Abstract
Although genome-wide association studies have identified several susceptibility loci for adult glioma, little is known regarding the potential contribution of genetic variation in the human leukocyte antigen (HLA) region to glioma risk. HLA associations have been reported for various malignancies, with many studies investigating selected candidate HLA polymorphisms. However, no systematic analysis has been conducted in glioma patients, and no investigation into potential non-additive effects has been described. We conducted comprehensive genetic analyses of HLA variants among 1,746 adult glioma patients and 2,312 controls of European-ancestry from the GliomaScan Consortium. Genotype data were generated with the Illumina 660-Quad array, and we imputed HLA alleles using a reference panel of 5,225 individuals in the Type 1 Diabetes Genetics Consortium who underwent high-resolution HLA typing via next-generation sequencing. Case-control comparisons were adjusted for population stratification using ancestry-informative principal components. Because alleles in different loci across the HLA region are linked, we created multigene haplotypes consisting of the genes DRB1, DQA1, and DQB1. Although none of the haplotypes were associated with glioma in additive models, inclusion of a dominance term significantly improved the model for multigene haplotype HLA-DRB1*1501-DQA1*0102-DQB1*0602 (P=0.002). Heterozygous carriers of the haplotype had an increased risk of glioma (odds ratio (OR)=1.23; 95% confidence interval (CI) 1.01–1.49), while homozygous carriers were at decreased risk compared with non-carriers (OR=0.64; 95% CI 0.40–1.01). Our results suggest that the DRB1*1501-DQA1*0102-DQB1*0602 haplotype may contribute to the risk of glioma in a non-additive manner, with the positive dominance effect partly explained by an epistatic interaction with HLA-DRB1*0401-DQA1*0301-DQB1*0301.
Keywords: Glioma, brain tumor, HLA, MHC, non-additive effects, epistasis, interactions
Introduction
Glioma accounts for ~80% of all primary malignant brain tumors and ~13,000 deaths in the United States annually[1]. With ionizing radiation being the only preventable exposure strongly linked to gliomagenesis, the etiology of glioma is largely unknown[2]. Epidemiologic studies have described potential links between glioma and risk of allergies, rosacea, and autoimmune disorders[3–6], leading to the hypothesis that genetic variants in immunologic pathways also contribute to glioma susceptibility. The above-mentioned immune disorders have also been associated with genetic variation in the major histocompatibility complex (MHC) region[7–9], which contains human leukocyte antigen (HLA) genes encoding cell-surface molecules responsible for antigen presentation and immune response modulation. HLA variation has also been associated with risk of several cancers, including chronic lymphocytic leukemia, lung squamous cell carcinoma, ovarian, breast, cervical, and nasopharyngeal cancers[10]. In view of these prior findings, we hypothesized that variants in the HLA region may also potentially be associated with glioma risk.
Genome-wide association studies (GWAS) have identified several susceptibility loci for glioma[11–13], although none were in the HLA region on chromosome 6. However, genetic variation and haplotype structure in HLA genes is not well-captured by standard genome-wide SNP arrays due to the unique characteristics of sequence and copy-number variation across the MHC region. Furthermore, glioma GWAS to-date have only examined additive models, which assume a linear increase in effect per risk allele on the log-odds scale and therefore have limited power to detect non-additive effects deviating from this linear relationship. Given the observation of widespread non-additive and epistatic effects within the HLA region for a number of autoimmune diseases[14–16], non-additive effects in this gene-dense region of highly polymorphic, multi-allelic variants may also play an important role in glioma risk.
Several previous studies have examined HLA with respect to glioma risk, yielding inconsistent results across both HLA class I and II genes[17–22]. However, prior analyses were generally limited in size (with the largest study involving 255 cases) and scope (selective candidate genotyping was commonly performed, resulting in a limited number of variants being interrogated). Furthermore, earlier studies were often limited by the resolution of HLA genotyping, assessing two-digit resolution that only uniquely identifies serological groups rather than at the four-digit resolution that identifies polymorphisms at the amino acid level. Long-range HLA supertypes (extended haplotypes) have also been ignored. Given that specific amino acid polymorphisms within the same serological group can have differing associations with the same autoimmune diseases, as observed in Type 1 diabetes[23], multiple sclerosis[24] and others, a more comprehensive investigation of HLA variants using high-resolution genotyping data may improve the detection of glioma susceptibility variants. We therefore examined associations between HLA polymorphisms and glioma risk under both additive and non-additive models in a large case-control study of glioma patients with high-resolution HLA imputation data.
