Summary
Non-Hodgkin lymphomas (NHL) represent a frequent complication of human immunodeficiency virus (HIV) infection. To elucidate HIV-NHL pathogenesis, we performed a genome-wide DNA profiling based on a single nucleotide polymorphism-based microarray comparative genomic hybridization in 57 HIV-lymphomas and, for comparison, in 105 immunocompetent diffuse large B-cell lymphomas (IC-DLBCL). Genomic complexity varied across HIV-NHL subtypes. HIV-Burkitt lymphoma showed a significantly lower number of lesions than HIV-DLBCL (p=0.032), whereas the median number of copy number changes was significantly higher in Epstein-Barr virus negative (EBV−) HIV-DLBCL (42.5, range 8-153) compared to EBV+ cases (22; range 3-41; p=0.029). Compared to IC-DLBCL, HIV-DLBCL displayed a distinct genomic profile with no gains of 18q and specific genetic lesions. Fragile sites-associated genes, including FHIT (FRA3B), WWOX (FRA16D), DCC (FRA18B) and PARK2 (FRA6E) were frequently inactivated in HIV-NHL by interstitial deletions, and a significantly higher prevalence of FHIT alterations was observed in HIV-DLBCL compared to IC-DLBCL. The same genes involved by fragile site deletions were also frequently affected by aberrant methylation of regulative regions.
Keywords: Acquired immunodeficiency syndrome (AIDS), FHIT, WWOX, diffuse large B-cell lymphoma, Affymetrix
Non-Hodgkin lymphomas (NHL) represent a major cause of morbidity and mortality among individuals infected by the human immunodeficiency virus (HIV) (Grogg, et al 2007, Navarro and Kaplan 2006).
The vast majority of HIV-related NHL (HIV-NHL) is represented by aggressive monoclonal B-cell lymphomas displaying distinctive clinical features, including widespread disease at presentation, poor prognosis and frequent involvement of extranodal sites. The pathological spectrum of the disease includes systemic HIV-NHL, primary central nervous system lymphoma (HIV-PCNSL), primary effusion lymphoma (HIV-PEL) and plasmablastic lymphoma of the oral cavity ’(Carbone, et al 2009, Carbone, et al 2001a, Grogg, et al 2007, Navarro and Kaplan 2006, Swerdlow, et al 2008). Systemic HIV-NHL comprise HIV-related Burkitt (HIV-BL) and HIV-related diffuse large B-cell lymphoma (HIV-DLBCL). The vast majority of HIV-PCNSL is histologically represented by DLBCL.
Despite their clinico-pathological heterogeneity, the vast majority of HIV-NHL have been demonstrated to derive molecularly from B cells that have experienced the germinal centre (GC) reaction (Capello, et al 2008, Carbone, et al 2001b). In particular, HIV-BL and a fraction of HIV-DLBCL express markers typical of GC B cells, whereas the remaining HIV-DLBCL and HIV-PEL are represented by post-GC, pre-terminally differentiated B-cells.
The pathogenesis of HIV-NHL is a multistep process involving factors provided by the host, as well as alterations intrinsic to the tumor clone (Capello, et al 2008, Carbone, et al 2009, Carbone, et al 2001b, Moir and Fauci 2009). Screening of known genetic lesions has revealed that the molecular pathogenesis of HIV-NHL is characterized by molecular heterogeneity and involves distinct pathogenetic pathways that preferentially associate with specific clinico-pathological entities (Carbone, et al 2009, Carbone, et al 2001a, Jenner, et al 2003, Klein, et al 2003, Nair, et al 2006, Navarro and Kaplan 2006).
In order to further elucidate the molecular pathogenesis of HIV-NHL and identify genetic lesions specifically associated with HIV infection, we: i) performed genome-wide DNA profiling based on single nucleotide polymorphism (SNP)-based microarray comparative genomic hybridization (a-CGH) in 43 HIV-NHL clinical specimens and 14 HIV-NHL cell lines; ii) compared the genomic profiles of HIV-DLBCL versus 105 immunocompetent DLBCL (IC-DLBCL); iii) evaluated the prognostic impact of genomic aberrations.
Methods
Tumour panel
Forty-three HIV-NHL clinical specimens formed the basis of this study. Based upon the 2001 World Health Organization Classification (Jaffe, et al 2001), HIV-NHL cases were centrally reviewed and histologically classified as HIV-BL (11 cases), HIV-DLBCL (28 cases) and HIV-PCNSL (four cases). Eight HIV-PEL (CRO-AP/2, CRO-AP/3, CRO-AP/5, CRO-AP/6, BCBL1, BC2, BC3 and HBL6) and six HIV-BL (PA-682, LAM-C3+, HBL2, HBL4, BRG-IgM and AS283A) cell lines were also analyzed. For comparative purposes, 105 IC-DLBCL clinical specimens were also investigated. IC-DLBCL were diagnosed by expert pathologists. All clinical specimens were derived from involved sites and were obtained in the course of routine diagnostic procedures before therapy. Cases were selected for the study based upon the availability of frozen material and for having a fraction of malignant cells in the pathological specimen representing >70% of overall cellularity as determined by morphological and immunophenotypic studies. All samples were studied according to a protocol approved by the Bellinzona Institutional Review Board.
DNA extraction, Epstein-Barr virus (EBV) detection and a-CGH
DNA was extracted and its integrity was verified as previously described (Capello, et al 2003, Forconi, et al 2008). Infection by EBV was investigated by Epstein-Barr encoded RNA (EBER) in situ hybridization (ISH) and polymerase chain reaction (PCR) analysis as previously reported (Carbone, et al 2001b). Genome-wide DNA profiles were obtained using the GeneChip Human Mapping 250K NspI (Affymetrix, Santa Clara, CA, USA), as previously described (Forconi, et al 2008). The Human Mapping 250K NspI has probesets targeting over 250,000 single nucleotide polymorphisms (SNPs), and is capable of providing copy number (CN) and genotyping information with a median physical distance between SNPs of 5 kb and an average distance between SNPs of approximately 10 kb.
Data mining
Data acquisition was performed using the GeneChip Operating Software (GCOS) v1.4 and Genotyping Analysis Software (GTYPE) v4.1. Genotype calls were calculated using the BRLMM algorithm (Bayesian Robust Linear Model with Mahalanobis distance classifier). Mapping data for probes were derived from the National Center for Biotechnology Information (NCBI) Human Genome Build 36, as provided by Affymetrix. The modified Bayesian Piecewise Regression (mBPCR) method (Rancoita, et al 2009) was used to estimate the copy number (CN) starting from the log2-ratios of raw CN values obtained with Affymetrix CNAT 4.01 using a Gaussian bandwidth of 0 Kb and 46 Caucasian normal female samples of the HapMap Project as reference samples. Afterwards, each genomic profile was normalized to a median log2-ratio of zero (i.e. CN of two). Thresholds for loss and gain were −0.08 and +0.08, respectively, corresponding to six times the median absolute deviation symmetrically around zero with an associated p-value lower than <0.001 after Bonferoni multiple test correction. Thresholds for homozygous deletion and amplification were defined at −0.6 and +0.6, respectively. Loss of heterozygosity (LOH) profiles were obtained applying the method with haplotype correction for tumour-only LOH inference available in the dChip software (Beroukhim, et al 2006), using as reference the 60 Centre d’Etude du Polymorphisme Humain parents of the HapMap Project and computing the allele A frequency from the data.
