Abstract
Migration of stem cells underpins the physiology of metazoan animals. For tissues to be maintained, stem cells and their progeny must migrate and differentiate in the correct positions. This need is even more acute after tissue damage by wounding or pathogenic infection. Inappropriate migration also underpins metastasis. Despite this, few mechanistic studies address stem cell migration during repair or homeostasis in adult tissues. Here, we present a shielded X-ray irradiation assay that allows us to follow stem cell migration in planarians. We demonstrate the use of this system to study the molecular control of stem cell migration and show that snail-1, snail-2 and zeb-1 EMT transcription factor homologs are necessary for cell migration to wound sites and for the establishment of migratory cell morphology. We also observed that stem cells undergo homeostatic migration to anterior regions that lack local stem cells, in the absence of injury, maintaining tissue homeostasis. This requires the polarity determinant notum. Our work establishes planarians as a suitable model for further in-depth study of the processes controlling stem cell migration in vivo.
KEY WORDS: EMT, Migration, Planarian, Pluripotency, Snail, Wounding, Schmidtea mediterranea
Highlighted Article: snail-1, snail-2 and zeb-1 control precise homing of stem cells and their progeny to wound sites, whereas the polarity determinant notum controls migration in the absence of wounding to maintain anterior tissue homeostasis.
INTRODUCTION
Regeneration and tissue homeostasis in multicellular animals are a result of stem cell activity. Most animal adult life histories include some potential to regenerate tissues and organs but the efficiency and extent of the regenerative process vary greatly among species. Many invertebrates, such as cnidarians, flatworms and annelids, are capable of whole-body regeneration and some of these are now available as experimentally tractable models for studying regeneration and homeostasis (Galliot, 2012; Gehrke and Srivastava, 2016; Tanaka and Reddien, 2011). Studies of the stem cells that contribute to regeneration and homeostasis can inform us about the origins of key stem cell properties. Few studies in regenerative models have investigated cell migration in vivo in adult animals, even though migration to sites of injury or homeostatic activity is necessary for regeneration and repair, and has important biomedical applications (Bradshaw et al., 2015; Guedelhoefer and Sánchez Alvarado, 2012b; Reig et al., 2014).
Overmigration leads to tumor tissue invasion and the pathology caused by cancers (Friedl and Gilmour, 2009; Friedl et al., 2012), and defects in stem cell migration are likely to contribute to aging. Many studies have revealed common mechanisms that drive cell migration in different contexts (Friedl and Alexander, 2011; Friedl et al., 2012; Goichberg, 2016; Ridley et al., 2003). However, studying cell migration in vivo is technically challenging, and a simple model might have much to offer. For example, in vivo studies in both Drosophila and C. elegans during embryogenesis and larval development have proven useful for unveiling fundamental molecular mechanisms (Geisbrecht and Montell, 2002; Hagedorn et al., 2013; Montell, 2003; Reig et al., 2014; Sato et al., 2015). The planarian system, in which pluripotent adult stem cells [known as neoblasts (NBs)] and their progeny can be studied, is another potentially tractable system for studying cell migration (Guedelhoefer and Sánchez Alvarado, 2012a).
Here, we establish new methods to study cell migration and show that NB and progeny migration utilize epithelial-mesenchymal transition (EMT)-related mechanisms in response to tissue damage. To date, relatively little focus has been given to stem cell migration in planarians (Guedelhoefer and Sánchez Alvarado, 2012b; Saló and Baguñà, 1985), although it is a necessary component of a successful regenerative outcome. We designed an assay to allow observation of cell migration and describe several phenomena within the planarian system, including the formation of extended processes by migrating NBs. Using markers of the epidermal lineage we uncover that cells at some stages of differentiation are more migratory than other cells that are at other stages of differentiation. RNAi of Smed-MMPa (mmpa) and of an ortholog of beta-integrin, Smed-β1-integrin (β1-integrin), disrupt cell migration and the formation of extended processes, providing proof of principle for this approach (Bonar and Petersen, 2017; Isolani et al., 2013; Seebeck et al., 2017). Using RNAi we also show that the polarity determinant Smed-notum (notum) is necessary for the homeostatic anterior migration of cells in unwounded animals, but not for cells to form processes or to migrate in response to wounding (Petersen and Reddien, 2011). Observation of migratory behavior and morphology suggested that EMT-related mechanisms control cell migration in planarians. We investigated three planarian orthologs of EMT transcription factors (EMT-TFs), namely snail-1, snail-2 and zeb-1, and found that they were all required for migration.
Our work establishes the conservation of EMT mechanisms controlling cell migration across the breadth of bilaterians and further establishes the use of Schmidtea mediterranea as an effective model system to study the migration of stem cells and their progeny in a regenerative context.
RESULTS
Establishment of an X-ray-shielded irradiation assay
The sensitivity of planarian regenerative properties to high doses of ionizing radiation was established over a century ago (Bardeen and Baetjer, 1904). This was subsequently attributed to the fact that NBs were killed by irradiation (Wolff, 1962). Partially exposing planarians to ionizing radiation, through use of a lead shield, was shown to slow down regenerative ability and suggested the possibility that NBs could move to exposed regions and restore regenerative ability (Dubois, 1949). Recently established methods for tracking cell migration in planarians have revisited shielding or involved transplanting tissue with stem cells into lethally irradiated hosts (Guedelhoefer and Sánchez Alvarado, 2012b; Tasaki et al., 2016). These methods clearly show movement of NBs and their progeny. There is also evidence for the migration of eye progenitors (Lapan and Reddien, 2011) and anterior pole cell progenitors (Oderberg et al., 2017) in regenerating animals. We set out with the goal of adapting the shielding approach to establish a practical assay for studying the molecular control of cell migration.
We designed an approach in which multiple animals can be uniformly irradiated with X-rays, except for a thin strip in a predetermined position along their body axis. This is achieved by placing the animals directly above a 0.8 mm strip of lead (6.1 mm thick) to significantly attenuate the X-rays in the region just above the lead to less than 5% of the dose applied to the rest of the animal (Fig. 1A-C, Fig. S1A-C).
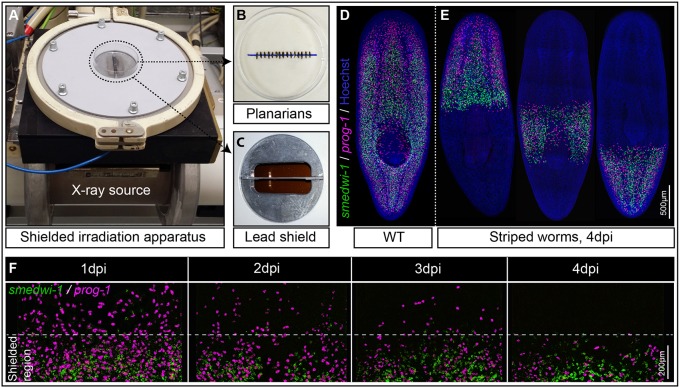
Fig. 1.
The shielded irradiation assay. (A-C) Point source X-ray irradiator (A) passing through a lead shield (C) with aligned Schmidtea mediterranea worms (B) that have been anesthetized in 0.2% chloretone. (D) Wild-type (WT) unirradiated planarians showing distribution of NBs (green) and their early progeny (magenta). (E) Striped planarians at 4 days post irradiation (dpi) showing bands of stem cells (green) and early progeny (magenta) restricted to the irradiation-protected region. (F) Loss of NBs (green) and early progeny (magenta) in the non-shielded region after 1, 2, 3 or 4 dpi (n=10), and maintenance within the shielded region. See also Fig. S1.
Our final version of the apparatus is designed to fit a standard 60 mm Petri dish, with the lead shield lying below the diameter (Fig. 1A, Fig. S1A,B). Anesthetized planarians are aligned across the diameter in preparation for X-ray exposure (Fig. 1A-C). We could then expose up to 20 ∼2-5 mm long worms simultaneously to a normally lethal 30 Gy X-ray dose with the shielded region receiving <1.5 Gy. This allows for some precision in controlling the position of a surviving band of NBs (Fig. 1D,E).
We performed whole-mount fluorescent in situ hybridization (WFISH) to assay the effectiveness of the shield. With the smedwi-1 NB marker we confirmed that all NBs (smedwi-1+) outside the shielded region disappear by 24 h post irradiation. With the early epidermal lineage marker prog-1 we confirmed that stem cell progeny (prog-1+) outside the shielded region have differentiated by 4 days post irradiation (dpi) as no NBs were present to renew the prog-1+ population (Fig. 1E,F). We observed that cells within the shield have a density equivalent to that of wild-type animals not subjected to shielded irradiation, suggesting that the shield is effective at protecting cells (Fig. 1E,F; see Fig. 2D for quantification). There is no cell migration from the shielded region during this time (Fig. 1E,F). These data established that any observation of migrating NBs and progeny should ideally occur after 4 dpi.
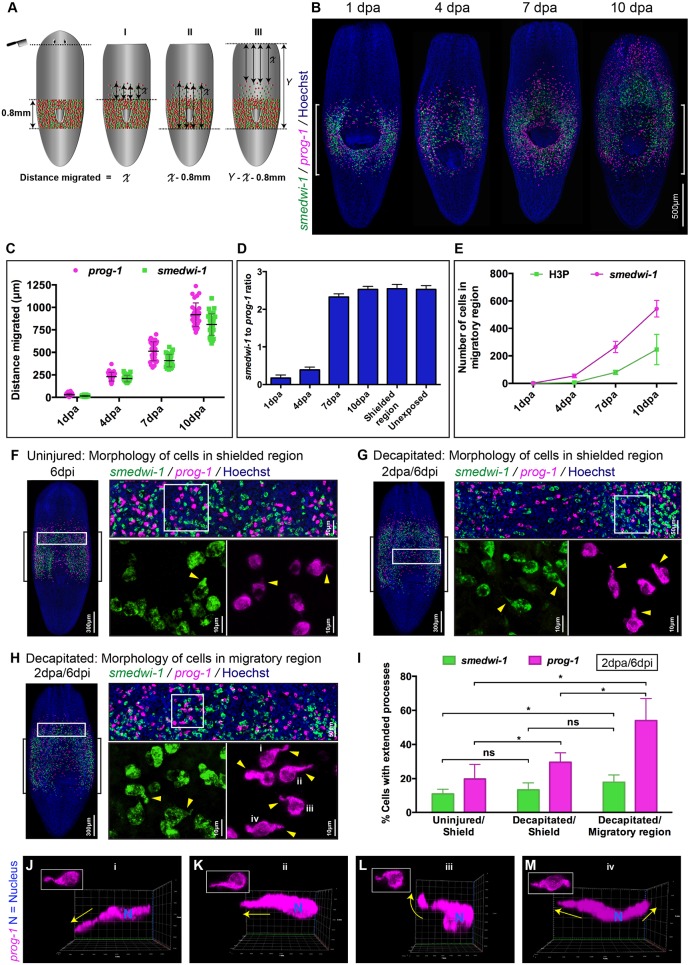
Fig. 2.