Materials and methods
Samples
GliomaScan is a consortium of studies comprising 1,856 adult glioma patients and 4,955 controls, with study periods ranging from 1974 to the present. Individual-level genotype and phenotype data were downloaded from the Database of Genotypes and Phenotypes (dbGaP, Study Accession phs000652.v1.p1) after review and approval by the NCI Data Access Committee. Information on case ascertainment, genotyping, and quality control are described in the Supplementary Materials.
Imputation of HLA genotypes
Four-digit classical HLA alleles were imputed in the three main class I genes (HLA-A, HLA-B, and HLA-C) and in five class II genes (HLA-DRB1, -DPA1, -DPB1, -DQA1, and -DQB1) with SNP2HLA[25], using a reference panel of 5,225 individuals of European ancestry from the Type 1 Diabetes Genetics Consortium who underwent high-resolution HLA typing via next-generation sequencing[26].
Imputation of loci outside the HLA region was carried out as described in the Supplemental Materials.
Selection of alleles and individuals for analysis
For the MHC-wide association analyses, we restricted to the commonly observed (i.e. frequency ≥5% of the total study population) classic four-digit HLA class I and II alleles among 1,746 cases and 2,312 controls of European ancestry. For the non-additive effect analyses, we created multigene haplotypes by combining phased four-digit classical alleles from genes DRB1, DQA1, and DQB1, chosen because their alleles are in strong linkage disequilibrium, allowing associations for multiple loci to be tested without loss of power due multiple test corrections. For the multigene haplotype analyses of DRB1-DQA1-DQB1 haplotypes, we restricted to individuals who had two best-guess haplotype variants at each of the three genes, and haplotypes with a frequency ≥5% of the total study population. Supplementary Figure 1 illustrates the selection of HLA variants and individuals in this study.
Statistical analyses
To analyze the additive effects of individual class I and II HLA variants on glioma risk, we performed MHC-wide logistic regression analyses using the SNP2HLA imputed allele dosage data, and the “logistic” command in PLINK 1.9[27], adjusting for the first five ancestry-informative principal components generated by Eigenstrat[28]. Additive effects of multigene haplotypes on glioma risk were estimated using the best-guess multigene haplotypes, and the glm command in R.
To investigate non-additive associations in class I and II alleles, and multigene haplotypes, we used dominance-effect models; in addition to the additive term capturing the dosage effect of a haplotype (i.e. 0,1,2 copies), a dominant term was included to capture any deviation from the additive scenario. The dominance model was compared against the baseline additive model for each haplotype by calculating the change in deviance (−2 × the difference in log likelihood) between models, which follows a chi-square distribution with 1 degree of freedom. The P-value for the chi-square test represents the significance in improvement of fit of the model after including the dominance term for a given haplotype. Bonferroni-adjusted significance thresholds were used to correct for multiple haplotype tests. All analyses were performed for individual genotypes and repeated for the multigene haplotypes. We also tested for general non-additive effects at the DRB1-DQA1-DQB1 multigene locus by simultaneously including the additive and dominance terms for all DRB1-DQA1-DQB1 variants in a global regression model (Supplementary Materials).
We examined the glioma risk of carriers of each haplotype compared to non-carriers, stratified by homozygous and heterozygous status for each haplotype. Under an additive model of association, the log-odds ratio of being a carrier of a given haplotype compared to non-carriers in heterozygotes should be half of that in homozygotes (i.e. the odds ratio of heterozygous carriers should be the square root of that of homozygous carriers). Deviation from this linear relationship can indicate the presence of non-additivity.
We tested for interaction effects as potential explanations for any observed departures from additivity in the multigene association analyses (Supplementary Materials).
For haplotypes with effects observed in the non-additive analyses, we performed conditional analyses further adjusting for 37 previously observed GWAS risk to assess any attenuation of non-additive effects.[12]
Additional sex-stratified analyses were performed because sex-specific differences were previously observed for the incidence of both glioma[1] and autoimmune disorders[29]. Stratified analyses by age of diagnosis (<50, ≥60), and carrier status of the risk variant (G) for SNP rs55705857 were performed because of their previously observed associations with tumor grade and IDH-status[30,32], allowing these variables to serve as proxies for lower-grade glioma. The age stratification cutoffs were chosen based on available data on age groups and to maximize the prediction of tumor grade by age[32].
Equations for the association analyses using the additive, dominance, and interaction effect models are described in detail in the Supplementary Materials.
RESULTS
MHC-wide allele association with glioma risk
Among the 20 class I alleles and 29 class II alleles tested, three HLA variants: HLA-B*4001, HLA-DRB1*1302 and HLA-DPB1*0101 showed nominally significant protective effects against glioma (P<0.05) under an additive model, but no associations were detected after Bonferroni correction (Supplementary Table 1).
