Abstract
Background
The Piezo1 protein ion channel is a novel mechanical activated ion channel which is related to mechanical signal transduction. However, the function of the mechanically activated ion channel Piezo1 had not been explored. In this study, we explored the function of the Piezo1 ion channel in human osteosarcoma (OS) cells related to apoptosis, invasion, and the cell proliferation.
Material/Methods
Reverse transcription polymerase chain reaction (RT-PCR) and western-blotting were used to detect the expression of the Piezo1 protein. CCK-8, Transwell experiments and AV-PI were used to detected cell proliferation, cell invasion and cell apoptosis.
Results
The Piezo1 protein ion channel was highly expressed in human OS cells. The Piezo1-shRNA inhibited the expression of the Piezo1. We explored whether LV3-PIEZO1-homo-3201 could act as Piezo1-shRNA, which could then be an inhibitor of Piezo1. The expression of Piezo1 in the 2-hour stretch group were slightly higher than the 0-hour stretch group, and the difference was not statistically significant (n=3, p>0.05, one-way ANOVA). The apoptotic gene such as the Bax, BAD, caspase-3, and caspase-9 had the same characteristics as the Piezo1 expression under the stretch force. We also explored the invasion of Piezo1 in vivo using nude mice, and found that Piezo1-shRNA could inhibit the invasion of the OS cells.
Conclusions
The Piezo1 protein may be a novel, potential therapeutic target for OS.
MeSH Keywords: Apoptosis, Cell Proliferation, Osteosarcoma
Background
Osteosarcoma (OS) is an aggressive bone neoplasm and occurs mainly in children and young adults, and it ranks as the second most prevalent leading cause of cancer-related death [1]. Surgical treatment combined with multidrug chemotherapy were mainly used to deal with this cancer, however, the five-year survival rate and recurrence risk for OS patients has not been significantly improved due to the local relapse and metastases of the tumor [2]. Many molecular alterations are involved in critical signal transduction pathways that contribute to OS pathogenesis via promoting key molecular targets, thus identifying the key molecular targets is important for the diagnosis and therapy of OS patients [3]. Therefore, in this study, we explored the novel mechanical activated ion channel Piezo1 protein as the key molecular target of OS patients under stretch force.
Recently, a novel mechanically activated (MA) cation channel known as the Piezo protein was identified by Coste et al. [4,5]. It is part of an evolutionarily conserved ion channel family of cation-permeable proteins involved in mechano-transduction that is closely related to the cytoskeleton. The hPiezo protein has been reported to play a key role in cellular response to mechanical stimuli in human erythrocyte membranes and bladder urothelium, and mutations in the hPiezo protein are associated with anemia, hereditary xerocytosis, and distal arthrogryposis type 5 [6–12]. hPiezo1 and hPiezo2 have been identified as ion channels involved in mechano-sensation, suggesting that they have the ability to sense mechanical signals to maintain cell volume. OS is associated with abnormal mechanical loading, thus, we hypothesized that the MA ion channel Piezo1 plays an important role in OS cells proliferation, migration, and apoptosis.
The caspases comprise a class of cysteine proteases involved in apoptosis, and they convey the apoptotic signal, such as mechanical stretch and inflammation, into the proteolytic cascade, with caspases cleaving and activating other caspases that subsequently degrade cellular targets to induce apoptosis. The activating caspases include caspase-9, caspase-12 and caspase-3 [13]. Caspase-9 can be activated by cytochrome C from the chondriosome, which can activate caspase-3 and trigger apoptosis. Caspase-12 is a marker of ER stress, and it can induce apoptosis by activating apoptotic genes in the nucleus. B cell lymphoma/leukemia-2 (Bcl-2), Bcl-associated X protein (Bax) and Bcl-2-associated death promoter (BAD) act in the apoptosis cascades and are closely related to apoptosis [14–17]. Bcl-2 is an anti-apoptosis signaling factor, and it promotes cell proliferation and inhibits apoptosis through complex pathways [15]. BAD is an important apoptosis factor, and its homology with Bcl-2 is restricted to the BH1 and BH2 domains [16]. Bcl-xl can activate BAD to lead to cell apoptosis by suppressing the Bcl-2 family, which has similar functions to Bax. In this study, we explored the connection between the apoptotic family and the mechanical stretch which was induced by the novel mechanical activated ion channel Piezo1 protein.
Material and Methods
Cell culture and treatment
Human OS cell line MG63 and U2 cell line were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). All OS cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 15% of heat-inactivated fetal bovine serum (FBS; Life Technologies); 100 U/mL of penicillin and 100 μg/mL of streptomycin (Pen Strep, Life Technologies); as well as L-glutamine (Life Technologies). All cells were cultured in humidified 5% CO2 incubator at 37°C and incubated for three days until confluence was reached. Subculture cells were treated with 0.05% trypsin solution (Sigma-Aldrich, St. Louis, MO, USA) and seeded in BioFlex6 culture plates (BioFlex, NC, USA).
Construction of the Piezo1-shRNA Lentivirus Vector
The designed shRNA template was cloned into a self-inactivating lentivirus that harbors a ZsGreen reporter and a PGK promoter upstream of the insertion sites. The cloned oligonucleotides can then produce in vitro shRNA namely pHBLV-U6-ZsGreen-PGK-Puro vector. The coding regions in the human Piezo1 gene were selected to design shRNAs. The designed olig sequence of the h-Piezo1 ix shown in Table 1. The successful construction of shRNA cassettes was determined by restriction mapping and DNA sequencing. The infectious lentivirus vectors were generated. The infectious lentivirus vectors were collected at 48 hours post-transduction and subjected to centrifugation to remove cell debris. The hole-by-dilution titer assay was used to determine the virus titer (~5.0×107 TU/mL)
Table 1.