The recurrent minimal common regions (MCR) were defined using the algorithm by Lenz et al. (2008a). MCR were calculated for DNA gains, losses, LOH (at least 5% of cases), homozygous deletions and amplifications (at least two samples). To calculate the frequency of each individual MCR, all cases bearing the specific lesion were included; i.e., a case with a gain of a whole chromosome was also counted as positive for a SRA within that chromosome. MCR were compared with the Database of Genomic Variants (http://projects.tcag.ca/variation/).
For MCR occurring in at least 15% of cases, differences in frequencies between HIV-DLBCL and IC-DLBCL were evaluated using a Fisher’s exact test followed by multiple test correction. A false discovery rate (FDR) ≤ 0.1 was considered as significant.
Genomic complexity and differences in the median number of DNA gains, losses and LOH among the different HIV-NHL subtypes were calculated using the Wilcoxon rank-sum test (Stata Corporation, College Station, TX, USA). The p-value for significance was ≤ 0.05.
Methylation analysis
The methylation status of Fragile HIstidine Triad (FHIT), WW domain-containing Oxidoreductase (WWOX), Deleted in Colon Cancer (DCC) and Parkinson disease (autosomal recessive, juvenile) 2, parkin (PARK2) CpG islands was investigated by methylation-specific polymerase chain reaction (MSP), as previously reported (Agirre, et al 2006, Hussain, et al 2004, Iliopoulos, et al 2005, Miyamoto, et al 2005). DNA used as a template was chemically modified by bisulfite treatment (EpiTect Bisulfite Kit, QIAGEN Milan, Italy). MSP was performed with primers specific for methylated and unmethylated sequences on 50 ng of bisulfite-treated DNA in a Verity thermal cycler (Applied Biosystems, Milan, Italy). All MSP reactions were performed with positive and negative controls for both unmethylated and methylated alleles. In vitro methylated DNA from normal peripheral blood mononuclear cells (PBMC) was used as positive control for methylated allele-specific reactions. DNA from normal PBMC was used as negative control for methylated allele-specific reactions and as positive control for unmethylated allele specific reactions. PCR products were separated by agarose gel electrophoresis and visualized under UV illumination. Dilution curve experiments showed that MSP assay enabled the detection of 1 cell carrying methylated alleles out of 100 cells.
Real-time quantitative reverse transcription polymerase chain reaction (RT-PCR)
Real-time RT-PCR for FHIT, WWOX and for the housekeeping gene β-glucuronidase (GUSB) were performed on a StepOnePlus Real-Time PCR System (Applied Biosystems) using TaqMan Inventoried Gene Expression Assays (Applied Biosystems), according to the manufacturer’s protocol. Linearity of PCR amplification and equal efficiency for primer/probe systems was demonstrated for all genes. The (2-ΔΔCt) algorithm was used to determine the relative expression of FHIT and WWOX mRNA in methylated and unmethylated cases.
Results
The degree of genomic complexity differs among HIV-NHL subtypes
We analyzed a total of 57 HIV-NHL including 43 primary samples and 14 cell lines by high-resolution SNP-based a-CGH. The level of genomic complexity varied among the different HIV-NHL subtypes. In particular, the median number of CN changes was lower in HIV-PCNSL (7; range, 1-29) and HIV-BL primary samples (17; range, 1-64), than in HIV-PEL (28; range, 6-149) and HIV-DLBCL (31.5; range, 3-153). When compared to systemic HIV-DLBCL, HIV-BL primary samples showed a significantly lower number of lesions (p=0.032). HIV-BL cell lines were characterized by a very unstable genomic profile (median number of CN changes 132.5; range, 20-176), and were therefore considered separately from HIV-BL primary samples in any further analysis.
Among HIV-DLBCL, genomic complexity varied according to EBV infection status, since the median number of CN changes was significantly higher in the 15 EBV-negative HIV-DLBCL (42.5, range 8-153) than in the eight EBV-positive cases (22; range 3-41; p=0.029).
HIV-NHL subtypes are characterized by distinct recurrent alterations
Figs 1 and S1 show the frequencies of DNA CN changes and of LOH, respectively, in each HIV-NHL subgroup. To have a more precise representation of the lesions characterizing these disorders, we determined the MCR, which are supposed to contain loci relevant for the tumour (Table SI). Four types of MCR were defined: abnormal chromosome arm (ACA), abnormal whole chromosome (AWC), short recurrent abnormality (SRA) and long recurrent abnormality (LRA) (Lenz, et al 2008a).
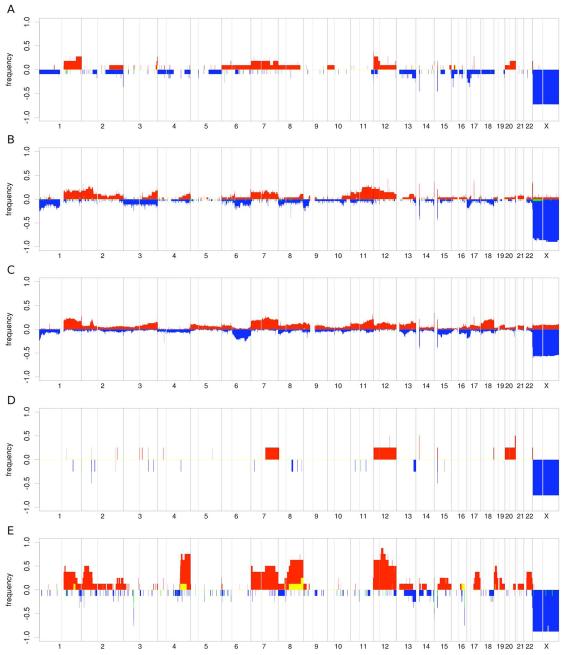
Fig 1.
Frequency of DNA gains (up) and losses (down) observed in 11 HIV-BL (A), 25 HIV-DLBCL (B), 105 IC-DLBCL (C), four HIV-PCNSL (D) and eight HIV-PEL (E). X-axis, chromosome localization and physical mapping; Y-axis, percentage of cases showing the aberrations.
ACA, detected in more than 15% HIV-BL primary samples, included gains of 1q (18%), 7p (18%), 12p (27%), 20q (18%), losses of 16p (18%) and 17p (27%, TP53), and LOH of 15q (18%) and 17q (18%) (Figs 1A and S1A, Table S1). SRA and LRA mapped at 1q, 3q, 7p, 7q, 12p, 12q, 15q, 17q (gains), 4q, 5p, 13q, 16p, 17p (losses) and 1q, 2p, 3q, 4p, 5q, 6q, 7p, 8p, 13q, 14q, 17q (LOH).
