ABSTRACT
Switch/sucrose non-fermentable (SWI/SNF) chromatin remodeling complexes are mutated in many human cancers. In this article, we make use of a Drosophila genetic model for epithelial tumor formation to explore the tumor suppressive role of SWI/SNF complex proteins. Members of the BAP complex exhibit tumor suppressor activity in tissue overexpressing the Yorkie (Yki) proto-oncogene, but not in tissue overexpressing epidermal growth factor receptor (EGFR). The Brahma-associated protein (BAP) complex has been reported to serve as a Yki-binding cofactor to support Yki target expression. However, we observed that depletion of BAP leads to ectopic expression of Yki targets both autonomously and non-autonomously, suggesting additional indirect effects. We provide evidence that BAP complex depletion causes upregulation of the Wingless (Wg) and Decapentaplegic (Dpp) morphogens to promote tumor formation in cooperation with Yki.
KEY WORDS: SWI/SNF, Hippo pathway, Yorkie, Oncogenic cooperation, Cancer, Drosophila
Summary: Using a Drosophila model of epithelial transformation, the authors identify the SWI/SNF BAP complex as an element that represses Yorkie activity and cooperates with the Yorkie protein in malignant tumor formation.
INTRODUCTION
Tumors accumulate multiple genetic and epigenetic modifications, and mutations in epigenetic regulators are associated with several types of human cancer. Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ) are the nuclear effectors of the Hippo pathway, regulate organ growth and are potent drivers of tumor formation (Harvey et al., 2013; Johnson and Halder, 2014; Piccolo et al., 2014; Yu et al., 2015). Even though activation of YAP and TAZ are widespread in human cancers, much remains to be learned about other factors that may cooperate with these oncogenes to promote malignant tumor formation.
Genes encoding Switch/sucrose non-fermentable (SWI/SNF) chromatin remodeling complex proteins are among the most commonly mutated in human cancer and have a crucial role in tumor suppression (Kadoch et al., 2013; Wilson and Roberts, 2011). However, specific mechanisms by which SWI/SNF complexes suppress tumor formation remain poorly understood. Using simple genetic tumor models provides a means to explore these mechanisms. Subunits of the SWI/SNF complex have been shown to play a tumor suppressive role in Drosophila (Eroglu et al., 2014; Koe et al., 2014; Xie et al., 2017).
Carcinomas originating in epithelial tissues are among the most common human cancers. The Drosophila imaginal discs are epithelial monolayers that proliferate actively during larval development, and have proven to be a useful model to study epithelial tumor formation (reviewed in Herranz et al., 2016). Overexpression of Yorkie (Yki), the fly ortholog of the YAP oncoprotein, leads to benign epithelial hyperplasia without driving the tissue into neoplasia (Dong et al., 2007; Herranz et al., 2012a; Huang et al., 2005). We made use of the wing imaginal disc of Drosophila to identify genes cooperating with Yki in malignant tumor formation.
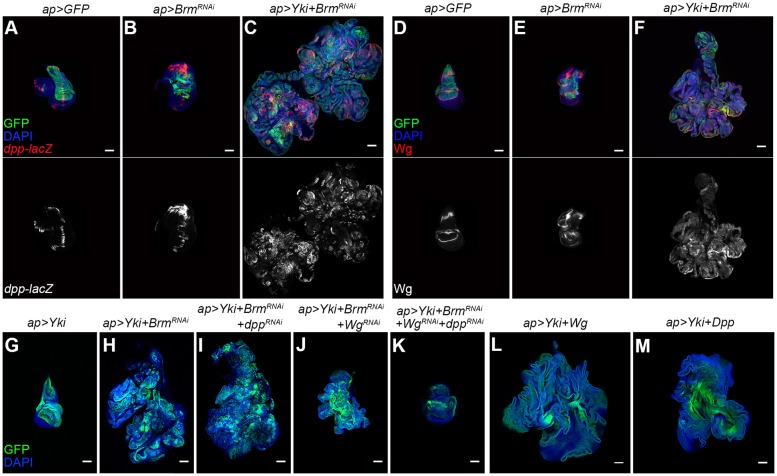
In this study, we identify the SWI/SNF Brahma-associated protein (BAP) complex as a suppressor of Yki-induced tumor formation. Although depletion of the BAP remodeling complex has been shown to reduce expression of Yki target genes in the wing pouch (Oh et al., 2013; Zhu et al., 2015), we observed upregulation of Yki targets in other regions of the wing imaginal disc. We also provide evidence for an indirect effect of BAP complex depletion mediated by upregulation of the signaling proteins Wingless (Wg) and Decapentaplegic (Dpp). Ectopic Wg and, to a lesser extent, Dpp expression contributes to the tumor suppressive effect of the BAP complex in the context of excessive Yki activity.
RESULTS
Synergistic interaction between Yki and the SWI/SNF BAP complex
To identify genes cooperating with Yki in malignant tumor formation, we made use of apterous-Gal4 (apGal4) to direct the expression of UAS-transgenes in the dorsal compartment of the wing imaginal disc epithelium; the ventral compartment serves as an internal control (Fig. 1A). Expression of a UAS-Yki transgene caused overgrowth of the dorsal compartment of the disc (Fig. 1B). The larvae pupariated, but died as pupae.
Fig. 1.
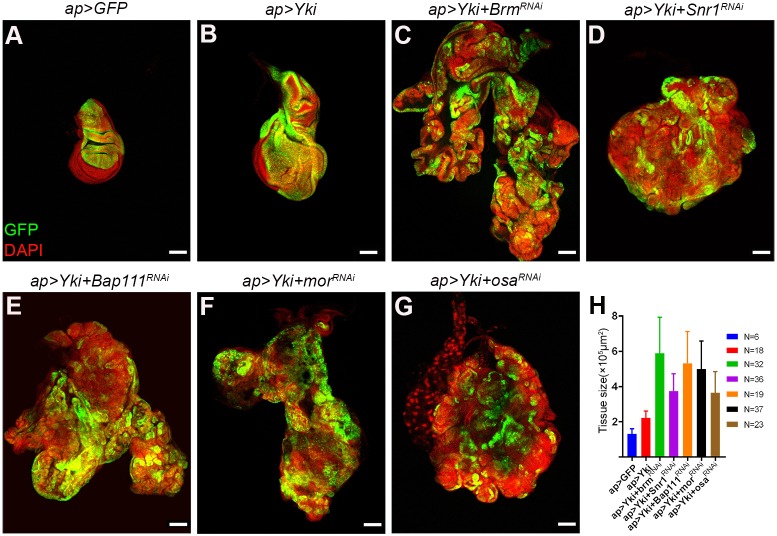
Depletion of BAP complex subunits enhances Yki-induced hyperplasia. Confocal micrographs of wing imaginal discs expressing combinations of UAS-transgenes under apGal4 control. Discs were labeled with UAS-GFP to mark the transgene-expressing tissue (green) and with DAPI to label nuclei (red). (A) Control wing imaginal disc expressing GFP. (B) Wing imaginal disc expressing Yki. (C-G) Wing imaginal discs co-expressing UAS-Yki with UAS-RNAi transgenes to deplete expression of BAP complex subunit genes. Scale bars: 100 μm. (H) Quantification of tissue size (ap>Yki versus ap>Yki+BAPRNAis, unpaired t-test, P<0.0001 for all).