The oligo sequence of the three Piezo1-shRNAs.
| Oligo name | Oligo sequence |
|---|---|
| LV3-PIEZO1-homo-2844 | 5′-GATCCGCGTCATCATCGTGTGTAAGATTCAAGAGATCTTACACACGATGATGACGCTTTTTTG-3′ |
| 3′-AATTCAAAAAAGCGTCATCATCGTGTGTAAGATCTCTTGAATCTTACACACGATGATGACGCG-5′ | |
| LV3-PIEZO1-homo-3201 | 5′-GATCCGCCTCAAGTACTTCATCAACTTTCAAGAGAAGTTGATGAAGTACTTGAGGCTTTTTTG-3′ |
| 3′-AATTCAAAAAAGCCTCAAGTACTTCATCAACTTCTCTTGAAAGTTGATGAAGTACTTGAGGCG-5′ | |
| LV3-PIEZO1-homo-4155 | 5′-GATCCGCGTCTTCCTTAGCCATTACTTTCAAGAGAAGTAATGGCTAAGGAAGACGCTTTTTTG-3′ |
| 3′-AATTCAAAAAAGCGTCTTCCTTAGCCATTACTTCTCTTGAAAGTAATGGCTAAGGAAGACGCG-5′ | |
| Negative Sh control | 5′-GATAGCGTCCATCATCGTGTGTAAGATTCAAGAGATCTTACACACGATGATGACGCTTTTTTG-3′ |
| 3′-AATAGCGTCAAGCGTCATCATCGTGTGTAAGATCTCTTGAATCTTACACACGATGATGACGCG-5′ |
RT-qPCR
Total RNAs were extracted with RNAiso kit (TaKaRa, Japan) after 2, 12, 24, and 48 hours of compressive stress, respectively. The concentration and purity of the total RNA were evaluated with a spectrophotometer. Reverse transcription-quantitative chain reaction (RT-qPCR) was performed and analyzed to assess the expression of hPiezo1 and hGAPDH with the SYBR Premix Ex Taq II kit (Perfect Real-Time, TaKaRa, Japan) on a FTC-3000 RT-qPCR System (Funglyn Biotech Inc., Canada) according to the manufacturer’s instructions. The PCR primers (synthesized by SangonBiotech, Shanghai, China) were used to amplify the genes (Table 2). The housekeeping gene GADPH levels were normalized to the threshold cycle of target genes. To evaluate hPiezo1, hcaspase-3, hcaspase-9, hBAD, hBcl-2, hBax, and hGAPDH expression, the relative expressions were analyzed by the comparative 2−ΔΔCT method.
Table 2.
Oligo sequences of the target genes.
| Oligo name | Oligo sequence | |
|---|---|---|
| hPiezo1 | Forward primer | 5′-CATCTTGGTGGTCTCCTCTGTCT-3′ |
| Reverse primer | 5′-CTGGCATCCACATCCCTCTCATC-3′ | |
| hCaspase3 | Forward primer | 5′-GGGTGCTATTGTGAGGCG-3′ |
| Reverse primer | 5′-CGCCTCACAATAGCACCC-3′ | |
| hCaspase9 | Forward primer | 5′-GGCTGTCTACGGCACAGATGGA-3′ |
| Reverse primer | 5′-CTGGCTCGGGGTTACTGCCAG-3′ | |
| hBAD | Forward primer | 5′-CCGGAGGATGAGTGACGAGT-3′ |
| Reverse primer | 5′-CCGATCCCACCAGGACTG-3′ | |
| hBcl-2 | Forward primer | 5′-TGGGATGCCTTTGTGGAACT-3′ |
| Reverse primer | 5′-GAGACAGCCAGGAGAAATCAAAC-3′ | |
| hBax | Forward primer | 5′-CCTTTTGCTTCAGGGTTTCAT-3′ |
| Reverse primer | 5′-GAGACACTCGCTCAGCTTCTTG-3′ | |
| hGAPDH | Forward primer | 5′-GCACCGTCAAGGCTGAGAAC-3′ |
| Reverse primer | 5′-TGGTGAAGACGCCAGTGGA-3′ |
Western blotting
After treatment with mechanical stretch force for 2, 12, 24, or 48 hours, the cells were harvested from the Flexcell 6-well plates. A 10 nM Tris-HCl buffer (pH 8.0) plus 1 mM PMSF was used to suspend the mitochondrial fractions. Cytosolic fractions were obtained by centrifuging for 1×105 rpm at 4°C for one hour. A Bradford protein assay kit (Beyotime, Shanghai, China) was used to measure the protein content. Briefly, the proteins were subjected to SDS-PAGE, followed by transfer to PVDF membranes. The samples were incubated with the Piezo1 primary antibody (Novus Biologicals, Littleton, CO, USA) at 4°C overnight. AlexaFluor 488 goat anti-rabbit IgG (diluted 1: 2,000; Invitrogen, Carlsbad, CA, USA) was used as a secondary antibody. After incubation with an anti-rabbit secondary antibody, the membrane was treated with a chemiluminescence detection kit and exposed to the VILBER Fusion FX5 automatic gel imaging analysis system. Protein band signal intensities were quantified using the GS-800 Calibrated Densitometer (Leica, Heidelberg, Germany) and Bio-Rad analysis software.
Application of cyclic stretch
OS MG63 cells were seeded in GM at a concentration of 3×106 cells/well on six-well collagen-coated BioFlex plates containing a flexible silicone elastomer substratum and were grown to 80% confluence under non-stretch conditions for 3–5 days. BioFlex plates were then mounted in a Flexercell Strain Unit (Flexercell International; McKeesport, PA, USA) and subjected to 20% surface elongation at a frequency of six cycles per minute. Each cycle consisted of a three second stretch alternating with three seconds of relaxation regulated by a computer-controlled vacuum stretch apparatus (FX-4000T Tension Plus System; FlexCell International Corporation, USA). All cells were divided into 0-hour stretch group (control group), 2-hour stretch group, 12-hour stretch group, 24-hour stretch group, and 48-hour stretch group under the treatment of the duration stretch force with Flexcell FX5000 Tension system (FlexCell International Corporation, USA). The observed time point and deformation rate were on the basis of our preliminary experiment.
Immunofluorescence
After mechanical stimulation for 24 hours, the cells were seeded into 24-well plates with glass coverslips. After two rinses in PBS, the cells were fixed with 4% paraformaldehyde (Hyclone, Logan, UT, USA) and permeabilized with 0.2% Triton X-100 (MP Biomedicals, Santa Ana, CA, USA) for 10 minutes at room temperature. A solution of 5% BSA in PBS was used as a blocking solution to prevent nonspecific binding for one hour. The coverslips were incubated with the Piezo1 primary antibody (Novus Biologicals, Littleton, CO, USA) at 4°C overnight. AlexaFluor 488 Goat Anti-Rabbit IgG (diluted 1: 2,000; Invitrogen, Carlsbad, CA, USA) was used as a secondary antibody. The coverslips were stained with Hoechst 33342 to visualize nuclei (Thermo Scientific, Shanghai, China). A Zeiss LSM 510 confocal microscope (Zeiss, Jena, Germany) was used to observe Piezo1 protein localization.
Morphology of apoptotic OS cells by Hoechst staining
OS cells nuclei were stained with Hoechst to observe morphological changes under stretch force. After treatment with mechanical stretch force for 2, 12, 24, or 48 hours, the cells were collected from the Flexcell 6-well plates using 0.25% trypsin. The OS cells were then fixed with 4% paraformaldehyde (MP Biomedicals, Santa Ana, CA, USA), and permeabilized with 0.2% Triton X-100 (MP Biomedicals, Santa Ana, CA, USA) for 10 minutes at room temperature. Cells were then stained with Hoechst for 10 minutes. A Leica TCS SP5 fluorescence microscope (Leica, Heidelberg, Germany) was used to observe nuclear fragmentation.
