ABSTRACT
Vinexin, c-Cbl associated protein (CAP) and Arg-binding protein 2 (ArgBP2) constitute an adaptor protein family called the vinexin (SORBS) family that is targeted to focal adhesions (FAs). Although numerous studies have focused on each of the SORBS proteins and partially elucidated their involvement in mechanotransduction, a comparative analysis of their function has not been well addressed. Here, we established mouse embryonic fibroblasts that individually expressed SORBS proteins and analysed their functions in an identical cell context. Both vinexin-α and CAP co-localized with vinculin at FAs and promoted the appearance of vinculin-rich FAs, whereas ArgBP2 co-localized with α-actinin at the proximal end of FAs and punctate structures on actin stress fibers (SFs), and induced paxillin-rich FAs. Furthermore, both vinexin-α and CAP contributed to extracellular matrix stiffness-dependent vinculin behaviors, while ArgBP2 stabilized α-actinin on SFs and enhanced intracellular contractile forces. These results demonstrate the differential roles of SORBS proteins in mechanotransduction.
KEY WORDS: Vinexin, CAP, ArgBP2, Focal adhesion, Actin cytoskeleton, Mechanotransduction
Summary: Comparing the functions of vinexin family (SORBS) proteins reveals that vinexin-α and CAP regulate vinculin behavior depending on the stiffness of the extracellular matrix, while ArgBP2 plays a role in generating greater contractile forces.
INTRODUCTION
Extracellular matrix (ECM) stiffness is a critical determinant of cell fates, such as migration (Pelham and Wang, 1997; Lo et al., 2000; Peyton and Putnam, 2005), proliferation (Wang et al., 2000; Klein et al., 2009) and differentiation (Engler et al., 2004, 2006). Understanding the mechanisms underlying mechanotransduction, by which cells convert mechanical properties into biochemical signals, has become important in tumour therapies (Paszek et al., 2005; Ulrich et al., 2009) and tissue engineering (Engler et al., 2006).
Cell–ECM adhesions, called focal adhesions (FAs), contain ECM receptor integrins and cytoplasmic scaffolding proteins and serve as the mechanical linkage between the ECM and force-generating actin stress fibers (SFs) (Geiger et al., 2001). This mechanical linkage acts as a ‘molecular clutch’ to transmit the force derived from non-muscle myosin-II-dependent contraction to the ECM. Cells on more rigid substrates exert greater contractile forces than those on soft substrates (Hoffman et al., 2011; Roca-Cusachs et al., 2012; LaCroix et al., 2015). These alterations can lead to stiffness-dependent biochemical signals.
Among the numerous FA scaffolding proteins, vinculin is one of the main ‘clutch’ molecules that can regulate force transmission. Vinculin consists of an N-terminal head region and a C-terminal tail region separated by a flexible proline-rich linker region (Bakolitsa et al., 2004; Borgon et al., 2004). Disruption of the head–tail intramolecular interaction induces a conformational change in vinculin to an active form exhibiting a high affinity for F-actin (Johnson and Craig, 1995; Cohen et al., 2005; Chen et al., 2006). As vinculin binds simultaneously to both the talin–integrin complex and F-actin, this protein plays a key role in regulating FA formation (Humphries et al., 2007) and generating traction force (Thievessen et al., 2013). Conversely, myosin II activity or force promotes vinculin recruitment to FAs and immobilization at FAs, suggesting that vinculin harbours force-dependent binding to its partners (Pasapera et al., 2010; Kuo et al., 2011; Wolfenson et al., 2011; Carisey et al., 2013; Dumbauld et al., 2013). In our previous study, we showed that myosin II activity and rigid substrates promote the interaction of the vinculin linker region with another FA protein vinexin-α (also known as SORBS3), which is indispensable for vinculin resistance to cytoskeleton stabilization (CSK) buffer treatment and vinculin immobilization at FAs. In addition, vinexin-α induces a conformational change in vinculin in vitro and is required for the stiffness-dependent regulation of cell migration, indicating that the vinculin–vinexin-α interaction functions as a mechanosensor of ECM stiffness (Yamashita et al., 2014).
Vinexin has several splice variants: vinexin-α contains a sorbin homology (SoHo) domain and three Src homology 3 (SH3) domains, whereas vinexin-β contains only three SH3 domains. Vinexin-α but not -β accumulates F-actin at FAs and functions as a mechanosensor (Kioka et al., 1999; Takahashi et al., 2005; Yamashita et al., 2014). Despite these functions, vinexin knockout (KO) mice merely exhibit a delay in cutaneous wound healing (Kioka et al., 2010) and an increased sensitivity to cardiac hypertrophy (Chen et al., 2013) without other severe phenotypes, suggesting compensatory mechanisms for the loss of vinexin expression. Vinexin and two other SORBS proteins, c-Cbl-associated protein (CAP)/ponsin (also known as SORBS1) and Arg-binding protein 2 (ArgBP2) (also known as SORBS2), constitute an adaptor protein family, also known as the vinexin (SORBS) family (Kioka et al., 2002). These proteins contain the same domain structures (Fig. 1A). Each of the SORBS proteins localizes at FAs in a cell context-dependent manner (Mandai et al., 1999; Cestra et al., 2005; Rönty et al., 2005), whereas CAP and ArgBP2 localize on actin SFs (Wang et al., 1997; Ribon et al., 1998a). Recent advances in proteomics have revealed that both vinexin-α and CAP are consensus adhesome proteins, while ArgBP2 is a conditional adhesome protein among more than 2400 proteins (Horton et al., 2015). These three SORBS proteins share binding partners, including vinculin (Kioka et al., 1999; Mandai et al., 1999; Cestra et al., 2005), the tyrosine kinase c-Abl (Wang et al., 1997; Lin et al., 2001; Mitsushima et al., 2006b), the E3 ubiquitin-protein ligase c-Cbl (Ribon et al., 1998b; Soubeyran et al., 2003; Mitsushima et al., 2006c) and the lipid raft protein flotillin (Kimura et al., 2001; Haglund et al., 2004). Among the vinexin family, only ArgBP2 is reported to interact with the actin crosslinking protein α-actinin (Rönty et al., 2005; Anekal et al., 2015). In addition to the function of vinexin-α as a mechanosensor, several studies have also demonstrated that SORBS proteins play roles in mechanotransduction: ArgBP2 increases phosphorylation of myosin regulatory light chain II (MRLC) (Martin et al., 2013), and the only orthologue in Drosophila, dCAP, regulates the assembly and function of tension-sensitive organs (Bharadwaj et al., 2013). However, few studies have analysed comparatively the mammalian SORBS proteins.
Fig. 1.

Establishment of SORBS-re-expressing TKO MEFs. (A) Schematic diagram of the domain structure of SORBS proteins (vinexin-α, CAP and ArgBP2). SORBS proteins share one SoHo domain and three SH3 domains. (B) TKO MEFs re-expressing each SORBS protein were lysed and subjected to SDS-PAGE. Re-expression of SORBS proteins in TKO MEFs was examined by immunoblotting using the indicated antibodies. Vinculin was used as a loading control. The lysate of astrocytes served as a reference sample, which expressed all SORBS proteins. (C–E) SORBS-re-expressing cells were cultured on coverslips for 24 h. Cell membranes were stained with CellMask Orange and photographed (C). Cell area (D) and aspect ratio (E) were quantified from images of ninety cells from three separate experiments. N.S., not significant in Kruskal–Wallis ANOVA. Scale bars: 50 µm.
In the present study, we establish mouse embryonic fibroblasts (MEFs), in which all SORBS proteins are depleted, followed by re-expressing each SORBS protein to compare the function of individual SORBS proteins. We show that stiffness-dependent behaviors of vinculin require either vinexin-α or CAP but not ArgBP2. Meanwhile, ArgBP2 immobilizes α-actinin on SFs and enhances the contractile force but neither vinexin-α nor CAP does. Altogether, these results suggest that SORBS proteins play distinct roles in sensing ECM stiffness and contractile force generation.
RESULTS
Establishment of SORBS-re-expressing cells
All cells express two or more SORBS proteins accompanied by multiple splice variants, making it difficult to compare the unique functions of the SORBS proteins. We first examined the expression of each SORBS protein in wild-type (WT) MEFs and found that two splice variants of vinexin (vinexin-α and β) and CAP were expressed, but ArgBP2 was not detected in WT MEFs (Fig. 1B; Fig. S1A). To establish SORBS triple-depleted cells, we first established vinexin/CAP double-depleted cells by generating vinexin/CAP double KO mice, which were viable and fertile. Spontaneously immortalized embryonic fibroblast cells (referred to as DKO MEFs) from double KO mice expressed neither vinexins nor CAP (Fig. S1A). As the plasticity of ArgBP2 expression has been reported (Anekal et al., 2015), it remained possible that there was a small population of DKO MEFs expressing ArgBP2, even though ArgBP2 expression was not detected by immunoblotting. To overcome this possibility, a lentivirus expressing shRNA against ArgBP2 was transduced into DKO MEFs to stably knock down residual ArgBP2. Because ArgBP2 was undetectable in ArgBP2 knockdown DKO MEFs (Fig. S1A), we tested the effectiveness of our designed shRNA against ArgBP2 using the mouse mesenchymal cell line ATDC5. We differentiated the ATDC5 cells into chondrocytes, which expressed ArgBP2. We found that our shRNA successfully knocked down endogenous ArgBP2 expression (Fig. S1B). We designated the ArgBP2 knockdown DKO MEFs as SORBS triple-depleted cells and refer to them as TKO MEFs.
Cells re-expressing each one of the SORBS proteins were then established from TKO MEFs, and stable expression of the individual SORBS proteins in the TKO MEFs was confirmed by immunoblotting (Fig. 1B). TKO MEF/vinexin-α expressed levels of vinexin-α comparable to WT MEFs. The expression level of CAP in TKO MEF/CAP was higher than that of WT MEFs, yet comparable to that of rat primary astrocytes. The level of ArgBP2 in TKO MEF/ArgBP2 was also comparable to that of astrocytes. These results indicate that the expression levels of SORBS proteins are within the physiological range. Hereafter, these four stable cell lines, TKO MEF/mock, TKO MEF/vinexin-α, TKO MEF/CAP and TKO MEF/ArgBP2, are collectively referred to as SORBS-re-expressing cells.
We investigated the effects of SORBS proteins on cell spreading by labelling the cell surface membrane with CellMask Orange (Fig. 1C). No significant differences in cell area (Fig. 1D) and aspect ratio (Fig. 1E) were found among these cells, suggesting that SORBS proteins do not affect cell spreading in MEFs.
