A globally distributed yet previously overlooked marine nitrite oxidizer can reduce nitrate, produce N2O, and oxidize sulfide.
Abstract
Nitrite-oxidizing bacteria (NOB) have conventionally been regarded as a highly specialized functional group responsible for the production of nitrate in the environment. However, recent culture-based studies suggest that they have the capacity to lead alternative lifestyles, but direct environmental evidence for the contribution of marine nitrite oxidizers to other processes has been lacking to date. We report on the alternative biogeochemical functions, worldwide distribution, and sometimes high abundance of the marine NOB Nitrococcus. These largely overlooked bacteria are capable of not only oxidizing nitrite but also reducing nitrate and producing nitrous oxide, an ozone-depleting agent and greenhouse gas. Furthermore, Nitrococcus can aerobically oxidize sulfide, thereby also engaging in the sulfur cycle. In the currently fast-changing global oceans, these findings highlight the potential functional switches these ubiquitous bacteria can perform in various biogeochemical cycles, each with distinct or even contrasting consequences.
INTRODUCTION
Nitrite oxidation is the major pathway that generates nitrate, the dominant form of biologically available nitrogen and often a limiting nutrient to biological production in surface oceans. The prevalence of nitrate below surface, however, attests to the substantial occurrence of nitrite oxidation in the ocean interior, which represents a key conduit to resupply nitrogen to the surface ocean and is thus essential to the global nitrogen cycle (1). Nevertheless, current knowledge of the physiology of marine nitrite-oxidizing bacteria (NOB) remains poor, with only a few species described to date that belong to the genera Nitrobacter, Nitrospira, Nitrospina, Candidatus Nitromaritima, or Nitrococcus (2, 3). Although several studies have challenged the conventional understanding of nitrate production as the only relevant contribution of NOB to global biogeochemical cycling (4–7), no direct environmental evidence of marine NOB participating in elemental cycles other than nitrogen and carbon has been reported to date.
Most recent research on marine nitrite oxidizers has been heavily biased toward Nitrospina, which can be found in a wide range of habitats, including open ocean (8), sediments (9, 10), and suboxic water columns (11, 12). Experimental and genome analyses of the two cultured Nitrospina strains, Nitrospina gracilis and Nitrospina watsonii, strongly suggest that these organisms are chemolithoautotrophs dependent on nitrite oxidation for energy conservation (13–15). In comparison, Nitrococcus, with only one species known to date, Nitrococcus mobilis, has had its presence reported only twice in the 1980s in the Pacific (16, 17) and once in a high-salinity pond (18) since its first isolation (13); its presence has not been reported again until its recently recorded abundance in the Namibian oxygen minimum zone (OMZ) where they even outnumbered Nitrospina (11). In that study, Nitrococcus and Nitrospina were the only NOB genera detectable via catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) and together comprised almost 10% of the total microbial community. Although Nitrospina use a putatively high-affinity cbb3-type terminal oxidase, which should allow them to catalyze nitrite oxidation even under hypoxic conditions (14), the adaptation strategy of Nitrococcus to oxygen-deficient conditions remains unclear. Nitrospina preferred residing in the oxycline and the upper OMZ, whereas Nitrococcus, in contrast, became more abundant in the lower OMZ (4.9% of total microbial abundance) where oxygen levels were minimal (11). Here, a similar trend was again observed for October 2011, with Nitrospina contributing up to 1.3% of total microbial community in the upper OMZ (65 m, 95 μM O2), whereas Nitrococcus displayed maximal abundance (1.3%) at 119 m (16 μM O2) (Fig. 1A). Overall, Nitrococcus showed a moderate yet significant negative correlation with oxygen (Spearman correlation = 0.44, P < 0.01) and was undetectable at oxygen concentrations >20 μM (fig. S1A).
Fig. 1. Occurrence and distribution of Nitrococcus in a marine OMZ and the global oceans.
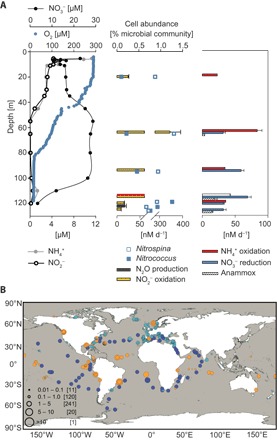
(A) Water column chemistry and nitrogen cycling rates in the Namibian OMZ. Left: Vertical distribution of oxygen, NO2−, NH4+, and NO3− at station 2. Middle: Nitrite oxidation rates, N2O production rates, and cellular abundance of Nitrococcus and Nitrospina. Right: Rates of ammonia oxidation, nitrate reduction, and anammox measured with 15N-labeling experiments. Error bars show SDs derived from linear regression analyses. (B) Worldwide distribution of Nitrococcus mobilis and Nitrococcus-like phylotypes in the oceans. Combined abundance of all four phylotypes in different metagenomic data sets is denoted in orange (MG-RAST), turquoise (OSD), and blue (Tara Oceans). Circle size indicates percentage of 16S rRNA gene reads relative to all bacterial and archaeal 16S rRNA gene reads within each sample.
To investigate whether Nitrococcus is restricted to certain environmental settings, we searched for Nitrococcus-like sequences in publicly available databases. Three novel Nitrococcus-like phylotypes, in addition to N. mobilis, were identified in the SILVA 16S ribosomal RNA (rRNA) database (19, 20), sharing 97.8 to 99.2%, 96.5 to 97.8%, and 96.6 to 97.3% sequence identities with N. mobilis (fig. S2 and table S1). Subsequent searches for all phylotypes’ 16S rRNA genes in marine amplicon and metagenomic data sets from the MetaGenome Rapid Annotation using Subsystems Technology (MG-RAST) server (21), Ocean Sampling Day 2014 (OSD) (22), and Tara Oceans (23) revealed the presence of N. mobilis and Nitrococcus-like phylotypes in 69% of all data sets examined. Among the data sets that carry geographical information, N. mobilis and Nitrococcus-like phylotypes were found to be globally distributed from coastal upwelling regions to oligotrophic open oceans, from tropical to temperate and polar latitudes, and from epipelagic (surface sunlit layer, ~0 to 100 m) to mesopelagic (twilight zone, ~100 to 1000 m) ocean waters (Fig. 1B and tables S2 and S3). Considering only metagenomic data sets (table S2) and excluding the more bias-prone amplicon sequencing data (table S3), higher contributions of N. mobilis and Nitrococcus-like phylotypes to total microbial community (>5%, up to 17%) were nearly exclusively associated with highly productive environments, such as the Peruvian OMZ and Guanabara Bay or with hypersaline lagoons, indicating an ecological advantage for them under these conditions (Fig. 1B and table S2). On the basis of Tara Oceans metagenomic data alone, which is currently the most comprehensive data set with consistent sequencing depths and covering the widest geographic range of global oceans to date, 16S rRNA genes affiliated with Nitrococcus were present in all samples and represented 1.2 to 5.1% (mean, 2.3%) of the microbial community (table S2).
We analyzed the genome of N. mobilis Nb-231 to identify potential functional roles Nitrococcus may play in such diverse environments. The resulting genome-derived hypotheses were tested in incubation experiments with both the pure culture isolate N. mobilis Nb-231 and Namibian OMZ seawater samples. Nitrococcus only encodes a putatively low-affinity aa3-type cytochrome c oxidase (Fig. 2 and table S4), thus ruling out a specialization on microaerobic respiration as suggested for Nitrospina (14). Instead, its abundance in OMZ waters indicates the utilization of alternative anaerobic metabolisms. In agreement with this hypothesis, Nitrococcus was abundant even in waters where no nitrite oxidation was detectable in the Namibian OMZ, and no correlation with nitrite oxidation rates was observed (Spearman correlation = 0.05, P > 0.05, n = 35) (Fig. 1A and fig. S1B). These findings suggest that NOB other than Nitrococcus, such as Nitrospina, were responsible for most of the nitrite oxidation detected in the Namibian OMZ (Fig. 1A).
