp-In T-REx 293 cells transiently expressing the model ERAD substrate NHK-HA were performed as described previously (Christianson et al., 2008). Briefly, cells were starved in DMEM lacking methionine and cysteine (Lonza)+10% dialysed FBS for 10 min, metabolically labelled by supplementing starvation medium with 35S-methionine/cysteine (EXPRE35S35S Protein Labelling Mix (PerkinElmer), 80 µCi/6 cm plate) for 15 min, rinsed three times in Dulbecco's PBS, and chased for indicated time points in DMEM supplemented with methionine and cysteine (50 mM each). Cells were lysed in buffer containing 1% Triton X-100 as above, and the detergent-soluble, post-nuclear lysates pre-cleared using mouse IgG conjugated to agarose beads followed by immunoprecipitation with anti-HA-agarose beads (Sigma). Bead-bound radiolabelled substrates were resuspended in Laemmli buffer+20 mM DTT, separated by SDS-PAGE and band intensities quantified by phosphorimager and QuantityOne software (Bio-Rad). After background subtraction, half-life (t1/2) values were calculated (GraphPad Prism 7) using data from at least three independent experiments, from which the average and s.e.m. values for each time point were determined. Significance was determined by Students t-test.
Bioinformatics analysis
Evolutionary conservation of Hrd1, FAM8A1 and Herp proteins were determined by obtaining a phylogenetic tree of organismal orthologs via TreeFam9 (www.treefam.org; Schreiber et al., 2014) and maximal likelihood parameters. The resulting multiple sequence alignment was analysed by ConSurf (http://consurf.tau.ac.il/2016/; Ashkenazy et al., 2016) and a conservation score determined for each amino acid position (value=1 to 10). A sliding window average over 10 amino acids was calculated and the resulting values divided by 10 to facilitate plotting on a 0 to 1 scale. Values for the intrinsic disorder of Hrd1, FAM8A1 and Herp were predicted using the Metadisorder analysis package (http://genesilico.pl/metadisorder/; Kozlowski and Bujnicki, 2012) and the MetadisorderMD values plotted. Predictions of α-helical content of protein sequences were made using the Phyre2 protein fold recognition algorithm (www.sbg.bio.ic.ac.uk/phyre2; Kelley et al., 2015).
Worm strains
C. elegans nematodes were grown according to standard protocols at 20°C. The Bristol strain N2 was used as wild-type (WT). Mutants and transgenic animals used in this study are listed as follows: N2; hhIs113 [Pnhx-2::cpl 1W32AY35A::YFP; Pmyo-2::mCherry]I cup-2&tag-353(gk443)I (3×outcrossed against WT), sel-11 (nDf59) V.sqt-3(sc8); sel-1 (e1948)V; F48B9.8 (gk272969) (4× outcrossed against WT).
Worm RNAi
RNAi was performed using nematode growth medium (NGM)-RNAi plates containing 2 mM isopropyl-b-D-thiogalactopyranoside (IPTG) seeded with the appropriate HT115 RNAi bacteria obtained from the Ahringer or ORFeome WS112 libraries (Geneservice Ltd, available via Source BioScience, UK, cat. # 3318 and 3320). The empty feeding vector pL4440 was used as control. Synchronised L1 larvae were fed with indicated RNAi for 72 h at 20°C.
Microscopy
Microscopy of RNAi-treated worms was performed using a Leica M80 Stereomicroscope. Processing of selected pictures was done with Adobe Photoshop CS4.
Preparation of worm lysates and immunoblots
Whole-worm proteins were extracted by sonication in denaturing SDS buffer, separated with SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked with Roti-Block (Roth) and incubated with primary antibodies against tubulin (DM1A; Sigma) or GFP (Clontech). Donkey anti-mouse IR-Dye 800 (LI-COR) was used as secondary antibody. Detection was carried out using a LI-COR Odyssey infrared imaging system.
Lifespan analysis
Adult lifespan analysis was performed at 20°C on E. coli OP50; the young adult stage was defined as day 0; 43-50 animals were used per strain and replication and scored every day. Worms that had undergone vulval bursting, internal hatching or crawling off the plates were censored. Four replications for each strain were performed. Statistical analyses were performed using GraphPad Prism 6 with the log-rank (Mantel–Cox) method.
