Abstract
HDL cholesterol (HDL-C) remains a superior biochemical predictor of CVD risk, but its genetic basis is incompletely defined. In patients with extreme HDL-C concentrations, we concurrently evaluated the contributions of multiple large- and small-effect genetic variants. In a discovery cohort of 255 unrelated lipid clinic patients with extreme HDL-C levels, we used a targeted next-generation sequencing panel to evaluate rare variants in known HDL metabolism genes, simultaneously with common variants bundled into a polygenic trait score. Two additional cohorts were used for validation and included 1,746 individuals from the Montréal Heart Institute Biobank and 1,048 individuals from the University of Pennsylvania. Findings were consistent between cohorts: we found rare heterozygous large-effect variants in 18.7% and 10.9% of low- and high-HDL-C patients, respectively. We also found common variant accumulation, indicated by extreme polygenic trait scores, in an additional 12.8% and 19.3% of overall cases of low- and high-HDL-C extremes, respectively. Thus, the genetic basis of extreme HDL-C concentrations encountered clinically is frequently polygenic, with contributions from both rare large-effect and common small-effect variants. Multiple types of genetic variants should be considered as contributing factors in patients with extreme dyslipidemia.
Keywords: genetics, genomics, dyslipidemias, genes in lipid dysfunction, complex trait, rare variants, common variants, next-generation sequencing, polygenic risk score
Despite apprehension over its direct causal role in atherogenesis and its value as a drug target (1), HDL cholesterol (HDL-C) remains a valid biochemical predictor of CVD risk (2–4). Understanding the full range of factors that determine plasma HDL-C concentrations, including genetics, still has relevance for epidemiology and risk projection (5). Furthermore, specific etiologies of extreme perturbations of HDL-C may have clinical importance in terms of diagnosis and directed therapies (1, 6).
Multiple genetic factors could be present in an individual, creating a polygenic network of influential determinants on HDL-C (7–9). These determinants include monogenic disorders (10, 11), such as extremely low or absent HDL-C due to rare homozygous mutations in ABCA1, LCAT, and APOA1 (12–15), and extremely elevated HDL-C due to rare homozygous mutations in CETP, LIPC, and SCARB1 (16–18). In contrast, the potential role of other genetic determinants in extreme HDL-C phenotypes, namely common SNPs (1), has not been systematically evaluated.
Polygenic factors, assessed by tallying SNPs with small phenotypic effects to derive “genetic risk scores” or “polygenic trait scores” (PTSs), contribute to numerous medical conditions, including coronary artery disease (19) and diabetes (20). Among dyslipidemias, polygenic factors play a substantial role in familial hypercholesterolemia (FH) (21), which was previously considered an archetypal “monogenic” disorder. For instance, in patients referred with extremely elevated LDL cholesterol (LDL-C), targeted next-generation sequencing demonstrated that ∼50% of individuals had rare heterozygous large-effect variants, whereas another ∼16% had an accumulation of common small-effect variants (i.e., SNPs) identified previously from genome-wide association studies (GWASs) as determinants of LDL-C (22). Although earlier sequencing experiments indicate that 11%–35% of patients with extremely low HDL-C and 5%–20% of patients with extremely high HDL-C have rare heterozygous large-effect variants (8, 23–28) driving the phenotypes, the proportion of such patients with excessive GWAS-identified SNPs, as quantified using PTSs, is unknown.
Here we used targeted next-generation sequencing to robustly characterize the genetic determinants influencing HDL-C levels in patients with extremely low and high HDL-C. This allowed us to concurrently evaluate the burden of rare large-effect variants and common small-effect GWAS variants, the latter bundled into a PTS. We saw that ∼30% of individuals at each HDL-C extreme had an identifiable genetic determinant, with the PTS explaining a marked increment above simple tallies of heterozygous large-effect variants. Our findings illustrate that both types of determinants are enriched in individuals with extremely high and low HDL-C levels.
MATERIALS AND METHODS
Study subjects
Two hundred and fifty-five unrelated patients with extreme HDL-C levels were selected for study from the Lipid Genetics Clinic at the London Health Sciences Centre, University Hospital (London, Ontario, Canada). Extremely low HDL-C was defined as ≤0.8 mmol/l (30.9 mg/dl) and ≤1.0 mmol/l (38.7 mg/dl) in males and females, respectively (N = 136). Extremely high HDL-C was defined as ≥1.4 mmol/l (54.1 mg/dl) and ≥1.8 mmol/l (69.6 mg/dl) in males and females, respectively (N = 119). These thresholds adhere closely to the top and bottom 10th percentiles of HDL-C levels in a North American population (29). The two patient exclusion criteria were triglyceride levels of ≥3.37 mmol/l (298.5 mg/dl) (as low HDL-C can simply be secondary to elevated triglycerides, which have their own distinct determinants) and diagnosis of clinical syndromes of extreme HDL-C (e.g., Tangier disease). All patients provided signed informed consent with approval from the Western University ethics review board (no. 07290E). As a reference group for the PTS analysis, the European subgroup of the 1000 Genomes Project (1KG; N = 503) was assumed to model the normal distribution of HDL-C levels among primarily normolipidemic individuals from the general population. A validation cohort from the Montréal Heart Institute (MHI) Biobank, ascertained as previously described (30), included individuals with extremely low HDL-C (N = 201), individuals with high HDL-C (N = 347), and normolipidemic controls (N = 1,198). A second validation cohort from the University of Pennsylvania (UPenn), ascertained as previously described (18, 31), included individuals with extremely low HDL-C (N = 349) as well as high HDL-C (N = 699). The studies in all clinic patients in this study were in compliance with the Declaration of Helsinki.
DNA processing and sequencing
Genomic DNA from the Lipid Genetics Clinic were isolated with the Puregene® DNA Blood Kit (Gentra Systems, Qiagen Inc., Mississauga, Ontario, Canada) (cat. no. 158389). Samples were indexed and pooled with the Nextera® Rapid Capture Custom Enrichment Kit (cat. no. FC-140-1009) “LipidSeq” design (32). Genomic libraries of our enriched samples were sequenced with an Illumina MiSeq personal sequencer (Illumina, San Diego, CA). Sequencing and genotyping methods performed at the MHI Biobank (30) and UPenn (18) have been described in detail previously.
Annotation of genetic variants
FASTQ files were generated for each patient and imported into CLC Bio Genomics Workbench (version 7.5; CLC Bio, Aarhus, Denmark) for read alignment against the human reference genome (build hg19), variant calling, and coverage statistics for targeted regions. Variants were annotated with customized ANNOVAR annotation scripts (33). Annotation methods performed at the MHI Biobank (30) and UPenn (18) have been described in detail previously.