Gains of 1q, 6p LOH, SRA and LRA at 1q, 6p, 15q (gains), 2p, and 4q (losses) and 6p (LOH) were the most common alterations observed in HIV-BL cell lines (Fig S2, Table SI).
ACA occurring in more than 15% HIV-DLBCL were represented by gains of 1q (18%), 2p (18%, comprising BCL11A and REL), 11q (25%), and 12p (21%), losses of 17p (18%, TP53), and LOH of 17q (Fig 1B and S1B). SRA and LRA detected in more than 15% HIV-DLBCL are shown in Table I.
Table I.
MCR (small and long recurrent aberrations) occurring in more than 15% HIV-DLBCL.
| Region | Frequency | Start | End | Size | Type | Candidate genes |
Notes | |
|---|---|---|---|---|---|---|---|---|
| Gains | 1q25.1 | 21% | 171,652,096 | 173,071,745 | 1,419,649 | SRA | ||
| 1q31.3 | 18% | 192,469,924 | 192,743,596 | 273,672 | SRA | ^ | ||
| 1q32.2 | 21% | 206,380,943 | 206,520,994 | 140,051 | SRA | * ^ | ||
| 1q44 | 18% | 245,702,638 | 247,110,269 | 1,407,631 | SRA | * | ||
| 2p15-p14 | 25% | 61,329,962 | 64,146,492 | 2,816,530 | SRA |
BCL11A, REL |
^ | |
| 3q29 | 18% | 194,627,100 | 199,318,155 | 4,691,055 | SRA | |||
| 6q27 | 18% | 169,778,234 | 170,716,684 | 938,450 | SRA | * | ||
| 7q11.22 | 18% | 68,070,037 | 70,003,179 | 1,933,142 | SRA | |||
| 7q31.33 | 18% | 126,322,656 | 126,846,369 | 523,713 | SRA | |||
| 11q22.3-q23.2 | 25% | 105,659,503 | 114,937,935 | 9,278,432 | SRA | ^ | ||
| 11q24.2-q25 | 21% | 125,356,265 | 134,439,182 | 9,082,917 | SRA | ^ | ||
| 12p13.33- p13.32 |
25% | 61,880 | 5,110,487 | 5,048,607 | SRA | CCND2 | ^ | |
| 12q21.31 | 43% | 83,148,358 | 83,183,479 | 35,121 | SRA | |||
| 14q11.2 | 32% | 19,336,854 | 19,489,991 | 153,137 | SRA | * | ||
| 14q32.33 | 36% | 106,043,812 | 106,356,482 | 312,670 | SRA | * | ||
| 15q11.2 | 21% | 18,427,103 | 20,335,459 | 1,908,356 | SRA | * | ||
| 17q21.31 | 32% | 41,474,846 | 41,707,706 | 232,860 | SRA | * | ||
| Losses | 1p36.33- p36.23 |
25% | 2,292,771 | 7,811,025 | 5,518,254 | SRA | TP73 | ^ |
| 1p36.22 | 21% | 11,399,128 | 11,540,865 | 141,737 | SRA | PTCHD2 | ||
| 1p34.2 | 18% | 41,635,248 | 41,911,980 | 276,732 | SRA | |||
| 3p21.31 | 18% | 47,933,041 | 49,649,347 | 1,716,306 | SRA | |||
| 3p14.2 | 25% | 60,401,632 | 60,564,495 | 162,863 | SRA | FHIT | ||
| 3q21.1-q21.2 | 21% | 124,994,673 | 126,226,160 | 1,231,487 | SRA | |||
| 6q15 | 21% | 87,940,789 | 88,118,556 | 177,767 | SRA | |||
| 6q26 | 18% | 161,993,412 | 162,532,109 | 538,697 | SRA | PARK2 | * | |
| 9p23 | 25% | 11,978,659 | 12,149,128 | 170,469 | SRA | * | ||
| 14q11.2 | 36% | 19,336,854 | 19,489,770 | 152,916 | SRA | * | ||
| 14q11.2 | 43% | 21,466,983 | 22,045,540 | 578,557 | SRA | * | ||
| 15q11.2 | 39% | 18,427,103 | 19,988,041 | 1,560,938 | SRA | * | ||
| LOH | 1p36.32- p36.23 |
18% | 2,713,412 | 7,263,431 | 4,550,019 | SRA | TP73 | |
| 1p13.2 | 18% | 113,591,971 | 114,671,830 | 1,079,859 | SRA | * | ||
| 1q24.1-q24.2 | 18% | 164,176,790 | 168,975,819 | 4,799,029 | SRA | |||
| 6p22.2 | 18% | 25,207,095 | 26,036,355 | 829,260 | SRA | |||
| 6p21.33 | 21% | 31,052,961 | 31,528,666 | 475,705 | SRA |
HLA-B,HLA- C |
||
| 17p13.3-p12 | 18% | 18,901 | 12,900,792 | 12,881,89 1 |
SRA | TP53 | ||
| 17q12-q25.3 | 18% | 32,281,667 | 78,599,918 | 46,318,25 1 |
LRA | |||
| 17q22 | 21% | 50,314,685 | 50,956,167 | 641,482 | SRA | |||
| 17q24.2-q25.1 | 18% | 64,593,327 | 69,907,679 | 5,314,352 | SRA |
SRA, small recurrent aberration; LRA, long recurrent aberration;
more than 92% of the probes of the region overlap with known copy number variations;
this SRA is included in a LRA, occurring in more than 15% of the cases, and not shown on this table
As mentioned above, HIV-PCNSL had very few genomic aberrations, with no recurrent lesions (Figs 1D and S1D).
HIV-PEL had common aberrations of whole chromosomes or chromosome arms: gains of 2p (29%,BCL11A, REL), 7p (29%), 7q (43%), whole 7 (29%), 8q (57%), 12p (57%), 12q (43%), whole 12 (29%), 19p (29%), 22q (43%) and LOH of 12q (29%) (Figs 1E and S1E). Table SI shows the SRA and LRA detected in at least 15% of HIV-PEL, respectively.
Occurrence of copy neutral LOH in HIV-NHL
The high density SNP array used in this study allowed the concomitant identification of CN changes and genotyping. As expected, the detection of DNA losses vastly overlapped with the presence of LOH. However, LOH occurred also in regions without DNA losses (Bacolod, et al 2008, Rinaldi, et al 2006), suggesting the presence of copy neutral LOH (uniparental disomy, UD). The incidence of UD was heterogeneous among different HIV-NHL subtypes. The proportion of probes mapping in regions of UD was relatively higher in HIV-PEL (median 4.6%; range, 0.2%-13.1%) and HIV-BL (2.31%; range, 0.14%-9.15%) compared to HIV-DLBCL (0.8%; 0%-9.15%), and HIV-PCNSL (0.8%; 0.15%-10.18%). Moreover, UD of at least 1 region longer than 5Mb (less likely due to the accidental presence of stretches of homozygous SNPs) (Forconi, et al 2008), was more frequently observed in HIV-BL (7/8; 88%) and PEL (8/11; 73%) than in HIV-DLBCL (15/28, 53%). The regions most frequently targeted by UD were 1q (HIV-BL and HIV-DLBCL), 3q (HIV-DLBCL), 6p (HIV-DLBCL), 10q (HIV-DLBCL), 15q (HIV-BL) and 17q (HIV-BL and HIV-DLBCL).