The invasive assay by the Transwell experiment
All cells including OS MG63 and OS U2 cells were treated with stretch force and the OS MG63 and OS U2 cells with Piezo1-shRNA were inoculated in the polycarbonate microporous membrane of Transwell system with a pore size of 10 μm at 37°C for 30 minutes. The upper chamber of the Transwell system was loaded with 100 μL of the cells, and the lower chamber of the Transwell system with 600 μL of the culture supernatant per well and then incubated in an atmosphere of 5% CO2 at 37°C for 48 hours. Then the cells were removed with cotton swabs, fixed and stained with Gimsa solution. The microscope was used to count of the number of invasive cells in five random visual fields. Experimental progress was done in triplicate in each group.
Cell counting Kit-8 (CCK-8)
All cells including OS MG63 and OS U2 cells treated with stretch force and the OS MG63 and OS U2 cells with Piezo1-shRNA were inoculated in a 96-well plate. After incubated in the 37°C CO2 incubator, the 10 μL of CCK-8 reagent was added into the plates. The micro plate reader (Bio-Rad) was used to measuring the optical density (OD) value at a wave length setting of 450 nm. Cell proliferation rate of the cells was calculated with the equation: proliferation rate (%)=OD value in experimental wells–OD value of control well/OD value of control well ×100%. Three duplicated wells were set for each experiment.
Analysis of apoptosis
Annexin V binding and propidium iodide staining were used to analyze human OS cell apoptosis. The cells were harvested and centrifuged after being exposed to continuous stretching for 0, 2, 12, 24, or 48 hours. The same conditions were applied to cells in the group treated with Piezo1-shRNA. Cells were stained with FITC-conjugated Annexin V and propidium iodide (PI) according to the manufacturer’s instructions for the Apoptosis Detection Kit (R&D Systems, MN, USA). Extra binding buffer was added to the control group. Flow cytometry (Epics XL, Beckman-Coulter, Kreefeld, Germany) was used to collect the data. GraphPad software (CA, USA) was used to analyze the number of cells in the early stage, in the late stage, and during total apoptosis.
Subcutaneous tumor formation assay in nude mice
A total of 18 three-week-old female BALB/c nude mice weighting between 10–12 g with congenital cellular immune defect were obtained from Shanghai Animal Experimental Center of Chinese Academy of Science. The nude mice were divided into three groups as following blank control group (BC group) containing serum-free and antibiotic-free DMEM without shRNA; negative control group (NC group) containing Piezo1-shRNA lentivirus vector without any target gene; and the Piezo1-shRNA group. All cells including OS MG63 and OS U2 cells, and the OS MG63 and OS U2 cells with Piezo1-shRNA, were subcutaneously injected into the right axilla of nude mice. 100 μL of cells with an average density of 3×107/mL were selected and transferred to a sterile Eppendorf tube (EP) tube before the injection. The tumor long and short diameter were measured every week and the tumor volume in each group was calculated according to the formula: V=1/6Πab2. The tumor inhibited rate (%)=(1-mean tumor volume in experimental group/mean tumor volume in BC group) ×100%.
Statistical analysis
The data were expressed as mean ± standard deviation (SD) of separate experiments. The unpaired t-test was used to value the difference between groups. Statistical significance was set at p value less than 0.05. Analysis was performed using SPSS version 13 (SPSS Inc., Chicago, IL).
Results
Detection of lentivirus transfection
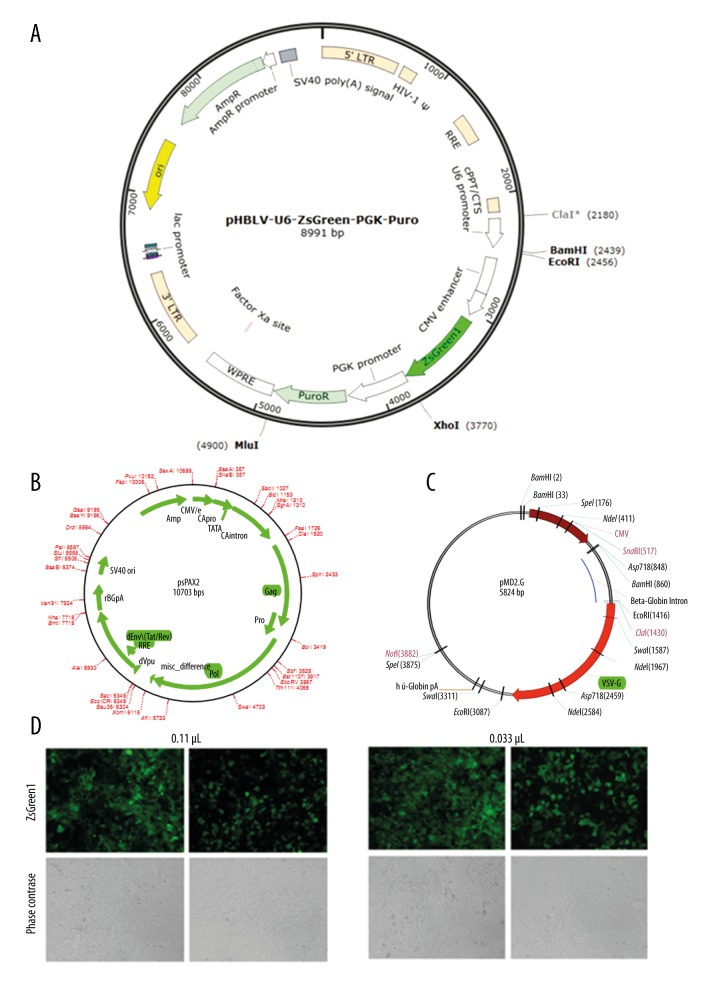
The construction of the Piezo1-sh RNA lentivirus vector is shown in Figure 1A. And the lentivirus packaging was according as the Figure 1B and 1C. The dilution method of counting was adopted as the lentivirus titer detection as shown Figure 1D and 1E. The titer of the virus (TU/mL)=cell counting × percentage of fluorescence × MOI × virus dilution factor ×103. The titer of the negative control group=4×104×10% x MOI(1)×30×103=1×108 TU/mL. And the virus titer of the Piezo1-shRNA group=4×104×20% × MOI(1)×30×103=2×108 TU/mL.
Figure 1.