Identification of rare large-effect variants
Variants likely to produce extreme HDL-C phenotypes were identified using a specific set of criteria for both the Lipid Genetics Clinic and MHI cohorts. Definite causative variants previously reported as being phenotype-inducing in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/all.php) (34) were identified immediately. Of the remaining coding and splice-site variants, filters were applied for a minor allele frequency of <1% or missing according to the 1KG (http://browser.1000genomes.org/index.html) (35), Exome Sequencing Project (http://evs.gs.washington.edu/EVS/), and the Exome Aggregation Consortium (http://exac.broadinstitute.org/) (36) databases. Of the remaining coding variants, in silico predictions of deleterious or damaging outcomes were required for half of the available prediction tools, including Polymorphism Phenotyping (version 2; PolyPhen2; http://genetics.bwh.harvard.edu/pph2/) (37), Sorting Intolerant From Tolerant (SIFT; http://sift.jcvi.org/) (38), MutationTaster (http://www.mutationtaster.org/), or Combined Annotation Dependent Depletion (CADD; http://cadd.gs.washington.edu/score) (39). Concordance for pathogenic outcomes for Splicing Based Analysis of Variants (http://tools.genes.toronto.edu/) (40) and Automated Splice Site and Exon Definition Analyses (http://splice.uwo.ca/) (41) were necessary to identify splice-site variant candidates.
Of the variants meeting the above criteria, those within lipid-associated genes with direct (primary) and indirect (secondary) HDL-C-altering effects (supplemental Table S1) were designated as large-effect mutations; our use of the term “mutation” here is relatively colloquial, because it connotes the likely pathogenic nature of the large-effect variants.
It is important to note that because the UPenn cohort comes from an established on-going study (the UPenn High HDL-C Study), the criteria used in identifying large-effect variants differ slightly from what was considered here (18, 31). As such, carrier patients with rare large-effect variants found through that project were excluded from our study to ensure consistent results for subsequent analyses.
Polygenic trait score
Between the discovery cohort and 1KG cohort, genotype data for 34 HDL-C-associated SNPs were available for study; these SNPs were selected from the most recent GWAS meta-analyses on blood lipids and lipoproteins, published by the Global Lipids Genetics Consortium (42). A PTS encompassing all available SNPs was calculated for all patients. In the interest of future application and usability, smaller SNP sets of 10 or fewer were tested and compared with the original set of 34; the aim was to select a smaller number of SNPs that were just as informative as was the full set of 34.
Scores were calculated by using a weighted approach; the number of HDL-C alleles associated with high HDL-C at a locus (0, 1, or 2) were summed and multiplied by the reported effect size for the high HDL-C-associated allele. The SNP products for each locus were totaled to provide the overall PTS for an individual. The underlying assumption when calculating the PTS was that each allele had an additive effect on their respective HDL-C phenotypes. Higher scores indicated that individuals carried a greater number of high HDL-C-associated alleles, and lower scores indicated that individuals carried fewer alleles associated with high HDL-C and therefore carried a greater number of low HDL-C-associated alleles.
Statistical analysis
Differences between groups for mean PTS were evaluated with one-tailed, unpaired Wilcoxon rank-sum tests assuming unequal variances and are reported as the mean ± SD. Odds ratios (ORs) were derived using two-by-two contingency tables. Statistical analyses were conducted using SAS (version 9.3; SAS Institute, Cary, NC). Statistical significance was defined as P < 0.05 for the mean PTS comparisons and ORs.
RESULTS
Subject characteristics
Clinical and demographic information for the patients from the Lipid Genetics Clinic, the MHI Biobank, and UPenn are summarized in Table 1; the UPenn cohort has also been described in detail previously (31). In the Lipid Genetics Clinic, the majority of individuals reported European descent, with <3% of individuals reporting Asian or African ancestry. In the MHI Biobank cohort, all individuals are of French-Canadian ancestry.
TABLE 1.
Demographics of extreme HDL-C patient cohorts
| Lipid Genetics Clinic | Montréal Heart Institute Biobank | University of Pennsylvania | ||||||||||
| Low HDL-C | High HDL-C | Low HDL-C | High HDL-C | Low HDL-C | High HDL-C | |||||||
| Males | Females | Males | Females | Males | Females | Males | Females | Males | Females | Males | Females | |
| N | 90 | 46 | 60 | 59 | 131 | 70 | 280 | 67 | 202 | 147 | 217 | 482 |
| Age | 48.1 ± 16.8a | 45.4 ± 12.5a | 58.5 ± 14.2 | 58.6 ± 10.5 | 64.4 ± 10.4 | 68.9 ± 8.4 | 65.6 ± 10.1 | 71.2 ± 7.2 | 56.0 ± 12.2 | 53.2 ± 15.0 | 58.7 ± 14.9 | 58.2 ± 11.7 |
| BMI | 29.0 ± 5.6a | 28.8 ± 6.0a | 26.5 ± 3.7 | 25.3 ± 3.5a | 31.0 ± 5.2a | 31.4 ± 6.4a | 26.9 ± 4.5a | 26.4 ± 6.0a | 32.4 ± 5.0a | 34.5 ± 7.4a | 29.0 ± 5.0a | 27.2 ± 7.0a |
| TC | 4.2 ± 1.4 | 5.8 ± 2.3 | 5.7 ± 1.4 | 6.9 ± 1.5 | 3.4 ± 1.1 | 3.7 ± 1.0 | 4.5 ± 1.0a | 5.2 ± 1.1 | 4.0 ± 1.1 | 4.5 ± 1.3 | 6.5 ± 1.6 | 6.4 ± 1.2 |
| HDL-C | 0.6 ± 0.2 | 0.8 ± 0.2 | 2.1 ± 0.5 | 2.7 ± 0.7 | 0.7 ± 0.1 | 0.9 ± 0.1 | 1.7 ± 0.2 | 2.1 ± 0.3 | 0.8 ± 0.2 | 0.9 ± 0.2 | 2.5 ± 0.5 | 2.9 ± 0.5 |
| LDL-C | 2.7 ± 1.3 | 4.0 ± 2.3 | 3.2 ± 1.4 | 3.7 ± 1.5 | 2.2 ± 1.0 | 2.2 ± 0.9 | 2.4 ± 0.9a | 2.5 ± 0.9 | 2.7 ± 1.3 | 3.2 ± 1.7 | 3.0 ± 1.2 | 3.6 ± 2.0 |
| TG | 2.2 ± 1.3 | 2.0 ± 1.1 | 1.0 ± 0.5 | 1.2 ± 0.6 | 2.2 ± 0.7 | 2.3 ± 0.7 | 1.4 ± 0.6 | 1.3 ± 0.5 | 1.8 ± 0.7 | 1.6 ± 0.6 | 0.9 ± 0.4 | 0.9 ± 0.4 |
| CVD Hx (%) | 45.2a | 21.9a | 29.8a | 16.7a | 60.3 | 67.1 | 40.7 | 31.3 | 11.9a | 12.9a | 6.0a | 4.1a |
Values are indicative of the mean ± SD. Lipid values are in millimoles per liter. CVD Hx, CVD history; TC, total cholesterol.
