Abstract
In this study, we aimed to examine the expression of SLC4A11 in ovarian cancer and in normal ovarian tissues, its prognostic value and the possible mechanism of its dysregulation. Bioinformatic analysis was performed by using data from the GEO datasets, the Cancer Genome Atlas-Ovarian Cancer (TCGA-OV) and the Human Protein Atlas (HPA). Results showed that SLC4A11 was upregulated in ovarian cancer compared with normal ovarian epithelial tissues. In patients with primary serous ovarian cancer in TCGA-OV, the cases with lymphatic invasion (N = 133) had significantly higher SLC4A11 expression than those without lymphatic invasion (N = 77) (p = 0.0069). High SLC4A11 expression was consistently associated with worse overall survival (OS). Univariate and multivariate analysis confirmed that high SLC4A11 expression was an independent prognostic factor for poor OS in grade 3/4 (G3/G4) tumors (HR = 1.416, 95%CI: 1.098–1.824, p = 0.007). 320 out of 578 (55.4%) ovarian cancer cases had SLC4A11 amplification. High methylation group had a significantly lower level of SLC4A11 expression. Based on these findings, we infer that high SLC4A11 expression is an independent predictor for poor OS in grade 3/4 serous ovarian cancer. Both DNA amplification and hypomethylation contribute to its upregulation in ovarian cancer.
Introduction
Intracellular and extracellular pH (pHi and pHe) homeostasis is an important constitution of cellular microenvironment and is a prerequisite for normal cell function [1]. Studies in the past decade revealed that pHi homeostasis is often dramatically altered in cancer [2, 3]. Cancer cells usually keep a pHi that is equal to or even more alkaline than the surrounding normal counterparts, suggesting that they upregulate net acid extrusion [2, 3]. This alteration leads to at least two fundamental differences to normal physiology: firstly, it is supportive to maintain pHi homeostasis by eliminating excessive production of acid equivalents in cancer cells due to hypoxia and/or oncogene-induced changes in glycolytic metabolism [4]; secondly, prolonged extracellular acidification (pHe), which favors tumor invasion and metastasis [5, 6].
Solute linked cotransporter 4 (SLC4) family is comprised of ten members (SLC4A1-5; SLC4A7-11), which have critical roles in pHi buffering [7, 8]. SLC4A1-3 are Cl−/HCO3− exchangers and function as cellular acid loaders [9]. SLC4A4, −4A5, −4A7, −4A8 and −4A10 are Na+-coupled HCO3− transporters and function as cellular acid extruders [9]. SLC4A11 is the most divergent member of this family and has recently been characterized as a Na+/OH− and NH4+ transporter [10]. Therefore, SLC4A11 can mediate H+ efflux and can be considered as a cellular acid extruder. Dysregulated SLC4 family members have been implied in pathological development of some cancers. For example, SLC4A7 regulates pHi and tumor cell progression in breast cancers [11, 12]. Upregulated SLC4A4 contributes to growth and migration of colon and breast cancer cells [13]. SLC4A9 disruption by either genetic or pharmaceutical approaches results in pHi acidification and reduced cell growth of breast cancer and glioma cells [14].
Ovarian cancer cells have an elevated H+ efflux compared with non-tumor cells [15]. One recent study found that the basal pHi is higher in ovarian cancer A2780 cells than in normal ovarian HOSE cells [16]. SLC9A1 amplification is a mechanism of the high basal PHi and is associated with unfavorable overall patient survival [16]. These findings suggest that pHi regulation is closely related to ovarian cancer cell behaviors and prognosis of the patients.
In this study, we examined the expression of SLC4A11 in ovarian cancer/normal ovarian tissues and further assessed its prognostic value and the possible mechanism of its dysregulation.
Materials and methods
Bioinformatic data mining in GEO
The normalized data of one previous microarray (GDS3592) [17] that explored the dysregulated genes in ovarian cancer epithelial cells (CEPIs) from patients with primary serous ovarian cancer compared with normal ovarian surface epithelia (OSE) was downloaded from GEO dataset for secondary analysis.
Data mining in the Human Protein Atlas
SLC4A11 expression at the protein level in normal ovarian tissues and in serous ovarian cancer tissues was compared by using the immunohistochemistry (IHC) staining data provided by the Human Protein Atlas (HPA) (http://www.proteinatlas.org/) [18, 19].