Construction of the Piezo1-shRNA lentivirus vector. (A) The mapping of the pHBLV-U6-ZsGreen-PGK-Puro vector. (B, C) The mapping of the construction of the Piezo1-ShPiezo1 lentivirus vector. (D) The virus titer of the negative control group at 0.11 μL and 0.033 μL (100×). (E) The virus titer of the Piezo1-shRNA group at 0.11 μL and 0.033 μL (100×).
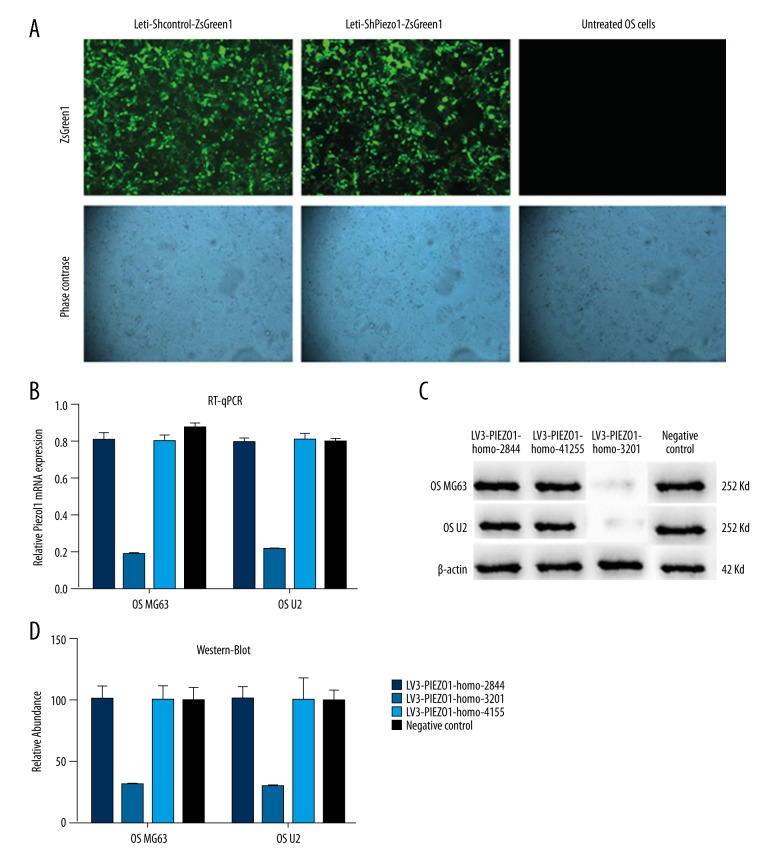
The selection of the effective Piezo1-shRNA
The result of the detection of the three designed group as LV3-PIEZO1-homo-2844, LV3-PIEZO1-homo-3201, LV3-PIEZO1-homo-4155 and the negative control group are shown in Figure 2. From the result of the RT-qPCR (Figure 2B.) and the western blotting (Figure 2C, 2D), we explored whether LV3-PIEZO1-homo-3201 could act as Piezo1-shRNA which could be an inhibitor of the Piezo1. So in the following experiment, we selected the LV3-PIEZO1-homo-3201 as the sh-Piezo1 group (Tables 3, 4).
Figure 2.
The selection of the effective Piezo1-shRNA (A) The letivirus effection of the osteosarcoma (OS) cells which divided into sh-control group and the sh-Piezo1 group. (B) The detection of the RT-qPCR of the three groups and the negative control group in OS MG63 and OS U2 cell line. (C) The detection of the western-blot of the three groups and the negative control group in OS MG63 and OS U2 cell line. (D) The relative abundance of the western blotting in the three groups and the negative control group.
Table 3.
The Expression of the three group of the Piezo1 with RT-PCR (χ̄±s, n=3).
| Groups | OS MG63 | OS U2 |
|---|---|---|
| LV3-PIEZO1-homo-2844 | 0.795±0.008 | 0.782±0.003 |
| LV3-PIEZO1-homo-3201 | 0.179±0.001 | 0.235±0.004 |
| LV3-PIEZO1-homo-4155 | 0.812±0.004 | 0.812±0.007 |
| Negative control | 0.915±0.003 | 0.773±0.002 |
Table 4.
The expression of the three groups of Piezo1 with western-blot (χ̄±s, n=3).
| Groups | OS MG63 | OS U2 |
|---|---|---|
| LV3-PIEZO1-homo-2844 | 100.23±1.112 | 100.14±1.321 |
| LV3-PIEZO1-homo-3201 | 25.79±2.279a | 23.25±2.492a |
| LV3-PIEZO1-homo-4155 | 102.12±2.324 | 102.82±4.327 |
| Negative control | 95.15±3.311 | 100.53±6.002 |
Indicates p<0.05, compared with other groups in the OS MG63 and the OS U2 cells.
The result of the RT-qPCR and the western blotting under stretch force
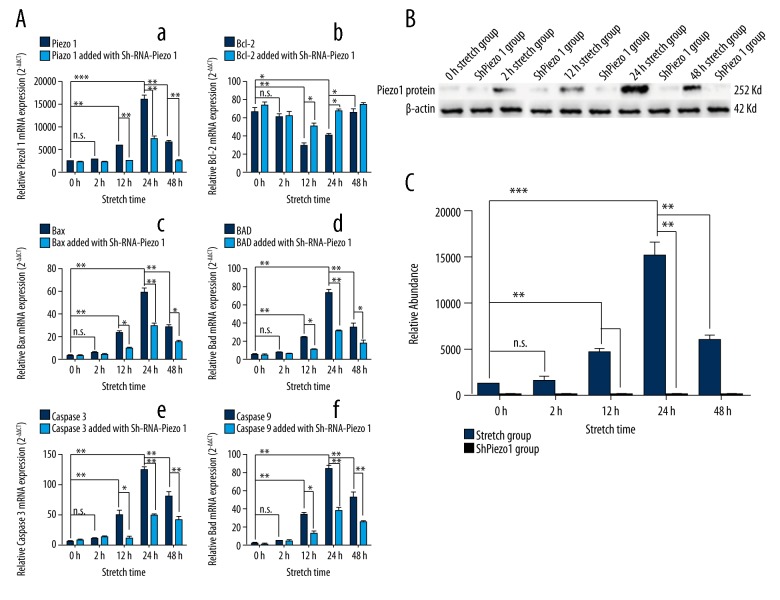
The expression of the Piezo1 was closely related with the stretch, and Piezo1 in the 12-hour stretch group was highly expressed than the 0-hour stretch group (n=3, p<0.05, one-way ANOVA). While the 24-hour stretch group was highly than the 12-hour stretch group (n=3, p<0.05, one-way ANOVA) and the 48-hour stretch group (n=3, p<0.05, one-way ANOVA) (Figure 3Aa). However, the expression of the Piezo1 in the 2-hour stretch group was slightly higher than the 0-hour stretch group, and the difference was not statistically significant (n=3, p>0.05, one-way ANOVA). The apoptotic genes, such as the Bax, BAD, caspase-3, and caspase-9, had the same characteristics as the Piezo1 expression under the stretch force (Figure 3Ac–3Af). However the Bcl-2, acted as the anti-apoptosis gene was the opposite expression as shown in Figure 3Ab as well as Figure 3B, and 3C. The result of the western blotting also showed the same trend as the result of the RT-qPCR.
