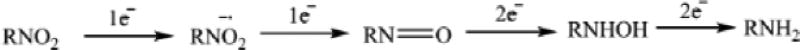Abstract
The existence of a substrate-sensitive equilibrium between high spin (S=5/2) and low spin (S=1/2) ferric iron is a well-established phenomenon in the cytochrome P450 (CYP) superfamily, although its origins are still a subject of discussion. A series of mutations that strongly perturb the spin state equilibrium in the camphor hydroxylase CYP101A1 were recently described (Colthart et al., Sci. Rep. 6, 22035 (2016)). Wild type CYP101A1 as well as some CYP101A1 mutants are herein shown to be capable of catalyzing the reduction of nitroacetophenones by NADH to the corresponding anilino compounds (nitroreductase or NRase activity). The distinguishing characteristic between those mutants that catalyze the reduction and those that cannot appears to be the extent to which residual high spin form exists in the absence of the native substrate d-camphor, with those showing the largest spin state shifts upon camphor binding also exhibiting NRase activity. Optical and EPR spectroscopy was used to further examine these phenomena. These results suggest that reduction of nitroaromatics may provide a useful probe of residual high spin states in the CYP superfamily.
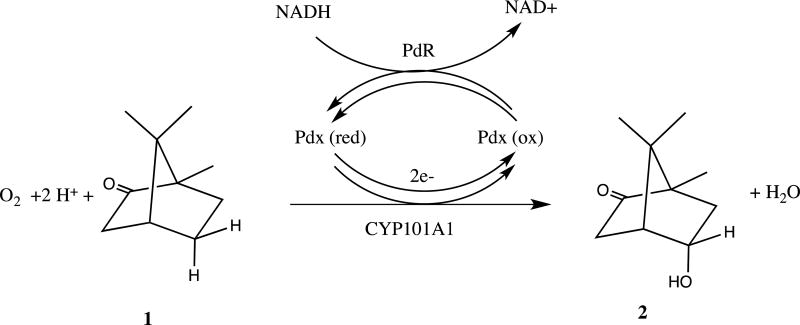
Cytochrome P450cam (CYP101A1) is a heme-containing monooxygenase that catalyzes the stereospecific 5-exo hydroxylation of the substrate camphor by molecular oxygen, O2 (Scheme I). The overall reaction involves insertion of one oxygen atom into the 5-exo C-H bond of camphor 1 to form 5-exo-hydroxycamphor 2, with concomitant reduction of the other oxygen atom to water. The oxygen reduction requires two electrons, provided by the iron-sulfur protein putidaredoxin (Pdx) via two separate one-electron shuttles from the flavoenzyme putidaredoxin reductase (PdR). PdR catalyzes the two-electron oxidation of NADH, the ultimate source of reducing equivalents in the P450 reaction cycle.
Scheme I.
Reaction stoichiometry of the P450cam CYP101A1 hydroxylase.
CYP101A1 has long provided the paradigm for understanding the mechanisms of P450 catalysis. Landmark publications in the 1970s identified a substrate-induced spin state shift for the heme iron that is required for efficient transfer of the first electron from Pdx to CYP101A1 [1, 2]. At ambient temperatures, the heme iron in substrate-free CYP101A1 exists predominantly in the low-spin state (Fe3+, S = 1/2, λmax = 418 nm), but upon binding of the native substrate d-camphor 1 (Scheme I), the equilibrium shifts to > 95% high-spin state (S = 5/2, λmax = 391 nm. The spin state change causes an upshift in reduction potential for the heme iron from −303 mV for substrate-free to −173 mV for substrate-bound CYP101A1, poising the electron transfer from reduced Pdx (reduction potential = −240 mV) to be nearly isoenergetic (Keq ~ 2.3), thereby lowering the activation barrier for optimal electron transfer rates [1]. The spin state change thus acts as an “on-off switch, essentially requiring substrate to be bound in the P450 active site before the first electron transfer occurs, thereby making unproductive oxygen activation that could lead to peroxide or superoxide formation thermodynamically unfavorable. Furthermore, there was an apparent relationship between the extent of the spin state shift and the “appropriateness” of the bound substrate. The native substrate of CYP101A1, d-camphor 1, induces the greatest spin state change, while non-native substrates induce stabilization of the high-spin ferric state to a lesser extent. For example, upon binding norcamphor 3 produces ~ 40% high-spin state, while adamantanone 4 induces ~90% high-spin state.