Subcellular localization of SORBS proteins in MEFs
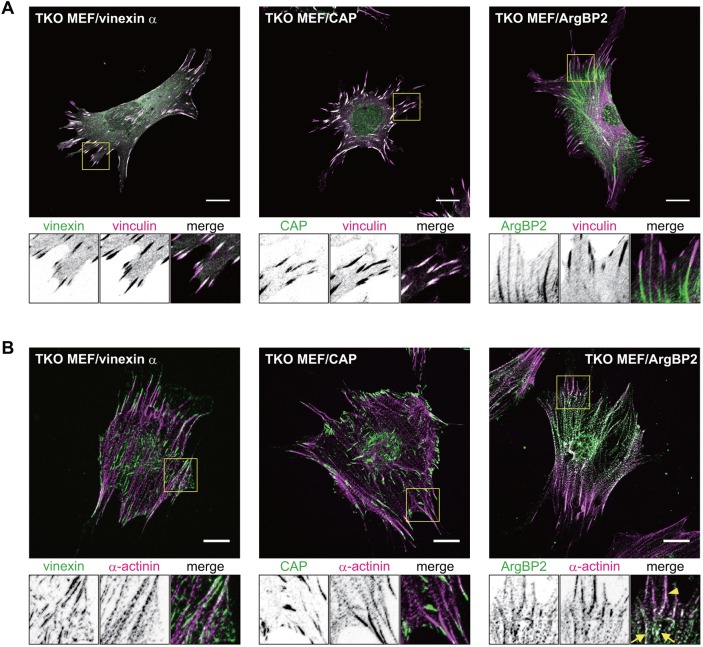
Several studies have suggested that subcellular localization patterns of SORBS proteins depend on cell type. We examined the localization of SORBS proteins in MEFs by immunostaining. Each of the antibodies specifically recognized a target SORBS protein (Fig. 2A; Fig. S1C-E). Both vinexin-α and CAP were enriched at FAs and clearly co-localized with vinculin, whereas ArgBP2 was enriched at the proximal end of FAs and slightly co-localized with vinculin (Fig. 2A). ArgBP2 has been reported to be co-localized with α-actinin along actin SFs in non-muscle cells (Anekal et al., 2015), prompting us to test for co-localization with α-actinin. As expected, ArgBP2 co-localized with α-actinin at the proximal end of FAs and the punctate structures on SFs, whereas vinexin-α or CAP hardly co-localized with α-actinin (Fig. 2B). To confirm further the localization of SORBS proteins, Green Fluorescent Protein (GFP)-tagged SORBS proteins were also stably transduced into TKO MEFs and observed using 3D-structured illumination microscopy (SIM) (Fig. S2). A maximum intensity projection provided images similar to those acquired by conventional confocal microscopy (Fig. 2; Fig. S2A-C), indicating a localization pattern for vinexin-α and CAP distinct from that observed for ArgBP2. Interestingly, both vinexin-α and CAP co-localized with the dorsal fraction of vinculin at FAs by 3D volume visualization (Fig. S2A′-C′), suggesting regulation of the nanoscale localization of vinexin-α or CAP.
Fig. 2.
Localization of SORBS proteins in re-expressing cells. (A,B) SORBS-re-expressing cells cultured on coverslips for 24 h were immunostained using the indicated antibodies including anti-vinculin (A) or anti-α-actinin antibodies (B). Representative images are displayed from three independent experiments. The FA-containing areas indicated by yellow boxes are cropped and displayed at the bottom of the figure. ArgBP2 and α-actinin were co-localized at FAs (arrowhead) and on SFs (arrow). Scale bars: 20 µm. See also Figs S1C-E and S2.
SORBS proteins differently affect the molecular composition of FAs
Our previous study showed that vinexin-α alters the molecular composition of FAs (Yamashita et al., 2014). Therefore, we investigated the effect of SORBS proteins on the distribution of FA proteins by immunostaining with anti-vinculin or anti-paxillin antibodies (Fig. 3A,C). Both vinculin and paxillin were observed at FAs in all SORBS-re-expressing cells. Interestingly, the quantification of images showed that while both vinexin-α and CAP significantly increased the number of vinculin-positive FAs per cell compared with the mock control, ArgBP2 only had a minor effect (Fig. 3B). A similar trend was observed in the integrated density, the product of area and mean intensity (Fig. S3A). Conversely, vinexin-α and CAP decreased both the number and the integrated densities of paxillin-positive FAs per cell, whereas ArgBP2 slightly increased these structures (Fig. 3D; Fig. S3B). Nevertheless, immunoblotting showed that total expression levels of vinculin and paxillin were comparable among SORBS-re-expressing cells (Fig. S1F). To corroborate that vinexin-α and CAP modulate the composition of FA, paxillin or talin was simultaneously immunostained with vinculin and the ratios to vinculin in individual FAs were determined (Fig. 3E-H). Consistent with the number and integrated density of paxillin-positive FA per cell, vinexin-α and CAP decreased the ratio of paxillin to vinculin of individual FAs, whereas they did not show a significant effect on the ratio of talin to vinculin. ArgBP2 slightly decreased the ratio of paxillin to vinculin in contrast to the number of paxillin-positive FAs. Collectively, these findings demonstrate that SORBS proteins differentially regulate FA protein composition; both vinexin-α and CAP increase the number of vinculin-positive FAs and make them paxillin-poor, but ArgBP2 has only a minor effect.
Fig. 3.
Both vinexin-α and CAP induce vinculin-rich FAs, whereas ArgBP2 induces paxillin-rich FAs. SORBS-re-expressing cells cultured on coverslips were immunostained using antibodies indicated as follows: anti-vinculin (hVIN) (A,B), anti-paxillin (C–F), anti-talin (G,H) and anti-vinculin (ab73412) (E–H). Ninety (A–D) or thirty (E–H) individual cells from three separate experiments were photographed for each condition. Scale bars: 20 µm. The number of vinculin-positive FAs and paxillin-positive FAs per cell were quantified from the images in A and C, respectively (B,D). The intensity ratio of paxillin and talin to vinculin within vinculin-positive FAs was quantified from the images in E and G, respectively (F,H); P-value calculated by Mann–Whitney U-tests; n.s., not significant. See also Fig. S1F.
The ECM stiffness-dependent CSK resistance of vinculin requires either vinexin-α or CAP
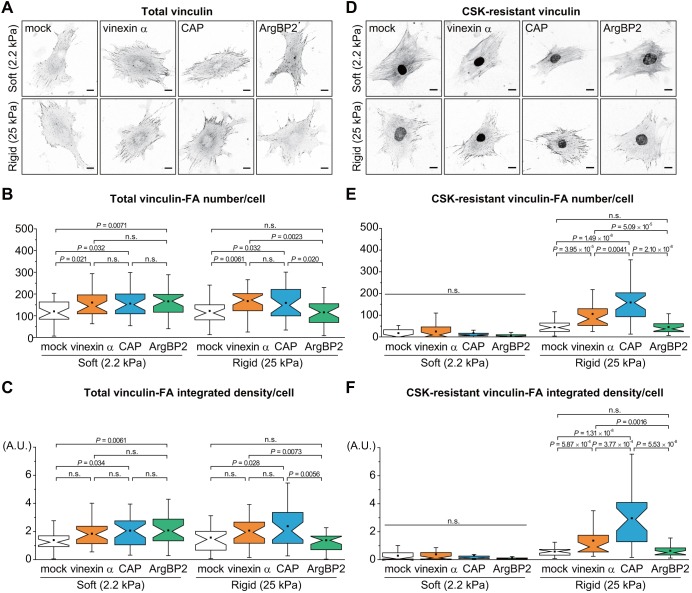
We next examined the effect of SORBS proteins on the ECM stiffness-dependent localization of vinculin by immunostaining cells cultured on polyacrylamide gel substrates with different levels of stiffness (Fig. 4A). Vinculin was similarly localized at FAs both on soft (2.2 kPa) and rigid (25 kPa) substrates in mock control cells. All SORBS proteins promoted vinculin localization at FAs on soft substrates (Fig. 4B,C), whereas vinexin-α and CAP but not ArgBP2 prompted vinculin to target to FAs on rigid substrates (25 kPa). This finding is similar to results obtained from cells on extremely rigid (glass) substrates (Fig. 3B,C). Interestingly, ArgBP2 increased vinculin localization at FAs in cells treated with low concentration of blebbistatin even on rigid substrates (glass) (Fig. S3M). These results suggest that rigid substrates or high contractile force mask the effect of ArgBP2 on recruiting vinculin to FAs and further that CAP may regulate vinculin behaviors in a manner similar to vinexin-α.
Fig. 4.
Both vinexin-α and CAP but not ArgBP2 contribute to CSK-resistant vinculin on rigid substrates. SORBS-re-expressing cells were cultured on polyacrylamide gel substrates with 2.2 kPa (soft) and 25 kPa (rigid) stiffness. Cells were treated without (A–C) or with (D–F) CSK, followed by immunostaining using an anti-vinculin antibody. Thirty individual cells from three separate experiments were photographed for each condition (A,D). Scale bars: 20 µm. The number (B,E) and integrated density (C,F) of total or CSK-resistant vinculin-positive FAs per cell were quantified from the images. Note that non-specific staining was observed in the nucleus after CSK treatment. n=30; P-value was calculated by Mann–Whitney U-test; n.s., not significant. See also Fig. S3C-E.
The cytoskeleton stabilization buffer (CSK)-resistant vinculin, which represents the fraction of vinculin tightly bound to the cytoskeleton, is induced by rigid substrates and its interaction with vinexin-α (Yamashita et al., 2014). To investigate the effects of SORBS proteins on CSK-resistant vinculin, SORBS-re-expressing cells were treated with CSK followed by immunostaining (Fig. 4D). CSK-resistant vinculin was hardly detected in any cells on soft substrates, suggesting that the vinculin localized at FAs on soft substrates are not tightly bound to the cytoskeleton regardless of the expression of SORBS proteins. In contrast, both vinexin-α and CAP but not ArgBP2 increased the fraction of CSK-resistant vinculin on rigid substrates. The effect of CAP on the CSK resistance of vinculin was greater than that observed for vinexin-α (Fig. 4E,F). To confirm further the effects of SORBS proteins on the CSK-resistance of vinculin, we tested SORBS-re-expressing cells cultured on extremely rigid glass substrates (Fig. S3C). Consistent with the results from cells on rigid gel substrates, CAP and vinexin-α but not ArgBP2 increased the CSK-resistant vinculin compared with the mock control on glass substrates (Fig. S3D,E). As observed above, CAP was more effective at producing CSK resistance than vinexin-α. These results demonstrate that vinexin-α and CAP regulate ECM stiffness-dependent vinculin status, a process in which ArgBP2 is not involved.