Wound-induced cell migration and characteristic extended morphology of migrating stem cells and stem cell progeny. (A) Model demonstrating the position of the wound and three (I, II and III) independent methods for measuring migration distances. (B) Representative WFISH showing NBs (green) and progeny (magenta). Brackets indicate the shielded area. (C) Distances migrated during migration and repopulation of NBs (green) and early progeny (magenta) after shielding across the pharynx at 1, 4, 7 and 10 days post injury (dpa). Each dot represents the average distance migrated by the ten most distal cells in each animal (n=20 per time point). (D) NB to early progeny ratio in the migratory region at 1, 4, 7 and 10 dpa (decapitation) (n=20 per time point). Ratio of cells in the shielded region and in unexposed worms is used as a control. Mean±s.d. (E) Quantification of NBs (magenta) and mitotic cells (green) in the migratory region following decapitation at 1, 4, 7 or 10 days (n=20 per time point). Mean±s.d. H3P, H3ser10p. (F-H) Morphology of cells within the shielded region in an uninjured worm (F), within the shielded region in a decapitated worm (G) and within the migratory region in the decapitated worm (H) shows NBs (green) and early progeny cells (magenta) with and without extended cytoplasmic projections (n=20 in each condition). Brackets indicate the shielded area. Arrowheads indicate examples of extended processes. (I) Quantification of cells with processes shows an increase in the number of NBs (green) and early progeny (magenta) with extended processes within the decapitated/migratory region as well as the decapitated/shielded region compared with the uninjured/shielded region (n=20 per condition). Mean±s.d. Student's t-test, *P<0.05; ns, not significant. (J-M) Early progeny cells (magenta) within the migratory region in decapitated worms (i-iv in H, boxed) show extended processes in various directions relative to the wound. Arrows indicate the direction of extended processes. Relative position of wound to cells is to the top. J-M and i-iv are the same cells: i-iv, top views; J-M, side views. See also Fig. S2.
In summary, our X-ray-shielded assay allows convenient and precise observation of NB and progeny behavior over time post-irradiation, and in animals of a size and number suitable for functional studies.
Features of planarian cell migration after wounding
We next employed the assay system to describe the movement of NBs and progeny. Cycling NBs in S. mediterranea are normally present throughout the body but absent from the region in front of the photoreceptors and the centrally positioned pharynx and are not detectable within early regenerative blastema (Fig. S2A,B). In normal animals: (1) NBs do not normally migrate far, as they are located relatively close to where they are required, except for the anterior region and the pharynx; (2) early in regeneration, progeny migrate to establish the blastema tissue before NBs; and (3) for the pharynx and the most anterior tissue, homeostasis is achieved by migration of postmitotic progeny, and not NBs. This led us to speculate that stem cell progeny have migratory properties distinct from those of NBs.
We shielded animals over the pharynx (Fig. 2A,B) and made anterior wounds by decapitation just under the photoreceptors, at 4 dpi (Fig. 1F). Using WFISH over a 10 day timecourse after wounding, we observed that NBs and progeny migrated anteriorly towards the wound, but not in a posterior direction (Fig. 2B). We used the lack of posterior migration in this experimental design to facilitate accurate measurements of individual cell migration distances over time (Fig. 2A). Quantifying smedwi-1+ NBs, prog-1+ progeny and mitotic cells in the migratory region allowed us to develop a detailed overview of the migration process (Fig. 2B-E).
While the most advanced smedwi-1+ cells can match the extent of migration of the most advanced prog-1+ cells, we found that many more prog-1+ cells enter the migratory region than smedwi-1+ cells over the first 4 days post amputation (dpa) (Fig. 2B-D). By 7 dpa, although the density of NBs and of progeny in the migratory region just anterior to the shield are still lower than in unexposed animals, homeostatic ratios of NBs and progeny are restored (Fig. 2D). We observed cells in M phase within the field of migrating cells, the numbers of which increased in proportion with the numbers of migrating smedwi-1+ NBs over time (Fig. S2C,D, Fig. 2E). This pattern of proliferation in the migratory region is consistent with the homeostatic ratio of NBs and progeny being restored by increased NB division and further migration from the shielded region (Fig. 2C-E). From this we deduce that increases in the number of NBs and progeny outside of the shielded region are fueled initially by migration, but then by both migration and proliferation of NBs.
prog-1+ progeny that reach the wound site at 10 dpa can only have arisen from division of NBs at 6 dpa or later, as 4 days is the maximum time before they differentiate further and stop expressing the prog-1 marker (Eisenhoffer et al., 2008). Given the NB migration speeds we observe (Fig. 2C), these prog-1+ cells must be the progeny of NBs that have themselves already migrated beyond the shielded region. Taken together, these data suggest that migrating smedwi-1+ NBs undergo cell divisions that increase both the number of smedwi-1+ cells and prog-1+ cells, importantly providing a source of stem cell progeny that do not derive from the shielded region. These dynamics are similar to those of regeneration, where stem cell progeny form the initial regeneration blastema and NBs follow later.
We performed single-poke wounds at the midline or notches confined to one side of the animal (Fig. S2E,F) to see how precise the homing of migrating cells to wounds could be. Even small injuries in relatively close proximity promoted distinct migratory responses around each wound site, indicating that migrating cells home with precision (Fig. S2E,F). Despite the absence of NBs and progeny in the anterior tissue field, the stem cell progeny only migrate and collect around the wound, and do not sense the absence of NBs and progeny elsewhere (Fig. S2E,F). We also observed as a general feature of migration towards the wound site that dorsal prog-1+ cells migrate more rapidly than ventral cells (Fig. S2G,H), and that dorsal smedwi-1+ cells migrate centrally whereas ventral stem cells migrate across the width of animals (Fig. S2I).
Migrating planarian cells have extended cell processes
We next investigated migrating cells in more detail to understand how they move. We imaged migrating cells compared with static cells in the shielded region. We observed a significantly higher frequency of NBs and progeny with extended cell processes in migratory regions of injured animals than in the shielded region (Fig. 2F-I; see Fig. S2J,K for different cell morphologies). We did not observe connections or alignment between cells, and cells appeared to migrate independently rather than by any mechanism involving collective cell movement (Friedl and Alexander, 2011; Friedl et al., 2012). This suggests that migration involves cellular mechanisms similar to those used during EMT (Kalluri and Weinberg, 2009; Lamouille et al., 2014). Although net movement is towards the wound site, cell processes can extend in all directions, not just towards the wound (Fig. 2J-M). Taken together, these data indicate that NBs and progeny respond to wounds with directional precision and by extending cell processes.
The order and extent of cell migration recapitulate the cell lineage
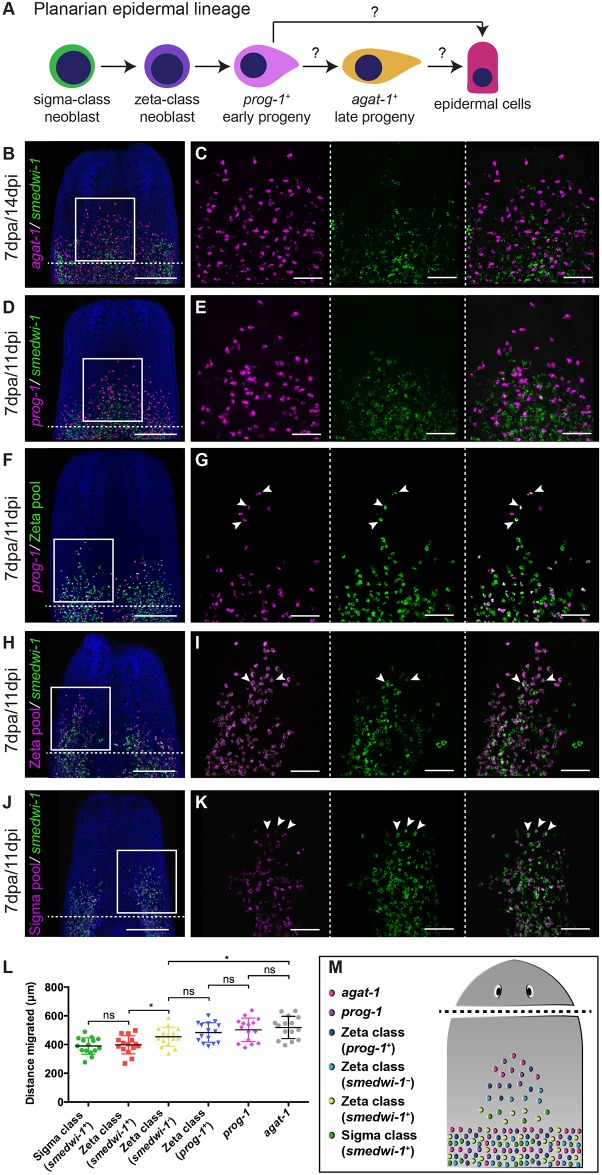
Details of planarian NBs and the epidermal progeny lineage allows detailed tracking of differentiation fates (Eisenhoffer et al., 2008; Tu et al., 2015; van Wolfswinkel et al., 2014). We used the cell-type markers from these studies to label different populations of NBs and progeny (Fig. 3A). We investigated expression of these markers in migrating cells using overlapping double WFISH experiments, allowing us to observe the relationship between migration and differentiation (Fig. 3B-M). We observed that the greatest increase in migration distances between cells occurred upon exit from the smedwi-1+ state. We saw a significant difference in the extent of migration between smedwi-1+ zeta+ NBs and smedwi-1− zeta+ progeny (Fig. 3H,I,L). These data suggest that very early postmitotic progeny might have the highest migratory potential in the epidermal cell lineage. Again, we note that this pattern of differentiation and migration recapitulates early regeneration.
