Abstract
PAR proteins constitute a highly conserved network of scaffolding proteins, adaptors and enzymes that form and stabilize cortical asymmetries in response to diverse inputs. They function throughout development and across the metazoa to regulate cell polarity. In recent years, traditional approaches to identifying and characterizing molecular players and interactions in the PAR network have begun to merge with biophysical, theoretical and computational efforts to understand the network as a pattern-forming biochemical circuit. Here, we summarize recent progress in the field, focusing on recent studies that have characterized the core molecular circuitry, circuit design and spatiotemporal dynamics. We also consider some of the ways in which the PAR network has evolved to polarize cells in different contexts and in response to different cues and functional constraints.
KEY WORDS: PAR, Polarity, C. elegans, Drosophila
Summary: This Review discusses studies that have revealed how the PAR protein network can form and stabilize the asymmetric distribution of its components.
Introduction
PAR proteins constitute a highly conserved network of scaffolds, adaptors and enzymes that control cell polarity in a wide variety of developmental and physiological contexts across the metazoa (Goldstein and Macara, 2007; Suzuki and Ohno, 2006). A key feature of the PAR network is that many of its members are asymmetrically localized in polarized cells. PAR asymmetries are established in response to many different kinds of polarizing cues; they are reinforced by complex networks of mutually supportive and antagonistic interactions, and they feed forward to control the localization and activities of downstream targets to elaborate polarized phenotypes.
In the past few decades, we have made substantial progress in identifying and characterizing the core molecular players and interactions that govern the formation and stabilization of PAR polarity. Advances in live imaging and biophysical analyses have allowed us to probe PAR protein dynamics in living cells. This has set the stage for using mathematical models to frame and test hypotheses for how specific interactions lead to robust and dynamically stable PAR polarity. Although much of the key work to define core mechanisms has been performed in a few model organisms and in a few specific developmental contexts, comparisons across these different developmental and organismal contexts are beginning to reveal a broader view of how the same core circuitry/mechanisms can be tuned differently to perform different tasks.
Here, we review recent progress in the field, emphasizing an emerging view of the PAR network as a dynamic pattern-forming module, the circuit logic and molecular interactions of which endow it with the ability to form and stabilize asymmetric distributions of its components. We focus on studies in the C. elegans zygote, where the most comprehensive picture of mechanism has emerged. However, we also discuss how core elements of this circuit are modified and/or extended in other contexts to achieve functional variants of the same core mechanism.
PAR proteins form a highly conserved, but versatile, polarity module
PAR proteins were first discovered in screens for mutations that affect asymmetric cell division in the C. elegans zygote (Kemphues et al., 1988; Tabuse et al., 1998; Watts et al., 1996). During polarization of the C. elegans zygote, a sperm-derived cue induces redistribution of cell fate determinants along the future anterior-posterior (AP) axis that are then inherited unequally during cell division (Rose and Gönczy, 2014). In seminal studies, Ken Kemphues and colleagues showed that PAR genes are required both for the initial segregation of cell fate determinants and to position the cleavage plane so these determinants are partitioned correctly into the anterior and posterior daughters of the zygote (Guo and Kemphues, 1996). They showed that, during polarization, a subset of PAR proteins becomes asymmetrically enriched at the cell cortex within complementary anterior and posterior domains, and that these asymmetries are controlled by mutual antagonism between anterior and posterior PAR proteins (Boyd et al., 1996; Etemad-Moghadam et al., 1995; Guo and Kemphues, 1995; Tabuse et al., 1998; Watts et al., 1996). Subsequent work showed that PAR protein homologues are asymmetrically localized in many other organisms and cell types (Denker et al., 2013; Nakaya et al., 2000; Tomancak et al., 2000; Wodarz et al., 2008). Complimentary PAR domains are associated with axis formation before and during fertilization in some oocytes, with contact-dependent polarities in early embryonic cells, with asymmetric cell divisions, and with apico-basal and planar polarities in embryonic epithelia. In other cells, such as neurons and neuroblast stem cells, subsets of PAR proteins can localize in a unipolar fashion, without an opposing domain. The polarizing inputs and functional outputs of PAR asymmetry are strikingly different in these different contexts. However, what appears to be most highly conserved is the core set of molecular interactions by which PAR proteins promote or inhibit the localization or activities of one another to convert transient polarizing inputs into stable cell polarity.
An overview of polarization in the C. elegans zygote
The most complete understanding of how PAR proteins mediate the establishment and maintenance of cortical polarity comes from studies in the C. elegans zygote. The core players in this system include the original proteins discovered by Kemphues et al. (Kemphues et al., 1988) plus later additions (Beatty et al., 2010; Gotta et al., 2001; Hoege et al., 2010; Kumfer et al., 2010; Tabuse et al., 1998); we refer to these collectively as PAR proteins (Table 1). Anterior PAR proteins (aPARs) include the oligomeric scaffold PAR-3, the adaptor PAR-6, the kinase PKC-3 and the small GTPase CDC-42. Posterior PAR proteins (pPARs) include the kinase PAR-1, the RING domain protein PAR-2, the tumor suppressor LGL-1 and a putative GTPase-activating protein (GAP) for CDC-42, called CHIN-1. Two additional proteins (the kinase PAR-4 and the 14-3-3 protein PAR-5) are not asymmetrically localized but control asymmetries of the other PARs. All of these proteins are highly conserved across the metazoa, with the exception of PAR-2, but even PAR-2 may have functional analogues in other organisms
Table 1.
The PAR proteins
The C. elegans zygote polarizes in two distinct phases, referred to as establishment and maintenance phases, which coincide with mitotic interphase and M phase, respectively (Fig. 1). Before polarity establishment, the aPARs are enriched throughout the cortex while pPARs are uniformly cytoplasmic. This symmetry is broken in response to multiple signals from a transient centrosome/microtubule-organizing center (the sperm MTOC) that forms near the site of sperm entry (Bienkowska and Cowan, 2012; Cowan and Hyman, 2004; Hamill et al., 2002; O'Connell et al., 2000). One of these signals, still poorly defined, acts locally to inhibit RhoA-dependent cortical actomyosin contractility (Motegi and Sugimoto, 2006; Munro et al., 2004). This creates a spatial gradient of contractility and anterior-directed cortical flows that segregate PAR-3, PAR-6 and PKC-3 towards the anterior pole, allowing PAR-1, PAR-2 and LGL-1 to accumulate on the posterior cortex (Cheeks et al., 2004; Goehring et al., 2011b; Mayer et al., 2010; Munro et al., 2004). A second signal from the sperm MTOC can induce symmetry-breaking in the absence of contractility (Motegi et al., 2011); during this event, microtubules associated with the sperm MTOC act locally to promote PAR-2 association with the posterior cortex, and PAR-2 in turn recruits PAR-1, which phosphorylates and promotes the dissociation of PAR-3.
Fig. 1.
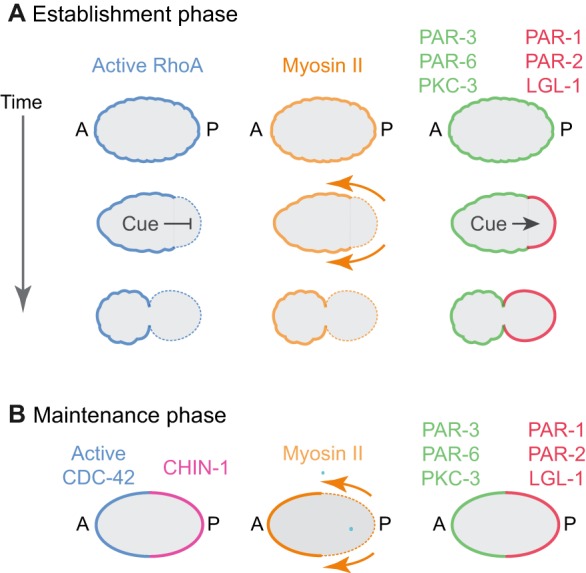
Overview of polarization in the C. elegans zygote. Polarization of the C. elegans zygote involves two distinct phases: establishment phase (A) and maintenance phase (B). (A) Before polarity establishment, anterior PAR proteins (green), active RhoA and Myosin II are uniformly enriched at the cell cortex. During polarity establishment, a transient sperm-derived cue acts locally to inhibit RhoA activity and induce actomyosin-based cortical flows that segregate anterior PAR proteins towards the anterior pole, and to promote local accumulation of posterior PAR proteins (red) on the posterior cortex where they act to inhibit local accumulation of anterior PAR proteins. (B) During maintenance phase, complementary distributions of anterior PAR proteins and posterior PAR proteins are maintained in the absence of a cue. RhoA activity is low, and CDC-42 activity becomes enriched at the anterior cortex, while the CDC-42 GAP CHIN-1 becomes enriched on the posterior cortex. CDC-42 acts through the kinase MRCK-1 to activate Myosin II on the anterior cortex, leading to persistent cortical flows.
These asymmetries persist during maintenance phase as the centrosomes, (i.e. the original cue) move towards the cell center. During this time, several new asymmetries appear. Active CDC-42 becomes enriched with other aPARs at the anterior pole (Kumfer et al., 2010). At the same time, CHIN-1 becomes highly enriched in a complementary posterior domain. In addition to interacting directly with other PAR proteins, CDC-42 acts through the conserved kinase MRCK-1 to activate myosin II, creating a gradient of contractility that drives anterior-directed cortical flows (Kumfer et al., 2010; Sailer et al., 2015). As discussed below, both flow-dependent and -independent mechanisms contribute to positioning and stabilizing the boundary between anterior and posterior PAR domains during maintenance phase.
The molecular basis for complementary PAR domains
Biophysical studies, using fluorescence recovery after photobleaching (FRAP) (Cheeks et al., 2004; Goehring et al., 2011a; Nakayama et al., 2009) or single-molecule imaging (Arata et al., 2016; Robin et al., 2014; Sailer et al., 2015), have shown that PAR proteins exchange dynamically between the cytoplasm and the cell surface, where they move locally by diffusion or transport. At the same time, genetic and biochemical studies have revealed a complex web of local interactions through which PAR proteins modulate the recruitment or dissociation of one another (see below). Together, these data support a conceptual model for dynamic stabilization of cell polarity in which PAR proteins exchange locally with the cell surface, and either synergize to promote the recruitment of one another or compete to promote the dissociation of one another. Below, we review the mechanisms that mediate local recruitment of anterior and posterior PARs, and then discuss how mutual interactions between aPARs and pPARs reinforce their enrichment within complementary domains.
