Abstract
Cell fate is a concept used to describe the differentiation and development of a cell in its organismal context over time. It is important in the field of regenerative medicine, where stem cell therapy holds much promise but is limited by our ability to assess its efficacy, which is mainly due to the inability to monitor what happens to the cells upon engraftment to the damaged tissue. Currently, several imaging modalities can be used to track cells in the clinical setting; however, they do not satisfy many of the criteria necessary to accurately assess several aspects of cell fate. In recent years, reporter genes have become a popular option for tracking transplanted cells, via various imaging modalities in small mammalian animal models. This review article examines the reporter gene strategies used in imaging modalities such as MRI, SPECT/PET, Optoacoustic and Bioluminescence Imaging. Strengths and limitations of the use of reporter genes in each modality are discussed.
Keywords: Molecular imaging, Stem cell engraftment, Stem cell tracking, Cell imaging modalities
Introduction
Regenerative medicine involving the use of stem cells for therapeutic purposes is a highly promising field with various applications across areas of medicine. The objective of cellular regenerative therapy is to regenerate damaged tissue, promote normal tissue function using cells that are capable of producing biologically active molecules, and redirect any aberrant processes associated with the medical condition [1]. To date, we have seen encouraging results obtained with stem cells in various diseases including diabetes as well as cardiovascular and orthopedic diseases [1–6]. To continue to advance successful translation of stem cell-based therapeutic approaches, it is essential to understand the behavior and functional outcome of stem cells to demonstrate their beneficial effect in a clinical setting [4].
Cell fate is a term used to describe the differentiation and development of a cell over time. It is a concept necessary and pertinent to studying stem cells and is closely related to a vast array of processes such as stem cell homeostasis, cell division, differentiation, as well as migration and engraftment of cells to damaged tissue. As cells progress down a specific developmental path, they undergo differential gene expression leading to irreversible acquisition of unique characteristics that contribute to cell fate determination. Since cells may undergo conditional or autonomous specification, they may appear nearly identical to their neighbor cells [7]. Consequently, the long-term and systemic effects of stem cell therapy are difficult to assess [3].
Tracking cell fate is, therefore, a critical need in the field of stem cell research and regenerative medicine as it provides insights into stem cell engraftment and enables evaluation of the success of stem cells as a therapeutic modality. With technological advances in imaging approaches, they have become instrumental in studying cell fate of stem cells in terms of temporal localization, functionality, and viability. There are challenges, however, and while whole-body imaging modalities such as magnetic resonance imaging (MRI), single-photon emission computed tomography (SPECT), and positive emission tomography (PET) can be utilized to track stem cells or immune cells labeled with exogenous contrast agents, several limitations exist that restrict their application [6]. The main limitations are that the in vitro-labeled cells lose the contrast agent over every successive cellular division, eventually leading to an absence of detectable signal. Ideally, the label should be retained by cells over prolonged periods of time, able to give an accurate quantitative measure of cell count, cleared rapidly after cell death, and non-toxic to the host [1, 6, 7]. One way to accomplish this is with the use of DNA sequences—known as reporter genes—that code for proteins able to generate contrast. The signal is not lost when the cells divide since the reporter gene is replicated with the rest of the genome. The intensity of the signal or the degree of light generated correlates well with the number of cells, so quantification of cell count is possible. This self-renewing and non-toxic mode of imaging can be used to provide crucial data on stem cell fate [8]. A common limitation, however, is that reporter gene expression becomes silenced after several cell divisions [9].
Here, we will discuss the strengths and limitations of reporter gene-based cell tracking approaches in preclinical settings. We will present important factors such as the biological distribution of the gene, availability of the probes, and the effect of the reporter gene on cell function. A brief review of the various current labeling techniques in the imaging modalities will be provided, while the strength and limitations of each modality in the context of stem cell tracking research are compared in Table 1.
Table 1.
Comparison of strengths and limitations of reporter gene strategies in various imaging modalities [10–24, 27–37, 40, 42, 46, 55, 56, 63, 65–67, 69–71, 93]
| Imaging modality | Mechanism | Strengths | Limitations |
|---|---|---|---|
| MRI (General) | Genes code for proteins that generate contrast upon the application of pulse sequences T1 and T2 |
High resolution Scans large segments of body No radiation exposure |
Expensive Strict selection criteria, many contraindications |
| Iron homeostasis reporter proteins |
Proteins increase expression of transferrin receptor or ferritin, leading to increased intracellular iron stores, which generate contrast when pulse sequences are applied |
Both endogenous genes; therefore, there is no worry about immunogenicity Additional contrast agent is not necessary although it may be used to gain higher resolution |
Weaker contrast than other methods Potential for augmenting Fenton reaction, leading to generation of free radicals |
| Reporter enzymes and proteins | Enzymes catalyze reactions and yield products capable of generating contrast or expressing surface proteins that bind to exogenously administered contrast agents |
Contrast generated by select cells For surface proteins, no internalization of contrast agent or any particles in general Can control the amount of resolution by giving more contrast agent |
Need to administer contrast agent or precursor contrast agent exogenously Potential for toxicity of exogenous agent Contrast agent may not reach low perfused areas |
| CEST | CEST proteins with saturated protons interact with surrounding water molecules, leading to chemical shifts |
Wide variety of CEST reporter genes available and theoretically feasible to use In theory, able to monitor multiple CEST reporter genes in one imaging session |
In some cases, an exogenous agent needs to be administered Some CEST reporter genes yield low contrast to noise ratio Few in vivo studies |
| PET (general) | PET scanner recognizes tracers that emit positrons, which are trapped, transported inside, or adhere to target cell via the reporter gene |
High sensitivity, contrast, and spatial resolution Can quantify cell number |
Very expensive Exogenous, radioactive tracer required |
| Reporter enzyme | Enzyme acts on exogenously administered tracer, allowing it to be trapped intracellularly and tracked over time | Uses a non-human kinase which allows selective trapping of exogenous tracers in only the cells that express the reporter gene |
Uses HSV1 TK, a non-human gene (potential for adverse immune reaction) Trapping of radioactive particles inside cells |
| Reporter receptor | Receptors allow radioactive ligand to bind to cell surface |
No intracellular accumulation of radioactive tracer Uses a human protein, substantially lowering chance of adverse immune reaction |
Radioactive tracers still inside the body, even if not intracellularly |
| Reporter transport protein | Membrane transporter allows selective intake of exogenously administered tracer |
Uses a human protein, substantially lowering chance of adverse immune reaction If imaging is done quickly after administration of tracer, high sensitivity can be achieved |
Accumulation of radioactive tracer inside cell Relatively fast efflux of tracer out of cell |
| SPECT (general) | Use gamma rays to detect radioactive isotopes |
Relatively high sensitivity and resolution Lower cost than PET |
Lower sensitivity than PET Need to administer radioactive isotopes |
| Reporter transport proteins | Express human transporter protein which allows for radioactive isotopes to accumulate intracellularly | Reporter protein is human; thus, there is less chance of adverse immune reaction | Accumulation of radioactive isotopes intracellularly |
| Bioluminescence imaging (BLI) | |||
| Reporter enzyme | Luciferin enzyme converts exogenously administered d-luciferin to yield the optically active metabolite oxyluciferin |
Low cost No radiation Lots of signal from small number of cells |
Small resolution Minimal tissue penetration Need to inject contrast agent |
| Fluorescence imaging | |||
| Reporter protein | Exogenous excitation source causes protein to emit light of a different wavelength | Similar to BLI |
Low resolution Minimal tissue penetration |
MRI reporter genes
MRI is an imaging technology that uses magnets to polarize water molecules in human tissue to come up with a high-resolution, multi-dimensional image. Because this modality does not involve ionizing radiation, it can be performed serially over time. Various labels such as iron oxide nanoparticles, Gd chelates, microcapsules with fluorine sodium carbon, ferritin and lysine-rich protein protamine can be used [6]. Traditionally, MRI images are generated using chemical agents that produce contrast through specific pulse sequences, most notably T1 and T2. Gadolinium-diethylenetriaminepentaacetic acid (DTPA), a T1-shortening contrast agent, is used to track cells in the context of microphage and fibroblast migration [9]. Additionally, iron oxide nanoparticles can be used to de-phase the surrounding magnetic field of the tissue, leading to shortened T2 relaxation times and thus allowing one to visualize the cell [10]. While the use of such agents has been wide and frequent, many limitations restrain their use in clinical research with stem cells. Upon division, cells lose or dilute the labeling agent, leading to diminished signaling after successive generations of the cell cycle. There is also the possibility of these agents’ altering the function of the cell. In the case of iron particles, they remain in the tissue after the death of labeled cells, which poses a significant barrier to cell tracking in the context of regenerative medicine [11].
The use of MRI reporter genes to visualize in vivo cell fate has been increasingly employed over the last few years. Overall, certain MRI reporter gene strategies overcome some of the caveats that limit traditional probes in their use in cell fate tracking. As a whole, MRI reporter genes can be divided into three main classes: iron homeostasis proteins, reporter enzymes, and reporter genes capable of achieving chemical exchange saturation transfer [11]. In general, the expression of the various classes of reporter genes does not appear to alter the cell function [12]. Additionally, certain methods may utilize iron oxide nanoparticles in addition to the reporter gene to obtain an image with enhanced contrast.