Bioinformatic analysis using data from TCGA-Ovarian Cancer (TCGA-OV)
SLC4A11 expression, its copy number alteration and its DNA methylation in patients with ovarian cancer were examined using data from TCGA-OV. Original data downloaded from the UCSC Xena Browser (https://xenabrowser.net/) were given in S1 Table. The association between SLC4A11 expression and overall survival (OS) or recurrence-free survival (RFS) was assessed by generating Kaplan-Meier survival curves, with median SLC4A11 expression as the cutoff. The analysis was performed using the UCSC Xena Browser or by using GraphPad Prism 6.0. Among 595 patients with primary serous ovarian cancer in TCGA-OV, 540 patients had SLC4A11 expression measured by AgilentG4502A_07_3. This proportion of patients was included in the univariate and multivariate analysis of the association between SLC4A11 expression and OS/RFS.
Statistical analysis
The association between clinicopathological characteristics and SLC4A11 expression was assessed by using χ2 tests. The significance of the difference between the survival curves was assessed by log-rank test. Univariate and multivariate Cox regression models were used to evaluate prognostic significance. Welch’s t-test was conducted to compare SLC4A11 expression between patients with or without lymphatic invasion and between groups with high/low methylation. p<0.05 was considered statistically significant.
Results
SLC4A11 is upregulated in ovarian cancer compared with normal ovarian epithelial tissues
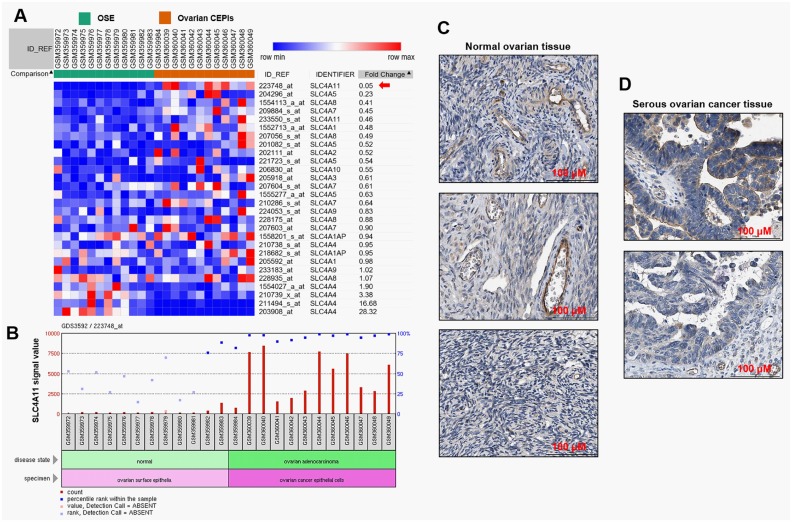
By re-analysis of the normalized data of GDS3592, we examined the expression of SLC4A family members in CEPIs and OSE (Fig 1A). SLC4A11 was the most significantly upregulated gene among the 10 SLC4A family members (Fig 1A, red arrow). The bar chart of the array signaling value further indicated that SLC4A11 was significantly upregulated in the CEPI samples compared with the OSE samples (Fig 1B). By data mining in the HPA, we found that in normal ovarian tissues, follicle cells had medium SLC4A11 staining (Fig 1C). But SLC4A11 expression was not detectable in ovarian stroma cells (Fig 1C). In comparison, the serous ovarian cancer tissues usually had low to medium SLC4A11 staining in both cytoplasma and cell membrane (Fig 1D).
Fig 1. SLC4A11 is upregulated in ovarian cancer compared with normal ovarian epithelial tissues.
A. Heat map of the expression of SLC4A family members in 12 cases of ovarian CEPIs compared with 12 cases of OSE. Red: up-regulation; Blue: down-regulation. The image was generated by re-analysis of the raw microarray data of GDS3592. B. SLC4A11 microarray signal values in 12 CEPIs and 12 OSE cases. Data were analyzed by using the tool provided by GEO datasets. C-D. Representative images of IHC staining of SLC4A11 in normal ovarian tissues (C) and serous ovarian cancer tissues (D). Data were obtained from the HPA: http://www.proteinatlas.org/ENSG00000088836-SLC4A11/pathology/tissue/ovarian+cancer.