The hierarchical recruitment of anterior PARs
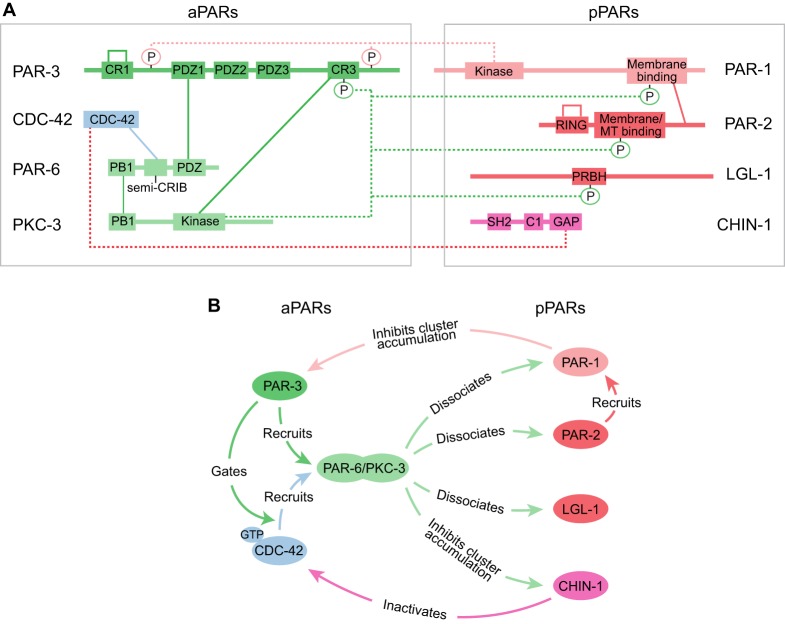
Cortical recruitment of aPARs is governed by a complex hierarchy of protein-protein interactions in which PAR-3 and CDC-42 bind independently to the cortex and synergize to recruit PAR-6 and PKC-3 (Fig. 2) (Kumfer et al., 2010; Sailer et al., 2015). Structure/function studies in C. elegans, Drosophila and vertebrate cells suggest that PAR-3 recruitment depends on direct binding to membrane phospholipids, self-oligomerization and specific protein/protein interactions (Feng et al., 2007; Li et al., 2010a; McKinley et al., 2012; Mizuno et al., 2003; Wu et al., 2007; Yu and Harris, 2012). PAR-3 contains several conserved functional domains, including an N-terminal (CR1) oligomerization domain, three PDZ domains and a C-terminal (CR3) domain that interacts with PKC-3. Specific domains/residues that mediate membrane binding have been characterized in flies (Krahn et al., 2010b; McKinley et al., 2012), mammalian cells (Mizuno et al., 2003; Wu et al., 2007) and to a lesser extent in C. elegans (Li et al., 2010a), but do not appear to be tightly conserved. In C. elegans zygotes, both C-terminal sequences and PDZ2 are required for normal membrane association (Li et al., 2010a). Accumulation of PAR-3 also depends crucially on its ability to assemble oligomers via its CR1 domain. Live imaging studies show that PAR-3 accumulates dynamically at the membrane in small clusters of indeterminant sizes (Achilleos et al., 2010; Beers and Kemphues, 2006; Dickinson et al., 2017; Hung and Kemphues, 1999; McGill et al., 2009; Rodriguez et al., 2017; Sailer et al., 2015; Wang et al., 2017). The CR1 domain of PAR-3 assembles into helical filaments in vitro (Feng et al., 2007; Zhang et al., 2013). Deleting the CR1 domain or introducing point mutations that abolish oligomerization in vitro (Zhang et al., 2013) strongly attenuates membrane accumulation of PAR-3 in the C. elegans zygote (Dickinson et al., 2017; Li et al., 2010a; Rodriguez et al., 2017), as in Drosophila or mammalian epithelial cells (Benton and St Johnston, 2003a; Feng et al., 2007; McKinley et al., 2012; Mizuno et al., 2003). These data suggest that PAR-3 monomers can bind weakly to the membrane, but that increased binding avidity due to oligomerization is essential for strong accumulation.
Fig. 2.
Core molecular interactions that underlie the dynamic stabilization of PAR asymmetries. (A) A schematic view of the PAR network indicating key domains and phosphorylation sites involved in protein-protein interactions. Solid lines indicate direct binding interactions, whereas dotted lines terminating in circles represent enzymatic action, either phosphorylation or GAP activity. In the case of PAR-3 and PAR-2, self-connecting loops indicate oligomerization. (B) A functional view of the same circuit emphasizing the consequences of protein-protein interactions. For clarity, some interactions documented in other contexts (e.g. inhibition of aPKC by LGL or by PAR-3) have been omitted here.
In C. elegans zygotes, fluorescently tagged CDC-42 transgenes localize to the cortex throughout the cell cycle, independently of other aPARs (Aceto et al., 2006; Nakayama et al., 2009; Schonegg and Hyman, 2006). As in other cells, only a subset of membrane-localized CDC-42 is likely to be in a GTP-bound active state. Analysis of a biosensor for CDC-42 activity suggests that active CDC-42 is enriched at the posterior pole during polarity establishment and at the anterior pole during polarity maintenance (Kumfer et al., 2010; Sailer et al., 2015). CDC-42 activation requires the GEF cGEF-1, which is uniformly enriched at the cortex of zygotes (Chan and Nance, 2013; Kumfer et al., 2010), while spatial restriction of CDC-42 activity during maintenance phase depends on its local inhibition by CHIN-1 (Beatty et al., 2013; Kumfer et al., 2010; Sailer et al., 2015) (see below). The mechanism(s) that localize CDC-42 activity during polarity establishment remain unclear.
PAR-6 and PKC-3 form heterodimers through conserved PB1 domain interactions (Hirano et al., 2005), and depend on one another for cortical accumulation in polarizing zygotes (Hung and Kemphues, 1999; Tabuse et al., 1998). Normal PAR-6/PKC-3 accumulation also depends on both PAR-3 and active CDC-42. Both PAR-6 and PKC-3 bind directly to PAR-3 (Joberty et al., 2000; Li et al., 2010b; Lin et al., 2000); however, direct binding of PAR-6 to PAR-3 is not required for PAR-6/PKC-3 accumulation in C. elegans (Li et al., 2010b). PAR-6/PKC-3 can also bind active CDC-42 through the conserved semi-CRIB domain of PAR-6 (Aceto et al., 2006). In the zygote, PAR-6/PKC-3 accumulate in two distinct pools: a punctate pool that requires PAR-3, but not CDC-42 (Aceto et al., 2006); and a diffuse pool that requires both PAR-3 and CDC-42 (Beers and Kemphues, 2006; Hung and Kemphues, 1999; Motegi and Sugimoto, 2006; Tabuse et al., 1998). PAR-6/PKC-3 punctae colocalize extensively with PAR-3 oligomers (Beers and Kemphues, 2006; Hung and Kemphues, 1999; Li et al., 2010b; Tabuse et al., 1998); they segregate to the anterior with cortical flow during polarity establishment and then decay during polarity maintenance (Motegi and Sugimoto, 2006; Munro et al., 2004). A more diffuse pool of PAR-6/PKC-3 becomes progressively enriched on the anterior cortex during polarity establishment and persists through maintenance phase. Recent work shows that PAR-6/PKC-3 punctae represent PAR-6/PKC-3 bound directly to oligomeric PAR-3 (Dickinson et al., 2017), which can be blocked by genetic or pharmacological inhibition of PKC-3 activity(Rodriguez et al., 2017). In contrast, the diffuse accumulation of PAR-6/PKC-3 requires direct binding of PAR-6 to CDC-42 (Aceto et al., 2006). Interestingly, inhibition of PKC-3 or depletion of the cytoplasmic chaperone CDC-37, allows CDC-42 to recruit PAR-6/PKC-3 independently of PAR-3 (Beers and Kemphues, 2006). These and other observations suggest that PAR-3 may promote the association of PAR-6/PKC-3 with CDC-42 by competing with CDC-37 (and/or other cytoplasmic factors) for an active (or activatable) form of PKC-3 (Beers and Kemphues, 2006; Rodriguez et al., 2017).
The hierarchical recruitment of posterior PARs
The mechanisms that govern pPAR recruitment have also been extensively characterized (Fig. 2). PAR-2 binding involves three distinct domains. A membrane localization domain binds to phosphoinositide lipids, while a microtubule-binding domain facilitates PAR-2 loading onto the posterior cortex by the sperm MTOC during polarity establishment. This is achieved through a mechanism in which microtubule binding protects PAR-2 from phosphorylation by PKC-3 (Motegi et al., 2011). Finally, a RING domain stabilizes PAR-2 binding to the cortex and mediates positive feedback in which membrane-localized PAR-2 facilitates recruitment of cytoplasmic PAR-2 (Hao et al., 2006; Motegi et al., 2011). Recent single molecule imaging studies indicate that PAR-2 forms oligomers at the membrane that appear to undergo size-dependent dissociation (Arata et al., 2016). Because oligomerization is common to many other RING domains (Deshaies and Joazeiro, 2009), RING domain-mediated oligomerization of PAR-2 may both stabilize PAR-2 binding and potentiate recruitment of cytoplasmic PAR-2.
PAR-1 can associate with the membrane via a conserved C-terminal domain that binds acidic phospholipids (Griffin et al., 2011; Moravcevic et al., 2010). The binding of PAR-2 to this domain is both necessary and sufficient for PAR-2 to promote PAR-1 recruitment during polarity establishment (Motegi et al., 2011), and contributes to recruiting PAR-1 during polarity maintenance (Boyd et al., 1996; Griffin et al., 2011).
The mechanism(s) that govern localization of LGL-1 in the C. elegans zygote remain unclear. Both vertebrate and invertebrate orthologues of LGL-1 bind non-muscle Myosin II (Betschinger et al., 2005; Dahan et al., 2014, 2012; Strand et al., 1994, 1995). Cortical localization of Lgl in Drosophila neuroblasts or S2 cells is myosin-dependent, but may not require direct binding to Myosin II (Barros et al., 2003; Betschinger et al., 2005). More recent studies of Drosophila Lgl identified a short conserved phospho-regulated polybasic and hydrophobic (PRBH) motif that is conserved in C. elegans LGL-1, binds negatively charged phospholipids, and is necessary and partially sufficient for cortical localization (Bailey and Prehoda, 2015; Dong et al., 2015). Thus, normal cortical recruitment of LGL-1 is likely to be mediated by direct binding to membranes and at least one other cortical target.
CHIN-1 is largely absent from the cortex during polarity establishment but accumulates in the form of larger clusters in the posterior cortex during polarity maintenance (Kumfer et al., 2010; Sailer et al., 2015). The mechanisms for cortical accumulation and cluster assembly remain unclear, although it is known that cortical accumulation does not require any of the other pPARs (Kumfer et al., 2010; Sailer et al., 2015) and that the spatial distribution of clusters is determined through local inhibition by aPARs (discussed below).
The molecular basis for mutual antagonism
In the C. elegans zygote, as in many other contexts, PKC-3 plays a dominant role in enforcing anterior cortex identity by phosphorylating and excluding pPARs (Fig. 2). Phosphorylation of PAR-1, PAR-2 and LGL-1 by PKC-3 is both necessary and sufficient to prevent them from binding to the anterior cortex (Hao et al., 2006; Hoege et al., 2010; Motegi et al., 2011). In all three cases, PKC-3 phosphorylates residues within the motifs that are known to mediate membrane binding (Bailey and Prehoda, 2015; Hao et al., 2006; Hoege et al., 2010; Motegi et al., 2011). PAR-6/PKC-3 also restrict local accumulation of CHIN-1 clusters to the posterior pole (Kumfer et al., 2010; Sailer et al., 2015), although the molecular basis for this restriction remains to be determined (discussed below).
The pPARs act in multiple ways to prevent posterior accumulation of PAR-6/PKC-3. Early studies assigned a key role for PAR-2 in promoting local dissociation of PAR-6/PKC-3, because loss of PAR-2 leads to rapid posterior accumulation of PAR-6/PKC-3 during maintenance phase, whereas loss of other pPARs does not (Beatty et al., 2010; Hoege et al., 2010). However, recent single molecule imaging studies have shown that during maintenance phase, PAR-6/PKC-3 asymmetries are determined by faster anterior association, rather than by faster posterior dissociation (Robin et al., 2014; Sailer et al., 2015). Thus, PAR-2 does not act by promoting posterior dissociation of PAR-6/PKC-3. Instead, live imaging studies show that the loss of asymmetry in par-2 mutants is due to the redistribution of PAR-6/PKC-3 by ectopic posterior-directed cortical flows, which can be reversed by inactivating Myosin II during maintenance phase (Beatty et al., 2010; Munro et al., 2004; Sailer et al., 2015). Interestingly, overexpressing LGL-1 rescues loss of polarity and embryonic lethality in par-2 mutants, suggesting that LGL-1 can substitute functionally for PAR-2 (Beatty et al., 2010; Hoege et al., 2010).
In contrast, PAR-1 and CHIN-1 act redundantly to exclude aPARs during maintenance phase through mechanisms that do not involve local inhibition of actomyosin contractility. Depletion of CHIN-1 causes uniform activation of CDC-42 during maintenance phase (Beatty et al., 2013; Kumfer et al., 2010; Sailer et al., 2015), but PAR-3, PAR-6 and PKC-3 are still excluded from the posterior cortex (Sailer et al., 2015). Depletion of PAR-1 causes weak posterior accumulation of PAR-3 clusters, and a slight increase in posterior association of PAR-6/PKC-3, but has no effect on active CDC-42 (Kumfer et al., 2010). However, depletion of both PAR-1 and CHIN-1 leads to uniform CDC-42 activation and weak posterior accumulation of PAR-3 clusters, and this is accompanied by rapid posterior association of PAR-6/PKC-3 and complete loss of PAR-6/PKC-3 asymmetry.