Multigene haplotype association with glioma risk
Nine HLA-DRB1-DQA1-DQB1 haplotypes had frequencies ≥5%, consisting of 99% of the multigene haplotypes in the study population (among individuals with allele frequencies ≥5% for DRB1, DQA1, and DQB1). A total of 887 cases and 1,211 controls had common diploid multigene haplotype information fulfilling the inclusion criteria. None of the haplotypes were associated with glioma risk under the additive model for individually modeled haplotype variants, nor under the global additive model for all haplotype variants. Association estimates and haplotype frequencies for the multigene haplotypes are summarized in Supplementary Table 2.
Dominance model analyses
The inclusion of the dominance term improved the fit of the model for HLA-DRB1*1501-DQA1*0102-DQB1*0602, Pimprove=0.002, with the dominance term estimate of OR=1.54 (95% confidence interval (CI) 1.16–2.03), i.e. heterozygous carriers have an approximately 50% greater risk than homozygous carriers. This improvement in model fit remained significant after Bonferroni correction for the nine HLA-DRB1-DQA1-DQB1 haplotypes under study (0.05/9=0.006). Estimates for this haplotype, and all other haplotypes are found in Table 1, and Supplementary Table 3, respectively. Estimates for the individual genotypes are summarized in Supplementary Table 4. The model fit was not improved when modeling the additive and dominance terms of all HLA-DRB1-DQA1-DQB1 haplotype variants simultaneously in global regression models (data not shown). We observed no substantial attenuation of the non-additive effect when adjusting for known glioma risk SNPs identified by previous GWAS (data not shown).
Table 1.
Additive and non-additive effect sizes of glioma risk for haplotype DRB1*1501-DQA1*0102-DQB1*0602.
| Model | Effect | OR (95% CI) | Pimprovementa |
|---|---|---|---|
| Additive | Additive | 1.03 (0.88–1.20) | 2.5×10−3 |
| Dominance | Additive | 0.80 (0.63–1.01) | |
| Dominance | 1.53 (1.16–2.03) |
P-value from likelihood ratio test of improvement in fit when including the dominance term.
Association analyses comparing homozygous vs heterozygous carriers
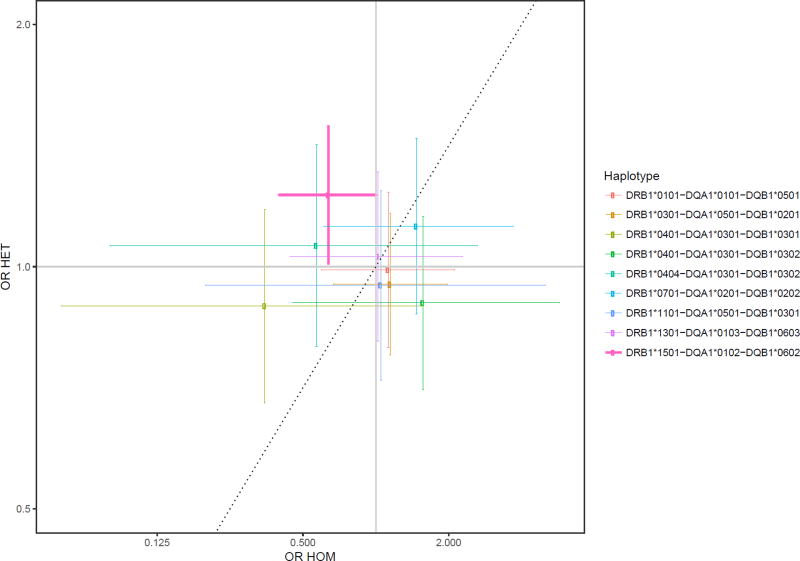
Individuals that were heterozygous for the HLA-DRB1*1501-DQA1*0102-DQB1*0602 multigene haplotype were at increased risk of glioma compared with non-carriers (Table 1), with an adjusted odds ratio (OR) of 1.23 (95% CI 1.01–1.49; P=0.040). Homozygous carriers of the haplotype were at modestly decreased risk of glioma compared with non-carriers (OR 0.64; 95% CI 0.40–1.01; P=0.057). These estimates were not significant after Bonferroni correction (P<0.006), although they were consistent with the estimates from the non-additive analysis of the DRB1*1501-DQA1*0102-DQB1*0602 haplotype using the chi-square tests of improvement of fit. Frequency counts showing the enrichment of glioma cases for heterozygous carriers of the HLA-DRB1*1501-DQA1*0102-DQB1*0602 haplotype (Table 2). Associations for this haplotype, along with all other multigene haplotypes, are displayed in Figure 1 as a scatterplot, and in Supplementary Figure 2 as forest plots. Associations for the individual genotypes show similar estimates for the linked DRB1*1501, DQA1*0102, and DQB1*0602 alleles separately (Supplementary Figure 3). No differences were observed between homozygous and heterozygous carriers of HLA class I alleles (data not shown).