CAP, as well as vinexin-α, immobilizes vinculin within FAs
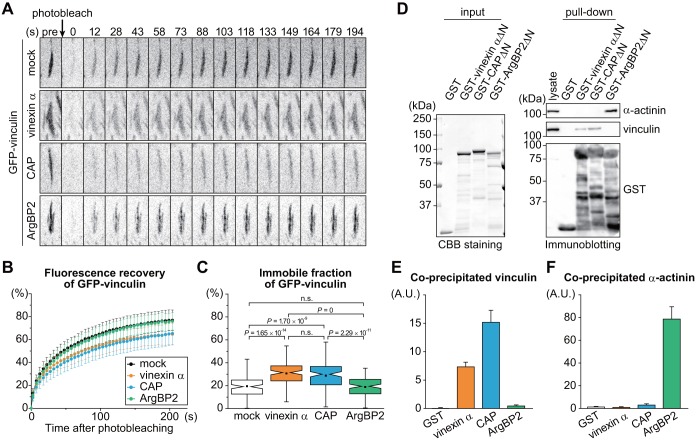
The molecular immobility within FAs is another ECM stiffness-dependent feature of the cellular effects of vinculin, which also requires its interaction with vinexin-α (Yamashita et al., 2014). To investigate the effects of SORBS proteins on vinculin immobility, fluorescence recovery after photobleaching (FRAP) studies were conducted using SORBS-re-expressing cells that express GFP–vinculin on rigid glass substrates (Fig. 5A,B; Movie 1). The immobile fraction and the time of half-life for recovery (t½) were calculated by curve-fitting the data (Fig. 5C; Fig. S3F). However, because our previous data have shown that ECM stiffness affects the immobile fraction of vinculin but not t½, we focused on the immobile fractions in the following experiments. The immobile fractions of vinculin in TKO MEF/mock, TKO MEF/vinexin-α, TKO MEF/CAP and TKO MEF/ArgBP2 were 20±0.9, 31±0.9, 29±1.1 and 19±0.8%, respectively (means±s.e.m.). These results indicate that both vinexin-α and CAP contribute to the immobility of vinculin within FAs, whereas ArgBP2 is dispensable. Note that both vinexin-α and CAP increase the immobile fraction of vinculin to the same extent, unlike our findings for CSK resistance, indicating that these two properties are not entirely coupled.
Fig. 5.
Both vinexin-α and CAP but not ArgBP2 immobilize vinculin at FAs and bind to vinculin. (A–C) SORBS-re-expressing cells stably expressing EGFP–vinculin were cultured on glass-based dishes and subjected to FRAP analysis. (A) More than 111 FAs containing GFP–vinculin from thirty individual cells in three independent experiments were photobleached for each condition, and the representative time-lapse montages are displayed. (B) The normalized fluorescence recovery of EGFP–vinculin at FAs in TKO MEF/mock (n=111), TKO MEF/vinexin-α (n=118), TKO MEF/CAP (n=116) or TKO MEF/ArgBP2 (n=115) was plotted as the mean±s.d. (C) The immobile fraction was calculated by curve-fitting the data. P-value was calculated by Mann–Whitney U-test; n.s., not significant. See also Fig. S3F and Movie 1. (D–F) Pulldown assay using GST–vinexin-αΔN, GST–CAPΔN and GST–ArgBP2ΔN. TKO MEFs were lysed and subjected to a pulldown assay. Input purified proteins were visualized by CBB staining (left panels). Co-precipitated proteins were visualized by immunoblotting using the indicated antibodies (right panels) (D). The amount of co-precipitated vinculin (E) or α-actinin (F) with GST-tagged proteins was quantified by Fiji software. The values represent the means±s.e.m. from three independent experiments.
Vinexin-α and CAP bind to vinculin, whereas ArgBP2 binds to α-actinin
To interpret these different effects of SORBS proteins on vinculin status, we investigated the binding abilities of SORBS proteins to vinculin. GST-tagged SORBSΔN proteins that lack the N-terminal intrinsically disordered region were purified and used for a pulldown assay instead of wild-type proteins to prevent degradation in E. coli (Yamashita et al., 2014). Vinculin was co-precipitated with GST–vinexin-αΔN or GST–CAPΔN but not GST–ArgBP2ΔN, suggesting that ArgBP2 has extremely low affinity for vinculin despite the similar domain structures among the SORBS proteins (Fig. 5D,E). Meanwhile, consistent with previous reports (Rönty et al., 2005; Anekal et al., 2015), α-actinin was co-precipitated with ArgBP2 but neither vinexin-α nor CAP (Fig. 5D,F). Collectively, these results demonstrate that ArgBP2 has unique binding selectivity, which differentiates it from vinexin-α and CAP.
The central region of vinexin-α and CAP play a critical role in binding to vinculin and inducing CSK resistance of vinculin
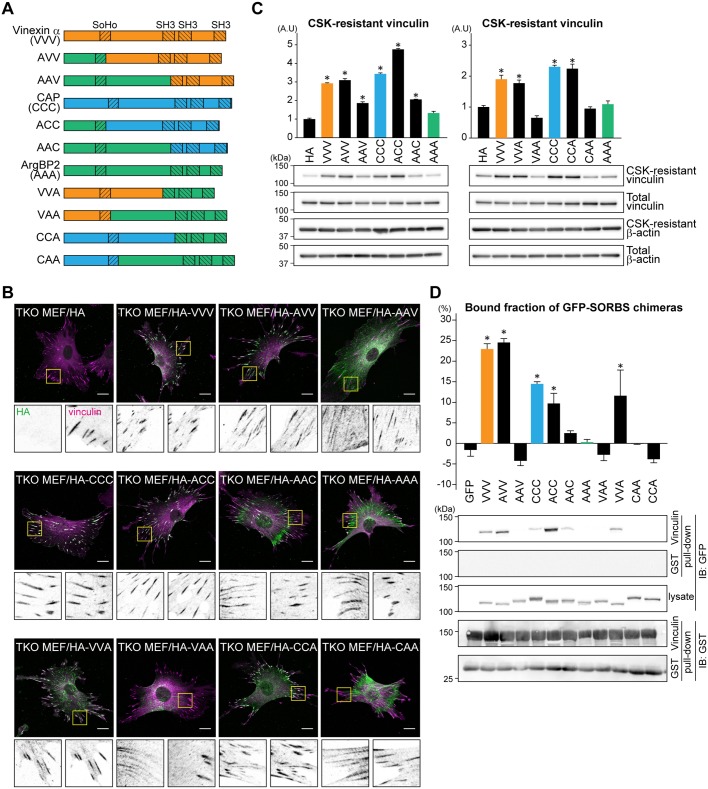
SH3 domains in SORBS proteins have been reported to mediate the binding to vinculin, while a central region of ArgBP2 mediates the binding to α-actinin (Anekal et al., 2015). To determine which regions are involved in the subcellular localization of SORBS protein and CSK resistance of vinculin, we divided each SORBS protein into three parts [N-terminal parts (N-terminus to SoHo domain), central parts (between SoHo domain and first SH3 domain), and C-terminal parts (first SH3 domain to C-terminus)] and constructed chimeras (Fig. 6A). TKO MEFs stably expressing each chimera were established, and the expressions were confirmed (Fig. S3G). Immunostaining analysis indicated that chimeras containing the central region of vinexin-α or CAP (AVV, VVA, ACC and CCA) clearly co-localized with vinculin, even though they have the N-terminal or C-terminal part of ArgBP2 (Fig. 6B). In contrast, chimeras containing the central region of ArgBP2 (AAV, VAA, AAC and CAA) did not show the co-localization with vinculin, but did show a similar subcellular localization to ArgBP2, suggesting that the central region of vinexin-α and CAP, as well as ArgBP2, plays a critical role in directing subcellular localization.
Fig. 6.
The central region of vinexin-α and CAP play critical roles in regulating vinculin. (A) Schematic diagram of the domain structure of chimeric proteins between SORBS proteins. Each SORBS protein was divided into three parts: the N-terminal part containing one SoHo domain, the C-terminal part containing three SH3 domains and the central part not containing any domain structure. (B) HA-tagged chimeric protein-expressing cells cultured on coverslips were immunostained using anti-vinculin and anti-HA antibodies. The FA-containing areas indicated by yellow boxes are cropped and displayed at the bottom of the figure. Scale bars: 20 µm. (C) Cells cultured on collagen-coated dishes were treated with or without CSK and then lysed with 1% SDS, followed by immunoblotting using the indicated antibodies (lower panels). The ratio of CSK-resistant vinculin to total vinculin was quantified by Fiji software. The values represent the means±s.e.m. from three independent experiments. *P<0.05 was compared with HA-expressing samples using an unpaired Student's t-test. (D) GFP–depletion assay using purified GST–vinculin. 293T cells expressing GFP–chimeric proteins were subjected to a pulldown assay. Intensity of the unbound fraction was measured by spectrofluorimeter and the bound fraction was calculated (upper panel). Co-precipitated chimeric proteins were visualized by immunoblotting using the indicated antibodies (lower panels). The values represent the means±s.e.m. from three independent experiments. *P<0.05 was compared with GFP-expressing samples using an unpaired Student's t-test.
We next examined the effect of these chimeras on CSK resistance of vinculin. Cells were treated with CSK buffer, and the remaining CSK-resistant proteins were solubilized and examined by immunoblotting using an anti-vinculin antibody. As shown in Fig. 6C, chimeras having the central region of vinexin-α or CAP significantly increased the CSK-resistant vinculin. In contrast, chimeras containing the central region of ArgBP2 showed no or moderate effects on the CSK-resistant vinculin. Finally, we tested whether these functions reflect the interaction with vinculin. GFP-tagged chimeras expressed in 293T cells were pulled down using GST–vinculin, and then the depletion of GFP fluorescence in supernatant was determined as proteins binding to vinculin. As expected, GFP-tagged vinexin-α or CAP, but not ArgBP2, was efficiently depleted by GST–vinculin (Fig. 6D). Replacement of a central region of ArgBP2 significantly decreased the depletion. Taken together, these observations using chimeras indicate a critical role of the central region of vinexin-α and CAP in binding to vinculin and inducing CSK resistance of vinculin.