Figure 3.
The result of the RT-qPCR and the western blotting under stretch force. (A) The detection of the RT-qPCR about the expression of the Piezo1 and the apoptosis gene such as Bcl-2, Bax, BAD, caspase-3, and caspase-9 under stretch force. (B) The detection of the western-Blot about the expression of the Piezo1 under stretch force. (C) The relative abundance of the western blotting. The results are presented as the means ±SD, n=3; ns means p>0.05 versus the control group; * p<0.05 versus the control group; ** p<0.01 versus the control group; one-way ANOVA.
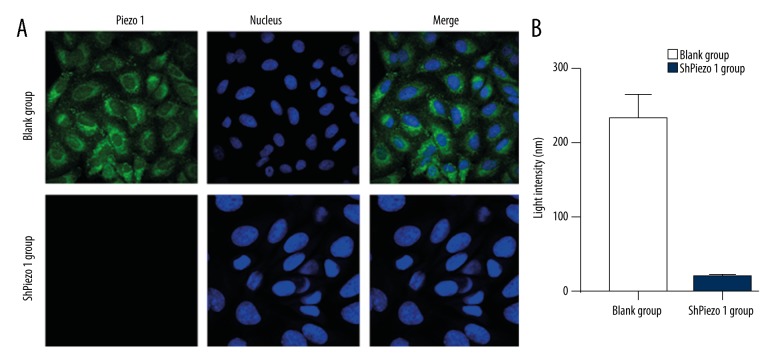
The novel MA ion channel Piezo1 protein immunofluorescence in OS cells
We performed immunofluorescence analysis to examine the expression and localization of the MA ion channel Piezo1 protein (Figure 4A). Piezo1 was expressed in human OS cells, and it localized in the cell membrane. The detection of the express of the Piezo1 with the immunofluorescence had shown that the Piezo1-sh-RNA group was lower than the blank group (n=3, p<0.05, Student’s t-test) (Figure 4B).
Figure 4.
The novel MA ion channel Piezo1 protein immunofluorescence in osteosarcoma (OS) cells. (A) The location of the novel MA ion channel Piezo1 protein-specific immunolabelling in the OS cells (600×). The nucleus was stained by DAPI (blue). Piezo1 localized in the cytoplasm of the OS cells. (A, B) The light intensity of the immunolabelling of the Piezo1 protein. The express of the Piezo1 with the Immunofluorescence had shown that the Piezo1-sh-RNA group was lower than the blank (BC) group. The results are presented as the means ±SD, n=3; ns means p>0.05 versus the control group; * p<0.05 versus the control group; ** p<0.01 versus the control group; one-way ANOVA.
The invasion assay by Transwell experiment
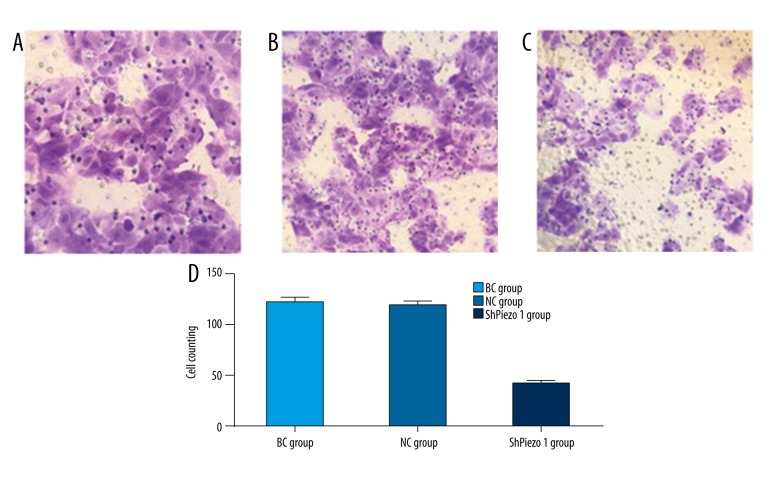
The cell counting of the OS cells to penetrate the membrane in Piezo1-shRNA group was significantly reduced compared with the blank group (n=3, p<0.05, Student’s t-test) and the negative control group (n=3, p<0.05, Student’s t-test) after virus transfection (Figure 5). However, the number of the cells to penetrate the membrane in the blank group was slightly increased compared to the negative control group, and there was no statistical difference (n=3, p>0.05, Student’s t-test).
Figure 5.
The result of the Transwell experiment. (A) The blank group (BC group) (200×). (B) The negative control group (NC group) (200×). (C) The shPiezo1 group (200×). (D) The cell counts of the cells penetrating the membrane of the three groups. The results are presented as the means ±SD, n=3; ns means p>0.05 versus the control group; * p<0.05 versus the control group; ** p<0.01 versus the control group; one-way ANOVA.
CCK-8 assay
The cell proliferation rate of the OS cells in the 12-hour stretch group was highly than the 0-hour stretch group (n=3, p<0.05, one-way ANOVA). While the 24-hour stretch group was highly than the 12-hour stretch group (n=3, p<0.05, one-way ANOVA) and the 48-hour stretch group (n=3, p<0.05, one-way ANOVA) (Table 5, Figure 6). However, the OS cells proliferation rate in the 2-hour stretch group were slightly higher than the 0-hour stretch group, and the difference was not statistically significant (n=3, p>0.05, one-way ANOVA). And in the 12-hour stretch group, the cell proliferation rate of the cells was highly than the blank group (n=3, p<0.05, Student’s t-test); in the 24-hour stretch group, the cell proliferation rate of the cells was highly than the blank group (n=3, p<0.05, Student’s t-test)
Table 5.
The cell proliferation rate of the groups (χ̄±s, n=3, %).
| Groups | Blank group | shPiezo1 group |
|---|---|---|
| 0-hour stretch group | 124.43±10.12 | 131.32±34.39 |
| 2-hour stretch group | 125.78±12.29 | 123.95±12.41 |
| 12-hour stretch group | 42.32±9.47a | 103.82±19.32 |
| 24-hour stretch group | 28.35±3.31a | 101.23±6.82 |
| 48-hour stretch group | 38.76±2.94a | 103.234±10.11 |
Indicates p < 0.05 compared with other groups at the same time point.