These observations imply that active CDC-42 is not sufficient to recruit PAR-6/PKC-3 to the cortex, but in the presence of active CDC-42, a small amount of PAR-3 is both necessary and sufficient to achieve maximum rates of PAR-6/PKC-3 recruitment. Importantly, most of the diffuse pool of PAR-6/PKC-3 that accumulates on the posterior pole in embryos depleted of CHIN-1 and PAR-1 cannot be directly associated with the few PAR-3 clusters that accumulate there. Thus, cortical PAR-3 somehow gates the local association of PAR-6/PKC-3 with active CDC-42. How this works remains an interesting puzzle. An attractive hypothesis is that cortical PAR-3 competes with cytoplasmic factors such as CDC-37 to bind PAR-6/PKC-3 dimers, then transfers those dimers to CDC-42. In other systems, PAR-3, CDC-42 and PAR-6/aPKC can form a quaternary complex (Joberty et al., 2000; Lin et al., 2000), PAR-3 can bind and inhibit the active form of aPKC, and this inhibitory bond can be released by inducing phosphorylation of PAR-3 by aPKC (Morais-de-Sá et al., 2010; Soriano et al., 2016; Walther and Pichaud, 2010). Thus, one possible scenario is that PAR-6/aPKC dock initially on PAR-3, followed by formation of a quaternary complex, and then phosphorylation of PAR-3 by aPKC releases CDC-42/PAR6/aPKC.
Regardless of the exact mechanism, the requirement for PAR-3 to gate the local association of PAR-6/PKC-3 with CDC-42 plays a key role in shaping the spatial gradient of active PKC-3 in the polarized zygote. Older work (Aceto et al., 2006; Gotta et al., 2001; Kay and Hunter, 2001; Schonegg and Hyman, 2006) and an elegant recent study (Rodriguez et al., 2017) suggest that the pool of PAR-6/PKC-3 that associates with PAR-3 is inactive, whereas the diffuse CDC-42-bound pool of PAR-6/PKC-3 is the active form that phosphorylates and excludes posterior PARs. Single-molecule imaging and particle-tracking analyses suggest that PAR-3/PAR-6/PKC-3-containing clusters couple tightly to the actin cortex and are efficiently segregated by cortical flow (Dickinson et al., 2017; Motegi and Sugimoto, 2006; Munro et al., 2004; Rodriguez et al., 2017; Sailer et al., 2015; Wang et al., 2017), whereas the pool of PAR-6/PKC-3 that associates with CDC-42 diffuses freely and rapidly (Robin et al., 2014). Thus, the gradient of active PKC-3 is likely shaped by two mechanisms: the distribution of relatively immobile PAR-3 defines the local source for PKC-3 recruitment, while diffusion and dissociation shape a broader gradient of active PKC-3 that acts upon its posterior targets.
A circuit-level view of PAR polarity
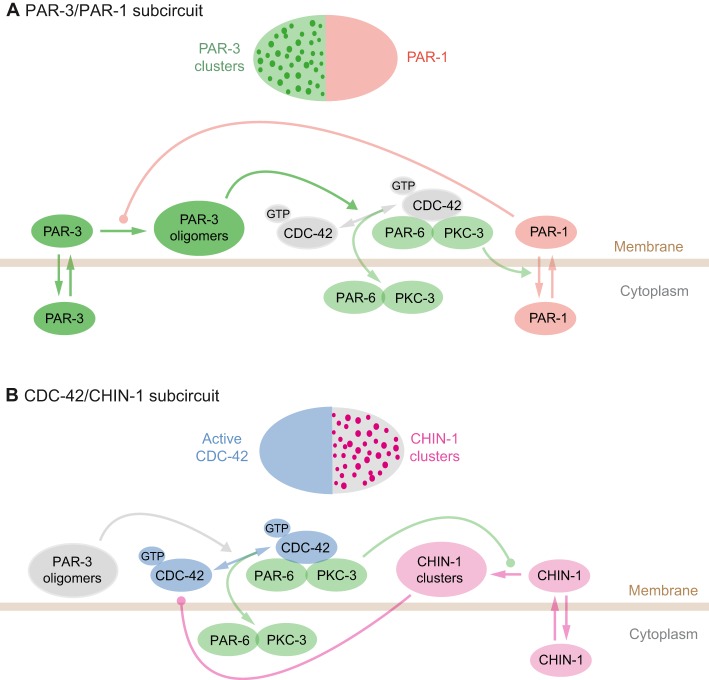
Although many important molecular and biochemical questions remain to be answered, the above studies support a model in which two distinct subcircuits act redundantly to stabilize cortical polarity (Fig. 3). In the PAR-1/PAR-3 subcircuit, PAR-3 promotes PAR-6/PKC-3 association with CDC-42, PKC-3 phosphorylates and promotes dissociation of PAR-1 (Sailer et al., 2015), while PAR-1 phosphorylates and promotes the dissociation of PAR-3. In a second CDC-42/CHIN-1 subcircuit, active CDC-42 recruits PAR-6/PKC-3, which locally inhibits the accumulation of CHIN-1 clusters (Beatty et al., 2013; Sailer et al., 2015), while CHIN-1 inactivates CDC-42. The PAR-1/PAR-3 and CDC-42/CHIN-1 subcircuits are coupled by their shared use of PKC-3 to phosphorylate and exclude pPARs, and by the dual requirement for PAR-3 and CDC-42 to recruit PAR-6/PKC-3, which allows either PAR-1 or CHIN-1 to prevent posterior accumulation of PAR-6/PKC-3.
Fig. 3.
Decomposition of the PAR network into two smaller, mutually inhibitory subcircuits. (A) The PAR-3/PAR-1 subcircuit involves mutual antagonism between PAR-3 oligomers and PAR-1. PAR-3 promotes local association of PAR-6/PKC-3 with active CDC-42; PKC-3 phosphorylates and inhibits membrane association of PAR-1; PAR-1 phosphorylates PAR-3 and inhibits membrane association and/or oligomerization. (B) The CDC-42/CHIN-1 subcircuit involves mutual antagonism between active CDC-42 and clustered CHIN-1. Active CDC-42 binds PAR-6/PKC-3 in the presence of PAR-3; PAR-6/PKC-3 inhibits local growth and accumulation of CHIN-1 clusters; CHIN-1 inactivates CDC-42.
In addition to preventing active redistribution of aPARs by cortical flow, PAR-2 plays an integral role in the PAR-3/PAR-1 subcircuit through its ability to recruit PAR-1, and positive feedback on PAR-2 recruitment can further reinforce posterior identity (Arata et al., 2016; Motegi and Seydoux, 2013) (and see below). Likewise, LGL-1 may contribute to both subcircuits by limiting the availability of PAR-6/PKC-3, either by reducing their total abundancies (Beatty et al., 2013) or through sequestration of cytoplasmic PAR-6/PKC-3 (Hoege et al., 2010; Wirtz-Peitz et al., 2008). Both PAR-2 and LGL-1 could also contribute by inhibiting the actions of PKC-3 towards other posterior targets (Hao et al., 2006; Soriano et al., 2016; Wirtz-Peitz et al., 2008). Finally, other uncharacterized feedback mechanisms must contribute to promoting local accumulation of PAR-3, because PAR-3 remains stably asymmetric during maintenance phase in embryos co-depleted of CHIN-1 and PAR-1, when all the other PARs are absent or uniformly distributed (Sailer et al., 2015).
Why all this redundancy? Theoretical studies and experimental analyses of synthetic circuits suggest that positive feedback and mutual inhibition are both in principle sufficient to stabilize cortical polarity, but the combination of both circuit motifs makes the process more robust with respect to variations in protein abundancies, diffusivities or interaction strengths (Chau et al., 2012). Redundancy may also have evolved to allow the same network to respond to a larger range of symmetry-breaking cues. For example, as discussed above, PAR asymmetries can be established in the C. elegans zygote by active transport of aPARs, or through local exclusion of PAR-3 by PAR-2/PAR-1, both directed by the sperm MTOC (Motegi et al., 2011). However, similar PAR asymmetries also form later in development but they do so from different starting points and in response to different cues. For example, in C. elegans P2 cells, extracellular signals from an adjacent cell (the EMS cell), mediated by SRC-1 and MES-1, bias PAR-2 recruitment to the EMS/P2 boundary to induce symmetry breaking (Arata et al., 2010; Seirin Lee, 2016). In contrast, somatic C. elegans blastomeres exhibit contact-dependent radial polarity in which pPARs are localized to cell-cell contacts and aPARs are localized to contact-free zones. In this case, the cue is mediated by junctional proteins that accumulate at cell-cell contacts, and recruit the GAP PAC-1 to inactivate CDC-42 (Anderson et al., 2008; Klompstra et al., 2015). Finally, redundancy may provide a way to partition specific polarity functions across different phases of the cell cycle, while maintaining overall continuity of cell polarity. For example, the PAR-3/PAR-1 circuit plays a key role in responding to polarity cues during interphase, while the CDC-42/CHIN-1 circuit may have distinct M-phase-specific roles.
From biochemical circuits to cell polarity
Genetic and biochemical approaches have revealed much of the core biochemical circuitry that mediates the formation and stabilization of PAR domains. However, deeper dynamic and mechanistic insights into how stable polarity emerges from circuit logic and pairwise molecular interactions has come from mathematical models, backed by quantitative imaging and biophysical analysis. Thus far, most models for Par polarity have focused on simplified versions of the polarity circuit in which anterior and/or posterior PARs are lumped together and assumed to exchange, diffuse and act locally to promote the dissociation of one another (Blanchoud et al., 2015; Dawes and Munro, 2011; Goehring et al., 2011b; Seirin Lee and Shibata, 2015). Several key insights have emerged from these studies. First, to stabilize complementary domains through mutual inhibition, the PAR circuit must exhibit locally bistable dynamics. That is, in the absence of diffusion, it must be able to stabilize either of two states in which aPARs win or pPARs win. Second, to achieve bistable dynamics, the strengths of opposing interactions must be sufficiently balanced and the effective exchange rates of one or more factors must have sigmoidal (S-shaped) or ‘ultrasensitive’ dependence on the concentrations of reactants (Blanchoud et al., 2015; Dawes and Munro, 2011; Goehring et al., 2011b; Seirin Lee and Shibata, 2015). Finally, given locally bistable dynamics, additional mechanisms are required to ensure a stably positioned AP boundary in the face of diffusion or transport by cortical flow (Dawes and Munro, 2011; Goehring et al., 2011b; Mori et al., 2008). Below, we discuss sources of ultrasensitivity and bistability in the PAR circuit and then discuss mechanisms that help to stabilize boundary position.
Sources of ultra-sensitivity and bistability in the PAR circuit
Ultrasensitivity underlies bistable dynamics in many biochemical control circuits and can arise in many different ways, e.g. through multi-site phosphorylation, stochiometric inhibition or in phosphorylation/dephosphorylation reactions, when kinase and phosphatase operate near saturation (Bryant and Mostov, 2008; Ferrell and Ha, 2014). All of these mechanisms are potentially relevant to generating bistable dynamics in the PAR circuit. For example, multi-site phosphorylation is common to many targets of aPKC, including LGL (Graybill and Prehoda, 2014) and PAR-2 (Hao et al., 2006). PAR-3, LGL and likely PAR-2 can act as stoichiometric inhibitors of PKC-3 (Hao et al., 2006; Soriano et al., 2016; Wirtz-Peitz et al., 2008), but whether these interactions are tuned to yield ultrasensitive responses has not been explored.
One source of ultrasensitivity that is particularly relevant to stabilizing PAR asymmetries emerges from the observation that multiple PARs, including PAR-2 (Arata et al., 2016), PAR-3 (Li et al., 2010a; Sailer et al., 2015), PAR-6/PKC-3 (Hung and Kemphues, 1999) and CHIN-1 (Kumfer et al., 2010; Sailer et al., 2015), form oligomers at the membrane. Oligomers will bind more avidly to the membrane than single monomers (Lemmon, 2008), and this can lead to ultrasensitive dependence of effective dissociation rates on the local concentration of the oligomerizing species. Indeed, a simple model of the PAR circuit suggests that oliogomerization of PAR-3 could endow this circuit with bistable dynamics (Dawes and Munro, 2011).