Fig. 2. Cell metabolic cartoon based on the annotation of the N. mobilis Nb-231 genome.
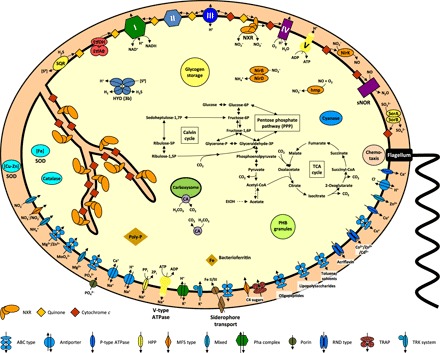
ETF, electron-transport flavoprotein; hmp, NO dioxygenase; HYD, hydrogenase; NirBD, assimilatory nitrite reductase; NirK, copper-containing nitrite reductase (NO-forming); NXR, nitrite oxidoreductase; sNOR, NO reductase; PHB, polyhydroxybuterate; poly-P, polyphosphates; SOR, sulfite dehydrogenase; SQR, sulfide/quinone oxidoreductase; PPP, pentose phosphate pathway; TRAP, tripartite ATP-independent periplasmic transporter (TRAP-T); TRK, K+ transporter; RND, resistance-nodulation-division; ATPase, adenosine triphosphatase; ABC, ATP-binding cassette. Enzyme complexes of the electron transport chain are labeled by Roman numerals. Red and orange diamonds represent cytochrome c proteins and quinones, respectively. The legend indicates transport protein classifications.
On the other hand, Nitrococcus abundance was significantly correlated with nitrate reduction rates (fig. S1C) (Spearman correlation = 0.54, P < 0.05), suggesting a contribution to this anaerobic process. The key enzyme for nitrite oxidation, nitrite oxidoreductase (NXR), is reversible, and nitrate reduction activities have previously been observed in Nitrobacter (24, 25) and Nitrospira (6). When the N. mobilis pure culture was subjected to anoxic conditions in the presence of formate and acetate, substrates that have previously been reported to support mixotrophic or heterotrophic growth in NOB (6, 25), NO3− was reduced at a rate of 3.66 ± 0.32 fmol per cell per day (±SD) within the first 230 hours of incubation (Fig. 3A). Because the N. mobilis genome does not encode any other known nitrate-reducing enzyme, the NXR of Nitrococcus is most likely the candidate for nitrate reduction (Fig. 2). Furthermore, N. mobilis encodes duplicated versions of NXR alpha subunit (NxrA), the substrate-binding catalytic subunit of the enzyme complex (fig. S3) (26, 27). NxrA1 is encoded in one genomic region together with genes for the NXR beta, delta, and gamma subunits. The duplicated subunit NxrA2 (fig. S3), with an amino acid identity of 89% to NxrA1, is not clustered with any other genes known to be involved in nitrite oxidation. This duplication of the catalytic NxrA subunit alone has also been observed in all sequenced Nitrobacter genomes and in Nitrolancea hollandica (28, 29). It is tempting to speculate that Nitrococcus uses one NxrA paralog for nitrite oxidation and the other for the reduction of nitrate, but biochemical verification will be necessary.
Fig. 3. Denitrifying potential of Nitrococcus mobilis Nb-231.
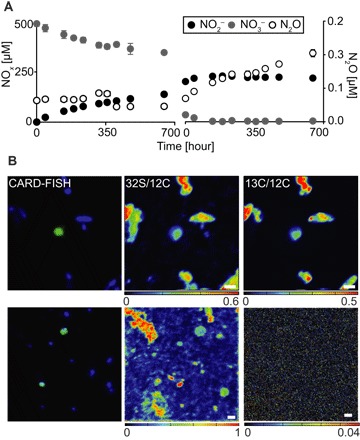
(A) Anaerobic nitrogen dissimilation under (left) organoheterotrophic conditions (500 μM 15NO3−, 250 μM acetate, and 250 μM formate, anoxic) and (right) without the addition of electron donor (200 μM 15NO2−, anoxic). See the main text for more details. Error bars indicate SD of three biological replicates. (B) FISH-SIMS analyses of single Nitrococcus cells in Namibian OMZ bottom water samples (1.20 m above seafloor), incubated with (top) 13C-labeled dissolved organic matter (DOM) or (bottom) 13C-labeled bicarbonate under hypoxic conditions. Left: Nitrococcus cells (green) targeted by CARD-FISH among other microbial cells (blue). Middle: 32S/12C of the corresponding cells. Right: 13C/12C ratio. Scale bars, 1 μm.
The observed switch to NO3− reduction with formate and acetate indicates that Nitrococcus can use organic matter as energy and carbon source. Although whether this truly translates to growth remains to be empirically verified, chemoorganoheterotrophic and autotrophic potentials of Nitrococcus are corroborated by the presence of all genes for the oxidative TCA (tricarboxylic acid) cycle, glycolysis, PPP (pentose phosphate pathway), and the Calvin-Benson-Bassham cycle in the genome (Fig. 2 and table S4). Genome annotation also revealed genes for respiratory complexes I to V (Fig. 2 and table S4). Complexes I to III are not required for energy conservation during nitrite oxidation, unlike during the oxidation of organic substrates, but they do mediate the reverse electron flow from nitrite to NAD+ during chemolithoautotrophic growth. An ETF (electron-transport flavoprotein) complex and an ETF/quinone oxidoreductase probably couple β-oxidation of fatty acids to the respiratory chain.
Consistent with the pure culture experiments and genome analyses, uptake of organic carbon was also demonstrated by single-cell analyses of uncultured Nitrococcus in hypoxic Namibian OMZ seawater samples (O2 < 2 μM, NO3− > 15 μM). Upon incubation with 13C-labeled algal DOM, fluorescence in situ hybridization–secondary ion mass spectrometry (FISH-SIMS) analyses (30) of individual Nitrococcus cells showed substantial uptake of 13C-DOM after 29 hours (0.3 to 48.1 atomic % 13C) (Fig. 3B and table S5). The possibility of 13C enrichment as a result of rapid remineralization of 13C-DOM by other organisms followed by lithoautotrophic uptake of 13C-HCO3− by Nitrococcus can be largely excluded, as no detectable 13C enrichment was observed in Nitrococcus cells in parallel anoxic incubations with 13C-HCO3− and NO2− (Fig. 3C). The high 13C enrichment in combination with our findings from genome analyses and pure culture experiments strongly indicate a chemoorganoheterotrophic potential of Nitrococcus under NO3− reducing conditions. The broad range of 13C enrichment observed in our incubations (table S5) can likely be attributed to the presence of different Nitrococcus phylotypes or phenotypic heterogeneity of individual cells within a population (31).
When exposed to elevated nitrite levels (~200 μM), pure cultures of N. mobilis continuously produced small but significant amounts of N2O during anoxic incubations [3.2 (±0.48) × 10−3 fmol per cell per day during the first 230 hours] (Fig. 3B and fig. S4). No organic substrates were amended in these incubations, and denitrification was likely driven by the oxidation of dissolved organic carbon present in the sterilized seawater used for medium preparation (table S6) and/or the oxidation of intracellular storage compounds such as glycogen (Fig. 2) (13). N. mobilis encodes a copper-containing nitrite reductase (NirK) (table S4), likely facilitating nitrite reduction to nitric oxide (NO). None of the enzymes known to be responsible for the reduction of NO to N2O—cytochrome c or quinone-dependent nitric oxide reductase (cNOR or qNOR, respectively)—were identified in the genome. However, the Nb-231 genome carries the genes for a two-subunit oxidase that belongs to a distinct phylogenetic cluster within the haem-copper oxidase family (table S4 and fig. S5). This enzyme group is designated the sNOR family and likely functions as NO reductases to detoxify NOx intermediates produced in the periplasm, as proposed for ammonia-oxidizing bacteria (32). To our knowledge, this is the first time that partial denitrification (NO3− or NO2− reduction to N2O) has been shown for an organism apparently containing this type of NOR only, thus giving indirect evidence that this enzyme family functions as NOR and might be used for energy conservation under denitrifying conditions. It should, however, be noted that the N. mobilis Nb-231 genome is not closed such that the definite absence of a canonical NOR is yet to be confirmed. Although we can exclude strictly abiotic N2O production based on our abiotic control incubation (fig. S4), we cannot rule out abiotic-biotic hybrid N2O formation, in which the NO formed from NirK-mediated NO2− reduction may react abiotically with medium components to give N2O. This has recently been reported to be the main N2O production pathway by ammonia-oxidizing archaea that also lack a known NOR (33). A nitrous oxide reductase (for reduction of N2O to N2) has not been identified in the genome, which is consistent with the lack of detectable N2 production in our pure-culture experiments.