ArgBP2 immobilizes α-actinin on actin SFs but not within FAs
Although ArgBP2 has been shown to interact with α-actinin, the effect of the interaction on α-actinin remains unclear. As both vinexin-α and CAP immobilized vinculin within FAs (Fig. 5A–C), we hypothesized that ArgBP2 may immobilize α-actinin. FRAP studies were thus conducted using SORBS-re-expressing cells that express GFP–α-actinin. The fluorescence recovery curves of GFP–α-actinin at the proximal end of FAs from all SORBS-re-expressing cells were similar (Fig. 7A,B; Movie 2), and no significant differences were observed in the immobile fractions (Fig. 7C): 48±1.2% (mean±s.e.m.) in TKO MEF/mock, 51±1.0% in TKO MEF/vinexin-α, 46±1.1% in TKO MEF/CAP, and 49±0.9% in TKO MEF/ArgBP2. In contrast, the maximum fluorescence recovery of GFP–α-actinin on punctate structures along SFs was suppressed (Fig. 7D,E; Movie 3), and the immobile fraction was consequently increased by ArgBP2 (Fig. 7F): 18±1.7% in TKO MEF/mock, 24±1.6% in TKO MEF/vinexin-α, 23±1.9% in TKO MEF/CAP and 33±2.0% in TKO MEF/ArgBP2. Both vinexin-α and CAP slightly increased the immobile fraction, but not significantly. These results suggest that ArgBP2 immobilizes α-actinin on SFs specifically.
Fig. 7.
ArgBP2 but neither vinexin-α nor CAP immobilizes α-actinin on SFs. SORBS-re-expressing cells stably expressing EGFP–α-actinin were cultured on glass-based dishes and subjected to FRAP analysis. (A–C) More than 78 FAs containing GFP–α-actinin from twenty individual cells in three independent experiments were photobleached for each condition, and the representative time-lapse montages are displayed (A). The normalized fluorescence recovery after photobleaching of EGFP–α-actinin at FAs in TKO MEF/mock (n=79), TKO MEF/vinexin-α (n=78), TKO MEF/CAP (n=79) or TKO MEF/ArgBP2 (n=79) was plotted as the mean±s.d. (B) The immobile fraction was calculated by curve-fitting the data (C); N.S., non-significant in Kruskal–Wallis ANOVA. See also Fig. S3H and Movie 2. (D–F) More than 25 cells were selected in three independent experiments, and one circular region along SFs containing GFP–α-actinin in each cell were photobleached for each condition (D). The normalized fluorescence recovery after photobleaching of EGFP–α-actinin at SFs in TKO MEF/mock (n=26), TKO MEF/vinexin-α (n=26), TKO MEF/CAP (n=25) or TKO MEF/ArgBP2 (n=25) was plotted as the mean±s.d. (E). The immobile fraction was calculated by curve-fitting the data (F). P-value was calculated by Tukey's post hoc test; n.s., not significant. See also Fig. S3I and Movie 3.
ArgBP2 is involved in the generation of cellular contractile forces
An α-actinin mutant with a high affinity for actin, which has a larger immobile fraction than wild type, is involved in control of cellular contractile forces (Ehrlicher et al., 2015). Additionally, the over-expression of ArgBP2 enhances phosphorylation of MRLC in vascular endothelial cells (Martin et al., 2013). These reports prompted us to investigate the effects of SORBS proteins, especially ArgBP2, on the generation of contractile forces. Contractile forces were visualized by wrinkle formation on silicone substrates. Every test was conducted as a paired experiment to determine the relative contributions of SORBS proteins to the generation of cellular contractile forces. As contractile forces within a cell correlate with the length of wrinkles (Burton and Taylor, 1997) (Fig. S4A), a single slice image containing an individual cell accompanied by the maximum length of wrinkles was extracted from 2 h time series data in order to evaluate the maximum force (Fig. 8A–F). We quantified the maximum length of wrinkles per cell in each combination between SORBS-re-expressing cells. Both vinexin-α and CAP did not affect wrinkle formation (Fig. 8A–C), but ArgBP2 significantly increased wrinkle formation compared with the other conditions (Fig. 8D–F). ArgBP2 also increased the averaged wrinkle length per cell (Fig. S3N). These results suggest that ArgBP2 specifically promotes the generation of contractile force in MEFs.
Fig. 8.
ArgBP2 generates the greatest contractile force in the wrinkle formation assay. (A–F) SORBS-re-expressing cells were cultured on type I collagen-coated silicone substrates and subjected to the cell contraction assay. Left panels: more than twenty individual cells were observed for 2 h and four independent experiments were performed for each combination. Representative images of cells that showed maximum wrinkle length during the 2 h observation in each combination are displayed. Scale bars: 50 µm. Right panels: the total length of wrinkles per cell at the time of maximum was quantified from images in the representative experiment with a custom-made program written in Fiji software. See also Fig. S4B. The number of cells is as follows: (A) n=42 (TKO MEF/mock) and n=30 (TKO MEF/vinexin-α), (B) n=27 (TKO MEF/mock) and n=22 (TKO MEF/CAP), (C) n=42 (TKO MEF/vinexin-α) and n=28 (TKO MEF/CAP), (D) n=47 (TKO MEF/mock) and n=39 (TKO MEF/ArgBP2), (E) n=40 (TKO MEF/vinexin-α) and n=32 (TKO MEF/ArgBP2), (F) n=24 (TKO MEF/CAP) and n=24 (TKO MEF/ArgBP2). P-value was calculated by Mann–Whitney U-test. (G,H) SORBS-re-expressing cells cultured on coverslips for 24 h were immunostained using anti-diphosphorylated (T18/S19) MRLC antibody with phalloidin staining (G). Scale bars: 20 µm. The integrated density of diphosphorylated MRLC per cell was quantified from images of ninety individual cells from three separate experiments (H). n=90; P-value calculated by Mann–Whitney U-tests; n.s., not significant.
To corroborate the marked contribution of ArgBP2 to force generation, we examined the effects of SORBS proteins on the phosphorylation of MRLC, which is necessary for myosin-dependent contraction. Immunostaining revealed that ArgBP2 considerably increased both mono- and diphosphorylated MRLC compared with the mock control (Fig. 8G,H; Fig. S3J,K). Both vinexin-α and CAP increased phospho-MRLC significantly, but less than that observed with ArgBP2, indicating that the increased levels induced by these proteins are insufficient to enhance further wrinkle formation. Interestingly, the amount of diphosphorylated MRLC estimated by immunoblotting was comparable among SORBS-re-expressing cells (Fig. S3L), suggesting that ArgBP2 expression modulates the subcellular distribution of phospho-MRLC to accumulate it on stress fibers, but does not affect the levels of phosphorylation of MRLC itself. Taken together, these results demonstrate that ArgBP2 but neither vinexin-α nor CAP is adequate for generation of contractile forces in MEFs.
DISCUSSION
SORBS proteins have received considerable attention related to the cellular mechanisms of mechanotransduction. We have established MEFs that individually express each of the SORBS proteins and investigated their functions in mechanotransduction using SORBS-re-expressing cells. CAP, as well as vinexin-α, contributes to the stiffness-dependent CSK resistance of vinculin and immobilizes vinculin at FAs. Meanwhile, ArgBP2 immobilizes α-actinin on SFs and markedly increases the contractile forces. These observations indicate the distinct roles of SORBS proteins in mechanotransduction.
SORBS proteins share domain structures and binding partners, mainly through their SH3 domains. Numerous reports have focused on each individual SORBS protein and have shown that their subcellular localization and function depend on cellular context. However, no study has comparatively analysed the localization and function of all SORBS proteins. Here, we shed light on the functional redundancy between vinexin-α and CAP by using cells re-expressing single SORBS proteins. This finding suggests that the mild phenotype in both vinexin and CAP KO mice (Lesniewski et al., 2007; Kioka et al., 2010; Chen et al., 2013) may be due to compensation by the other proteins. Interestingly, the EMBL-EBI Clustal Omega program demonstrates the highest sequence identity between CAP and ArgBP2 of all the combinations: 33.8% between vinexin-α and CAP, 33.7% between vinexin-α and ArgBP2, and 41.7% between CAP and ArgBP2. In addition, loss of CAP expression causes enhanced expression of ArgBP2 for compensation in myotubes (Hallock et al., 2015). These observations suggest that a compensatory mechanism may work in the case of cells or animals bearing a knockout of a single SORBS protein. Therefore, TKO MEFs and SORBS-re-expressing cells are good model cells to analyse the functions of SORBS proteins.
We showed that both purified vinexin-α and CAP but not ArgBP2 pulled down endogenous vinculin in TKO MEFs. In contrast, ArgBP2 but neither vinexin-α nor CAP pulled down endogenous α-actinin in TKO MEFs. However, several studies have reported that the first two SH3 domains of vinexin-α (Kioka et al., 1999; Takahashi et al., 2005), CAP (Mandai et al., 1999; Zhang et al., 2006) and ArgBP2 (Cestra et al., 2005) share binding ability to vinculin in vitro and are highly homologous (Kioka et al., 2002). Thus we further constructed chimeric proteins and showed that central region of vinexin-α and CAP are necessary for the co-precipitation with vinculin. It remains unclear how the central regions of vinexin-α and CAP promote the interaction with vinculin. It is possible that central regions in SORBS proteins would contain the additional binding sites for target proteins and contribute to the interaction along with SH3 domains. Indeed, ArgBP2 includes an α-actinin binding sequence in the central region (Anekal et al., 2015). Alternatively, the central region of vinexin-α and CAP might regulate the SH3 function sterically or allosterically. Most importantly, we also showed that chimeras having the central region of vinexin-α or CAP co-localized with vinculin and induced the CSK resistance of vinculin in TKO cells, whereas chimeras having those of ArgBP2 did not. These observations suggest that the different binding selectivity of SORBS proteins results in different localization and functions in mechanotransduction.