However, as more P450 enzymes are characterized, it has become clear that CYP101A1 is somewhat of an outlier in terms of tight coupling between the spin state equilibrium, substrate binding and/or reactivity towards a particular substrate. Many P450 enzymes are capable of efficient substrate turnover in the absence of large substrate-dependent induced spin shifts, suggesting that only the presence of the high-spin state (and not the extent of the equilibrium) is critical. Sigmen et al. noted that substrate deoxycorticosterone binding to CYP106A2 is not detectable by UV-visible spectroscopy, but evidenced by perturbations of heme-connected infrared absorbances [3]. The presence of an isosbestic point at 406 nm for the spin state changes indicates that a two-state equilibrium is operant in all cases [4]. The link, however, between coupling efficiency towards a given substrate (that is, substrate turnover relative to consumption of reducing equivalents) and spin state is not always straightforward, even for the case of CYP101A1.
Indeed, Wong et al. noted that while α-pinene 5 binding to WT CYP101A1 results in ~85% high spin, the observed coupling efficiency in turnover assays was only 23%. Also, mutations that induced a lower fraction of the high spin state upon binding of 5 were in some cases more efficient towards substrate turnover [5]. We also recently described a series of mutations in CYP101A1, all of which affected substrate specificity, turnover rates and spin equilibrium to different extents. However, again, we could not draw a significant correlation between enzyme efficiency, substrate specificity and spin equilibrium perturbations [6].
Nevertheless, for efficient electron transfer from physiological redox partners, the presence of a detectable high-spin P450 population would seem to be required. The high-spin ferric state required for the catalytic cycle would be replenished as reduction occurs by Le Chatelier’s principle, thereby maintaining catalytic turnover. In other words, simply the presence of a high-spin form, rather than the extent to which it is stabilized at equilibrium, would be the critical factor for substrate turnover.
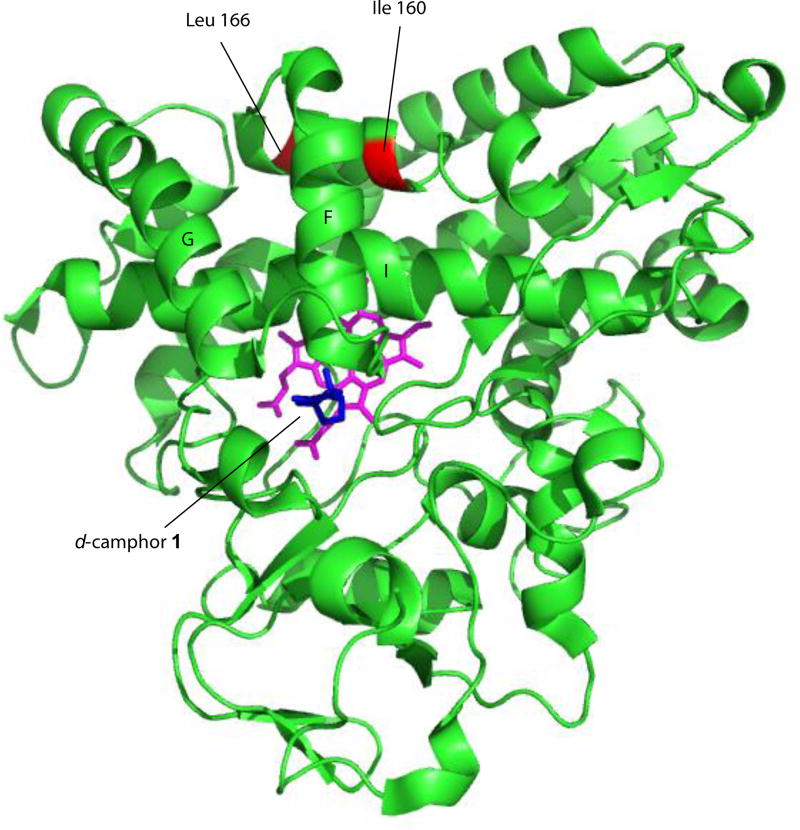
We have recently found that wild-type (WT) and selected mutant CYP101A1 enzymes can catalyze an unusual reaction, the reduction of nitroacetophenones to the corresponding anilinoacetophenones by NADH. Nitroreductase (NRase) activity by a P450 enzyme is not unprecedented, with NAD(P)H-supported NRase activity towards nitrobenzene reported for rat liver microsomal P450s [7, 8]. Those studies also reported that nitrobenzene and nitrosobenzene strongly inhibit O-demethylation reactions typically catalyzed by microsomal P450s (in particular, CYP2D6 [9]), indicating a direct interaction between the P450 and the nitroaromatic compound. Interestingly, P450nor (nitric oxide reductase), which catalyzes the NADH-supported reduction of NO to N2O, is isostructural with the P450 superfamily, and directly binds NADH in the cleft between the F and G helices (see Figure 1 for the location of these features) [10].
Figure 1.
Solution structure of CYP101A1 (PDB entry 2L8M, ref. [13]) with positions of mutations described in text identified, along with secondary structural features discussed in the text. Heme is shown in magenta, and substrate camphor in the active site in blue. Structural features are labeled according to Poulos et al. [24].