Fig. 3.
Epidermal lineage cell migration. (A) Current model of planarian epidermal lineage differentiation. Question marks indicate that currently there is no direct evidence demonstrating direct transitions from one cell type to another. (B-K) WFISH showing migration of the epidermal lineage at 7 dpa. (B,C) agat-1+ cells (magenta) and smedwi-1+ cells (green). (D,E) prog-1+ cells (magenta) and smedwi-1+ cells. (F,G) prog-1+ cells (magenta), zeta class cells (green) and prog-1+/zeta class double-positive cells (white). (H,I) smedwi-1− zeta class cells (magenta) and smedwi-1+ zeta stem cells (white) and smedwi-1+ cells (green). (J,K) smedwi-1+ sigma stem cells (white) and smedwi-1+ cells (green) migrate. Arrowheads indicate examples of double-positive cells. The shielded region is beneath the dotted line. Scale bars: 300 μm in B,D,F,H,J; 100 μm for C,E,G,I,K. (L) Distance traveled by the ten most distal cells in each population (smedwi-1+ sigma class stem cells, smedwi-1+ zeta class stem cells, smedwi-1− zeta class cells, prog-1+/zeta class double-positive cells, prog-1+ cells and agat-1+ cells) in decapitated worms at 7 dpa. n=15 per condition. Student's t-test, *P<0.05. (M) Summary of migration and differentiation data after wounding.
A matrix metalloprotease and beta-integrin are both required for cell migration to wound sites
We next tested whether we could study gene function in the context of migration. We considered candidate genes that might be required for cell migration based on previous work, and selected mmpa and β1-integrin as strong candidates for proof-of-principle experiments.
Previous research had attempted to implicate mmpa, one of four matrix metalloprotease enzymes identifiable in the S. mediterranea genome, as having a role in cell migration (Isolani et al., 2013). We first performed RNAi in the context of normal regeneration and observed that mmpa(RNAi) animals showed regeneration defects as previously described, with failure to correctly regenerate anterior or posterior tissues (Fig. S3A). RNAi in the context of our assay revealed that anterior tissues regressed and that animals failed to regenerate (Fig. S3B). We used WFISH to monitor the movement of smedwi-1+ NBs and prog-1+ stem cell progeny after mmpa RNAi, and observed almost no migration of cells compared with control gfp(RNAi) worms (Fig. 4A,D,M; see also Fig. S3M,N). Additionally, we examined the morphology of NBs and progeny and observed reduced numbers of cells with extended processes compared with migrating cells in the gfp(RNAi) control animals (Fig. 4B,C,E,F,N). These results confirm that this matrix metalloprotease enzyme is required to facilitate cell migration in planarians. We found that mmpa is only expressed at relatively low levels in stem cells and in stem cell progeny, with the bulk of its expression being in differentiated radiation-insensitive cells (Fig. S3C-E) (Kao et al., 2017 preprint). We also did not detect mmpa expression in migrating cells (Fig. S3F,G), suggesting also that it might be produced by differentiated cells and might be required in the extracellular matrix to allow cell extensions to form and permit migration.
Fig. 4.
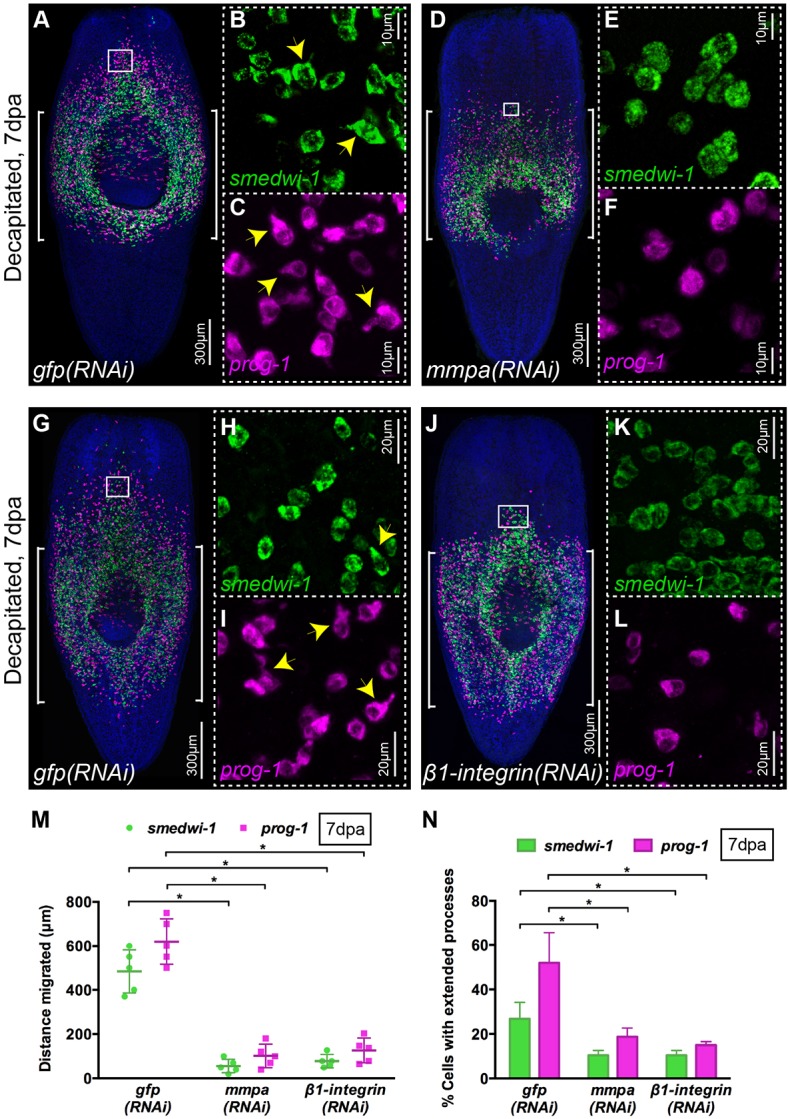
mmpa and β1-integrin are essential for migration and cell extension formation. (A-L) WFISH shows migration of NBs (green) and early progeny (magenta) at 7 dpa in control gfp(RNAi) (A-C,G-I) and lack of migration in mmpa(RNAi) (D-F) and β1-integrin(RNAi) (J-L) animals. Insets show the presence of NBs and early progeny with extended cytoplasmic projections in the migratory region of gfp(RNAi) worms (B,C,H,I, arrowheads) that are almost absent in mmpa(RNAi) (E,F) and β1-integrin(RNAi) (K,L) worms (n=5). Brackets indicate the shielded area. (M) Distance migrated by NBs (green) and early progeny (magenta) at 7 dpa in mmpa(RNAi) and β1-integrin(RNAi) animals compared with control gfp(RNAi) worms (n=5). Each dot represents the average distance migrated by the ten most distal cells from each animal. Mean±s.d. Student's t-test, *P<0.05. (N) Quantification of NBs (green) and early progeny (magenta) with extended processes in mmpa(RNAi), β1-integrin(RNAi) and control gfp(RNAi) animals at 7 dpa (n=5). Mean±s.d. Student's t-test, *P<0.05. See also Fig. S3.
We next investigated whether β1-integrin also has a conserved role in allowing cell migration. Integrins have conserved roles in orchestrating cell migration, providing a connection between physical actions of the actin cytoskeleton and signaling mechanisms instructing migratory activity (Mogilner and Keren, 2009; Vicente-Manzanares et al., 2009). The recently published regenerative phenotypes for planarian β1-integrin suggested to us that the cellular disorganization observed in these studies could be due to failures in migratory activity (Bonar and Petersen, 2017; Seebeck et al., 2017). We observed that β1-integrin transcript is expressed in nearly all smedwi-1+ NBs and in about a third of migrating progeny in the migration region of wild-type animals in our assay (Fig. S3H-L). We performed β1-integrin RNAi and found that cell migration was greatly impaired compared with gfp(RNAi) controls (Fig. 4G-N, Fig. S3M,N). Cell process formation in NBs and progeny was also disrupted (Fig. 4K,L,N). These data confirm a conserved role for β1-integrin in NB and progeny cell migration in planarians and, along with the mmpa(RNAi) phenotype, confirm that our assay can be combined with RNAi-based loss-of-function studies.