Iron homeostasis proteins
The two main reporter genes for MRI in the iron homeostasis mechanism of action are the transferrin receptor and ferritin, the iron storage protein. The mechanism behind increased transferrin receptor expression and cell fate detection is that as a transporter protein, the receptor will cause more iron to accumulate intracellularly as ferritin (Fig. 1). The increase in iron concentration will enable the genetically engineered cell to have an increase in R2 relaxation and thus generate a detectable change in contrast [11]. The gene can be manipulated so that it lacks the iron-regulatory region and mRNA destabilization motif in the 3′ untranslated region, leading to constitutive expression of the receptor [12]. One major limitation hindering its widespread use as a reporter gene is the weak change in image contrast that is produced. Incidentally, one can overcome this by administering transferrin conjugated to iron oxide nanoparticles [11]. However, using these nanoparticles with reporter genes brings about several limitations. For long-term imaging, residual iron oxide nanoparticles left over from previous imaging sessions may interfere with the signal, leading to inferior contrast-to-noise ratio [13, 14]. On the other hand, if we use techniques that utilize reporter genes without the administration of iron oxide nanoparticles, the resolution obtained by the MRI scanner is low [13]. The rise in iron stores poses another issue—augmenting Fenton reaction, leading to toxicity [11, 15].
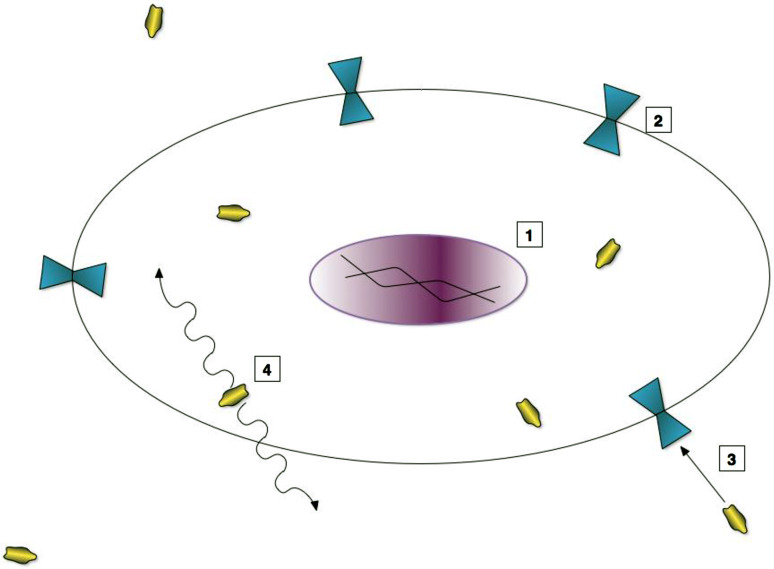
Fig. 1.
Mechanism of action of cell surface transporter reporter proteins. 1 Reporter gene is integrated within stem cell’s genome. 2 Protein is expressed and synthesized within the cell. Subsequently, the protein gets anchored as a cell membrane receptor. 3 An exogenously administered contrast agent is transported inside the cell via the reporter protein. 4 Contrast agent makes detection of cell possible using various imaging modalities, like MRI or PET
Ferritin itself is one of the most commonly used MRI reporter genes. Ferritin is a 450-kDa globular protein consisting of multiple heavy and light protein subunits. Elevation of the heavy chain, or in combination with the light chain, causes an increase in intracellular iron stores [11]. Ferritin overexpression inside the cell leads to iron accumulation, which in turn allows for precise tracking of the stem cells in vivo. The disruption in the magnetic field caused by the iron accumulation is picked up by measuring myocardial T2-star relaxation parameter, which appears as areas of hypointensity on the image. The benefits of this technique are vast. For one, increasing ferritin expression protects the cells from the Fenton reaction-mediated oxidative damage. Second, the contrast generated from the ferritin upregulation is sufficient without exogenous sources of iron. Nonetheless, the sensitivity of this technique, especially in the in vivo context, remains limited and significantly less clear contrast is generated when compared to techniques that utilize exogenous particles [11].
Interestingly, several studies have shown that the cells overexpressing ferritin do not have to be cultured in an iron-rich medium for them to generate sufficient contrast [11]. However, recent data demonstrated that stem cells with overexpressed ferritin heavy chain-1 showed a noticeable change in morphology and rate of proliferation, which was reversed when cultured in iron-rich media [16]. In addition, associations have been made between increased ferritin expression and neurodegeneration [17]. Thus, the chronic overexpression of ferritin as a reporter gene could have cytotoxic effects. A study showed that one way to address this issue is through the use of an inducible reporter gene system, such as MagA with the Tet-on expression system [13]. The function of the gene is transportation of iron and intracellular formation of magnetite crystals causing a significant reduction in signal on T2-weighted MRI images [15]. The magnetite crystals have similar properties as SPIO nanoparticles and thus are excellent as T2-contrast agents. Another benefit of utilizing this approach is that levels of MagA can be controlled via administration of a tetracycline antibiotic. Having an inducible reporter system leads to a reduced accumulation of MagA and iron limiting cytotoxicity and thus permits longitudinal imaging [13]. Although this technique can potentially overcome the limitation that ferritin overexpression induces, MagA is a non-human gene, which could lead to adverse immune responses.
Reporter enzymes
Several different kinds of reporter enzymes with varying mechanisms can be used for MRI reporter imaging. The divalent metal transporter (DMT1) can be used to allow increased manganese uptake, resulting in altered T1-relaxation properties that, in comparison to the iron-regulatory proteins, provides enhanced sensitivity. A major drawback of this method is the possibility of adverse reactions to high manganese levels. Toxicity aside, many other cell types are capable of manganese import, thus contributing to a lot of noise on an MRI [18]. Alternatively, overexpression of tyrosinase, an enzyme responsible for the production of melanin, can be used to create a surplus of melanin inside cells to sequester paramagnetic ions and generate contrast on an MRI [11].
Artificially expressed plasma membrane peptides and antigens as reporters have also been used, in the context of cancer cell proliferation imaging. Experiments using this method were conducted by implanting embryonic stem cells that were engineered to express the fusion reporter protein myelocytomatosis (myc) and hemagglutinin (HA), which anchors to the plasma membrane. Upon intravenous administration of contrast agents such as super-paramagnetic iron oxide particles conjugated to either anti-HA or anti-myc monoclonal antibodies that bind to such reporter surface proteins, T2-weighted images can be generated [12]. Since exogenous agents need to be administered for the reporter genes to work, several limitations arise from this requirement. There is the potential of the contrast agent having a toxic effect on the cells or the organism as a whole. Additionally, the contrast agent may have difficulty reaching areas that have low perfusion or areas of the body that are hard to reach in general, such as the central nervous system with its blood–brain barrier [11].
Genes capable of achieving chemical exchange saturation transfer (CEST)
The third strategy of utilizing MRI reporter genes is through a process called chemical exchange saturation transfer, or CEST. Generation of contrast in vivo is possible through overexpression of lysine-rich protein (LRP), whose amide protons exchange with water protons. The first step in achieving CEST is the irradiation of protons of the reporter protein with a resonance frequency that causes protons to be exchanged [19]. When the irradiated protons are exchanged with those found in the surrounding water molecules, saturation is transferred and the said water molecules generate a reduced signal that is seen on MRI [19, 20]. This reduction in signal corresponds to achieving contrast (Fig. 2) [11]. Alternatively, Bar-Shir et al. have shown that a supercharged green fluorescent protein (GFP) mutant could also be used as a CEST agent [21]. A limitation of utilizing LRP or GFP is that they are synthetic genes and thus increase the potential for an adverse immune reaction when transduced into human cells. One potential alternative is to utilize a biocompatible MRI reporter gene based on a human gene, the human protamine-1 (hPRM1), which is arginine rich and thus would yield high contrast using the CEST mechanism [22]. However, imaging of this reporter was only done in vitro and its efficacy in vivo is yet to be determined.
Fig. 2.
Mechanism behind MRI CEST. 1 Reporter gene, such as lysine-rich protein (LRP), is integrated within stem cell’s genome. 2 LRP is expressed and synthesized inside the cell. 3 Amide protons in LRP are irradiated, causing them to be switched with the surrounding ones in the water molecule. 4–5 Protons are transferred and the new water molecule generates reduced signaling on MRI, which corresponds to a change in contrast
A second mechanism for generating CEST-MRI images is through expression of an enzyme that catalyzes the exchange of an amide proton from an exogenously administered probe with the surrounding water molecules. By expressing the enzyme Herpes Simplex Virus 1-thymidine kinase (HSV1-tk), Bar-Shir et al. successfully generated CEST contrast by having the kinase phosphorylate an exogenous CEST-MRI reporter probe, 5-methyl-5,6-dihydrothymidine (5-MDHT), and trapping it intracellularly. The increase in concentration of 5-MDHT within cells expressing HSV1-tk allowed it to be visualized using the CEST-MRI method, which relies on the exchange of protons on the reporter probe with the surrounding water protons [23].
In theory, one of the major benefits of this CEST-MRI is that multiple reporter genes responding to varying resonant frequencies can be used. Thus, investigators can selectively activate various CEST agents, enabling the visualization of multiple CEST targets within the same organism in one imaging session. In studies where cells were electroporated to allow intake of exogenous CEST probes, the tracking of two different probes simultaneously in one imaging session was possible [24].
Lastly, Shapiro et al. recently reported the first genetically encoded reporter for hyperpolarized 129Xe MRI [25]. These expressible reporters are based on gas vesicles, gas-binding protein nanostructures expressed by certain buoyant microorganisms. The same group also demonstrated that aquaporin-1 (AQP1) can be used as a genetically encoded reporter for diffusion-weighted MRI [26].