In our hands, the NRase reaction proceeds readily for WT and mutant CYP101A1 enzymes that exhibit a significant fraction (>50%) of the high-spin ferric form in the presence of d-camphor 1. Those variants that exhibit smaller (or undetectable) camphor-induced spin state shifts do not catalyze the reduction to any detectable extent. These results suggested to us that the ability of the WT or variant P450s to catalyze the reduction of nitroacetophenones by NADH is contingent on the presence of some residual high-spin ferric state in the absence of native substrate. Because the high-spin form is required for efficient electron transfer from Pdx, NRase activity could thus be valuable for directly probing the presence of residual high-spin forms in P450 enzymes.
Results
Identification of NRase activity in CYP101A1
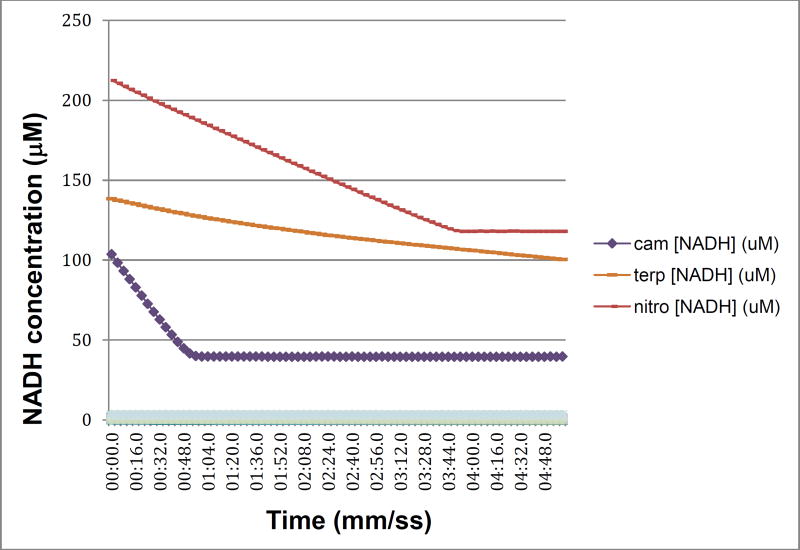
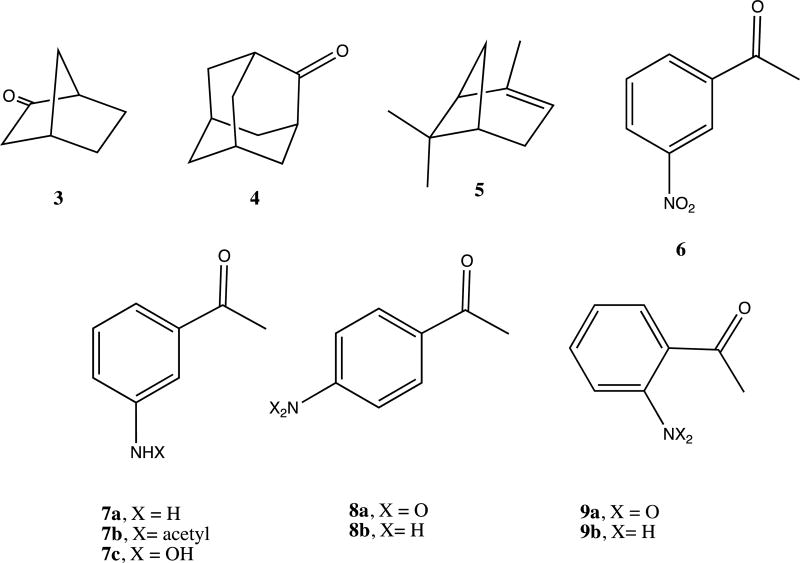
In a recent publication, we described a series of mutations in CYP101A1 that impact a variety of enzymatic and spectroscopic parameters, including turnover rates, substrate selectivity, coupling efficiency, and the position of the substrate-induced spin state equilibrium [6]. The sites for mutation were chosen based on conformational and dynamic perturbations that occur upon substrate binding as detected by multidimensional nuclear magnetic resonance (NMR) [6]. In particular, substitutions at Ile 160 and Leu 166 on the E helix showed significant perturbations in substrate-dependent spin state equilibria (Figure 1). As part of that work, we screened the WT and variant CYP101A1s against a variety of substrates testing for modified substrate selectivity and catalytic function. Catalysis by WT and mutant CYP101A1s was monitored by following the time-dependent decay of the characteristic λmax = 340 nm absorption of NADH in the reconstituted CYP101A1 activity assay including the Pdx/PdR redox relay. Product mixtures were then analyzed by GC/MS for determining coupling efficiencies and product distributions. Screened compounds included a variety of standard terpene and terpenoid compounds 1, 3 and 4 [6] as well as m-nitroacetophenone 6. The WT and some CYP101A1 variants exhibited activity towards 6, with the WT CYP101A1 showing NADH consumption rates that exceeded that of any other substrate except for the native substrate 1 (Figure 2). The results of the terpene oxidations were described in the previous work, but the activity towards 6 was not, as we did not have an explanation for the observations.