Table 2.
Proportion of cases and controls with 0, 1, or 2 copies of haplotype DRB1*1501-DQA1*0102-DQB1*0602, and overall haplotype frequencies between cases and controls.
| Number of copies | Cases (% with haplotype) | Controls (% with haplotype) |
|---|---|---|
| 0 | 539 (60.8%) | 754 (62.3%) |
| 1 | 318 (35.9%) | 385 (31.8%) |
| 2 | 30 (3.4%) | 72 (5.9%) |
|
| ||
| Haplotype frequency | 378 (21.3%)a | 529 (21.8%)a |
Refers to haplotype frequencies and percentages in cases and controls, where each individual contributes two haplotypes to the total count
Figure 1.
Scatter plot of glioma risk for homozygous haplotype carriers plotted on the x-axis, and heterozygous haplotype carriers plotted on the y-axis. Associations are shown as the odds ratios for glioma risk in carriers compared to non-carriers of the haplotype. The dashed line represents a purely additive relationship, in which heterozygotes have half the risk of homozygotes (on a log-odds scale). The haplotype showing a non-additive effect is bolded.
Interaction model analyses
We examined interaction models of the HLA-DRB1*1501-DQA1*0102-DQB1*0602 haplotype paired with each of the remaining eight haplotypes (Table 3). The inclusion of the interaction term with HLA-DRB1*0401-DQA1*0301-DQB1*0301 improved the fit of the model, P=0.009, although the significance level was above the Bonferroni-adjusted cutoff (0.05/8=0.006). The interaction term estimate was OR=2.28 (95% CI 1.23–4.26), meaning that carriers of the HLA-DRB1*1501-DQA1*0102-DQB1*0602 haplotype had an approximately 2.3-fold greater risk when their second haplotype was HLA-DRB1*0401-DQA1*0301-DQB1*0301.
Table 3.
Additive odds ratios for each haplotype, and fold changes in odds ratios due to the inclusion of interaction effects between DRB1*1501-DQA1*0102-DQB1*0602 and each haplotype. The reference group for each analysis is all other haplotypes combined.
| Haplotype | Case/ Control |
Additive OR (95% CI) |
Interaction term OR |
Interaction P-value |
|---|---|---|---|---|
| DRB1*0101-DQA1*0101-DQB1*0501 | 49/50 | 0.99 (0.79, 1.24) | 1.01 | 0.97 |
| DRB1*0301-DQA1*0501-DQB1*0201 | 88/105 | 0.98 (0.79, 1.21) | 1.26 | 0.23 |
| DRB1*0401-DQA1*0301-DQB1*0301 | 31/33 | 0.85 (0.63, 1.13) | 2.28 | 0.009 |
| DRB1*0401-DQA1*0301-DQB1*0302 | 31/43 | 0.93 (0.72, 1.21) | 1.00 | 0.99 |
| DRB1*0404-DQA1*0301-DQB1*0302 | 17/35 | 1.01 (0.75, 1.35) | 0.84 | 0.63 |
| DRB1*0701-DQA1*0201-DQB1*0202 | 42/37 | 1.11 (0.86, 1.44) | 1.18 | 0.54 |
| DRB1*1101-DQA1*0501-DQB1*0301 | 30/29 | 0.94 (0.71, 1.26) | 1.57 | 0.15 |
| DRB1*1301-DQA1*0103-DQB1*0603 | 30/43 | 1.01 (0.79, 1.28) | 1.22 | 0.5 |
Stratified analyses
No differences were observed when analyses were stratified by sex (data not shown).
In the age-stratified analyses, inclusion of the dominance term improved the model fit for HLA-DRB1*1501-DQA1*0102-DQB1*0602 for younger subjects (Pimprove=0.05) but not older subjects (Pimprove=0.35). However, only 4 controls and 0 cases were homozygous carriers of the haplotype among younger subjects, resulting in unstable point estimates of the additive and dominance terms due to collinearity.
In the rs55705857-stratified analyses, inclusion of the dominance term improved the model fit for both low-grade variant carriers (Pimprove=0.021) and non-carriers (Pimprove=0.009), with a larger dominance term estimate of carriers (OR=2.00, 95% CI 1.10–4.26) compared to non-carriers (OR=1.54, 95% CI 1.16–2.03).