HIV-DLBCL and IC-DLBCL are characterized by similar degrees of genomic complexity but by distinct genomic profiles
With the aim of evaluating genomic differences between HIV-DLBCL and IC-DLBCL, we compared the genomic profiles of 28 HIV-DLBCL and 105 IC-DLBCL obtained with the same SNP array (Figs 1F and S1F). The genomic complexity of IC-DLBCL, including CN changes (median 31.5; range, 2-154) and the proportion of probes mapping in UD regions (median 1.15%, range 0%-12.9%), was similar to that observed in HIV-DLBCL. Nevertheless, HIV- and IC-DLBCL showed a different distribution of the most common MCR and a comparison of the single genomic alterations led to the identification of statistically significant differences between the two groups of lymphomas. HIV-DLBCL cases more commonly presented deletion of 3p14.3 (7/28, 25%, vs 5/105, 5%; p-value=0.003, FDR 0.1), which contains the fragile site FRA3B and FHIT, and gains of 12q21.31 (12/28, 43%, vs 18/105, 17%; p-value=0.007, FDR 0.1). Conversely, IC-DLBCL more commonly had gains of 18q (21/105, 19%, vs 0/28, 0%; p-value=0.007, FDR 0.1), containing among others BCL2 and NFATC1. As 49/105 (47%) of the IC-DLBCL had been previously characterized for their cell of origin using gene expression profiling (Lenz, et al 2008b), we also compared HIV-DLBCL versus 30 GC-DLBCL and 19 activated B-cell like (ABC)-DLBC. HIV-DLBCL did not show any significant difference versus GC-DLBCL whereas again, they had less chromosome 18 gains (0/28, 0%, vs 6/19, 32%; p-value=0.002, FDR=0.068) and more gains of 12q21.31 (12/28, 43%, vs 1/19, 5%; p-value=0.007, FDR 0.082), when compared to ABC-DLBCL.
Genes spanning fragile sites are preferentially targeted by genomic deletions in HIV-NHL
As shown before, the loss of chromosomal region 3p14, that includes the fragile site FRA3B and the FHIT gene, was the most significant difference between HIV-DLBCL and IC-DLBCL. The region was also frequently affected in HIV-PEL (6/8, 75%), but less in HIV-BL (1/11, 9%). Interestingly, nine HIV-NHL with deletion of FHIT showed the concomitant alteration of the fragile site FRA16D, localized at 16q23.1 and involving the WWOX gene.
The observation that 3p14, 16q23.1 and also 6q26, another known fragile site containing region (PARK2), were included among the MCR of HIV-DLBCL prompted thte screening of 39 genes mapping to well characterized fragile sites (Smith, et al 2007, Smith, et al 2006) both in HIV-NHL and in IC-DLBCL (Table SII). Among the loci analyzed, 13 genes, including FHIT and WWOX, were frequent targets of alterations in HIV-NHL and were deleted in more than 10% HIV-DLBCL. Besides the significantly higher prevalence of FHIT alterations in HIV-DLBCL compared to IC-DLBCL, also WWOX, DCC, and DAB1 were also more frequently deleted in HIV-DLBCL than in IC-DLBCL, although the differences were not statistically significant. Importantly, HIV-DLBCL and IC-DLBCL differed not only for the frequency of inactivation of genes associated to fragile sites, but also for the pattern of deletions. Whereas most IC-DLBCL showed deletions affecting a wide chromosomal region, including the entire fragile site, HIV-DLBCL were characterized by short, interstitial, deletions localized within the fragile site associated gene. In particular, FHIT was mainly affected in the proximity of exon 5, whereas WWOX was preferentially targeted within the last intron of its longest splice variant. The same pattern of interstitial deletions of fragile site genes observed in HIV-DLBCL recurred also in other HIV-NHL subtypes.
In HIV-NHL, FHIT, WWOX, DCC and PARK2 are inactivated by deletion and/or methylation
FHIT, WWOX, DCC and PARK2 are considered tumor suppressor genes that may be inactivated by deletion and/or methylation in many tumors (Agirre, et al 2006, Aqeilan and Croce 2007, Arakawa 2004, Bednarek, et al 2001, Cesari, et al 2003, Ishii, et al 2003, O’Keefe and Richards 2006).
The methylation status of FHIT, WWOX, DCC and PARK2 in HIV-NHL was evaluated by MSP and results were compared to array-CGH data. Aberrant methylation of FHIT intron 1 was detected in 8/47 (17%) HIV-NHL, including 6/28 (21%) HIV-DLBCL, 2/11 (18%) HIV-BL, whereas it was absent in HIV-PEL (0/8). Aberrant methylation of the promoter region of WWOX was observed in 24/47 (51%) HIV-NHL, including 6/8 (75%) HIV-PEL, 15/28 (54%) HIV-DLBCL, and 3/11 (27%) HIV-BL. In HIV-NHL, the mechanisms of inactivation of FHIT by deletion or methylation were mutually exclusive, as none of the cases with aberrant methylation of FHIT showed concomitant deletion of the gene. On the contrary, all HIV-NHL with WWOX deletion showed the presence of WWOX methylated alleles.
To verify the association between FHIT and WWOX epigenetic and genetic alterations and gene expression, we analyzed FHIT and WWOX mRNA levels by real-time quantitative RT-PCR in a representative panel of HIV-NHL (Fig 2).
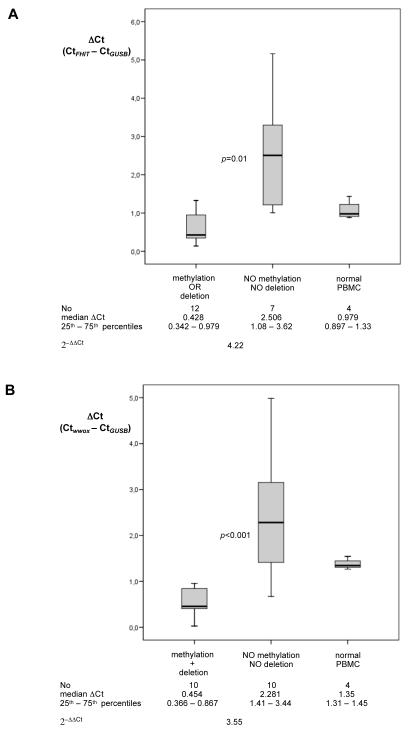
Fig 2.