Figure 2.
NADH consumption as a function of time for WT CYP101A1/Pdx/PdR in the presence of native substrate camphor 1 (cam, purple diamonds), poor substrate α-terpineol (terp- orange dots) and m-nitroacetophenone 6 (nitro- red dots). Assay conditions are as described in Materials and Methods. X-axis is time in min:sec.0, y-axis is concentration of NADH calculated from absorbance at λ= 340 nm. The offset of the nitroacetophenone trace is due to the significant absorbance shoulder of product 7a at 340 nm.
We originally hypothesized that hydroxylation of the aromatic ring of 6 was occurring, which would introduce a relatively acidic phenolic OH group. We thus extracted the assay solution with aqueous base, and indeed observed a yellow color in the assay solution upon basification, which is typical of nitrophenolate anions. This color change was not observed in assays with CYP101A1 variants that failed to exhibit NADH consumption in the presence of 6, strongly suggesting that the color observed in our assays was indeed due to a product of a catalytic process. The enzymatic reaction was then run on a sufficiently large scale so that product could be isolated and characterized, and the reaction mixture extracted with methylene chloride. The crude extract was further purified by flash chromatography over activated silica using a hexane:ethyl acetate eluent, and both the crude extract and the purified material were analyzed by GC/MS and 1H NMR.
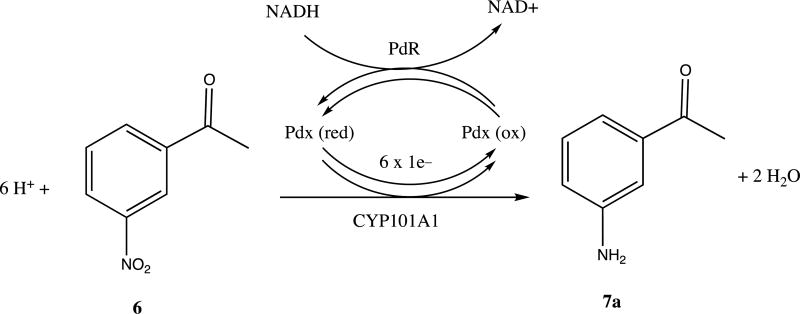
To our surprise, neither the crude extract nor the chromatographically purified material gave parent ion expected for a singly hydroxylated nitroacetophenone (m/z = 181). Furthermore, the apparent M+ ion (m/z = 135) from the crude extract was different from that obtained after chromatography (m/z = 177), with the increase in mass being consistent with that of the addition of an acetyl group. Mass spectral library matching identified the pre-column product as m-aminoacetophenone 7a and the post-column product as m-(acetylamino)acetophenone 7b (see Supplementary Information for details). Apparently, in the presence of ethyl acetate the activated silica gel column catalyzes the acetylation of the aniline amino group. In any case, our results are thus consistent with the idea that CYP101A1 acts as a reductase, catalyzing the six-electron reduction of the aromatic nitro group to the aniline with the overall stoichiometry shown in Scheme III. The origin of the yellow color upon basification is still unclear, but may be the result of the presence of a hydroxylamine intermediate in the reduction, 7c, which has an expected pKa of 8.66 [7, 11]. Scheme IV illustrates a plausible pathway for the reduction that includes nitroso and hydroxylamine intermediates. If a hydroxlamine intermediate is responsible for the yellow color, this implies that the intermediates can be released from the active site. It was pointed out by a reviewer of the original version of this manuscript that under basic conditions, condensation reactions might occur involving products of the reduction that could also impart a yellow color to the workup.
Scheme III.
Reaction stoichiometry of NRase activity of CYP101A1.
Scheme IV.
Possible nitroso and hydroxylamine intermediates in nitroaromatic reduction by CYP101A1.
The reduction of 6 is observed only when all the components of the standard assay (CYP101A1, Pdx or PdR) are included, indicating that the complete electron transport system, as well as the P450, are required in the present case (Scheme III).
The reductive capabilities of the WT and mutant CYP101A1 enzymes toward m-nitroacetophenone, along with camphor-induced spin state equilibria, are summarized in Table I. As can be seen, only those mutants (and WT) that display appreciable accumulation (>50%) of the high-spin ferric state at room temperature in the presence of d-camphor 1 catalyze the reduction of 6. The assay was also carried out using p- and o-nitroacetophenones 8a and 9a, with both of these isomers being efficiently reduced under the same conditions by the same mutant/wild-type set of enzymes, confirming that the reduction is not specific for the meta isomer. The product of the o-nitroacetophenone reduction, 2-aminoacetophenone 9b, is found in red wine and is used as an artificial grape oderant, and when isolated from our reaction mixture by thin layer chromatography exhibits a strong grape odor. Mass spectra of this compound after purification did not yield the expected parent ion at m/z = 135, but rather a peak at m/z = 133 (loss of H2) consistent with a facile gas phase cyclization of the radical cation to the hydroxyindole.