Anterior migration of stem cells and stem cell progeny in the absence of wounding
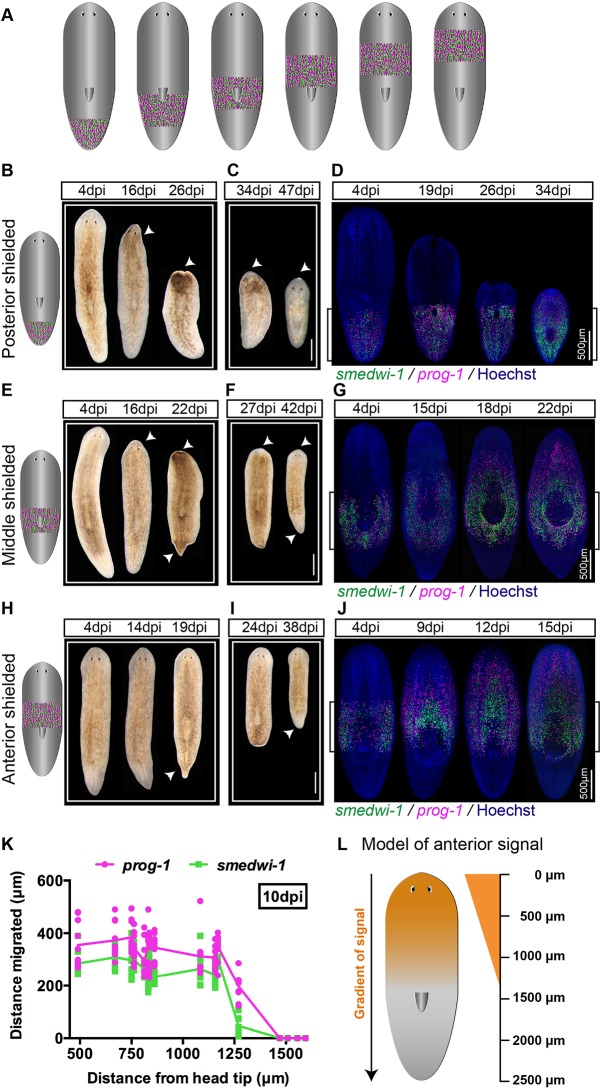
While wounding will trigger migration and, in fact, precise homing of NBs and progeny (Fig. S2E,F), we wanted to observe what happens in the absence of wounding. We shielded animals of equal size at different positions along the anteroposterior (AP) axis and irradiated them (Fig. 5A). When the shield was placed in the posterior region of worms we observed tissue death and regression from the anterior towards the shield (Fig. 5B). Subsequently, we observed blastema formation and normal regeneration that took up to 50 dpi (Fig. 5C). Using WFISH we were able to observe that NBs and progeny did not migrate until the regressing anterior tissue boundary was relatively close to the anterior of the shielded region (Fig. 5D). When animals were shielded in mid-body regions with the top of the shield level with the most anterior region of the pharynx, we observed regression of the anterior and posterior tissue (Fig. 5E). We subsequently observed blastema formation and regeneration that took up to 45 dpi (Fig. 5F). WFISH revealed that in these animals NBs and progeny migrate towards the anterior (Fig. 5G) and later towards the posterior once regressing tissue is close to the shielded regions. These data suggested that remaining NBs maintain local tissue homeostasis, and remain stationary within the shielded region until regressing tissue boundaries are close enough to trigger migration.
Fig. 5.
NBs and their progeny migrate anteriorly in the absence of injury. (A) Strategy of shielding worms at various places along the AP axis. (B,C,E,F,H,I) Bright-field images of worms shielded at three different places, namely posterior (B,C), middle (E,F) and anterior (H,I), showing regression and recovery over time (n=20 per time point). Arrowheads indicate regressed (B,E,H) and regenerated (C,F,I) regions. Scale bars: 500 μm. (D,G,J) WFISH showing no migration of NBs (green) and early progeny (magenta) in posteriorly shielded worms until the anterior tissue regresses close enough to the shielded region (D). By contrast, NBs and early progeny migrate after failure in anterior tissue integrity in middle shielded worms (G). In anteriorly shielded worms, cells migrate without a visible loss of anterior integrity (J). n=20 per time point. Brackets indicate the shielded area. (K) Distance migrated by NBs (green) and early progeny (magenta) in worms shielded at different places along the AP axis in the absence of anterior wound. Each dot represents the average distance migrated by the ten most distal cells in each animal (n=6). (L) Model showing a gradient of signal (orange) from head tip to up to ∼1.3 mm towards posterior in ∼2.5 mm-long worms.
By contrast, when the posterior of the shield was positioned level with the anterior of the pharynx the worms displayed posterior regression but not anterior regression (Fig. 5H,I). The heads of these animals never regressed, whereas tails regressed and then regenerated over several weeks (Fig. 5I). WFISH subsequently revealed that NBs and progeny could migrate towards the anterior in the absence of wounding or loss of tissue homeostasis (Fig. 5J). These results suggest that leaving a stripe of more anteriorly positioned cells is somehow sufficient to trigger anterior migration and maintain anterior tissue homeostasis.
To investigate this phenomenon further we irradiated animals with shields positioned at different points along the AP axis and performed WFISH to observe NB and stem cell progeny migration at different time points. We were able to observe migration of cells towards the anterior in the absence of wounding as long as the shield was within a set distance of the anterior tip (≤1.2 mm in animals of 2.5 mm in length; Fig. 5K,L).
These data add to previous work that described that migration only occurs after wounding or when tissue homeostasis fails and tissue regression reaches remaining stem cells (Guedelhoefer and Sánchez Alvarado, 2012b). We find that stem cells and stem cell progeny in the pre-pharyngeal anterior region can migrate to the anterior in the absence of wounding. This migratory activity suggests the presence of anterior signals that can call NBs and progeny into the brain and anterior structures over a restricted range. These observations suggest that an anterior signal exists for encouraging cell migration in intact animals that acts at least over the brain region (Fig. 5L).
notum is required for anterior cell migration in intact animals, but not after wounding
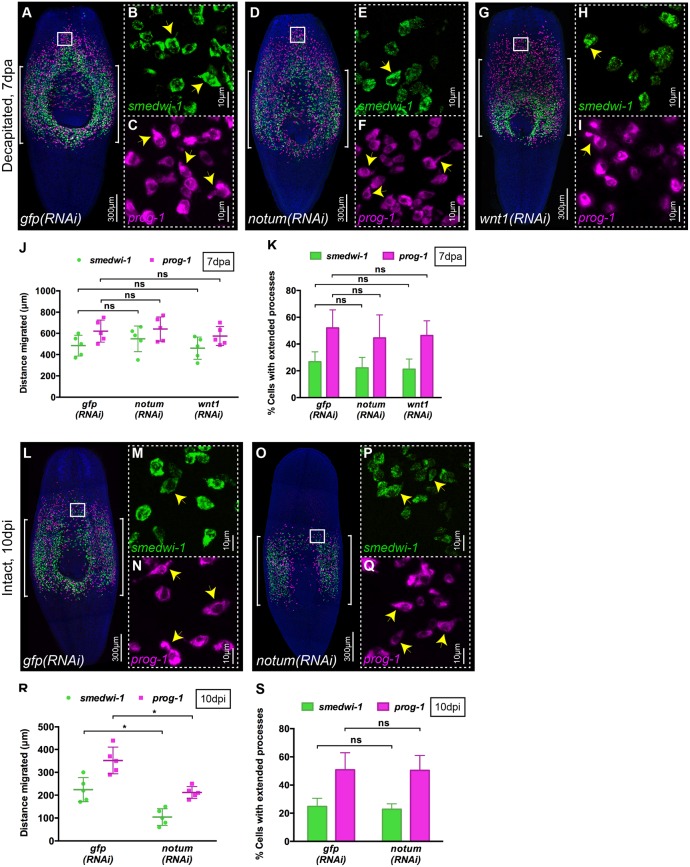
There are a large number of conserved candidate signaling pathways that could be involved in promoting cell migration. We chose to study two candidates, namely Smed-wnt1 (wnt1) and notum, which are both upregulated at anterior wounds in planarians (Petersen and Reddien, 2009). In addition, notum is also expressed at the anterior medial tip of intact animals (Petersen and Reddien, 2011) and is therefore also a candidate for controlling anterior migration in the absence of wounding.
It has previously been shown that wounding results in wnt1 in muscle cells at the wound site (Witchley et al., 2013). Given that Wnt signaling has a role in regulating cell migration elsewhere (Mayor and Theveneau, 2014), wound-induced Wnt1 expression could be required for cell migration to the wound in planarians. We performed wnt1 RNAi and observed full penetrance of the tailless phenotype previously described for these animals (Fig. S4A) (Petersen and Reddien, 2009). After shielded irradiation we also observed that wnt1(RNAi) animals were able to regenerate anterior structures completely (Fig. S4B). Using WFISH we observed no effects on either NB or progeny migration after wounding, and both cell populations formed cell extensions to a similar extent to control gfp(RNAi) animals, suggesting that Wnt1 has no essential role in the migration process (Fig. 6A-C,G-K).
Fig. 6.
notum is required for migration in the absence of wounding. (A-I) WFISH showing migration of NBs (green) and early progeny (magenta) at 7 dpa in (A-C) control gfp(RNAi), (D-F) notum(RNAi) and (G-I) wnt1(RNAi) animals. Brackets indicate the shielded area. Insets show that NBs and early progeny in the migratory region from (B,C) gfp(RNAi), (E,F) notum(RNAi) and (H,I) wnt1(RNAi) animals are able to form extended processes (arrowheads). (J) Distances migrated by NBs (green) and early progeny (magenta) at 7 dpa in gfp(RNAi), notum(RNAi) and wnt1(RNAi) animals are equal (n=5). Each dot represents the average distance migrated by the ten most distal cells from each animal. Mean±s.d. Student's t-test. (K) Number of NBs and early progeny with extended processes in notum(RNAi), wnt1(RNAi) and gfp(RNAi) animals (n=5). Mean±s.d. Student's t-test. (L-Q) WFISH showing migration of NBs (green) and early progeny (magenta) at 10 dpi in intact (O-Q) notum(RNAi) animals compared with intact (L-N) gfp(RNAi) animals. Brackets indicate the shielded area. (M,N,P,Q) The morphology of NBs and early progeny in the migratory region. Arrowheads indicate examples of cells with extended processes. (R) Distance migrated by NBs (green) and early progeny (magenta) at 10 dpi in notum(RNAi) animals compared with gfp(RNAi) animals (n=5). Each dot represents the average distance migrated by the ten most distal cells from each animal. Mean±s.d. Student's t-test, *P<0.05. (S) Quantification of extended processes of NBs and early progeny in notum(RNAi) compared with gfp(RNAi) animals (n=5). Mean±s.d. Student's t-test. See also Fig. S4.