An incomplete dataset was used for these calculations.
Rare large-effect variants in HDL-C-altering genes
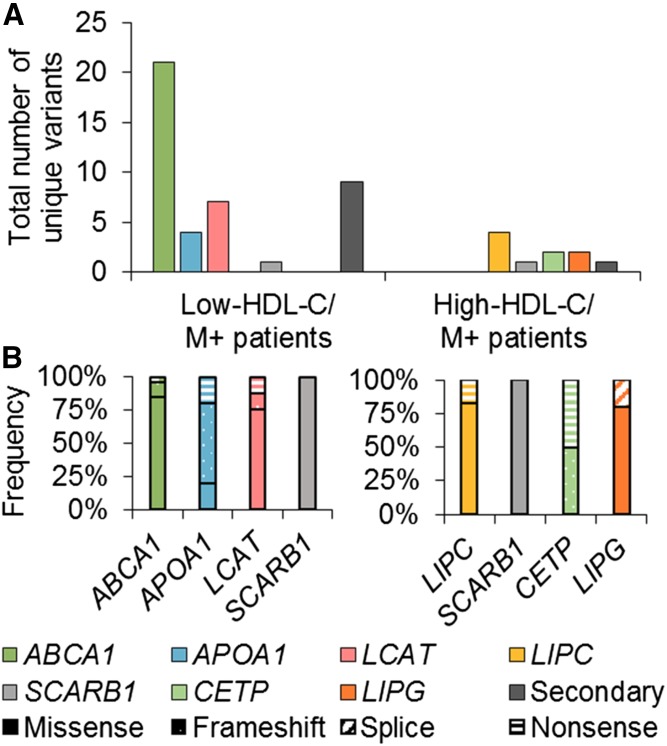
Considering the primary genes associated with low HDL-C (i.e., ABCA1, APOA1, LCAT) and high HDL-C (i.e., LIPC, SCARB1, CETP, and LIPG), we identified 43 unique large-effect variants that are either likely or definite causes of either extreme (Fig. 1A and supplemental Table S2). When considering variant types, 72.1% were missense, 4.7% were splicing, 14.0% were frameshift, and 9.3% were nonsense variants (Fig. 1B). One individual was homozygous for ABCA1 p.G851R, and one individual was a compound heterozygote for ABCA1 p.W590C and p.W590L. A single individual carried rare heterozygous variants in both a low and a high HDL-C-associated gene, that is, ABCA1 and SCARB1, and presented with a low HDL-C phenotype.
Fig. 1.
Summary of rare large-effect variant types in the Lipid Genetics Clinic cohort. Forty-three unique variants were identified in primary genes, and 10 unique variants were identified in secondary genes; there were 68 variants total. A: Total number of unique variants per gene per patient group. B: Frequency of variant type for each unique variant for the low-HDL-C M+ patients (left) and high-HDL-C M+ patients (right).
Only a few large-effect variants were identified in secondary HDL-C-altering genes (supplemental Table S2). In nine low-HDL-C patients, missense variants were identified in LPL, APOA5, LMF1, GPD1, and APOE. In two high-HDL-C patients, the same splicing variant was identified in APOC3. All variants in the secondary genes were heterozygous.
Overall, 30.1% and 12.6% of low- and high-HDL-C patients, respectively, carried at least one variant that could explain their phenotypes and were labeled “mutation positive” (M+). Conversely, patients who were noncarriers were labeled “mutation negative” (M−). Patients were grouped by HDL-C phenotype and mutation status (i.e., M+ or M−) (supplemental Fig. S1).
In the MHI validation cohort, 10.9% of patients with low HDL-C and 10.4% of patients with high HDL-C carried large-effect variants, respectively. The same distinctions between phenotype and M+ or M− status were applied. In the UPenn validation cohort, because different criteria were used in large-effect variant identification, only M− patients were considered for this study.
Measuring accumulation of common small-effect variants using a polygenic trait score
A set of nine SNPs produced results that most closely matched the results from the original 34-SNP score and was used as the primary score in this study. The nine SNPs were in linkage equilibrium and showed significant primary associations with plasma levels of HDL-C; some of the loci were previously implicated either directly or indirectly to HDL metabolism (Table 2). Each SNP was selected on the basis of their reported β coefficients, or “effect sizes,” reported P values, and frequency within the general population. The allele associated with higher HDL-C levels was taken as the primary variable.
TABLE 2.
Nine SNPs used in polygenic trait score
| Chr:Position | rsID | Gene | High HDL-C-Associated Allele | Effect Size | Relation with HDL-C or HDL Metabolism |
| 1:182199750 | rs1689800 | ZNF648 | A | 0.034 | Mechanism underlying association is poorly characterized. |
| 1:230159944 | rs4846914 | GALNT2 | A | 0.048 | Recently confirmed as an important determinant of HDL-C (43) |
| 9:104902020 | rs1883025 | ABCA1 | C | 0.07 | Causative gene for Tangier disease (6) |
| 12:109562388 | rs7134594 | MVK | T | 0.035 | MVK encodes mevalonate kinase, which is involved in biosynthesis of cholesterol and isoprenoids (44), although the closely linked MMAB gene encoding cob(I)alamin adenosyltransferase may actually underlie the HDL-C association at this locus (45). |
| 12:124777047 | rs838880 | SCARB1 | C | 0.048 | Causative gene for scavenger receptor B1 deficiency |
| 15:58391167 | rs1532085 | LIPC | A | 0.107 | Causative gene for hepatic lipase deficiency |
| 16:56959412 | rs3764261 | CETP | A | 0.241 | Causative gene for cholesteryl ester transfer protein deficiency |
| 16:81501185 | rs2925979 | CMIP | C | 0.035 | Mechanism underlying association is poorly characterized. |
| 19:8368312 | rs7255436 | ANGPTL4 | A | 0.032 | Regulates LPL with reciprocal effects on triglycerides and HDL-C (46) |
SNP information from this table has been extracted from (42). Chr, chromosome; rsID, reference SNP cluster ID (accession number).