Ultrasensitive dependence of CHIN-1 cluster accumulation on PAR-6/PKC-3 may account for bistable dynamics in the CHIN-1/CDC-42 subcircuit (Kumfer et al., 2010; Sailer et al., 2015). CHIN-1 clusters nucleate and grow on the posterior cortex during polarity maintenance (Kumfer et al., 2010; Sailer et al., 2015). Strikingly, cluster growth has threshold dependence on local concentration of PAR-6/PKC-3: below a certain concentration of PAR-6/PKC-3, clusters grow, whereas above that concentration, they shrink. This threshold dependence converts a shallow gradient of PAR-6/PKC-3 into a sharp step change in the local density of CHIN-1 clusters. Incorporating these findings into a detailed mathematical model of the CDC-42/CHIN-1 subcircuit, in which exchange rates and interactions strengths are constrained by experiments, shows that threshold-dependence of CHIN-1 cluster growth of PAR-6/ PKC-3 endows this circuit with locally bistable dynamics (Sailer et al., 2015). While the mechanisms that govern CHIN-1 cluster growth and its inhibition by PAR-6/PKC-3 remain unknown, threshold dependence of cluster growth on PKC-3 emerges naturally from a model in which CHIN-1 clusters (and/or factors that recruit CHIN-1) are dynamic polymers that assemble through reversible monomer addition. A characteristic feature of polymer assembly is the existence of a critical monomer concentration, above which polymers grow and below which they shrink (Edelstein-Keshet and Ermentrout, 1998). Threshold dependence of polymer growth on kinase activity can emerge if the kinase modulates either the availability of monomers relative to a fixed critical concentration or the critical concentration relative to a fixed-size pool of available monomers.
A similar mechanism may also lead to ultrasensitive dependence of PAR-3 accumulation on PAR-1. The PAR-3 boundary is exceedingly sharp relative to the opposing gradient of PAR-1 (Beers and Kemphues, 2006; Motegi et al., 2011; Sailer et al., 2015). In flies and vertebrate cells, PAR-1 phosphorylates conserved N-terminal residues adjacent to the CR1 domain of PAR-3 to inhibit oligomerization (Benton and St Johnston, 2003a; Mizuno et al., 2003). This is consistent with the possibility that PAR-1 could modulate the concentration of monomers that are competent to oligomerize, and thus the effective critical concentration for PAR-3 oligomer assembly. Whether it does so during polarization of the C. elegans zygote remains to be seen. As discussed above, additional mechanisms must shape PAR-3 asymmetries because PAR-3 remains asymmetric in embryos lacking PAR-1.
Stabilizing the position of the polarity boundary
Once polarity is established, the zygote can maintain a normally positioned boundary in the absence of contractility and flow (Kumfer et al., 2010; Sailer et al., 2015) or when actin filaments are depolymerized by acute drug treatments (Goehring et al., 2011a). However, wild-type embryos maintain the same boundary position in the face of persistent contractile asymmetries and redistribution of polarity proteins by cortical flow. Moreover, in embryos that fail to establish normal asymmetries during interphase, cortical flows can restore normal polarity and a normally positioned boundary during maintenance phase (Zonies et al., 2010). Thus, there appear to be both flow-dependent and -independent mechanisms for stabilizing the same boundary position.
Mathematical models suggest one possible mechanism for stabilizing boundaries in the absence of flow (Mori et al., 2008). In a bistable reaction-diffusion system based on mutual inhibition, if the reactants are abundant, then one domain will tend to spread at the expense of the other, unless the competition between domains is finely tuned (Dawes and Munro, 2011; Goehring et al., 2011b; Mori et al., 2008). However, if the total abundance of one or more factors that ‘feed’ the expanding domain is limiting, then depletion of those factors as the domain expands can slow and stall domain expansion, ‘pinning’ the boundary at a characteristic position (Dawes and Munro, 2011; Goehring et al., 2011b; Mori et al., 2008). A number of observations are consistent with this mechanism operating in the C. elegans zygote: the AP boundary position shifts when aPARs or pPARs are overexpressed (Goehring et al., 2011b; Kumfer et al., 2010; Sailer et al., 2015; Zonies et al., 2010); when the PAR boundary is mis-positioned during polarity establishment by increasing or decreasing RhoA activity, it corrects rapidly during maintenance phase, as predicted by a wave-pinning model (Spiga et al., 2013; Zonies et al., 2010); and the progressive depletion of cytoplasmic PAR-2 as the cortical PAR-2 domain expands during polarity establishment is consistent with the idea that PAR-2 could act as a limiting factor (Goehring et al., 2011b). However, in embryos that lack maintenance phase contractility, boundary position is insensitive to posterior PAR depletion (Sailer et al., 2015), and boundary correction during maintenance depends on actomyosin contractility (Zonies et al., 2010). Thus, whether and how individual PAR proteins act as limiting components to stabilize boundary position in the absence of actomyosin contractility remains to be determined.
Indeed, a growing body of data suggests that boundary position is strongly shaped by feedback loops in which cortical flow redistributes PAR proteins, while PAR proteins modulate contractility and cortical flow. Particle tracking analyses reveal strong dependencies of PAR-3 and CHIN-1 cluster mobilities on actin assembly and cluster size, and strong correlations between cluster movement and cortical flow (Dickinson et al., 2017; Rodriguez et al., 2017; Sailer et al., 2015; Wang et al., 2017). Local gating of PAR-6/PKC-3 by PAR-3 and local inhibition of CDC-42 by CHIN-1 ensure tight coupling between movements of PAR-3 and CHIN-1 clusters and movements of the polarity boundary. Thus, local clustering of polarity proteins plays a central role in coupling PAR asymmetries to cortical flow. Localizing cluster assembly to the cell surface may also provide a way to avoid counter-transport of polarity proteins by cytoplasmic flows.
During polarity establishment, PAR proteins modulate a gradient of RhoA-dependent actomyosin contractility induced by the sperm cue (Motegi and Sugimoto, 2006; Schmutz et al., 2007; Schonegg and Hyman, 2006; Tse et al., 2012). PAR-3, PAR-6 and PKC-3 promote anterior contraction and thus their own segregation, while PAR-2 inhibits posterior contractility to promote segregation of anterior PARs. The molecular mechanisms remain unclear, although recent work suggests that PAR-3 may promote contractility by inhibiting the TAO kinase KIN-18, a putative negative regulator of RhoA activity (Spiga et al., 2013).
During polarity maintenance, actomyosin contractility is primarily controlled by CDC-42 through MRCK-1 (Kumfer et al., 2010). Anterior restriction of CDC-42 activity by CHIN-1 biases Myosin II activation at the anterior cortex. In addition, PAR-2 (and possibly LGL-1) act downstream of CDC-42 to inhibit posterior accumulation and/or activation of Myosin II (Beatty et al., 2010; Munro et al., 2004; Sailer et al., 2015). PAR-3/PAR-6/PKC-3 indirectly promote anterior contractility by locally inhibiting PAR-2 and CHIN-1 accumulation as discussed above (Beatty et al., 2010; Munro et al., 2004; Sailer et al., 2015). Whether they also act more directly to promote contractility during polarity maintenance remains unclear (Beatty et al., 2013; Zonies et al., 2010).
Regardless of the molecular mechanism(s), local activation of Myosin II by anterior PARs and local inhibition by posterior PARs, will synergize to promote anterior-directed cortical flows and anterior movement of the PAR boundary. This positive-feedback loop could help to explain the ability of the zygote to amplify an initially weak PAR asymmetry, and hence rescue polarity during maintenance phase, in embryos that lack RhoA activity (Zonies et al., 2010). However, it does not explain why the boundary stabilizes at a unique position in the face of persistent contractile asymmetry and cortical flow. Additional mechanisms must exist to counteract the anterior movement of the PAR boundary. One theoretical possibility (Bois et al., 2011) is that anterior transport of some factors, such as PAR-3 and CHIN-1 clusters, could be balanced by diffusive spread of other factors, such as a CDC-42/PAR-6/PKC-3 complex. Indeed quantitative measurements and mathematical modeling suggest that the counterbalance of directed and diffusive transport could mediate boundary stabilization by the CDC-42/CHIN-1 subcircuit when PAR-1 is missing (Sailer et al., 2015). However, it remains to be explained how this balance is achieved, in the face of persistent contractile asymmetries, at a unique AP position. Understanding how a stable boundary position is determined by the dynamic interplay of PAR proteins and contractility remains a challenging problem for future studies.
A comparison with other systems
To what extent are the mechanisms that operate in C. elegans embryos more widely conserved? Studies in Drosophila oocytes, in epithelia, and in neural precursors and stem cells (Prehoda, 2009; St Johnston and Ahringer, 2010; Tepass, 2012) reveal conservation of many core molecular interactions. At the same time, these studies highlight how the core circuitry has been modified in different contexts to serve distinct functional requirements of different cells or tissues.
PAR asymmetries in Drosophila oocytes
In Drosophila oocytes, complementary domains are formed and stabilized through mutual antagonism between anterior (Par-3, Cdc-42, Par-6/aPKC) and posterior (Par-1 and Lgl) PAR proteins. As in C. elegans, both Par-3 and Cdc-42 are required to recruit and activate Par-6/aPKC (Doerflinger et al., 2010; Leibfried et al., 2013); aPKC phosphorylates and excludes both Par-1 and Lgl (Doerflinger et al., 2010; Tian and Deng, 2008), and both Par-1 and Lgl contribute to excluding Par-3 and Par-6/aPKC from the posterior pole. Par-1 plays the dominant role, whereas Lgl plays a smaller role that can be enhanced by overexpression (Morais-de-Sá et al., 2014; Tian and Deng, 2008). While Drosophila lack a homologue of PAR-2, an E3 ubiquitin ligase called Slmb may play an analogous role in the sense that Slmb is required for posterior accumulation of Par-1 and the establishment of oocyte polarity, and loss of Slmb can be rescued by overexpressing Lgl (Morais-de-Sá et al., 2014; Skwarek et al., 2014). Whether posterior inhibition of Cdc-42 activity is also involved remains unclear. However, unlike the situation in C. elegans, actomyosin contractility and cortical flow do not appear to be involved in either establishing or maintaining Drosophila oocyte polarity (Doerflinger et al., 2010). Instead, local signals from posterior follicle cells are thought to break symmetry by displacing Par-6/aPKC from the posterior cortex, allowing local recruitment of Par-1 and local displacement of Par-3 (Doerflinger et al., 2010; Morais-de-Sá et al., 2014). Thus, while Drosophila oocytes appear to rely heavily on a variant of the Par-3/Par-1 subcircuit, some of the mechanisms that operate redundantly to establish and maintain PAR asymmetries in C. elegans zygotes may be missing, and additional factors (e.g. Slmb) may fulfill analogous roles.
PAR asymmetries in epithelia
Epithelial cells partition themselves into three domains: apical, junctional and basolateral. The apical domain is defined by co-enrichment of Cdc-42, Par-6/aPKC and members of the highly conserved Crumbs (Crb) complex, which contains Crumbs (Crb), Stardust (Std) and PatJ. The junctional domain is enriched in Par-3 and overlaps with adherens and tight or septate junctions. The basolateral domain is enriched in Par-1 and members of the Scribble polarity module, comprising Scribble (Scrib), Discs Large (Dlg) and Lgl. Not surprisingly, therefore, the molecular circuitry in epithelial cells is more complex than that in C. elegans, although the circuit design principles are strikingly analogous (Laprise and Tepass, 2011; Rodriguez-Boulan and Macara, 2014; Tepass, 2012).
During epithelial polarization in vivo, Par-3 serves as a crucial landmark that localizes early to the apical margin of cell-cell contacts, where it recruits components of adherens junctions and acts as a transient docking station for Par-6/aPKC and other members of the apical Crb/Cdc-42/Par-6/aPKC complex to facilitate their association with the apical membrane (Achilleos et al., 2010; Franz and Riechmann, 2010; Harris and Peifer, 2005; Horikoshi et al., 2009; Krahn et al., 2010a; Mccaffrey and Macara, 2009; McKinley et al., 2012). As in other contexts, Par-1 phosphorylates and excludes Par-3 from lateral membranes (Bayraktar et al., 2006; Benton and St Johnston, 2003b; McKinley et al., 2012). Par-3 is also excluded from the apical surface through competition with the apical Crumbs complex for binding to Par-6/aPKC (Morais-de-Sá et al., 2010; Soriano et al., 2016; Tepass, 2012; Walther and Pichaud, 2010). Binding to the conserved CR3 domain of Par-3 can transiently trap Par-6/aPKC in a high-affinity inhibited state (Soriano et al., 2016). However, unknown input(s) stimulate aPKC to phosphorylate Par-3, reducing their mutual affinity, and allowing Crumbs to out-compete Par-3 for binding to Par-6/aPKC (Morais-de-Sá et al., 2010; Walther and Pichaud, 2010). Cdc-42 may promote this segregation by binding Par-6 and increasing its affinity for Crumbs (Whitney et al., 2016). Together, these interactions promote the accumulation of Par-6/aPKC with Crb and Cdc-42 on the apical surface, while restricting Par-3 to adherens junctions.