Discussion
This is the most comprehensive study to date of the association between HLA variants and glioma risk, and the first to systematically compare additive and non-additive associations, which enabled the identification of effects not previously described. We observed no additive effect, and a non-additive dominance effect for HLA-DRB1*1501-DQA1*0102-DQB1*0602, a multigene haplotype present in ~40% of all study subjects. The non-additive effect was not attenuated in conditional analyses adjusted for known risk SNPs identified in prior GWAS, suggesting that the effect is independent of previously described genetic associations. Our association estimates suggest that being a homozygous carrier of HLA-DRB1*1501-DQA1*0102-DQB1*0602 is protective against glioma, and that being a heterozygous carrier confers increased risk, with the positive dominance effect at least in part explained by the interaction with HLA-DRB1*0401-DQA1*0301-DQB1*0301.
Among our three nominally significant MHC-wide associations, two have been previously described in association with glioma at the two-digit resolution level. Namely, our finding that HLA-B*4001 and HLA-DRB1*1302 were protective against glioma was consistent with prior observations that HLA-B*40 and HLA-DRB1*13 were enriched among the control group[17,18,21], although the association was not significant in those prior studies and the opposite direction of effect was observed in another study[22]. Although a previous GWAS found that rosacea (recently linked to glioma risk in a large Danish cohort[5]) is associated with three HLA alleles found to be in high linkage disequilibrium in our study (DRB1*0301, DQA1*0501, and DQB1*0201)[8], we did not observe these alleles to be associated with glioma risk, suggesting that the glioma-rosacea link is not likely due to shared HLA risk variants with pleiotropic effects.
HLA-DRB1*1501-DQA1*0102-DQB1*0602 is sometimes more simply referred to as the DR15 haplotype, a long-range HLA haplotype frequently observed in Northern Europeans (also known as the ancestral DR2 haplotype)[33]. The Northern European origin of this haplotype is intriguing, as its distribution may contribute to the lower incidence of gliomas observed in individuals of African ancestry[1]. Previous disease associations observed with this haplotype (and components of the haplotype) include several autoimmune disorders such as narcolepsy, systemic lupus erythematosus, type 1 diabetes, and multiple sclerosis[34–37]. Additionally, the DRB1*1501-DQB1*0602 haplotype has been previously implicated in cervical neoplasia risk, with heterogeneity of effect by human papillomavirus infection type[38]. Prior literature describing the role of viral infections, such as human cytomegalovirus[39] and varicella-zoster[40], on the pathogenesis of gliomas further highlights the relevance of investigating HLA variants in association with glioma.
Our primary finding that multigene haplotype HLA-DRB1*1501-DQA1*0102-DQB1*0602 was associated with glioma risk in a non-additive manner may, in part, explain prior conflicting reports on the association between glioma risk and DRB1*15. Namely, our finding of the increased risk for glioma in heterozygous carriers of HLA-DRB1*1501-DQA1*0102-DQB1*0602 is consistent with studies reporting increased glioma risk associated with DRB1*15[18,22], with the multigene haplotypes including both DRB1*15 and DQB1*06 also conferring increased glioma risk. Other studies detected no association between DRB1*15 and glioma risk[17,19,20]. The previously reported association between increased glioma risk and DQB1*06[18,19,22] is consistent with our finding that DQB1*0602 (in linkage disequilibrium with DRB1*1501) increased glioma risk in heterozygous carriers. We posit that the method of comparing allele frequency distribution between cases and controls used in prior studies is not as well powered to detect non-additive effects, particularly in light of the possible opposing effect directions we observed in homozygous and heterozygous carriers, contributing to the inconsistent evidence in the prior literature. Notably, we would also have detected no association had we chosen to analyze the data simply as a χ2 test of haplotype variant frequency distributions between cases and controls (P=0.8239), due to the misspecification of the model. Our findings reinforce the idea that non-additive effects in the MHC region are an important area of investigation, particularly as necessary functional redundancy in the immune system is hypothesized as a driver of non-additive effects[41], and such effects have been observed in many autoimmune-related disorders previously linked to glioma risk[14,15].
Our observation of a synergistic increase in glioma risk associated with the interaction between HLA-DRB1*1501-DQA1*0102-DQB1*0602 and HLA-DRB1*0401-DQA1*0301-DQB1*0301 may be partially driven by the dominance effect detected in the dominance model. However, this diplotype does not constitute a large portion of the heterozygous individuals, so it is not likely to be a main driver of the observed dominance effect. While it is not apparent why heterozygosity of the HLA-DRB1*1501-DQA1*0102-DQB1*0602 haplotype would increase glioma risk, prior association studies of HLA haplotypes in autoimmune disorders such as Type 1 diabetes, lupus, narcolepsy, ulcerative colitis, celiac disease, rheumatoid arthritis, type 1 diabetes, and psoriasis show both increased or decreased risk in homozygotes compared to heterozygotes[14,15,34–36], suggesting that the exact nature of non-additivity is allele- and disease-specific.