Expression analysis of FHIT and WWOX mRNA by real-time quantitative RT-PCR. Levels of FHIT (A) and WWOX (B) mRNA in HIV-NHL samples with and without genetic and/or epigenetic alterations of the gene were normalized to the endogenous reference GUSB mRNA to obtain the normalized threshold cycle (ΔCt) for each sample. The median ΔCt value was set to 1. The ΔCt value of FHIT (A) and WWOX (B) of cases without alterations of the gene was related to the ΔCt value of FHIT (A) and WWOX (B) of cases displaying genetic and/or epigenetic alterations to obtain the relative threshold cycle (ΔΔCt). The (2−ΔΔCt) algorithm was used to determine the relative expression of FHIT and WWOX mRNA. Statistical analysis, performed by the non parametric Mann-Whitney test with Bonferroni adjustment, revealed a significant difference in FHIT and WWOX expression between samples without genetic and/or epigenetic alterations and HIV-NHL cases displaying alterations of the gene (p=0.01 for FHIT, and p<0.001 for WWOX).
Relative expression levels of FHIT were evaluated, comparing samples with methylation or deletions (n=12) to samples without genetic or epigenetic alterations (n=7) (Fig 2A). Compared to samples without genetic or epigenetic alterations, expression of FHIT mRNA was 4.22-fold (p=0.01) lower in HIV-NHL displaying alterations of the gene.
Relative expression levels of WWOX were evaluated comparing ten samples with deletion and/or methylation of the gene with ten samples without alterations. (Fig 2B). Compared to samples without genetic and/or epigenetic alterations, expression of WWOX mRNA was 3.55-fold (p<0.001) lower in HIV-NHL displaying alterations of the gene.
DCC methylation was detected in 16/47 (34%) HIV-NHL, and was more frequent in HIV-DLBCL (12/28; 43%) compared to HIV-BL (3/11; 27%), and HIV-PEL (1/8; 13%). PARK2 methylation was observed in 12/47 (25%) HIV-NHL , including 7/28 (25%) HIV-DLBCL, 4/11 (36%) HIV-BL, and 1/8 (12.5%) HIV-PEL.
Discussion
This study reports the largest series of HIV-NHL so far analyzed using a high-density genome wide SNP-based microarray analysis. Overall, the level of genomic complexity varied across HIV-NHL subtypes. In particular, HIV-BL primary samples were characterized by a more stable profile when compared to both HIV-DLBCL and IC-DLBCL. The lower genomic complexity of BL compared to DLBCL has also been observed in the context of lymphomas arising after solid organ transplantation (Rinaldi, et al 2006) and in immunocompetent hosts (Hummel, et al 2006), and therefore may represent an intrinsic feature of BL not related to the host immune status.
Among systemic HIV-DLBCL, genomic complexity varied according to EBV infection status, because EBV-positive HIV-DLBCL showed a lower genomic complexity than EBV-negative cases. Notably, HIV-PCNSL, which carry EBV in virtually all cases, were also characterized by the presence of a very low number of genomic alterations. This observation corroborates previous studies (Capello, et al 2008, Vaghefi, et al 2006) and may be ascribed to a direct EBV transforming effect, which, at least in specific cellular contexts, can replace the need of additional chromosomal aberrations.
In our study, HIV-PEL showed a high prevalence of recurrent lesions. Overall, the HIV-PEL genomic profile, in accordance with a previous CGH study (Nair, et al 2006), is different from that of HIV-DLBCL or IC-DLBCL and multiple myeloma (Carrasco, et al 2006). The observed genomic profile of PEL suggests that the pathogenesis of this lymphoma also involves CN changes, in addition to human herpes virus 8 (HHV8) infection and miRNA deregulation (Carbone, et al 2009, O’Hara, et al 2008). Intriguingly, the regions recurrently gained by PEL contain some genes previously shown to be overexpressed in PEL cells, including Vitamin D3 receptor (VDR, 12q13.11) and interleukin 2 receptor beta (IL2RB, 22q13.1). Moreover, the presence of the gene encoding the interleukin 6 (IL6) receptor within the gained 1q31.1 region suggests a possible autocrine loop with the viral IL6 homologue encoded by HHV8.
Although HIV-DLBCL and IC-DLBCL were characterized by an overall similar quantitative genomic complexity, HIV-DLBCL showed peculiar genomic alterations. Comparison of the genomic profiles of HIV-DLBCL with the profiles of 105 de novo IC-DLBCL and their GC- and post GC-subtypes, suggests that HIV-DLBCL are intermediate between post GC- and GCB-DLBCL, with more similarities with GCB-DLBCL. In immunocompetent hosts, de novo post GC-cases are characterized by deletions of 6q (PRDM1 and TNFAIP3), 9p (CDKN2A), trisomy of chromosome 3, gain/amplifications of 3p (FOXP1, NFKBIZ), 3q, 18q (BCL2) and 19q (SPIB) (Bea, et al 2005, Chen, et al 2006, Compagno, et al 2009, Lenz, et al 2008a, Tagawa, et al 2004). On the contrary, GCB-DLBCL display more commonly gains of 1q, 2p (REL), 7q, 9q, 12q (MDM2), 13q (MIR17HG), and deletions of 1p (TP73), 10q (PTEN) and 13q (ING1) (Bea, et al 2005, Chen, et al 2006, Lenz, et al 2008a, Tagawa, et al 2004). Analogous to GCB-DLBCL, HIV-DLBCL clearly showed recurrent gains of 2p (REL), 7q and 12q, as well as losses of 1p. Also, lesions that characterize post GC-DLBCL, such as gains of 18q (BCL2), were not detected among HIV-DLBCL. However, at variance with IC GCB-DLBCL, HIV-DLBCL displayed gains in 3q, an abnormality that is absent or extremely rare among immunocompetent patients (Lenz, et al 2008a), and did not show preferential deletion of PTEN, which has been reported as specific in this NHL subset (Lenz, et al 2008a).
When compared to IC-DLBCL, HIV-DLBCL showed a higher incidence of interstitial deletions affecting genes overlapping fragile sites. Fragile sites are regions of profound genomic instability distributed throughout the genome and are often the site of DNA breakage in cancers (Freudenreich 2007, Smith, et al 2007). The high frequency of deletions in fragile site-associated genes was not limited to HIV-DLBCL, but was common to other HIV-NHL. FHIT (FRA3B), WWOX (FRA16D), DCC (FRA18B) and PARK2 (FRA6E) were the fragile site-associated genes more frequently affected in HIV-NHL, and were characterized by interstitial deletions possibly imputable to fragile site instability.
In HIV-NHL, FHIT, WWOX, PARK2 and DCC were frequently inactivated not only by interstitial deletion, but also by gene methylation. Inactivation of tumor suppressor genes by deletion, mutation or methylation is a common finding in human neoplasia and tumor suppressor genes spanning fragile sites are expected to be inactivated by similar mechanisms.
FHIT and WWOX are well-known tumor suppressor genes frequently and coordinately inactivated in a variety of human malignancies, including hematopoietic neoplasia (Ishii, et al 2003, Kameoka, et al 2004, Ludes-Meyers, et al 2007). The pathogenetic relevance of FHIT and WWOX for lymphomagenesis is documented by the observation that FHIT- and WWOX-deficient mice have a higher incidence of lymphoma (Ludes-Meyers, et al 2007) (Aqeilan, et al 2007, Roz, et al 2002).