PET reporter genes
Positive emission tomography is a tomographic technique that produces images of functional processes in the body via detection of biologically active radiotracers. Common tags include: fluorodeoxyglucose (FDG), 9-(4-(18)F-fluoro-3-[hydroxymethyl]butyl)guanine (FHBG), and 18F-fluorodopamine (FDOPA) [27–30]. It is useful in providing an image of general distribution of the labeled target. It works by allowing one to precisely localize and quantify molecular events in vivo by detecting the annihilation of positrons and photons. While PET imaging has poor spatial resolution, with poor anatomic information, a clinically valuable optical image can be obtained by co-registering the PET image with anatomical computer tomography (CT) image data [27]. The main issue with PET imaging is that of endurance; the tags steadily decay in the body and thus prevent imaging over the course of a few weeks [6].
Reporter genes utilized in PET techniques are classified into three general categories based on how the protein products interact with the PET reporter probes (PRP). The groupings are: PET reporter genes encoding enzymes that phosphorylate its target PRPs, leading to their intracellular retention and accumulation; PET reporter genes encoding proteins that act as receptors for the PRP; and finally genes that translate into membrane transporters for the specific PRPs [28]. These PET probes allow for high spatial resolution as well as sensitivity that is higher than what can be achieved with MRI [29].
An example of a PET reporter gene with enzymatic function is HSV-1 TK. Various tracers can be used in combination with HSV1-TK, including thymidine radionucleotides. One study created such a reporter gene for the purpose of assessing gene therapy results. The researchers engineered a reporter gene that allowed for highly sensitive imaging of liver cells in rats in vivo. They utilized an 18F-labeled tracer, 18F-FHBG, which was acted upon by the reporter enzyme, allowing the reporter probe to be trapped within cells expressing HSV1-tk intracellularly [30]. It is very likely that such a procedure could be replicable in the setting of stem cell tracking. While the use of non-human genes as reporter genes may have disadvantages in terms of immunogenicity, one benefit is that the non-human kinases will not get trapped inside cells that do not express them which would lead to non-targeted cells being visualized in the PET image [28].
Reporter genes that act as membrane transporters for PRPs have been heavily utilized in various fields of research (Fig. 1). A prime example is the human sodium iodide symporter (hNIS), which is capable of transporting radioactively labeled forms of iodide. One study used hNIS to examine the ability of mesenchymal stem cells to differentiate into various lineages in vivo. To generate the image, Wolfs et al. injected mice with 124I, which was taken up by the mesenchymal cells via the hNIS. Since a human protein was utilized in this genetic construct, there was reduced incidence of adverse immunological responses. One observable drawback to this specific technique, however, was a substantial efflux of the tracer molecule from the cells. Nonetheless, 31% of the tracer remained trapped intracellularly after 3 h, and thus detection of the mesenchymal stem cells was still possible [27].
The third PET reporter gene strategy involves utilizing transgenic receptors to allow the binding of a radioactive ligand. One such example is the use of the dopamine D2 receptor and a high-affinity radioactively labeled D2 antagonist, [11C]FLB 457. Other studies have used ligands such as 18F-fallypride and 1C-(1)-4-propyl-9-hydroxynaphthoxazine, with each probe having varying degrees of uptake in different tissue types [29, 31]. The binding is accomplished though transfection of a cell line with a vector containing a promoter to drive the expression of D2R. Binding of the antagonist does not elicit any specific responses in the cell, and the ligand–receptor complex can be recognized using PET imaging [32]. Overall, the mutated D2 receptor is nonfunctional and it can be construed that it will have a low potential for malignant transformation or for starting adverse or unnecessary signaling cascades [29]. An advantage of this method is that the radioactive tracer does not make it inside the cell.
SPECT reporter genes
SPECT uses gamma rays emitted from radioactive isotopes such as Tc-99m and In-111. The isotope Tc-99m can even be used to detect apoptotic cells, which is important to determine the viability of transplanted stem cells and as such can be used to detect early rejection in heart transplant recipients [6].
Reporter genes for SPECT imaging are quite similar to the ones utilized for PET. Overall, however, SPECT images have lower sensitivity and resolution [33]. For in vivo imaging of implanted cells (both mesenchymal and immunologic) in small mammals, reporter systems that act as (ion)channels have been used most heavily. The two most notable reporter systems are the human norepinephrine transporter (hNET) and the human sodium iodide symporter (hNIS); both genes are introduced to the host cells via retroviral vectors. The proliferation of cells after expression of hNIS is unaffected [34].
hNET works by transporting norepinephrine analogues into cells. The contrast agent used with hNET to generate the image in SPECT is [123I]metaiodobenzylguanidine ([123I]MIBG), which happens to be a clinically approved probe. Once hNET is introduced into T cells, the cells can be injected into tumor sites and upon introduction of the radiolabel probe, the biodistribution, growth, and activity of the cells can be monitored for up to 28 days [34].
The other human protein that has been used in SPECT, hNIS, is capable of delivering the SPECT tracer 99mTc-pertechnetate as well as iodide intracellularly, which can then be used as a tracer for SPECT analysis. In one study, the hNIS was introduced into cardiac-derived stem cells that were then injected into rat myocardium after an infarction. The reporter gene stayed in the cell for up to 2 weeks, with increasing visualization of the cells from days 1–3 and a gradual decrease in signal by day 12 [35]. In another study, the gene was used to study transplanted cells in the brain over time, since it is not naturally expressed in the brain. The spatial resolution of the probe system was sufficient to determine the grafting of cells in the brain, allowing for differentiation between regions of varying stem cell density. Some key features made this reporter probe a good choice for cell fate tracking: the radio probes for the transporter are readily available in many clinics and their clearance and metabolism in the body are well documented. However, one limitation was the inability of the probe to cross the blood–brain barrier, and thus the probe had to be delivered by an intracranial injection [36, 37].
The benefit of using such reporter genes is the same as with other human proteins; the chance of an adverse immunogenic response to a xenogeneic reporter construct is reduced. Similar to PET, SPECT does not generate anatomically relevant data on its own and thus the imaging modality can be combined with CT scans to allow precise localization of the labeled cells [34]. In addition, while it is believed that PET is a more sensitive imaging modality, studies utilizing reporter genes for tracking cell fate have shown that SPECT shows resolution on par with that of PET [34, 35]. Considering the difference in costs utilizing PET and SPECT, these findings could be significant in a clinical context [34, 35, 37].
Reporter genes for bioluminescence imaging
Another reporter gene method widely used in preclinical studies is in vivo bioluminescence imaging (BLI) [38, 39]. For BLI, a gene expressing an enzyme, such as firefly luciferase, oxidizes a bioluminescent substrate and thus induces its light emission is transduced for stable or inducible expression (e.g., with a retroviral vector) into cells of interest. d-Luciferin, the substrate most commonly used for the firefly luciferase enzyme, is administered exogenously (i.e., by intravenous or intraperitoneal injection) and is converted by the luciferase into its optically active metabolite, oxyluciferin. After substrate administration, the animal is anesthetized and placed into an imaging chamber equipped with a charge-coupled device (CCD) camera to non-invasively capture bioluminescent photons [38–40].
Modifications of various luciferases have been developed to advance in vivo cell tracking via BLI [41], the most common being the modified firefly luciferase Luc2 optimized for expression in mammalian cells [42].
BLI, in contrast to fluorescence imaging, suffers only from minimal background signals emanating from the animal’s tissues. Therefore, relatively weak signals from few cells can be detected with a high signal-to-background ratio [43]. However, a major limitation of BLI has been the relatively low spatial resolution and tissue penetration as compared to MRI and PET/SPECT. This is due to the low energy of the produced photons that are scattered or absorbed by myoglobin and hemoglobin, the two main endogenous absorbers of bioluminescent light in tissue, in particular in areas with a high blood-to-tissue ratio [15, 44]. Since levels of light emitted by the native firefly luciferase drop exponentially by ~90% for every centimeter of tissue penetrated, the utility of BLI in animals larger than rabbits is limited and has been rarely explored. To avoid the absorption of light below 600 nm and improve in vivo detection, a number of researchers have developed luciferases and/or substrates, as well as fluorescent proteins that emit light at longer wavelengths [45].
Red-shifted luciferases
Shifting the emitted bioluminescent light to the red spectrum has by now been achieved in a number of ways. Genetically engineered luciferase variants, for example based on the luc2 gene, such as Ppy RE9, emit light that is shifted towards the red spectrum and hence create a narrower BLI signal without undesirable tissue absorbance. The major drawback of Ppy RE9 appears to be that cells expressing it showed four to five times lower luminescence intensity compared to cells expressing Luc2 [42]. Mezzanotte et al. have studied the use of two luciferases simultaneously, a red light-emitting codon-optimized Photinus pyralis luciferase mutant, Ppy RE8, and the green click beetle luciferase, CBG99. The described d-luciferin-dependent red/green couplet allows for quantitative gene expression studies in vivo, thus enabling simultaneous tracking of different populations of stem cells [46].
Rumyantsev et al. constructed chimeric reporters, in which an intramolecular bioluminescence resonance energy transfer (BRET) occurs between an enhanced Renilla luciferase variant RLuc8 and two bacterial phytochrome-based near-infrared (NIR) fluorescent proteins iRFP670 or iRFP720. These chimeric proteins exhibit NIR bioluminescence with maxima at 670 and 720 nm, respectively [47]. The 50 nm spectral shift between emissions of the two iRFP chimeras enables combined multicolor imaging, while the iRFPs can also be detected by multicolor fluorescence imaging (FLI); Conley et al. synthesized a selenium analogue of amino-d-luciferin, aminoseleno-d-luciferin, that served as a competent substrate for the firefly luciferase enzyme and exhibited red-shifted bioluminescence emission peaking at 600 nm [48].