Supplementary Material
Acknowledgements
We would like to thank the following individuals for the careful reading and commenting on the manuscript: James Olzmann, Skirmantas Kriaucionis, Mary Muers and Federica Lari.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: T.H., J.C.C.; Methodology: J.S., J.C.C.; Validation: J.S., D.A., J.C.C.; Formal analysis: D.A., M.A.Q., A.G., L.-S.D., J.C.C.; Investigation: J.S., D.A., M.A.Q., A.G., L.-S.D., E.J.F., N.V., Y.H., J.C.C.; Resources: J.C.; Data curation: J.S., J.C.C.; Writing - original draft: L.-S.D., J.C.C.; Writing - review & editing: D.A., E.J.F., T.H., J.C.C.; Visualization: J.S., D.A., M.A.Q., A.G., L.-S.D., J.C.C.; Supervision: T.H., J.C.C.; Project administration: T.H., J.C.C.; Funding acquisition: T.H., J.C.C.
Funding
D.A. is supported by a European Molecular Biology Organization (EMBO) Long Term Fellowship (ALTF 60-2015). J.C.C. is supported by a grant from the Medical Research Council (MR/L001209/1) and by the Ludwig Institute for Cancer Research. T.H. is supported by funding from the Deutsche Forschungsgemeinschaft (CECAD, SFB635 and DIP8 grant 2014376) and the European Research Council (consolidator grant 616499). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.206847.supplemental
References
- Amano T., Yamasaki S., Yagishita N., Tsuchimochi K., Shin H., Kawahara K.-I., Aratani S., Fujita H., Zhang L., Ikeda R. et al. (2003). Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel pathogenic factor for arthropathy. Genes Dev. 17, 2436-2449. 10.1101/gad.1096603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T. and Ben-Tal N. (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344-W350. 10.1093/nar/gkw408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. T., Catanzariti A.-M., Karunasekara Y., Soboleva T. A., Sharwood R., Whitney S. and Board P. G. (2005). Using deubiquitylating enzymes as research tools. Meth. Enzymol. 398, 540-554. 10.1016/S0076-6879(05)98044-0 [DOI] [PubMed] [Google Scholar]
- Baldridge R. D. and Rapoport T. A. (2016). Autoubiquitination of the Hrd1 ligase triggers protein retrotranslocation in ERAD. Cell 166, 394-407. 10.1016/j.cell.2016.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell J. C. A. and Jakob U. (2012). Conditional disorder in chaperone action. Trends Biochem. Sci. 37, 517-525. 10.1016/j.tibs.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R., Pertel T., Luban J. and Molinari M. (2008). A dual task for the Xbp1-responsive OS-9 variants in the mammalian endoplasmic reticulum: inhibiting secretion of misfolded protein conformers and enhancing their disposal. J. Biol. Chem. 283, 16446-16454. 10.1074/jbc.M802272200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma W., Nielsen S. V., Lindorff-Larsen K., Hartmann-Petersen R. and Ellgaard L. (2016). Bioinformatics analysis identifies several intrinsically disordered human E3 ubiquitin-protein ligases. PeerJ 4, e1725 10.7717/peerj.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr M. L., Cano F., Svobodova S., Boyle L. H., Boname J. M. and Lehner P. J. (2011). HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. USA 108, 2034-2039. 10.1073/pnas.1016229108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr M. L., van den Boomen D. J. H., Bye H., Antrobus R., Wiertz E. J. and Lehner P. J. (2013). MHC class I molecules are preferentially ubiquitinated on endoplasmic reticulum luminal residues during HRD1 ubiquitin E3 ligase-mediated dislocation. Proc. Natl. Acad. Sci. USA 110, 14290-14295. 10.1073/pnas.1303380110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. M. and Hampton R. Y. (2010). Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase. J. Biol. Chem. 285, 5146-5156. 10.1074/jbc.M109.067876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V. and Rapoport T. A. (2006). Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126, 361-373. 10.1016/j.cell.2006.05.043 [DOI] [PubMed] [Google Scholar]
- Carvalho P., Stanley A. M. and Rapoport T. A. (2010). Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143, 579-591. 10.1016/j.cell.2010.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zhong Y., Wu Y., Liu L., Wang P., Liu R., Cui F., Li Q., Yang X., Fang S. et al. (2016). HRD1-mediated ERAD tuning of ER-bound E2 is conserved between plants and mammals. Nat. Plants 2, 16094 10.1038/nplants.2016.94 [DOI] [PubMed] [Google Scholar]
