Abstract
Filamentous fungi and bacteria form mixed-species biofilms in nature and diverse clinical contexts (Frey-Klett et al., 2011; Peleg et al., 2010). The interactions between fungi and bacteria, often mediated by secreted metabolites, have important ramifications for the biology of the interacting partners (Frey-Klett et al., 2011). This is particularly true for the bacterium Pseudomonas aeruginosa (P. aeruginosa) and the fungus Aspergillus fumigatus (A. fumigatus) which often reside in the same niche such as lungs of cystic fibrosis (CF) patients. Some studies have reported that co-infection with P. aeruginosa and A. fumigatus could lead to a decrease in lung function relative to their respective single species infection (Amin et al., 2010; Peleg et al., 2010). Metabolite extraction and analysis allow for the characterization of specific microbial metabolites in the polymicrobial biofilm. This protocol describes how to prepare the Pseudomonas-Aspergillus co-culture biofilm on solid medium in preparation for metabolite extraction.
Materials and Reagents
Plastic spreader (Excel Scientific, catalog number: 5PR-L-510)
Falcon tubes (15 ml) (Corning Inc., catalog number: 352196)
Petri dish (100 × 15 mm) (Corning Inc., catalog number: 351029)
Petri dish (60 × 15 mm) (Corning Inc., catalog number: 351007)
Inoculating needle (VWR International, catalog number: 12000-816)
Glass vial (10 ml) (VWR International, catalog number: SC66022-300)
Aluminum foil
Pseudomonas aeruginosa culture
Aspergillus fumigatus culture
D-glucose (Thermo Fisher Scientific, Alfa Aesar, catalog number: A1682836)
Anhydrous dextrose (VWR International, BDH, catalog number: BDH9230500G)
Yeast extract (BD, Bacto™, catalog number: 288620)
Agar-agar (Merck Millipore Corporation, catalog number: EM1.01614.0500)
Peptone (BD, Bacto™, catalog number: 212120)
Luria-Bertani (LB) dehydrated medium (Merck Millipore Corporation, catalog number: 1.10285.5000)
Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: S5881)
GR ACS Sodium Nitrate (NaNO3) (Merck Millipore Corporation, catalog number: SX06551)
NaCl
Potassium Chloride (KCl) (VWR International, BDH, catalog number: 0395VBD500G)
Potassium dihydrogen phosphate (KH2PO4) (Thermo Fisher Scientific, Alfa Aesar, catalog number: 1159436)
Boric acid (H3BO3) (Sigma-Aldrich, catalog number: B6768)
Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: M1880)
Copper(II) sulfate pentahydrate (CuSO4·5H2O) (Sigma-Aldrich, catalog number: C8027)
Zinc sulfate heptahydrate (ZnSO4·7H2O) (Sigma-Aldrich, catalog number: Z0251)
Ammonium molybdate tetrahydrate [(NH4)6Mo7O24·4H2O] (Sigma-Aldrich, catalog number: 431346)
Manganese(II) sulfate monohydrate (MnSO4·H2O) (Sigma-Aldrich, catalog number: M7634)
Iron(II) sulfate heptahydrate (FeSO4·7H2O) (Sigma-Aldrich, catalog number: F8633)
Ethylenediaminetetraacetic acid disodium salt dehydrate (Na2EDTA·2H2O) (Sigma-Aldrich, catalog number: E4884)
Cobalt(II) chloride hexahydrate (CoCl2·6H2O) (Sigma-Aldrich, catalog number: 255599)
Tween 20 solution (Sigma-Aldrich, catalog number: P2287)
Chloroform (VWR International, BDH, catalog number: PL049ZA4)
Acetonitrile (Honeywell International Inc., catalog number: AH015-4)
PBS-0.01 % Tween 20 buffer (see Recipes)
20× sodium nitrate salts solution (see Recipes)
Trace elements (see Recipes)
-
Medium recipes (see Recipes)
Solid Luria-Bertani (LB) dehydrated medium (see Recipes)
Solid yeast extract-peptone-dextrose medium (YPD) (see Recipes)
Equipment
Incubator (VWR International, model: 1535)
Orbital shaker (Eppendorf AG, New Brunswick Scientific, model: I26)
Centrifuge (Beckman, model: J20) with rotor (Beckman, model: JLA-10.500)
Microscope (Celestron, model: 44345)
Hemocytometer (Hausser Scientific, catalog number: 3500)
Steel corer (38.1 mm in diameter) (Figure 1)
Steel blade (VWR International, catalog number: 55411-050)
Steel pestle (VWR International, catalog number: 62400-336)
0.2 µm-pore-size hydrophilic filter (VWR International, catalog number: 28145-477)
Glass culture tube (15 ml) (VWR International, catalog number: 47729-583)
Glass centrifuge tube (15 ml) (SP Industries, Wilmad-LabGlass, catalog number: C-8060-15)
Nitrogen gas cylinder
Figure 1.