Red-shifted luciferase substrates
In recent years, several red-shifted luciferase substrates have been developed, with some of them possessing unique properties. Mofford et al. created synthetic aminoluciferin analogues that increase near-infrared photon flux more than tenfold over that of d-luciferin in luciferase-expressing cells [49]. Furthermore, they found a mutated firefly luciferase that can accept and utilize rigid aminoluciferins with high activity, while exhibiting virtually no light emission with the natural d-luciferin substrate. Jathoul et al. synthesized a dual-color, far-red to near-infrared (nIR)-emitting analogue of beetle luciferin, which, akin to natural luciferin, exhibits pH-dependent fluorescence spectra and emits bioluminescence of different colors with different engineered Fluc enzymes. The analogue produces different far-red to nIR emission maxima up to λ max = 706 nm with different Fluc mutants. This emission is the most red-shifted bioluminescence reported without using a resonance energy transfer acceptor [50]. Steinhardt et al. generated an alkyne-modified d-luciferin that exhibited a red-shifted emission spectrum compared to the parent substrate [51].
AkaLumine-HCl, a luciferin analogue synthesized by Kuchimaru et al., produces bioluminescence in reactions with native firefly luciferase in the near-infrared wavelength ranges (λ max = 677 nm), and significantly increases target-detection sensitivity from deep tissues, as compared with d-luciferin [52, 53]. A series of N-cycloalkylaminoluciferins (cyaLucs) containing lipophilic N-cycloalkylated substitutions were developed by Wu et al. and are effective substrates for native FLuc. Importantly, they can produce elevated bioluminescent signals in vitro in cells, and in vivo, where 0.01% of the standard dose of d-luciferin (dLuc) used in mouse imaging radiated 20-fold more bioluminescent light than d-luciferin (dLuc) or aminoluciferin (aLuc) at the same concentration [54].
Thus, chemical modification of the luciferin substrate together with mutant luciferases has extended the capabilities of bioluminescent reporters by offering high photon flux in the red and NIR spectrum. Since the substrate for firefly luciferase, luciferin, can undergo strenuous chemical modification and still be acted on by the enzyme in its wild-type or mutant form, the generation of targeted substrates suitable for specific conditions and situations can further broaden the scope and range of applications of BLI [55].
Besides firefly and click beetle luciferase, light-emitting enzymes have also been isolated from other species like the sea pansy Renilla reniformis and the crustacean Gaussia princeps that utilize coelenterazine as a substrate. Their reactions do not require ATP and oxygen, but are limited by substrate penetration and light absorption. Thus, unfortunately, they are rarely utilized for in vivo studies [55, 56]. To improve the in vivo imaging performance of Renilla luciferases Loening et al. and Rahnama et al. have engineered variants with a red-shifted spectrum (peak emission of 556 and 540 nm), of which a substantial portion is above 600 nm [57, 58].
GLuc is the smallest luciferase cloned (18 kDa) with several advantages over other commonly used reporters as it is over 2000-fold more sensitive than firefly or Renilla luciferase. GLuc is naturally secreted and, therefore, permits the real-time monitoring of biological processes and reaction kinetics in culture. Luker et al. fused complementary GLuc protein fragments to ligand–receptor pairs to study ligand–receptor binding in vitro and in vivo. Specifically, they quantified the binding of chemokine (C-X-C motif) ligand 12 (CXCL12) to chemokine (C-X-C motif) receptors 4 (CXCR4) and 7 (CXCR7). BLI showed CXCL12–CXCR7 binding in primary and metastatic tumors in a mouse model of breast cancer and enabled them to quantify the drug-mediated inhibition of CXCL12–CXCR4 binding in living mice [59].
NanoLuc is an ATP-independent luciferase enzyme from the deep-sea shrimp Oplophorus gracilirostris that uses furimazine, an analogue of coelenterazine optimized as a substrate for NanoLuc. It is the brightest luciferase and produces approximately two orders of magnitude more bioluminescence than GLuc [60]. Chu et al. engineered CyOFP1, a bright, orange-red FP excitable by cyan light. CyOFP1 serves as an efficient acceptor for resonance energy transfer from the blue-emitting NanoLuc. An optimized fusion of CyOFP1 and NanoLuc, called Antares, produces substantially brighter signals in vitro and in vivo from deep tissues than firefly luciferase and other bioluminescent proteins [61]. Suzuki et al. reported five new spectral variants of a bright luminescent protein, enhanced Nano-lantern (eNL), made by concatenation of NanoLuc with various color variants of fluorescent proteins [62]. eNLs allow five-color live-cell imaging, as well as detection of single protein complexes and even single molecules. An eNL-based Ca2+ sensor can image spontaneous Ca2+ dynamics in cardiomyocytes and neural cell models responding with a fivefold signal change.
Aequorin is a calcium-dependent, blue light-emitting luciferase from the jellyfish (hydrozoan) Aequorea victoria. It utilizes coelenterazine, too, and is expressed together with the green fluorescent protein to produce green light via resonant energy transfer. A red fluorescent protein–aequorin fusion has been developed by Bakayan et al. and can serve as a Ca2+ sensor yielding improved bioluminescence images from single cells and in vivo [63]. Grinstead et al. modified aequorin using coelenterazine analogues and genetic engineering with non-canonical amino acids to shift peak emission from 472 nm for the native enzyme to 526 nm of the most red-shifted variant, thereby increasing the portion of emitted light above 600 nm [64].
BLI has been applied to track cell fate in various preclinical applications such as monitoring the behavior and localization of adipose tissue-derived progenitor cells in a rodent model of myocardial infarction, tracking the viability and distribution of injected mesenchymal stem cells in a partially pancreatectomized mouse, and tracking the activation process of injected stem cells as they graft and differentiate into the neuronal lineage [65–67].
Finally, the multiplexing of bioluminescence imaging is being pursued to increase the number of cell types that can be tracked in vivo. For this, Jones et al. custom-synthesized sterically modified luciferins and by screening libraries of mutant luciferases with them they identified orthogonal enzyme–substrate pairs that specifically interacted and produced light in vitro and in cell culture. The resulting reporter–substrate pairs allow simultaneous multicomponent imaging, e.g., of different cell types labeled with different luciferase mutants and visualized with orthogonal luciferin derivatives, thus enabling the direct interrogation of cell networks in vivo [68]. Alternatively, double- and even triple-luciferase systems, in which enzymes oxidizing different substrates can be multiplexed together in vivo, can further expand the utility of BLI for stem cell and other in vivo research [46, 69].
Reporter genes for fluorescence imaging
Fluorescence imaging (FLI) uses nearly the same principles and equipment as BLI, in which an enzyme acts on a delivered substrate to create photon emission from within, whereas the green and red fluorescent proteins utilized in fluorescence imaging contain an internal fluorophore that needs to be excited by an external light source [39, 70]. The main advantage of fluorescence imaging is that fluorescent molecules can be brighter than their counterpart BLI enzymes since many photons can be generated from a single fluorophore through continuous application of excitation light [71]. The main limitations for fluorescent reporter genes are: (1) the possibility of the fluorescent protein to cause potential toxic effects (due to singlet oxygen generation during protein maturation); (2) the low depth of penetration of light through tissue where both excitation and emitted light are distorted through scattering and non-specific absorption by overlying tissue; and (3) the high autofluorescence of cells and tissue, predominantly in the blue-green part of the light spectrum. Like BLI, light absorbance from hemoglobin is a problem, decreasing signal to noise ratio [2, 45, 72], hence fluorescent protein variants have been developed that emit at longer wavelength, with peak emission wavelength of 592, 635, 646, and 675 nm [73–78]. Rodriguez et al. developed a novel RFP from an allophycocyanin α-subunit (APCα), small ultra-red FP (smURFP), which covalently attaches a biliverdin (BV) chromophore without help from a lyase, and has a 670-nm excitation–emission peak [79]. Shu et al. engineered IFP1.1, a bacterial phytochrome protein from Deinococcus radiodurans that incorporates biliverdin as the chromophore and fluoresces with peak emission at 708 nm that is well expressed in mammalian cells and in mice for whole-body imaging. [80]. The second-generation IFP2.0, has an emission maximum of 711 nm, is significantly brighter when expressed in mammalian cells (HEK293) in the absence of exogenous biliverdin and in cells low in biliverdin, such as neuronal cells, its brightness can be augmented by coexpression of heme oxygenase-1 [81].
An alternative option for in vivo FLI is red and NIR dyes. Xie et al. used a combination of activatable and targeting NIR fluorescent (NIRF) probes to detected luciferase-expressing 4T1-luc2 mouse breast cancer cells in vitro. In vivo, 4T1-luc2 cells orthotopically implanted in nude mice could be followed and tumor progression monitored longitudinally both by BLI and dual-wavelength FLI [82]. Thanks to the development of far-red or NIR proteins and dyes, FLI now offers several advantages over non-optical imaging modalities: low cost and ease of use, relatively high resolution and sensitivity, as well as the ability to detect cells of interest in vivo with intravital two-photon microscopy and ex vivo by flow cytometry and fluorescence microscopy.
Reporter genes for (opto)acoustic imaging
In the emerging field optoacoustics, chromophores in the tissue absorb light and release this energy as acoustic waves that can be imaged using ultrasound detectors. This hybrid imaging technique offers several advantages over strictly optical methods of imaging. The acoustic waves scatter significantly less than photons in tissue, eliminating the depth and spatial restrictions found in strictly optical techniques [83].