Smed-notum is also expressed in muscle cells on wounding, but only at anterior-facing wounds where it is required to ensure the proper specification of anterior fates, probably by repressing Wnt signaling (Petersen and Reddien, 2011). Additionally, notum has a homeostatic expression pattern at the anterior margin and has previously been shown to promote the homeostasis and correct size of the brain in combination with the activity of a wnt11-6 gene expressed in posterior brain regions (Hill and Petersen, 2015). On this basis, notum represents a candidate for both wound-induced migration and migration of cells towards anterior regions in uninjured animals. We performed notum RNAi and observed full penetrance of the double-tailed phenotype previously described for these animals in a standard regeneration assay (Fig. S4A) (Petersen and Reddien, 2011). After shielded irradiation and wounding we observed that although notum(RNAi) animals failed to regenerate normal anterior structures compared with controls, there were no differences in the migration of cells or migrating cell morphology compared with control gfp(RNAi) animals as assessed by WFISH (Fig. 6A-F,J,K). However, when using an anteriorly positioned shield, which led to anterior migration of cells in control intact unwounded gfp(RNAi) animals, there was a significant reduction in anterior migration in notum(RNAi) animals (Fig. 6L-S, Fig. S4C-E). This reduction in migration was not accompanied by a difference in the number of cells with cell extensions (Fig. 6S), suggesting that notum might act by contributing a directional signal rather than by controlling cellular migratory behavior of anteriorly positioned NBs and progeny. These data suggest that notum is not essential for wound-induced cell migration but is required for the homeostatic anterior migration in intact animals that we uncovered in this work. It seems likely that an earlier description of a notum/wnt11-6 regulatory circuit involved in homeostatic regulation of brain size may also have a broader role in the homeostatic maintenance of anterior regions that do not normally contain NBs (Hill and Petersen, 2015).
Conserved EMT-TFs regulate cell migration in planarians
We next considered if we could establish a broad comparative context for the control of cell migration in planarians and migration in other systems, including mammals. Our observation that NBs and progeny appear to migrate individually, using cell extensions to interact with the extracellular matrix and non-migratory neighboring differentiated cells, suggested that they might use similar mechanisms to those attributed to EMT (Thiery and Sleeman, 2006). EMT in different contexts requires the activity of a restricted group of transcription factors (EMT-TFs) (Batlle et al., 2000; Cano et al., 2000; Colvin Wanshura et al., 2011; Lamouille et al., 2014). We identified two members of the snail transcription factor family (snail-1 and snail-2) of EMT-TFs and an ortholog of the zinc-finger E-box-binding homeobox 1 (zeb-1) EMT-TF.
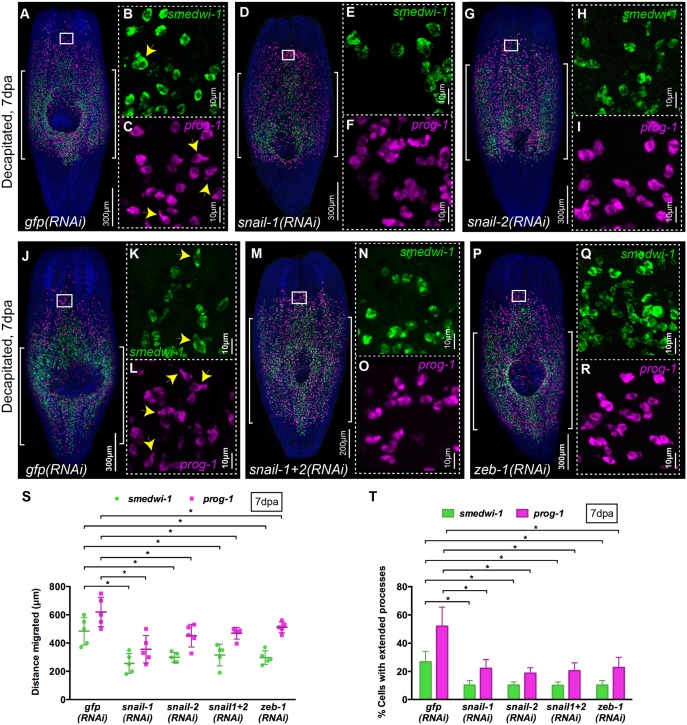
We tested whether any of these conserved EMT-TF genes were involved in cell migration in planarians. Previously, the snail family transcription factor snail-2 was reported as being expressed in collagen-positive muscle cells, in a small percentage of G2/M NBs before wounding and in ∼35% of G2/M NBs after wounding (Scimone et al., 2014). To our knowledge no phenotype has been reported for a snail family gene in planarians and when we assessed snail-1(RNAi) or snail-2(RNAi) animals in a standard regenerative assay they all regenerated normally (Fig. S5A).
However, in the context of our migration assay, snail-1(RNAi) or snail-2(RNAi) animals failed to regenerate after wounding suggesting a defect in cell migration (Fig. S5B). In WFISH experiments we observed a clear decrease in the extent of cell migration compared with gfp(RNAi) control animals (Fig. 7A,D,G,S, Fig. S5M,N). This defect in migration of both NBs and progeny was accompanied by a decrease in the number of cells with cell extensions (Fig. 7B,C,E,F,H,I,T). Performing double snail-1/2 RNAi did not lead to a stronger phenotype (Fig. 7M,N,O,S).
Fig. 7.
Snail family genes control migration of stem cells and their progeny. (A-R) WFISH shows migration of NBs (green) and early progeny (magenta) at 7 dpa in (A-C,J-L) gfp(RNAi), (D-F) snail-1(RNAi), (G-I) snail-2(RNAi), (M-O) snail-1+2(RNAi) and (P-R) zeb-1(RNAi) animals. Brackets indicate the shielded area. Insets show the presence of NBs and early progeny with extended cytoplasmic projections in migratory regions (arrowheads). n=5 per condition. (S) Measurements of distance migrated by NBs (green) and early progeny (magenta) at 7 dpa in snail-1(RNAi), snail-2(RNAi), snail-1+2(RNAi) and zeb-1(RNAi) animals compared with control gfp(RNAi) (n=5). Each dot represents the average distance migrated by the ten most distal cells from each animal. Mean±s.d. Student's t-test, *P<0.05. (T) Quantification of NBs and early progeny with extended processes in snail-1(RNAi), snail-2(RNAi), snail-1+2(RNAi) and zeb-1(RNAi) compared with control gfp(RNAi) animals at 7 dpa (n=5). Mean±s.d. Student's t-test, *P<0.05. See also Figs S5 and S6.
We found that both snail-1 and snail-2 were expressed in smedwi-1+ NBs in the migratory region after wounding (87% and 93%, respectively) (Fig. S5F,K). This expression pattern suggests that these EMT-TFs have a cell-autonomous role in controlling NB migration. Taken together, our data suggest that cell-autonomous migratory mechanisms are affected in snail-1(RNAi) and snail-2(RNAi) animals and establish that snail EMT-TFs in planarians have a conserved role in regulating cell migration in response to wound signals.
We also investigated the role of zeb-1 and, similar to our observations for snail genes, no defects were observed in zeb-1(RNAi) animals in a normal regeneration assay (Fig. S6A). zeb-1 RNAi also led to a failure to regenerate correctly in our migration assay (Fig. S6B). WFISH experiments revealed clear defects in cell migration and cell process formation, very similar to those observed for both snail TFs (Fig. 7P-T, Fig. S6H,I). We could only detect zeb-1 using WISH in relatively few migrating smedwi-1+ NBs (8%, Fig. S6C-F). This is in agreement with the relatively low level of zeb-1 expression observed across planarian RNAseq data (Fig. S6C-F) (Kao et al., 2017 preprint).
Taken together, our data establish that conserved EMT-TFs are required for NB and progeny migration in planarians, establishing conservation of this regulatory circuit across bilaterians.
DISCUSSION
An X-ray-shielded assay allows precise observation of cell migration and application of functional genomic approaches
During homeostasis as well as standard regeneration experiments, NBs and stem cell progeny are always close to where they are required. Nonetheless, NBs and progeny must still move into the correct functional positions in tissues and organs. Precise monitoring of this process is difficult as the migratory distances involved are short, and so confidently inferring changes in migratory behavior as opposed to changes in differentiation, for example, is not possible. Our X-ray-shielded assay creates a ‘blank canvas’ into which migrating stem cells and stem cell progeny move and we can accurately assign relationships between migration, differentiation and proliferation of groups of these cells over time. NBs and progeny are capable of restoring full tissue and organ function by migrating from the small shielded region. The innovations we have made here allow for a thinner shield, smaller worms to be irradiated and technical consistency over relatively large numbers of worms.
A detailed description of migratory behavior in a regenerative context
We have revealed a number of detailed properties of cell migration in planarians that can be used to help unpick the underlying control mechanisms. We have shown that migration occurs in response to wounding or damaged tissue as previously described (Guedelhoefer and Sánchez Alvarado, 2012b), but we also find that migration can occur without wounding or failure in tissue homeostasis in the case of anteriorly positioned stem cells and stem cell progeny. This observation tallies with the absence of NBs in anterior regions and the brain in intact animals, which suggests that a mechanism for encouraging homeostatic cell migration must exist. Migrating cells home precisely to wounds without recognizing other tissue regions that lack NBs and progeny. In regions containing moving cells we can see a clear increase in the number of cells with pronounced cell extensions. Migrating cells are unconnected to other migrating cells. Together, these observations give an EMT-like characteristic to planarian cell migration, as opposed to other mechanisms involving collective cell migration. Overall, we establish a set of basic phenotypic criteria that can be used to study the genetic and molecular control of cell migration.
The relationship between stem cell migration, proliferation and differentiation
Stem cell migration during normal healthy tissue homeostasis must be intricately linked to cell division, differentiation and the integration of new cells. Highly regenerative animal models provide an opportunity to study these processes. Thus, perhaps the most important observations facilitated by our assay are those concerning the relationships between migration, proliferation and differentiation.