Although we were unable to directly assess the observed non-additive effect separately among glioma histological subtypes (an important clarification given known differences between subtypes with regards to incidence, biology, natural history and association with genetic variants[12,13,43]), we used age of diagnosis and rs55705857 genotype as proxies of tumor grade and histology[30,32,33]. Both stratified analyses suggest that the non-additive effect of HLA-DRB1*1501-DQA1*0102-DQB1*0602 is stronger in low-grade glioma, although several caveats merit mentioning. The age-stratified analysis was limited by the smaller sample size of the younger age group (n=119 vs n=637 among haplotype carriers with available age of diagnosis data). Furthermore, although the magnitude of the dominance term estimate suggested a stronger association in carriers of the low-grade variant, we observed a more significant Pimprove in non-carriers. This discrepancy in effect size versus statistical significance is likely due to the larger sample size of cases not carrying the low-grade variant (n=717 vs. n=170 carriers). This observation suggesting differential association with immune-related variants by glioma type is consistent with a prior study observing stronger allergy associations in non-glioblastoma tumors[43], although other studies have also suggested no difference by subtype[6], and slightly stronger effects in high-grade gliomas[4]. Future studies incorporating histopathology and tumor molecular markers (e.g. IDH and 1p/19q co-deletion status) are needed to clarify any subtype-specificity of HLA risk variants.
Our study is limited by its focus on subjects of European ancestry, and as such may not be generalizable to subjects with other ancestral backgrounds. More work is necessary to replicate our findings, and extend the investigations to non-European subjects. GliomaScan did not include information on other disease status such as diagnoses of atopic and autoimmune diseases, which would have allowed further clarification of whether the association between glioma and immune-related diseases have a shared genetic basis in the HLA region. Finally, additional studies of potential interactions between HLA variants and viral infection status on conferring glioma risk are warranted to expand this line of inquiry.
In summary, we observed a dominance effect on glioma risk for a common HLA class II multigene haplotype with a frequency of ~40% of our study subjects. Future studies are needed to validate the significance of this haplotype association and to identify the causal mechanisms underlying the apparent non-additive effects.
Supplementary Material
Supplementary Figure 1: Flowchart of HLA allele and individual inclusions.
Supplementary Figure 2: Forest plots of glioma risk for a) homozygous haplotype carriers, and b) heterozygous haplotype carriers. Associations are shown as the odds ratios for glioma risk in carriers compared to non-carriers of the haplotype.
Supplementary Figure 3: Scatter plot of glioma risk for homozygous haplotype carriers plotted on the x-axis, and heterozygous haplotype carriers plotted on the y-axis, for a) DRB1, b) DQA1, and c) DQB1. Associations are shown as the odds ratios for glioma risk in carriers compared to non-carriers of the haplotype. The dashed line represents a purely additive relationship, in which heterozygotes have exactly half the risk of homozygotes (on a log-odds scale).
Supplementary Table 1: Glioma risk estimates for nominally significant (P<0.05) HLA alleles
Supplementary Table 2: Glioma risk estimates for multigene haplotype associations based on additive models.
Supplementary Table 3: Additive and non-additive effect sizes of glioma risk for multigene haplotypes across the DRB1-DQA1-DQB1 loci.
Supplementary Table 4: Additive and non-additive effect sizes of glioma risk for DRB1*1501, DQA1*0102, and DQB1*0602.
Acknowledgments
Funding: This work was supported by the National Institutes of Health T32CA151022-06 (C.Z.), R25T CA112355 (J.S.W.), and The Sontag Foundation (K.M.W.)
The results published here are, in part, based upon data obtained from dbGaP Study Accession phs000652.v1.p1: “Cohort-based Genome-Wide Association Study of Glioma (GliomaScan)” which was supported by intramural funds from the NCI and federal funds from the NCI under contract N01-CO-12400. The authors additionally acknowledge use of the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. This research uses resources provided by the Type 1 Diabetes Genetics Consortium (T1DGC); a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK); National Institute of Allergy and Infectious Diseases (NIAID); National Human Genome Research Institute (NHGRI); National Institute of Child Health and Human Development; Juvenile Diabetes Research Foundation International (JDRF), supported by U01 DK062418.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent: Informed consent was obtained from all individual participants included in the study.