As in C. elegans, multiple forms of feedback contribute to reinforcing apical versus basaolateral identities (Rodriguez-Boulan and Macara, 2014; Tepass, 2012). For example, aPKC enforces apical identity by phosphorylating and displacing Par-1, Lgl and other basolateral polarity factors (Gamblin et al., 2014; Suzuki and Ohno, 2006). In addition, Cdc-42/Par-6/aPKC can inhibit the endocytosis of Crb and other apical factors to reinforce their apical accumulation (Fletcher et al., 2012; Harris and Tepass, 2008). If left unchecked, this positive feedback can lead to apical expansion (Fletcher et al., 2012). However, multiple basolateral factors, including members of the Scribble module (Bilder and Perrimon, 2000; Bilder et al., 2003; Laprise et al., 2009; Tanentzapf and Tepass, 2003) and a later acting Yurt/Coracle module (Laprise et al., 2009), oppose expansion. The molecular basis for this opposition remains unclear, but several basolateral factors, including Lgl and Yurt, appear to play dual roles as substrates and inhibitors of aPKC. (Wirtz-Peitz et al., 2008; Yamanaka et al., 2006). Local inhibition of aPKC activity could indirectly prevent local accumulation of aPKC by inhibiting aPKC-dependent positive feedback (Fletcher et al., 2012) or by allowing the accumulation of aPKC targets that cross-inhibit aPKC accumulation.
Finally, a growing body of work has begun to reveal additional feedback interactions involving PAR proteins, the Crumbs and Scribble polarity modules, phosphoinositide lipids and polarized membrane traffic that reinforce apical versus basolateral identities in epithelia (Eaton and Martin-Belmonte, 2014; Rodriguez-Boulan and Macara, 2014; Shewan et al., 2011). In many epithelia, local enrichment of PIP2 [PtdIns(4,5)P2] and PIP3 [PtdIns(3,4,5)P3] play key roles in defining, respectively, apical and basolateral identities (Claret et al., 2014; Gassama-Diagne et al., 2006; Martin-Belmonte and Mostov, 2008; Martin-Belmonte et al., 2007). Polarity proteins can modulate the relative levels of PIP2 and PIP3 by promoting or inhibiting the local recruitment of lipid kinases (e.g. PI3 kinase or PPK-5) or phosphatases (e.g. PTEN) (Chartier et al., 2011; Claret et al., 2014; Martin-Belmonte et al., 2007; von Stein et al., 2005), and PIP2 and PIP3 can in turn bias the recruitment of polarity proteins, either by direct binding or by targeting secretion to apical versus basolateral domains (reviewed by Eaton and Martin-Belmonte, 2014). These additional slower forms of feedback may be essential to maintain compartmental identities on longer timescales against the continuous insertion and removal of membrane proteins that elaborates compartment identities, and during the continuous and active remodeling of epithelia during tissue morphogenesis. Indeed, membrane recycling appears to play a much smaller role in the C. elegans zygote where cortical polarities are formed and maintained on much faster timescales (Nakayama et al., 2009; Shivas and Skop, 2012).
Unipolar PAR asymmetries in neuroblast stem cells
Studies of Drosophila neural stem cells (neuroblasts) highlight another interesting variation on PAR-mediated polarity. Embryonic and larval neuroblasts delaminate from a surface epithelium and then execute a sequence of asymmetric divisions, each producing a small ganglion mother cell and a self-renewed neuroblast (Hartenstein and Wodarz, 2013). As in epithelia, Par-3 forms an essential landmark that is enriched in an apical cap from late interphase through to the end of mitosis. During mitosis, Par-3 cooperates with Cdc-42 to recruit and activate Par-6/aPKC; aPKC in turn phosphorylates and displaces basal determinants from the apical cortex (Atwood and Prehoda, 2009; Atwood et al., 2007; Petronczki and Knoblich, 2001; Wodarz et al., 2000).
Neuroblasts inherit an apical Par-3 cap when they first delaminate. This cap disappears as neuroblasts divide then reform in daughter neuroblast cells during late interphase in response to spatial cues from the overlying epithelium (in embryos) (Siegrist and Doe, 2006) or from an apical centrosome that ‘remembers’ the position of the previous apical cap (in larvae) (Januschke and Gonzalez, 2010; Januschke et al., 2013; Rusan and Peifer, 2007). In cells that lack these spatial cues, Par-3 caps form later in mitosis, with random orientation, but can still direct a normally asymmetric division (Januschke and Gonzalez, 2010; Siegrist and Doe, 2006).
Interestingly, apical polarity in neuroblasts does not appear to require basolateral inhibition. Phosphorylation by Par-1 is not required to restrict Par-3 to a polar cap, although Par-1 may help to align Par-3 caps with polarity cues (Krahn et al., 2009). Members of the Scribble polarity module are co-enriched with Par-3 at the apical cortex of neuroblasts, and mutating Dlg, Scrib or Lgl does not cause basal accumulation of apical factors or loss of Par-3 asymmetry (Lee et al., 2006; Ohshiro et al., 2000; Peng et al., 2000; Siegrist and Doe, 2005). So how do neuroblasts form and stabilize an apical PAR domain? Par-3 binds the adapter Inscuteable (Insc) (Schober et al., 1999; Wodarz et al., 1999) to localize a conserved Pins/Ga/Dlg complex to the apical cap (Betschinger and Knoblich, 2004). This complex plays a key role in aligning the mitotic spindle with the polarity axis to ensure correct partitioning of cell fate determinants between daughter cells (reviewed by Lu and Johnston, 2013). In Insc mutants, Par-3 caps are lost, but a normal Pins cap forms late in mitosis at a random position relative to apical cues, possibly via positive feedback involving microtubules and the kinesin Khc-73 (Siegrist and Doe, 2005). On the other hand, in Pins or Ga mutants, Par-3 caps are positioned normally with respect to apical cues but are attenuated or lost later in the cell cycle (Cai et al., 2003; Schaefer et al., 2001, 2000; Yu et al., 2000). Thus, Pins feedback may reinforce the Par-3 cap that forms during interphase at a position dictated by apical cues; but in the absence of a polarizing cue, a Pins cap can form de novo at a random position and recruit Par-3 through the adaptor Insc. Whether the weaker Par-3 caps that forms in neuroblasts lacking Pins depends upon a polarizing cue, or reflects additional mechanism(s) for spontaneous formation of PAR-3 caps, remains unclear. Interestingly, Par-3 can spontaneously form polar caps during mitosis in Drosophila S2 cells, even in the absence of Insc (Wodarz et al., 1999).
Examples that do not involve mutual antagonism
While much of this Review has emphasized an essential role for mutual antagonism in reinforcing complementary PAR protein distributions, there are many examples in which aPARs localize to discrete subcellular domains in the apparent absence of mutual antagonism. A classic example comes from studies of hippocampal neurons in vitro, in which Par-3/Par-6/aPKC localize to the growing tips of multiple individual neurites, before becoming restricted to the tip of a single fast-growing axon (Shi et al., 2003). This localization is thought to be sustained in part by positive feedback in which Par-3 is delivered to neurite tips by microtubule-based transport (Nishimura et al., 2004; Shi et al., 2004), while oligomeric Par-3 feeds back by binding, bundling and stabilizing microtubules (Chen et al., 2013).
Discrete patches of Par-3 also associate with centrosomes in a variety of different cells, including Drosophila neuroblasts (Januschke and Gonzalez, 2010; Rebollo et al., 2007; Rusan and Peifer, 2007), C. elegans intestinal epithelia (Feldman and Priess, 2012) and Drosophila male germline stem cells (Inaba et al., 2015; Yuan et al., 2012). While the underlying mechanisms remain unknown, studies of Par-3 localization in Drosophila aPKC mutant epithelia have provided some insights (Harris and Peifer, 2007; Jiang et al., 2015). In wild-type epithelia, Par-3 is uniformly distributed along apical cell-cell junctions. In aPKC mutant epithelia, Par-1 is no longer excluded from the apical surface and Par-3 is restricted to discrete patches that colocalize with apical centrosomes in aPKC mutants. This restriction appears to be governed by a positive-feedback loop between centrosomes and Par-3 (Jiang et al., 2015). Strikingly, Par-1 promotes this positive feedback by phosphorylating Par-3 (and also through its actions on centrosomal microtubule dynamics) (Jiang et al., 2015). This suggests that the Par-1/Par-3 interaction can undergo a context-dependent switch between mutual antagonism and synergism that supports positive feedback and the formation of discrete unipolar patches of Par-3. In the embryonic epithelium, aPKC enforces mutual antagonism by phosphorylating and excluding Par-1 from the apical surface. However, the synergistic mode is likely to play a functional role in other contexts, where Par-3 patches colocalize with centrosomes (Feldman and Priess, 2012; Inaba et al., 2015).
Conclusions
In the past decade, we have made rapid progress in understanding how cells form and stabilize PAR asymmetries. Molecular, genetic and biochemical approaches continue to supply new information about individual polarity proteins and their pairwise interactions. Mechanistic insights into the dynamics of polarization have come from biophysical and imaging approaches that allow one to quantify PAR protein dynamics in living cells, and from mathematical models that attempt to explain these dynamics in terms of known interactions. Looking forward, the system is now ripe for approaches from optogenetics, synthetic biology and biochemical reconstitution (Chau et al., 2012; Loose et al., 2017; Witte et al., 2017).
Despite (and because of) this progress, there is still much to learn from the study of traditional model systems. Recent work highlights key processes involved in the clustering of polarity proteins and raises many new questions. How does clustering shape the local recruitment of polarity proteins, their physical coupling to the cytoskeleton and their biochemical activities? How is this modulated through cell cycle control and developmental progression? Another pressing challenge is to uncover the molecular mechanisms by which PAR proteins modulate cytoskeletal dynamics and motor-based transport, how these in turn feed back to govern local recruitment and redistribution of PAR proteins, and how these feedback loops are integrated to ensure robust self-stabilizing polarity. The overarching question that remains to be addressed is how cells stabilize uniquely positioned boundaries between complementary domains (or a unique size and position for unipolar domains) through a dynamic balance of local exchange, assembly/disassembly, diffusion/transport and mechanical force.
Finding answers to these questions in simple and tractable model systems will provide a blueprint for understanding how the PAR network carries out its functions in many other contexts. Indeed, rapid advances in microscopy and genome editing make it increasingly possible to characterize the dynamic behaviors of PAR proteins in many other developmental and organismal contexts. Using our increasingly deep knowledge of individual systems to ground comparative analyses, we are now poised to gain deep insights into how this highly conserved and yet versatile circuit has evolved to do so many different variants of its core job in so many different contexts.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Work in E.M.'s lab is funded bythe National Institutes of Health (R01 HD088831). Additionally, C.F.L. is supported by the National Institutes of Health (T32 GM007197). Deposited in PMC for release after 12 months.