High SLC4A11 expression is associated with lymphatic invasion
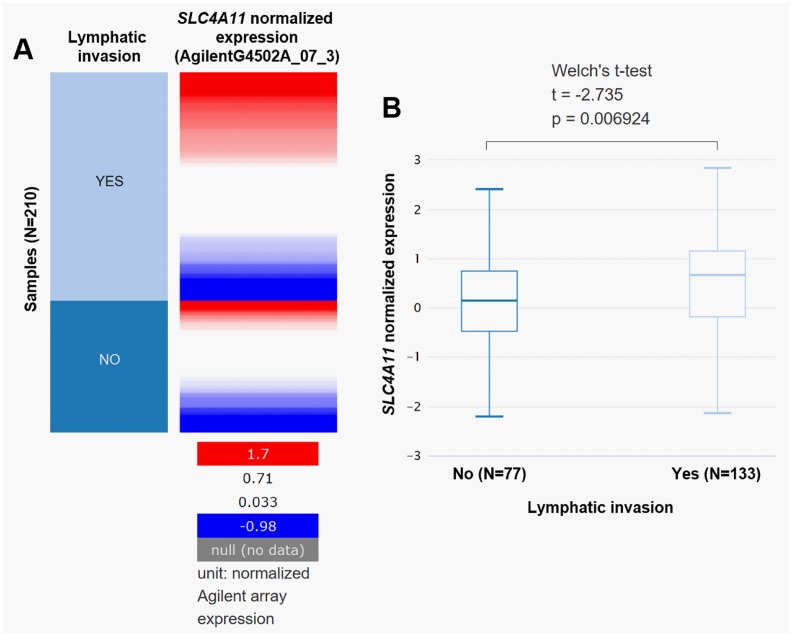
Using data from TCGA-OV, we compared SLC4A11 expression in ovarian cancer patients with or without lymphatic invasion. Results revealed that the cases with lymphatic invasion (N = 133) had significantly higher SLC4A11 expression (p = 0.0069) than those without lymphatic invasion (N = 77) (Fig 2A and 2B).
Fig 2. High SLC4A11 expression is associated with lymphatic invasion.
A-B. Heat map (A) and box plots (B) of SLC4A11 expression in patients with or without lymphatic invasion.
High SLC4A11 expression is an independent predictor of poor OS in ovarian cancer patients
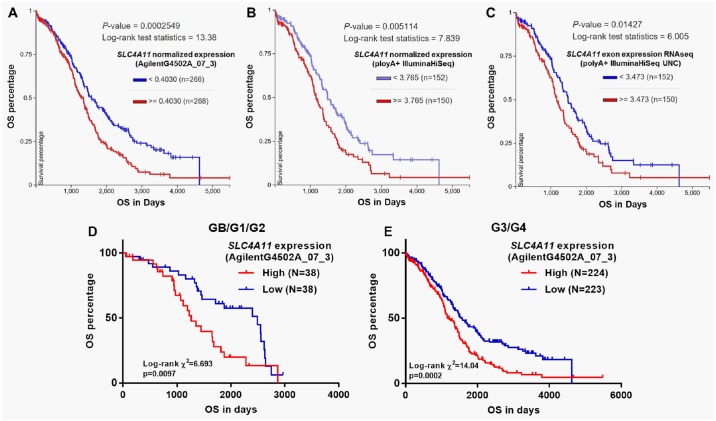
The association between clinicopathological features and SLC4A11 expression in patients with primary serous ovarian cancer was summarized in Table 1. High SLC4A11 expression was significantly associated with lower grade tumors (GB/G1/G2) (p = 0.038), and a larger proportion of lymphatic invasion (p = 0.027) and deceased cases (p = 0.036) (Table 1). In addition, the high SLC4A11 expression group also had a higher ratio of tumor residual disease at the margin level of significance (p = 0.050) (Table 1). However, no significant difference was observed in recurrence rate between the two groups (Table 1). Then, Kaplan-Meier survival analysis was performed between the high and low SLC4A11 expression groups in TCGA-OV. In the database, SLC4A11 expression was quantified by RNAseq (AgilentG4502A_07_3 and IlluminaHiSeq respectively) and exon RNAseq (polyA+ IlluminaHiSeq). Among 540 patients with SLC4A11 measured by AgilentG4502A_07_3, 534 patients had OS data. Both IlluminaHiSeq and RNAseq data included 302 patients with OS data (Fig 2A–2C). Log-rank test showed that in all measurements, high SLC4A11 expression was consistently associated with worse OS compared with low SLC4A11 expression (p = 0.00025, 0.0051 and 0.014 respectively, Fig 2A–2C). Recent studies suggest that GB/G1/G2 carcinoma belong to type I, while G3/G4 carcinomas belong to type II tumors, which have significant clinicopathologic and molecular differences [20, 21]. We then performed subgroup analysis in GB/G1/G2 and G3/G4 tumors respectively. In both groups, we found that high SLC4A11 expression was associated with unfavorable OS (Fig 3D and 3E). In univariate analysis, high SLC4A11 expression was associated with poor prognosis in terms of OS in both GB/G1/G2 and G3/G4 tumors (p = 0.011 and p<0.001 respectively) (Table 2). However, following multivariate analysis only confirmed the independent prognostic value of high SLC4A11 expression in G3/G4 tumors (HR = 1.416, 95%CI: 1.098–1.824, p = 0.007; Table 2), but not in GB/G1/G2 tumors (HR = 1.832, 95%CI: 0.987–3.402, p = 0.055; Table 2). Due to insufficient data of recurrence, we only assessed the association between SLC4A11 expression and RFS in patients with G3/G4 diseases. No independent prognostic value of SLC4A11 expression was observed in terms of RFS (Table 2).