OMZs are a major global source of oceanic N2O (25 to 59%) (34), a potent greenhouse gas and ozone-depleting agent that has usually been attributed to denitrifying and ammonia-oxidizing microorganisms (35, 36). In the Namibian OMZ, N2O production (~17 nM day−1) was detected particularly in the bottom waters where Nitrococcus abundance was maximal and nitrite oxidation was minimal (Fig. 1A), strongly indicating a contribution from Nitrococcus. Considering the worldwide distribution and high abundance of N. mobilis and Nitrococcus-like phylotypes especially within productive coastal ecosystems, Nitrococcus might represent a previously overlooked yet significant source of oceanic N2O. Because OMZs are expanding and intensifying globally (37), Nitrococcus may switch from a nitrogen-preserving (NO2−-oxidizing) mode to one that facilitates nitrogen loss (NO3−/NO2− reduction to N2O).
Apart from their importance in nitrogen cycling, Nitrococcus may also affect sulfur cycling. The N. mobilis genome harbors several key sulfur-metabolizing enzymes, including a sulfide/quinone oxidoreductase (sqr) that might oxidize hydrogen sulfide (H2S) to elemental sulfur or polysulfide (S0) in the periplasmic space with concomitant reduction of quinone (Fig. 2 and table S4). In addition, a periplasmic sulfite dehydrogenase (sor) may oxidize sulfite to sulfate and transfer the electrons onto cytochrome c. Last, a cytoplasmic type 3b bidirectional hydrogenase (38) is present (Fig. 2 and table S4). This so-called sulfhydrogenase can catalyze the reversible oxidation of H2 with NAD(P)+ (39) and has been shown to reduce S0 to H2S in the hyperthermophilic archaeon Pyrococcus furiosus (40).
The ability of Nitrococcus to catalyze sulfide oxidation in the presence of O2 was confirmed with N. mobilis pure cultures, which consumed sulfide at a rate of 43 nmol S per microgram of protein per day, whereas the disappearance of sulfide in the abiotic control was negligible even during 5 hours of incubation (Fig. 4B). To our knowledge, this is the first time that capability for sulfur oxidation has ever been reported in association with nitrifying organisms, adding another remarkable facet to the versatile metabolic repertoire already known in NOB.
Fig. 4. Sulfide oxidation potential of N. mobilis Nb-231.
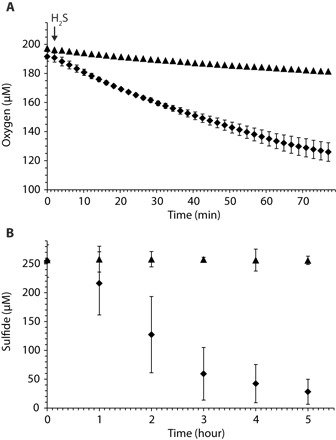
(A) Oxygen consumption in the presence (diamonds; mean of three replicates) and absence (triangles; single measurement) of Na2S. Only one measurement every 2 min is shown. The arrow indicates the time point of substrate addition. (B) Sulfide consumption by N. mobilis Nb-231 (diamonds) and abiotic control (triangles). The curves represent the mean of three biological replicates, and error bars indicate the SD of three technical replicates.
Among 20 screened marine environmental metagenomes, 50% were found to contain sqr genes with high sequence identities to the N. mobilis sequence, indicating the prevalent potential of Nitrococcus to oxidize reduced S-compounds in the oceans. More than half of the metagenomes from mesopelagic habitats contain N. mobilis–like sqr, but none do from the deep chlorophyll maxima and epipelagic ocean (fig. S6 and table S7). Hence, sulfide-oxidizing capacity seems to present an ecological advantage for Nitrococcus in subsurface waters: This could mean a direct coupling with growth, or facilitation of sulfide detoxification and/or preservation of biologically available sulfur (41, 42).
The prevalence of this functional trait of Nitrococcus in the mesopelagic is consistent with the recently reported widespread occurrence of sulfide-oxidizing enzyme complexes in the mesopelagic ocean and within the Global Ocean Sampling (GOS) metagenomic data sets, despite the apparent lack of detectable sulfide (42–44). On the basis of the 138 Tara Oceans data sets that distinguish mesopelagic from surface and deep chlorophyll maximum (DCM) samples, N. mobilis and Nitrococcus-like phylotypes appeared to be particularly abundant in the mesopelagic ocean (mean, 3.3%, versus 2 and 2.1% for surface and DCM, respectively) (table S2). Particulate organic matter, both sinking and suspended, serves as a major carbon source for heterotrophic communities in the mesopelagic ocean. Although Nitrococcus does not necessarily rely on these particles for their carbon demand, active remineralization by other microbes may create oxygen-deficient microniches and facilitate anaerobic processes within marine snow (45–47). Moreover, remineralization processes release inorganic and organic nutrients, including reduced sulfur compounds, proteins, or osmolytes such as dimethylsulfoniopropionate that may further be metabolized to release sulfide (42, 43). Nitrococcus, sometimes found enriched on particles (11), could then use these reduced sulfur compounds to support sulfide oxidation. The actual occurrence and ecological significance, however, remain to be determined in situ.
In coastal OMZs associated with highly productive upwelling areas, such as the Namibian and Peruvian OMZs, sulfide sometimes accumulates to levels that are detrimental to marine life and fisheries. Previous studies have shown that sulfide-oxidizing bacteria affiliated with Gammaproteobacteria and Epsilonproteobacteria participate in detoxifying the harmful sulfide in these oxygen-deficient zones (48, 49). The relatively high abundance of 16S rRNA genes affiliated with N. mobilis and Nitrococcus-like phylotypes (up to 9.2%; table S2) in Peruvian OMZ metagenomes during a sulfidic event (49) indicates that they are well adapted to such episodic sulfidic conditions and may hint toward a contribution to sulfide detoxification in these waters (48, 49).
NOB are important preservers of biologically available nitrogen in global oceans. Their significance in OMZs, specifically in counteracting nitrogen loss through anammox and denitrification, which both reduce NO2− to N2, has been increasingly recognized in recent years through direct activity measurements, molecular surveys, and natural stable isotope analyses (11, 12, 50). Thus far, nitrite oxidation is postulated to have prevented oxygen-deficient water bodies from turning into active nitrogen-loss zones (51). This nitrogen conservation role is most certainly becoming more crucial as global ocean deoxygenation continues (37). Nonetheless, our findings of relatively high abundance of Nitrococcus in these waters and of the diverse alternative biogeochemical functions Nitrococcus may perform suggest that they may be on the verge of reversing the NOB community’s usual role to instead reduce nitrate and produce N2O, thus promoting nitrogen loss and exacerbating the global greenhouse effect.