- Christensen L. C., Jensen N. W., Vala A., Kamarauskaite J., Johansson L., Winther J. R., Hofmann K., Teilum K. and Ellgaard L. (2012). The Human Selenoprotein VCP-interacting Membrane Protein (VIMP) Is Non-globular and Harbors a Reductase Function in an Intrinsically Disordered Region. J. Biol. Chem. 287, 26388-26399. 10.1074/jbc.M112.346775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J. C. and Ye Y. (2014). Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat. Struct. Mol. Biol. 21, 325-335. 10.1038/nsmb.2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J. C., Shaler T. A., Tyler R. E. and Kopito R. R. (2008). OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10, 272-282. 10.1038/ncb1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J. C., Olzmann J. A., Shaler T. A., Sowa M. E., Bennett E. J., Richter C. M., Tyler R. E., Greenblatt E. J., Harper J. W. and Kopito R. R. (2012). Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 14, 93-105. 10.1038/ncb2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen J. H. L., Kundrat L. and Ploegh H. L. (2012). Protein quality control in the ER: balancing the ubiquitin checkbook. Trends Cell Biol. 22, 22-32. 10.1016/j.tcb.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier J. H., Tamura T., Sunryd J. C. and Hebert D. N. (2009). EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex. Mol. Cell 34, 627-633. 10.1016/j.molcel.2009.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J. and Joazeiro C. A. P. (2009). RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399-434. 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- Eura Y., Yanamoto H., Arai Y., Okuda T., Miyata T. and Kokame K. (2012). Derlin-1 deficiency is embryonic lethal, Derlin-3 deficiency appears normal, and Herp deficiency is intolerant to glucose load and ischemia in mice. PLoS ONE 7, e34298 10.1371/journal.pone.0034298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco A. B., Singh R., Li S., Vani A. K., Yang L., Munroe R. J., Diaferia G., Cardano M., Biunno I., Qi L. et al. (2010). Deficiency of suppressor enhancer Lin12 1 like (SEL1L) in mice leads to systemic endoplasmic reticulum stress and embryonic lethality. J. Biol. Chem. 285, 13694-13703. 10.1074/jbc.M109.085340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco A. B., Singh R., Sha H., Yan X., Qi L., Lei X. and Long Q. (2011). Haploid insufficiency of suppressor enhancer Lin12 1-like (SEL1L) protein predisposes mice to high fat diet-induced hyperglycemia. J. Biol. Chem. 286, 22275-22282. 10.1074/jbc.M111.239418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. G., Swarbrick G. M., Bays N. W., Cronin S. R., Wilhovsky S., Seelig L., Kim C. and Hampton R. Y. (2000). Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 151, 69-82. 10.1083/jcb.151.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R., Sommer T. and Jarosch E. (2006). The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 25, 1827-1835. 10.1038/sj.emboj.7601088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt E. J., Olzmann J. A. and Kopito R. R. (2011). Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat. Struct. Mol. Biol. 18, 1147-1152. 10.1038/nsmb.2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotzke J. E., Lu Q. and Cresswell P. (2013). Deglycosylation-dependent fluorescent proteins provide unique tools for the study of ER-associated degradation. Proc. Natl. Acad. Sci. USA 110, 3393-3398. 10.1073/pnas.1300328110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R. Y., Gardner R. G. and Rine J. (1996). Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell 7, 2029-2044. 10.1091/mbc.7.12.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S. C., Hanna J., Hirsch C., Volkwein C., Schütz A., Heinemann U., Sommer T. and Jarosch E. (2009). Usa1 Functions as a Scaffold of the HRD-Ubiquitin Ligase. Mol. Cell 36, 782-793. 10.1016/j.molcel.2009.10.015 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Wada I., Okawa K. and Nagata K. (2008). Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 283, 20914-20924. 10.1074/jbc.M709336200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.-H., Chu Y.-R., Ye Y. and Chen X. (2014). Role of HERP and a HERP-related protein in HRD1-dependent protein degradation at the endoplasmic reticulum. J. Biol. Chem. 289, 4444-4454. 10.1074/jbc.M113.519561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J., Walczak C. P., Shaler T. A., Olzmann J. A., Zhang L., Elias J. E. and Kopito R. R. (2017). Characterization of protein complexes of the endoplasmic reticulum-associated degradation E3 ubiquitin ligase Hrd1. J. Biol. Chem. 292, 9104-9116. 10.1074/jbc.M117.785055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Sim H. J., Song E. K., Lee H., Ha S. C., Jun Y., Park T. J. and Lee C. (2016). Crystal structure of SEL1L: Insight into the roles of SLR motifs in ERAD pathway. Sci. Rep. 6, 20261 10.1038/srep20261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K., Xie W. and Ng D. T. W. (2010). Modularity of the Hrd1 ERAD complex underlies its diverse client range. J. Cell Biol. 188, 707-716. 10.1083/jcb.200907055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M., Ishiguro M., Niinuma Y., Uesugi M. and Nomura Y. (2002). Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 532, 147-152. 10.1016/S0014-5793(02)03660-8 [DOI] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N. and Sternberg M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845-858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-Y., Kim E., Yoon S. K. and Yoon J.-B. (2008). Herp enhances ER-associated protein degradation by recruiting ubiquilins. Biochem. Biophys. Res. Commun. 369, 741-746. 10.1016/j.bbrc.2008.02.086 [DOI] [PubMed] [Google Scholar]