References
- 1.Ostrom QT, Gittleman H, Farah P, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2006–2010. Neuro-Oncol. 2013;15:ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro-Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiemels JL, Wilson D, Patel C, et al. IgE, Allergy, and Risk of Glioma: Update from the San Francisco Bay Area Adult Glioma Study in the Temozolomide Era. Int J Cancer J Int Cancer. 2009;125:680–687. doi: 10.1002/ijc.24369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amirian ES, Zhou R, Wrensch MR, et al. Approaching a Scientific Consensus on the Association between Allergies and Glioma Risk: A Report from the Glioma International Case-Control Study. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2016;25:282–290. doi: 10.1158/1055-9965.EPI-15-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egeberg A, Hansen PR, Gislason GH, Thyssen JP. Association of Rosacea With Risk for Glioma in a Danish Nationwide Cohort Study. JAMA Dermatol. 2016;152:541–545. doi: 10.1001/jamadermatol.2015.5549. [DOI] [PubMed] [Google Scholar]
- 6.Brenner AV, Linet MS, Fine HA, et al. History of allergies and autoimmune diseases and risk of brain tumors in adults. Int J Cancer. 2002;99:252–259. doi: 10.1002/ijc.10320. [DOI] [PubMed] [Google Scholar]
- 7.Hinds DA, McMahon G, Kiefer AK, et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 2013;45:907–911. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang ALS, Raber I, Xu J, et al. Assessment of the genetic basis of rosacea by genome-wide association study. J Invest Dermatol. 2015;135:1548–1555. doi: 10.1038/jid.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bateman AC, Howell WM. Human leukocyte antigens and cancer: is it in our genes? J Pathol. 1999;188:231–236. doi: 10.1002/(SICI)1096-9896(199907)188:3<231::AID-PATH325>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 11.Walsh KM, Codd V, Smirnov IV, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melin BS, Barnholtz-Sloan JS, Wrensch MR, et al. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49:789–794. doi: 10.1038/ng.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajaraman P, Melin BS, Wang Z, et al. Genome-wide association study of glioma and meta-analysis. Hum Genet. 2012;131:1877–1888. doi: 10.1007/s00439-012-1212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenz TL, Deutsch AJ, Han B, et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat Genet. 2015;47:1085–1090. doi: 10.1038/ng.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goyette P, Boucher G, Mallon D, et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet. 2015;47:172–179. doi: 10.1038/ng.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.the International Multiple Sclerosis Genetics Consortium. Class II HLA interactions modulate genetic risk for multiple sclerosis. Nat Genet. 2015;47:1107–1113. doi: 10.1038/ng.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerini FR, Agliardi C, Zanzottera M, et al. Human leukocyte antigen distribution analysis in North Italian brain Glioma patients:an association with HLA-DRB1*14. J Neurooncol. 2005;77:213–217. doi: 10.1007/s11060-005-9032-x. [DOI] [PubMed] [Google Scholar]
- 18.Machulla HKG, Steinborn F, Schaaf A, et al. Brain Glioma and Human Leukocyte Antigens (HLA) – is There an Association. J Neurooncol. 2001;52:253–261. doi: 10.1023/A:1010612327647. [DOI] [PubMed] [Google Scholar]
- 19.La Torre D, Maugeri R, Angileri FF, et al. Human leukocyte antigen frequency in human high-grade gliomas: a case-control study in Sicily. Neurosurgery. 2009;64:1082–8. doi: 10.1227/01.NEU.0000345946.35786.92. discussion 1088–1089. [DOI] [PubMed] [Google Scholar]
- 20.Tang J, Shao W, Dorak MT, et al. Positive and Negative Associations of Human Leukocyte Antigen Variants with the Onset and Prognosis of Adult Glioblastoma Multiforme. Cancer Epidemiol Biomark Amp Prev. 2005;14:2040. doi: 10.1158/1055-9965.EPI-05-0136. [DOI] [PubMed] [Google Scholar]
- 21.Song W, Ruder AM, Hu L, et al. Genetic Epidemiology of Glioblastoma Multiforme: Confirmatory and New Findings from Analyses of Human Leukocyte Antigen Alleles and Motifs. PLOS ONE. 2009;4:e7157. doi: 10.1371/journal.pone.0007157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassig BA, Inskip PD, Burdette L, et al. Selected human leukocyte antigen class II polymorphisms and risk of adult glioma. J Neuroimmunol. 2011;233:185–191. doi: 10.1016/j.jneuroim.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guja C, Guja L, Nutland S, et al. Type 1 diabetes genetic susceptibility encoded by HLA DQB1 genes in Romania. J Cell Mol Med. 2004;8:249–256. doi: 10.1111/j.1582-4934.2004.tb00280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laaksonen M, Pastinen T, Sjoroos M, et al. HLA class II associated risk and protection against multiple sclerosis-a Finnish family study. J Neuroimmunol. 2002;122:140–145. doi: 10.1016/s0165-5728(01)00456-8. [DOI] [PubMed] [Google Scholar]