Steel corer
Procedure
-
Biofilm co-culture of Pseudomonas and Aspergillus
-
Pseudomonas aeruginosa culture preparation
Streak-inoculate Pseudomonas aeruginosa PA14 strain from −80 °C glycerol stock onto solid LB medium in a 100 × 15 mm petri dish and incubate at 37 °C for 24 h.
Immediately prior to use for co-culture experiments, inoculate 5 ml of liquid LB medium in a 15 ml glass culture tube with a single colony from PA14 LB solid medium culture in step A1a and incubate at 37 °C, 225 rpm for 14 h to early stationary phase.
-
Aspergillus fumigatus conidia preparation
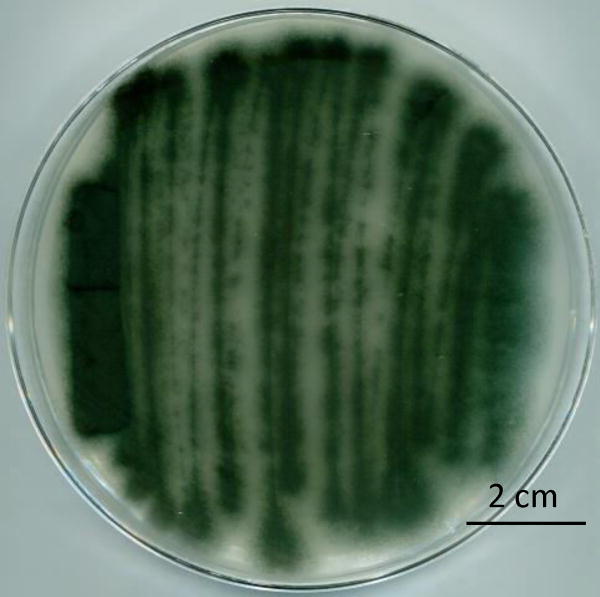
Take a ~10 µl plug from the −80 °C glycerol stock of Aspergillus fumigatus AF293 using a sterile inoculating needle and streak onto solid glucose minimal medium (GMM) in a 100 × 15 mm petri dish and incubate at 37 °C for 5 days (Figure 2).
Immediately prior to use, add 5 ml of phosphate buffered saline (PBS) −0.01% Tween 20 buffer (pH 7.2) to the fungal plate and gently scrap the surface of fungal colony with a sterile, plastic spreader.
Collect the conidia suspension from petri dish and transfer into a 15 ml Falcon tube.
Centrifuge the conidia suspension at 5,000 × g for 5 min. Discard supernatant and re-suspend conidia pellet with 5 ml of PBS-0.01% Tween 20 buffer.
Dilute the conidia suspension with PBS-0.01% Tween 20 buffer to a final concentration between 1 to 2 × 106 conidia/ml. Quantify the number of conidia using a hemocytometer (Figure 3).
-
Pseudomonas and Aspergillus co-culture biofilm preparation
-
Pour 10 ml of autoclaved YPD solid medium (1.6% agar) (bottom layer) into a 60 × 15 mm petri dish and solidify at room temperature.
Note: Prepare multiple YPD agar plates for different co-culture time points and replicates.
Autoclave YPD solid medium (0.5% agar) (top layer) and cool in 50 °C water bath for 15 min.
After YPD top layer is cooled, inoculate with freshly prepared and vortex-mixed Aspergillus conidia suspension to a final concentration of 3.33 × 105 conidia/ml.
-
Mix the YPD top layer containing conidia thoroughly and pour 3 ml of the mixed top layer onto the solidified bottom layer.
Note: High temperature of autoclaved agar will kill Aspergillus conidia. Thus, it is critical to ensure that the temperature of YPD top layer is cooled enough to the extent that medium bottle can be held with hands. In the meanwhile, agar will solidify if the temperature drops too much. Steps A3c-d should be done as quickly as possible.
-
Once the top layer is solidified, incubate the plate (fungal biofilm facing up) at 25 °C for 12 h.
Note: A. fumigatus grows slower than P. aeruginosa. 12 h pre-culture is necessary to avoid overwhelming bacterial growth.
Spot-inoculate 10 µl aliquots of freshly prepared early stationary phase P. aeruginosa PA14 LB liquid culture in the center of the performed fungal lawn.