- Klemm E. J., Spooner E. and Ploegh H. L. (2011). Dual role of ancient ubiquitous protein 1 (AUP1) in lipid droplet accumulation and endoplasmic reticulum (ER) protein quality control. J. Biol. Chem. 286, 37602-37614. 10.1074/jbc.M111.284794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Finger A., Braun T., Hellmuth K. and Wolf D. H. (1996). Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 15, 753-763. [PMC free article] [PubMed] [Google Scholar]
- Kny M., Standera S., Hartmann-Petersen R., Kloetzel P.-M. and Seeger M. (2011). Herp Regulates Hrd1-mediated Ubiquitylation in a Ubiquitin-like Domain-dependent Manner. J. Biol. Chem. 286, 5151-5156. 10.1074/jbc.M110.134551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokame K., Agarwala K. L., Kato H. and Miyata T. (2000). Herp, a new ubiquitin-like membrane protein induced by endoplasmic reticulum stress. J. Biol. Chem. 275, 32846-32853. 10.1074/jbc.M002063200 [DOI] [PubMed] [Google Scholar]
- Kozlowski L. P. and Bujnicki J. M. (2012). MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics 13, 111 10.1186/1471-2105-13-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan M. N., Ng A., Sukumaran B., Gilfoy F. D., Uchil P. D., Sultana H., Brass A. L., Adametz R., Tsui M., Qian F. et al. (2008). RNA interference screen for human genes associated with West Nile virus infection. Nature 455, 242-245. 10.1038/nature07207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley B. N. and Ploegh H. L. (2005). Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA 102, 14296-14301. 10.1073/pnas.0505014102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard J. R., Schulman B. A. and Harper J. W. (2013). Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 14, 1050-1061. 10.1038/embor.2013.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Dang Y., Wu Y., Jia G., Anaya E., Zhang J., Abraham S., Choi J.-G., Shi G., Qi L. et al. (2015). A CRISPR-based screen identifies genes essential for west-nile-virus-induced cell death. Cell Rep. 12, 673-683. 10.1016/j.celrep.2015.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert M., Sommer T. and Jarosch E. (2014). Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat. Cell Biol. 16, 77-86. 10.1038/ncb2882 [DOI] [PubMed] [Google Scholar]
- Menon M. B., Tiedje C., Lafera J., Ronkina N., Konen T., Kotlyarov A. and Gaestel M. (2013). Endoplasmic reticulum-associated ubiquitin-conjugating enzyme Ube2j1 is a novel substrate of MK2 (MAPKAP kinase-2) involved in MK2-mediated TNFα production. Biochem. J. 456, 163-172. 10.1042/BJ20130755 [DOI] [PubMed] [Google Scholar]
- Morreale G., Conforti L., Coadwell J., Wilbrey A. L. and Coleman M. P. (2009). Evolutionary divergence of valosin-containing protein/cell division cycle protein 48 binding interactions among endoplasmic reticulum-associated degradation proteins. FEBS J. 276, 1208-1220. 10.1111/j.1742-4658.2008.06858.x [DOI] [PubMed] [Google Scholar]
- Mueller B., Lilley B. N. and Ploegh H. L. (2006). SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J. Cell Biol. 175, 261-270. 10.1083/jcb.200605196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B., Klemm E. J., Spooner E., Claessen J. H. and Ploegh H. L. (2008). SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. USA 105, 12325-12330. 10.1073/pnas.0805371105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Piffanelli P., Devoto A., Miklis M., Elliott C., Ortmann B., Schulze-Lefert P. and Panstruga R. (2005). Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell 17, 149-163. 10.1105/tpc.104.026625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutzner A., Neutzner M., Benischke A.-S., Ryu S.-W., Frank S., Youle R. J. and Karbowski M. (2011). A systematic search for endoplasmic reticulum (ER) membrane-associated RING finger proteins identifies Nixin/ZNRF4 as a regulator of calnexin stability and ER homeostasis. J. Biol. Chem. 286, 8633-8643. 10.1074/jbc.M110.197459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh R. S., Bai X. and Rommens J. M. (2006). Human homologs of Ubc6p ubiquitin-conjugating enzyme and phosphorylation of HsUbc6e in response to endoplasmic reticulum stress. J. Biol. Chem. 281, 21480-21490. 10.1074/jbc.M601843200 [DOI] [PubMed] [Google Scholar]