Tyrosinase, the enzyme responsible for the synthesis of melanin, has found use as a reporter gene for optoacoustics. Human melanin pigment exhibits high photostability and strong broadband optical absorption, which even though its peak absorption occurs around 335 nm, extends to wavelengths beyond that of hemoglobin [84, 85]. Hence, melanin efficiently absorbs photons generated by the excitation lasers used for optoacoustic imaging. It was recently shown that transgenic expression of tyrosinase enabled longitudinal cell tracking of cancer dissemination at tissue depths up to 10 mm [86].
Other chromophores commonly used for optoacoustics include fluorescent proteins, such as bacteriophytochrome-based near-infrared fluorescent protein (iRFP). It is stable, non-toxic, and capable of producing a signal in cells, tissues, and whole animals at low concentrations [87]. Liu et al. expressed the far-red fluorescent protein (FP) E2-Crimson as a transgene in the exocrine pancreas of adult zebrafish and non-invasively mapped it in 3D in vivo using photoacoustic tomography [88].
The Shapiro group, which uses bacterial-derived gas vesicles as reporters, showed that by engineering genetic variants of the proteins that self-assemble to form these gas vesicles, they were able to generate gas vesicles with different mechanical properties. As such, the different gas vesicles mechanically resisted distinct acoustic pressures, which were used for pressure unmixing to obtain multiplexed images using ultrasound [89].
Multimodal imaging with reporter genes
The various imaging modalities that can be used with reporter genes have their own strengths and weaknesses such as signal generation, clinical relevance, degree of contrast generated, and tissue distribution. To overcome limitations of certain modalities and exploit the strengths of others, multimodal imaging techniques have been developed [26, 27, 90–95]. For example, the DMT1 reporter gene discussed in the MRI reporter gene section has been shown to be useful for tracking neural stem cells in a rat brain using a dual-modality PET and MRI manganese-based imaging approach [91]. Another reporter gene that has been used in multimodal imaging is a triple fusion reporter consisting of firefly luciferase, a red or green fluorescent protein and a herpes simplex virus type 1 thymidine kinase (HSV1 TK), which allows one to track cell fate using bioluminescent, fluorescent, and PET imaging [26, 79, 92, 93]. Another previously mentioned reporter gene, hNIS, can be used for multimodal imaging using PET and Cerenkov Luminescence Imaging. Cerenkov luminescence imaging (CLI) utilizes Cerenkov radiation, a form of electromagnetic radiation emitted when a particle exceeds the speed of light while traveling in a medium. As the moving particle displaces electrons in the medium, they eventually return to their ground state and emit photons that can be detected by a CCD camera [13]. A major limitation of this method is that the wavelength of light-emitted peaks is around 450 nm and the signal emitted is weak requiring long exposure times to obtain a quality image [9], which is not ideal for in vivo experiments.
Promoter selection
Gene expression is regulated by upstream promoter sequences, which contain canonical binding sites for transcription factors and which are often controlled by enhancer elements acting at large distances either up- or downstream [96]. The expression of genes that specify cell type identity and function is associated with densely spaced clusters of active enhancers known as ‘super-enhancers’. Lineage progression involves chromatin remodeling where super-enhancers and their dense clusters (‘epicenters’) of transcription factor-binding sites are switched on or off [97]. Lineage-determining transcription factors (LDTFs) and collaborating transcription factors (CTFs) bind to enhancers prior to signal-dependent activation. Such “primed” enhancers may exhibit basal enhancer activity for promoter binding and are activated by broadly expressed signal-dependent transcription factors (SDTFs) in a cell type-specific manner [98].
The functions of enhancers and super-enhancers are influenced by, and affect, higher order genomic organization [98]. A pivotal role in the control of gene expression is played by cytosine–guanosine dinucleotide (CpG) sites contained in mammalian gene promoters. Methylation of the cytosine residues at CpG sites regulates transcription directly by inhibiting the binding of specific transcription factors, and indirectly by recruiting methyl-CpG-binding proteins that repress chromatin remodeling and transcription. Thus, epigenetic modifications are responsible for the modulation of developmentally regulated and tissue-specific gene expression.
Therefore, the promoter portion for reporter constructs is selected depending on the cell type, the conditions of expression, and the differentiation pathway(s) of interest [99]. The selected promoter sequence, with or without an enhancer sequence, is normally placed at the 5′-region of the reporter gene where it can be switched on in the presence of transcription factors governing cell fate and state leading to the expression of the reporter protein. When choosing promoters, the key factors are achieving the best possible sensitivity, determined by the signal-to-noise ratio (SNR) between basal and induced expression in the cell, and the highest specificity of the promoter, i.e., a high SNR for the cell type of interest vs. off-target cells.
For monitoring cell survival and migration over long periods of time alone, constitutively active promoters are employed that are expressed across broad spectrum of cell types, most commonly (listed in order of expression levels) the CMV enhancer fused to the chicken beta-actin promoter, the core elongation factor 1 alpha promoter, and the human Ubiquitin C promoter. Viral promoters, while producing high transient levels of expression, are to be advised against in long-term tracking studies since they become methylated and silenced over time [100].
A variety of well-characterized promoters are available for monitoring tissue development and cell differentiation [6]. A prominent example is the promoter for pancreatic beta cell-specific expression of insulin located within approximately 400 nucleotides upstream of the transcription start site. It directs both tissue-specific and metabolic-responsive transcription of the insulin gene [101–103]. Differentiation protocols have been developed to generate beta cells from stem or progenitor cells [104, 105]. The resulting cells may only be a first approximation, however, since recently emerging evidence suggests that beta cells are not homogenous but consist of heterogeneous subpopulations exhibiting morphological, functional and gene expression differences [106].
Multiplexing of reporters can be facilitated using bidirectional promoters, short (<1 kbp) intergenetic regions of DNA that regulate expression of two adjacent genes encoded on opposite strands and whose 5′ ends are oriented toward one another. As the two genes are often functionally related, they can be co-regulated by modification of their shared promoter region. Bidirectional promoters are a common feature of mammalian genomes, with ~11% of human genes being bidirectionally paired.
Overcoming limitations of reporter genes
One major complication of using reporter genes in the clinical setting to track therapeutic stem cells is safety concerns associated with gene modification of cells via viral vectors. In the past, reports have been published where physicians attempted to treat two male patients with X-linked severe combined immunodeficiency disease by integrating retroviruses and as a result of the genetic manipulation the patients developed clonal T cell expansion that is seen in acute leukemia [5, 6]. Theoretically, the integration of foreign DNA into a host’s genome could occur at any site and impede the function of an endogenous gene. Seeing how a pseudo-cancer syndrome could arise from such transfections, it might be worthwhile to utilize an alternate technique to drive reporter gene expression in cells that will be transplanted into humans for therapeutic purposes, one that does not integrate into the cells’ genomes and is capable of autonomous replication [8].
Over the years, non-integrating vector systems have been developed, which include human artificial chromosomes, viral vectors with replication origins that require transactivating factors, and scaffold/matrix attachment region (S/MAR) vectors. The main issue with artificial chromosomes is the difficulty in introducing this large piece of DNA into cells, whereas vectors with viral replication origins need viral-based proteins to initiate transformation, which would be difficult to employ in a clinical setting [8].
There are several characteristics of S/MAR vectors that make them promising candidates for being utilized as non-integrative inducers of reporter gene expression in the context of cell fate tracking in a clinical setting. They utilize their host’s genetic machinery to replicate and are thus capable of being passed down from progeny to progeny. Because of their size, they are readily taken up by cells and their unique sequence allows them to remain episomal. Originally, S/MAR vectors contained prokaryotic components; however, studies have shown that they can be removed, resulting in the creation of S/MAR Minicircles (MCs) that do not require antibiotic selection and have a greater tendency to resist integration into the host’s genome [8].
It has been shown that S/MAR MCs can be inserted into cancer cells and injected into animals to track cell fate in vivo over successive generations. In this case, the MC vector was created to express the gene firefly luciferase, which as mentioned previously can be tracked using bioluminescent imaging upon injection of d-luciferin. The viability of the MCs allowed them to produce a bioluminescent-detectable signal for a time greater than 40 days [8].
Despite the numerous advantages it has to offer, this technology is not devoid of limitations. The establishment rate of the vectors within cells is only about 5%, meaning the creation of trackable stem cells for in vivo imaging containing MCs can be difficult. To address this issue, methods have been developed to improve transfection rates, such as histone hyperacetylation. In addition, the S/MAR MC’s ability to remain episomal has a time frame; it has been shown that integration and vector defragmentation occur after prolonged periods of time in culture, with an onset of 3 weeks after transfection and with the majority of the DNA being defragmented or integrated by week 21 [107].
Conclusion
The use of stem cells for therapeutic purposes holds much potential in various fields of medicine. However, despite the growing evidence of the high utility of stem cells, many hurdles need to be overcome before the cells can be used to treat disease. Reporter genes have shown much promise when utilized to track cell fate in vivo in preclinical disease models. Many different probes have been utilized with various imaging modalities, such as MRI, SPECT/PET, and BLI. In addition, multimodal imaging enables the research to combine the strengths of several reporter genes to overcome the weaknesses of a single reporter gene, resulting in methods to track stem cells using whole-body imaging approaches using MRI/SPECT or PET, but also providing high-resolution imaging capabilities down to the cellular level using optical reporters. The selection of appropriate promoter sequences for the insertion of said reporter genes is another factor to take into consideration as it can aid in the efficacy of such constructs in monitoring cell fate. Recent advances in molecular biology have led to the discovery of replicating minicircles, which could help overcome the current issue of potential adverse reactions from the integration of foreign DNA into human cells, paving a viable pathway to clinical translation of the discussed approaches.
Acknowledgements
This study was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748.