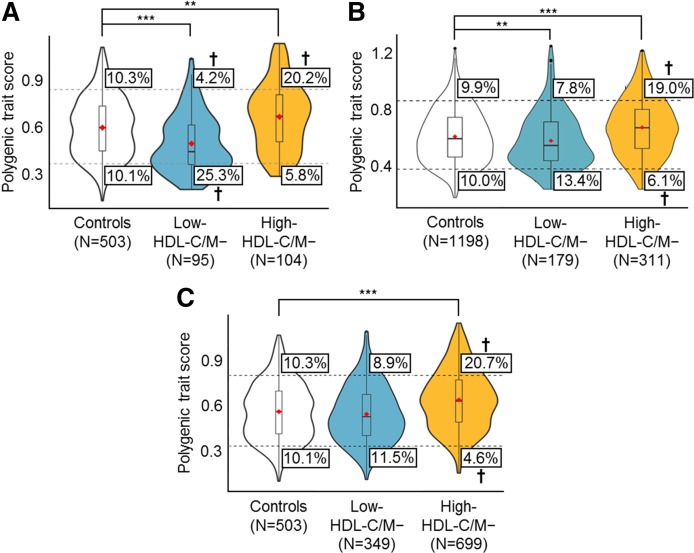
The PTS results for the Lipid Genetics Clinic cohort were analyzed by phenotype and mutation status rather than on an individual level and were visualized with violin plots (Fig. 2A), which illustrate the distribution of PTSs within reference individuals (i.e., the European subgroup of the 1KG population) and low- or high-HDL-C patients. Neither the low- nor high-HDL-C/M+ group had a mean PTS that was significantly different from the reference population.
Fig. 2.
PTS analysis for low- and high-HDL-C/M− patients. Violin plots (similar to box plots, except that they also show the probability density of the data at different values) illustrate the distribution of polygenic scores for individuals in the control, low-HDL-C/M−, and high-HDL-C/M− groups in the Lipid Genetics Clinic cohort (A), the MHI Biobank cohort (B), and UPenn cohort (C). Red diamonds mark the mean PTS of the group. The total area of a single plot represents 100% of individuals in that group; wider sections of each plot represent an increased number of individuals with scores at that point, and narrower sections represent a decreased number of individuals. The top dashed line and bottom dashed line represent the threshold for the top 10th and bottom 10th percentiles of PTSs in the control population, respectively. The boxes indicate the percentage of individuals falling above or below the percentile thresholds. †Percentile groups with significant ORs (Tables 3, 4); **P < 0.01; ***P < 0.0001.
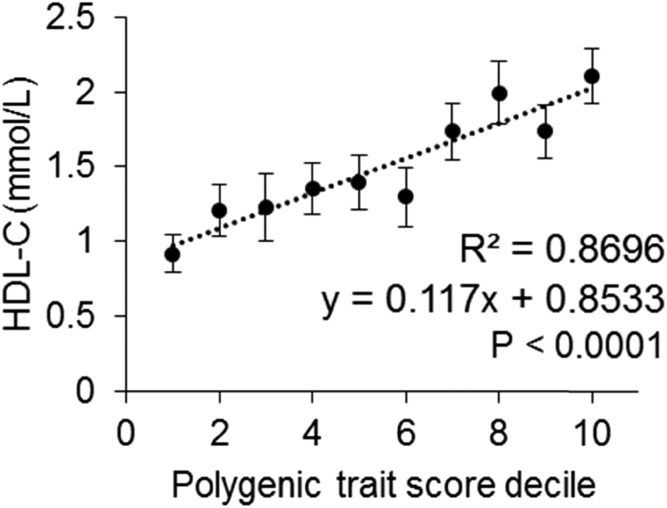
However, the mean PTSs for the low-HDL-C/M− patients (0.48 ± 0.18, unpaired Wilcoxon rank-sum, P < 0.0001) and the high-HDL-C/M− patients (0.65 ± 0.21, unpaired Wilcoxon rank-sum, P = 0.0015) were significantly less than and greater than the reference population (0.58 ± 0.19), respectively. In addition, 25.3% of low-HDL-C/M− patients fell below the bottom 10th percentile in comparison with 10.1% of the reference population (OR: 3.00 [95% CI: 1.67–5.35], P < 0.0001), whereas 20.2% of high-HDL-C/M− patients fell above the top 10th percentile in comparison with 10.3% of the reference population (OR: 2.19 [95% CI: 1.21–3.96], P = 0.006). A primarily polygenic basis for extreme HDL-C was considered for M− patients with an “extreme” PTS, below the bottom 10th or above the top 10th percentile for low- or high-HDL-C phenotypes, respectively (supplemental Fig. S2). When patients are grouped by PTS decile, there is a strong linear relationship between increasing PTS and HDL-C level (Fig. 3).
Fig. 3.
Mean HDL-C level in low- and high-HDL-C/M− patients from the Lipid Genetics Clinic by PTS decile. There is a strong linear relationship between increasing PTSs and HDL-C levels, as is indicated by the R2 value of 0.8696 (P < 0.0001). Vertical bars indicate standard errors of the mean.
External validation of polygenic trait score
Results from the MHI validation cohort were similar to those of the Lipid Genetics Clinic cohort. Groups with a mean PTS that was significantly different from the reference population (0.58 ± 0.19) were the low-HDL-C/M− patients (0.55 ± 0.20, P = 0.007) and the high-HDL-C/M− patients (0.64 ± 0.20, P < 0.0001) (Fig. 2B). Additionally, only the high-HDL-C/M− patients showed a significant OR: 19.0% of patients fell above the top 10th percentile in comparison with 9.9% of reference individuals (OR: 2.12 [95% CI: 1.48–3.02], P < 0.0001) (Tables 3, 4). From the UPenn validation cohort, only the high-HDL-C/M− individuals (0.66 ± 0.20, P < 0.0001) had a mean PTS that was significantly different from that of the reference population (0.58 ± 0.19) (Fig. 2C). Similarly, only the high-HDL-C/M− patients showed a significant OR: 20.7% of patients fell above the top 10th percentile in comparison with 10.3% of reference individuals (OR: 2.27 [95% CI: 1.59–3.24], P < 0.0001) (Tables 3, 4).
TABLE 3.
PTS comparison of patients based on low HDL-C levels and mutation status
| Top 10th Percentile of PTS | Bottom 10th Percentile of PTS | |||||||
| Control | M+ | M− | OR (95% CI, P) | Control | M+ | M− | OR (95% CI, P) | |
| Lipid Genetics Clinic | 52/503 | 4/41 | 4/95 | M+: 0.94 (0.27–2.90, 0.583) | 51/503 | 8/41 | 24/95 | M+: 2.15 (0.86–5.19, 0.063) |
| M−: 0.38 (0.11–1.13, 0.038) | M−: 3.00 (1.67–5.35, <0.0001) | |||||||
| MHI Biobank | 119/1,198 | 3/22 | 14/179 | M+: 1.43 (0.33–5.21, 0.380) | 120/1,198 | 1/22 | 24/179 | M+: 0.43 (0.02–3.03, 0.341) |
| M−: 0.77 (0.41–1.41, 0.228) | M−: 1.39 (0.85–2.27, 0.107) | |||||||
| UPenn | 52/503 | NA | 31/349 | M+: NA | 51/503 | NA | 40/349 | M+: NA |
| M−: 0.85 (0.52–1.38, 0.280) | M−: 1.15 (0.72–1.82, 0.307) | |||||||
| Overall | 171/1,701a | 7/63 | 49/623 | M+: 1.12 (0.46–2.60, 0.455) | 171/1,701a | 9/63 | 88/623 | M+: 1.49 (0.67–3.20, 0.186) |
| M−: 0.76 (0.54–1.08, 0.063) | M−: 1.47 (1.11–1.96, <0.01) | |||||||
Whole numbers represent patient counts, with the numerator indicating the number of individuals with a PTS in the top 10th or bottom 10th percentile and the denominator indicating the total number of individuals in the patient subgroup. ORs were calculated with a confidence level of 95%. Left-tailed and right-tailed tests of significance were considered for the top 10th and bottom 10th percentiles, respectively. NA, not available. Boldface type indicates statistically significant ORs.