-
Incubate the co-culture biofilm (co-culture biofilm facing up) at 25 °C for an additional 7 days.
Notes:
See Figure 5 for an example of the co-culture biofilm.
It is critical to synchronize the growth of P. aeruginosa (24 h) and A. fumigatus (5 d) prior to the preparation of co-culture biofilm.
-
-
-
Metabolite extraction
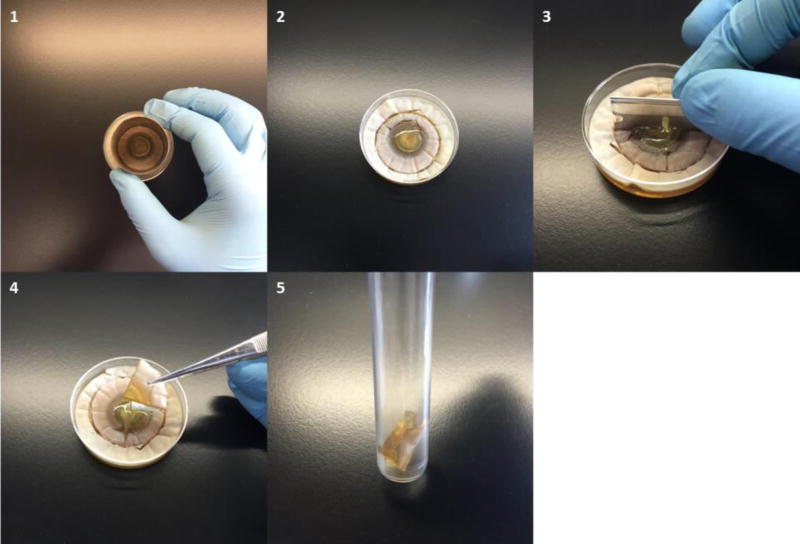
After different periods of growth (for example, every two days after inoculation), cut the co-culture biofilm with a sterile steel corer (38.1 mm in diameter), which encompasses the co-culture biofilm.
Cut the 38.1 mm diameter extraction core into four, equal quarters using a sterile blade.
-
Transfer one quarter (about 285 mm2 of surface) to a glass culture tube.
Note: Transfer both co-culture biofilm and supporting agar (Figure 4).
Add 4 ml milli-Q water and homogenize using a pestle.
Add 4 ml chloroform, cap the tube, and vortex vigorously for 15 min.
Transfer the mixture to a 15 ml glass centrifuge tube.
Centrifuge at 500 × g for 10 min.
Collect water phase supernatant and filter through a 0.2 µm-pore-size hydrophilic filter.
Collect 1.5 ml of chloroform phase to a 10 ml glass vial.
Evaporate chloroform to dryness by using a gentle flow of nitrogen.
-
Re-dissolve the dried extract with 1.5 ml mixture of 50/50% acetonitrile/water solution.
Note: This dissolution solvent can be used for the assessment of metabolites that have similar chemical property as Pseudomonas phenazines, which were analyzed in Zheng et al. (2015).
-
If extracted sample contains metabolites that are unstable in acetonitrile such as 5-methoxy-phenazine-1-carboxylic acid (5-Me-PCA), collect another 1.5 ml aliquot of chloroform phase after step B9, evaporate to dryness under a gentle flow of nitrogen and re-dissolve in 1.5 ml milli-Q water.
Note: Most bacterial and fungal metabolites are light sensitive, thus, it is strongly suggested to protect extracts from light by wrapping all containers with aluminum foil.
Figure 2.
Example of A. fumigatus AF293 streak culture after 5 days of incubation at 37 °C
Figure 3.
Example of A. fumigatus conidia under microscope
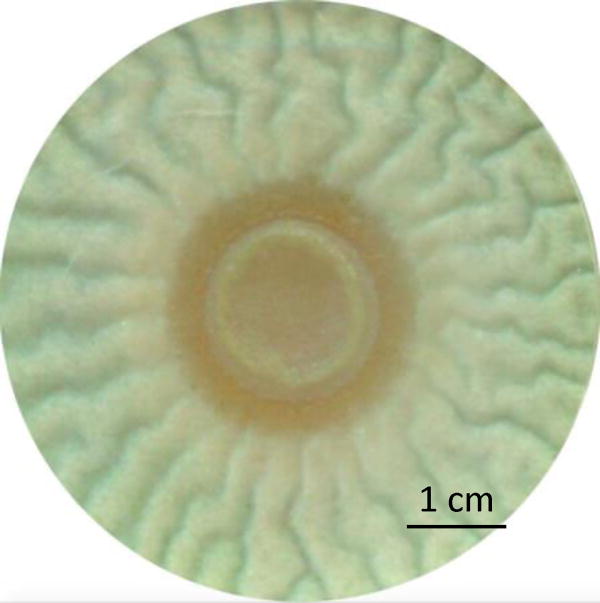
Figure 5.