References
- Aceto D., Beers M. and Kemphues K. J. (2006). Interaction of PAR-6 with CDC-42 is required for maintenance but not establishment of PAR asymmetry in C. elegans. Dev. Biol. 299, 386-397. 10.1016/j.ydbio.2006.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilleos A., Wehman A. M. and Nance J. (2010). PAR-3 mediates the initial clustering and apical localization of junction and polarity proteins during C. elegans intestinal epithelial cell polarization. Development 137, 1833-1842. 10.1242/dev.047647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. C., Gill J. S., Cinalli R. M. and Nance J. (2008). Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science 320, 1771-1774. 10.1126/science.1156063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arata Y., Lee J.-Y., Goldstein B. and Sawa H. (2010). Extracellular control of PAR protein localization during asymmetric cell division in the C. elegans embryo. Development 137, 3337-3345. 10.1242/dev.054742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arata Y., Hiroshima M., Pack C.-G., Ramanujam R., Motegi F., Nakazato K., Shindo Y., Wiseman P. W., Sawa H., Kobayashi T. J. et al. (2016). Cortical polarity of the RING protein PAR-2 is maintained by exchange rate kinetics at the cortical-cytoplasmic boundary. Cell Rep. 16, 2156-2168. 10.1016/j.celrep.2016.07.047 [DOI] [PubMed] [Google Scholar]
- Atwood S. X. and Prehoda K. E. (2009). aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr. Biol. 19, 723-729. 10.1016/j.cub.2009.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood S. X., Chabu C., Penkert R. R., Doe C. Q. and Prehoda K. E. (2007). Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J. Cell Sci. 120, 3200-3206. 10.1242/jcs.014902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M. J. and Prehoda K. E. (2015). Establishment of par-polarized cortical domains via phosphoregulated membrane motifs. Dev. Cell 35, 199-210. 10.1016/j.devcel.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros C. S., Phelps C. B. and Brand A. H. (2003). Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev. Cell 5, 829-840. 10.1016/S1534-5807(03)00359-9 [DOI] [PubMed] [Google Scholar]
- Bayraktar J., Zygmunt D. and Carthew R. W. (2006). Par-1 kinase establishes cell polarity and functions in Notch signaling in the Drosophila embryo. J. Cell Sci. 119, 711-721. 10.1242/jcs.02789 [DOI] [PubMed] [Google Scholar]
- Beatty A., Morton D. and Kemphues K. (2010). The C. elegans homolog of Drosophila Lethal giant larvae functions redundantly with PAR-2 to maintain polarity in the early embryo. Development 137, 3995-4004. 10.1242/dev.056028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty A., Morton D. G. and Kemphues K. (2013). PAR-2, LGL-1 and the CDC-42 GAP CHIN-1 act in distinct pathways to maintain polarity in the C. elegans embryo. Development 140, 2005-2014. 10.1242/dev.088310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers M. and Kemphues K. (2006). Depletion of the co-chaperone CDC-37 reveals two modes of PAR-6 cortical association in C. elegans embryos. Development 133, 3745-3754. 10.1242/dev.02544 [DOI] [PubMed] [Google Scholar]
- Benton R. and St Johnston D. (2003a). A conserved oligomerization domain in drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr. Biol. 13, 1330-1334. 10.1016/S0960-9822(03)00508-6 [DOI] [PubMed] [Google Scholar]
- Benton R. and St Johnston D. (2003b). Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell 115, 691-704. 10.1016/S0092-8674(03)00938-3 [DOI] [PubMed] [Google Scholar]
- Betschinger J. and Knoblich J. A. (2004). Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr. Biol. 14, R674-R685. 10.1016/j.cub.2004.08.017 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Eisenhaber F. and Knoblich J. A. (2005). Phosphorylation-induced autoinhibition regulates the cytoskeletal protein Lethal (2) giant larvae. Curr. Biol. 15, 276-282. 10.1016/j.cub.2005.01.012 [DOI] [PubMed] [Google Scholar]
- Bienkowska D. and Cowan C. R. (2012). Centrosomes can initiate a polarity axis from any position within one-cell C. elegans embryos. Curr. Biol. 22, 583-589. 10.1016/j.cub.2012.01.064 [DOI] [PubMed] [Google Scholar]
- Bilder D. and Perrimon N. (2000). Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature 403, 676-680. 10.1038/35001108 [DOI] [PubMed] [Google Scholar]
- Bilder D., Schober M. and Perrimon N. (2003). Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat. Cell Biol. 5, 53-58. 10.1038/ncb897 [DOI] [PubMed] [Google Scholar]
- Blanchoud S., Busso C., Naef F. and Gönczy P. (2015). Quantitative analysis and modeling probe polarity establishment in C. elegans embryos. Biophys. J. 108, 799-809. 10.1016/j.bpj.2014.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bois J. S., Jülicher F. and Grill S. W. (2011). Pattern formation in active fluids. Phys. Rev. Lett. 106, 028103 10.1103/PhysRevLett.106.028103 [DOI] [PubMed] [Google Scholar]
- Boyd L., Guo S., Levitan D. and Stinchcomb D. T. (1996). PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos . Development 122, 3075-3084. [DOI] [PubMed] [Google Scholar]
- Bryant D. M. and Mostov K. E. (2008). From cells to organs: building polarized tissue. Nat. Rev. Mol. Cell Biol. 9, 887-901. 10.1038/nrm2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Yu F., Lin S., Chia W. and Yang X. (2003). Apical complex genes control mitotic spindle geometry and relative size of daughter cells in Drosophila neuroblast and pI asymmetric divisions. Cell 112, 51-62. 10.1016/S0092-8674(02)01170-4 [DOI] [PubMed] [Google Scholar]
- Chan E. and Nance J. (2013). Mechanisms of CDC-42 activation during contact-induced cell polarization. J. Cell Sci. 126, 1692-1702. 10.1242/jcs.124594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier F. J.-M., Hardy É. J.-L. and Laprise P. (2011). Crumbs controls epithelial integrity by inhibiting Rac1 and PI3K. J. Cell Sci. 124, 3393-3398. 10.1242/jcs.092601 [DOI] [PubMed] [Google Scholar]
- Chau A. H., Walter J. M., Gerardin J., Tang C. and Lim W. A. (2012). Designing synthetic regulatory networks capable of self-organizing cell polarization. Cell 151, 320-332. 10.1016/j.cell.2012.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeks R. J., Canman J. C., Gabriel W. N., Meyer N., Strome S. and Goldstein B. (2004). C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. 14, 851-862. 10.1016/j.cub.2004.05.022 [DOI] [PubMed] [Google Scholar]
- Chen S., Chen J., Shi H., Wei M., Castaneda-Castellanos D. R., Bultje R. S., Pei X., Kriegstein A. R., Zhang M. and Shi S.-H. (2013). Regulation of microtubule stability and organization by mammalian Par3 in specifying neuronal polarity. Dev. Cell 24, 26-40. 10.1016/j.devcel.2012.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret S., Jouette J., Benoit B., Legent K. and Guichet A. (2014). PI(4,5)P2 produced by the PI4P5K SKTL controls apical size by tethering PAR-3 in Drosophila epithelial cells. Curr. Biol. 24, 1071-1079. 10.1016/j.cub.2014.03.056 [DOI] [PubMed] [Google Scholar]
- Cowan C. R. and Hyman A. A. (2004). Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature 431, 92-96. 10.1038/nature02825 [DOI] [PubMed] [Google Scholar]
- Dahan I., Yearim A., Touboul Y. and Ravid S. (2012). The tumor suppressor Lgl1 regulates NMII-A cellular distribution and focal adhesion morphology to optimize cell migration. Mol. Biol. Cell 23, 591-601. 10.1091/mbc.E11-01-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan I., Petrov D., Cohen-Kfir E. and Ravid S. (2014). The tumor suppressor Lgl1 forms discrete complexes with NMII-A and Par6α-aPKCζ that are affected by Lgl1 phosphorylation. J. Cell Sci. 127, 295-304. 10.1242/jcs.127357 [DOI] [PubMed] [Google Scholar]
- Dawes A. T. and Munro E. M. (2011). PAR-3 oligomerization may provide an actin-independent mechanism to maintain distinct par protein domains in the early caenorhabditis elegans embryo. Biophys. J. 101, 1412-1422. 10.1016/j.bpj.2011.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker E., Bocina I. and Jiang D. (2013). Tubulogenesis in a simple cell cord requires the formation of bi-apical cells through two discrete Par domains. Development 140, 2985-2996. 10.1242/dev.092387 [DOI] [PubMed] [Google Scholar]
- Deshaies R. J. and Joazeiro C. A. P. (2009). RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399-434. 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Schwager F., Pintard L., Gotta M. and Goldstein B. (2017). A single-cell biochemistry approach reveals PAR complex dynamics during cell polarization. Dev. Cell. 42, 416-434. 10.1016/j.devcel.2017.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger H., Vogt N., Torres I. L., Mirouse V., Koch I., Nüsslein-Volhard C. and St Johnston D. (2010). Bazooka is required for polarisation of the Drosophila anterior-posterior axis. Development 137, 1765-1773. 10.1242/dev.045807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W., Zhang X., Liu W., Chen Y.-J., Huang J., Austin E., Celotto A. M., Jiang W. Z., Palladino M. J., Jiang Y. et al. (2015). A conserved polybasic domain mediates plasma membrane targeting of Lgl and its regulation by hypoxia. J. Cell Biol. 211, 273-286. 10.1083/jcb.201503067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S. and Martin-Belmonte F. (2014). Cargo sorting in the endocytic pathway: a key regulator of cell polarity and tissue dynamics. Cold Spring Harbor Perspect. Biol. 6, a016899-a016899 10.1101/cshperspect.a016899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein-Keshet L. and Ermentrout G. B. (1998). Models for the length distributions of actin filaments: I. Simple polymerization and fragmentation. Bull. Math. Biol. 60, 449-475. 10.1006/bulm.1997.0011 [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B., Guo S. and Kemphues K. J. (1995). Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell 83, 743-752. 10.1016/0092-8674(95)90187-6 [DOI] [PubMed] [Google Scholar]
- Feldman J. L. and Priess J. R. (2012). A role for the centrosome and PAR-3 in the hand-off of MTOC function during epithelial polarization. Curr. Biol. 22, 575-582. 10.1016/j.cub.2012.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Wu H., Chan L.-N. and Zhang M. (2007). The Par-3 NTD adopts a PB1-like structure required for Par-3 oligomerization and membrane localization. EMBO J. 26, 2786-2796. 10.1038/sj.emboj.7601702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J. E. and Ha S. H. (2014). Ultrasensitivity part II: multisite phosphorylation, stoichiometric inhibitors, and positive feedback. Trends Biochem. Sci. 39, 556-569. 10.1016/j.tibs.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher G. C., Lucas E. P., Brain R., Tournier A. and Thompson B. J. (2012). Positive feedback and mutual antagonism combine to polarize Crumbs in the Drosophila follicle cell epithelium. Curr. Biol. 22, 1116-1122. 10.1016/j.cub.2012.04.020 [DOI] [PubMed] [Google Scholar]
- Franz A. and Riechmann V. (2010). Stepwise polarisation of the Drosophila follicular epithelium. Dev. Biol. 338, 136-147. 10.1016/j.ydbio.2009.11.027 [DOI] [PubMed] [Google Scholar]
- Gamblin C. L., Hardy É. J.-L., Chartier F. J.-M., Bisson N. and Laprise P. (2014). A bidirectional antagonism between aPKC and Yurt regulates epithelial cell polarity. J. Cell Biol. 204, 487-495. 10.1083/jcb.201308032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassama-Diagne A., Yu W., ter Beest M., Martin-Belmonte F., Kierbel A., Engel J. and Mostov K. (2006). Phosphatidylinositol-3,4,5-trisphosphate regulates the formation of the basolateral plasma membrane in epithelial cells. Nat. Cell Biol. 8, 963-970. 10.1038/ncb1461 [DOI] [PubMed] [Google Scholar]
- Goehring N. W., Hoege C., Grill S. W. and Hyman A. A. (2011a). PAR proteins diffuse freely across the anterior-posterior boundary in polarized C. elegans embryos. J. Cell Biol. 193, 583-594. 10.1083/jcb.201011094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring N. W., Trong P. K., Bois J. S., Chowdhury D., Nicola E. M., Hyman A. A. and Grill S. W. (2011b). Polarization of PAR proteins by advective triggering of a pattern-forming system. Science 334, 1137-1141. 10.1126/science.1208619 [DOI] [PubMed] [Google Scholar]
- Goldstein B. and Macara I. G. (2007). The PAR proteins: fundamental players in animal cell polarization. Dev. Cell 13, 609-622. 10.1016/j.devcel.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Abraham M. C. and Ahringer J. (2001). CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr. Biol. 11, 482-488. 10.1016/S0960-9822(01)00142-7 [DOI] [PubMed] [Google Scholar]