In a wider context, the remarkably versatile and sometimes contrasting functional capacities of Nitrococcus unveiled in our study—nitrate reduction versus conventional nitrite oxidation, heterotrophy (degradation of organic carbon releasing CO2) versus autotrophy (CO2 fixation into organic carbon), along with N2O production and sulfide oxidation—have conferred on these organisms appreciable adaptability and robustness toward dynamic, changing environmental conditions. Their success is shown by their worldwide distribution across diverse habitats, with particular prominence in dynamic settings such as coastal upwelling, or the vast mesopelagic ocean where resources likely come in pulses. Hence, potential occurrence of any functional switches of Nitrococcus could extend beyond oxygen-deficient waters to the wider ocean and may substantially affect the cycling and budgets of nitrogen, carbon, oxygen, and sulfur in global oceans as a result. In the face of rapid global environmental changes, the key regulatory controls and the tipping points of contrasting biogeochemical functions associated with the ubiquitous Nitrococcus thus urgently need to be determined.
MATERIALS AND METHODS
Sample collection
Water sampling was conducted on board the R/V Maria S. Merian in October 2011 (MSM19/1c) over the Namibian shelf between 21°59.9′S/13°40.92′E and 28°30′S/15°45.01′E. Salinity, temperature, dissolved oxygen, and chlorophyll a fluorescence were measured with a conductivity-temperature-depth (CTD) system, equipped with an oxygen sensor and a fluorometer (Sea-Bird Electronics). Water samples were collected with 12-liter Niskin bottles attached to the CTD system. Oxygen data were calibrated against Winkler titration. The benthic boundary layer (BBL) was sampled at three depths from 30 cm to 2 m above seafloor using a bottom water sampler (52). NO2− and NH4+ were measured on board spectrophotometrically (53) and fluorometrically (54). Water samples were frozen for later NO3− and PO43− analyses with an autoanalyzer in a shore-based laboratory (TrAAcs 800, Bran & Luebbe). Detection limits for NH4+, NO2−, NO3−, and PO43− were 0.01, 0.01, 0.1, and 0.1 μM, respectively.
Incubation experiments with environmental samples
Incubation experiments were conducted with water collected from two stations in the Namibian OMZ, located at 22°S/13°4′E and 22°6′S/16°6′E. Water samples (250 ml) were collected from five to six depths with the CTD system or the bottom water sampler for samples from the BBL. Different combinations of 15N and 14N substrates were added for rate determinations of ammonia and nitrite oxidation as well as nitrate reduction (table S8). Samples were purged with helium for 15 min to eliminate any O2 contamination from the CTD system (55). For NH3 and NO2− oxidation, a second set of incubation experiments without helium purging was conducted. Each 15N(/14N)-amended sample was immediately transferred into five 12-ml Exetainer vials (Labco). These samples were incubated for up to 48 hours in the dark at in situ temperatures. At each time interval (approximately 0, 6, 12, 24, and 48 hours), incubation in one Exetainer was terminated by adding 100 ml of saturated mercuric chloride solution to stop biological activities. Samples were stored upside down in the dark at room temperature until further processing in a shore-based laboratory [for a more detailed description, see the study of Holtappels et al. (56)].
Stable isotopic analyses of N2
Nitrogen-stable isotopic ratios of N2 were determined by gas chromatography–isotope ratio mass spectrometry (GC-IRMS; VG Optima). Ammonia oxidation rates and nitrate reduction rates were determined as the 15NO2− production over time from incubations amended with 15NH4+/14NO2− or 15NO3−/14NO2− via the conversion of NO2− by sulfamic acid (57). Nitrite oxidation rates were determined in incubations amended with 15NO2−/14NO3−. After the removal of residual NO2−, NO3− was reduced to NO2− with cadmium and subsequently to N2 via sulfamic acid, as previously described (11). Process rates were calculated from the slopes of linear regressions with 15N2 production as a function of time, and only when the production was instantaneous. All rates were calculated as net rates from five samples obtained over the course of incubation (0 to 48 hours) (P < 0.05) and have been corrected for the 15N-labeling percentages of initial substrate pools.
CARD-FISH
Water samples were fixed with 2% (final concentration) paraformaldehyde (PFA) in phosphate-buffered saline solution for 8 to 12 hours at 4°C, followed by filtration onto polycarbonate membrane filters (GTTP, 0.22 μM pore size, 47 mm diameter; Millipore). CARD-FISH was performed according to the protocol by Pernthaler et al. (58). First, cells were immobilized on the GTTP filters by embedding in 0.2% agarose, followed by permeabilization in lysozyme (10 mg/ml) in 50 mM EDTA and 100 mM tris-HCl. Labeled oligonuctleotide probes (Biomers) were added and allowed to hybridize for 2 hours at 46°C at varying formamide concentrations, depending on the oligonucleotide applied (table S9). This was followed by tyramide signal amplification for 15 min at 46°C and subsequent staining of cells with DAPI (4′,6′-diamidino-2-phenylindole). DAPI-stained cells and positive hybridization signals were enumerated with epifluorescence microscopy (Axioplan 2, Zeiss).
Pure culture experiments
N. mobilis strain Nb-231 was grown in aerobic batch cultures that were moderately stirred for 32 days in 3.5 liters of marine NOB medium (table S6). NO2− was added to a final concentration of 3 mM and resupplied when consumed. The cultures had reached stationary phase when the various incubation experiments were conducted.
For anoxic incubation experiments, cultures were harvested by centrifugation and transferred into 2 liters of anaerobic marine NOB medium, including the various amendments for experiments 1 to 8 as listed in table S10. Subsequently, each experimental step was conducted in an anaerobic chamber (90% N2/10% CO2 atmosphere) to avoid oxygen contamination. One hundred fifty milliliters of culture per incubation was transferred into 250-ml serum bottles. All experiments were conducted in triplicate. Control incubations with autoclaved culture were included to test for abiotic reactions (fig. S5B). Through the duration of 28 days, a total of 10 subsamples consisting of 3-ml culture and 2-ml headspace were obtained every 2 to 3 days. The sampling volume was replaced with 90% N2/10% CO2 from the anaerobic chamber to avoid underpressure in the incubation vessels. Sulfide-dependent oxygen consumption was measured by using a respiration cell RC-350 (Warner Instruments), equipped with a Clark-type electrode (model 1302) connected to a picoammeter PA2000 (Unisense). Dissolved oxygen concentration was recorded in SensorTrace Basic version 3.0.2 (Unisense), with one measurement every 2 s. Measurements were performed by using approximately 25× concentrated N. mobilis Nb-231 biomass from actively nitrite-oxidizing cultures. Biomass was harvested by centrifugation (4000g for 15 min), washed twice, and resuspended in marine NOB medium. For measurements, the cell chamber was filled with 2 ml of biomass suspension and closed with the electrode inserted into the general electrode holder EH-100 (Warner Instruments) without enclosing air bubbles. Measurements were performed at 28°C. Na2S was added to an end concentration of 5 mM with a 250-μl glass syringe (Hamilton Gastight, #1725). Biomass without substrate was used as a negative control to measure the electrode drift.
To determine sulfide oxidation in aerobic batch incubation assays, 200 ml of actively nitrite-oxidizing N. mobilis Nb-231 culture was harvested and washed, as described above, and resuspended in 100 ml of marine NOB medium. Ten milliliters of nitrite-free culture per incubation was transferred to 30-ml serum bottles, and 250 μM Na2S was added. Overpressure in the bottles was ensured by adding 5-ml sterile air to each incubation. Incubations were performed in a shaking incubator at 28°C. Cell-free medium was used as a negative control. One milliliter of headspace was sampled every hour. Sulfide concentrations were determined by gas chromatography. One milliliter of approximately 557× diluted headspace samples was injected in triplicate into a gas chromatographer [Agilent 7890B, equipped with a Chrompack ML 848 glass column (2 m × 4 mm) with Carbopack B HT 100 40 to 60 mesh packing, at 80°C; Agilent Technologies] combined with a flame photometric detector (Agilent Technologies).