References
- 1.Aarntzen EH, Srinivas M, Walczak P, Janowski M, Heerschap A, de Vries IJ, Figdor CG, Bulte JW, Oyen WJ. In vivo tracking techniques for cellular regeneration, replacement, and redirection. J Nucl Med. 2012;53(12):1825–1828. doi: 10.2967/jnumed.112.106146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leahy M, Thompson K, Zafar H, Alexandrov S, Foley M, O’Flatharta C, Dockery P. Functional imaging for regenerative medicine. Stem Cell Res Ther. 2016;7(1):57. doi: 10.1186/s13287-016-0315-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su W, Leng L, Han Z, He Z, Li Z. Bioluminescence imaging of human embryonic stem cell-derived endothelial cells for treatment of myocardial infarction. Methods Mol Biol. 2013;1052:203–215. doi: 10.1007/7651_2013_15. [DOI] [PubMed] [Google Scholar]
- 4.Yu Q, Fan W, Cao F. Mechanistic molecular imaging of cardiac cell therapy for ischemic heart disease. Am J Physiol Heart Circ Physiol. 2013;305(7):H947–H959. doi: 10.1152/ajpheart.00092.2013. [DOI] [PubMed] [Google Scholar]
- 5.Chen IY, Wu JC. Molecular imaging: the key to advancing cardiac stem cell therapy. Trends Cardiovasc Med. 2013;23(6):201–210. doi: 10.1016/j.tcm.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naumova AV, Modo M, Moore A, Murry CE, Frank JA. Clinical imaging in regenerative medicine. Nat Biotechnol. 2014;32(8):804–818. doi: 10.1038/nbt.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nolta JA. New advances in understanding stem cell fate and function. Stem Cells. 2015;33(2):313–315. doi: 10.1002/stem.1905. [DOI] [PubMed] [Google Scholar]
- 8.Ronald JA, Cusso L, Chuang HY, Yan X, Dragulescu-Andrasi A, Gambhir SS. Development and validation of non-integrative, self-limited, and replicating minicircles for safe reporter gene imaging of cell-based therapies. PLoS One. 2013;8(8):e73138. doi: 10.1371/journal.pone.0073138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Jokerst JV. Stem cell imaging: tools to improve cell delivery and viability. Stem Cells Int. 2016;2016:9240652. doi: 10.1155/2016/9240652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cromer Berman SM, Walczak P, Bulte JW. Tracking stem cells using magnetic nanoparticles. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2011;3(4):343–355. doi: 10.1002/wnan.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vande Velde G, Himmelreich U, Neeman M. Reporter gene approaches for mapping cell fate decisions by MRI: promises and pitfalls. Contrast Media Mol Imaging. 2013;8(6):424–431. doi: 10.1002/cmmi.1590. [DOI] [PubMed] [Google Scholar]
- 12.Vandsburger M. Cardiac cell tracking with MRI reporter genes: welcoming a new field. Curr Cardiovasc Imaging Rep. 2014;7:9250. doi: 10.1007/s12410-013-9250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho IK, Moran SP, Paudyal R, Piotrowska-Nitsche K, Cheng PH, Zhang X, Mao H, Chan AW. Longitudinal monitoring of stem cell grafts in vivo using magnetic resonance imaging with inducible maga as a genetic reporter. Theranostics. 2014;4(10):972–989. doi: 10.7150/thno.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung J, Kee K, Barral JK, Dash R, Kosuge H, Wang X, Weissman I, Robbins RC, Nishimura D, Quertermous T, Reijo-Pera RA, Yang PC. In vivo molecular MRI of cell survival and teratoma formation following embryonic stem cell transplantation into the injured murine myocardium. Mag Reson Med. 2011;66(5):1374–1381. doi: 10.1002/mrm.22929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivastava AK, Bulte JW. Seeing stem cells at work in vivo. Stem Cell Rev. 2014;10(1):127–144. doi: 10.1007/s12015-013-9468-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira SM, Moss D, Williams SR, Murray P, Taylor A. Overexpression of the MRI reporter genes ferritin and transferrin receptor affect iron homeostasis and produce limited contrast in mesenchymal stem cells. Int J Mol Sci. 2015;16(7):15481–15496. doi: 10.3390/ijms160715481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur D, Rajagopalan S, Chinta S, Kumar J, Di Monte D, Cherny RA, Andersen JK. Chronic ferritin expression within murine dopaminergic midbrain neurons results in a progressive age-related neurodegeneration. Brain Res. 2007;1140:188–194. doi: 10.1016/j.brainres.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Bartelle BB, Szulc KU, Suero-Abreu GA, Rodriguez JJ, Turnbull DH. Divalent metal transporter, DMT1: a novel MRI reporter protein. Magn Reson Med. 2013;70(3):842–850. doi: 10.1002/mrm.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilad A, McMahon M, Walczak P, Winnard P, Raman V, van Laarhoven H, Skoglund C, Bulte J, van Zijl P. Artificial reporter gene providing MRI contrast based on proton exchange. Nat Biotechnol. 2007;25(2):217–219. doi: 10.1038/nbt1277. [DOI] [PubMed] [Google Scholar]
- 20.Airan RD, Bar-Shir A, Liu G, Pelled G, McMahon MT, Zijl PCMV, Bulte JWM, Gilad AA. MRI biosensor for protein kinase A encoded by a single synthetic gene. Magn Reson Med. 2012;68(6):1919–1923. doi: 10.1002/mrm.24483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bar-Shir A, Liang Y, Chan KW, Gilad AA, Bulte JW. Supercharged green fluorescent proteins as bimodal reporter genes for CEST MRI and optical imaging. Chem Commun (Camb) 2015;51(23):4869–4871. doi: 10.1039/C4CC10195B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bar-Shir A, Liu G, Chan KW, Oskolkov N, Song X, Yadav NN, Walczak P, McMahon MT, van Zijl PC, Bulte JW, Gilad AA. Human protamine-1 as an MRI reporter gene based on chemical exchange. ACS Chem Biol. 2014;9(1):134–138. doi: 10.1021/cb400617q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bar-Shir A, Liu G, Greenberg MM, Bulte JW, Gilad AA. Synthesis of a probe for monitoring HSV1-tk reporter gene expression using chemical exchange saturation transfer MRI. Nat Protoc. 2013;8(12):2380–2391. doi: 10.1038/nprot.2013.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrauto G, Castelli DD, Terreno E, Aime S. In vivo MRI visualization of different cell populations labeled with PARACEST agents. Magn Reson Med. 2013;69(6):1703–1711. doi: 10.1002/mrm.24411. [DOI] [PubMed] [Google Scholar]
- 25.Shapiro MG, Ramirez RM, Sperling LJ, Sun G, Sun J, Pines A, Schaffer DV, Bajaj VS. Genetically encoded reporters for hyperpolarized xenon magnetic resonance imaging. Nat Chem. 2014;6(7):629–634. doi: 10.1038/nchem.1934. [DOI] [PubMed] [Google Scholar]
- 26.Kedziorek DA, Solaiyappan M, Walczak P, Ehtiati T, Fu Y, Bulte JW, Shea SM, Brost A, Wacker FK, Kraitchman DL. Using C-arm X-ray imaging to guide local reporter probe delivery for tracking stem cell engraftment. Theranostics. 2013;3(11):916–926. doi: 10.7150/thno.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfs E, Holvoet B, Gijsbers R, Casteels C, Roberts SJ, Struys T, Maris M, Ibrahimi A, Debyser Z, Van Laere K, Verfaillie CM, Deroose CM. Optimization of multimodal imaging of mesenchymal stem cells using the human sodium iodide symporter for PET and Cerenkov luminescence imaging. PLoS One. 2014;9(4):e94833. doi: 10.1371/journal.pone.0094833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yaghoubi SS, Campbell DO, Radu CG, Czernin J. Positron emission tomography reporter genes and reporter probes: gene and cell therapy applications. Theranostics. 2012;2(4):374–391. doi: 10.7150/thno.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schönitzer V, Haasters F, Käsbauer S, Ulrich V, Mille E, Gildehaus FJ, Carlsen J, Pape M, Beck R, Delker A, Böning G, Mutschler W, Böcker W, Schieker M, Bartenstein P. In vivo mesenchymal stem cell tracking with PET using the dopamine type 2 receptor and 18F-fallypride. J Nucl Med. 2014;55(8):1342–1347. doi: 10.2967/jnumed.113.134775. [DOI] [PubMed] [Google Scholar]
- 30.Muñoz-Álvarez KA, Altomonte J, Laitinen I, Ziegler S, Steiger K, Esposito I, Schmid RM, Ebert O. PET imaging of oncolytic VSV expressing the mutant HSV-1 thymidine kinase transgene in a preclinical HCC rat model. Mol Ther. 2015;23(4):728–736. doi: 10.1038/mt.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallezot JD, Beaver JD, Gunn RN, Nabulsi N, Weinzimmer D, Singhal T, Slifstein M, Fowles K, Ding YS, Huang Y, Laruelle M, Carson RE, Rabiner EA. Affinity and selectivity of [11C]-(+)-PHNO for the D3 and D2 receptors in the rhesus monkey brain in vivo. Synapse. 2012;66(6):489–500. doi: 10.1002/syn.21535. [DOI] [PubMed] [Google Scholar]
- 32.Aung W, Okauchi T, Sato M, Saito T, Nakagawa H, Ishihara H, Ikota N, Suhara T, Anzai K. In-vivo PET imaging of inducible D2R reporter transgene expression using [11C]FLB 457 as reporter probe in living rats. Nucl Med Commun. 2005;26(3):259–268. doi: 10.1097/00006231-200503000-00011. [DOI] [PubMed] [Google Scholar]