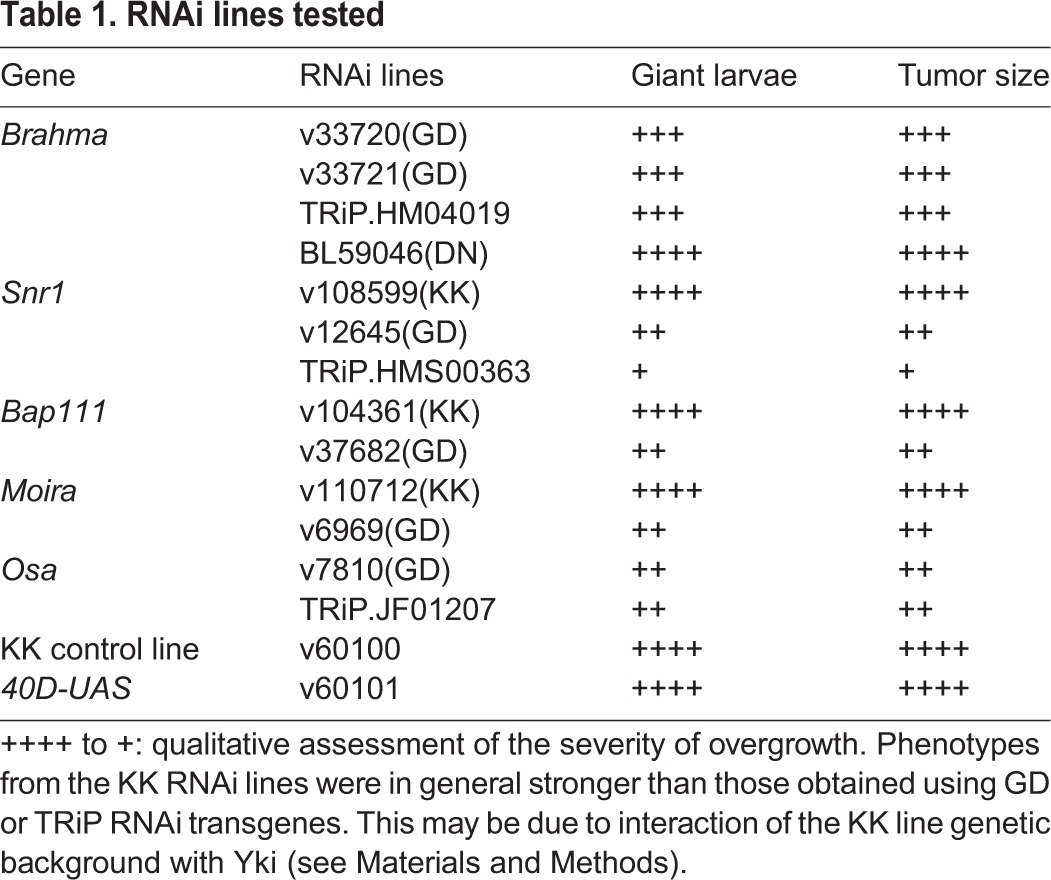
To look for cooperating factors that drive neoplasia in the context of increased Yki activity, we coexpressed UAS-RNAi transgenes directed against a variety of epigenetic regulators together with UAS-Yki. Under these conditions, depletion of Brahma (Brm), a subunit of the SWI/SNF chromatin remodeling complex, led to massive overgrowth of the disc (Fig. 1C). Inactivating mutations in several SWI/SNF subunits have been identified at a high frequency in a variety of cancers (Wilson and Roberts, 2011). We asked whether other SWI/SNF components cooperated with Yki in the formation of tumors. Drosophila has two distinct SWI/SNF complexes: BAP and Polybromo-BAP (PBAP) (Mohrmann and Verrijzer, 2005; Moshkin et al., 2007). Both share a common multi-subunit core, comprising the Brm ATPase, Snr1, Bap111 and Moira (Mor) proteins. This core associates with Osa protein, to form the BAP complex, or with Polybromo, Bap170 and SAYP proteins to form the PBAP complex (Chalkley et al., 2008; Mohrmann and Verrijzer, 2005; Moshkin et al., 2007). As with Brm depletion, co-expressing Yki with UAS-RNAi transgenes against the other common core elements (Snr1, Bap111 and Mor), as well as depletion of Osa, led to massive overgrowth (Fig. 1D-G). The resulting tissues were significantly larger than that produced by expression of UAS-Yki alone (Fig. 1H). At least two independent RNAi transgenes were tested for each gene, with comparable results (Table 1). Introducing UAS-RNAi transgenes targeting the PBAP-specific subunits Polybromo, Bap170 or SAYP did not lead to synergistic overgrowth when combined with UAS-Yki (Fig. S1). These observations suggest that the BAP SWI/SNF complex in some way limits the growth promoting effects of Yki overexpression, whereas the PBAP complex does not have this effect.
Table 1.
RNAi lines tested
Yki+BAPRNAi tumors exhibit malignant characteristics
Larvae co-expressing UAS-Yki and UAS-RNAi transgenes targeting BAP complex subunits under the control of the apGal4 driver carried large tumorous discs (Fig. 2A-F; we refer to these as Yki+BAPRNAi larvae). Some Yki+BAPRNAi larvae did not pupate and instead continued to grow to form giant larvae, in some cases growing for more than 20 days before eventually dying. The ‘giant larva’ phenotype is characteristic of larvae with malignant tumors (Bilder, 2004). In some cases, we observed GFP-expressing tissue growths at a distance from the main tumorous mass (Fig. 2F). Ectopically located GFP-expressing tissue was never observed in larvae overexpressing Yki on its own (Fig. 2A). These ectopic GFP masses are likely to reflect metastasis from the overgrown tumorous disc.
Fig. 2.