The 1KG group was counted only once, given that it was used in both the Lipid Genetics Clinic and UPenn cohorts.
TABLE 4.
PTS comparison of patients based on high HDL-C levels and mutation status
| Top 10th Percentile of PTS | Bottom 10th Percentile of PTS | |||||||
| Control | M+ | M− | OR (95% CI, P) | Control | M+ | M− | OR (95% CI, P) | |
| Lipid Genetics Clinic | 52/503 | 2/15 | 21/104 | M+: 1.33 (0.20–6.47, 0.476) | 51/503 | 0/15 | 6/104 | M+: 0 (0–3.11, 0.207) |
| M−: 2.19 (1.21–3.96, <0.01) | M−: 0.54 (0.20–1.36, 0.110) | |||||||
| MHI Biobank | 119/1,198 | 4/36 | 59/311 | M+: 1.13 (0.33–3.44, 0.490) | 120/1,198 | 2/36 | 19/311 | M+: 0.53 (0.09–2.29, 0.291) |
| M−: 2.12 (1.49–3.03, <0.0001) | M−: 0.59 (0.34–0.99, 0.019) | |||||||
| UPenn | 52/503 | NA | 145/699 | M+: NA | 51/503 | NA | 32/699 | M+: NA |
| M−: 2.27 (1.59–3.24, <0.0001) | M−: 0.43 (0.26–0.69, <0.0001) | |||||||
| Overall | 171/1,701a | 6/51 | 225/1,114 | M+: 1.19 (0.45–2.97, 0.412) | 171/1,701a | 2/51 | 57/1,114 | M+: 0.37 (0.6–1.55, 0.106) |
| M−: 2.27 (1.82–2.83, <0.0001) | M−: 0.48 (0.35–0.67, <0.0001) | |||||||
Whole numbers represent patient counts, with the numerator indicating the number of individuals with a PTS in the top 10th or bottom 10th percentile and the denominator indicating the total number of individuals in the patient subgroup. ORs were calculated with a confidence level of 95%. Right-tailed and left-tailed tests of significance were considered for the top 10th and bottom 10th percentiles, respectively. Boldface type indicates statistically significant ORs.
The 1KG group was counted only once, given that it was used in both the Lipid Genetics Clinic and UPenn cohorts.
DISCUSSION
We report a polygenic trait score for HDL-C that expands the proportion of individuals with extreme levels that can be explained genetically. In our subjects, we first confirmed an excess of rare heterozygous large-effect variants in ABCA1, LCAT, and APOA1, and in CETP, LIPC, LIPG, and SCARB1 among individuals with extremely low and high HDL-C, respectively. Overall, 18.7% of low- and 10.9% of high-HDL-C patients carried at least one such variant. Then, among remaining individuals with extreme levels and without such rare variants, we showed an ∼1.5- to 2-fold increased risk of having an extreme PTS due to multiple common small-effect variants known to influence HDL-C levels from earlier GWAS (P < 0.01 for each extreme). Overall, 12.8% of low-HDL-C patients and 19.3% of high-HDL-C patients had an extreme PTS. Cumulatively, >30% of individuals had either a rare large-effect variant or a bundle of common small-effect variants associated with their respective extreme HDL-C phenotype. Our study highlights the importance of polygenic effects as determinants of extreme HDL-C and reinforces the polygenic nature of this complex trait.
In Lipid Genetics Clinic patients, 47.7% and 30.2% of low- and high-HDL-C patients, respectively, had identifiable genetic contributors to their extreme phenotypes. The prevalence of M+ patients in the low-HDL-C subgroup was higher than was the proportion of M+ patients in the MHI cohort, perhaps reflecting ascertainment bias. Mean HDL-C levels were markedly lower in the clinically ascertained low-HDL-C extreme subgroup than in the MHI and UPenn cohorts; rare large-effect variants may be more important determinants of the phenotype. Furthermore, there was no excess of extreme PTSs among M+ individuals with both extremes of HDL-C in both study cohorts, suggesting that when a large-effect variant was present, it was the main determinant of the extreme HDL-C phenotype, overriding small polygenic effects.
In contrast, among clinically ascertained M− individuals with low HDL-C, a large excess had low PTSs (OR: 3.00 [95% CI: 1.67–5.35], P < 0.0001). There were nonsignificant trends to low PTSs among M− patients from MHI and UPenn, leading to an excess risk for a low PTS in the overall low-HDL-C/M− sample (OR: 1.47 [95% CI: 1.11–1.96], P < 0.01). This pattern was mirrored by respective deficits of high PTSs in the same subgroups (Table 3). This demonstrates that individuals with low HDL-C and no large-effect variants have a significant polygenic contribution of small-effect variants. In the Lipid Genetics Clinic, MHI, and UPenn cohorts, among clinically ascertained M− individuals with high HDL-C, many had a high PTS (overall OR: 2.27 [95% CI: 1.82–2.83], P < 0.0001). This pattern was mirrored by deficits of low PTSs in the same subgroups (Table 4). This demonstrates that among individuals with high HDL-C and no large-effect variants, there was a significant polygenic contribution from small-effect variants.
We also found that M+ individuals with the respective phenotype and concurrent excess PTS did not have HDL-C levels that were significantly different from those of M+ individuals with normal PTSs (data not shown). This suggests that rare large-effect variants and polygenic determinants are independent and, when present together, are not necessarily additive: rare large-effect variants appear to predominantly determine the HDL-C phenotype. This contrasts with conclusions derived from a whole-genome sequence analysis of individuals with less extreme phenotypes, in whom common variants were determined to be the predominant determinants of HDL-C (47). Of course, our cohorts were still relatively small: a possible additive or synergistic relationship between large- and small-effect variants will require evaluation in much larger samples of such extreme individuals.
Application of PTSs, or “genetic risk scores,” is becoming popular in the area of cardiovascular health and related complex traits (48). Mendelian randomization studies have previously evaluated genetic risk scores to infer a causal role of HDL-C in CVD outcomes (49). However, until now there has been minimal to no evaluation of polygenic scores in individuals selected for extremes of HDL-C levels.