Table I.
Summary of high-spin (HS) ferric heme iron detected by optical spectroscopy and NRase activity towards m-nitroacetophenone 6 for WT and mutant CYP101A1 enzymes. <5%, (n.d., not detected), indicates that a shoulder at 390 nm could not be distinguished in optical spectra in the presence of d-camphor 1. All six enzymes catalyze the hydroxylation of 1 to 2, although with differences in rate and efficiency (see ref. [6] for details).
| CYP101A1 | % high-spin w/ camphor 1 bound |
NRase activity |
|---|---|---|
| WT | >95% | Y |
| I160L | ~35% | N |
| I160L/L166A | ~80% | Y |
| I160L/L166F | <5% (n.d.) | N |
| I160L/L166T | ~75% | Y |
| I160L/L166V | ~50% | Y |
Optical spectroscopy and activity assays on WT and variant CYP101A1s
Ile 160 (Cα-Fe distance = 14.9 Å) and Leu 166 (Cα-Fe distance = 15.9 Å) are located on the E helix distal to the I helix that borders the active site (Fig. 1). Despite their relative remoteness from the heme, substitutions at these positions resulted in significant displacement of the position of equilibrium for the substrate-induced spin equilibrium from the wild-type enzyme (> 95% high-spin state with substrate 1 bound). The optical spectra of these variants in the presence of d-camphor are shown in Figure 3, in which it is also indicated whether the variant could successfully reduce nitrobenzophenones. The low-spin state is observed spectroscopically at ~418 nm, and the high-spin form at ~390 nm with an isosbestic point at 406 nm. Despite the absence of a large spin state change upon camphor binding, most of the mutants could turn over camphor with good efficiency (i.e., little waste of reducing equivalents in the production of reactive oxygen species) although initial velocities of NADH consumption were slower in most variants when compared to that of the WT enzyme [6].
Figure 3.
Optical spectra of d-camphor (1)-bound WT CYP101A (top), I160L/L166A (second down), I160L (third down) and I160L/L166F. Horizontal axes are wavelengths (nm), and vertical axes are absorptions. The top two (WT and I160L/L166A) both reduce nitrobenzophenone 6 to 7a (+NRase) and have a significant high-spin component (λmax= 390 nm) with camphor bound. The bottom two exhibit little or no high-spin state with camphor bound (low-spin (λmax = 390 nm) and neither show reductase activity towards 6 (-NRase).
We suspected that in the cases in which the variants turned over 1 efficiently without a large substrate-induced spin state shift, binding of 6 either induced a small spin state shift not readily detectable by optical means, or that there was sufficient residual high-spin ferric state in the absence of substrate to allow for efficient electron transfer from Pdx. If this were the case, then the ability of WT and some mutant P450s to reduce 6 should be contingent on the presence of residual high-spin state, either in the absence of 1 or in the presence of a non-specific substrate such as 6.
EPR characterization of the WT and I160L CYP101A1
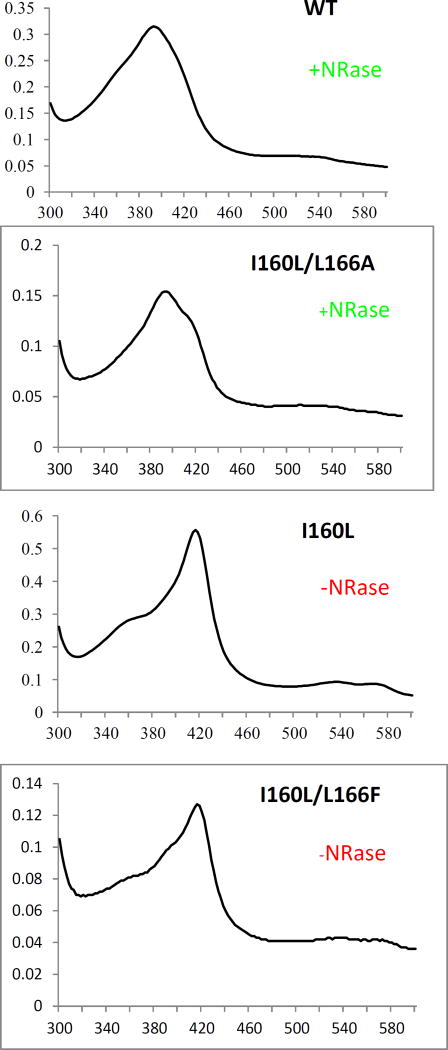