Figure 6.
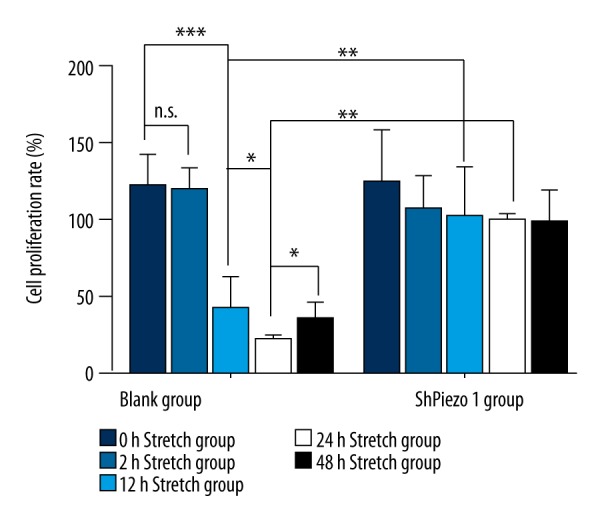
The result of the CCK-8. The cell proliferation rate of the osteosarcoma cells in the 0-hour stretch group, 2-hour stretch group, 12-hour stretch group, 24-hour stretch group, and the 48-hour stretch group. The results are presented as the means ±SD, n=3; ns means p>0.05 versus the control group; * p<0.05 versus the control group; ** p<0.01 versus the control group; one-way ANOVA.
OS cell apoptosis by Hoechst staining and Annexin V-PI staining
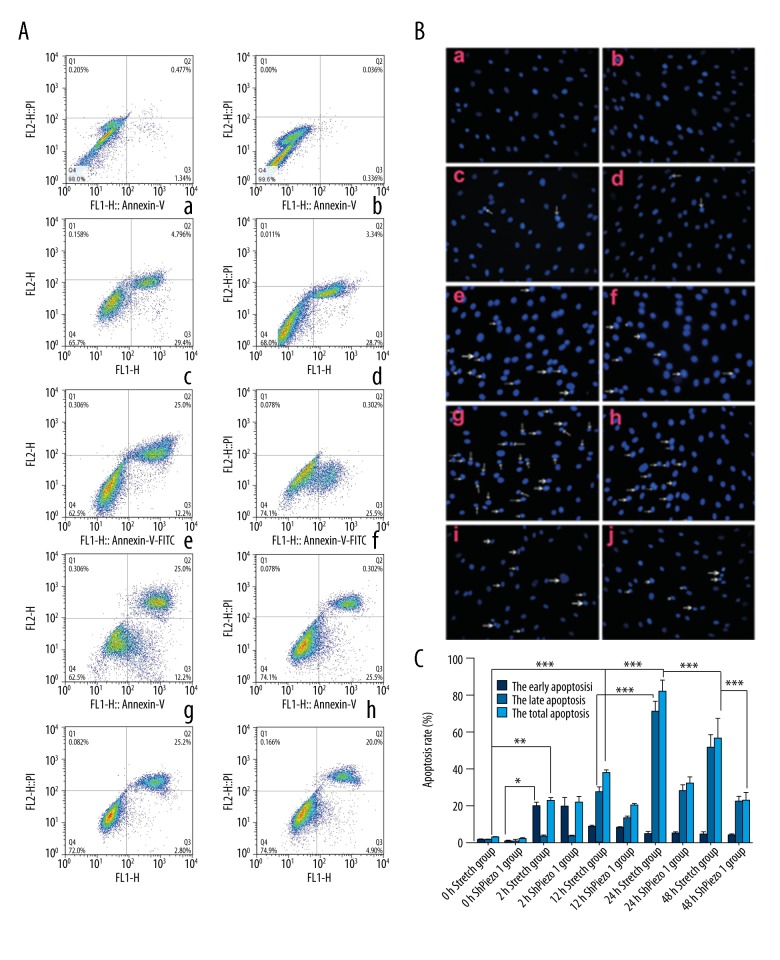
We performed Annexin V binding, PI staining and flow cytometry to analyze OS cell apoptosis. The results showed a time-dependent shift in the apoptotic response to mechanical stretch (Figure 7A). The amount of apoptosis was lower after 48 hours compared to 24 hours. We observed a time-dependent shift in apoptosis. Treatment with shPiezo1RNA could block cell apoptosis. We used GraphPad Prism 5.0 to analyze apoptosis. The results showed that the 2-hour group was characterized by early stages of apoptosis, with few cells in late apoptosis (n=3, p>0.05, Student’s t-test) (Figure 7Ac). The highest rate of apoptosis appeared in the 24-hour group (n=3, p<0.05, Student’s t-test). Meanwhile, shPiezo1RNA inhibited late stage apoptosis in the 24-hour group and the 12-hour group (n=3, p<0.05, Student’s t-test).
Figure 7.
(A) Images of nuclei shrinking and evident nuclear condensation of osteosarcoma (OS) cells (shown by arrowhead) after mechanical stretch forces of 0, 2, 12, 24, and 48 hours. Notes: (a) 0-hour group 200×. (b) 0-hour plus shPiezo1RNA group 200x. (c) 2-hour group 200×. (d) 2-hour plus shPiezo1RNA group 200×. (e) 12-hour group 200×. (f) 12-hour plus shPiezo1RNA group 200×. (g) 24-hour group 200×. (h) 24-hour plus shPiezo1RNA group 200×. (i) 48-hour group 200×. (j) 48-hour plus shPiezo1RNA group 200×. (B) Results of flow cytometry detection of 0S cell apoptosis in 0, 2, 12, 24, and 48 hours and shPiezo1RNA groups. Q1: (AnnexinV-FITC)−/PI+, necrotic cells. Q2: (AnnexinV-FITC)+/PI+, late stage apoptotic cells. Q3: (AnnexinV-FITC)+/PI−, early stage apoptotic cells. Q4: (AnnexinV-FITC)−/PI−, live cells. Notes: (a) 0-hour group. (b) 0-hour group plus shPiezo1RNA. (c) 2-hour group. (d) 2-hour group plus shPiezo1RNA. (e) 12-hour group. (f) 12-hour group plus shPiezo1RNA. (g) 24-hour group. (h) 24-hour group plus shPiezo1RNA. (i) 48-hour group. (j) 48-hour group plus shPiezo1RNA. (C) Early stage apoptosis of human OS cells in 0, 2, 12, 24, 48 hors and shPiezo1RNA groups. After a 2 hour mechanical stretch, the rate of early stage apoptosis significantly increased. After a 12 hour mechanical stretch, the late stage apoptotic rate increased and peaked in the 24-hour group. The late stage apoptotic rate was lower in the 48-hour group compared to the 24-hour group. The results are presented as the means ±SD, n=3; ns means p>0.05 versus the control group; * p<0.05 versus the control group; ** p<0.01 versus the control group; one-way ANOVA.