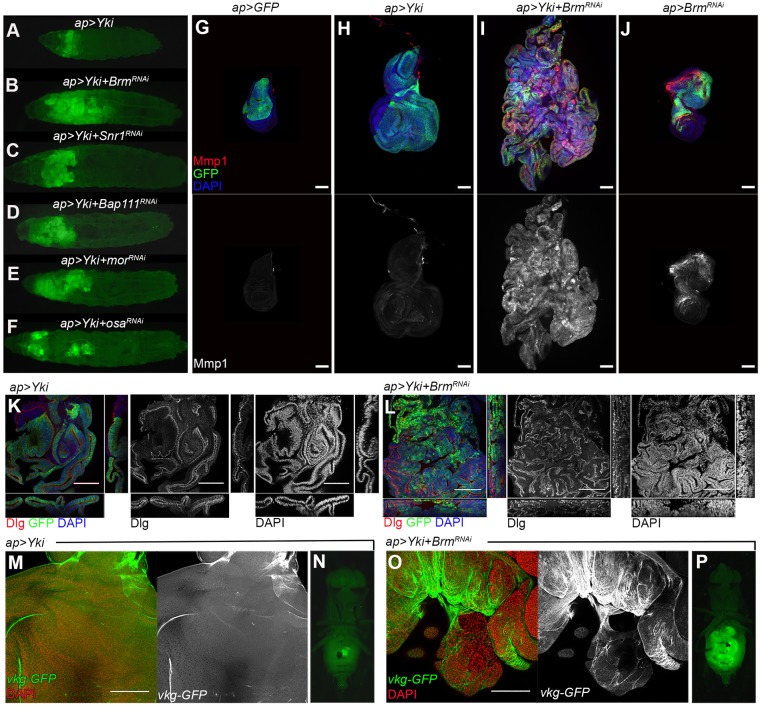
Tumor formation by BAP-complex-depleted tissue. (A-F) Low-magnification views of whole larvae showing the imaginal disc overgrowth for the indicated transgene combinations. GFP-expressing tissue expands massively to fill the anterior of the animal. (G-J) Confocal micrographs of wing discs expressing the indicated combinations of UAS-transgenes. Discs were labeled with antibody to Matrix metalloprotease 1 (Mmp1, red) as well as UAS-GFP to mark the Yki-expressing tissue (green) and DAPI (blue) to outline the tissue. Mmp1 channel shown separately in gray below. (K,L) Confocal micrographs of wing discs expressing the indicated combinations of UAS-transgenes. xy section is shown in the central picture; xz section is shown in the bottom; and yz section is shown in the right side. Note that the tissue in K maintains the normal epithelial organization whereas the tumor in L is highly disorganized. Discs large (Dlg) is show in red and gray. DAPI labels the nuclei and is shown in blue and gray. GFP is shown in green. (M,O) Confocal micrographs of wing discs expressing the indicated combinations of UAS-transgenes. Viking (vkg)-GFP was used to label the basement membrane and is shown in green and gray. The basement membrane appears as a continuous layer in the disc in M, whereas it is more disorganized and in some parts has been degraded in the tumor in O. (N,P) Fluorescent images of whole flies, showing GFP-expressing allograft tissue in the host abdomen. Fragments of imaginal discs were implanted in the abdomen of a host female and allowed to grow for 2 weeks. (N) apGal4, UAS-Yki+UAS-GFP, 0/12 showed overgrowth; (P) apGal4, UAS-Yki+UAS-BrmRNAi, 16/19 showed overgrowth. Scale bars: 100 μm.
Malignant fly tumors express the secreted matrix metalloproteinase 1 (Mmp1). Mmp1 degrades the basement membrane of the imaginal disc, allowing tumor cell migration and invasion (Beaucher et al., 2007; Uhlirova and Bohmann, 2006). In normal discs, Mmp1 is expressed in the trachea but is not detected in the proliferating epithelium (Fig. 2G). We did not detect Mmp1 expression in discs expressing UAS-Yki (Fig. 2H), but the Yki+BAPRNAi tumors had high levels of Mmp1 (Fig. 2I and Fig. S2A-D). Mmp1 expression correlated with degradation of basement membranes, visualized using Viking-GFP (vkg-GFP) (Fig. 2O; compare with the continuous basement membrane layer in Yki control discs, Fig. 2M). Interestingly, Mmp1 expression was observed when BAP complex transcripts were depleted in the absence of Yki overexpression (Fig. 2J and Fig. S2E-H). Carcinomas show defects in polarity as they evolve towards malignancy. Yki expression results in tissue overgrowth but those discs maintained normal epithelial polarity, as shown by localized expression of the apical polarity marker Discs large (Dlg, Fig. 2K). In contrast, epithelial polarity was disrupted in the Yki+BAPRNAi tumors (Fig. 2L).
Transplantation of imaginal disc fragments into the abdomen of adult hosts provides an in vivo assay system for tumor formation (Caussinus and Gonzalez, 2005). To assess the growth potential of the Yki+BAPRNAi tumors, we injected fragments of Yki+GFP-expressing discs, and Yki+BrmRNAi discs. Discs fragments expressing Yki alone with GFP survived in the abdomen of adult hosts 2 weeks after tumor injection, but they did not grow to form tumors (Fig. 2N). In contrast, fragments of Yki+BrmRNAi discs grew rapidly to fill much of the host abdomen and killed the hosts by 2 weeks (Fig. 2P). Taken together, these observations suggest that the Yki+BAPRNAi tumors have acquired malignant features.
BAP complex depletion does not synergize with EGFR
Activating mutations in epidermal growth factor receptors (EGFRs) and downstream effectors in the Ras/MAPK pathway are frequent in human cancer (Kandoth et al., 2013). EGFR behaves as an oncogene in Drosophila, and EGFR overexpression results in activation of the Ras/MAPK pathway and tissue hyperplasia (Herranz et al., 2012a). EGFR expression results in benign tissue overgrowth, resembling that produced by Yki overexpression (Fig. S3). As with Yki, combining EGFR with other factors can drive tumor formation (Eichenlaub et al., 2016; Herranz et al., 2012b, 2014). However, depletion of the BAP complex transcripts did not show synergistic interaction with EGFR. Growth of the tissue was similar to that produced by EGFR alone (Fig. 3A-F). These observations suggest that the BAP complex does not simply limit tissue overgrowth by any growth driver. Instead, there appears to be a specific interaction in Yki-expressing tissue.
Fig. 3.
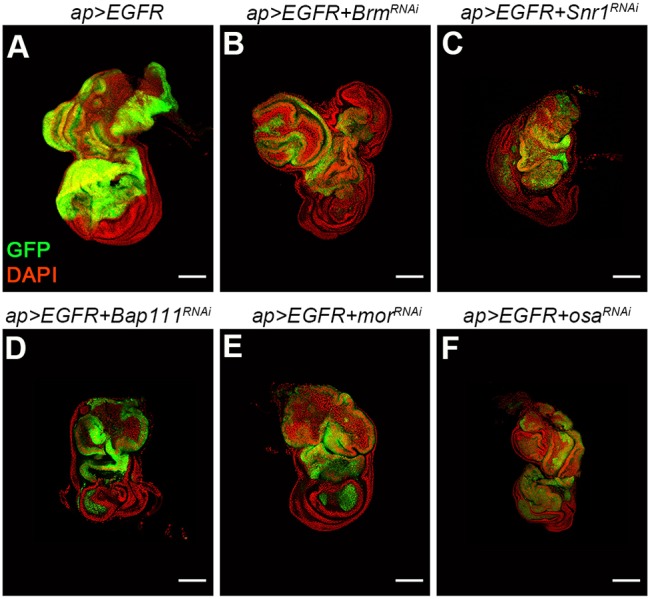
Depletion of BAP complex subunits does not enhance EGFR-induced hyperplasia. Confocal micrographs of wing discs expressing the indicated combinations of UAS-transgenes. Discs were labeled with UAS-GFP to mark the transgene-expressing tissue (green) and DAPI (red) to outline the tissue. Scale bars: 100 μm.