The resolution of the UV/VIS spectra does not allow for an unambiguous determination of small amounts of residual high-spin ferric state, because of significant band overlap. To overcome this limitation, we performed EPR spectroscopy on samples of the WT CYP101A1 (which catalyzes the reduction), and the I160L variant (which does not). The EPR spectrum of substrate-free WT CYP101A1 recorded at 10 K (Figure 4) shows an appreciable amount of high-spin Fe3+ state (S = 5/2) as well as low-spin Fe3+ state (S = 1/2). The high-spin ferric signal is characterized by three effective g-values: 7.74, 3.96, and 1.75, which are similar to the values reported previously, and are consistent with zero-field parameters of E/D = 0.088 and D = 4 cm−1. The signal corresponding to the low-spin form in the absence of a substrate analog has effective g-values of 2.44, 2.25 and 1.89. The ratios of the high-spin (HS) and low-spin (LS) configurations were determined by comparing the numerical simulations of their respective signals, and in the case of the WT these were 1.4:1 HS:LS. Upon addition of d-camphor, the equilibrium shifts largely to the high-spin state configuration, with their ratio now being 4:1, whereas upon addition of 6, the ratio hardly changes. These results are collected in Table 2, confirming that the WT enzyme contains a considerable fraction of high spin Fe(III) at 10 K both in the absence of substrate 1 and in the presence of m-nitroacetophenone 6 (Figure 4). We also note that excess of 6 appears to suppress the high spin form (Figure 4). This suggests that any residual high-spin present in 6-bound CYP101A1 is intrinsic, and not induced by the binding of 6. Interestingly, the spectrum of the low-spin component is not perturbed by addition of 6 in either the WT or I160L enzymes, in contrast to the binding of 1, which does perturb the low-spin spectrum (see Table II and Supplementary Information).
Figure 4.
CW EPR spectra of WT CYP101A1 under different conditions. (A) 337 µM substrate free WT CYP101A1 (red trace “as isolated”), (B) 327 µM of WT CYP101A1 with 2 mM d-camphor 1 (blue trace), bottom, (C) 331 µM of WT CYP101A1 with 2 mM nitroacetophenone 6 (black trace), (D) 320 µM of WT CYP101A1 with 6 mM nitroacetophenone 6 (green trace). Experimental conditions: temperature 10 K, microwave power = 2 mW, microwave frequency = 9.384 GHz, modulation amplitude = 1 mT.
Table II.
Principal components of the g-tensor for the low-spin forms of CYP101A1. Related EPR spectra can be seen in the Supplementary Information.
| Species | g1 | g2 | g3 |
|---|---|---|---|
| WT CYP101A1 | 2.44 | 2.25 | 1.89 |
| WT CYP101A1+camphor 1 | 2.46 (2.41) | 2.25 | 1.89 |
| WT CYP101A1+ 6 | 2.44 | 2.25 | 1.90 |
| I160L CYP101A1 | 2.45 | 2.25 | 1.91 |
| I160L CYP101A1 + 6 | 2.45 | 2.25 | 1.91 |
| 2.49 (shoulder) | 2.25 | 1.89 (shoulder) |
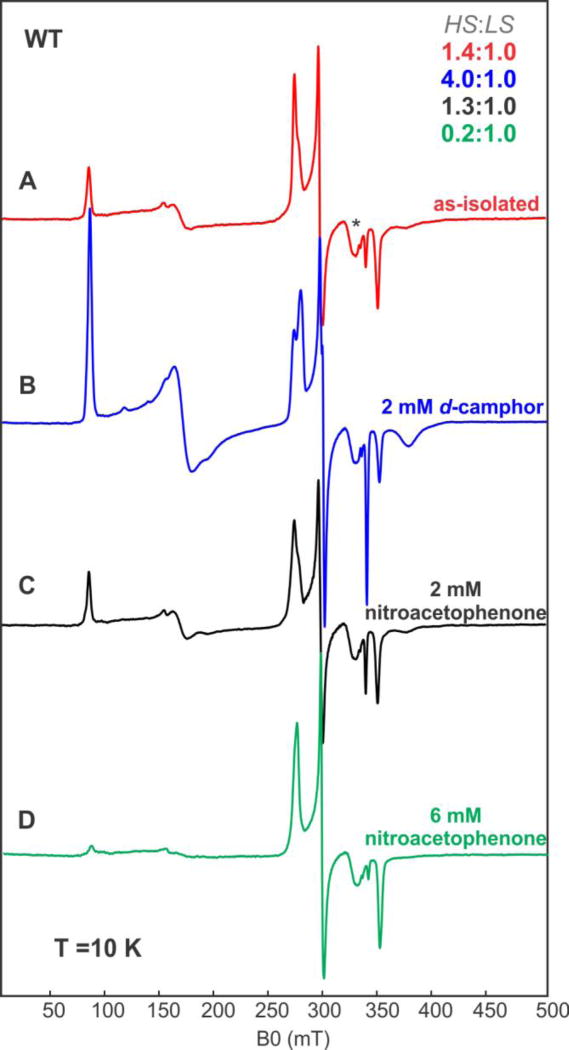
In contrast to the WT, the I160L variant was unable to carry out the catalytic reduction of 6, and, based on optical spectroscopy, the I160L mutant does not accumulate > 35% high-spin ferric form with 1 bound. The EPR spectrum of the I160L variant in the absence of 1 lacks any detectable EPR signals characteristic of the high-spin configuration (Figure 5). Nor does addition of 6 result in a detectable spin state change, with the low-spin state being the sole detectable EPR-active component (Figure 5). These results are thus consistent with the hypothesis that the inability of I160L to exhibit NRase activity towards 6 is due to the absence of significant residual high-spin ferric state.