Table 1. Demographic and clinicopathological parameters of patients with primary ovarian cancer in TCGA-OV.
| Parameters | SLC4A11 expression RNAseq | χ2 | p Value | ||
|---|---|---|---|---|---|
| High (N = 270) | Low (N = 270) | ||||
| Age (Mean ± SD) | ≥60 | 131 | 122 | 0.54 | 0.46 |
| <60 | 139 | 147 | |||
| Null | 0 | 1 | |||
| Grade | GB/G1/G2 | 46 | 30 | 4.30 | 0.038 |
| G3/G4 | 215 | 236 | |||
| GX + Null | 9 | 4 | |||
| Clinical stage | I/II | 18 | 24 | 0.93 | 0.33 |
| III/IV | 249 | 243 | |||
| Null | 3 | 3 | |||
| Venous invasion | No | 30 | 37 | 2.05 | 0.15 |
| Yes | 48 | 37 | |||
| Null | 192 | 196 | |||
| Lymphatic invasion | No | 33 | 44 | 4.88 | 0.027 |
| Yes | 78 | 55 | |||
| Null | 159 | 171 | |||
| Tumor residual disease | No | 44 | 62 | 3.83 | 0.050 |
| Yes | 194 | 177 | |||
| Null | 32 | 31 | |||
| Recurrence status | No | 5 | 10 | 0.24 | 0.62 |
| Yes | 21 | 31 | |||
| Null | 244 | 229 | |||
| Living Status | Living | 99 | 122 | 4.38 | 0.036 |
| Dead | 169 | 144 | |||
| Null | 2 | 4 | |||
GB: Border line malignancy; G1: Well differentiated; G2: Moderately differentiated; G3-G4: Poorly differentiated or Undifferentiated; GX: Grade cannot be assessed. Null: no data.
Fig 3. High SLC4A11 expression is associated with poor survivals in ovarian cancer patients.
A-E. Kaplan-Meier curves of OS in ovarian cancer patients with high and low SLC4A11 expression. Data were obtained from TCGA-OV. SLC4A11 expression was quantified by RNAseq (AgilentG4502A_07_3 (A) and IlluminaHiSeq respectively (B)) and exon RNAseq (polyA+ IlluminaHiSeq) (C). Kaplan-Meier curves of OS in GB/G1/G2 patients (D) and in G3/G4 patients (E).
Table 2. Univariate and multivariate analyses of OS/RFS in patients with primary ovarian cancer in TCGA-OV.