Progeny migrate in large numbers in an initial response to wounding, and proliferating NBs accompany them in smaller numbers. In response to both wounding and homeostatic signals we observe that NBs can divide as they migrate, and that the new progeny differentiate while they migrate. Our observations broadly recapitulate cell behavior observed during regeneration, in which progeny migrate to form the regeneration blastema where they complete differentiation, and cycling cells follow later. We observe significant differences in migration between smedwi-1+ cells and zeta class/smedwi-1− cells, which we interpret as suggesting that nascent progeny migrate ahead of cycling NBs as they do in blastema formation. NBs may migrate more slowly on average as they stop to divide, or because they require the presence of progeny at a certain density before they can be healthily maintained in a repopulating tissue region, or perhaps simply because they are slower due to having smaller cell extensions. Based on these observations our assay will provide an alternate method of assessing cell lineage relationships with WFISH approaches and, when combined with RNAi, it allows the molecular processes controlling the interplay between migration, proliferation and differentiation to be studied.
As we have established that migration homes precisely to wound sites we can also now study whether differentiation programs show specificity to the type of wound depending on which cell types are damaged. Recently, it was shown that the production of photoreceptor precursors and cells is independent of whether eyes are present or not (LoCascio et al., 2017), suggesting that for some organs the differentiation programs are independent of the state of the target tissue. Combining our assay with experimental paradigms that damage one or a few defined cell types will help to answer how demands for new cells are regulated and how stem cells and their progeny sense and adjust to these demands.
A role for notum in homeostatic migration of stem cells and stem cell progeny
Two genes that have already been shown to have complementary roles in controlling the polarity of planarian regeneration, wnt1 and notum, are both known to be wound induced (Petersen and Reddien, 2011) and represented good candidates for potential roles in cell migration after wounding. In addition, homeostatic expression of notum was recently shown to be involved in regulating planarian brain size in combination with wnt11-6, specifically ensuring that sufficient neural precursors are produced to maintain correct brain size (Hill and Petersen, 2015). These observations made notum a candidate for involvement in the homeostatic cell migration that we described in intact animals in anterior regions.
Using RNAi we found no role for either wnt1 or notum in wound-induced migration; however, we found that notum is required for the homeostatic anterior migration. Given the homeostatic expression of notum transcript and the observation that cells migrate homeostatically within a certain distance from the anterior tip of the animal, we propose that a gradient of notum somehow provides directional cues to migrating cells. We note that the formation of cell extensions is not affected by notum RNAi, suggesting that other signals might be responsible for this aspect of migratory behavior, while notum activity provides a directional cue. In planarians, mammals and flies notum has been implicated as a Wnt signaling inhibitor (Kakugawa et al., 2015; Traister et al., 2008; Zhang et al., 2015), so it is possible that inhibition of local homeostatic levels of Wnt signaling, specifically of anteriorly expressed planarian Wnts (wnt11-6 and wnt5), might then allow homeostatic migration. Future work with our assay will aim to understand the mechanism by which notum facilitates homeostatic migration and wound-induced migration.
Conservation of EMT-TF function and the potential to study processes relevant to tumor invasion
The fact that cells appear to migrate individually and that in migratory regions increased numbers of cells have extended cell processes suggested that molecular mechanisms associated with EMT might regulate migration. In order to begin to test this possibility, we investigated the function of two planarian snail family transcription factors and a planarian ortholog of zeb-1, as these are conserved positive regulators of cell migration during EMT, required to downregulate the expression of genes that encode proteins that maintain cell-cell contacts, such as E-cadherin (Thiery and Sleeman, 2006). Enhanced Snail gene expression has been reported in several cancer types, including ovarian carcinoma (Davidson et al., 2012), breast tumors (Blanco et al., 2002; Elloul et al., 2005), gastric cancers (Peng et al., 2014; Rosivatz et al., 2002), hepatocellular carcinomas (Miyoshi et al., 2005; Sugimachi et al., 2003), colon cancers (Pálmer et al., 2004) and synovial sarcomas (Saito et al., 2004). Overexpression or downregulation of Snail has been shown to modulate invasiveness and metastasis in in vitro cancer cell culture studies (Adhikary et al., 2014; Belgiovine et al., 2016; Fan et al., 2012; Horvay et al., 2015; Sharili et al., 2013; Smith et al., 2014; Villarejo et al., 2015). Similarly, zeb-1 overactivity has also been implicated in tumorigenesis. These reports clearly suggest that EMT-TFs are key players in cancer invasion and metastasis.
Within the context of our assay, RNAi of all three of these genes led to failure in cell migration and we were able to clearly observe a decrease in cells showing the extended cell processes that are indicative of migratory morphology. Our data confirm the role of EMT-TFs in controlling migration in the context of our assay and suggest that we can use this as a basis for studying EMT-related processes in planarians. By combining functional approaches with expression screens starting with planarian homologs of EMT-related transcription factor regulators and known upstream EMT regulatory signals, we will be able to discover more about EMT in the context of tissue homeostasis, regeneration and adult stem cell activity.
MATERIALS AND METHODS
Planarian culture
A Schmidtea mediterranea asexual strain was cultured and maintained in 0.5% Instant Ocean water in the dark at 20°C. Animals were starved for at least 7 days before experiments.
X-ray irradiation and design of shield
Irradiations were performed using a Comet MXR-321 X-ray set operated at 225 kVp and 17 mA with a 0.5 mm aluminium filter. The X-ray field is collimated to 40×20 mm with a 6.1 mm thick lead disc positioned centrally, directly above the X-ray tube focal spot and supported within an aluminium frame. The removable central shielded area is achieved using a 0.8 mm wide, 6.1 mm thick lead strip spanning the long axis of the collimated field; this sits slightly proud of the main lead collimator so that it is in contact with the base of the Petri dish. When in position, the worms are irradiated at a dose rate of 23 Gy/min, reducing to ∼1 Gy/min underneath the shielded region. The variation in dose distribution across the strip is shown in Fig. S1C. The circular hole in the top aluminium plate corresponds to the outside diameter of the Petri dish and enables dishes to be positioned quickly and reproducibly. Thin strips of materials such as tungsten or tantalum could be used to replace the lead strip to achieve thinner shielded regions if required.
Dosimetry
Dose rate measurements and spatial characterization of the treatment field were performed using Gafchromic EBT3 film (International Specialty Products, Wayne, NJ, USA) placed in the base of an empty 60 mm Petri dish. Twenty-four hours following exposure the EBT3 film was scanned in transmission mode at 48 bit RGB (16 bits per color) with 300 dots per inch resolution using a flatbed scanner (Epson Expression 10000XL). A template was used to position the film within the scanner and the scanning direction was kept constant with respect to the film orientation, as recommended in the manufacturer's guidelines. The dose was calculated using the optical density of the red channel and corrected using the optical density of the blue channel in order to compensate for small non-uniformities in the film, which cause false apparent variations in dose (as described by the manufacturer; http://www.veritastk.co.jp/attached/2062/GAFCHROMICEBT2TechnicalBrief-Rev1.pdf). The batch of EBT3 film was calibrated following the recommendations of the report of AAPM Task Group 61 (Ma et al., 2001).
Shielded irradiation
Up to 20 size-matched planarians (3-4 mm) were anesthetized in ice-cold 0.2% chloretone and aligned on a 60 mm Petri dish (Guedelhoefer and Sánchez Alvarado, 2012a). The anterior tip of each worm was perfectly aligned in order to keep the absolute migratory distance (distance between tip of the head and shielded region) fixed. The Petri dish was pre-marked with a line at the bottom denoting the placement and dimensions (length and thickness) of the shield strip. Excess liquid was removed to minimize movement of worms during irradiation. The Petri dish containing worms was then placed on to the shield of a bottom-source X-ray irradiator. Care is taken to perfectly match the position of the shield strip and the line marked on the Petri dish to ascertain the exact region of the worm to be shielded. 30 Gy X-ray (225 kV for 1 min 18 s) was used for irradiation. Planarians were then immediately washed with 0.5% Instant Ocean water and transferred into fresh 0.5% Instant Ocean water and incubated in the dark at 20°C.
WFISH, immunostaining and imaging
Whole-mount fluorescent in situ hybridization (WFISH) was performed as described previously (Currie et al., 2016; King and Newmark, 2013). H3ser10p rabbit monoclonal antibody (Millipore, 04-817; 1:100) was used for immunostaining (Felix and Aboobaker, 2010). Confocal imaging was performed with Olympus FV1000 and Zeiss 880 Airyscan microscopes. Bright-field images were taken with a Zeiss Discovery V8 using a Canon 1200D camera. Images were processed with Fiji (Schindelin et al., 2012) and Adobe Photoshop. ZEN 2.1 (blue edition) software (Zeiss) was used to construct 3D images of cells. All measurements and quantifications were performed with Fiji and Adobe Photoshop. Significance was determined by unpaired two-tailed Student's t-test.
Measurement of cell migration
Lack of posterior migration of cells allowed us to define the posterior boundary of the shield. This posterior boundary was used as reference for all distance measurements. Usually, cells in the posterior boundary are confined in a straight lateral line. Next, we defined the anterior boundary of the shield at 0.8 mm in front of the posterior boundary (thickness of shield). By knowing both the posterior and anterior boundary of the shielded region we moved on to measure distances migrated by individual cells by following three independent methods (see Fig. 2A). (1) The distance between cells and the anterior boundary gave a direct measurement of the distance migrated by cells. (2) The distance between cells and the posterior boundary of the shield minus 0.8 mm (thickness of shielded region) gave another measurement of the distance migrated by cells. (3) The distance between the posterior boundary of the shield and the anterior boundary of the worm (site of amputation), denoted as Υ; as well as the distance between cells and the site of amputation, denoted as X. Then, Υ−X−0.8 mm will give the distance migrated by cells. A requirement for consistency between the methods reduced the chance of measurement error in the migration distance of a cell.