Among extreme dyslipidemias, PTSs have been well studied in cohorts of patients with extremely high LDL-C levels, particularly FH. In fact, the genetic architecture is analogous among individuals with FH and those with extreme HDL-C studied here. For instance, among clinically ascertained individuals with likely FH, 50%–80% have a rare heterozygous large-effect variant in LDLR, APOB, or PCSK9, whereas another 15%–20% have an extreme PTS comprising small-effect SNPs for LDL-C (21, 22). The exact proportions of individuals with large- and small-effect variants differ in our cohorts with extreme HDL-C levels, but the overall pattern of genetic contributors to both complex lipoprotein traits is similar. One possible difference is that syndromic FH was intentionally enriched in the extreme LDL-C studies, whereas we excluded patients with known clinical syndromes of extreme HDL-C.
Also, for LDL-C, only individuals with extremely high levels are typically studied. In contrast, our current study assessed individuals with both very low and very high HDL-C extremes. The fact that our PTS was directionally associated with both extremes of HDL-C (i.e., excessive high and low PTS among individuals with high and low HDL-C phenotypes, respectively) indicates that this score applies bidirectionally for HDL-C and is thus relatively unique among such scores evaluated for most quantitative metabolic traits.
There may be clinical relevance in knowing the genetic basis of a patient’s extreme HDL-C level. For instance, in patients with high LDL-C, the CVD risk compared to normolipidemic individuals was ∼22-fold higher in those who carried a rare heterozygous large-effect variant in comparison with ∼6-fold higher among those who did not (50). Although polygenic effects were not evaluated, extreme LDL-C in at least some individuals in the latter subgroup likely had a polygenic basis. Although both groups are at high risk having such patient substrata, it generates hypotheses for different interventions under the framework of precision medicine. For instance, prospective randomized studies may show that among individuals with extremely high LDL-C, carriers of a rare variant may benefit relatively more from certain treatments, such as PCSK9 inhibitors, than might individuals with a polygenic basis (51). By analogy, individuals with extremely low HDL-C who carry a rare variant versus those who have a high polygenic burden can be studied to determine whether there are differential effects of therapies targeted toward raising HDL-C (52).
This study has some limitations. First, patient ascertainment differed among the three cohorts: Lipid Genetics Clinic patients were referred because of abnormal lipid profiles; MHI Biobank participants were recruited on the basis of cardiovascular health; and though UPenn patients also came from lipid referrals, there was more focus on high-HDL-C phenotypes. This may explain why the low-HDL-C patients from the discovery cohort had a greater burden of rare variants: these individuals’ HDL-C phenotypes were more pronounced and perhaps more likely to have a genetic basis. In contrast, because CVD was of primary interest at the MHI, abnormal HDL-C profiles were less extreme and perhaps more often secondary to other, nongenetic health issues. Testing the PTS in additional cohorts with more closely matched patient-ascertainment parameters would not only increase the power of our study but also alleviate these biases. Second, application of the PTS assumes that each allele has a linearly additive effect, with no epistatic interactions. Modeling epistasis could improve PTS accuracy and comprehension. Third, the PTS was tested largely in individuals of European ancestry and may not be generalizable to other ethnic groups. Also, we did not evaluate other factors, such as epigenetic regulators or large copy-number variations, as possible explanations for their extreme phenotypes. Additionally, some important variants may have been overlooked, because only specific known genes associated with HDL-C were screened and only a subset of HDL-C-associated SNPs were considered; this could have led to a skew in the percentage of M+ patients or patients with an extreme accumulation of polygenic SNPs. Finally, given that low-pass whole-genome sequencing was used to genetically characterize participants from the MHI Biobank, it is possible that rare variants in the HDL-C candidate genes may have been missed. Despite these limitations, we have for the first time demonstrated the genetic complexity underlying extreme HDL-C phenotypes by considering both rare variants and the accumulation of common SNPs simultaneously.
In summary, we concurrently detected both rare large-effect and common small-effect variants using our next-generation sequencing platform. In patients with both low- and high- HDL-C extremes, we confirmed the enrichment of rare large-effect variants, and simultaneously detected individuals with extreme PTSs. This substantially expanded the number of individuals with a genetic basis for their phenotype: about one-sixth of patients with extreme HDL-C had an extreme PTS. Loci for rare and common variants contributing to extreme HDL-C levels encode products acting at all stages of the HDL lifecycle; we suggest that both rare and common variants be considered concurrently for understanding extreme HDL-C. The large proportion of individuals still unaccounted for can be studied for additional mechanisms, such as possible new genes, gene-gene or gene-environment interactions, and nonmendelian influences, including mitochondrial or epigenetic effects. In addition to acquiring a more complete genetic picture of patients with extreme dyslipidemia, stratifying them genetically may help evaluate interindividual differences in their clinical course or responses to interventions.