| Parameters | Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| p | HR | 95%CI (lower/upper) | p | HR | 95%CI (lower/upper) | |||
|
OS (GB/G1/G2) Age ≥60 vs.< 60 |
0.011 | 2.171 | 1.192 | 3.956 | 0.999 | 1.00 | 0.549 | 1.824 |
|
Clinical stage III/IV vs. I/II |
0.018 | 5.557 | 1.338 | 23.07 | 0.0367 | 4.63 | 1.099 | 19.504 |
|
Venous invasion No vs. Yes |
0.112 | 0.392 | 0.123 | 1.244 | ||||
|
Lymphatic invasion No vs. Yes |
0.999 | 1.001 | 0.27 | 3.712 | ||||
|
Tumor residual disease No vs. Yes |
0.208 | 1.693 | 0.746 | 3.841 | ||||
|
SLC4A11 expression High vs. Low |
0.011 | 2.171 | 1.192 | 3.956 | 0.055 | 1.832 | 0.987 | 3.402 |
|
OS (G3/G4) Age ≥60 vs.< 60 |
<0.001 | 1.616 | 1.266 | 2.062 | 0.001 | 1.516 | 1.187 | 1.937 |
|
Clinical stage III/IV vs. I/II |
0.123 | 1.743 | 0.861 | 3.529 | ||||
|
Venous invasion No vs. Yes |
0.533 | 1.2 | 0.676 | 2.132 | ||||
|
Lymphatic invasion No vs. Yes |
0.055 | 0.613 | 0.373 | 1.01 | 0.563 | 0.859 | 0.514 | 1.437 |
|
Tumor residual disease No vs. Yes |
<0.001 | 2.46 | 1.647 | 3.675 | <0.001 | 2.252 | 1.497 | 3.39 |
|
SLC4A11 expression High vs. Low |
<0.001 | 1.595 | 1.247 | 2.04 | 0.007 | 1.416 | 1.098 | 1.824 |
|
RFS (G3/G4) Age ≥60 vs.< 60 |
0.3004 | 1.366 | 0.757 | 2.465 | ||||
|
Clinical stage III/IV vs. I/II |
0.146 | 0.22 | 0.029 | 1.692 | ||||
|
Tumor residual disease No vs. Yes |
0.017 | 2.421 | 1.175 | 4.989 | 0.041 | 2.167 | 1.031 | 4.553 |
|
SLC4A11 expression High vs. Low |
0.052 | 1.769 | 0.996 | 3.14 | 0.179 | 1.511 | 0.828 | 2.757 |
GB: Border line malignancy; G1: Well differentiated; G2: Moderately differentiated; G3-G4: Poorly differentiated or Undifferentiated. OS: overall survival; RFS: recurrence-free survival.
SLC4A11 expression is regulated by DNA amplification and methylation in ovarian cancer
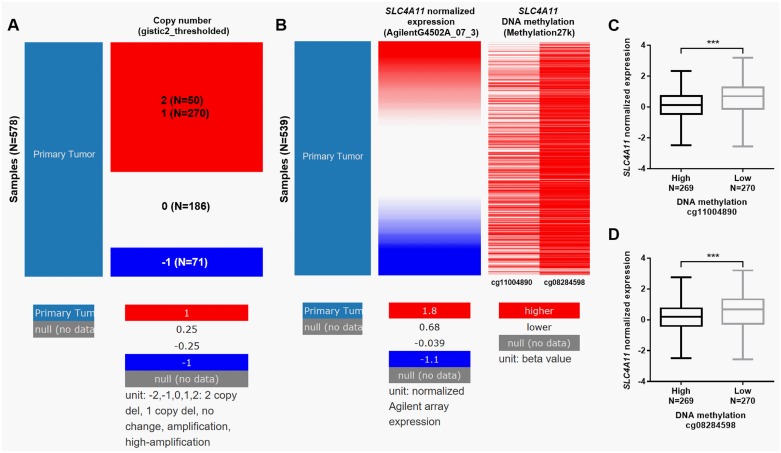
Via analyzing the deep sequencing data in TCGA-OV, we further explored the mechanisms of SLC4A11 dysregulation. Among 578 patients with copy number measured, 320 (55.4%) cases had DNA amplification (Fig 4A). By comparing SLC4A11 expression and its DNA methylation status, we also confirmed that the high methylation group had a significantly lower level of SLC4A11 expression (Fig 4B–4D).
Fig 4. SLC4A11 expression is regulated by DNA amplification and methylation in ovarian cancer.
A. Heat map of SLC4A11 copy number changes in TCGA-OV. -1: deletion; 0: no change; 1: amplification; 2: high-amplification. B-D. Heat map (B) and box plots (C-D) of SLC4A11 expression and DNA methylation in TCGA-OV. Patients were divided into high and low methylation groups according to the median value of the two probes (cg11004890 and cg08284598) respectively.