The nuclei of the apoptotic OS cells appeared shrunken and condensed (Figure 7B). In the 12-hour group and the 24-hour group, the number of apoptotic OS cells was higher compared to the 0-hour group, but the amount of apoptosis was lower in the 48-hour group compared to the 24-hour group.
In vivo anti-tumor experiment
OS cells were successfully inoculated in all of the study nude mice, and subcutaneous tumors formed and gradually increased. After treatment, tumor growth was marked in the BC and NC group; no mice died in either group. After four weeks of treatment, tumor growth was significantly inhibited in the Piezo1-shRNA group compared with the BC group (n=3, p<0.05, Student’s t-test) and the NC group (n=3, p<0.05, Student’s t-test) (Table 6). However, during the early stage of the treatment, tumor growth was not significantly inhibited in the Piezo1-shRNA group compared with the BC group (n=3, p>0.05, Student’s t-test) and the NC group (n=3, p>0.05, Student’s t-test) (Figure 7C).
Table 6.
The tumor size in the nude mice.
| 2w | 3w | 4w | 5w | 6w | |
|---|---|---|---|---|---|
| BC group | 0.0742±0.0100 | 0.1242±0.0089 | 0.6753±0.0351 | 1.0021±0.0029 | 1.1766±0.3109 |
| NC group | 0.0669±0.0129 | 0.1288±0.0091 | 0.6942±0.0308 | 0.9792±0.0031 | 0.0742±0.0311 |
| Piezo1-shRNA group | 0.0710±0.0098 | 0.1319±0.0073 | 0.1739±0.0009a | 0.1841±0.0056a | 0.2081±0.0077a |
Indicates p < 0.05, compared with other groups at the same time point.
BC (blank control group containing serum-free and antibiotic-free DMEM without shRNA) group, NC (negative control group containing shRNA plasmid without any target gene) group. The results are presented as the means ±SD, n=3; ns means p>0.05 versus the control group;
p<0.05 versus the control group;
p<0.01 versus the control group; one-way ANOVA.
Discussion
Piez1 ion channel is a novel mechanical activated ion channel, which was discovery recently. Current research has focused on the structure of the Piezo1 protein. In this study, we explored the function of the Piezo1 ion channel in human OS cells. We found that the Piezo1 protein could act as the promoter in the overload mechanical stretch induced apoptosis of the OS cells in in vitro experiments and played the role of the inhibitor in the cell proliferation of the OS cells. Also, in the in vivo experiment, we find that the special inhibitor of the Piezo1, and Piezo1-shRNA could promote tumor growth in nude mice.
Previous studies [18–22] have identified the exact structure, ion permeation, and mechano-transduction of the Piezo1 ion channel in various physiological and pathophysiological processes in mammals. The Piezo1 protein has been shown to form a remarkable three-bladed, propeller-shaped homotrimeric channel complex comprising a separable ion-conducting pore module and mechano-transduction modules. In this study, we focused on the function about the Piezo1 in human OS cells. Through the FX-4000T Tension Plus System, we built the mechanical stretch model in vitro, and the result of the RT-qPCR and western-blot showed that Piezo1 was closely related with the stretch, and Piezo1 in the 12-hour stretch group was highly expressed compared to the 0-hour stretch group (n=3, p<0.05, one-way ANOVA). While the 24-hour stretch group was higher compared to the 12-hour stretch group (n=3, p<0.05, one-way ANOVA) and the 48-hour stretch group (n=3, p<0.05, one-way ANOVA) (Figure 3Aa). However, the expression of the Piezo1 in the 2-hour stretch group were slightly higher than the 0-hour stretch group, and the difference was not statistically significant (n=3, p>0.05, one-way ANOVA). The apoptotic genes, such as the Bax, BAD, caspase-3 and caspase-9, had the same characteristics as the Piezo1 expression under the stretch force (Figure 3Ac–3Af). However, Bcl-2 acted as the anti-apoptosis gene and had the opposite expression as shown in Figure 3Ab. The result of the western blotting also showed the same trend as the result of the RT-qPCR. In addition, we also explored the connection between the result of the western blotting and the RT-PCR using Spearman’s correlation analysis; the results showed that the RT-PCR results were correlated with the results of the western blotting (r=0.441, p≤0.05). From this result, we concluded that Piezo1 was closely related to the mechanical force in human OS cells, and with the apoptosis of the OS cells.
Some studies [23,24] have shown that Piezo1 could act as a promoter in the mechanical-reduce over apoptosis of the chondrocytes and the human OS cells. In this study, we also explored the connection between Piezo1 and the OS cells proliferation, invasion and apoptosis. The result of the CCK-8 analysis showed the cell proliferation rate of the OS cells in the 12-hour stretch group was highly than the 0-hour stretch group (n=3, p<0.05, one-way ANOVA). While the 24-hour stretch group was highly than the 12-hour stretch group (n=3, p<0.05, one-way ANOVA) and the 48-hour stretch group (n=3, p<0.05, one-way ANOVA) (Figure 6). However, the OS cells proliferation rate in the 2-hour stretch group were slightly higher than the 0-hour stretch group, and the difference was not statistically significant (n=3, p>0.05, one-way ANOVA). And in the 12-hour stretch group, the cell proliferation rate of the cells was highly than the BC group (n=3, p<0.05, Student’s t-test); in the 24-hour stretch group, the cell proliferation rate of the cells was higher than the BC group (n=3, p<0.05, Student’s t-test). The Transwell experiment showed that Piezo1-shRNA group was significantly reduced compared with the BC group (n=3, p<0.05, Student’s t-test) and the negative control group (n=3, p<0.05, Student’s t-test) after virus transfection. However, the number of the cells to penetrate the membrane in the blank group was slightly increased compared to the NC group, there was no statistical difference (n=3, p>0.05, Student’s t-test). In addition, the results showed a time-dependent shift in the apoptotic response to mechanical stretch (Figure 7A). The amount of apoptosis was lower after 48 hours compared to 24 hours. We observed a time-dependent shift in apoptosis. Treatment with shPiezo1RNA could block cell apoptosis. We used GraphPad Prism 5.0 to analyze apoptosis. The results showed that the 2-hour group was characterized by early stages of apoptosis, with few cells in late apoptosis (n=3, p>0.05, Student’s t-test) (Figure 7Ac). The highest rate of apoptosis appeared in the 24-hour group (n=3, p<0.05, Student’s t-test). Meanwhile, shPiezo1RNA inhibited late stage apoptosis in the 24-hour group and 12-hour group (n=3, P<0.05, Student’s t-test). The nuclei of the apoptotic OS cells appeared shrunken and condensed (Figure 7B). In the 12-hour group and the 24-hour group, the number of apoptotic OS cells was higher compared to the 0-hour group, but the amount of apoptosis was lower in the 48-hour group compared to the 24-hour group. From these results, we concluded that the novel ion channel Piezo1 protein could promote the invasion and apoptosis of OS cells and inhibitor the proliferation of OS cells, especially after the 24-hour stretch mechanical force treatment.