Although vinexin-α or CAP were required for stiffness-dependent CSK resistant vinculin that is tightly associated with the actin cytoskeleton, CAP enhanced CSK resistance more dramatically than vinexin-α on rigid gel (25 kPa) and glass (∼GPa) substrates. We also showed that CAP more effectively co-precipitated vinculin than vinexin-α. These results suggest that CAP has a higher affinity for vinculin compared with vinexin-α, and that CAP forms a ternary complex with vinculin and F-actin more effectively than vinexin-α. A recent report has shown that vinexin-α and CAP are necessary for myogenic differentiation on 15 kPa gels and osteogenic differentiation on 42 kPa gels in human mesenchymal stem cells, respectively (Holle et al., 2016). In addition, vinculin is also necessary for stiffness-dependent myogenesis and, to a lesser extent, involved in osteogenesis (Holle et al., 2013, 2016). We have also clarified that vinculin regulates stiffness-dependent adipogenesis of the mouse mesenchymal stem cell line ST2 cells (Kuroda et al., 2017). These observations raise the possibility that the formation of the vinexin-α–vinculin and CAP–vinculin complexes have a different optimum range of ECM stiffness and regulate stiffness-dependent cell differentiation. However, we cannot exclude the possibility that different expression levels of vinexin-α and CAP affect the substantial influence of CAP on CSK-resistant vinculin. As we have re-expressed SORBS proteins to the levels observed in native proteins, and these re-expressed proteins do not contain any epitope tags, it is difficult to compare the precise expression levels of vinexin-α and CAP directly. Future studies should examine these possibilities.
In the present study, CAP as well as vinexin-α, but not ArgBP2, increased the immobile fraction of vinculin. The vinculin immobile fraction is increased by both rigid ECM (Yamashita et al., 2014) and a vinculin conformational change triggered by disruption of the head–tail interaction (Cohen et al., 2006; Auernheimer et al., 2015; Liu et al., 2016). Although myosin II activity and force have been reported to be required for the vinculin immobile fraction (Wolfenson et al., 2011; Dumbauld et al., 2013), a remarkable increase in contractile force or accumulation of phosphorylated myosin II induced by ArgBP2 did not cause an increase in the vinculin immobile fraction. These results indicate that increases in contractile force are insufficient to enhance immobilization of vinculin. Rather, vinculin binding to vinexin-α or CAP seems necessary for vinculin immobilization at FAs.
We showed that both vinexin-α and CAP were not fully co-localized with the entirety of vinculin but were slightly positioned at the proximal side and dorsal layer of the vinculin localization area. An elegant study focusing on nanoscale architecture of FAs has reported that vinculin activation correlates with the vinculin vertical position within FAs: inactive vinculin localizes in the ventral layer close to integrins by binding to phosphorylated paxillin, whereas activated vinculin moves to the dorsal layer close to F-actin by binding to talin (Case et al., 2015). We previously reported that vinexin-α induces a conformational change in vinculin that results in its activated form (Yamashita et al., 2014). These observations suggest that vinexin-α and CAP activate vinculin to move to the dorsal layer and simultaneously co-localize with activated vinculin.
We found that ArgBP2 immobilized α-actinin on SFs but not at FAs. Anekal et al. reported that PKA phosphorylation of ArgBP2 disrupts its binding to α-actinin, leading to loss of ArgBP2 from SFs, but does not affect the FA localization of ArgBP2 (Anekal et al., 2015). This finding indicates that ArgBP2 targeting to FAs is independent of the interaction with α-actinin despite their clear co-localization. Thus differences in the ArgBP2 interaction with α-actinin may explain the varied effects on immobilization of α-actinin on SFs and FAs. The lack of effects of vinexin-α or CAP, both of which did not associate with α-actinin, on the immobilization of α-actinin supports this idea. It is worth noting that ArgBP2 as well as vinexin-β forms a complex with WAVE2, which works as a PKA anchoring protein in tumour cells (Cestra et al., 2005; Mitsushima et al., 2006a; Yamashita et al., 2011). Future studies will be necessary to clarify the involvement of PKA in these regulatory events.
In this study, we showed that ArgBP2 generated greater cellular contractile forces in a wrinkle formation assay. The mechanism of this effect is not completely understood, but α-actinin seems to be involved in this process. α-Actinin is one of the ‘clutch’ molecules that transmits intracellular forces to FAs (Roca-Cusachs et al., 2013) and assembles SFs with short spacing in response to contractile forces (Aratyn-Schaus et al., 2011). Reciprocally, an enhancement of α-actinin affinity for F-actin increases its immobile fraction, which gives rise to greater contractile forces (Ehrlicher et al., 2015; Schiffhauer et al., 2016). The immobile fraction of α-actinin on SFs estimated by FRAP analysis was increased by ArgBP2 in our study. These observations suggest that α-actinin immobilization in SFs by ArgBP2 contributes to greater contractile forces.
In summary, we conclude that vinexin-α and CAP redundantly regulate vinculin behaviors depending on ECM stiffness, whereas ArgBP2 enhances contractile forces in collaboration with α-actinin. Every SORBS protein has been recently reported as a tumour suppressor: vinexin-α in hepatocarcinoma (Ploeger et al., 2016), CAP in breast cancer (Song et al., 2016) and ArgBP2 in pancreatic cancer (Taieb et al., 2008) and gastric cancer (Tong et al., 2015). Further investigation of the SORBS proteins using cancer cells and stem cells may provide insight into ECM stiffness-dependent tumour malignancies and tissue development, respectively.
MATERIALS AND METHODS
Plasmid construction
The shRNA for ArgBP2 (5′-GGATGGTTTGTGGGAACTTCA-3′) was subcloned into the lentiviral transfer vector pLKO.1-Puro from Open Biosystems (Huntsville, AL, USA). The lentiviral transfer vector pCDH-EF1-IRES-Puro from System Biosciences (Mountain View, CA, USA) was modified into pCDH-EF1-IRES-Blast or -Hygro. The cDNA encoding mouse vinexin-α and vinexin-αΔN (157–733 a.a.) were described previously (Yamashita et al., 2014). The cDNA encoding mouse CAP and mouse ArgBP2 were identified from WT MEFs, and their sequences have been registered in the DNA Data Bank of Japan (DDBJ) (accession numbers: LC200510 and LC200511, respectively). The cDNA encoding shRNA-resistant ArgBP2 was generated by site-directed mutagenesis using an In-Fusion HD cloning kit (Clontech, Mountain View, CA, USA). The full-length vinexin-α, CAP and shRNA-resistant ArgBP2 were subcloned into pCDH-EF1-IRES-Blast, while monomeric GFP-tagged vinculin and α-actinin-1 (kindly provided by Dr Seiji Tadokoro, Osaka University, Japan) were subcloned into pCDH-EF1-IRES-Hygro. The cDNA encoding chimeric proteins of SORBS proteins were generated using an In-Fusion HD cloning kit, then HA-tagged or GFP-tagged chimeras were subcloned into pCDH-EF1-IRES-Blast. The full-length of vinculin, N-terminal region deleted CAP (CAPΔN; 202–760 a.a.) and ArgBP2 (ArgBP2ΔN; 143–718 a.a.) were generated by PCR and then subcloned into pColdI-ΔHis-GST vector for protein purification. The ArgBP2 fragment (392–535 a.a.) was subcloned into pColdI-GST vector for antigen production.
Antibodies and reagents
Mouse monoclonal anti-vinculin (hVIN-1, V9131, 1:10,000 for western blotting, 1:400 for immunostaining), anti-α-actinin (BM-75.2, A5044, 1:10,000/1:100), anti-talin (8d4, T3287, -/1:100) and rat monoclonal anti-HA (3F10, 11867423001, 1:1000/-) antibodies were purchased from Sigma Aldrich (St Louis, MO, USA). Mouse monoclonal anti-paxillin (5H11, AHO0492, 1:10,000/1:200) antibody was from Invitrogen/Thermo Fisher Scientific (Carlsbad, CA, USA). Rabbit monoclonal anti-MLC2 (D18E2, 8505, 1:1000/-), rabbit polyclonal anti-phospho-MLC2 (Ser19, 3671, 1:1000/1:100) and anti-phospho-MLC2 (Thr18/Ser19, 3674, 1:1000/1:100) antibodies were from Cell Signaling Technology (Danvers, MA, USA). Rabbit polyclonal anti-vinculin antibody (ab73412, -/1:25) was from Abcam (Cambridge, UK). Rabbit polyclonal anti-vinexin (1:7000/1:100) antibody was described previously (Kioka et al., 1999). Rabbit polyclonal anti-CAP antibody (06-994, 1:1000/1:50) was from Upstate/Merck Millipore (Darmstadt, Germany). Rabbit polyclonal anti-ArgBP2 antibody (1:2000/1:50) was produced as described below. His–GST–ArgBP2 (392–535 a.a.) was purified for antigen. His–GST tags were cleaved by HRV3C protease (Novagen/Merck Millipore) and removed by HiTrap SP HP column (GE Healthcare). The purified ArgBP2 fragment was injected into two Japanese White rabbits, and their antiserum was collected by Medical & Biological Laboratories CO., LTD. (Nagoya, Japan). Polyclonal antibodies were affinity-purified using Affi-Gel 10 conjugated with ArgBP2 fragment (Bio-Rad). Alexa Fluor 488 goat anti-mouse IgG antibody, Alexa 555 goat anti-rabbit IgG antibodies, Alexa Fluor 488 phalloidin and CellMask Orange were from Thermo Fisher Scientific (Waltham, MA, USA). Type I collagen was from Nitta Gelatin (Osaka, Japan). Puromycin, Blasticidin S and Hygromycin B were from Sigma Aldrich, Kaken Pharmaceutical (Tokyo, Japan) and Wako Pure Chemical Industries (Osaka, Japan), respectively.
Generation of vinexin/CAP double knockout (DKO) mice and isolation of DKO MEFs
Mice were housed in a specific pathogen-free facility with a 12 h:12 h light:dark cycle and given free access to food and water, except when food was restricted during fasting. All animal use was in compliance with the Institute of Laboratory Animal Research Guide for the Care and Use of Laboratory Animals and approved by the University Committee on Use and Care of Animals at the University of Michigan. Vinexin/CAP double hetero knockout (DKO) mice were generated using vinexin KO mice (Kioka et al., 2010) and CAP KO mice (Lesniewski et al., 2007). These DKO mice were viable and fertile. Thus DKO mice were mated, and primary DKO MEFs were isolated.