N2O and N2 measurements
The gas samples were injected into sterile water–filled 3-ml Exetainers, and the water was displaced via a compensation needle. The concentration of N2O in the headspace was measured by a gas chromatograph with a 63Ni electron capture detector (Shimadzu, GC-8A). The linear increase over time was used to calculate N2O production rates by Nitrococcus. Nitrogen-stable isotopic ratios of N2 (15N15N/14N14N and 15N14N/14N14N) were determined by GC-IRMS (VG Optima). Rates were calculated from the slopes of linear regressions with 29N2/30N2 and N2O production as a function of time.
NanoSIMS analyses
During the cruise to the Namibian OMZ, 250-ml water samples were amended with either 13C-labeled DOM (13C-DOM) or 5 μM 15NO2− and 200 μM 13C-HCO13−, alongside incubations for rate measurements. After ~29 hours of incubation, samples were fixed with 1% PFA (final concentration) for 8 to 12 hours at 4°C. Subsequently, 11 ml of the sample was filtered onto gold-palladium precoated polycarbonate filters (GTTP, 0.2 μm pore size, 25 mm diameter; Millipore) and stored at −80°C until further analyses in a shore-based laboratory.
Nitrococcus cells were hybridized on a filter with Nitrococcus-specific CARD-FISH probes as described previously (11) and analyzed with a NanoSIMS 50L (Cameca). To obtain a stable ion emission rate and to clean the sample from any contamination, we presputtered the area of interest with a Cs+ primary ion beam of 100 pA. The cells were analyzed by rastering a primary Cs+ ion beam with a beam current of 0.8 to 1 pA and a beam diameter of <100 nm. Secondary ion images of 12C−, 13C−, 19F−, 12C14N−, and 32S− were recorded in parallel. The analyzed areas ranged from 10 × 10 μm to 20 × 20 μm with an image size of 256 × 256 pixels and a dwell time of 1 ms per pixel. Tuning of the instrument for high mass resolution (~7000 mass resolving power) reduced the interference for 13C. Data analyses were performed with the freeware Look@NanoSIMS (59).
Genome annotation of N. mobilis
The genome of N. mobilis Nb-231 was sequenced by the J. Craig Venter Institute in the framework of the Gordon and Betty Moore Foundation Marine Microbiology Initiative. The sequence was deposited in the GenBank database in 2006 and consists of 43 assembled contigs.
The draft genome of N. mobilis Nb-231 (GenBank project PRJNA13475) was integrated into the MicroScope annotation platform (60). After automated prediction and annotation of coding sequences (CDS), the annotation of all CDS in key pathways, including those for nitrite oxidation, respiration, and carbon fixation, was manually refined by using the respective tools of MaGe (60), as described in detail elsewhere (27).
Phylogenetic analyses
16S rRNA sequences classified as Nitrococcus were identified within the SILVA small subunit rRNA database release 119 (20). Affiliation with the genus was confirmed by phylogenetic analyses and by selecting for sequence identities ≥94.5% (61). The three N. mobilis 16S rRNA gene sequences (HM038001, L35510, and AAOF01000010) originate from the N. mobilis–type strain and the genome sequence. Nitrococcus-like phylotypes were defined on the basis of a 98.7% sequence identity threshold (19, 61). Only sequences ≥1300 bases were used for phylogenetic tree reconstruction. Shorter sequences were added to the tree using the parsimony insertion tool in ARB (62). NxrA and heme-copper oxidase (HCO) family member proteins were imported in custom-made databases.
Sequence alignments for all data sets were generated and manually refined using the sequence editor in ARB. Phylogenetic trees were calculated using RAxML version 7.0.4 (63), with 100 (NxrA and HCO) or 1000 (16S rRNA) bootstraps. To test for tree stability, other treeing methods (neighbor joining, maximum parsimony, and Bayesian interference) were used but yielded almost identical branching patterns. For 16S rRNA analyses, the GTRGAMMA substitution model and a 50% conservation filter resulting in 1515 valid alignment positions were used. NxrA and HCO trees were calculated using the PROTMIX rate distribution and the WAG substitution model, with a 10% conservation filter for NxrA and without a conservation filter for HCO, resulting in 1198 and 1367 distinct alignment positions, respectively.
Metagenomic data analyses
In total, 253 metagenomic and 422 amplicon data sets of marine origin were downloaded from MG-RAST (21), 150 metagenomic and 155 amplicon data sets were obtained from OSD 2014, as detailed in https://github.com/MicroB3-IS/osd-analysis/wiki/Guide-to-OSD-2014-data, and 138 samples of 16S rDNA miTags Illumina sequence data were obtained from http://ocean-microbiome.embl.de/companion.html as fasta files from the Tara Oceans project. Metadata for the data sets are provided in table S1. All sequence reads were processed with the next-generation sequencing analysis pipeline of the SILVA project (available at www.arb-silva.de/ngs). A cutoff of (BLASTN percent query coverage + BLASTN percent alignment identity)/2 > 97% was used to assign Nitrococcus-like phylotypes to a read. Of the 1118 data sets, 772 were found to contain at least one N. mobilis or Nitrococcus-like phylotype (tables S1 and S2). Of these, only metagenomic data sets (Fig. 1B) were used to create a world map showing the global distribution and relative abundance of N. mobilis or Nitrococcus-like phylotypes.
Selected OSD metagenomes, Tara, and MG-RAST samples were mapped to the N. mobilis Nb-231 genome (accession no. gi|211606481|ref|NZ_CH672427.1) using FR-HIT version 0.7 with the following parameters: -e 0.001 -u 0 -p 8 -c 80 -m (dynamically set to 40% of average read length in a sample). Hits for nitrogen and sulfur cycling genes of interest were considered significant when the e value was <10−5, and percent sequence identity was >80%. The results were evaluated on the basis of presence/absence of genes, rather than quantitatively, as the Nitrococcus species had rather low relative abundance in most data sets.
Supplementary Material
Acknowledgments
We thank M. Zabel (chief scientist; Marum Bremen), the crew of the R/V Maria S. Merian (MSM19/1c), K. Emeis (cruise leader; University of Hamburg), and the crew of the R/V Meteor (M76/2) for their support at sea. We also thank G. Klockgether, D. Tienken, D. Nini, A. Ellrott, and V. Meyer (Max Planck Institute for Marine Microbiology) for conscientious technical and analytical assistance. LABGeM and the national infrastructure “France Genomique” are acknowledged for providing access to the MaGe annotation system. Funding: This work was supported by the Max-Planck-Gesellschaft, the Deutsche Forschungsgemeinschaft, Bundesministerium für Bildung und Forschung (for cruises M76/2 and MSM 19/1c), the Vienna Science and Technology Fund (grant LS09-40), the Austrian Science Fund (grant P25231-B21), the Netherlands Organization for Scientific Research (VENI grant 863.14.019), and the U.K. Natural Environment Research Council (SONiC grant NE/N003187/1). Author contributions: J.F., S.L., P.Y., B.N., H.D., E.S., M.M.M.K., and P.L. designed the study. J.F. and P.F.H. collected environmental samples. J.F., S.L., B.N., M.A.H.J.v.K., J.B., and P.B. conducted pure culture experiments. J.F. was responsible for sample analyses. S.L. performed NanoSIMS analyses. S.L. annotated and analyzed the N. mobilis Nb-231 genome. P.Y. was responsible for analyses of environmental amplicon and metagenome data. J.F., S.L., P.Y., and P.L. wrote the manuscript. All authors revised the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/11/e1700807/DC1
Supplementary Text
fig. S1. Abundance of Nitrococcus-affiliated cells in the Namibian OMZ based on CARD-FISH counts.
fig. S2. 16S rRNA gene-based phylogenetic tree visualizing the relation of four Nitrococcus phylotypes.