Representative image of the co-culture biofilm of Pseudomonas aeruginosa PA14 wild-type (center colony) and Aspergillus fumigatus AF293 wild-type (wrinkled biofilm surrounding bacterial colony) after 6 days of incubation at 25 °C
Figure 4.
Example of cutting co-culture plate for extraction
Representative data
Notes
It is critical to keep the same pre-culture condition for both bacteria and fungi before co-culture inoculation. For fungi, you need to ensure the same pre-culture time. For bacteria, early stationary phase culture is recommended in this experiment. OD600 can be measured to check bacteria growth. If you use different fungal and/or bacterial strains, it is strongly recommended to do a series of co-cultures with different pre-culture time to determine a condition that gives the best result.
Co-culture biofilms can grow in various petri dishes of different sizes. You can scale up or down the dimension of co-cultures based on your research need. Note that the co-culture biofilm growth may vary in different types of petri dishes.
Recipes
-
PBS-0.01% Tween 20 buffer (1 L)
NaCl 8 g KCl 0.2 g Na2HPO4 1.42 g KH2PO4 0.24 g Tween 20 solution 0.1 ml Mix in about 900 ml milli-Q water and adjust pH to 7.2 with 5 M NaOH solution
Adjust volume with milli-Q water to 1 L and autoclave
-
20× sodium nitrate salts solution
NaNO3 120 g KCl 10.4 g MgSO4·7H2O 10.4 g KH2PO4 30.4 g Add milli-Q water up to 1 L
Autoclave and store at room temperature
-
Trace elements (100 ml)
ZnSO4·7H2O 2.2 g H3BO3 1.1 g MnSO4·H2O 0.43 g FeSO4·7H2O 0.5 g CoCl2·6H2O 0.17 g CuSO4·5H2O 0.16 g (NH4)6Mo7O24·4H2O 0.11g Na4EDTA·2H2O 4.9 g Add solids into 80 ml milli-Q water, dissolving each completely before adding the next
Add 20 ml of 1 M NaOH solution to adjust the pH to 6.5
Mix solution until solids are completely dissolved
-
Medium recipes
-
Solid glucose minimal medium (GMM) (1 L)
20× sodium nitrate salts 50 ml Trace elements (mix before using) 1 ml D-glucose 10 g Agar-agar 16 g Mix in 900 ml milli-Q water and adjust pH to 6.5 with 5 M NaOH solution
Adjust volume with Milli-Q water to 1 L
-
Solid yeast extract-peptone-dextrose medium (YPD) (1 L)
Anhydrate dextrose 20 g Yeast extract 10 g Bacto peptone 20 g Agar-agar 16 g Trace elements (mix before using) 1 ml Mix in 900 ml milli-Q water, adjust volume with milli-Q water to 1 L, and autoclave
-
Solid Luria-Bertani (LB) dehydrated medium (1 L)
Luria-Bertani medium powder 25 g *Agar-agar 16 g Mix in 900 ml milli-Q water, adjust volume with milli-Q water to 1 L, and autoclave
Note: *Do not add agar when preparing liquid LB medium.
-
Acknowledgments
This work was supported by startup and ISEN funding from Northwestern University (to Y. W.), NIH Grant R01 GM 067725 (to N. L. K.), and National Science Foundation Grant Emerging Frontiers in Research and Innovation 1136903 (to N. P. K.). Protocol was adapted from Zheng et al. (2015).
References
- 1.Amin R, Dupuis A, Aaron SD, Ratjen F. The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in patients with cysticfibrosis. Chest. 2010;137(1):171–176. doi: 10.1378/chest.09-1103. [DOI] [PubMed] [Google Scholar]
- 2.Frey-Klett P, Burlinson P, Deveau A, Barret M, Tarkka M, Sarniguet A. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev. 2011;75(4):583–609. doi: 10.1128/MMBR.00020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nat Rev Microbiol. 2010;8(5):340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 4.Shimizu K, Keller NP. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics. 2001;157(2):591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng H, Kim J, Liew M, Yan JK, Herrera O, Bok JW, Kelleher NL, Keller NP, Wang Y. Redox metabolites signal polymicrobial biofilm development via the NapA oxidative stress cascade in Aspergillus. Curr Biol. 2015;25(1):29–37. doi: 10.1016/j.cub.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]