Discussion
As a Na+-coupled base transporter capable of Na+-H+ exchange in mammalian cells, SLC4A11 mutation is associated with a series of corneal endothelial diseases characterized by dysfunction of the endothelial cells in the inner surface of the cornea [22–24]. These diseases finally result in the apoptosis of corneal endothelial cells, and subsequent edema and impaired vision [25]. In fact, the normal corneal endothelial function is dependent on the balanced regulation of bicarbonate concertation and pHi/pHe. Based on these findings, we infer that the putative bicarbonate transport or Na+-H+ exchange activity of SLC4A11 is essential for maintaining normal cellular physiological functions.
In this study, we found that SLC4A11 expression is significantly upregulated in ovarian cancer tissues than in normal tissues. The alkaline intracellular environment of cancer cells is usually associated with the upregulation of the expression and/or activity of acid extruders or downregulation of the expression and/or activity of acid loading transporters [1]. Given the pHi regulatory activity demonstrated in previous studies [10, 26], SLC4A11 naturally owns a strong cellular buffering capacity that enables the highly glycolytic cancer cells to remove lactic acid rapidly. Its upregulation can be considered as an adaptive response of the cancer cells to metabolic changes. In addition, upregulation of the acid extruders leads to extracellular acidification, thereby generating a favorable environment for tumor invasion and metastasis [5, 6]. In this study, we observed that the patients with metastasis had significantly higher SLC4A11 expression than their counterparts without metastasis. Therefore, we infer that SLC4A11 upregulation is an important mechanism leading to malignant cellular behaviors in ovarian cancer. By comparing the clinicopathological features between high and low SLC4A11 expression groups, we found that the high SLC4A11 expression group had a significantly higher ratio of lymphatic invasion and deceased cases. However, we also observed that the high SLC4A11 expression group had a substantially larger proportion of low-grade tumors (GB/G1/G2) (46/261, 17.6% vs. 30/261, 11.3%) compared with the low SLC4A11 group. Actually, G1/G2 serous ovarian carcinomas belong to type I tumors, while G3/G4 serous ovarian carcinomas belong to type II tumors, which have different molecular and biological profiles [20, 21]. We hypothesized that the difference in tumor grade might be related to the genetic difference between type I and type II tumors. However, more studies are required to confirm this hypothesis.
Currently, a series of predictors such as clinical stage, CA125 levels, age, response to chemotherapy and residual disease after debulking surgery are used to predict prognosis of ovarian cancer patients. However, there is still some discrepancy in the prognosis of similar tumors characterized by these clinical variables [27]. Ovarian cancers actually are a heterogeneous group of neoplasias derived from the ovarian surface epithelium, inclusion cysts, or the fallopian tube [28, 29]. The differences at the molecular level may be important sources of the variations. Therefore, it is necessary to explore other molecular parameters for better prediction of prognosis. In this study, based on the survival data of patients in TCGA-OV, we found that high SLC4A11 expression was robustly associated with poor OS. Following univariate and multivariate analysis confirmed that high SLC4A11 expression was an independent prognostic factor for poor OS in G3/G4 tumors. These findings suggest that SLC4A11 might be a potential clinical marker in this group of patients. Although the association between SLC4A11 expression and OS in GB/G1/G2 patients was not significant, it showed a trend toward significance (p = 0.055). Considering the small number of GB/G1/G2 patients included in this study (N = 76), it is meaningful to further explore its prognostic value with a large sample base in the future.
SLC4A11 locates in chromosome 20p in the human genome, a region with a relatively high frequency of genetic amplification in ovarian cancer [30, 31]. By using deep sequencing data in TCGA-OV, we found that 320 out of 578 ovarian cancer cases had SLC4A11 amplification. Therefore, DNA amplification is one important mechanism of upregulated SLC4A11 in ovarian cancer. Epigenetic regulation, such as methylation and histone modification are also important mechanisms of dysregulated genes in ovarian cancer [32, 33]. Thus, we also investigated whether methylation status influences SLC4A11 expression. By using the results from Illumina 27k methylation array, we found that the group with high DNA methylation had significantly lower SLC4A11 expression, suggesting that epigenetic alterations also contribute to SLC4A11 upregulation in ovarian cancer.
Conclusion
Based on findings above, we infer that high SLC4A11 expression is an independent predictor for poor OS in grade 3/4 serous ovarian cancer. Both DNA amplification and hypomethylation contribute to its upregulation in ovarian cancer.