- Okuda-Shimizu Y. and Hendershot L. M. (2007). Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol. Cell 28, 544-554. 10.1016/j.molcel.2007.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann J. A., Kopito R. R. and Christianson J. C. (2013a). The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb. Perspect. Biol. 5, a013185-a013185 10.1101/cshperspect.a013185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann J. A., Richter C. M. and Kopito R. R. (2013b). Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc. Natl. Acad. Sci. USA 110, 1345-1350. 10.1073/pnas.1213738110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T., Kaneko M., Okuma Y., Orba Y., Nagashima K., Takahashi R., Fujitani N., Matsumura S., Hata A., Kubota K. et al. (2006). A ubiquitin ligase HRD1 promotes the degradation of Pael receptor, a substrate of Parkin. J. Neurochem. 99, 1456-1469. 10.1111/j.1471-4159.2006.04155.x [DOI] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A. and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281-2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer J., Appenzeller-Herzog C., Johansson L., Bodenmiller B., Hartmann-Petersen R. and Ellgaard L. (2009). A luminal flavoprotein in endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. USA 106, 14831-14836. 10.1073/pnas.0900742106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer J., Hansen H. G., Appenzeller-Herzog C., Johansson L. and Ellgaard L. (2011). Identification of the PDI-family member ERp90 as an interaction partner of ERFAD. PLoS ONE 6, e17037 10.1371/journal.pone.0017037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J. C., Fredrickson E. K., Oeser M. L., Garrett-Engele C. M., Locke M. N., Richardson L. A., Nelson Z. W., Hetrick E. D., Milac T. I., Gottschling D. E. et al. (2011). Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol. Cell 41, 93-106. 10.1016/j.molcel.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoebel S., Mi W., Stein A., Ovchinnikov S., Pavlovicz R., DiMaio F., Baker D., Chambers M. G., Su H., Li D. et al. (2017). Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature 548, 352-355. 10.1038/nature23314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber F., Patricio M., Muffato M., Pignatelli M. and Bateman A. (2014). TreeFam v9: a new website, more species and orthology-on-the-fly. Nucleic Acids Res. 42, D922-D925. 10.1093/nar/gkt1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A., Standera S., Buerger E., Kikkert M., van Voorden S., Wiertz E., Koning F., Kloetzel P.-M. and Seeger M. (2005). The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J. Mol. Biol. 354, 1021-1027. 10.1016/j.jmb.2005.10.020 [DOI] [PubMed] [Google Scholar]
- Stein A., Ruggiano A., Carvalho P. and Rapoport T. A. (2014). Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell 158, 1375-1388. 10.1016/j.cell.2014.07.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Shi G., Han X., Francisco A. B., Ji Y., Mendonça N., Liu X., Locasale J. W., Simpson K. W., Duhamel G. E. et al. (2014). Sel1L is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. Proc. Natl. Acad. Sci. USA 111, E582-E591. 10.1073/pnas.1318114111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler R. E., Pearce M. M. P., Shaler T. A., Olzmann J. A., Greenblatt E. J. and Kopito R. R. (2012). Unassembled CD147 is an endogenous endoplasmic reticulum-associated degradation substrate. Mol. Biol. Cell 23, 4668-4678. 10.1091/mbc.E12-06-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y. and Nagata K. (2008). ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321, 569-572. 10.1126/science.1159293 [DOI] [PubMed] [Google Scholar]
- Yagishita N., Ohneda K., Amano T., Yamasaki S., Sugiura A., Tsuchimochi K., Shin H., Kawahara K.-I., Ohneda O., Ohta T. et al. (2005). Essential role of synoviolin in embryogenesis. J. Biol. Chem. 280, 7909-7916. 10.1074/jbc.M410863200 [DOI] [PubMed] [Google Scholar]
- Yamasaki S., Yagishita N., Sasaki T., Nakazawa M., Kato Y., Yamadera T., Bae E., Toriyama S., Ikeda R., Zhang L. et al. (2006). Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase “Synoviolin”. EMBO J. 26, 113-122. 10.1038/sj.emboj.7601490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Liu W., Zhang H., Liu C., Shang Y., Ye Y., Zhang X. and Li W. (2014). Ube2g2-gp78-mediated HERP polyubiquitylation is involved in ER stress recovery. J. Cell Sci. 127, 1417-1427. 10.1242/jcs.135293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Meyer H. H. and Rapoport T. A. (2001). The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652-656. 10.1038/414652a [DOI] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E. and Rapoport T. A. (2005). Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA 102, 14132-14138. 10.1073/pnas.0505006102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.