Depletion of BAP induces reaper expression and apoptosis
We next examined the consequences of depleting BAP complex subunits on their own in the developing wing epithelium. Discs depleted for different members of the BAP complex showed morphological defects and appeared moderately overgrown (Fig. 4A-D and Fig. S4A-J). These discs contained pyknotic nuclei, which are a sign of DNA fragmentation in apoptotic cells.
Fig. 4.
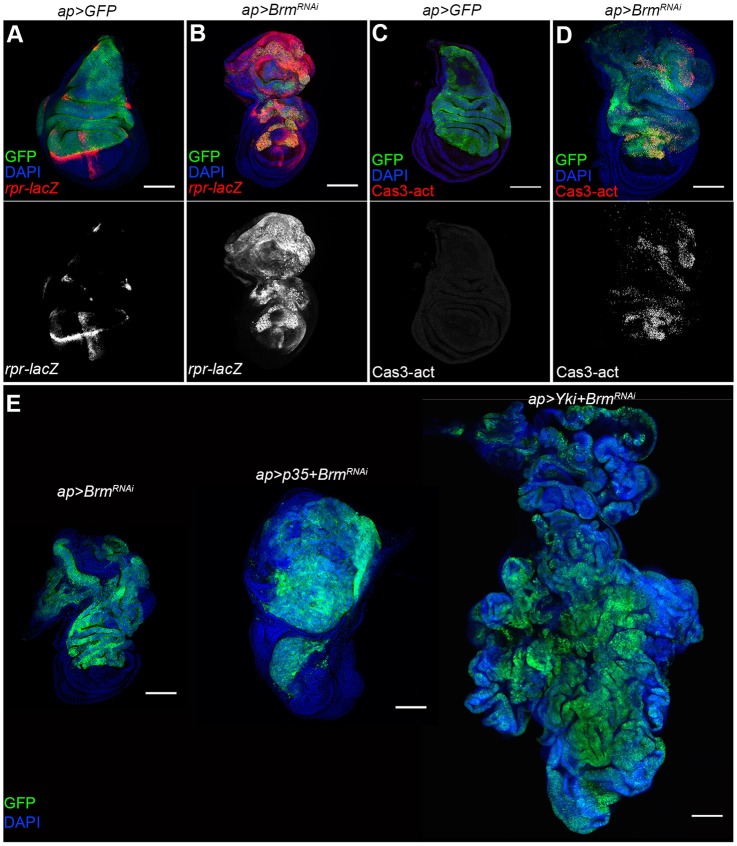
Apoptosis in BAP-complex-depleted tissue. Confocal micrographs of wing discs expressing the indicated combinations of UAS-transgenes. Discs were labeled with UAS-GFP to mark the transgene-expressing tissue (green) and DAPI (blue) to outline the tissue. (A,B) A lacZ transgene insertion at the reaper (rpr) locus was used to visualize reaper expression (red). (C,D) Discs were labeled with antibody to the activated form of Caspase-3 to visualize apoptotic cells (red). (E) Comparison of the effect of blocking apoptosis by expression on baculovirus p35 protein or UAS-Yki with UAS-BrmRNAi. Examples of other transgene combinations are shown in Fig. S4. Scale bars: 100 μm.
The Drosophila proapototic gene reaper is transcriptionally activated in response to a number of different signals (Brodsky et al., 2000; Lohmann et al., 2002; Nordstrom et al., 1996). Reaper induces apoptosis by regulating Drosophila Death-associated inhibitor of apoptosis protein 1 (DIAP1) (Goyal et al., 2000; Lisi et al., 2000; Yoo et al., 2002). We monitored reaper expression using a lacZ transgene inserted at the reaper locus. In normal discs, reaper-lacZ is expressed along the dorso-ventral and anterior-posterior compartment boundaries (Fig. 4A). We found that depletion of BAP components resulted in substantial ectopic reaper-lacZ expression (Fig. 4B and Fig. S4A-E). We also used an antibody against the activated form of Caspase-3 to visualize apoptotic cells. Apoptosis is normally undetectable in control wing discs (Fig. 4C), but many Caspase-3-positive cells were found in discs depleted of the BAP complex proteins (Fig. 4D and Fig. S4F-J).
Apoptosis is a tumor suppressor mechanism. Suppression of apoptosis can induce cell proliferation in Drosophila and, under some conditions, can also induce tumor formation (Huh et al., 2004; Perez-Garijo et al., 2004; Ryoo et al., 2004). To investigate whether the interaction of BAP components with Yki could be explained by suppression of apoptosis, we blocked apoptosis with the baculovirus protein p35. In discs depleted for BAP complex transcripts, co-expression of p35 led to robust overgrowth (Fig. 4E), although not as much as with the combination of p35 and Yki+BrmRNAi (Fig. 4E and Fig. S4K-O).
BAP inhibits Yki activity in the hinge and notum regions of the wing disc
The BAP subunits Brm and Mor have been identified as Yki-associated proteins, and depletion of these subunits has been reported to downregulate Yki target genes in the wing pouch region of the wing disc (Oh et al., 2013; Zhu et al., 2015). Based on this, we should expect Brm depletion to lower Yki activity, perhaps limiting the effects of Yki-induced growth. However, we found that Brm depletion enhanced Yki-induced tissue overgrowth.