- Graybill C. and Prehoda K. E. (2014). Ordered multisite phosphorylation of lethal giant larvae by atypical protein kinase C. Biochemistry 53, 4931-4937. 10.1021/bi500748w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin E. E., Odde D. J. and Seydoux G. (2011). Regulation of the MEX-5 gradient by a spatially segregated kinase/phosphatase cycle. Cell 146, 955-968. 10.1016/j.cell.2011.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S. and Kemphues K. J. (1995). par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81, 611-620. 10.1016/0092-8674(95)90082-9 [DOI] [PubMed] [Google Scholar]
- Guo S. and Kemphues K. J. (1996). Molecular genetics of asymmetric cleavage in the early Caenorhabditis elegans embryo. Curr. Opin. Genet. Dev. 6, 408-415. 10.1016/S0959-437X(96)80061-X [DOI] [PubMed] [Google Scholar]
- Hamill D. R., Severson A. F., Carter J. C. and Bowerman B. (2002). Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev. Cell 3, 673-684. 10.1016/S1534-5807(02)00327-1 [DOI] [PubMed] [Google Scholar]
- Hao Y., Boyd L. and Seydoux G. (2006). Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev. Cell 10, 199-208. 10.1016/j.devcel.2005.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. J. C. and Peifer M. (2005). The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J. Cell Biol. 170, 813-823. 10.1083/jcb.200505127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. J. C. and Peifer M. (2007). aPKC controls microtubule organization to balance adherens junction symmetry and planar polarity during development. Dev. Cell 12, 727-738. 10.1016/j.devcel.2007.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K. P. and Tepass U. (2008). Cdc42 and Par proteins stabilize dynamic adherens junctions in the Drosophila neuroectoderm through regulation of apical endocytosis. J. Cell Biol. 183, 1129-1143. 10.1083/jcb.200807020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V. and Wodarz A. (2013). Initial neurogenesis in Drosophila. Wiley Interdiscip Rev. Dev. Biol. 2, 701-721. 10.1002/wdev.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y., Yoshinaga S., Takeya R., Suzuki N. N., Horiuchi M., Kohjima M., Sumimoto H. and Inagaki F. (2005). Structure of a cell polarity regulator, a complex between atypical PKC and Par6 PB1 domains. J. Biol. Chem. 280, 9653-9661. 10.1074/jbc.M409823200 [DOI] [PubMed] [Google Scholar]
- Hoege C., Constantinescu A.-T., Schwager A., Goehring N. W., Kumar P. and Hyman A. A. (2010). LGL can partition the cortex of one-cell Caenorhabditis elegans embryos into two domains. Curr. Biol. 20, 1296-1303. 10.1016/j.cub.2010.05.061 [DOI] [PubMed] [Google Scholar]
- Horikoshi Y., Suzuki A., Yamanaka T., Sasaki K., Mizuno K., Sawada H., Yonemura S. and Ohno S. (2009). Interaction between PAR-3 and the aPKC-PAR-6 complex is indispensable for apical domain development of epithelial cells. J. Cell Sci. 122, 1595-1606. 10.1242/jcs.043174 [DOI] [PubMed] [Google Scholar]
- Hung T. J. and Kemphues K. J. (1999). PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development 126, 127-135. [DOI] [PubMed] [Google Scholar]
- Inaba M., Venkei Z. G. and Yamashita Y. M. (2015). The polarity protein Baz forms a platform for the centrosome orientation during asymmetric stem cell division in the Drosophila male germline. Elife 4, 659 10.7554/eLife.04960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J. and Gonzalez C. (2010). The interphase microtubule aster is a determinant of asymmetric division orientation in Drosophila neuroblasts. J. Cell Biol. 188, 693-706. 10.1083/jcb.200905024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Reina J., Llamazares S., Bertran T., Rossi F., Roig J. and Gonzalez C. (2013). Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 15, 241-248. 10.1038/ncb2671 [DOI] [PubMed] [Google Scholar]
- Jiang T., McKinley R. F. A., McGill M. A., Angers S. and Harris T. J. C. (2015). A Par-1-Par-3-centrosome cell polarity pathway and its tuning for isotropic cell adhesion. Curr. Biol. 25, 2701-2708. 10.1016/j.cub.2015.08.063 [DOI] [PubMed] [Google Scholar]
- Joberty G., Petersen C., Gao L. and Macara I. G. (2000). The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2, 531-539. 10.1038/35019573 [DOI] [PubMed] [Google Scholar]
- Kay A. J. and Hunter C. P. (2001). CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Curr. Biol. 11, 474-481. 10.1016/S0960-9822(01)00141-5 [DOI] [PubMed] [Google Scholar]
- Kemphues K. J., Priess J. R., Morton D. G. and Cheng N. S. (1988). Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52, 311-320. 10.1016/S0092-8674(88)80024-2 [DOI] [PubMed] [Google Scholar]
- Klompstra D., Anderson D. C., Yeh J. Y., Zilberman Y. and Nance J. (2015). An instructive role for C. elegans E-cadherin in translating cell contact cues into cortical polarity. Nat. Cell Biol. 17, 726-735. 10.1038/ncb3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn M. P., Egger-Adam D. and Wodarz A. (2009). PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical-basal polarity in dividing embryonic neuroblasts. Dev. Cell 16, 901-908. 10.1016/j.devcel.2009.04.011 [DOI] [PubMed] [Google Scholar]
- Krahn M. P., Bückers J., Kastrup L. and Wodarz A. (2010a). Formation of a Bazooka-Stardust complex is essential for plasma membrane polarity in epithelia. J. Cell Biol. 190, 751-760. 10.1083/jcb.201006029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn M. P., Klopfenstein D. R., Fischer N. and Wodarz A. (2010b). Membrane targeting of Bazooka/PAR-3 is mediated by direct binding to phosphoinositide lipids. Curr. Biol. 20, 636-642. 10.1016/j.cub.2010.01.065 [DOI] [PubMed] [Google Scholar]
- Kumfer K. T., Cook S. J., Squirrell J. M., Eliceiri K. W., Peel N., O'Connell K. F. and White J. G. (2010). CGEF-1 and CHIN-1 regulate CDC-42 activity during asymmetric division in the Caenorhabditis elegans embryo. Mol. Biol. Cell 21, 266-277. 10.1091/mbc.E09-01-0060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprise P. and Tepass U. (2011). Novel insights into epithelial polarity proteins in Drosophila. Trends Cell Biol. 21, 401-408. 10.1016/j.tcb.2011.03.005 [DOI] [PubMed] [Google Scholar]
- Laprise P., Lau K. M., Harris K. P., Silva-Gagliardi N. F., Paul S. M., Beronja S., Beitel G. J., McGlade C. J. and Tepass U. (2009). Yurt, Coracle, Neurexin IV and the Na(+),K(+)-ATPase form a novel group of epithelial polarity proteins. Nature 459, 1141-1145. 10.1038/nature08067 [DOI] [PubMed] [Google Scholar]
- Lee C.-Y., Robinson K. J. and Doe C. Q. (2006). Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature 439, 594-598. 10.1038/nature04299 [DOI] [PubMed] [Google Scholar]
- Leibfried A., Müller S. and Ephrussi A. (2013). A Cdc42-regulated actin cytoskeleton mediates Drosophila oocyte polarization. Development 140, 362-371. 10.1242/dev.089250 [DOI] [PubMed] [Google Scholar]
- Lemmon M. A. (2008). Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99-111. 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- Li B., Kim H., Beers M. and Kemphues K. (2010a). Different domains of C. elegans PAR-3 are required at different times in development. Dev. Biol. 344, 745-757. 10.1016/j.ydbio.2010.05.506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Kim H., Aceto D. G., Hung J., Aono S. and Kemphues K. J. (2010b). Binding to PKC-3, but not to PAR-3 or to a conventional PDZ domain ligand, is required for PAR-6 function in C. elegans. Dev. Biol. 340, 88-98. 10.1016/j.ydbio.2010.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., Edwards A. S., Fawcett J. P., Mbamalu G., Scott J. D. and Pawson T. (2000). A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2, 540-547. 10.1038/35019582 [DOI] [PubMed] [Google Scholar]
- Loose M., Zieske K. and Schwille P. (2017). Reconstitution of protein dynamics involved in bacterial cell division. Subcell. Biochem. 84, 419-444. 10.1007/978-3-319-53047-5_15 [DOI] [PubMed] [Google Scholar]
- Lu M. S. and Johnston C. A. (2013). Molecular pathways regulating mitotic spindle orientation in animal cells. Development 140, 1843-1856. 10.1242/dev.087627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Belmonte F. and Mostov K. (2008). Regulation of cell polarity during epithelial morphogenesis. Curr. Opin. Cell Biol. 20, 227-234. 10.1016/j.ceb.2008.01.001 [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F., Gassama A., Datta A., Yu W., Rescher U., Gerke V. and Mostov K. (2007). PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell 128, 383-397. 10.1016/j.cell.2006.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M., Depken M., Bois J. S., Jülicher F. and Grill S. W. (2010). Anisotropies in cortical tension reveal the physical basis of polarizing cortical flows. Nature 467, 617-621. 10.1038/nature09376 [DOI] [PubMed] [Google Scholar]
- Mccaffrey L. M. and Macara I. G. (2009). The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev. 23, 1450-1460. 10.1101/gad.1795909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M. A., McKinley R. F. A. and Harris T. J. C. (2009). Independent cadherin-catenin and Bazooka clusters interact to assemble adherens junctions. J. Cell Biol. 185, 787-796. 10.1083/jcb.200812146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley R. F. A., Yu C. G. and Harris T. J. C. (2012). Assembly of Bazooka polarity landmarks through a multifaceted membrane-association mechanism. J. Cell Sci. 125, 1177-1190. 10.1242/jcs.091884 [DOI] [PubMed] [Google Scholar]
- Mizuno K., Suzuki A., Hirose T., Kitamura K., Kutsuzawa K., Futaki M., Amano Y. and Ohno S. (2003). Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J. Biol. Chem. 278, 31240-31250. 10.1074/jbc.M303593200 [DOI] [PubMed] [Google Scholar]
- Morais-de-Sá E., Mirouse V. and St Johnston D. (2010). aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell 141, 509-523. 10.1016/j.cell.2010.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais-de-Sá E., Mukherjee A., Lowe N. and St Johnston D. (2014). Slmb antagonises the aPKC/Par-6 complex to control oocyte and epithelial polarity. Development 141, 2984-2992. 10.1242/dev.109827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K., Mendrola J. M., Schmitz K. R., Wang Y.-H., Slochower D., Janmey P. A. and Lemmon M. A. (2010). Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell 143, 966-977. 10.1016/j.cell.2010.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y., Jilkine A. and Edelstein-Keshet L. (2008). Wave-pinning and cell polarity from a bistable reaction-diffusion system. Biophys. J. 94, 3684-3697. 10.1529/biophysj.107.120824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi F. and Seydoux G. (2013). The PAR network: redundancy and robustness in a symmetry-breaking system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20130010-20130010 10.1098/rstb.2013.0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi F. and Sugimoto A. (2006). Sequential functioning of the ECT-2 RhoGEF, RHO-1 and CDC-42 establishes cell polarity in Caenorhabditis elegans embryos. Nat. Cell Biol. 8, 978-985. 10.1038/ncb1459 [DOI] [PubMed] [Google Scholar]
- Motegi F., Zonies S., Hao Y., Cuenca A. A., Griffin E. and Seydoux G. (2011). Microtubules induce self-organization of polarized PAR domains in Caenorhabditis elegans zygotes. Nat. Cell Biol. 13, 1361-1367. 10.1038/ncb2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E., Nance J. and Priess J. R. (2004). Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell 7, 413-424. 10.1016/j.devcel.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Nakaya M., Fukui A., Izumi Y., Akimoto K., Asashima M. and Ohno S. (2000). Meiotic maturation induces animal-vegetal asymmetric distribution of aPKC and ASIP/PAR-3 in Xenopus oocytes. Development 127, 5021-5031. [DOI] [PubMed] [Google Scholar]
- Nakayama Y., Shivas J. M., Poole D. S., Squirrell J. M., Kulkoski J. M., Schleede J. B. and Skop A. R. (2009). Dynamin participates in the maintenance of anterior polarity in the Caenorhabditis elegans embryo. Dev. Cell 16, 889-900. 10.1016/j.devcel.2009.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Kato K., Yamaguchi T., Fukata Y., Ohno S. and Kaibuchi K. (2004). Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat. Cell Biol. 6, 328-334. 10.1038/ncb1118 [DOI] [PubMed] [Google Scholar]