Gene cloning and RNAi
Planarian genes were cloned into the pPR-T4P plasmid vector, which contains opposable T7 RNA polymerase promoters (kind gift from Jochen Rink, MPI-CBG, Dresden, Germany). The constructs were then used for in vitro double-stranded (ds) RNA synthesis and probe synthesis as described previously (King and Newmark, 2013; Rouhana et al., 2013). The primers used to generate dsRNA template from genes were as follows (5′-3′, forward and reverse): mmpa (GenBank HE577120.1), ATCCTGATTACGGCTCCAA and TTTATTGGGGGTGCAACTGT; β1-integrin (GenBank KU961518.1), GAACTCAACACACAACGCCC and TCTCGACAGGGAACAATGGC; snail-1 (GenBank KY964486), AGCAATCAATCCTAAAGTCG and CGATAGATTCTTCCACGGAG; snail-2 (GenBank KJ934814.1), GTTATCAAGCCAGACCTTCA and GTTTGACTTGTGAATGGGTC; zeb-1 (GenBank, KY992929), TCGTACCCTCATCTACCGCA and GGGTTTCTCTCCGCTGTGAA.
Previously described sequence regions were used for dsRNA synthesis of wnt1 (Petersen and Reddien, 2009) and notum (Petersen and Reddien, 2011). Reported sequences were used for riboprobe synthesis of smedwi-1 (Reddien et al., 2005), prog-1 (Eisenhoffer et al., 2008), agat-1 (Eisenhoffer et al., 2008), zeta pool (van Wolfswinkel et al., 2014) and sigma pool (van Wolfswinkel et al., 2014). To generate probes for mmpa, β1-integrin, snail-1, snail-2 and zeb-1 the same regions of their respective dsRNAs were used. For knockdown of genes, animals were injected with 3×32 nl dsRNA six times over 2 weeks. If worms needed to be used for shielded irradiation after RNAi, a 1 day gap was kept between the last RNAi injection and the shielded irradiation.
Acknowledgements
We thank all members of the A.A.A. lab past and present for discussions and reagent sharing.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: P.A., M.A.H., A.A.A.; Methodology: P.A., E.A., N.K., J.T., M.A.H., A.A.A.; Validation: P.A., J.T., M.A.H., A.A.A.; Formal analysis: P.A., J.T., M.A.H., A.A.A.; Investigation: P.A., E.A., N.K., A.A.A.; Resources: M.A.H., A.A.A.; Writing - original draft: P.A., A.A.A.; Writing - review & editing: J.T., M.A.H., A.A.A.; Supervision: M.A.H., A.A.A.; Project administration: A.A.A.; Funding acquisition: M.A.H., A.A.A.
Funding
The work of P.A., E.A., N.K. and A.A.A. is funded by the Medical Research Council (MRC) (MR/M000133/1), Biotechnology and Biological Sciences Research Council (BBSRC) (BB/K007564/1), the John Fell Fund, University of Oxford, Oxford University Press (OUP), and by a small grant from the Cancer Research UK (CRUK) Oxford Centre. N.K. is funded by a Marie Curie Sklodowska Fellowship funded by the Horizon 2020 Framework Programme. M.A.H. and J.T. acknowledge funding from the MRC Strategic Partnership Funding (MC-PC-12004) for the CRUK/MRC Oxford Institute for Radiation Oncology. Deposited in PMC for release after 6 months.
Data availability
New sequence data for the zeb-1 nucleotide sequence and the snail-1 nucleotide sequence have been deposited in GenBank under accession numbers KY992929 and KY964486, respectively.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.154971.supplemental
References
- Adhikary A., Chakraborty S., Mazumdar M., Ghosh S., Mukherjee S., Manna A., Mohanty S., Nakka K. K., Joshi S., De A. et al. (2014). Inhibition of epithelial to mesenchymal transition by E-cadherin up-regulation via repression of slug transcription and inhibition of E-cadherin degradation: dual role of scaffold/matrix attachment region-binding protein 1 (SMAR1) in breast cancer cells. J. Biol. Chem. 289, 25431-25444. 10.1074/jbc.M113.527267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeen C. R. and Baetjer F. H. (1904). The inhibitive action of the Roentgen rays on regeneration in planarians. J. Exp. Zool. 1, 191-195. 10.1002/jez.1400010107 [DOI] [Google Scholar]
- Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J. and García de Herreros A. (2000). The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2, 84-89. 10.1038/35000034 [DOI] [PubMed] [Google Scholar]
- Belgiovine C., Chiesa G., Chiodi I., Frapolli R., Bonezzi K., Taraboletti G., D'Incalci M. and Mondello C. (2016). Snail levels control the migration mechanism of mesenchymal tumor cells. Oncol. Lett. 12, 767-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M. J., Moreno-Bueno G., Sarrio D., Locascio A., Cano A., Palacios J. and Nieto M. A. (2002). Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene 21, 3241-3246. 10.1038/sj.onc.1205416 [DOI] [PubMed] [Google Scholar]
- Bonar N. A. and Petersen C. P. (2017). Integrin suppresses neurogenesis and regulates brain tissue assembly in planarian regeneration. Development 144, 784-794. 10.1242/dev.139964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw B., Thompson K. and Frank U. (2015). Distinct mechanisms underlie oral vs aboral regeneration in the cnidarian Hydractinia echinata. Elife 4, e05506 doi:10.7554/eLife.05506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano A., Pérez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F. and Nieto M. A. (2000). The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2, 76-83. 10.1038/35000025 [DOI] [PubMed] [Google Scholar]
- Colvin Wanshura L. E., Galvin K. E., Ye H., Fernandez-Zapico M. E. and Wetmore C. (2011). Sequential activation of Snail1 and N-Myc modulates sonic hedgehog-induced transformation of neural cells. Cancer Res. 71, 5336-5345. 10.1158/0008-5472.CAN-10-2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie K. W., Brown D. D. R., Zhu S., Xu C., Voisin V., Bader G. D. and Pearson B. J. (2016). HOX gene complement and expression in the planarian Schmidtea mediterranea. Evodevo 7, 7 10.1186/s13227-016-0044-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B., Tropé C. G. and Reich R. (2012). Epithelial-mesenchymal transition in ovarian carcinoma. Front. Oncol. 2, 33 10.3389/fonc.2012.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois F. (1949). Contribution á l ‘ètude de la migration des cellules de règènèration chez les Planaires dulcicoles. Bull. Biol. Fr. Belg. 83, 213-283. [Google Scholar]
- Eisenhoffer G. T., Kang H. and Sánchez Alvarado A. (2008). Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 3, 327-339. 10.1016/j.stem.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elloul S., Elstrand M. B., Nesland J. M., Tropé C. G., Kvalheim G., Goldberg I., Reich R. and Davidson B. (2005). Snail, Slug, and Smad-interacting protein 1 as novel parameters of disease aggressiveness in metastatic ovarian and breast carcinoma. Cancer 103, 1631-1643. 10.1002/cncr.20946 [DOI] [PubMed] [Google Scholar]
- Fan F., Samuel S., Evans K. W., Lu J., Xia L., Zhou Y., Sceusi E., Tozzi F., Ye X.-C., Mani S. A. et al. (2012). Overexpression of snail induces epithelial-mesenchymal transition and a cancer stem cell-like phenotype in human colorectal cancer cells. Cancer Med. 1, 5-16. 10.1002/cam4.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix D. A. and Aboobaker A. A. (2010). The TALE class homeobox gene Smed-prep defines the anterior compartment for head regeneration. PLoS Genet. 6, e1000915 10.1371/journal.pgen.1000915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P. and Alexander S. (2011). Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147, 992-1009. 10.1016/j.cell.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Friedl P. and Gilmour D. (2009). Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10, 445-457. 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- Friedl P., Locker J., Sahai E. and Segall J. E. (2012). Classifying collective cancer cell invasion. Nat. Cell Biol. 14, 777-783. 10.1038/ncb2548 [DOI] [PubMed] [Google Scholar]
- Galliot B. (2012). Hydra, a fruitful model system for 270 years. Int. J. Dev. Biol. 56, 411-423. 10.1387/ijdb.120086bg [DOI] [PubMed] [Google Scholar]
- Gehrke A. R. and Srivastava M. (2016). Neoblasts and the evolution of whole-body regeneration. Curr. Opin. Genet. Dev. 40, 131-137. 10.1016/j.gde.2016.07.009 [DOI] [PubMed] [Google Scholar]
- Geisbrecht E. R. and Montell D. J. (2002). Myosin VI is required for E-cadherin-mediated border cell migration. Nat. Cell Biol. 4, 616-620. 10.1038/ncb830 [DOI] [PubMed] [Google Scholar]
- Goichberg P. (2016). Current understanding of the pathways involved in adult stem and progenitor cell migration for tissue homeostasis and repair. Stem Cell Rev. 12, 421-437. 10.1007/s12015-016-9663-7 [DOI] [PubMed] [Google Scholar]
- Guedelhoefer O. C. and Sánchez Alvarado A. (2012a). Planarian immobilization, partial irradiation, and tissue transplantation. J. Vis. Exp. 66, pii: 4015 10.3791/4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedelhoefer O. C. and Sánchez Alvarado A. (2012b). Amputation induces stem cell mobilization to sites of injury during planarian regeneration. Development 139, 3510-3520. 10.1242/dev.082099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn E. J., Ziel J. W., Morrissey M. A., Linden L. M., Wang Z., Chi Q., Johnson S. A. and Sherwood D. R. (2013). The netrin receptor DCC focuses invadopodia-driven basement membrane transmigration in vivo. J. Cell Biol. 201, 903-913. 10.1083/jcb.201301091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E. M. and Petersen C. P. (2015). Wnt/Notum spatial feedback inhibition controls neoblast differentiation to regulate reversible growth of the planarian brain. Development 142, 4217.-. 10.1242/dev.123612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvay K., Jardé T., Casagranda F., Perreau V. M., Haigh K., Nefzger C. M., Akhtar R., Gridley T., Berx G., Haigh J. J. et al. (2015). Snai1 regulates cell lineage allocation and stem cell maintenance in the mouse intestinal epithelium. EMBO J. 34, 1319-1335. 10.15252/embj.201490881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isolani M. E., Abril J. F., Saló E., Deri P., Bianucci A. M. and Batistoni R. (2013). Planarians as a model to assess in vivo the role of matrix metalloproteinase genes during homeostasis and regeneration. PLoS ONE 8, e55649 10.1371/journal.pone.0055649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakugawa S., Langton P. F., Zebisch M., Howell S. A., Chang T.