Figure 5.
CW EPR spectra of I160L CYP101A1 under different conditions. (A) 300 µM of substrate-free I160L CYP101A1 (B) 300 µM uM of I160L CYP101A1 with 2 mM m-nitroacetophenone 6. Experimental conditions: temperature 10 K, microwave power = 2 mW, microwave frequency = 9.625 GHz, modulation amplitude = 1 mT.
Discussion
We are intrigued by the role that remote (second- and third-sphere) residues and secondary structural features play in substrate recognition, binding and spin state shifts in cytochromes P450. There are now close to one million non-redundant putative P450 sequences in various databases as the result of world-wide high-throughput genome sequencing efforts, and while doubtless many of these enzymes perform identical functions in different organisms, this number nevertheless represents a vast number of potential substrates for P450 enzymes, with each substrate presenting multiple possibilities for regio- and stereoselective oxidations. Yet despite the plethora of possible sequence-substrate-product combinations, the P450 fold is highly conserved. Every structurally characterized P450 exhibits an identical folding topology unique to the superfamily that is optimized for the safe activation of molecular oxygen [12]. We have used solution structural and dynamic nuclear magnetic resonance (NMR) methods to identify those structural features that are perturbed upon substrate binding, and in turn might be expected to exert influence on substrate selection and orientation in the active site [13, 14]. In the case of CYP101A1, we tested the feasibility of this approach by mutating residues in that enzyme that NMR results suggested are most likely to be involved in these functions. These results are detailed elsewhere [6], but it suffices to say that every mutation made as a result of our NMR observations perturbed some aspect of enzyme function, either substrate selectivity, efficiency (coupling) or spin state equilibrium, and in some cases all three.
The spin state equilibrium is a characteristic of the P450 superfamily that is instantly identifiable: While not all substrates induce a large or obvious spin state shift even in P450 enzymes that exhibit activity towards them, if the shift is observed, it varies little from enzyme to enzyme. Despite its ubiquity, the origin of the spin equilibrium is still a subject of discussion. The low-spin form is usually rationalized by the presence of a sixth ligand to the heme iron (water or hydroxide) in the active site that drives the 10Dq splitting of the iron 3d orbitals so as to favor the low-spin form (S = 1/2) [15]. Conversely, the absence of the axial ligand favors the high-spin state (S = 5/2). However, Poulos noted that the crystallographic structure of norcamphor 3-bound CYP101A1 has a high-occupancy water ligand present, yet the complex accumulates ~40% high-spin Fe3+ state in solution [16]. Furthermore, both water/OH− and thiolate (the axial Fe ligand on the proximal heme face originating from a conserved Cys residue) are classically considered “soft” ligands, that is, they are too polarizable to drive the formation of the low-spin form. Indeed, the heme Fe in met-aquo-myoglobin is almost completely in the high-spin configuration, yet has water and a “hard” imidazole nitrogen as axial ligands [17].
In the present study, we observed spin equilibrium perturbations that were particularly intriguing. Residue substitutions at the E helix positions of Ile 160 and Leu 166, both remote from the active site and changing little other than conformational flexibility of the I helix, are shown to have a large effect upon the spin state equilibrium in CYP101A1. Given that the I helix and substrate are the primary heme porphyrin contacts in the P450 active site, it may be that the spin equilibrium is as much a reflection of heme conformation as it is of the presence or absence of an axial water ligand. Indeed, the two may be interrelated, with the porphyrin conformation determining whether a water molecule in the active site is within bonding distance of the heme iron on not. Extensive DFT and NMR characterization of heme 13C chemical shifts showed conclusively that out-of-plane distortions affect redox potentials and efficiency of electron transfer in cytochrome c [18]. Champion and Sligar have reported that camphor binding to CYP101A1 results in activation of a heme vibrational mode associated with “doming”, that is, out-of-plane motion of the equatorial heme pyrrole nitrogen Fe ligands that is absent or weak in the substrate-free form. In the substrate-free form, instead there is an active “saddling” mode that would exclude the doming mode [19]. A study of the binding of a series of homologous tertiary amines in CYP2B4 showed conclusively that substrate size and shape have a significant effect on the position of the spin state equilibrium [20].