Cell culture and lentiviral transduction
Spontaneously immortalized vinexin/CAP double knockout MEFs (DKO MEFs) were generated with the 3T3 passaging method (Todaro and Green, 1963). Stable knockdown and expression were accomplished by lentiviral transduction as previously described (Yamashita et al., 2014), with slight modifications. Briefly, 293T cells were transiently transfected with lentiviral second generation vectors (i.e. pLKO.1, pMD2.G and psPAX2) or third generation vectors (i.e. pCDH, pMD2.G, pRSV-Rev and pMDLg/pRRE) using Lipofectamine LTX and PLUS Reagent (Thermo Fisher Scientific). All envelope and packaging vectors were from Addgene (Cambridge, MA, USA). After incubation for 48 h, the supernatants were collected, and the virus particles were concentrated using Lenti-X Concentrator (Clontech). The target cells were incubated in lentivirus-containing medium for 18 h, followed by an incubation in selection medium containing 1 µg/ml Puromysin, 3 µg/ml Blasticidin S or 500 µg/ml Hygromycin B for more than 7 days. All cells were grown in Dulbecco's modified Eagle's medium (DMEM; Nacalai tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum (FBS; Gibco/Thermo Fisher Scientific) at 37°C in a humidified atmosphere containing 5% CO2. Polyacrylamide (PAA) gel substrates were prepared as previously described (Yamashita et al., 2014). Both 2.2 kPa and 25 kPa PAA gels were used as soft and rigid substrates, respectively.
Immunofluorescence and confocal microscopy
Immunofluorescence was performed as previously described (Yamashita et al., 2014), with slight modification. Briefly, cells were cultured on 10 µg/ml type I collagen-coated no.1 coverslips or 200 µg/ml type I collagen-coated PAA gels, and subjected to immunostaining. To visualize CSK-resistant vinculin in cells on PAA gels, cells were treated with CSK buffer (0.5% Triton X-100, 10 mM PIPES, pH 6.8, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose) on ice for 1 min, followed by fixation with 4% (w/v) paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature. To visualize total FA protein in cells on coverslips, cells were fixed with 2% (w/v) paraformaldehyde in PBS for 20 min at room temperature. To visualize phosphorylated myosin regulatory light chain, PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9) was used instead of PBS. The fixed cells were then subjected to immunostaining. Cell spreading area was quantified using a plasma membrane staining reagent, CellMask Orange. Cells were incubated with 2 µg/ml CellMask Orange in DMEM without FBS for 5 min at 37°C, followed by fixation with 4% (w/v) paraformaldehyde in PBS for 10 min at 37°C. Images were acquired using an LSM700 laser scanning confocal microscope with a Plan-Apochromat 40×/1.3 NA oil immersion objective lens (Carl Zeiss, Oberkochen, Germany). Any image quantifications were performed using ImageJ-based Fiji software. Focal adhesions were classed as structures of 1–20 µm2 in the ‘Analyze Particles’ command. Gray images were treated with the ‘Enhance Contrast’ command and then inverted to increase their visibility.
Structured illumination microscopy
Cells were cultured on 10 µg/ml type I collagen-coated no.1S coverslips and subjected to immunostaining as described above. 3D-SIM acquisition was conducted on an N-SIM super-resolution microscope with an Apo TIRF 100×/1.49 NA oil immersion lens (Nikon, Tokyo, Japan), according to the manufacturer's instructions.
Fluorescence recovery after photobleaching assay and analysis
FRAP studies were performed as described previously (Yamashita et al., 2014), with slight modifications. Briefly, cells expressing GFP-tagged proteins were plated on 10 µg/ml type I collagen-coated glass-bottomed dishes and imaged at 37°C in an atmosphere of 5% CO2 in FluoroBrite DMEM (Thermo) supplemented with 10% FBS, sodium pyruvate and GlutaMAX (Thermo). FRAP experiments were conducted with an LSM700 microscope with a Plan-Apochromat 63×/1.4 NA oil immersion objective lens. In FRAP experiments focusing on FAs, four different FAs that did not show FA maturation or decay near the cell edge were selected, and each whole FA was manually encircled as a region of interest (ROI). In FRAP experiments focusing on SFs, SFs showing punctate localization of GFP–α-actinin were selected, and one circular region was defined as an ROI. Image acquisition started 10 s before photobleaching and continued every 5 s for 210 s. Fluorescence recovery within an FA was analysed by ZEN 2009 software (Carl Zeiss). Fluorescence recovery within punctate structures along SFs was analysed by Fiji software to remove noise. The average background intensity was subtracted from that of the photobleached ROI as well as the reference ROI. Subsequently, the photobleached ROI was normalized by the reference ROI. Every FRAP curve was fitted to a single exponential function, followed by the calculation of the immobile fraction and the time of half-life for recovery (t½), using KaleidaGraph software (Synergy software, Reading, PA, USA). To exclude the effect of protein diffusion, data points taken at <10 s after photobleaching were excluded from fitting. Data with a coefficient of determination less than 0.95 were eliminated. Ten random cells were analysed per experiment, and at least three independent experiments were performed.
Cell contraction assay
Cell contractility was evaluated using a modified version of a deformable silicone substrate technique (Sakane et al., 2016; Yokoyama et al., 2016). Silicone substrates were prepared as follows: parts A and B of CY 52-276 (Dow Corning Toray, Komatsu, Japan) were mixed at a weight ratio of 1.1:1. The mixture was coated onto 35 mm plastic dishes using a K-359S1 spin coater (Kyowa Riken, Tokyo, Japan) at 1500 rpm, then baked at 60°C for 20 h to create a cured gel. For hydrophilization, the gels were exposed to 4 mA oxygen plasma for 1 min at 10 Pa using an SEDE-GE plasma generator (Meiwafosis, Tokyo, Japan). The Young's modulus of the silicone substrates was evaluated with a method described elsewhere (Beningo et al., 2002) and was 17.2±0.7 kPa (mean±s.d.). The substrates were coated with 10 µg/ml type I collagen prior to seeding two different cell populations, one of which was stained by CellMask Orange. Cells were incubated at 37°C in an atmosphere of 5% CO2 in a chamber on an IX71 inverted microscope (Olympus, Tokyo, Japan). Twelve hours after cell seeding, wrinkle formation was photographed for 2 h with phase-contrast microscopy using a 10×/0.4 NA objective lens every 5 min. More than twenty random positions were observed per experiment with an electric motorized stage, and two independent experiments were performed in each permutation. Therefore, every combination was tested in quadruplicate.
The acquired images were analysed to automatically extract wrinkles on the silicone substrates using a custom-made program written in Fiji software (Fig. S4). Briefly, images were processed with a two-dimensional fast Fourier transformation (FFT), subjected to a band-pass filter to detect the wrinkles with a spatial period between 3 and 6 µm that are typical for the wrinkles generated by MEFs, and then an inverse FFT. The resulting images still contain geometric information on the contour of cells because the band-pass filter could not completely distinguish between the wrinkles and cell contour. To remove cell contour, the original images were processed with a command to eliminate high-intensity signals, followed by the same two-dimensional FFT, a band-pass filter, and inverse FFT. The resulting cell contour data were subtracted from the separately processed images to finally allow for integrating the total length of the wrinkles after skeletonization, i.e. line segmentation.
Protein purification
Protein purification was performed as described previously (Takahashi et al., 2005; Yamashita et al., 2014) with slight modifications. Briefly, the N-terminal regions of the SORBS proteins were deleted, because the full-length SORBS proteins showed low solubility in buffers and intensive degradation. GST–vinexin αΔN, GST–CAPΔN and GST–ArgBP2ΔN were individually expressed in E.coli Rosetta (DE3) strain (Novagen/Merck Millipore), and then lysed with purification buffer (50 mM Tris-HCl, 500 mM NaCl, 10% glycerol, pH 8.0) containing lysozyme. GST–vinculin was also expressed in E.coli, and then lysed by sonication in PBS. These proteins were purified using Glutathione Sepharose 4B, followed by size exclusion chromatography using Superdex 200 Increase 10/300 GL (GE Healthcare).
Pulldown assay and GFP-depletion assay
Pulldown assay was performed as described previously (Yamashita et al., 2014). Briefly, TKO MEFs were lysed with 1% Triton X-100 containing PBS. Cell lysates (500 µg) were incubated with 3 µg of purified GST or its equivalent moles of GST–tagged SORBSΔN at 4°C for 2 h, followed by further incubation with Glutathione Sepharose 4B beads. The beads were collected and washed with 0.1% Triton X-100 containing PBS three times. Co-precipitated proteins were separated by SDS-PAGE and subjected to immunoblotting with specific antibodies. For the GFP-depletion assay, 293T cells expressing GFP–SORBS chimeric proteins were lysed as described above. The initial GFP fluorescence in each lysate was measured by a Cytation 5 imaging plate reader (BioTek Japan, Tokyo, Japan) and then incubated with GST–vinculin at 4°C for 1 h, followed by further incubation with Glutathione Sepharose 4B beads. The unbound GFP–SORBS chimeric proteins were separated by centrifugation and then assayed again by plate reader. The bound fraction of SORBS chimeric proteins were calculated by subtraction and presented in the graph as the mean±s.e.m. from three independent experiments.
Statistical analysis
Data were presented as means±s.e.m., unless otherwise stated. For each dataset, the normality of the distribution was tested by the Shapiro–Wilk test. When the data followed a normal distribution, differences among compared groups were assessed by one-way ANOVA followed by multiple Tukey's post hoc tests. Otherwise, non-parametric Kruskal–Wallis ANOVA was used with multiple Mann–Whitney U-tests. P<0.05 was considered statistically significant. Box and whisker plots show the following: boxes represent the 25th to 75th percentile range, whiskers represent the 1.5× interquartile range, and notches represent the median with confidence intervals. All statistical analysis was performed using Origin 8.6J software.
Supplementary Material
Acknowledgements
We thank Ms Yuki Wantani for generating ArgBP2-knockdown ATDC5 cells. We thank Dr Beatrice Haimovich (University of Medicine and Dentistry of New Jersey) and Dr Seiji Tadokoro (Osaka University) for providing α-actinin-1 cDNA. We also thank Nikon Instech (Tokyo, Japan) for technical assistance with microscopy.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: T.I., N.K.; Methodology: Y.K.; Software: T.I., T.S.M., S.D.; Formal analysis: T.I., T.S.M., Y.K., K.U., S.D., A.S., N.K.; Investigation: T.I., M.K., T.S.M., A.I.N., T.A., S.-H.C., T.S., S.D.; Resources: A.S.; Writing - original draft: T.I.; Writing - review & editing: T.I., N.K.; Supervision: N.K.; Project administration: N.K.; Funding acquisition: T.I., T.S., K.U., A.S., N.K.