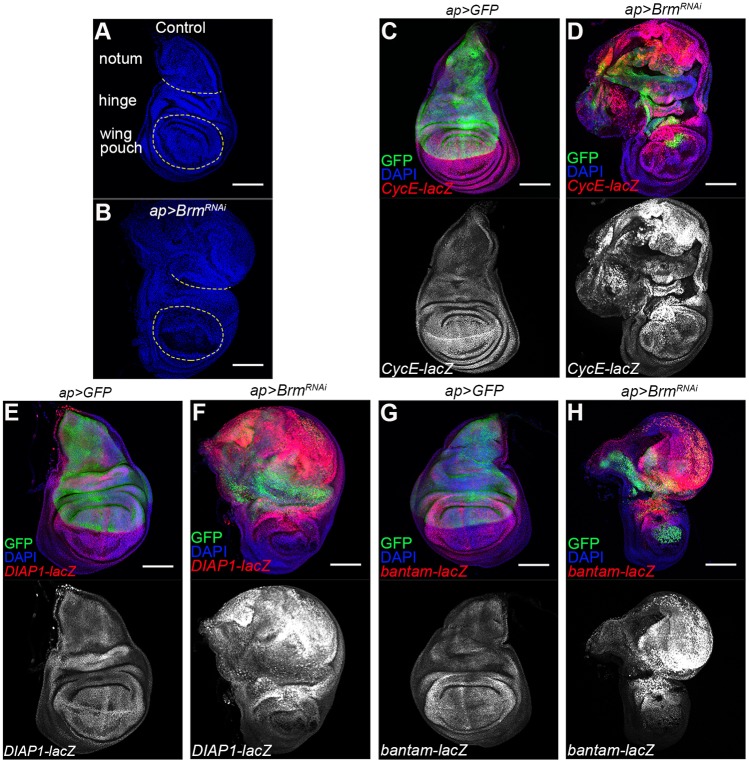
We noted that most of the tissue overgrowth in the imaginal discs depleted of the BAP complex subunits was in the region of the disc that corresponds to the wing hinge and to the presumptive dorsal thorax (notum; Fig. 5A,B), although ectopic cell death was observed throughout the apGal4 expression domain in the dorsal wing pouch, as well as in the hinge and notum areas (Fig. 4A-D). We therefore examined expression of the Yki target genes Cyclin E, the inhibitor of apoptosis DIAP1, and bantam microRNA (Huang et al., 2005; Nolo et al., 2006; Thompson and Cohen, 2006). Transgenic reporters for Cyclin E, DIAP1 and bantam showed strong ectopic expression in the overgrowing wing hinge and notum tissue in discs expressing the UAS-BAPRNAi transgenes compared to their expression levels in normal control discs, whereas expression in the wing pouch was normal to low (Fig. 5C-H and Fig. S5). Thus, the finding of BAP complex depletion causing high ectopic Yki target expression are consistent with the observed increase in tissue growth.
Fig. 5.
Yki target gene expression in BAP-complex-depleted wing discs. Confocal micrographs of wing discs expressing the indicated combinations of UAS-transgenes. Discs were labeled with DAPI (blue) to outline the tissue (A,B) and with UAS-GFP to mark the transgene-expressing tissue (green, C-H). (A,B) Schematic representation of regions of the wing disc to show that overgrowth due to UAS-BrmRNAi was mostly in the notum region. (C-H) Effects of Brm depletion on expression of Yki target genes. (C,D) CyclinE-lacZ transgene expression (red). (E,F) DIAP1-lacZ transgene expression (red). (G,H) bantam-lacZ transgene expression (red). Scale bars: 100 μm.
Depletion of the BAP complex leads to ectopic Wg and Dpp
The observed ectopic induction of Yki targets in BAP-depleted discs would be difficult to explain solely in terms of reduced Yki activity due to depletion of the BAP cofactor proteins. This led us to consider the possibility that the BAP complex might also affect other pathways that regulate growth control in the wing disc.
The secreted growth factors Dpp and Wg are important regulators of tissue growth and cell proliferation in the wing disc (Dekanty and Milán, 2011). Using a lacZ reporter, we monitored dpp expression in discs depleted for the BAP components. dpp-lacZ is normally expressed in a stripe close to the anterior-posterior compartment boundary (Fig. 6A). dpp-lacZ was ectopically expressed in the overgrowing tissue of the BAP-complex-depleted discs (Fig. 6B and Fig. S6A-E) and was ectopically expressed throughout the Yki+BAPRNAi tumorous discs (Fig. 6C and Fig. S6F-J). Wg regulates wing disc growth and patterning, and ectopic Wg induces the formation of ectopic wing structures (Ng et al., 1996). We observed ectopic expression of Wg protein in discs depleted for the BAP complex proteins (Fig. 6D,E and Fig. S7A-E). Ectopic Wg expression was extensive in the Yki+BAPRNAi tumorous discs (Fig. 6F and Fig. S7F-J). Consistent with the earlier work of Ng et al. (Ng et al., 1996), ectopic Wg expression led to the formation of ectopic wing pouch tissue in the notum region (marked by Nubbin expression; data not shown). However, our findings contrast in part with those of Collins and Treisman (Collins and Treisman, 2000), who reported that ectopic wing tissue was formed in osa mutants without induction of Wg expression.
Fig. 6.
Ectopic expression of Wg and Dpp in BAP-complex-depleted wing discs. Confocal micrographs of wing discs expressing the indicated combinations of UAS-transgenes. Discs were labeled with UAS-GFP to mark the transgene-expressing tissue (green) and DAPI (blue) to outline the tissue. (A-C) dpp-lacZ was used to visualize dpp expression (red). (D-F) Discs were labeled with antibody to Wingless (Wg) protein (red). (G-K) Effect of RNAi-mediated depletion of dpp or Wingless in the UAS-Yki+UAS-BrmRNAi background. (L) Coexpression of UAS-Wingless with UAS-Yki. (M) Coexpression of UAS-Dpp with UAS-Yki. Scale bars: 100 μm.
To assess the contributions of ectopic Wg and Dpp expression to tumor formation, we used RNAi-mediated depletion to limit their expression in Yki+BrmRNAi discs (Fig. 6I,J). Wg depletion strongly suppressed the growth of the Yki+BrmRNAi tumors, whereas depletion of Dpp on its own had a relatively limited effect. However, simultaneous depletion of Wg and Dpp was more effective than depletion of Wg alone (Fig. 6K), and coexpression of Yki with Wg or with Dpp proved to be sufficient to promote overgrowth (Fig. 6L,M). Thus, misexpression of Wg and Dpp each contribute to the formation of the Yki+BrmRNAi tumors.
DISCUSSION
Benign tumors accumulate mutations that enable them to progress to malignancy and metastasis. Although Yki overexpression promotes cell proliferation and inhibits apoptosis, Yki expression does not normally lead to the formation of malignant tumors in the Drosophila wing epithelia. Our findings show that inactivation of the BAP complex in discs expressing Yki results in the formation of giant larvae, a phenomenon characteristic of larvae with neoplastic tumors. The overgrown imaginal discs in these animals exhibit features of malignant transformation, including loss of epithelial polarity and expression of the proinvasive marker Mmp1. Moreover, when transplanted to a normal host, fragments of these discs produced tumors that grew and spread to kill the host.