- 33.Moroz MA, Zhang H, Lee J, Moroz E, Zurita J, Shenker L, Serganova I, Blasberg R, Ponomarev V. Comparative analysis of T cell imaging with human nuclear reporter genes. J Nucl Med. 2015;56(7):1055–1060. doi: 10.2967/jnumed.115.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doubrovin MM, Doubrovina ES, Zanzonico P, Sadelain M, Larson SM, O’Reilly RJ. In vivo imaging and quantitation of adoptively transferred human antigen-specific T cells transduced to express a human norepinephrine transporter gene. Cancer Res. 2007;67(24):11959–11969. doi: 10.1158/0008-5472.CAN-07-1250. [DOI] [PubMed] [Google Scholar]
- 35.Hu S, Cao W, Lan X, He Y, Lang J, Li C, Hu J, An R, Gao Z, Zhang Y. Comparison of rNIS and hNIS as reporter genes for noninvasive imaging of bone mesenchymal stem cells transplanted into infarcted rat myocardium. Mol Imaging. 2011;10(4):227–237. doi: 10.2310/7290.2010.00051. [DOI] [PubMed] [Google Scholar]
- 36.Micci MA, Boone DR, Parsley MA, Wei J, Patrikeev I, Motamedi M, Hellmich HL. Development of a novel imaging system for cell therapy in the brain. Stem Cell Res Ther. 2015;6:131. doi: 10.1186/s13287-015-0129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acton PD, Zhou R. Imaging reporter genes for cell tracking with PET and SPECT. Q J Nucl Med Mol Imaging. 2005;49(4):349–360. [PubMed] [Google Scholar]
- 38.Contag CH, Contag PR, Mullins JI, Spilman SD, Stevenson DK, Benaron DA. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 1995;18(4):593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- 39.Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- 40.Wilson K, Yu J, Lee A, Wu JC. In vitro and in vivo bioluminescence reporter gene imaging of human embryonic stem cells. J Vis Exp. 2008 doi: 10.3791/740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prescher JA, Contag CH. Guided by the light: visualizing biomolecular processes in living animals with bioluminescence. Curr Opin Chem Biol. 2010;14(1):80–89. doi: 10.1016/j.cbpa.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Liang Y, Walczak P, Bulte JW. Comparison of red-shifted firefly luciferase Ppy RE9 and conventional Luc2 as bioluminescence imaging reporter genes for in vivo imaging of stem cells. J Biomed Opt. 2012;17(1):016004. doi: 10.1117/1.JBO.17.1.016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JB, Urban K, Cochran E, Lee S, Ang A, Rice B, Bata A, Campbell K, Coffee R, Gorodinsky A, Lu Z, Zhou H, Kishimoto TK, Lassota P. Non-invasive detection of a small number of bioluminescent cancer cells in vivo. PLoS One. 2010;5(2):e9364. doi: 10.1371/journal.pone.0009364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J Biomed Opt. 2005;10(4):41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- 45.Rice BW, Contag CH. The importance of being red. Nat Biotechnol. 2009;27(7):624–625. doi: 10.1038/nbt0709-624. [DOI] [PubMed] [Google Scholar]
- 46.Mezzanotte L, Aswendt M, Tennstaedt A, Hoeben R, Hoehn M, Löwik C. Evaluating reporter genes of different luciferases for optimized in vivo bioluminescence imaging of transplanted neural stem cells in the brain. Contrast Media Mol Imaging. 2013;8(6):505–513. doi: 10.1002/cmmi.1549. [DOI] [PubMed] [Google Scholar]
- 47.Rumyantsev KA, Turoverov KK, Verkhusha VV. Near-infrared bioluminescent proteins for two-color multimodal imaging. Sci Rep. 2016;6:36588. doi: 10.1038/srep36588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conley NR, Dragulescu-Andrasi A, Rao J, Moerner WE. A selenium analogue of firefly d-luciferin with red-shifted bioluminescence emission. Angew Chem Int Ed Engl. 2012;51(14):3350–3353. doi: 10.1002/anie.201105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mofford DM, Reddy GR, Miller SC. Aminoluciferins extend firefly luciferase bioluminescence into the near-infrared and can be preferred substrates over d-luciferin. J Am Chem Soc. 2014;136(38):13277–13282. doi: 10.1021/ja505795s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jathoul AP, Grounds H, Anderson JC, Pule MA. A dual-color far-red to near-infrared firefly luciferin analogue designed for multiparametric bioluminescence imaging. Angew Chem Int Ed Engl. 2014;53(48):13059–13063. doi: 10.1002/anie.201405955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinhardt RC, O’Neill JM, Rathbun CM, McCutcheon DC, Paley MA, Prescher JA. Design and synthesis of an alkynyl luciferin analogue for bioluminescence imaging. Chemistry. 2016;22(11):3671–3675. doi: 10.1002/chem.201503944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuchimaru T, Iwano S, Kiyama M, Mitsumata S, Kadonosono T, Niwa H, Maki S, Kizaka-Kondoh S. A luciferin analogue generating near-infrared bioluminescence achieves highly sensitive deep-tissue imaging. Nat Commun. 2016;7:11856. doi: 10.1038/ncomms11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiyama M, Saito R, Iwano S, Obata R, Niwa H, Maki SA. Multicolor bioluminescence obtained using firefly luciferin. Curr Top Med Chem. 2016;16(24):2648–2655. doi: 10.2174/1568026616666160413135055. [DOI] [PubMed] [Google Scholar]
- 54.Wu W, Su J, Tang C, Bai H, Ma Z, Zhang T, Yuan Z, Li Z, Zhou W, Zhang H, Liu Z, Wang Y, Zhou Y, Du L, Gu L, Li M. cybLuc: an effective aminoluciferin derivative for deep bioluminescence imaging. Anal Chem. 2017;89(9):4808–4816. doi: 10.1021/acs.analchem.6b03510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams ST, Miller SC. Beyond D-luciferin: expanding the scope of bioluminescence imaging in vivo. Curr Opin Chem Biol. 2014;21:112–120. doi: 10.1016/j.cbpa.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim JE, Kalimuthu S, Ahn BC. In vivo cell tracking with bioluminescence imaging. Nucl Med Mol Imaging. 2015;49(1):3–10. doi: 10.1007/s13139-014-0309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loening AM, Dragulescu-Andrasi A, Gambhir SS. A red-shifted Renilla luciferase for transient reporter-gene expression. Nat Methods. 2010;7(1):5–6. doi: 10.1038/nmeth0110-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rahnama S, Saffar B, Kahrani ZF, Nazari M, Emamzadeh R. Super RLuc8: a novel engineered Renilla luciferase with a red-shifted spectrum and stable light emission. Enzyme Microb Technol. 2017;96:60–66. doi: 10.1016/j.enzmictec.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 59.Luker KE, Mihalko LA, Schmidt BT, Lewin SA, Ray P, Shcherbo D, Chudakov DM, Luker GD. In vivo imaging of ligand receptor binding with Gaussia luciferase complementation. Nat Med. 2011;18(1):172–177. doi: 10.1038/nm.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stacer AC, Nyati S, Moudgil P, Iyengar R, Luker KE, Rehemtulla A, Luker GD. NanoLuc reporter for dual luciferase imaging in living animals. Mol Imaging. 2013;12(7):1–13. [PMC free article] [PubMed] [Google Scholar]
- 61.Chu J, Oh Y, Sens A, Ataie N, Dana H, Macklin JJ, Laviv T, Welf ES, Dean KM, Zhang F, Kim BB, Tang CT, Hu M, Baird MA, Davidson MW, Kay MA, Fiolka R, Yasuda R, Kim DS, Ng HL, Lin MZ. A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat Biotechnol. 2016;34(7):760–767. doi: 10.1038/nbt.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suzuki K, Kimura T, Shinoda H, Bai G, Daniels MJ, Arai Y, Nakano M, Nagai T. Five colour variants of bright luminescent protein for real-time multicolour bioimaging. Nat Commun. 2016;7:13718. doi: 10.1038/ncomms13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bakayan A, Vaquero CF, Picazo F, Llopis J. Red fluorescent protein-aequorin fusions as improved bioluminescent Ca2+ reporters in single cells and mice. PLoS One. 2011;6(5):e19520. doi: 10.1371/journal.pone.0019520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grinstead KM, Rowe L, Ensor CM, Joel S, Daftarian P, Dikici E, Zingg JM, Daunert S. Red-shifted aequorin variants incorporating non-canonical amino acids: applications in in vivo imaging. PLoS One. 2016;11(7):e0158579. doi: 10.1371/journal.pone.0158579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bagó JR, Soler-Botija C, Casaní L, Aguilar E, Alieva M, Rubio N, Bayes-Genis A, Blanco J. Bioluminescence imaging of cardiomyogenic and vascular differentiation of cardiac and subcutaneous adipose tissue-derived progenitor cells in fibrin patches in a myocardium infarct model. Int J Cardiol. 2013;169(4):288–295. doi: 10.1016/j.ijcard.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 66.Lee S, Youn H, Chung T, Hwang DW, Oh SW, Kang KW, Chung JK, Lee DS. In vivo bioluminescence imaging of transplanted mesenchymal stem cells as a potential source for pancreatic regeneration. Mol Imaging. 2014 doi: 10.2310/7290.2014.00023. [DOI] [PubMed] [Google Scholar]
- 67.Oh HJ, Hwang DW, Youn H, Lee DS. In vivo bioluminescence reporter gene imaging for the activation of neuronal differentiation induced by the neuronal activator neurogenin 1 (Ngn1) in neuronal precursor cells. Eur J Nucl Med Mol Imaging. 2013;40(10):1607–1617. doi: 10.1007/s00259-013-2457-0. [DOI] [PubMed] [Google Scholar]