Conclusions
We explore the function of the novel MA ion channel Piezo1 ion channel in human OS cells, and we found that Piezo1 protein could be detected in human OS cells and that Piezo1-shRNA could inhibited the expression of the Piezo1. Also, Piezo1 could promote the invasion and apoptosis of OS cells and inhibit the proliferation of OS cells
Footnotes
Source of support: This work was supported by the National Natural Science Foundation of China (NO.81272056)
Conflict of interest
None.
References
- 1.Ene R, Sinescu RD, Ene P, et al. Proximal tibial osteosarcoma in young patients: Early diagnosis, modular reconstruction. Rom J Morphol Embryol. 2015;56:413–17. [PubMed] [Google Scholar]
- 2.Liebner DA. The indications and efficacy of conventional chemotherapy in primary and recurrent sarcoma. J Surg Oncol. 2015;111:622–31. doi: 10.1002/jso.23866. [DOI] [PubMed] [Google Scholar]
- 3.Durfee RA, Mohammed M, Luu HH. Review of osteosarcoma and current management. Rheumatol Ther. 2016;3:221–43. doi: 10.1007/s40744-016-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coste B, Mathur J, Schmidt M, et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coste B, Xiao B, Santos JS, et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature. 2012;483(7388):176–81. doi: 10.1038/nature10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarychanski R, Schulz VP, Houston BL, et al. Mutations in the mechanotransduction protein PIEZO1 are associated with hereditary xerocytosis. Blood. 2012;120(9):1908–15. doi: 10.1182/blood-2012-04-422253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyamoto TMH, Nakagomi S, Kira T, et al. Piezo1, a novel mechanosensor in the bladder urothelium, transmits signals of bladder sensation. Eur Urol. 2012;31(6):1015–17. [Google Scholar]
- 8.Gottlieb PA, Bae C, Gnanasambandam R. Piezo1 mutations identified in xerocytosis alter the inactivation rate. Biophys J. 2013;104(2):467A. [Google Scholar]
- 9.Demolombe S, Duprat F, Honoré E, Patel A. Slower Piezo1 inactivation in dehydrated hereditary stomatocytosis (xerocytosis) Biophysl J. 2013;105(4):833–34. doi: 10.1016/j.bpj.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zarychanski R, Schulz VP, Houston BL, et al. Multiple clinical forms of dehydrated hereditary stomatocytosis arise from mutations in PIEZO1. Blood. 2013;121(19):3925–35. S1–12. doi: 10.1182/blood-2013-02-482489. [DOI] [PubMed] [Google Scholar]
- 11.Coste B, Houge G, Murray MF, et al. Gain-of-function mutations in the mechanically activated ion channel PIEZO2 cause a subtype of Distal Arthrogryposis. PNAS. 2013;110(12):4667–72. doi: 10.1073/pnas.1221400110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMillin MJ, Beck AE, Chong JX, et al. Mutations in PIEZO2 cause Gordon syndrome, Marden-Walker syndrome, and distal arthrogryposis type 5. Am J Hum Genet. 2014;94(5):734–44. doi: 10.1016/j.ajhg.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green DR, Llambi F. Cell death signaling. Cold Spring Harb Perspect Biol. 2015;7(12) doi: 10.1101/cshperspect.a006080. pii: a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiren KM, Toombs AR, Semirale AA, Zhang X. Osteoblast and osteocyte apoptosis associated with androgen action in bone: Requirement of increased Bax/Bcl-2 ratio. Bone. 2006;38(5):637–51. doi: 10.1016/j.bone.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 15.Mocetti P, Silvestrini G, Ballanti P, et al. Bcl-2 and Bax expression in cartilage and bone cells after high-dose corticosterone treatment in rats. Tissue Cell. 2001;33(1):1–7. doi: 10.1054/tice.2000.0144. [DOI] [PubMed] [Google Scholar]
- 16.Yang E, Zha J, Jockel J, et al. Bad, a heterodimeric partner for Bc-lxL and Bc-l2, displacesBax and promotes cell death. Cell. 1995;80(2):285–91. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 17.Yi J, Wu L, Liu Z, et al. High-intensity focused ultrasound ablation induced apoptosis in human hepatocellular carcinoma. Hepatogastroenterology. 2014;61(136):2336–39. [PubMed] [Google Scholar]
- 18.Geng J, Zhao Q, Zhang T, Xiao B. In touch with the mechanosensitive Piezo channels: Structure, ion permeation, and mechanotransduction. Curr Top Membr. 2017;79:159–95. doi: 10.1016/bs.ctm.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Gao N, Yang M. The structural basis for sensing by the Piezo1 protein. Curr Top Membr. 2017;79:135–58. doi: 10.1016/bs.ctm.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Ge J, Li W, Zhao Q, et al. Architecture of the mammalian mechanosensitive Piezo1 channel. Nature. 2015;527(7576):64–69. doi: 10.1038/nature15247. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Q, Wu K, Chi S, et al. Heterologous expression of the Piezo1-ASIC1 chimera induces mechanosensitive currents with properties distinct from Piezo1. Neuron. 2017;94(2):274–77. doi: 10.1016/j.neuron.2017.03.040. [DOI] [PubMed] [Google Scholar]
- 22.Vora NL, Powell B, Brandt A, et al. Prenatal exome sequencing in anomalous fetuses: New opportunities and challenges. Genet Med. 2017 doi: 10.1038/gim.2017.33. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XF, Zhang Z, Chen ZK, et al. Piezo1 protein induces the apoptosis of human osteoarthritis-derived chondrocytes by activating caspase-12, the signaling marker of ER stress. Int J Mol Med. 2017;40(3):845–53. doi: 10.3892/ijmm.2017.3075. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Li XF, Leng P, Zhang Z, Zhang HN. The Piezo1 protein ion channel functions in human nucleus pulposus cell apoptosis by regulating mitochondrial dysfunction and the endoplasmic reticulum stress signal pathway. Exp Cell Res. 2017;358(2):377–89. doi: 10.1016/j.yexcr.2017.07.010. [DOI] [PubMed] [Google Scholar]