Supporting information
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Gorbatenko A, Olesen CW, Boedtkjer E, Pedersen SF. Regulation and roles of bicarbonate transporters in cancer. Front Physiol. 2014;5:130 doi: 10.3389/fphys.2014.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parks SK, Chiche J, Pouyssegur J. Disrupting proton dynamics and energy metabolism for cancer therapy. Nat Rev Cancer. 2013;13(9):611–23. doi: 10.1038/nrc3579 . [DOI] [PubMed] [Google Scholar]
- 3.Pedersen SF, Stock C. Ion channels and transporters in cancer: pathophysiology, regulation, and clinical potential. Cancer Res. 2013;73(6):1658–61. doi: 10.1158/0008-5472.CAN-12-4188 . [DOI] [PubMed] [Google Scholar]
- 4.Andersen AP, Moreira JM, Pedersen SF. Interactions of ion transporters and channels with cancer cell metabolism and the tumour microenvironment. Philos Trans R Soc Lond B Biol Sci. 2014;369(1638):20130098 doi: 10.1098/rstb.2013.0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estrella V, Chen T, Lloyd M, Wojtkowiak J, Cornnell HH, Ibrahim-Hashim A, et al. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 2013;73(5):1524–35. doi: 10.1158/0008-5472.CAN-12-2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B, Gillies RJ. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res. 2006;66(10):5216–23. doi: 10.1158/0008-5472.CAN-05-4193 . [DOI] [PubMed] [Google Scholar]
- 7.Romero MF, Chen AP, Parker MD, Boron WF. The SLC4 family of bicarbonate (HCO(3)(-)) transporters. Mol Aspects Med. 2013;34(2–3):159–82. doi: 10.1016/j.mam.2012.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thornell IM, Bevensee MO. Regulators of Slc4 bicarbonate transporter activity. Front Physiol. 2015;6:166 doi: 10.3389/fphys.2015.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shei W, Liu J, Htoon HM, Aung T, Vithana EN. Differential expression of the Slc4 bicarbonate transporter family in murine corneal endothelium and cell culture. Mol Vis. 2013;19:1096–106. [PMC free article] [PubMed] [Google Scholar]
- 10.Ogando DG, Jalimarada SS, Zhang W, Vithana EN, Bonanno JA. SLC4A11 is an EIPA-sensitive Na(+) permeable pHi regulator. Am J Physiol Cell Physiol. 2013;305(7):C716–27. doi: 10.1152/ajpcell.00056.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, Axelsen TV, Andersen AP, Vahl P, Pedersen SF, Boedtkjer E. Disrupting Na(+), HCO(3)(-)-cotransporter NBCn1 (Slc4a7) delays murine breast cancer development. Oncogene. 2016;35(16):2112–22. doi: 10.1038/onc.2015.273 . [DOI] [PubMed] [Google Scholar]
- 12.Boedtkjer E, Moreira JM, Mele M, Vahl P, Wielenga VT, Christiansen PM, et al. Contribution of Na+,HCO3(-)-cotransport to cellular pH control in human breast cancer: a role for the breast cancer susceptibility locus NBCn1 (SLC4A7). Int J Cancer. 2013;132(6):1288–99. doi: 10.1002/ijc.27782 . [DOI] [PubMed] [Google Scholar]
- 13.Parks SK, Pouyssegur J. The Na(+)/HCO3(-) Co-Transporter SLC4A4 Plays a Role in Growth and Migration of Colon and Breast Cancer Cells. J Cell Physiol. 2015;230(8):1954–63. doi: 10.1002/jcp.24930 . [DOI] [PubMed] [Google Scholar]
- 14.McIntyre A, Hulikova A, Ledaki I, Snell C, Singleton D, Steers G, et al. Disrupting Hypoxia-Induced Bicarbonate Transport Acidifies Tumor Cells and Suppresses Tumor Growth. Cancer Res. 2016;76(13):3744–55. doi: 10.1158/0008-5472.CAN-15-1862 . [DOI] [PubMed] [Google Scholar]
- 15.Sanhueza C, Araos J, Naranjo L, Villalobos R, Westermeier F, Salomon C, et al. Modulation of intracellular pH in human ovarian cancer. Curr Mol Med. 2016;16(1):23–32. . [DOI] [PubMed] [Google Scholar]
- 16.Sanhueza C, Araos J, Naranjo L, Toledo F, Beltran AR, Ramirez MA, et al. Sodium/proton exchanger isoform 1 regulates intracellular pH and cell proliferation in human ovarian cancer. Biochim Biophys Acta. 2017;1863(1):81–91. doi: 10.1016/j.bbadis.2016.10.013 . [DOI] [PubMed] [Google Scholar]