- O'Connell K. F., Maxwell K. N. and White J. G. (2000). The spd-2 gene is required for polarization of the anteroposterior axis and formation of the sperm asters in the Caenorhabditis elegans zygote. Dev. Biol. 222, 55-70. 10.1006/dbio.2000.9714 [DOI] [PubMed] [Google Scholar]
- Ohshiro T., Yagami T., Zhang C. and Matsuzaki F. (2000). Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature 408, 593-596. 10.1038/35046087 [DOI] [PubMed] [Google Scholar]
- Peng C.-Y., Manning L., Albertson R. and Doe C. Q. (2000). The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature 408, 596-600. 10.1038/35046094 [DOI] [PubMed] [Google Scholar]
- Petronczki M. and Knoblich J. A. (2001). DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 3, 43-49. 10.1038/35050550 [DOI] [PubMed] [Google Scholar]
- Prehoda K. E. (2009). Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harbor Perspect. Biol. 1, a001388-a001388 10.1101/cshperspect.a001388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E., Sampaio P., Januschke J., Llamazares S., Varmark H. and Gonzalez C. (2007). Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell 12, 467-474. 10.1016/j.devcel.2007.01.021 [DOI] [PubMed] [Google Scholar]
- Robin F. B., McFadden W. M., Yao B. and Munro E. M. (2014). Single-molecule analysis of cell surface dynamics in Caenorhabditis elegans embryos. Nat. Meth. 11, 677-682. 10.1038/nmeth.2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J., Peglion F., Martin J., Hubatsche L., Reich J., Hirani N., Gubieda A. G., Roffey J., Fernandez A. R., St Johnston D. et al. (2017). aPKC cycles between functionally distinct PAR protein assemblies to drive cell polarity. Dev. Cell 42, 400-415. 10.1016/j.devcel.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E. and Macara I. G. (2014). Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 15, 225-242. 10.1038/nrm3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose L. and Gönczy P. (2014). Polarity establishment, asymmetric division and segregation of fate determinants in early C. elegans embryos. WormBook 10.1895/wormbook.1.30.2 [DOI] [PubMed] [Google Scholar]
- Rusan N. M. and Peifer M. (2007). A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177, 13-20. 10.1083/jcb.200612140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer A., Anneken A., Li Y., Lee S. and Munro E. (2015). Dynamic opposition of clustered proteins stabilizes cortical polarity in the C. elegans Zygote. Dev. Cell 35, 131-142. 10.1016/j.devcel.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M., Shevchenko A. and Knoblich J. A. (2000). A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr. Biol. 10, 353-362. 10.1016/S0960-9822(00)00401-2 [DOI] [PubMed] [Google Scholar]
- Schaefer M., Petronczki M., Dorner D., Forte M. and Knoblich J. A. (2001). Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell 107, 183-194. 10.1016/S0092-8674(01)00521-9 [DOI] [PubMed] [Google Scholar]
- Schmutz C., Stevens J. and Spang A. (2007). Functions of the novel RhoGAP proteins RGA-3 and RGA-4 in the germ line and in the early embryo of C. elegans. Development 134, 3495-3505. 10.1242/dev.000802 [DOI] [PubMed] [Google Scholar]
- Schober M., Schaefer M. and Knoblich J. A. (1999). Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature 402, 548-551. 10.1038/990135 [DOI] [PubMed] [Google Scholar]
- Schonegg S. and Hyman A. A. (2006). CDC-42 and RHO-1 coordinate acto-myosin contractility and PAR protein localization during polarity establishment in C. elegans embryos. Development 133, 3507-3516. 10.1242/dev.02527 [DOI] [PubMed] [Google Scholar]
- Seirin Lee S. (2016). Positioning of polarity formation by extracellular signaling during asymmetric cell division. J. Theor. Biol. 400, 52-64. 10.1016/j.jtbi.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Seirin Lee S. and Shibata T. (2015). Self-organization and advective transport in the cell polarity formation for asymmetric cell division. J. Theor. Biol. 382, 1-14. 10.1016/j.jtbi.2015.06.032 [DOI] [PubMed] [Google Scholar]
- Shewan A., Eastburn D. J. and Mostov K. (2011). Phosphoinositides in cell architecture. Cold Spring Harbor Perspect. Biol. 3, a004796-a004796 10.1101/cshperspect.a004796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.-H., Jan L. Y. and Jan Y.-N. (2003). Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell 112, 63-75. 10.1016/S0092-8674(02)01249-7 [DOI] [PubMed] [Google Scholar]
- Shi S.-H., Cheng T., Jan L. Y. and Jan Y.-N. (2004). APC and GSK-3beta are involved in mPar3 targeting to the nascent axon and establishment of neuronal polarity. Curr. Biol. 14, 2025-2032. 10.1016/j.cub.2004.11.009 [DOI] [PubMed] [Google Scholar]
- Shivas J. M. and Skop A. R. (2012). Arp2/3 mediates early endosome dynamics necessary for the maintenance of PAR asymmetry in Caenorhabditis elegans. Mol. Biol. Cell 23, 1917-1927. 10.1091/mbc.E12-01-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist S. E. and Doe C. Q. (2005). Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell 123, 1323-1335. 10.1016/j.cell.2005.09.043 [DOI] [PubMed] [Google Scholar]
- Siegrist S. E. and Doe C. Q. (2006). Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development 133, 529-536. 10.1242/dev.02211 [DOI] [PubMed] [Google Scholar]
- Skwarek L. C., Windler S. L., de Vreede G., Rogers G. C. and Bilder D. (2014). The F-box protein Slmb restricts the activity of aPKC to polarize epithelial cells. Development 141, 2978-2983. 10.1242/dev.109694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano E. V., Ivanova M. E., Fletcher G., Riou P., Knowles P. P., Barnouin K., Purkiss A., Kostelecky B., Saiu P., Linch M. et al. (2016). aPKC inhibition by Par3 CR3 flanking regions controls substrate access and underpins apical-junctional polarization. Dev. Cell 38, 384-398. 10.1016/j.devcel.2016.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiga F. M., Prouteau M. and Gotta M. (2013). The TAO kinase KIN-18 regulates contractility and establishment of polarity in the C. elegans embryo. Dev. Biol. 373, 26-38. 10.1016/j.ydbio.2012.10.001 [DOI] [PubMed] [Google Scholar]
- St Johnston D. and Ahringer J. (2010). Cell polarity in eggs and epithelia: parallels and diversity. Cell 141, 757-774. 10.1016/j.cell.2010.05.011 [DOI] [PubMed] [Google Scholar]
- Strand D., Jakobs R., Merdes G., Neumann B., Kalmes A., Heid H. W., Husmann I. and Mechler B. M. (1994). The Drosophila lethal(2)giant larvae tumor suppressor protein forms homo-oligomers and is associated with nonmuscle myosin II heavy chain. J. Cell Biol. 127, 1361-1373. 10.1083/jcb.127.5.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand D., Unger S., Corvi R., Hartenstein K., Schenkel H., Kalmes A., Merdes G., Neumann B., Krieg-Schneider F. and Coy J. F. (1995). A human homologue of the Drosophila tumour suppressor gene l(2)gl maps to 17p11.2-12 and codes for a cytoskeletal protein that associates with nonmuscle myosin II heavy chain. Oncogene 11, 291-301. [PubMed] [Google Scholar]
- Suzuki A. and Ohno S. (2006). The PAR-aPKC system: lessons in polarity. J. Cell Sci. 119, 979-987. 10.1242/jcs.02898 [DOI] [PubMed] [Google Scholar]
- Tabuse Y., Izumi Y., Piano F., Kemphues K. J., Miwa J. and Ohno S. (1998). Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development 125, 3607-3614. [DOI] [PubMed] [Google Scholar]
- Tanentzapf G. and Tepass U. (2003). Interactions between the crumbs, lethal giant larvae and bazooka pathways in epithelial polarization. Nat. Cell Biol. 5, 46-52. 10.1038/ncb896 [DOI] [PubMed] [Google Scholar]
- Tepass U. (2012). The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell Dev. Biol. 28, 655-685. 10.1146/annurev-cellbio-092910-154033 [DOI] [PubMed] [Google Scholar]
- Tian A.-G. and Deng W.-M. (2008). Lgl and its phosphorylation by aPKC regulate oocyte polarity formation in Drosophila. Development 135, 463-471. 10.1242/dev.016253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P., Piano F., Riechmann V., Gunsalus K. C., Kemphues K. J. and Ephrussi A. (2000). A Drosophila melanogaster homologue of Caenorhabditis elegans par-1 acts at an early step in embryonic-axis formation. Nat. Cell Biol. 2, 458-460. 10.1038/35017101 [DOI] [PubMed] [Google Scholar]
- Tse Y. C., Werner M., Longhini K. M., Labbé J.-C., Goldstein B. and Glotzer M. (2012). RhoA activation during polarization and cytokinesis of the early Caenorhabditis elegans embryo is differentially dependent on NOP-1 and CYK-4. Mol. Biol. Cell 23, 4020-4031. 10.1091/mbc.E12-04-0268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stein W., Ramrath A., Grimm A., Müller-Borg M. and Wodarz A. (2005). Direct association of Bazooka/PAR-3 with the lipid phosphatase PTEN reveals a link between the PAR/aPKC complex and phosphoinositide signaling. Development 132, 1675-1686. 10.1242/dev.01720 [DOI] [PubMed] [Google Scholar]
- Walther R. F. and Pichaud F. (2010). Crumbs/DaPKC-dependent apical exclusion of Bazooka promotes photoreceptor polarity remodeling. Curr. Biol. 20, 1065-1074. 10.1016/j.cub.2010.04.049 [DOI] [PubMed] [Google Scholar]
- Wang S.-C., Low T. Y. F., Nishimura Y., Gole L., Yu W. and Motegi F. (2017). Cortical forces and CDC-42 control clustering of PAR proteins for Caenorhabditis elegans embryonic polarization. Nat. Cell Biol. 19, 988-995. 10.1038/ncb3577 [DOI] [PubMed] [Google Scholar]
- Watts J. L., Etemad-Moghadam B., Guo S., Boyd L., Draper B. W., Mello C. C., Priess J. R. and Kemphues K. J. (1996). par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development 122, 3133-3140. [DOI] [PubMed] [Google Scholar]
- Whitney D. S., Peterson F. C., Kittell A. W., Egner J. M., Prehoda K. E. and Volkman B. F. (2016). Binding of crumbs to the Par-6 CRIB-PDZ module is regulated by Cdc42. Biochemistry 55, 1455-1461. 10.1021/acs.biochem.5b01342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Nishimura T. and Knoblich J. A. (2008). Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell 135, 161-173. 10.1016/j.cell.2008.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte K. L., Strickland D. and Glotzer M. (2017). Cell cycle entry triggers a switch between two modes of Cdc42 activation during yeast polarization. Elife 6, e26722 10.7554/eLife.26722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Kuchinke U. and Knust E. (1999). Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature 402, 544-547. 10.1038/990128 [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Grimm A. and Knust E. (2000). Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 150, 1361-1374. 10.1083/jcb.150.6.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Kuchinke U. and Knust E. (2008). Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Curr. Biol. 18, 1639-1648. 10.1016/j.cub.2008.09.063 [DOI] [PubMed] [Google Scholar]
- Wu H., Feng W., Chen J., Chan L.-N., Huang S. and Zhang M. (2007). PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol. Cell 28, 886-898. 10.1016/j.molcel.2007.10.028 [DOI] [PubMed] [Google Scholar]
- Yamanaka T., Horikoshi Y., Izumi N., Suzuki A., Mizuno K. and Ohno S. (2006). Lgl mediates apical domain disassembly by suppressing the PAR-3-aPKC-PAR-6 complex to orient apical membrane polarity. J. Cell Sci. 119, 2107-2118. 10.1242/jcs.02938 [DOI] [PubMed] [Google Scholar]
- Yu C. G. and Harris T. J. C. (2012). Interactions between the PDZ domains of Bazooka (Par-3) and phosphatidic acid: in vitro characterization and role in epithelial development. Mol. Biol. Cell 23, 3743-3753. 10.1091/mbc.E12-03-0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Morin X., Cai Y., Yang X. and Chia W. (2000). Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100, 399-409. 10.1016/S0092-8674(00)80676-5 [DOI] [PubMed] [Google Scholar]
- Yuan H., Chiang C.-Y. A., Cheng J., Salzmann V. and Yamashita Y. M. (2012). Regulation of cyclin A localization downstream of Par-1 function is critical for the centrosome orientation checkpoint in Drosophila male germline stem cells. Dev. Biol. 361, 57-67. 10.1016/j.ydbio.2011.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang W., Chen J., Zhang K., Gao F., Gao B., Zhang S., Dong M., Besenbacher F., Gong W. et al. (2013). Structural insights into the intrinsic self-assembly of Par-3 N-terminal domain. Structure 21, 997-1006. 10.1016/j.str.2013.04.004 [DOI] [PubMed] [Google Scholar]
- Zonies S., Motegi F., Hao Y. and Seydoux G. (2010). Symmetry breaking and polarization of the C. elegans zygote by the polarity protein PAR-2. Development 137, 1669-1677. 10.1242/dev.045823 [DOI] [PMC free article] [PubMed] [Google Scholar]