The possible role of PARK2 inactivation in neoplasia is not known, although reported to be methylated in leukemias (Agirre, et al 2006). The gene encodes a RING finger E3 ubiquitin ligase, also expressed in normal B-cells, that regulates a variety of substrates, including HSP70 (Jiang, et al 2006, Moore, et al 2008). DCC is a known tumor suppressor gene that is inactivated in solid tumors (Arakawa 2004). DCC is expressed in GC B cells and encodes a dependence receptor that induces apoptosis after ligand withdrawal (Bredesen, et al 2005, Teyssier, et al 2001). DCC inactivation may protect B-cells bearing DNA damage from the killing induced by ligand deprivation at the exit from the GC (Tanikawa, et al 2003).
The apparent greater instability of fragile sites in HIV-DLBCL compared to IC-DLBCL suggests a possible contribution played by viral infections, within the context of the HIV-induced B-cell hyper-activation and dysfunction (Moir and Fauci 2009). In fact, fragile sites have been indicated as preferential integration sites for exogenous DNA, such as viruses (Feitelson and Lee 2007, Luo, et al 2004, Rassool, et al 1992, Thorland, et al 2003). EBV, Simian Virus 40 (SV40), Hepatitis B virus (HBV), herpes simplex virus types 1 and 2, adenovirus types 8 and 19, human papilloma virus 16 (HPV16), and HHV8 are only some of the viruses that could infect B-cells, integrate into the cellular genome and induce local genomic instability at the insertion sites (Rassool, et al 1992). In HIV-infected individuals, as a consequence of immunodeficiency, B-cells are expected to be more exposed to viral infections. We may hypothesize that viruses might contribute to the pathogenesis of HIV-NHL by a direct action at the genomic level, and that this transforming effect could be, at least in part, independent from the expression of viral oncogenes. Further experiments, such the demonstration of viral genomes within fragile sites, are needed to validate this hypothesis.
The results of this study expand our knowledge on the genomic heterogeneity of HIV-NHL and underlie the clinico-pathological diversity of this disease, revealing that HIV-DLBCL are characterized by distinct genomic lesions when compared to their counterpart arising in immunocompetent patients.
Supplementary Material
Acknowledgments
Work supported by: Oncosuisse grant OCS-1939-8-2006; Swiss National Science Foundation (grants 205321-112430, 205320-121886/1); Cantone Ticino (“Computational life science / Ticino in rete” program); Fondazione per la Ricerca e la Cura sui Linfomi (Lugano, Switzerland); Ricerca Sanitaria Finalizzata, Regione Piemonte, Torino, Italy; VI Programma Nazionale di Ricerca sull’AIDS, ISS, Rome, Italy; PRIN-MIUR 2006; Novara-AIL Onlus, Novara, Italy; Fondazione CRT, Torino, Italy; Alto Adige Bolzano-AIL Onlus; Cantone Ticino “Ticino in rete”; PHS grant, UO1 CA 114778.
Footnotes
Conflict of interest disclosure The authors declare no competing financial interests.
References
- Agirre X, Roman-Gomez J, Vazquez I, Jimenez-Velasco A, Garate L, Montiel-Duarte C, Artieda P, Cordeu L, Lahortiga I, Calasanz MJ, Heiniger A, Torres A, Minna JD, Prosper F. Abnormal methylation of the common PARK2 and PACRG promoter is associated with downregulation of gene expression in acute lymphoblastic leukemia and chronic myeloid leukemia. Int J Cancer. 2006;118:1945–1953. doi: 10.1002/ijc.21584. [DOI] [PubMed] [Google Scholar]
- Aqeilan RI, Croce CM. WWOX in biological control and tumorigenesis. J Cell Physiol. 2007;212:307–310. doi: 10.1002/jcp.21099. [DOI] [PubMed] [Google Scholar]
- Aqeilan RI, Trapasso F, Hussain S, Costinean S, Marshall D, Pekarsky Y, Hagan JP, Zanesi N, Kaou M, Stein GS, Lian JB, Croce CM. Targeted deletion of Wwox reveals a tumor suppressor function. Proc Natl Acad Sci U S A. 2007;104:3949–3954. doi: 10.1073/pnas.0609783104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H. Netrin-1 and its receptors in tumorigenesis. Nat Rev Cancer. 2004;4:978–987. doi: 10.1038/nrc1504. [DOI] [PubMed] [Google Scholar]
- Bacolod MD, Schemmann GS, Wang S, Shattock R, Giardina SF, Zeng Z, Shia J, Stengel RF, Gerry N, Hoh J, Kirchhoff T, Gold B, Christman MF, Offit K, Gerald WL, Notterman DA, Ott J, Paty PB, Barany F. The signatures of autozygosity among patients with colorectal cancer. Cancer Res. 2008;68:2610–2621. doi: 10.1158/0008-5472.CAN-07-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bea S, Zettl A, Wright G, Salaverria I, Jehn P, Moreno V, Burek C, Ott G, Puig X, Yang L, Lopez-Guillermo A, Chan WC, Weisenburger DD, Armitage JO, Gascoyne RD, Connors JM, Grogan TM, Braziel R, Fisher RI, Smeland EB, Kvaloy S, Delabie J, Simon R, Wilson WH, Jaffe ES, Montserrat E, Muller-Hermelink HK, Staudt LM, Campo E, Rosenwald A, Timothy GC, Harald H, John P. Diffuse Large B-Cell Lymphoma Subgroups Have Distinct Genetic Profiles that Influence Tumor Biology and Improve Gene Expression-Based Survival Prediction. Blood. 2005;106:3183–3190. doi: 10.1182/blood-2005-04-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61:8068–8073. [PubMed] [Google Scholar]
- Beroukhim R, Lin M, Park Y, Hao K, Zhao X, Garraway LA, Fox EA, Hochberg EP, Mellinghoff IK, Hofer MD, Descazeaud A, Rubin MA, Meyerson M, Wong WH, Sellers WR, Li C. Inferring loss-of-heterozygosity from unpaired tumors using high-density oligonucleotide SNP arrays. PLoS Comput Biol. 2006;2:e41. doi: 10.1371/journal.pcbi.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredesen DE, Mehlen P, Rabizadeh S. Receptors that mediate cellular dependence. Cell Death Differ. 2005;12:1031–1043. doi: 10.1038/sj.cdd.4401680. [DOI] [PubMed] [Google Scholar]
- Capello D, Cerri M, Muti G, Berra E, Oreste P, Deambrogi C, Rossi D, Dotti G, Conconi A, Vigano M, Magrini U, Ippoliti G, Morra E, Gloghini A, Rambaldi A, Paulli M, Carbone A, Gaidano G. Molecular histogenesis of posttransplantation lymphoproliferative disorders. Blood. 2003;102:3775–3785. doi: 10.1182/blood-2003-05-1683. [DOI] [PubMed] [Google Scholar]