fig. S3. Phylogenetic analysis of NxrA.
fig. S4. Incubation experiments with N. mobilis Nb-231.
fig. S5. Phylogenetic analysis of the sNOR.
fig. S6. Selection of Tara Oceans, MG-RAST, and OSD samples that were mapped to the N. mobilis Nb-231 genome.
fig. S7. Effect of reduced O2 and enhanced IO3 concentrations on nitrite oxidation rates.
table S1. Percent identity of 16S rRNA genes between N .mobilis and the newly identified Nitrococcus-like phylotype 1.
table S2. List of marine environmental metagenomes that contain at least one of the four Nitrococcus phylotypes.
table S3. List of marine amplicon sequencing data sets that contain at least one of the four Nitrococcus phylotypes.
table S4. N. mobilis strain Nb-231 proteins with predicted functions in key metabolic pathways.
table S5. 13C enrichment of single Nitrococcus cells from the Namibian OMZ.
table S6. Composition of marine NOB medium used to grow N. mobilis.
table S7. Functional genes associated with Nitrococcus spp. in selected Tara Oceans metagenomes.
table S8. Summary of stations, sampling depths, and 15N incubation experiments conducted.
table S9. NOB specific 16S rRNA-targeted oligonucleotide probes.
table S10. Substrate amendments for N. mobilis Nb-231 incubation experiments.
REFERENCES AND NOTES
- 1.Yool A., Martin A. P., Fernández C., Clark D. R., The significance of nitrification for oceanic new production. Nature 447, 999–1002 (2007). [DOI] [PubMed] [Google Scholar]
- 2.E. Spieck, E. Bock, The lithoautotrophic nitrite-oxidizing bacteria, in Bergey’s Manual of Systematic Bacteriology, G. Garrity, D. J. Brenner, N. R. Krieg, J. R. Staley, Eds. (Springer-Verlag, ed. 2, 2005), pp. 149–153. [Google Scholar]
- 3.Daims H., Lücker S., Wagner M., A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 24, 699–712 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koch H., Galushko A., Albertsen M., Schintlmeister A., Gruber-Dorninger C., Lücker S., Pelletier E., Le Paslier D., Spieck E., Richter A., Nielsen P. H., Wagner M., Daims H., Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science 345, 1052–1054 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Daims H., Lebedeva E. V., Pjevac P., Han P., Herbold C., Albertsen M., Jehmlich N., Palatinszky M., Vierheilig J., Bulaev A., Kirkegaard R. H., von Bergen M., Rattei T., Bendinger B., Nielsen P. H., Wagner M., Complete nitrification by Nitrospira bacteria. Nature 528, 504–509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koch H., Lücker S., Albertsen M., Kitzinger K., Herbold C., Spieck E., Nielsen P. H., Wagner M., Daims H., Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc. Natl. Acad. Sci. 112, 11371–11376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Kessel M. A. H. J., Speth D. R., Albertsen M., Nielsen P. H., Op den Camp H. J. M., Kartal B., Jetten M. S. M., Lücker S., Complete nitrification by a single microorganism. Nature 528, 555–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mincer T. J., Church M. J., Taylor L. T., Preston C., Karl D. M., DeLong E. F., Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9, 1162–1175 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Davis R. E., Stakes D. S., Wheat C. G., Moyer C. L., Bacterial variability within an iron-silica-manganese-rich hydrothermal mound located off-axis at the Cleft Segment, Juan de Fuca Ridge. Geomicrobiol. J. 26, 570–580 (2009). [Google Scholar]
- 10.Durbin A. M., Teske A., Microbial diversity and stratification of South Pacific abyssal marine sediments. Environ. Microbiol. 13, 3219–3234 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Füssel J., Lam P., Lavik G., Jensen M. M., Holtappels M., Günter M., Kuypers M. M. M., Nitrite oxidation in the Namibian oxygen minimum zone. ISME J. 6, 1200–1209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beman J. M., Leilei Shih J., Popp B. N., Nitrite oxidation in the upper water column and oxygen minimum zone of the eastern tropical North Pacific Ocean. ISME J. 7, 2192–2205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson S. W., Waterbury J. B., Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch. Mikrobiol. 77, 203–230 (1971). [Google Scholar]
- 14.Lücker S., Nowka B., Rattei T., Spieck E., Daims H., The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front. Microbiol. 4, 27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spieck E., Keuter S., Wenzel T., Bock E., Ludwig W., Characterization of a new marine nitrite oxidizing bacterium, Nitrospina watsonii sp. nov., a member of the newly proposed phylum “Nitrospinae”. Syst. Appl. Microbiol. 37, 170–176 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Ward B. B., Carlucci A. F., Marine ammonia- and nitrite-oxidizing bacteria: Serological diversity determined by immunofluorescence in culture and in the environment. Appl. Environ. Microbiol. 50, 194–201 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward B. B., Glover H. E., Lipschultz F., Chemoautotrophic activity and nitrification in the oxygen minimum zone off Peru. Deep-Sea Res. 36, 1031–1051 (1989). [Google Scholar]
- 18.Ghai R., Pašić L., Fernández A. B., Martin-Cuadrado A.-B., Mizuno C. M., McMahon K. D., Papke R. T., Stepanauskas R., Rodriguez-Brito B., Rohwer F., Sánchez-Porro C., Ventosa A., Rodríguez-Valera F., New abundant microbial groups in aquatic hypersaline environments. Sci. Rep. 1, 135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stackebrandt E., Ebers J., Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 33, 152–155 (2006). [Google Scholar]
- 20.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F. O., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer F., Paarmann D., D’Souza M., Olson R., Glass E. M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R. A., The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopf A., Bicak M., Kottmann R., Schnetzer J., Kostadinov I., Lehmann K., Fernandez-Guerra A., Jeanthon C., Rahav E., Ullrich M., Wichels A., Gerdts G., Polymenakou P., Kotoulas G., Siam R., Abdallah R. Z., Sonnenschein E. C., Cariou T., O’Gara F., Jackson S., Orlic S., Steinke M., Busch J., Duarte B., Caçador I., Canning-Clode J., Bobrova O., Marteinsson V., Reynisson E., Loureiro C. M., Luna G. M., Quero G. M., Löscher C. R., Kremp A., DeLorenzo M. E., Øvreås L., Tolman J., LaRoche J., Penna A., Frischer M., Davis T., Katherine B., Meyer C. P., Ramos S., Magalhães C., Jude-Lemeilleur F., Aguirre-Macedo M. L., Wang S., Poulton N., Jones S., Collin R., Fuhrman J. A., Conan P., Alonso C., Stambler N., Goodwin K., Yakimov M. M., Baltar F., Bodrossy L., Van De Kamp J., Frampton D. M., Ostrowski M., Van Ruth P., Malthouse P., Claus S., Deneudt K., Mortelmans J., Pitois S., Wallom D., Salter I., Costa R., Schroeder D. C., Kandil M. M., Amaral V., Biancalana F., Santana R., Pedrotti M. L., Yoshida T., Ogata H., Ingleton T., Munnik K., Rodriguez-Ezpeleta N., Berteaux-Lecellier V., Wecker P., Cancio I., Vaulot D., Bienhold C., Ghazal H., Chaouni B., Essayeh S., Ettamimi S., Zaid el H., Boukhatem N., Bouali A., Chahboune R., Barrijal S., Timinouni M., El Otmani F., Bennani M., Mea M., Todorova N., Karamfilov V., Ten Hoopen P., Cochrane G., L’Haridon S., Bizsel K. C., Vezzi A., Lauro F. M., Martin P., Jensen R. M., Hinks J., Gebbels S., Rosselli R., De Pascale F., Schiavon R., Dos Santos A., Villar E., Pesant S., Cataletto B., Malfatti F., Edirisinghe R., Silveira J. A., Barbier M., Turk V., Tinta T., Fuller W. J., Salihoglu I., Serakinci