The tumor suppressive role of the BAP complex appears to be context dependent. Overexpression of EGFR and Yki each results in tissue hyperplasia. Yki regulates cell proliferation and represses apoptosis by regulating target genes, including the cell cycle regulator Cyclin E, the inhibitor of apoptosis DIAP1 and the microRNA bantam (Huang et al., 2005; Nolo et al., 2006; Thompson and Cohen, 2006). Similarly, EGFR activates the Ras/MAPK pathway and induces cell proliferation by inducing Myc proto-oncogene expression (Prober and Edgar, 2000, 2002). EGFR signaling also represses apoptosis by inhibiting activity of the proapoptotic gene Hid (Bergmann et al., 1998; Kurada and White, 1998). Even though overexpression of EGFR and Yki resulted in a very similar growth phenotype, inactivation of BAP subunits drove tumor formation in discs expressing Yki but not in discs expressing EGFR.
Previous work has shown that some BAP subunits interact with Yki to regulate gene expression, and that Yki target gene expression was reduced following BAP complex depletion (Oh et al., 2013; Zhu et al., 2015). We were therefore surprised to find ectopic activation of Yki targets in the discs depleted for the BAP complex. Interestingly, we noted that BAP-depleted tissue was largely overgrown in the hinge and notum regions, where Yki target expression was elevated, and that the Yki targets were expressed at normal to low levels in the wing pouch in these discs (Fig. 5). Thus, there appears to be a region-specific difference in the response to BAP complex depletion. Yki regulates gene expression by interacting with a number of different DNA-binding transcription factors: Scalloped, Homothorax, Mad and Cabut (Goulev et al., 2008; Oh and Irvine, 2011; Ruiz-Romero et al., 2015; Wu et al., 2008; Zhang et al., 2008). Brm has been shown to interact with the Yki-Scalloped complex to regulate gene expression in the wing pouch (Zhu et al., 2015). Scalloped promotes wing blade development and shows high levels of expression in the wing pouch, whereas its expression levels are much lower in other regions of the wing disc (Halder et al., 1998; Simmonds et al., 1998). Homothorax is expressed in a pattern complementary to Scalloped, and acts in the hinge and notum regions of the disc (Azpiazu and Morata, 2000; Casares and Mann, 2000). It is possible that interaction of the BAP complex with the Yki-Homothorax complex might produce a different outcome with respect to Yki target gene expression than interaction with the Yki-Scalloped complex.
An alternative hypothesis is that induction of the Yki targets in the BAP-depleted notum tissue reflects an independent input. In support of this, we found that Wg and Dpp were ectopically expressed in the notum region in BAP-depleted discs as well as in the Yki+BAPRNAi discs, and that they contributed to formation of these tumors when co-expressed with Yki. Wg and Dpp are not direct targets of Yki activity but are required for normal growth of the wing imaginal disc, where they act as long-range signals to support cell survival and tissue growth. It may be of interest to explore how Wg and Dpp are ectopically induced. One possibility is that BAP complexes, acting as epigenetic factors, may normally suppress the expression of genes involved in wing pouch development in the notum region, including Wg and Dpp. This may be independent of their effects on Yki.
Another possibility involves indirect consequences of the cell death that results from BAP complex depletion. Previous reports have shown that dying cells in the wing imaginal disc produce Wg and Dpp and that blocking cell death allows for ongoing production of Wg and Dpp by the ‘undead’ cells, leading to overproliferation of the tissue (Huh et al., 2004; Perez-Garijo et al., 2004; Ryoo et al., 2004). Yki expression is anti-apoptotic, through induction of DIAP1 and bantam miRNA, and we observed many cells expressing reaper and showing Caspase-3 activation in the Yki+BAPRNAi tumorous discs. Whereas cell death seems to predominate in the tissue depleted for the BAP complex alone (despite some induction of Wg and Dpp), co-expression with Yki leads to tissue survival and extensive overgrowth. Ultimately, this leads to acquisition of tumorous features in the tissue, including the ability to make invasive malignant tumors that can kill a host animal in allograft experiments. However, it is important to note that blocking apoptosis was not sufficient to mimic the effects of Yki expression in the BAP-complex-depleted tissue, so other Yki targets must also be important.
MATERIALS AND METHODS
Drosophila genetics
Transgenes used: BrmRNAi (v37720, v37721 and BL31712), Snr1RNAi (v12645, v108599 and BL32372), osaRNAi (v7810 and BL31266), morRNAi (v6969 and v110712), Bap111RNAi (v38672 and v104361), Bap170RNAi (v34832), PolybromoRNAi (BL32840), SAYPRNAi (v38638), 40D-UAS (v60101), KK control (v60100), WgRNAi (v13351), UAS-Wg.HA (BL5918), UAS-dpp (BL1486), reaper-lacZ (BL58793), UAS-mCD8:GFP (BL5137), dppRNAi (BL25782), UAS-p35 (BL5072), bantam-lacZ (P{lacW}banL1170a), DIAP1-lacZ, CyclinE-lacZ, dpp-lacZ, viking-GFP (vkgG454) (Morin et al., 2001).
UAS-GFP was used to visualize the apGal4-expressing cells in all experiments. apGal4/Gal80ts was used to direct transgene expression in 3rd instar imaginal discs. Animals were reared at 18°C until early 3rd instar, and shifted to 29°C to induce transgene expression as described (Herranz et al., 2012b). Stocks used for these experiments were: (1) apGal4, UAS-mCD8:GFP/Cyo; tub-Gal80ts/TM6B; (2) apGal4, UAS-GFP/Cyo; UAS-Yki, tub-Gal80ts; and (3) apGal4, UAS-mCD8:GFP/Cyo; UAS-EGFR, tub-Gal80ts.
Interaction of KK RNAi lines with the Hippo pathway
It has been reported that ∼25% KK lines have insertions at both 30B and 40D, and that 40D insertions can affect expression of the nearby tiptop gene. This can result in false positives in screens based on sensitized Hippo pathway phenotypes (Green et al., 2014; Vissers et al., 2016). Among the KK lines used here, only v110712, targeting Bap111, has a 40D insertion (data not show). In each case the phenotypes obtained with KK transgenes were confirmed using independent GDP element insertion library stocks or Transgenic RNAi Project (TRiP) RNAi transgene lines (Table 1).
We tested for interaction in our screen with a known 40D-UAS integration site as well as with the background line, which contains the original landing sites used to produce the KK collection (VDRC line 60100). As expected, the 40D-UAS transgene showed interaction with UAS-Yki in tumor formation. We were surprised to find that this was also true of the original targeting line (Table 1). These tumors were qualitatively different from those resulting from BAP complex depletion (not shown). The experimental results presented in the figures were obtained with GD and TRiP RNAi transgenes, to avoid any potential bias due to cooperation with the KK line genetic background. A list of transgenes used for each figure is provided in Table S1.
Allograft transplantation
Wing disc tissue was removed from larvae in PBS. The discs were cut into small pieces using tweezers and sharp tungsten needles and transplanted by inserting glass capillary needles into the abdomens of 1-week-old w1118 female virgin host flies as described previously (Herranz et al., 2012b). The allografted flies were raised at 29°C to ensure ongoing transgene induction in the implant.