- 68.Jones KA, Porterfield WB, Rathbun CM, McCutcheon DC, Paley MA, Prescher JA. Orthogonal luciferase-luciferin pairs for bioluminescence imaging. J Am Chem Soc. 2017;139(6):2351–2358. doi: 10.1021/jacs.6b11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maguire CA, Bovenberg MS, Crommentuijn MH, Niers JM, Kerami M, Teng J, Sena-Esteves M, Badr CE, Tannous BA. Triple bioluminescence imaging for in vivo monitoring of cellular processes. Mol Ther Nucleic Acids. 2013;2:e99. doi: 10.1038/mtna.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aswendt M, Adamczak J, Tennstaedt A. A review of novel optical imaging strategies of the stroke pathology and stem cell therapy in stroke. Front Cell Neurosci. 2014;8:226. doi: 10.3389/fncel.2014.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tung JK, Berglund K, Gutekunst CA, Hochgeschwender U, Gross RE. Bioluminescence imaging in live cells and animals. Neurophotonics. 2016;3(2):025001. doi: 10.1117/1.NPh.3.2.025001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Subach FV, Piatkevich KD, Verkhusha VV. Directed molecular evolution to design advanced red fluorescent proteins. Nat Methods. 2011;8(12):1019–1026. doi: 10.1038/nmeth.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bajar BT, Wang ES, Lam AJ, Kim BB, Jacobs CL, Howe ES, Davidson MW, Lin MZ, Chu J. Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci Rep. 2016;6:20889. doi: 10.1038/srep20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4(9):741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 75.Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418(3):567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pandelieva AT, Baran MJ, Calderini GF, McCann JL, Tremblay V, Sarvan S, Davey JA, Couture JF, Chica RA. Brighter red fluorescent proteins by rational design of triple-decker motif. ACS Chem Biol. 2016;11(2):508–517. doi: 10.1021/acschembio.5b00774. [DOI] [PubMed] [Google Scholar]
- 77.Strack RL, Hein B, Bhattacharyya D, Hell SW, Keenan RJ, Glick BS. A rapidly maturing far-red derivative of DsRed-Express2 for whole-cell labeling. Biochemistry. 2009;48(35):8279–8281. doi: 10.1021/bi900870u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Piatkevich KD, Malashkevich VN, Morozova KS, Nemkovich NA, Almo SC, Verkhusha VV. Extended Stokes shift in fluorescent proteins: chromophore-protein interactions in a near-infrared TagRFP675 variant. Sci Rep. 2013;3:1847. doi: 10.1038/srep01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rodriguez EA, Tran GN, Gross LA, Crisp JL, Shu X, Ling JY, Tsien RY. A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat Methods. 2016;13:763–769. doi: 10.1038/nmeth.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324(5928):804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu D, Gustafson WC, Han C, Lafaye C, Noirclerc-Savoye M, Ge WP, Thayer DA, Huang H, Kornberg TB, Royant A, Jan LY, Jan YN, Weiss WA, Shu X. An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat Commun. 2014;5:3626. doi: 10.1038/ncomms4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie BW, Mol IM, Keereweer S, van Beek ER, Que I, Snoeks TJ, Chan A, Kaijzel EL, Löwik CW. Dual-wavelength imaging of tumor progression by activatable and targeting near-infrared fluorescent probes in a bioluminescent breast cancer model. PLoS One. 2012;7(2):e31875. doi: 10.1371/journal.pone.0031875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luke GP, Yeager D, Emelianov SY. Biomedical applications of photoacoustic imaging with exogenous contrast agents. Ann Biomed Eng. 2012;40(2):422–437. doi: 10.1007/s10439-011-0449-4. [DOI] [PubMed] [Google Scholar]
- 84.Kollias N, Baqer AH. Absorption mechanisms of human melanin in the visible, 400–720 nm. J Investig Dermatol. 1987;89(4):384–388. doi: 10.1111/1523-1747.ep12471764. [DOI] [PubMed] [Google Scholar]
- 85.Zonios G, Dimou A, Bassukas I, Galaris D, Tsolakidis A, Kaxiras E. Melanin absorption spectroscopy: new method for noninvasive skin investigation and melanoma detection. J Biomed Opt. 2008;13(1):014017. doi: 10.1117/1.2844710. [DOI] [PubMed] [Google Scholar]
- 86.Jathoul AP, Laufer J, Ogunlade O, Treeby B, Cox B, Zhang E, Johnson P, Rizzey AR, Philip B, Marafioti T, Lythgoe MF, Pedley RB, Pule MA, Beard P. Deep in vivo photoacoustic imaging of mammalian tissues using a tyrosinase-based genetic reporter. Nat Photonics. 2015;9:239–246. [Google Scholar]
- 87.Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29(8):757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu M, Schmitner N, Sandrian MG, Zabihian B, Hermann B, Salvenmoser W, Meyer D, Drexler W. In vivo three dimensional dual wavelength photoacoustic tomography imaging of the far red fluorescent protein E2-Crimson expressed in adult zebrafish. Biomed Opt Express. 2013;4(10):1846–1855. doi: 10.1364/BOE.4.001846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lakshmanan A, Farhadi A, Nety SP, Lee-Gosselin A, Bourdeau RW, Maresca D, Shapiro MG. Molecular engineering of acoustic protein nanostructures. ACS Nano. 2016;10(8):7314–7322. doi: 10.1021/acsnano.6b03364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ray P, Tsien R, Gambhir SS. Construction and validation of improved triple fusion reporter gene vectors for molecular imaging of living subjects. Cancer Res. 2007;67(7):3085–3093. doi: 10.1158/0008-5472.CAN-06-2402. [DOI] [PubMed] [Google Scholar]
- 91.Lewis CM, Graves SA, Hernandez R, Valdovinos HF, Barnhart TE, Cai W, Meyerand ME, Nickles RJ, Suzuki M. 52Mn production for PET/MRI tracking of human stem cells expressing divalent metal transporter 1 (DMT1) Theranostics. 2015;5(3):227–239. doi: 10.7150/thno.10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Love Z, Wang F, Dennis J, Awadallah A, Salem N, Lin Y, Weisenberger A, Majewski S, Gerson S, Lee Z. Imaging of mesenchymal stem cell transplant by bioluminescence and PET. J Nucl Med. 2007;48(12):2011–2020. doi: 10.2967/jnumed.107.043166. [DOI] [PubMed] [Google Scholar]
- 93.Pei Z, Lan X, Cheng Z, Qin C, Xia X, Yuan H, Ding Z, Zhang Y. Multimodality molecular imaging to monitor transplanted stem cells for the treatment of ischemic heart disease. PLoS One. 2014;9(3):e90543. doi: 10.1371/journal.pone.0090543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kircher MF, Gambhir SS, Grimm J. Noninvasive cell-tracking methods. Nat Rev Clin Oncol. 2011;8(11):677–688. doi: 10.1038/nrclinonc.2011.141. [DOI] [PubMed] [Google Scholar]
- 95.Mukherjee A, Wu D, Davis HC, Shapiro MG. A genetically encoded reporter for diffusion weighted magnetic resonance imaging. BioRxiv. 2016 [Google Scholar]
- 96.Yao L, Berman BP, Farnham PJ. Demystifying the secret mission of enhancers: linking distal regulatory elements to target genes. Crit Rev Biochem Mol Biol. 2015;50(6):550–573. doi: 10.3109/10409238.2015.1087961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, Fuchs E. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521(7552):366–370. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16(3):144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cherry AB, Daley GQ. Reprogrammed cells for disease modeling and regenerative medicine. Annu Rev Med. 2013;64:277–290. doi: 10.1146/annurev-med-050311-163324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garrison BS, Yant SR, Mikkelsen JG, Kay MA. Postintegrative gene silencing within the Sleeping Beauty transposition system. Mol Cell Biol. 2007;27(24):8824–8833. doi: 10.1128/MCB.00498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ohlsson H, Karlsson O, Edlund T. A beta-cell-specific protein binds to the two major regulatory sequences of the insulin gene enhancer. Proc Natl Acad Sci USA. 1988;85(12):4228–4231. doi: 10.1073/pnas.85.12.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hanahan D. Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985;315(6015):115–122. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- 103.Kuroda A, Rauch TA, Todorov I, Ku HT, Al-Abdullah IH, Kandeel F, Mullen Y, Pfeifer GP, Ferreri K. Insulin gene expression is regulated by DNA methylation. PLoS One. 2009;4(9):e6953. doi: 10.1371/journal.pone.0006953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pagliuca FW, Millman JR, Gürtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159(2):428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ameri J, Borup R, Prawiro C, Ramond C, Schachter KA, Scharfmann R, Semb H. Efficient generation of glucose-responsive beta cells from isolated GP2(+) human pancreatic progenitors. Cell Rep. 2017;19(1):36–49. doi: 10.1016/j.celrep.2017.03.032. [DOI] [PubMed] [Google Scholar]
- 106.Gutierrez GD, Gromada J, Sussel L. Heterogeneity of the pancreatic beta cell. Front Genet. 2017;8:22. doi: 10.3389/fgene.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Broll S, Oumard A, Hahn K, Schambach A, Bode J. Minicircle performance depending on S/MAR-nuclear matrix interactions. J Mol Biol. 2010;395(5):950–965. doi: 10.1016/j.jmb.2009.11.066. [DOI] [PubMed] [Google Scholar]