- 17.Bowen NJ, Walker LD, Matyunina LV, Logani S, Totten KA, Benigno BB, et al. Gene expression profiling supports the hypothesis that human ovarian surface epithelia are multipotent and capable of serving as ovarian cancer initiating cells. BMC Med Genomics. 2009;2:71 doi: 10.1186/1755-8794-2-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, et al. A subcellular map of the human proteome. Science. 2017;356(6340). doi: 10.1126/science.aal3321 . [DOI] [PubMed] [Google Scholar]
- 19.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–50. doi: 10.1038/nbt1210-1248 . [DOI] [PubMed] [Google Scholar]
- 20.Kurman RJ, Shih Ie M. The Dualistic Model of Ovarian Carcinogenesis: Revisited, Revised, and Expanded. Am J Pathol. 2016;186(4):733–47. doi: 10.1016/j.ajpath.2015.11.011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshiyama M, Matsumura N, Konishi I. Recent concepts of ovarian carcinogenesis: type I and type II. Biomed Res Int. 2014;2014:934261 doi: 10.1155/2014/934261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vithana EN, Morgan PE, Ramprasad V, Tan DT, Yong VH, Venkataraman D, et al. SLC4A11 mutations in Fuchs endothelial corneal dystrophy. Hum Mol Genet. 2008;17(5):656–66. doi: 10.1093/hmg/ddm337 . [DOI] [PubMed] [Google Scholar]
- 23.Vithana EN, Morgan P, Sundaresan P, Ebenezer ND, Tan DT, Mohamed MD, et al. Mutations in sodium-borate cotransporter SLC4A11 cause recessive congenital hereditary endothelial dystrophy (CHED2). Nat Genet. 2006;38(7):755–7. doi: 10.1038/ng1824 . [DOI] [PubMed] [Google Scholar]
- 24.Vilas GL, Loganathan SK, Liu J, Riau AK, Young JD, Mehta JS, et al. Transmembrane water-flux through SLC4A11: a route defective in genetic corneal diseases. Hum Mol Genet. 2013;22(22):4579–90. doi: 10.1093/hmg/ddt307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel SP, Parker MD. SLC4A11 and the Pathophysiology of Congenital Hereditary Endothelial Dystrophy. Biomed Res Int. 2015;2015:475392 doi: 10.1155/2015/475392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao L, Azimov R, Abuladze N, Newman D, Kurtz I. Human SLC4A11-C functions as a DIDS-stimulatable H(+)(OH(-)) permeation pathway: partial correction of R109H mutant transport. Am J Physiol Cell Physiol. 2015;308(2):C176–88. doi: 10.1152/ajpcell.00271.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimada T, Saito T, Shimokawa M, Shimamoto K, Matsushita S, Yamaguchi S, et al. Improvement in the prognosis of ovarian cancer in the era before addition of molecular targeting therapy. Jpn J Clin Oncol. 2017;47(6):494–8. doi: 10.1093/jjco/hyx026 . [DOI] [PubMed] [Google Scholar]
- 28.McCluggage WG. Morphological subtypes of ovarian carcinoma: a review with emphasis on new developments and pathogenesis. Pathology. 2011;43(5):420–32. doi: 10.1097/PAT.0b013e328348a6e7 . [DOI] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe T, Imoto I, Kosugi Y, Ishiwata I, Inoue S, Takayama M, et al. A novel amplification at 17q21-23 in ovarian cancer cell lines detected by comparative genomic hybridization. Gynecol Oncol. 2001;81(2):172–7. doi: 10.1006/gyno.2001.6132 . [DOI] [PubMed] [Google Scholar]
- 31.Lambros MB, Fiegler H, Jones A, Gorman P, Roylance RR, Carter NP, et al. Analysis of ovarian cancer cell lines using array-based comparative genomic hybridization. J Pathol. 2005;205(1):29–40. doi: 10.1002/path.1681 . [DOI] [PubMed] [Google Scholar]
- 32.Kwon MJ, Shin YK. Epigenetic regulation of cancer-associated genes in ovarian cancer. Int J Mol Sci. 2011;12(2):983–1008. doi: 10.3390/ijms12020983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asadollahi R, Hyde CA, Zhong XY. Epigenetics of ovarian cancer: from the lab to the clinic. Gynecol Oncol. 2010;118(1):81–7. doi: 10.1016/j.ygyno.2010.03.015 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.