Funding
This work was supported in part by The Naito Foundation, by the Asahi Glass Foundation, by Grants-in-Aid for Scientific Research (B) (JSPS KAKENHI grant numbers 20380186, 24380185) (N.K.), by a Grant-in-Aid for Challenging Exploratory Research (JSPS KAKENHI grant numbers 24658094, 26660291, 16K15090) (N.K.) and by Grants-in-Aid for JSPS Fellows (JSPS KAKENHI grant numbers 12J03633, 09J02878) (T.I. and T.S., respectively) from the Japan Society for the Promotion of Science, by a Grant-in-Aid for Scientific Research (S) (25221203) (K.U.) and by a Grant-in-Aid for Scientific Research on Innovative Areas (MEXT KAKENHI grant number 26112707) (N.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by the Advanced Research and Development Programs for Medical Innovation (N.K.) from the Japan Agency for Medical Research and Development. A.R.S. was supported by US National Institutes of Health grants DK60597 and 60591. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.200691.supplemental
References
- Anekal P. V., Yong J. and Manser E. (2015). Arg Kinase-binding Protein 2 (ArgBP2) interaction with α-actinin and actin stress fibers inhibits cell migration. J. Biol. Chem. 290, 2112-2125. 10.1074/jbc.M114.610725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aratyn-Schaus Y., Oakes P. W. and Gardel M. L. (2011). Dynamic and structural signatures of lamellar actomyosin force generation. Mol. Biol. Cell 22, 1330-1339. 10.1091/mbc.E10-11-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auernheimer V., Lautscham L. A., Leidenberger M., Friedrich O., Kappes B., Fabry B. and Goldmann W. H. (2015). Vinculin phosphorylation at residues Y100 and Y1065 is required for cellular force transmission. J. Cell Sci. 128, 3435-3443. 10.1242/jcs.172031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakolitsa C., Cohen D. M., Bankston L. A., Bobkov A. A., Cadwell G. W., Jennings L., Critchley D. R., Craig S. W. and Liddington R. C. (2004). Structural basis for vinculin activation at sites of cell adhesion. Nature 430, 583-586. 10.1038/nature02610 [DOI] [PubMed] [Google Scholar]
- Beningo K. A., Lo C.-M. and Wang Y.-L. (2002). Flexible polyacrylamide substrata for the analysis of mechanical interactions at cell-substratum adhesions. Methods Cell Biol. 69, 325-339. 10.1016/S0091-679X(02)69021-1 [DOI] [PubMed] [Google Scholar]
- Bharadwaj R., Roy M., Ohyama T., Sivan-Loukianova E., Delannoy M., Lloyd T. E., Zlatic M., Eberl D. F. and Kolodkin A. L. (2013). Cbl-associated protein regulates assembly and function of two tension-sensing structures in Drosophila. Development 140, 627-638. 10.1242/dev.085100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgon R. A., Vonrhein C., Bricogne G., Bois P. R. J. and Izard T. (2004). Crystal structure of human vinculin. Structure 12, 1189-1197. 10.1016/j.str.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Burton K. and Taylor D. L. (1997). Traction forces of cytokinesis measured with optically modified elastic substrata. Nature 385, 450-454. 10.1038/385450a0 [DOI] [PubMed] [Google Scholar]
- Carisey A., Tsang R., Greiner A. M., Nijenhuis N., Heath N., Nazgiewicz A., Kemkemer R., Derby B., Spatz J. and Ballestrem C. (2013). Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr. Biol. 23, 271-281. 10.1016/j.cub.2013.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case L. B., Baird M. A., Shtengel G., Campbell S. L., Hess H. F., Davidson M. W. and Waterman C. M. (2015). Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat. Cell Biol. 17, 880-892. 10.1038/ncb3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestra G., Toomre D., Chang S. and De Camilli P. (2005). The Abl/Arg substrate ArgBP2/nArgBP2 coordinates the function of multiple regulatory mechanisms converging on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 102, 1731-1736. 10.1073/pnas.0409376102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Choudhury D. M. and Craig S. W. (2006). Coincidence of actin filaments and Talin is required to activate Vinculin. J. Biol. Chem. 281, 40389-40398. 10.1074/jbc.M607324200 [DOI] [PubMed] [Google Scholar]
- Chen K., Gao L., Liu Y., Zhang Y., Jiang D.-S., Wei X., Zhu X. H., Zhang R., Chen Y., Yang Q. et al. (2013). Vinexin-β protects against cardiac hypertrophy by blocking the Akt-dependent signalling pathway. Basic Res. Cardiol. 108, 338 10.1007/s00395-013-0338-0 [DOI] [PubMed] [Google Scholar]
- Cohen D. M., Chen H., Johnson R. P., Choudhury B. and Craig S. W. (2005). Two distinct head-tail interfaces cooperate to suppress activation of Vinculin by Talin. J. Biol. Chem. 280, 17109-17117. 10.1074/jbc.M414704200 [DOI] [PubMed] [Google Scholar]
- Cohen D. M., Kutscher B., Chen H., Murphy D. B. and Craig S. W. (2006). A conformational switch in Vinculin drives formation and dynamics of a Talin-Vinculin complex at focal adhesions. J. Biol. Chem. 281, 16006-16015. 10.1074/jbc.M600738200 [DOI] [PubMed] [Google Scholar]
- Dumbauld D. W., Lee T. T., Singh A., Scrimgeour J., Gersbach C. A., Zamir E. A., Fu J., Chen C. S., Curtis J. E., Craig S. W. et al. (2013). How vinculin regulates force transmission. Proc. Natl. Acad. Sci. USA 110, 9788-9793. 10.1073/pnas.1216209110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlicher A. J., Krishnan R., Guo M., Bidan C. M., Weitz D. A. and Pollak M. R. (2015). Alpha-actinin binding kinetics modulate cellular dynamics and force generation. Proc. Natl. Acad. Sci. USA 112, 6619-6624. 10.1073/pnas.1505652112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A. J., Griffin M. A., Sen S., Bönnemann C. G., Sweeney H. L. and Discher D. E. (2004). Myotubes differentiate optimally on substrates with tissue-like stiffness. J. Cell Biol. 166, 877-887. 10.1083/jcb.200405004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A. J., Sen S., Sweeney H. L. and Discher D. E. (2006). Matrix elasticity directs stem cell lineage specification. Cell 126, 677-689. 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- Geiger B., Bershadsky A., Pankov R. and Yamada K. M. (2001). Transmembrane extracellular matrix– cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2, 793-805. 10.1038/35099066 [DOI] [PubMed] [Google Scholar]
- Haglund K., Ivankovic-Dikic I., Shimokawa N., Kruh G. D. and Dikic I. (2004). Recruitment of Pyk2 and Cbl to lipid rafts mediates signals important for actin reorganization in growing neurites. J. Cell Sci. 117, 2557-2568. 10.1242/jcs.01148 [DOI] [PubMed] [Google Scholar]
- Hallock P. T., Chin S., Blais S., Neubert T. A. and Glass D. J. (2015). Sorbs1 and 2 interact with Crk-L and are required for acetylcholine receptor cluster formation. Mol. Cell. Biol. 36, 262-270. 10.1128/MCB.00775-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B. D., Grashoff C. and Schwartz M. A. (2011). Dynamic molecular processes mediate cellular mechanotransduction. Nature 475, 316-323. 10.1038/nature10316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holle A. W., Tang X., Vijayraghavan D., Vincent L. G., Fuhrmann A., Choi Y. S., del Álamo J. C. and Engler A. J. (2013). In situ mechanotransduction via vinculin regulates stem cell differentiation. Stem Cells 31, 2467-2477. 10.1002/stem.1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holle A. W., McIntyre A. J., Kehe J., Wijesekara P., Young J. L., Vincent L. G. and Engler A. J. (2016). High content image analysis of focal adhesion-dependent mechanosensitive stem cell differentiation. Integr. Biol. 8, 1049-1058. 10.1039/C6IB00076B [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton E. R., Byron A., Askari J. A., Ng D. H. J., Millon-Frémillon A., Robertson J., Koper E. J., Paul N. R., Warwood S., Knight D. et al. (2015). Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat. Cell Biol. 17, 1577-1587. 10.1038/ncb3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries J. D., Wang P., Streuli C., Geiger B., Humphries M. J. and Ballestrem C. (2007). Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179, 1043-1057. 10.1083/jcb.200703036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. P. and Craig S. W. (1995). F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature 373, 261-264. 10.1038/373261a0 [DOI] [PubMed] [Google Scholar]
- Kimura A., Baumann C. A., Chiang S.-H. and Saltiel A. R. (2001). The sorbin homology domain: a motif for the targeting of proteins to lipid rafts. Proc. Natl. Acad. Sci. USA 98, 9098-9103. 10.1073/pnas.151252898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioka N., Sakata S., Kawauchi T., Amachi T., Akiyama S. K., Okazaki K., Yaen C., Yamada K. M. and Aota S. (1999). Vinexin: a novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J. Cell Biol. 144, 59-69. 10.1083/jcb.144.1.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioka N., Ueda K. and Amachi T. (2002). Vinexin, CAP/ponsin, ArgBP2: a novel adaptor protein family regulating cytoskeletal organization and signal transduction. Cell Struct. Funct. 27, 1-7. 10.1247/csf.27.1 [DOI] [PubMed] [Google Scholar]
- Kioka N., Ito T., Yamashita H., Uekawa N., Umemoto T., Motoyoshi S., Imai H., Takahashi K., Watanabe H. and Yamada M. (2010). Crucial role of vinexin for keratinocyte migration in vitro and epidermal wound healing in vivo. Exp. Cell Res. 316, 1728-1738. 10.1016/j.yexcr.2010.03.019 [DOI] [PubMed] [Google Scholar]
- Klein E. A., Yin L., Kothapalli D., Castagnino P., Byfield F. J., Xu T., Levental I., Hawthorne E., Janmey P. A. and Assoian R. K. (2009). Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr. Biol. 19, 1511-1518. 10.1016/j.cub.2009.07.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo J.-C., Han X., Hsiao C.-T., Yates J. R. III and Waterman C. M. (2011). Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 13, 383-393. 10.1038/ncb2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Wada H., Kimura Y., Ueda K. and Kioka N. (2017). Vinculin promotes nuclear localization of TAZ to inhibit ECM stiffness-dependent differentiation into adipocytes. J. Cell Sci. 130, 989-1002. 10.1242/jcs.194779 [DOI] [PubMed] [Google Scholar]