N., Ergoren M. C., Bresnan E., Iriberri J., Nyhus P. A., Bente E., Karlsen H. E., Golyshin P. N., Gasol J. M., Moncheva S., Dzhembekova N., Johnson Z., Sinigalliano C. D., Gidley M. L., Zingone A., Danovaro R., Tsiamis G., Clark M. S., Costa A. C., El Bour M., Martins A. M., Collins R. E., Ducluzeau A. L., Martinez J., Costello M. J., Amaral-Zettler L. A., Gilbert J. A., Davies N., Field D., Glöckner F. O., The ocean sampling day consortium. GigaScience 4, 1–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunagawa S., Coelho L. P., Chaffron S., Kultima J. R., Labadie K., Salazar G., Djahanschiri B., Zeller G., Mende D. R., Alberti A., Cornejo-Castillo F. M., Costea P. I., Cruaud C., d’Ovidio F., Engelen S., Ferrera I., Gasol J. M., Guidi L., Hildebrand F., Kokoszka F., Lepoivre C., Lima-Mendez G., Poulain J., Poulos B. T., Royo-Llonch M., Sarmento H., Vieira-Silva S., Dimier C., Picheral M., Searson S., Kandels-Lewis S. Tara Oceans coordinators, Bowler C., de Vargas C., Gorsky G., Grimsley N., Hingamp P., Iudicone D., Jaillon O., Not F., Ogata H., Pesant S., Speich S., Stemmann L., Sullivan M. B., Weissenbach J., Wincker P., Karsenti E., Raes J., Acinas S. G., Bork P., Structure and function of the global ocean microbiome. Science 348, 1261359 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Freitag A., Rudert M., Bock E., Growth of Nitrobacter by dissimilatoric nitrate reduction. FEMS Microbiol. Lett. 48, 105–109 (1987). [Google Scholar]
- 25.Bock E., Koops H.-P., Möller U. C., Rudert M., A new facultatively nitrite oxdizing bacterium, Nitrobacter vulgaris sp. nov. Arch. Microbiol. 153, 105–110 (1990). [Google Scholar]
- 26.Sundermeyer-Klinger H., Meyer W., Warninghoff B., Bock E., Membrane-bound nitrite oxidoreductase of Nitrobacter: Evidence for a nitrate reductase system. Arch. Microbiol. 140, 153–158 (1984). [Google Scholar]
- 27.Lücker S., Wagner M., Maixner F., Pelletier E., Koch H., Vacherie B., Rattei T., Damsté J. S. S., Spieck E., Le Paslier D., Daims H., A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 13479–13484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starkenburg S. R., Arp D. J., Bottomley P. J., Expression of a putative nitrite reductase and the reversible inhibition of nitrite-dependent respiration by nitric oxide in Nitrobacter winogradskyi Nb-255. Environ. Microbiol. 10, 3036–3042 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Sorokin D. Y., Lücker S., Vejmelkova D., Kostrikina N. A., Kleerebezem R., Rijpstra W. I. C., Damsté J. S. S., Le Paslier D., Muyzer G., Wagner M., van Loosdrecht M. C. M., Daims H., Nitrification expanded: Discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 6, 2245–2256 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musat N., Halm H., Winterholler B., Hoppe P., Peduzzi S., Hillion F., Horreard F., Amann R., Jørgensen B. B., Kuypers M. M. M., A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc. Natl. Acad. Sci. 105, 17861–17866 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ackermann M., A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol. 13, 497–508 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Stein L. Y., Arp D. J., Berube P. M., Chain P. S. G., Hauser L., Jetten M. S. M., Klotz M. G., Larimer F. W., Norton J. M., Op den Camp H. J. M., Shin M., Wei X., Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: Implications for niche adaptation. Environ. Microbiol. 9, 2993–3007 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Kozlowski J. A., Stieglmeier M., Schleper C., Klotz M. G., Stein L. Y., Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J. 10, 1836–1845 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suntharalingam P., Sarmiento J., Toggweiler J. R., Global significance of nitrous-oxide production and transport from oceanic low-oxygen zones: A modeling study. Global Biogeochem. Cycles 14, 1353–1370 (2000). [Google Scholar]
- 35.Goreau T. J., Kaplan W. A., Wofsy S. C., McElroy M. B., Valois F. W., Watson S. W., Production of NO2− and N2O by nitrifying bacteria at reduced concentrations of oxygen. Appl. Environ. Microbiol. 40, 526–532 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravishankara A. R., Daniel J. S., Portmann R. W., Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science 326, 123–125 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Stramma L., Johnson G. C., Sprintall J., Mohrholz V., Expanding oxygen-minimum zones in the tropical oceans. Science 320, 655–658 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Vignais P. M., Billoud B., Occurrence, classification, and biological function of hydrogenases: An overview. Chem. Rev. 107, 4206–4272 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Silva P. J., van den Ban E. C. D., Wassink H., Haaker H., de Castro B., Robb F. T., Hagen W. R., Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur. J. Biochem. 267, 6541–6551 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Ma K., Weiss R., Adams M. W. W., Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 182, 1864–1871 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luebke J. L., Shen J., Bruce K. E., Kehl-Fie T. E., Peng H., Skaar E. P., Giedroc D. P., The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol. Microbiol. 94, 1343–1360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia Y., Lü C., Hou N., Xin Y., Liu J., Liu H., Xun L., Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 10.1038/ismej.2017.125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swan B. K., Martinez-Garcia M., Preston C. M., Sczyrba A., Woyke T., Lamy D., Reinthaler T., Poulton N. J., Masland E. D. P., Gomez M. L., Sieracki M. E., DeLong E. F., Herndl G. J., Stepanauskas R., Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333, 1296–1300 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Murillo A. A., Ramírez-Flandes S., DeLong E. F., Ulloa O., Enhanced metabolic versatility of planktonic sulfur-oxidizing γ-proteobacteria in an oxygen-deficient coastal ecosystem. Front. Mar. Sci. 1, 18 (2014). [Google Scholar]
- 45.Karl D. M., Knauer G. A., Martin J. H., Ward B. B., Bacterial chemolithotrophy in the ocean is associated with sinking particles. Nature 309, 54–56 (1984). [Google Scholar]
- 46.Alldredge A. L., Cohen Y., Can microscale chemical patches persist in the sea? Microelectrode study of marine snow, fecal pellets. Science 235, 689–691 (1987). [DOI] [PubMed] [Google Scholar]
- 47.Stief P., Kamp A., Thamdrup B., Glud R. N., Anaerobic nitrogen turnover by sinking diatom aggregates at varying ambient oxygen levels. Front. Microbiol. 7, 98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lavik G., Stührmann T., Brüchert V., Van der Plas A., Mohrholz V., Lam P., Mußmann M., Fuchs B. M., Amann R., Lass U., Kuypers M. M. M., Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature 457, 581–584 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Schunck H., Lavik G., Desai D. K., Großkopf T., Kalvelage T., Löscher C. R., Paulmier A., Contreras S., Siegel H., Holtappels M., Rosenstiel P., Schilhabel M. B., Graco M., Schmitz R. A., Kuypers M. M. M., LaRoche J., Giant hydrogen sulfide plume in the oxygen minimum zone off Peru supports chemolithoautotrophy. PLOS ONE 8, e68661 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchwald C., Santoro A. E., Stanley R. H., Casciotti K. L., Nitrogen cycling in the secondary nitrite maximum of the eastern tropical North Pacific off Costa Rica. Global Biogeochem. Cycles 29, 2061–2081 (2015). [Google Scholar]