- 25.Jia X, Han B, Onengut-Gumuscu S, et al. Imputing Amino Acid Polymorphisms in Human Leukocyte Antigens. PLOS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rich SS, Concannon P, Erlich H, et al. The Type 1 Diabetes Genetics Consortium. Ann N Y Acad Sci. 2006;1079:1–8. doi: 10.1196/annals.1375.001. [DOI] [PubMed] [Google Scholar]
- 27.Chang CC, Chow CC, Tellier LC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 29.Whitacre CC. Sex differences in autoimmune disease. Nat Immunol. 2001;2:777–780. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 30.Jenkins RB, Xiao Y, Sicotte H, et al. A low-frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nat Genet. 2012;44:1122–1125. doi: 10.1038/ng.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grier JT, Batchelor T. Low-Grade Gliomas in Adults. The Oncologist. 2006;11:681–693. doi: 10.1634/theoncologist.11-6-681. [DOI] [PubMed] [Google Scholar]
- 32.Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Traherne JA. Human MHC architecture and evolution: implications for disease association studies. Int J Immunogenet. 2008;35:179–192. doi: 10.1111/j.1744-313X.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mignot E, Lin L, Rogers W, et al. Complex HLA-DR and -DQ Interactions Confer Risk of Narcolepsy-Cataplexy in Three Ethnic Groups. Am J Hum Genet. 2001;68:686–699. doi: 10.1086/318799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernando MMA, Stevens CR, Sabeti PC, et al. Identification of Two Independent Risk Factors for Lupus within the MHC in United Kingdom Families. PLOS Genet. 2007;3:e192. doi: 10.1371/journal.pgen.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erlich H, Valdes AM, Noble J, et al. HLA DR-DQ Haplotypes and Genotypes and Type 1 Diabetes Risk. Diabetes. 2008;57:1084. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt H, Williamson D, Ashley-Koch A. HLA-DR15 haplotype and multiple sclerosis: a HuGE review. Am J Epidemiol. 2007;165:1097–1109. doi: 10.1093/aje/kwk118. [DOI] [PubMed] [Google Scholar]
- 38.Hildesheim A, Schiffman M, Scott DR, et al. Human leukocyte antigen class I/II alleles and development of human papillomavirus-related cervical neoplasia: results from a case-control study conducted in the United States. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 1998;7:1035–1041. [PubMed] [Google Scholar]
- 39.Dziurzynski K, Chang SM, Heimberger AB, et al. Consensus on the role of human cytomegalovirus in glioblastoma. Neuro-Oncol. 2012;14:246–255. doi: 10.1093/neuonc/nor227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amirian ES, Scheurer ME, Zhou R, et al. History of chickenpox in glioma risk: a report from the glioma international case–control study (GICC) Cancer Med. 2016;5:1352–1358. doi: 10.1002/cam4.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose AM, Bell LCK. Epistasis and immunity: the role of genetic interactions in autoimmune diseases. Immunology. 2012;137:131–138. doi: 10.1111/j.1365-2567.2012.03623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinnersley B, Labussière M, Holroyd A, et al. Genome-wide association study identifies multiple susceptibility loci for glioma. Nat Commun. 2015;6:8559. doi: 10.1038/ncomms9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiemels JL, Wiencke JK, Sison JD, et al. History of allergies among adults with glioma and controls. Int J Cancer. 2002;98:609–615. doi: 10.1002/ijc.10239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Flowchart of HLA allele and individual inclusions.
Supplementary Figure 2: Forest plots of glioma risk for a) homozygous haplotype carriers, and b) heterozygous haplotype carriers. Associations are shown as the odds ratios for glioma risk in carriers compared to non-carriers of the haplotype.
Supplementary Figure 3: Scatter plot of glioma risk for homozygous haplotype carriers plotted on the x-axis, and heterozygous haplotype carriers plotted on the y-axis, for a) DRB1, b) DQA1, and c) DQB1. Associations are shown as the odds ratios for glioma risk in carriers compared to non-carriers of the haplotype. The dashed line represents a purely additive relationship, in which heterozygotes have exactly half the risk of homozygotes (on a log-odds scale).
Supplementary Table 1: Glioma risk estimates for nominally significant (P<0.05) HLA alleles
Supplementary Table 2: Glioma risk estimates for multigene haplotype associations based on additive models.
Supplementary Table 3: Additive and non-additive effect sizes of glioma risk for multigene haplotypes across the DRB1-DQA1-DQB1 loci.
Supplementary Table 4: Additive and non-additive effect sizes of glioma risk for DRB1*1501, DQA1*0102, and DQB1*0602.