Immunostaining and imaging
Primary antibodies used were: mouse anti-Wg [1:40, Developmental Studies Hybridoma Bank (DSHB), 4D4], mouse anti-βGal (1:50, DSHB, 40-1a), mouse anti-Mmp1 (1:10, DSHB, 3A6B4/5H7B11/3B8D12 were mixed in equal amounts), mouse anti-Dlg (1:200, DSHB, 4F3) and rabbit anti-cleaved Caspase-3 (1:500, Cell Signaling, 9661S). Samples were dissected in PBS, fixed in 4% formaldehyde for 20 min, washed for 3×10 min in PBX (1% Triton X-100 in PBS), blocked in PBX with 2% BSA (BBX) for 30 min, and incubated in BBX with primary antibody at 4°C overnight. Samples were then washed 4×30 min with BBX to remove primary antibody, and incubated in 200 µl BBX with 1 µl secondary antibody for 2 h. Alexa Fluor 555 series secondary antibodies from Thermo Fisher Scientific were used. After washing for 3×10 min in PBX (including DAPI for 1 wash), samples were mounted on glass slides. Images were taken with a Leica SP8 microscope. Image analysis was performed with Fiji software. Whole larva images were taken with a Leica Fluorescence Stereomicroscope.
Tissue size quantification and statistics
Tissue size measure was performed with Fiji software. Statistics was performed with Prism software.
Supplementary Material
Acknowledgements
We thank Qi Le for contributing to initial experiments that tested BAP complex depletion. Strains were obtained from the Bloomington Drosophila Stock Center and the Vienna Drosophila RNAi Center. We thank the Developmental Studies Hybridoma Bank for antibodies.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.S., H.H., S.M.C.; Methodology: S.S., H.H., S.M.C.; Formal analysis: S.S.; Investigation: S.S.; Resources: S.M.C.; Data curation: S.S., S.M.C.; Writing - original draft: S.S., H.H.; Writing - review & editing: H.H., S.M.C.; Supervision: H.H., S.M.C.; Project administration: H.H., S.M.C.; Funding acquisition: S.M.C.
Funding
This work was supported by the Danish Council for Strategic Research (DISC-B) and Novo Nordisk Foundation (NNF12OC0000552 to S.C).
Supplementary information
Supplementary information available online at http://dmm.biologists.org/lookup/doi/10.1242/dmm.030122.supplemental
References
- Azpiazu N. and Morata G. (2000). Function and regulation of homothorax in the wing imaginal disc of Drosophila. Development 127, 2685-2693. [DOI] [PubMed] [Google Scholar]
- Beaucher M., Hersperger E., Page-McCaw A. and Shearn A. (2007). Metastatic ability of Drosophila tumors depends on MMP activity. Dev. Biol. 303, 625-634. 10.1016/j.ydbio.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Bergmann A., Agapite J., McCall K. and Steller H. (1998). The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell 95, 331-341. 10.1016/S0092-8674(00)81765-1 [DOI] [PubMed] [Google Scholar]
- Bilder D. (2004). Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 18, 1909-1925. 10.1101/gad.1211604 [DOI] [PubMed] [Google Scholar]
- Brodsky M. H., Nordstrom W., Tsang G., Kwan E., Rubin G. M. and Abrams J. M. (2000). Drosophila p53 binds a damage response element at the reaper locus. Cell 101, 103-113. 10.1016/S0092-8674(00)80627-3 [DOI] [PubMed] [Google Scholar]
- Casares F. and Mann R. S. (2000). A dual role for homothorax in inhibiting wing blade development and specifying proximal wing identities in Drosophila. Development 127, 1499-1508. [DOI] [PubMed] [Google Scholar]
- Caussinus E. and Gonzalez C. (2005). Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 37, 1125-1129. 10.1038/ng1632 [DOI] [PubMed] [Google Scholar]
- Chalkley G. E., Moshkin Y. M., Langenberg K., Bezstarosti K., Blastyak A., Gyurkovics H., Demmers J. A. A. and Verrijzer C. P. (2008). The transcriptional coactivator SAYP is a trithorax group signature subunit of the PBAP chromatin remodeling complex. Mol. Cell. Biol. 28, 2920-2929. 10.1128/MCB.02217-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R. T. and Treisman J. E. (2000). Osa-containing Brahma chromatin remodeling complexes are required for the repression of wingless target genes. Genes Dev. 14, 3140-3152. 10.1101/gad.854300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekanty A. and Milán M. (2011). The interplay between morphogens and tissue growth. EMBO Rep. 12, 1003-1010. 10.1038/embor.2011.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A. and Pan D. (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120-1133. 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub T., Cohen S. M. and Herranz H. (2016). Cell competition drives the formation of metastatic tumors in a Drosophila model of epithelial tumor formation. Curr. Biol. 26, 419-427. 10.1016/j.cub.2015.12.042 [DOI] [PubMed] [Google Scholar]
- Eroglu E., Burkard T. R., Jiang Y., Saini N., Homem C. C. F., Reichert H. and Knoblich J. A. (2014). SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell 156, 1259-1273. 10.1016/j.cell.2014.01.053 [DOI] [PubMed] [Google Scholar]
- Goulev Y., Fauny J. D., Gonzalez-Marti B., Flagiello D., Silber J. and Zider A. (2008). SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 18, 435-441. 10.1016/j.cub.2008.02.034 [DOI] [PubMed] [Google Scholar]
- Goyal L., McCall K., Agapite J., Hartwieg E. and Steller H. (2000). Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19, 589-597. 10.1093/emboj/19.4.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E. W., Fedele G., Giorgini F. and Kyriacou C. P. (2014). A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat. Methods 11, 222-223. 10.1038/nmeth.2856 [DOI] [PubMed] [Google Scholar]
- Halder G., Polaczyk P., Kraus M. E., Hudson A., Kim J., Laughon A. and Carroll S. (1998). The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes Dev. 12, 3900-3909. 10.1101/gad.12.24.3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K. F., Zhang X. and Thomas D. M. (2013). The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246-257. 10.1038/nrc3458 [DOI] [PubMed] [Google Scholar]
- Herranz H., Hong X. and Cohen S. M. (2012a). Mutual repression by bantam miRNA and Capicua links the EGFR/MAPK and Hippo pathways in growth control. Curr. Biol. 22, 651-657. 10.1016/j.cub.2012.02.050 [DOI] [PubMed] [Google Scholar]
- Herranz H., Hong X., Hung N. T., Voorhoeve P. M. and Cohen S. M. (2012b). Oncogenic cooperation between SOCS family proteins and EGFR identified using a Drosophila epithelial transformation model. Genes Dev. 26, 1602-1611. 10.1101/gad.192021.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H., Weng R. and Cohen S. M. (2014). Crosstalk between epithelial and mesenchymal tissues in tumorigenesis and imaginal disc development. Curr. Biol. 24, 1476-1484. 10.1016/j.cub.2014.05.043 [DOI] [PubMed] [Google Scholar]
- Herranz H., Eichenlaub T. and Cohen S. M. (2016). Cancer in Drosophila: imaginal discs as a model for epithelial tumor formation. Curr. Top. Dev. Biol. 116, 181-199. 10.1016/bs.ctdb.2015.11.037 [DOI] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K. and Pan D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421-434. 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Huh J. R., Guo M. and Hay B. A. (2004). Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr. Biol. 14, 1262-1266. 10.1016/j.cub.2004.06.015 [DOI] [PubMed] [Google Scholar]