- 51.Bristow L. A., Callbeck C. M., Larsen M., Altabet M. A., Dekaezemacker J., Forth M., Gauns M., Glud R. N., Kuypers M. M. M., Lavik G., Milucka J., Naqvi S. W. A., Pratihary A., Revsbech N. P., Thamdrup B., Treusch A. H., Canfield D. E., N2 production rates limited by nitrite availability in the Bay of Bengal oxygen minimum zone. Nat. Geosci. 10, 24–29 (2017). [Google Scholar]
- 52.Holtappels M., Kuypers M. M. M., Schlüter M., Brüchert V., Measurement and interpretation of solute concentration gradients in the benthic boundary layer. Limnol. Oceanogr. Meth. 9, 1–13 (2011). [Google Scholar]
- 53.K. Grasshoff, M. Ehrhardt, K. Kremling, Methods of Seawater Analysis (Wiley-VCH, Weinheim, 1999). [Google Scholar]
- 54.Holmes R. M., Aminot A., Kérouel R., Hooker B. A., Peterson B. J., A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can. J. Fish. Aquat. Sci. 56, 1801–1808 (1999). [Google Scholar]
- 55.Dalsgaard T., Canfield D. E., Petersen J., Thamdrup B., Acuña-González J., N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature 422, 606–608 (2003). [DOI] [PubMed] [Google Scholar]
- 56.M. Holtappels, G. Lavik, M. M. Jensen, M. M. M. Kuypers, 15N-labelling experiments to dissect the contributions of heterotrophic denitrification and anammox to nitrogen removal in the OMZ waters of the ocean, in Methods in Enzymology: Research on Nitrification and Related Processes, Part A, M. Klotz, Ed. (Academic Press, ed. 1, 2011), vol. 486, pp. 223–251. [DOI] [PubMed] [Google Scholar]
- 57.Granger J., Sigman D. M., Removal of nitrite with sulfamic acid for nitrate N and O isotope analysis with the denitrifier method. Rapid Commun. Mass Spectrom. 23, 3753–3762 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Pernthaler A., Pernthaler J., Amann R., Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68, 3094–3101 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polerecky L., Adam B., Milucka J., Musat N., Vagner T., Kuypers M. M. M., Look@NanoSIMS—A tool for the analysis of nanoSIMS data in environmental microbiology. Environ. Microbiol. 14, 1009–1023 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Vallenet D., Labarre L., Rouy Z., Barbe V., Bocs S., Cruveiller S., Lajus A., Pascal G., Scarpelli C., Médigue C., MaGe: A microbial genome annotation system supported by synteny results. Nucleic Acids Res. 34, 53–65 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yarza P., Yilmaz P., Pruesse E., Glöckner F. O., Ludwig W., Schleifer K.-H., Whitman W. B., Euzéby J., Amann R., Rosselló-Móra R., Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Ludwig W., Strunk O., Westram R., Richter L., Meier H.,Yadhukumar, Buchner A., Lai T., Steppi S., Jobb G., Förster W., Brettske I., Gerber S., Ginhart A. W., Gross O., Grumann S., Hermann S., Jost R., König A., Liss T., Lüßmann R., May M., Nonhoff B., Reichel B., Strehlow R., Stamatakis A., Stuckmann N., Vilbig A., Lenke M., Ludwig T., Bode A., Schleifer K.-H., ARB: A software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thamdrup B., Fossing H., Jørgensen B. B., Manganese, iron and sulfur cycling in a coastal marine sediment, Aarhus bay, Denmark. Geochim. Cosmochim. Acta 58, 5115–5129 (1994). [Google Scholar]
- 65.Lees H., Simpson J. R., The biochemistry of the nitrifying organisms. 5. Nitrite oxidation by Nitrobacter. Biochem. J. 65, 297–305 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Farrenkopf A. M., Dollhopf M. E., Ní Chadhain S., Luther G. W. III, Nealson K. H., Reduction of iodate in seawater during Arabian Sea shipboard incubations and in laboratory cultures of the marine bacterium Shewanella putrefaciens strain MR-4. Mar. Chem. 57, 347–354 (1997). [Google Scholar]
- 67.Farrenkopf A. M., Luther G. W. III, Truesdale V. W., Van der Weijden C. H., Sub-surface iodide maxima: Evidence for biologically catalyzed redox cycling in Arabian Sea OMZ during the SW intermonsoon. Deep-Sea Res. Pt. II 44, 1391–1409 (1997). [Google Scholar]
- 68.Truesdale V. W., Bailey G. W., Iodine distribution in the Southern Benguela system during an upwelling episode. Cont. Shelf Res. 22, 39–49 (2002). [Google Scholar]
- 69.Sung Y.-C., Parsell D., Anderson P. M., Fuchs J. A., Identification, mapping, and cloning of the gene encoding cyanase in Escherichia coli K-12. J. Bacteriol. 169, 2639–2642 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meincke M., Bock E., Kastrau D., Kroneck P. M. H., Nitrite oxidoreductase from Nitrobacter hamburgensis: Redox centers and their catalytic role. Arch. Microbiol. 158, 127–131 (1992). [Google Scholar]
- 71.Rothery R. A., Workun G. J., Weiner J. H., The prokaryotic complex iron–sulfur molybdoenzyme family. Biochim. Biophys. Acta. 1778, 1897–1929 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Grimaldi S., Schoepp-Cothenet B., Ceccaldi P., Guigliarelli B., Magalon A., The prokaryotic Mo/W-bisPGD enzymes family: A catalytic workhorse in bioenergetic. Biochim. Biophys. Acta. 1827, 1048–1085 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Blasco F., Dos Santos J.-P., Magalon A., Frixon C., Guigliarelli B., Santini C.-L., Giordano G., NarJ is a specific chaperone required for molybdenum cofactor assembly in nitrate reductase A of Escherichia coli. Mol. Microbiol. 28, 435–447 (1998). [DOI] [PubMed] [Google Scholar]
- 74.Tabita F. R., Hanson T. E., Li H., Satagopan S., Singh J., Chan S., Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol. Mol. Biol. Rev. 71, 576−599 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yeates T. O., Kerfeld C. A., Heinhorst S., Cannon G. C., Shively J. M., Protein-based organelles in bacteria: Carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6, 681–691 (2008). [DOI] [PubMed] [Google Scholar]
- 76.Bowes G., Ogren W. L., Hageman R. H., Phosphoglycolate production catalyzed by ribulose diphosphate carboxylase. Biochem. Biophys. Res. Commun. 45, 716–722 (1971). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/11/e1700807/DC1
Supplementary Text
fig. S1. Abundance of Nitrococcus-affiliated cells in the Namibian OMZ based on CARD-FISH counts.
fig. S2. 16S rRNA gene-based phylogenetic tree visualizing the relation of four Nitrococcus phylotypes.
fig. S3. Phylogenetic analysis of NxrA.
fig. S4. Incubation experiments with N. mobilis Nb-231.
fig. S5. Phylogenetic analysis of the sNOR.
fig. S6. Selection of Tara Oceans, MG-RAST, and OSD samples that were mapped to the N. mobilis Nb-231 genome.
fig. S7. Effect of reduced O2 and enhanced IO3 concentrations on nitrite oxidation rates.
table S1. Percent identity of 16S rRNA genes between N .mobilis and the newly identified Nitrococcus-like phylotype 1.
table S2. List of marine environmental metagenomes that contain at least one of the four Nitrococcus phylotypes.
table S3. List of marine amplicon sequencing data sets that contain at least one of the four Nitrococcus phylotypes.
table S4. N. mobilis strain Nb-231 proteins with predicted functions in key metabolic pathways.
table S5. 13C enrichment of single Nitrococcus cells from the Namibian OMZ.
table S6. Composition of marine NOB medium used to grow N. mobilis.
table S7. Functional genes associated with Nitrococcus spp. in selected Tara Oceans metagenomes.
table S8. Summary of stations, sampling depths, and 15N incubation experiments conducted.
table S9. NOB specific 16S rRNA-targeted oligonucleotide probes.
table S10. Substrate amendments for N. mobilis Nb-231 incubation experiments.