- LaCroix A. S., Rothenberg K. E. and Hoffman B. D. (2015). Molecular-scale tools for studying mechanotransduction. Annu. Rev. Biomed. Eng. 17, 287-316. 10.1146/annurev-bioeng-071114-040531 [DOI] [PubMed] [Google Scholar]
- Lesniewski L. A., Hosch S. E., Neels J. G., de Luca C., Pashmforoush M., Lumeng C. N., Chiang S.-H., Scadeng M., Saltiel A. R. and Olefsky J. M. (2007). Bone marrow–specific Cap gene deletion protects against high-fat diet–induced insulin resistance. Nat. Med. 13, 455-462. 10.1038/nm1550 [DOI] [PubMed] [Google Scholar]
- Lin W.-H., Huang C.-J., Liu M.-W., Chang H.-M., Chen Y.-J., Tai T.-Y. and Chuang L.-M. (2001). Cloning, mapping, and characterization of the human sorbin and SH3 domain containing 1 (SORBS1) gene: a protein associated with c-Abl during insulin signaling in the hepatoma cell line Hep3B. Genomics 74, 12-20. 10.1006/geno.2001.6541 [DOI] [PubMed] [Google Scholar]
- Liu Z., Bun P., Audugé N., Coppey-Moisan M. and Borghi N. (2016). Vinculin head–tail interaction defines multiple early mechanisms for cell substrate rigidity sensing. Integr. Biol. 8, 693-703. 10.1039/C5IB00307E [DOI] [PubMed] [Google Scholar]
- Lo C.-M., Wang H.-B., Dembo M. and Wang Y. (2000). Cell movement is guided by the rigidity of the substrate. Biophys. J. 79, 144-152. 10.1016/S0006-3495(00)76279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai K., Nakanishi H., Satoh A., Takahashi K., Satoh K., Nishioka H., Mizoguchi A. and Takai Y. (1999). Ponsin/SH3P12: an l-Afadin- and vinculin-binding protein localized at cell–cell and cell–matrix adherens junctions. J. Cell Biol. 144, 1001-1018. 10.1083/jcb.144.5.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Geudens I., Bruyr J., Potente M., Bleuart A., Lebrun M., Simonis N., Deroanne C., Twizere J.-C., Soubeyran P. et al. (2013). PP2A regulatory subunit Bα controls endothelial contractility and vessel lumen integrity via regulation of HDAC7. EMBO J. 32, 2491-2503. 10.1038/emboj.2013.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsushima M., Sezaki T., Akahane R., Ueda K., Suetsugu S., Takenawa T. and Kioka N. (2006a). Protein kinase A-dependent increase in WAVE2 expression induced by the focal adhesion protein vinexin. Genes Cells 11, 281-292. 10.1111/j.1365-2443.2006.00932.x [DOI] [PubMed] [Google Scholar]
- Mitsushima M., Takahashi H., Shishido T., Ueda K. and Kioka N. (2006b). Abl kinase interacts with and phosphorylates vinexin. FEBS Lett. 580, 4288-4295. 10.1016/j.febslet.2006.06.072 [DOI] [PubMed] [Google Scholar]
- Mitsushima M., Ueda K. and Kioka N. (2006c). Vinexin β regulates the phosphorylation of epidermal growth factor receptor on the cell surface. Genes Cells 11, 971-982. 10.1111/j.1365-2443.2006.00995.x [DOI] [PubMed] [Google Scholar]
- Pasapera A. M., Schneider I. C., Rericha E., Schlaepfer D. D. and Waterman C. M. (2010). Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 188, 877-890. 10.1083/jcb.200906012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek M. J., Zahir N., Johnson K. R., Lakins J. N., Rozenberg G. I., Gefen A., Reinhart-King C. A., Margulies S. S., Dembo M., Boettiger D. et al. (2005). Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241-254. 10.1016/j.ccr.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Pelham R. J. Jr and Wang Y.-L. (1997). Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. 94, 13661-13665. 10.1073/pnas.94.25.13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyton S. R. and Putnam A. J. (2005). Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J. Cell. Physiol. 204, 198-209. 10.1002/jcp.20274 [DOI] [PubMed] [Google Scholar]
- Ploeger C., Waldburger N., Fraas A., Goeppert B., Pusch S., Breuhahn K., Wang X. W., Schirmacher P. and Roessler S. (2016). Chromosome 8p tumor suppressor genes SH2D4A and SORBS3 cooperate to inhibit interleukin-6 signaling in hepatocellular carcinoma. Hepatology 64, 828-842. 10.1002/hep.28684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribon V., Herrera R., Kay B. K. and Saltiel A. R. (1998a). A role for CAP, a novel, multifunctional Src homology 3 domain-containing protein in formation of actin stress fibers and focal adhesions. J. Biol. Chem. 273, 4073-4080. 10.1074/jbc.273.7.4073 [DOI] [PubMed] [Google Scholar]
- Ribon V., Printen J. A., Hoffman N. G., Kay B. K. and Saltiel A. R. (1998b). A novel, multifunctional c-Cbl binding protein in insulin receptor signaling in 3T3-L1 adipocytes. Mol. Cell. Biol. 18, 872-879. 10.1128/MCB.18.2.872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P., Iskratsch T. and Sheetz M. P. (2012). Finding the weakest link – exploring integrin-mediated mechanical molecular pathways. J. Cell Sci. 125, 3025-3038. 10.1242/jcs.095794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P., del Rio A., Puklin-Faucher E., Gauthier N. C., Biais N. and Sheetz M. P. (2013). Integrin-dependent force transmission to the extracellular matrix by α-actinin triggers adhesion maturation. Proc. Natl. Acad. Sci. USA 110, E1361-E1370. 10.1073/pnas.1220723110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönty M., Taivainen A., Moza M., Kruh G. D., Ehler E. and Carpen O. (2005). Involvement of palladin and α-actinin in targeting of the Abl/Arg kinase adaptor ArgBP2 to the actin cytoskeleton. Exp. Cell Res. 310, 88-98. 10.1016/j.yexcr.2005.06.026 [DOI] [PubMed] [Google Scholar]
- Sakane A., Yoshizawa S., Nishimura M., Tsuchiya Y., Matsushita N., Miyake K., Horikawa K., Imoto I., Mizuguchi C., Saito H. et al. (2016). Conformational plasticity of JRAB/MICAL-L2 provides ‘law and order’ in collective cell migration. Mol. Biol. Cell 27, 3095-3108. 10.1091/mbc.E16-05-0332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffhauer E. S., Luo T., Mohan K., Srivastava V., Qian X., Griffis E. R., Iglesias P. A. and Robinson D. N. (2016). Mechanoaccumulative elements of the mammalian actin cytoskeleton. Curr. Biol. 26, 1473-1479. 10.1016/j.cub.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Chang R., Dai C., Wu Y., Guo J., Qi M., Zhou W. and Zhan L. (2016). SORBS1 suppresses tumor metastasis and improves the sensitivity of cancer to chemotherapy drug. Oncotarget 8, 9108-9122. 10.18632/oncotarget.12851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubeyran P., Barac A., Szymkiewicz I. and Dikic I. (2003). Cbl-ArgBP2 complex mediates ubiquitination and degradation of c-Abl. Biochem. J. 370, 29-34. 10.1042/bj20021539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taieb D., Roignot J., Andre F., Garcia S., Masson B., Pierres A., Iovanna J.-L. and Soubeyran P. (2008). ArgBP2-dependent signaling regulates pancreatic cell migration, adhesion, and tumorigenicity. Cancer Res. 68, 4588-4596. 10.1158/0008-5472.CAN-08-0958 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Mitsushima M., Okada N., Ito T., Aizawa S., Akahane R., Umemoto T., Ueda K. and Kioka N. (2005). Role of interaction with vinculin in recruitment of vinexins to focal adhesions. Biochem. Biophys. Res. Commun. 336, 239-246. 10.1016/j.bbrc.2005.08.064 [DOI] [PubMed] [Google Scholar]
- Thievessen I., Thompson P. M., Berlemont S., Plevock K. M., Plotnikov S. V., Zemljic-Harpf A., Ross R. S., Davidson M. W., Danuser G., Campbell S. L. et al. (2013). Vinculin–actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 202, 163-177. 10.1083/jcb.201303129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro G. J. and Green H. (1963). Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17, 299-313. 10.1083/jcb.17.2.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y., Li Y., Gu H., Wang C., Liu F., Shao Y., Li J., Cao L. and Li F. (2015). Microchidia protein 2, MORC2, downregulates the cytoskeleton adapter protein, ArgBP2, via histone methylation in gastric cancer cells. Biochem. Biophys. Res. Commun. 467, 821-827. 10.1016/j.bbrc.2015.10.059 [DOI] [PubMed] [Google Scholar]
- Ulrich T. A., de Juan Pardo E. M. and Kumar S. (2009). The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 69, 4167-4174. 10.1158/0008-5472.CAN-08-4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Golemis E. A. and Kruh G. D. (1997). ArgBP2, a multiple Src homology 3 domain-containing, Arg/Abl-interacting protein, is phosphorylated in v-Abl-transformed cells and localized in stress fibers and cardiocyte Z-disks. J. Biol. Chem. 272, 17542-17550. 10.1074/jbc.272.28.17542 [DOI] [PubMed] [Google Scholar]
- Wang H. B., Dembo M. and Wang Y. L. (2000). Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am. J. Physiol. Cell Physiol. 279, C1345-C1350. [DOI] [PubMed] [Google Scholar]
- Wolfenson H., Bershadsky A., Henis Y. I. and Geiger B. (2011). Actomyosin-generated tension controls the molecular kinetics of focal adhesions. J. Cell Sci. 124, 1425-1432. 10.1242/jcs.077388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H., Ueda K. and Kioka N. (2011). WAVE2 forms a complex with PKA and is involved in PKA enhancement of membrane protrusions. J. Biol. Chem. 286, 3907-3914. 10.1074/jbc.M110.145409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H., Ichikawa T., Matsuyama D., Kimura Y., Ueda K., Craig S. W., Harada I. and Kioka N. (2014). The role of the interaction of the vinculin proline-rich linker region with vinexin α in sensing the stiffness of the extracellular matrix. J. Cell Sci. 127, 1875-1886. 10.1242/jcs.133645 [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Matsui T. S. and Deguchi S. (2016). New wrinkling substrate assay reveals traction force fields of leader and follower cells undergoing collective migration. Biochem. Biophys. Res. Commun. 482, 975-979. 10.1016/j.bbrc.2016.11.142 [DOI] [PubMed] [Google Scholar]
- Zhang M., Liu J., Cheng A., DeYoung S. M., Chen X., Dold L. H. and Saltiel A. R. (2006). CAP interacts with cytoskeletal proteins and regulates adhesion-mediated ERK activation and motility. EMBO J. 25, 5284-5293. 10.1038/sj.emboj.7601406 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.