Supplementary Material
Footnotes
Abbreviations:
- 1KG
- 1000 Genomes Project
- FH
- familial hypercholesterolemia
- GWAS
- genome-wide association study
- HDL-C
- HDL cholesterol
- LDL-C
- LDL cholesterol
- M+
- mutation positive
- M−
- mutation negative
- MHI
- Montréal Heart Institute
- OR
- odds ratio
- PTS
- polygenic trait score
- UPenn
- University of Pennsylvania
This work was supported by the Jacob J. Wolfe Distinguished Medical Research Chair, the Edith Schulich Vinet Research Chair in Human Genetics, and the Martha G. Blackburn Chair in Cardiovascular Research, as well as operating grants from the Canadian Institutes of Health Research (Foundation Award), the Heart and Stroke Foundation of Ontario (G-15-0009214), and Genome Canada, through Genome Quebec (award 4530) (R.A.H.). This work was also supported by the Montréal Heart Institute foundations and the Desmarais family, in support of Genome Quebec/Genome Canada. R.A.H. is a consultant and speakers’ bureau member for Aegerion, Amgen, Boston Heart Diagnostics, Cerenis, Eli Lilly, Gemphire, Pfizer, and Sanofi. The other authors have no disclosures.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Rosenson R. S. 2016. The high-density lipoprotein puzzle: why classic epidemiology, genetic epidemiology, and clinical trials conflict? Arterioscler. Thromb. Vasc. Biol. 36: 777–782. [DOI] [PubMed] [Google Scholar]
- 2.Di Angelantonio E., Gao P., Pennells L., Kaptoge S., Caslake M., Thompson A., Butterworth A. S., Sarwar N., Wormser D., Saleheen D., et al. 2012. Lipid-related markers and cardiovascular disease prediction. JAMA. 307: 2499–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perk J., De Backer G., Gohlke H., Graham I., Reiner Z., Verschuren M., Albus C., Benlian P., Boysen G., Cifkova R., et al. 2012. European guidelines on cardiovascular disease prevention in clinical practice (version 2012). The fifth joint task force of the European Society of Cardiology and other societies on cardiovascular disease prevention in clinical practice (constituted by representatives of nine societies and by invited experts). Eur. Heart J. 33: 1635–1701. [Erratum. 2012. Eur. Heart J. 33: 2126.] [DOI] [PubMed] [Google Scholar]
- 4.Parish S., Offer A., Clarke R., Hopewell J. C., Hill M. R., Otvos J. D., Armitage J., and Collins R.. 2012. Lipids and lipoproteins and risk of different vascular events in the MRC/BHF heart protection study. Circulation. 125: 2469–2478. [DOI] [PubMed] [Google Scholar]
- 5.Raffield L. M., Cox A. J., Hsu F. C., Ng M. C., Langefeld C. D., Carr J. J., Freedman B. I., and Bowden D. W.. 2013. Impact of HDL genetic risk scores on coronary artery calcified plaque and mortality in individuals with type 2 diabetes from the diabetes heart study. Cardiovasc. Diabetol. 12: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hovingh G. K., Rader D. J., and Hegele R. A.. 2015. HDL re-examined. Curr. Opin. Lipidol. 26: 127–132. [DOI] [PubMed] [Google Scholar]
- 7.Hegele R. A. 2009. Plasma lipoproteins: genetic influences and clinical implications. Nat. Rev. Genet. 10: 109–121. [DOI] [PubMed] [Google Scholar]
- 8.Cohen J. C., Kiss R. S., Pertsemlidis A., Marcel Y. L., McPherson R., and Hobbs H. H.. 2004. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 305: 869–872. [DOI] [PubMed] [Google Scholar]
- 9.Motazacker M. M., Peter J., Treskes M., Shoulders C. C., Kuivenhoven J. A., and Hovingh G. K.. 2013. Evidence of a polygenic origin of extreme high-density lipoprotein cholesterol levels. Arterioscler. Thromb. Vasc. Biol. 33: 1521–1528. [DOI] [PubMed] [Google Scholar]
- 10.Weissglas-Volkov D., and Pajukanta P.. 2010. Genetic causes of high and low serum HDL-cholesterol. J. Lipid Res. 51: 2032–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dron J. S., and Hegele R. A.. 2016. Genetics of lipid and lipoprotein disorders and traits. Curr. Genet. Med. Rep. 4: 130–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks-Wilson A., Marcil M., Clee S. M., Zhang L. H., Roomp K., van Dam M., Yu L., Brewer C., Collins J. A., Molhuizen H. O., et al. 1999. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat. Genet. 22: 336–345. [DOI] [PubMed] [Google Scholar]
- 13.Kuivenhoven J. A., Stalenhoef A. F., Hill J. S., Demacker P. N., Errami A., Kastelein J. J., and Pritchard P. H.. 1996. Two novel molecular defects in the lcat gene are associated with fish eye disease. Arterioscler. Thromb. Vasc. Biol. 16: 294–303. [DOI] [PubMed] [Google Scholar]
- 14.Ng D. S., Leiter L. A., Vezina C., Connelly P. W., and Hegele R. A.. 1994. Apolipoprotein A-I Q[-2]x causing isolated apolipoprotein A-I deficiency in a family with analphalipoproteinemia. J. Clin. Invest. 93: 223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaefer E. J., Anthanont P., Diffenderfer M. R., Polisecki E., and Asztalos B. F.. 2016. Diagnosis and treatment of high density lipoprotein deficiency. Prog. Cardiovasc. Dis. 59: 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegele R. A., Little J. A., Vezina C., Maguire G. F., Tu L., Wolever T. S., Jenkins D. J., and Connelly P. W.. 1993. Hepatic lipase deficiency. Clinical, biochemical, and molecular genetic characteristics. Arterioscler. Thromb. 13: 720–728. [DOI] [PubMed] [Google Scholar]
- 17.Inazu A., Brown M. L., Hesler C. B., Agellon L. B., Koizumi J., Takata K., Maruhama Y., Mabuchi H., and Tall A. R.. 1990. Increased high-density lipoprotein levels caused by a common cholesteryl-ester transfer protein gene mutation. N. Engl. J. Med. 323: 1234–1238. [DOI] [PubMed] [Google Scholar]
- 18.Zanoni P., Khetarpal S. A., Larach D. B., Hancock-Cerutti W. F., Millar J. S., Cuchel M., DerOhannessian S., Kontush A., Surendran P., Saleheen D., et al. 2016. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science. 351: 1166–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McPherson R., and Tybjaerg-Hansen A.. 2016. Genetics of coronary artery disease. Circ. Res. 118: 564–578. [DOI] [PubMed] [Google Scholar]
- 20.Bonnefond A., and Froguel P.. 2015. Rare and common genetic events in type 2 diabetes: what should biologists know? Cell Metab. 21: 357–368. [DOI] [PubMed] [Google Scholar]
- 21.Talmud P. J., Shah S., Whittall R., Futema M., Howard P., Cooper J. A., Harrison S. C., Li K., Drenos F., Karpe F., et al. 2013. Use of low-density lipoprotein cholesterol gene score to distinguish patients with polygenic and monogenic familial hypercholesterolaemia: a case-control study. Lancet. 381: 1293–1301. [DOI] [PubMed] [Google Scholar]
- 22.Wang J., Dron J. S., Ban M. R., Robinson J. F., McIntyre A. D., Alazzam M., Zhao P. J., Dilliott A. A., Cao H., Huff M. W., et al. 2016. Polygenic versus monogenic causes of hypercholesterolemia ascertained clinically. Arterioscler. Thromb. Vasc. Biol. 36: 2439–2445. [DOI] [PubMed] [Google Scholar]
- 23.Singaraja R. R., Sivapalaratnam S., Hovingh K., Dube M. P., Castro-Perez J., Collins H. L., Adelman S. J., Riwanto M., Manz J., Hubbard B., et al. 2013. The impact of partial and complete loss-of-function mutations in endothelial lipase on high-density lipoprotein levels and functionality in humans. Circ Cardiovasc Genet. 6: 54–62. [DOI] [PubMed] [Google Scholar]