- Johnson R. and Halder G. (2014). The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 13, 63-79. 10.1038/nrd4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C., Hargreaves D. C., Hodges C., Elias L., Ho L., Ranish J. and Crabtree G. R. (2013). Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592-601. 10.1038/ng.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C., McLellan M. D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J. F., Wyczalkowski M. A. et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature 502, 333-339. 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koe C. T., Li S., Rossi F., Wong J. J., Wang Y., Zhang Z., Chen K., Aw S. S., Richardson H. E., Robson P. et al. (2014). The Brm-HDAC3-Erm repressor complex suppresses dedifferentiation in Drosophila type II neuroblast lineages. Elife 3, e01906 10.7554/eLife.01906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurada P. and White K. (1998). Ras promotes cell survival in Drosophila by downregulating hid expression. Cell 95, 319-329. 10.1016/S0092-8674(00)81764-X [DOI] [PubMed] [Google Scholar]
- Lisi S., Mazzon I. and White K. (2000). Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics 154, 669-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann I., McGinnis N., Bodmer M. and McGinnis W. (2002). The Drosophila Hox gene deformed sculpts head morphology via direct regulation of the apoptosis activator reaper. Cell 110, 457-466. 10.1016/S0092-8674(02)00871-1 [DOI] [PubMed] [Google Scholar]
- Mohrmann L. and Verrijzer C. P. (2005). Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta 1681, 59-73. 10.1016/j.bbaexp.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Morin X., Daneman R., Zavortink M. and Chia W. (2001). A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98, 15050-15055. 10.1073/pnas.261408198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkin Y. M., Mohrmann L., van Ijcken W. F. J. and Verrijzer C. P. (2007). Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol. Cell. Biol. 27, 651-661. 10.1128/MCB.01257-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M., Diaz-Benjumea F. J., Vincent J.-P., Wu J. and Cohen S. M. (1996). Specification of the wing by localized expression of wingless protein. Nature 381, 316-318. 10.1038/381316a0 [DOI] [PubMed] [Google Scholar]
- Nolo R., Morrison C. M., Tao C., Zhang X. and Halder G. (2006). The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 16, 1895-1904. 10.1016/j.cub.2006.08.057 [DOI] [PubMed] [Google Scholar]
- Nordstrom W., Chen P., Steller H. and Abrams J. M. (1996). Activation of the reaper gene during ectopic cell killing in Drosophila. Dev. Biol. 180, 213-226. 10.1006/dbio.1996.0296 [DOI] [PubMed] [Google Scholar]
- Oh H. and Irvine K. D. (2011). Cooperative regulation of growth by Yorkie and Mad through bantam. Dev. Cell 20, 109-122. 10.1016/j.devcel.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Slattery M., Ma L., Crofts A., White K. P., Mann R. S. and Irvine K. D. (2013). Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep 3, 309-318. 10.1016/j.celrep.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Garijo A., Martin F. A. and Morata G. (2004). Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development 131, 5591-5598. 10.1242/dev.01432 [DOI] [PubMed] [Google Scholar]
- Piccolo S., Dupont S. and Cordenonsi M. (2014). The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev. 94, 1287-1312. 10.1152/physrev.00005.2014 [DOI] [PubMed] [Google Scholar]
- Prober D. A. and Edgar B. A. (2000). Ras1 promotes cellular growth in the Drosophila wing. Cell 100, 435-446. 10.1016/S0092-8674(00)80679-0 [DOI] [PubMed] [Google Scholar]
- Prober D. A. and Edgar B. A. (2002). Interactions between Ras1, dMyc, and dPI3K signaling in the developing Drosophila wing. Genes Dev. 16, 2286-2299. 10.1101/gad.991102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Romero M., Blanco E., Paricio N., Serras F. and Corominas M. (2015). Cabut/dTIEG associates with the transcription factor Yorkie for growth control. EMBO Rep. 16, 362-369. 10.15252/embr.201439193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo H. D., Gorenc T. and Steller H. (2004). Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev. Cell 7, 491-501. 10.1016/j.devcel.2004.08.019 [DOI] [PubMed] [Google Scholar]
- Simmonds A. J., Liu X., Soanes K. H., Krause H. M., Irvine K. D. and Bell J. B. (1998). Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes Dev. 12, 3815-3820. 10.1101/gad.12.24.3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J. and Cohen S. M. (2006). The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126, 767-774. 10.1016/j.cell.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Uhlirova M. and Bohmann D. (2006). JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO J. 25, 5294-5304. 10.1038/sj.emboj.7601401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers J. H. A., Manning S. A., Kulkarni A. and Harvey K. F. (2016). A Drosophila RNAi library modulates Hippo pathway-dependent tissue growth. Nat. Commun. 7, 10368 10.1038/ncomms10368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B. G. and Roberts C. W. M. (2011). SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481-492. 10.1038/nrc3068 [DOI] [PubMed] [Google Scholar]
- Wu S., Liu Y., Zheng Y., Dong J. and Pan D. (2008). The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388-398. 10.1016/j.devcel.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Xie G., Chen H., Jia D., Shu Z., Palmer W. H., Huang Y.-C., Zeng X., Hou S. X., Jiao R. and Deng W.-M. (2017). The SWI/SNF complex protein Snr1 is a tumor suppressor in Drosophila imaginal tissues. Cancer Res. 77, 862-873. 10.1158/0008-5472.CAN-16-0963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. J., Huh J. R., Muro I., Yu H., Wang L., Wang S. L., Feldman R. M., Clem R. J., Müller H.-A. J. and Hay B. A. (2002). Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 4, 416-424. 10.1038/ncb793 [DOI] [PubMed] [Google Scholar]
- Yu F.-X., Zhao B. and Guan K.-L. (2015). Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811-828. 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ren F., Zhang Q., Chen Y., Wang B. and Jiang J. (2008). The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377-387. 10.1016/j.devcel.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Li D., Wang Y., Pei C., Liu S., Zhang L., Yuan Z. and Zhang P. (2015). Brahma regulates the Hippo pathway activity through forming complex with Yki-Sd and regulating the transcription of Crumbs. Cell. Signal. 27, 606-613. 10.1016/j.cellsig.2014.12.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.