-H., Liu Y., Feizi T., Bineva G., O'Reilly N., Snijders A. P. et al. (2015). Notum deacylates Wnt proteins to suppress signalling activity. Nature 519, 187-192. 10.1038/nature14259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R. and Weinberg R. A. (2009). The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420-1428. 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao D., Mihaylova Y., Hughes S., Lai A. and Aboobaker A. (2017). Epigenetic analyses of the planarian genome reveals conservation of bivalent promoters in animal stem cells. bioRxiv, 122135 https://doi.org/10.1101/122135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R. S. and Newmark P. A. (2013). In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev. Biol. 13, 8 10.1186/1471-213X-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S., Xu J. and Derynck R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178-196. 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan S. W. and Reddien P. W. (2011). dlx and sp6-9 control optic Cup regeneration in a prototypic eye. PLoS Genet. 7, e1002226 10.1371/journal.pgen.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoCascio S. A., Lapan S. W. and Reddien P. W. (2017). Eye absence does not regulate planarian stem cells during eye regeneration. Dev. Cell 40, 381-391.e3. 10.1016/j.devcel.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.-M., Coffey C. W., DeWerd L. A., Liu C., Nath R., Seltzer S. M. and Seuntjens J. P., American Association of Physicists in Medicine (2001). AAPM protocol for 40-300 kV x-ray beam dosimetry in radiotherapy and radiobiology. Med. Phys. 28, 868-893. 10.1118/1.1374247 [DOI] [PubMed] [Google Scholar]
- Mayor R. and Theveneau E. (2014). The role of the non-canonical Wnt-planar cell polarity pathway in neural crest migration. Biochem. J. 457, 19-26. 10.1042/BJ20131182 [DOI] [PubMed] [Google Scholar]
- Miyoshi A., Kitajima Y., Kido S., Shimonishi T., Matsuyama S., Kitahara K. and Miyazaki K. (2005). Snail accelerates cancer invasion by upregulating MMP expression and is associated with poor prognosis of hepatocellular carcinoma. Br. J. Cancer 92, 252-258. 10.1038/sj.bjc.6602266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A. and Keren K. (2009). The shape of motile cells. Curr. Biol. 19, R762-R771. 10.1016/j.cub.2009.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell D. J. (2003). Border-cell migration: the race is on. Nat. Rev. Mol. Cell Biol. 4, 13-24. 10.1038/nrm1006 [DOI] [PubMed] [Google Scholar]
- Oderberg I. M., Li D. J., Scimone M. L., Gaviño M. A. and Reddien P. W. (2017). Landmarks in existing tissue at wounds are utilized to generate pattern in regenerating tissue. Curr. Biol. 27, 733-742. 10.1016/j.cub.2017.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pálmer H. G., Larriba M. J., García J. M., Ordóñez-Morán P., Peña C., Peiró S., Puig I., Rodríguez R., de la Fuente R., Bernad A. et al. (2004). The transcription factor SNAIL represses vitamin D receptor expression and responsiveness in human colon cancer. Nat. Med. 10, 917-919. 10.1038/nm1095 [DOI] [PubMed] [Google Scholar]
- Peng Z., Wang C.-X., Fang E.-H., Wang G.-B. and Tong Q. (2014). Role of epithelial-mesenchymal transition in gastric cancer initiation and progression. World J. Gastroenterol. 20, 5403-5410. 10.3748/wjg.v20.i18.5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. P. and Reddien P. W. (2009). A wound-induced Wnt expression program controls planarian regeneration polarity. Proc. Natl. Acad. Sci. USA 106, 17061-17066. 10.1073/pnas.0906823106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. P. and Reddien P. W. (2011). Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332, 852-855. 10.1126/science.1202143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Oviedo N. J., Jennings J. R., Jenkin J. C. and Sánchez Alvarado A. (2005). SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327-1330. 10.1126/science.1116110 [DOI] [PubMed] [Google Scholar]
- Reig G., Pulgar E. and Concha M. L. (2014). Cell migration: from tissue culture to embryos. Development 141, 1999-2013. 10.1242/dev.101451 [DOI] [PubMed] [Google Scholar]
- Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T. and Horwitz A. R. (2003). Cell migration: integrating signals from front to back. Science 302, 1704-1709. 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Rosivatz E., Becker I., Specht K., Fricke E., Luber B., Busch R., Höfler H. and Becker K.-F. (2002). Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am. J. Pathol. 161, 1881-1891. 10.1016/S0002-9440(10)64464-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L., Weiss J. A., Forsthoefel D. J., Lee H., King R. S., Inoue T., Shibata N., Agata K. and Newmark P. A. (2013). RNA interference by feeding in vitro-synthesized double-stranded RNA to planarians: methodology and dynamics. Dev. Dyn. 242, 718-730. 10.1002/dvdy.23950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Oda Y., Kawaguchi K., Sugimachi K., Yamamoto H., Tateishi N., Tanaka K., Matsuda S., Iwamoto Y., Ladanyi M. et al. (2004). E-cadherin mutation and Snail overexpression as alternative mechanisms of E-cadherin inactivation in synovial sarcoma. Oncogene 23, 8629-8638. 10.1038/sj.onc.1207960 [DOI] [PubMed] [Google Scholar]
- Saló E. and Baguñà J. (1985). Cell movement in intact and regenerating planarians. Quantitation using chromosomal, nuclear and cytoplasmic markers. J. Embryol. Exp. Morphol. 89, 57-70. [PubMed] [Google Scholar]
- Sato K., Hiraiwa T., Maekawa E., Isomura A., Shibata T. and Kuranaga E. (2015). Left-right asymmetric cell intercalation drives directional collective cell movement in epithelial morphogenesis. Nat. Commun. 6, 10074 10.1038/ncomms10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., and Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone M. L., Kravarik K. M., Lapan S. W. and Reddien P. W. (2014). Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Rep. 3, 339-352. 10.1016/j.stemcr.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebeck F., März M., Meyer A.-W., Reuter H., Vogg M. C., Stehling M., Mildner K., Zeuschner D., Rabert F. and Bartscherer K. (2017). Integrins are required for tissue organization and restriction of neurogenesis in regenerating planarians. Development 144, 795-807. 10.1242/dev.139774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharili A.-S., Allen S., Smith K., Price J. and McGonnell I. M. (2013). Snail2 promotes osteosarcoma cell motility through remodelling of the actin cytoskeleton and regulates tumor development. Cancer Lett. 333, 170-179. 10.1016/j.canlet.2013.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B. N., Burton L. J., Henderson V., Randle D. D., Morton D. J., Smith B. A., Taliaferro-Smith L., Nagappan P., Yates C., Zayzafoon M. et al. (2014). Snail promotes epithelial mesenchymal transition in breast cancer cells in part via activation of nuclear ERK2. PLoS ONE 9, e104987 10.1371/journal.pone.0104987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimachi K., Tanaka S., Kameyama T., Taguchi K., Aishima S., Shimada M., Sugimachi K. and Tsuneyoshi M. (2003). Transcriptional repressor snail and progression of human hepatocellular carcinoma. Clin. Cancer Res. 9, 2657-2664. [PubMed] [Google Scholar]
- Tanaka E. M. and Reddien P. W. (2011). The cellular basis for animal regeneration. Dev. Cell 21, 172-185. 10.1016/j.devcel.2011.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki J., Uchiyama-Tasaki C. and Rouhana L. (2016). Analysis of stem cell motility in vivo based on immunodetection of planarian neoblasts and tracing of BrdU-labeled cells after partial irradiation. Methods Mol. Biol. 1365, 323-338. 10.1007/978-1-4939-3124-8_18 [DOI] [PubMed] [Google Scholar]
- Thiery J. P. and Sleeman J. P. (2006). Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7, 131-142. 10.1038/nrm1835 [DOI] [PubMed] [Google Scholar]
- Traister A., Shi W. and Filmus J. (2008). Mammalian Notum induces the release of glypicans and other GPI-anchored proteins from the cell surface. Biochem. J. 410, 503-511. 10.1042/BJ20070511 [DOI] [PubMed] [Google Scholar]
- Tu K. C., Cheng L.-C., T K Vu H., Lange J. J., McKinney S. A., Seidel C. W. and Sánchez Alvarado A. (2015). Egr-5 is a post-mitotic regulator of planarian epidermal differentiation. Elife 4, e10501 10.7554/eLife.10501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel J. C., Wagner D. E. and Reddien P. W. (2014). Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 15, 326-339. 10.1016/j.stem.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Choi C. K. and Horwitz A. R. (2009). Integrins in cell migration--the actin connection. J. Cell Sci. 122, 199-206. 10.1242/jcs.018564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo A., Molina-Ortiz P., Montenegro Y., Moreno-Bueno G., Morales S., Santos V., Gridley T., Pérez-Moreno M. A., Peinado H., Portillo F. et al. (2015). Loss of Snail2 favors skin tumor progression by promoting the recruitment of myeloid progenitors. Carcinogenesis 36, 585-597. 10.1093/carcin/bgv021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchley J. N., Mayer M., Wagner D. E., Owen J. H. and Reddien P. W. (2013). Muscle cells provide instructions for planarian regeneration. Cell Rep. 4, 633-641. 10.1016/j.celrep.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff E. (1962). Recent researches on the regeneration of planaria. In Regeneration. 20th Growth Symposium (ed. Rudnick D.), pp. 53-84. New York: Ronald Press. [Google Scholar]
- Zhang X., Cheong S.-M., Amado N. G., Reis A. H., MacDonald B. T., Zebisch M., Jones E. Y., Abreu J. G. and He X. (2015). Notum is required for neural and head induction via Wnt deacylation, oxidation, and inactivation. Dev. Cell 32, 719-730. 10.1016/j.devcel.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]