- 24.Tietjen I., Hovingh G. K., Singaraja R. R., Radomski C., Barhdadi A., McEwen J., Chan E., Mattice M., Legendre A., Franchini P. L., et al. 2012. Segregation of LIPG, CETP, and GALNT2 mutations in Caucasian families with extremely high HDL cholesterol. PLoS One. 7: e37437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiss R. S., Kavaslar N., Okuhira K., Freeman M. W., Walter S., Milne R. W., McPherson R., and Marcel Y. L.. 2007. Genetic etiology of isolated low HDL syndrome: incidence and heterogeneity of efflux defects. Arterioscler. Thromb. Vasc. Biol. 27: 1139–1145. [DOI] [PubMed] [Google Scholar]
- 26.Holleboom A. G., Kuivenhoven J. A., Peelman F., Schimmel A. W., Peter J., Defesche J. C., Kastelein J. J., Hovingh G. K., Stroes E. S., and Motazacker M. M.. 2011. High prevalence of mutations in LCAT in patients with low HDL cholesterol levels in the Netherlands: identification and characterization of eight novel mutations. Hum. Mutat. 32: 1290–1298. [DOI] [PubMed] [Google Scholar]
- 27.Candini C., Schimmel A. W., Peter J., Bochem A. E., Holleboom A. G., Vergeer M., Dullaart R. P., Dallinga-Thie G. M., Hovingh G. K., Khoo K. L., et al. 2010. Identification and characterization of novel loss of function mutations in ATP-binding cassette transporter a1 in patients with low plasma high-density lipoprotein cholesterol. Atherosclerosis. 213: 492–498. [DOI] [PubMed] [Google Scholar]
- 28.Sadananda S. N., Foo J. N., Toh M. T., Cermakova L., Trigueros-Motos L., Chan T., Liany H., Collins J. A., Gerami S., Singaraja R. R., et al. 2015. Targeted next-generation sequencing to diagnose disorders of HDL cholesterol. J. Lipid Res. 56: 1993–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rifkind B. M., and Segal P.. 1983. Lipid research clinics program reference values for hyperlipidemia and hypolipidemia. JAMA. 250: 1869–1872. [PubMed] [Google Scholar]
- 30.Low-Kam C., Rhainds D., Lo K. S., Provost S., Mongrain I., Dubois A., Perreault S., Robinson J. F., Hegele R. A., Dube M. P., et al. 2016. Whole-genome sequencing in French Canadians from Quebec. Hum. Genet. 135: 1213–1221. [DOI] [PubMed] [Google Scholar]
- 31.Edmondson A. C., Brown R. J., Kathiresan S., Cupples L. A., Demissie S., Manning A. K., Jensen M. K., Rimm E. B., Wang J., Rodrigues A., et al. 2009. Loss-of-function variants in endothelial lipase are a cause of elevated HDL cholesterol in humans. J. Clin. Invest. 119: 1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johansen C. T., Dube J. B., Loyzer M. N., MacDonald A., Carter D. E., McIntyre A. D., Cao H., Wang J., Robinson J. F., and Hegele R. A.. 2014. Lipidseq: a next-generation clinical resequencing panel for monogenic dyslipidemias. J. Lipid Res. 55: 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K., Li M., and Hakonarson H.. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenson P. D., Mort M., Ball E. V., Shaw K., Phillips A., and Cooper D. N.. 2014. The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 133: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auton A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., Korbel J. O., Marchini J. L., McCarthy S., McVean G. A., and Abecasis G. R.. 2015. A global reference for human genetic variation. Nature. 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lek M., Karczewski K., Minikel E., Samocha K., Banks E., Fennell T., O’Donnell-Luria A., Ware J., Hill A., Cummings B., et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adzhubei I., Jordan D. M., and Sunyaev S. R.. 2013. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 7: 7.20.1–7.20.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar P., Henikoff S., and Ng P. C.. 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 39.Kircher M., Witten D. M., Jain P., O’Roak B. J., Cooper G. M., and Shendure J.. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong H. Y., Alipanahi B., Lee L. J., Bretschneider H., Merico D., Yuen R. K., Hua Y., Gueroussov S., Najafabadi H. S., Hughes T. R., et al. 2015. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 347: 1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mucaki E. J., Shirley B. C., and Rogan P. K.. 2013. Prediction of mutant mRNA splice isoforms by information theory-based exon definition. Hum. Mutat. 34: 557–565. [DOI] [PubMed] [Google Scholar]
- 42.Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M. L., Mora S., et al. 2013. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khetarpal S. A., Schjoldager K. T., Christoffersen C., Raghavan A., Edmondson A. C., Reutter H. M., Ahmed B., Ouazzani R., Peloso G. M., Vitali C., et al. 2016. Loss of function of GALNT2 lowers high-density lipoproteins in humans, nonhuman primates, and rodents. Cell Metab. 24: 234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Browne C., and Timson D. J.. 2015. In silico prediction of the effects of mutations in the human mevalonate kinase gene: towards a predictive framework for mevalonate kinase deficiency. Ann. Hum. Genet. 79: 451–459. [DOI] [PubMed] [Google Scholar]
- 45.Fogarty M. P., Xiao R., Prokunina-Olsson L., Scott L. J., and Mohlke K. L.. 2010. Allelic expression imbalance at high-density lipoprotein cholesterol locus MMAB-MVK. Hum. Mol. Genet. 19: 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dijk W., and Kersten S.. 2014. Regulation of lipoprotein lipase by ANGPTL4. Trends Endocrinol. Metab. 25: 146–155. [DOI] [PubMed] [Google Scholar]
- 47.Morrison A. C., Voorman A., Johnson A. D., Liu X., Yu J., Li A., Muzny D., Yu F., Rice K., Zhu C., et al. 2013. Whole-genome sequence-based analysis of high-density lipoprotein cholesterol. Nat. Genet. 45: 899–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith J. A., Ware E. B., Middha P., Beacher L., and Kardia S. L.. 2015. Current applications of genetic risk scores to cardiovascular outcomes and subclinical phenotypes. Curr. Epidemiol. Rep. 2: 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voight B. F., Peloso G. M., Orho-Melander M., Frikke-Schmidt R., Barbalic M., Jensen M. K., Hindy G., Holm H., Ding E. L., Johnson T., et al. 2012. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 380: 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khera A. V., Won H. H., Peloso G. M., Lawson K. S., Bartz T. M., Deng X., van Leeuwen E. M., Natarajan P., Emdin C. A., Bick A. G., et al. 2016. Diagnostic yield of sequencing familial hypercholesterolemia genes in patients with severe hypercholesterolemia. J. Am. Coll. Cardiol. 67: 2578–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santos R. D., Gidding S. S., Hegele R. A., Cuchel M. A., Barter P. J., Watts G. F., Baum S. J., Catapano A. L., Chapman M. J., Defesche J. C., et al. 2016. Defining severe familial hypercholesterolaemia and the implications for clinical management: a consensus statement from the International Atherosclerosis Society Severe Familial Hypercholesterolemia Panel. Lancet Diabetes Endocrinol. 4: 850–861. [DOI] [PubMed] [Google Scholar]
- 52.Zheng K. H., van der Valk F. M., Smits L. P., Sandberg M., Dasseux J. L., Baron R., Barbaras R., Keyserling C., Coolen B. F., Nederveen A. J., et al. 2016. HDL mimetic cer-001 targets atherosclerotic plaques in patients. Atherosclerosis. 251: 381–388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.