- Capello D, Martini M, Gloghini A, Cerri M, Rasi S, Deambrogi C, Rossi D, Spina M, Tirelli U, Larocca LM, Carbone A, Gaidano G. Molecular analysis of immunoglobulin variable genes in human immunodeficiency virus-related non-Hodgkin’s lymphoma reveals implications for disease pathogenesis and histogenesis. Haematologica. 2008;93:1178–1185. doi: 10.3324/haematol.12705. [DOI] [PubMed] [Google Scholar]
- Carbone A, Gloghini A, Capello D, Gaidano G. Genetic pathways and histogenetic models of AIDS-related lymphomas. Eur J Cancer. 2001a;37:1270–1275. doi: 10.1016/s0959-8049(01)00119-8. [DOI] [PubMed] [Google Scholar]
- Carbone A, Gloghini A, Larocca LM, Capello D, Pierconti F, Canzonieri V, Tirelli U, Dalla-Favera R, Gaidano G. Expression profile of MUM1/IRF4, BCL-6, and CD138/syndecan-1 defines novel histogenetic subsets of human immunodeficiency virus-related lymphomas. Blood. 2001b;97:744–751. doi: 10.1182/blood.v97.3.744. [DOI] [PubMed] [Google Scholar]
- Carbone A, Cesarman E, Spina M, Gloghini A, Schulz TF. HIV-associated lymphomas and gamma-herpesviruses. Blood. 2009;113:1213–1224. doi: 10.1182/blood-2008-09-180315. [DOI] [PubMed] [Google Scholar]
- Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, Stewart JP, Zhan F, Khatry D, Protopopova M, Protopopov A, Sukhdeo K, Hanamura I, Stephens O, Barlogie B, Anderson KC, Chin L, Shaughnessy JD, Jr., Brennan C, Depinho RA. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–325. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Cesari R, Martin ES, Calin GA, Pentimalli F, Bichi R, McAdams H, Trapasso F, Drusco A, Shimizu M, Masciullo V, D’Andrilli G, Scambia G, Picchio MC, Alder H, Godwin AK, Croce CM. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc Natl Acad Sci U S A. 2003;100:5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Houldsworth J, Olshen AB, Nanjangud G, Chaganti S, Venkatraman ES, Halaas J, Teruya-Feldstein J, Zelenetz AD, Chaganti RS. Array comparative genomic hybridization reveals genomic copy number changes associated with outcome in diffuse large B-cell lymphomas. Blood. 2006;107:2477–2485. doi: 10.1182/blood-2005-07-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Bertoni F, Ponzoni M, Scandurra M, Califano A, Bhagat G, Chadburn A, Favera R. Dalla, Pasqualucci L. Mutations at multiple genes cause deregulation of the NF-kB pathway in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitelson MA, Lee J. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 2007;252:157–170. doi: 10.1016/j.canlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Forconi F, Poretti G, Kwee I, Sozzi E, Rossi D, Rancoita PMV, Capello D, Rinaldi A, Zucca E, Raspadori D, Spina V, Lauria F, Gaidano G, Bertoni F. High density genome-wide DNA profiling reveals a remarkably stable profile in hairy cell leukaemia. Br J Haematol. 2008;141:622–630. doi: 10.1111/j.1365-2141.2008.07106.x. [DOI] [PubMed] [Google Scholar]
- Freudenreich CH. Chromosome fragility: molecular mechanisms and cellular consequences. Front Biosci. 2007;12:4911–4924. doi: 10.2741/2437. [DOI] [PubMed] [Google Scholar]
- Grogg KL, Miller RF, Dogan A. HIV infection and lymphoma. J Clin Pathol. 2007;60:1365–1372. doi: 10.1136/jcp.2007.051953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, Bernd HW, Cogliatti SB, Dierlamm J, Feller AC, Hansmann ML, Haralambieva E, Harder L, Hasenclever D, Kuhn M, Lenze D, Lichter P, Martin-Subero JI, Moller P, Muller-Hermelink HK, Ott G, Parwaresch RM, Pott C, Rosenwald A, Rosolowski M, Schwaenen C, Sturzenhofecker B, Szczepanowski M, Trautmann H, Wacker HH, Spang R, Loeffler M, Trumper L, Stein H, Siebert R. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- Hussain A, Gutierrez MI, Timson G, Siraj AK, Deambrogi C, Al-Rasheed M, Gaidano G, Magrath I, Bhatia K. Frequent silencing of fragile histidine triad gene (FHIT) in Burkitt’s lymphoma is associated with aberrant hypermethylation. Genes Chromosomes Cancer. 2004;41:321–329. doi: 10.1002/gcc.20099. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Guler G, Han SY, Johnston D, Druck T, McCorkell KA, Palazzo J, McCue PA, Baffa R, Huebner K. Fragile genes as biomarkers: epigenetic control of WWOX and FHIT in lung, breast and bladder cancer. Oncogene. 2005;24:1625–1633. doi: 10.1038/sj.onc.1208398. [DOI] [PubMed] [Google Scholar]
- Ishii H, Vecchione A, Furukawa Y, Sutheesophon K, Han SY, Druck T, Kuroki T, Trapasso F, Nishimura M, Saito Y, Ozawa K, Croce CM, Huebner K, Furukawa Y. Expression of FRA16D/WWOX and FRA3B/FHIT genes in hematopoietic malignancies. Mol Cancer Res. 2003;1:940–947. [PubMed] [Google Scholar]
- Ishii H, Wang Y, Huebner K. A Fhit-ing role in the DNA damage checkpoint response. Cell Cycle. 2007;6:1044–1048. doi: 10.4161/cc.6.9.4213. [DOI] [PubMed] [Google Scholar]
- Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. IARC Press; Lyon: 2001. [Google Scholar]
- Jenner RG, Maillard K, Cattini N, Weiss RA, Boshoff C, Wooster R, Kellam P. Kaposi’s sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc Natl Acad Sci U S A. 2003;100:10399–10404. doi: 10.1073/pnas.1630810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Jiang Q, Liu W, Feng J. Parkin suppresses the expression of monoamine oxidases. J Biol Chem. 2006;281:8591–8599. doi: 10.1074/jbc.M510926200. [DOI] [PubMed] [Google Scholar]
- Kameoka Y, Tagawa H, Tsuzuki S, Karnan S, Ota A, Suguro M, Suzuki R, Yamaguchi M, Morishima Y, Nakamura S, Seto M. Contig array CGH at 3p14.2 points to the FRA3B/FHIT common fragile region as the target gene in diffuse large B-cell lymphoma. Oncogene. 2004;23:9148–9154. doi: 10.1038/sj.onc.1208136. [DOI] [PubMed] [Google Scholar]
- Klein U, Gloghini A, Gaidano G, Chadburn A, Cesarman E, Dalla-Favera R, Carbone A. Gene expression profile analysis of AIDS-related primary effusion lymphoma (PEL) suggests a plasmablastic derivation and identifies PEL-specific transcripts. Blood. 2003;101:4115–4121. doi: 10.1182/blood-2002-10-3090. [DOI] [PubMed] [Google Scholar]
- Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, Powell J, Rosenwald A, Ott G, Muller-Hermelink HK, Gascoyne RD, Connors JM, Campo E, Jaffe ES, Delabie J, Smeland EB, Rimsza LM, Fisher RI, Weisenburger DD, Chan WC, Staudt LM. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008a;105:13520–13520. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, Xu W, Tan B, Goldschmidt N, Iqbal J, Vose J, Bast M, Fu K, Weisenburger DD, Greiner TC, Armitage JO, Kyle A, May L, Gascoyne RD, Connors JM, Troen G, Holte H, Kvaloy S, Dierickx D, Verhoef G, Delabie J, Smeland EB, Jares P, Martinez A, Lopez-Guillermo A, Montserrat E, Campo E, Braziel RM, Miller TP, Rimsza LM, Cook JR, Pohlman B, Sweetenham J, Tubbs RR, Fisher RI, Hartmann E, Rosenwald A, Ott G, Muller-Hermelink HK, Wrench D, Lister TA, Jaffe ES, Wilson WH, Chan WC, Staudt LM. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008b;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludes-Meyers JH, Kil H, Nunez MI, Conti CJ, Parker-Thornburg J, Bedford MT, Aldaz CM. WWOX hypomorphic mice display a higher incidence of B-cell lymphomas and develop testicular atrophy. Genes Chromosomes Cancer. 2007;46:1129–1136. doi: 10.1002/gcc.20497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo WJ, Takakuwa T, Ham MF, Wada N, Liu A, Fujita S, Sakane-Ishikawa E, Aozasa K. Epstein-Barr virus is integrated between REL and BCL-11A in American Burkitt lymphoma cell line (NAB-2) Lab Invest. 2004;84:1193–1199. doi: 10.1038/labinvest.3700152. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Fukutomi T, Akashi-Tanaka S, Hasegawa T, Asahara T, Sugimura T, Ushijima T. Identification of 20 genes aberrantly methylated in human breast cancers. Int J Cancer. 2005;116:407–414. doi: 10.1002/ijc.21054. [DOI] [PubMed] [Google Scholar]
- Moir S, Fauci AS. B cells in HIV infection and disease. Nat Rev Immunol. 2009;9:235–245. doi: 10.1038/nri2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DJ, West AB, Dikeman DA, Dawson VL, Dawson TM. Parkin mediates the degradation-independent ubiquitination of Hsp70. J Neurochem. 2008;105:1806–1819. doi: 10.1111/j.1471-4159.2008.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair P, Pan H, Stallings RL, Gao SJ. Recurrent genomic imbalances in primary effusion lymphomas. Cancer Genet Cytogenet. 2006;171:119–121. doi: 10.1016/j.cancergencyto.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro WH, Kaplan LD. AIDS-related lymphoproliferative disease. Blood. 2006;107:13–20. doi: 10.1182/blood-2004-11-4278. [DOI] [PubMed] [Google Scholar]
- O’Hara AJ, Vahrson W, Dittmer DP. Gene alteration and precursor and mature microRNA transcription changes contribute to the miRNA signature of primary effusion lymphoma. Blood. 2008;111:2347–2353. doi: 10.1182/blood-2007-08-104463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe LV, Richards RI. Common chromosomal fragile sites and cancer: focus on FRA16D. Cancer Lett. 2006;232:37–47. doi: 10.1016/j.canlet.2005.07.041. [DOI] [PubMed] [Google Scholar]
- Rancoita PM, Hutter M, Bertoni F, Kwee I. Bayesian DNA copy number analysis. BMC Bioinformatics. 2009;10:10. doi: 10.1186/1471-2105-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassool FV, Le Beau MM, Neilly ME, van Melle E, Espinosa R, 3rd, McKeithan TW. Increased genetic instability of the common fragile site at 3p14 after integration of exogenous DNA. Am J Hum Genet. 1992;50:1243–1251. [PMC free article] [PubMed] [Google Scholar]
- Rinaldi A, Kwee I, Poretti G, Mensah A, Pruneri G, Capello D, Rossi D, Zucca E, Ponzoni M, Catapano C, Tibiletti MG, Paulli M, Gaidano G, Bertoni F. Comparative genome wide profiling of post-transplant lymphoproliferative disorders and diffuse large B-cell lymphomas. Br J Haematol. 2006;134:27–36. doi: 10.1111/j.1365-2141.2006.06114.x. [DOI] [PubMed] [Google Scholar]
- Roz L, Gramegna M, Ishii H, Croce CM, Sozzi G. Restoration of fragile histidine triad (FHIT) expression induces apoptosis and suppresses tumorigenicity in lung and cervical cancer cell lines. Proc Natl Acad Sci U S A. 2002;99:3615–3620. doi: 10.1073/pnas.062030799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DI, McAvoy S, Zhu Y, Perez DS. Large common fragile site genes and cancer. Semin Cancer Biol. 2007;17:31–41. doi: 10.1016/j.semcancer.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Smith DI, Zhu Y, McAvoy S, Kuhn R. Common fragile sites, extremely large genes, neural development and cancer. Cancer Lett. 2006;232:48–57. doi: 10.1016/j.canlet.2005.06.049. [DOI] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri A, Stein H, Thiele J, Vardiman JW, editors. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. IARC Press; Lyon: 2008. [Google Scholar]
- Tagawa H, Tsuzuki S, Suzuki R, Karnan S, Ota A, Kameoka Y, Suguro M, Matsuo K, Yamaguchi M, Okamoto M, Morishima Y, Nakamura S, Seto M. Genome-wide array-based comparative genomic hybridization of diffuse large B-cell lymphoma: comparison between CD5-positive and CD5-negative cases. Cancer Res. 2004;64:5948–5955. doi: 10.1158/0008-5472.CAN-03-4056. [DOI] [PubMed] [Google Scholar]
- Tanikawa C, Matsuda K, Fukuda S, Nakamura Y, Arakawa H. p53RDL1 regulates p53-dependent apoptosis. Nat Cell Biol. 2003;5:216–223. doi: 10.1038/ncb943. [DOI] [PubMed] [Google Scholar]
- Teyssier JR, Rousset F, Garcia E, Cornillet P, Laubriet A. Upregulation of the netrin receptor (DCC) gene during activation of b lymphocytes and modulation by interleukins. Biochem Biophys Res Commun. 2001;283:1031–1036. doi: 10.1006/bbrc.2001.4902. [DOI] [PubMed] [Google Scholar]
- Thorland EC, Myers SL, Gostout BS, Smith DI. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene. 2003;22:1225–1237. doi: 10.1038/sj.onc.1206170. [DOI] [PubMed] [Google Scholar]
- Vaghefi P, Martin A, Prevot S, Charlotte F, Camilleri-Broet S, Barli E, Davi F, Gabarre J, Raphael M, Poirel HA. Genomic imbalances in AIDS-related lymphomas: relation with tumoral Epstein-Barr virus status. AIDS. 2006;20:2285–2291. doi: 10.1097/QAD.0b013e328010ac5b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.