The apparent sensitivity of the NRase activity of CYP101A1 to whether or not significant high-spin state accumulates in the presence of d-camphor may be useful in further investigations in this area. It is evident that the spin equilibrium is sensitive to relatively small changes in the conformational equilibria associated with substrate binding in CYP101A1. If the equilibrium is shifted by mutation towards those conformations favoring the low-spin state to the extent that binding of 1 only moves the equilibrium slightly to the right, this could be sufficient for the electron transfer and subsequent turnover of 1 observed with all of the mutants discussed here and elsewhere [6]. However, based on our EPR results, 6 does not shift the conformational equilibrium (at least at low temperatures), and even appears to suppress high-spin formation at higher concentrations (Figure 5). In this case, only if there is some residual high-spin form in the absence of 1 will reductase activity be seen. As such, nitroaromatic reduction by cytochromes P450 may be a sensitive probe for the existence of residual high-spin in these enzymes.
Experimental
Site directed mutagenesis, protein expression and purification
All mutations were made in the background of the C334A mutant of CYP101A1 (hereafter referred to as wild type, or WT). The C334A mutant has been found to be enzymatically and functionally identical to the wild-type enzyme, but does not dimerize at high concentrations [21]. Mutations were made via the QuikChange protocol (Stratagene) using the pDNC334A plasmid as template, with appropriate mutagenic oligonucleotides for primers (Eurofins MWG Operon, Huntsville, AL). After amplification, the original template was digested with DpnI, and the mutated plasmids transformed into XL1Blue E. coli for mini-prep of plasmid DNA. The presence of the appropriate mutation was confirmed by DNA sequencing (GeneWiz, South Plainfield, NJ). Plasmids were transformed into E coli strain NCM533 for overexpression. WT and mutant CYP101A1 enzymes, putidaredoxin (Pdx) and putidaredoxin reductase (PdR) were expressed and purified as described previously [22, 23]. Substrate d-camphor 1 was present at 2 mM in the purification buffers in all cases for stabilization. In order to remove d-camphor thoroughly prior to spectroscopy, samples were passed over a desalting column in a camphor-free buffer multiple times until no further change in the ratio of absorbances at 390 and 417 nm was observed.
Activity assays
Preliminary assays were performed using a 96-well spectroscopic plate reader (BioTek XS2) to monitor NADH consumption at 340 nm. Each well contained 0.5 µM CYP101A1 (WT or mutant), 5 µM Pdx, 0.2 µM PdR and 2 mM substrate in 50 mM Tris HCl pH 7.4, and 100 mM KCl. To start the reactions, NADH was added to a final concentration of 160 µM. If significant NADH consumption was observed, the reaction was run on a larger (1 mL) scale, and the products along with unreacted substrates were extracted with methylene chloride, the extracts passed over anhydrous MgSO4 to remove moisture, and analyzed by gas chromatography/mass spectrometry (GC/MS) on an Agilent (10 eV electron impact ionization, array ion current detector). Chromatographic peaks were identified by automated matching of MS fragmentation patterns against the ChemStation library. For chromatographic purification of the NRase products, the reaction was run on a 10 mL scale. Flash chromatography was conducted over activated silica using a 10:1 hexane:ethyl acetate eluent.
Optical spectroscopy
Optical spectra of WT and mutant CYP101A1 were obtained on a Agilent 8543 diode array spectrophotometer using 2 mL of solution containing 10 µM enzyme buffered with 50 mM Tris HCl pH 7.4, 100 mM KCl and saturating (2 mM) d-camphor 1.
Electron paramagnetic resonance (EPR) spectroscopy
Continuous wave EPR experiments at variable temperatures (5–70 K) were carried out at a Bruker ESP300 spectrometer equipped with a continuous flow cryostat (Oxford Instruments) and a Bruker ER 4116DM dual mode resonator, which operates both in the perpendicular ~ 9.6 GHz (TE102) and parallel ~ 9.4 GHz (TE012) microwave modes, respectively. The microwave frequency was measured with a 5350B Hewlett Packard frequency counter. For all experiments custom-made quartz tubes of the same inner and outer diameter were used (QSI). All quantifications were carried out using spectra recorded at T = 10 K. The first-derivative EPR spectra were simulated using the MATLAB (Mathworks)-based Easyspin simulation software.
Supplementary Material
Figure 6.
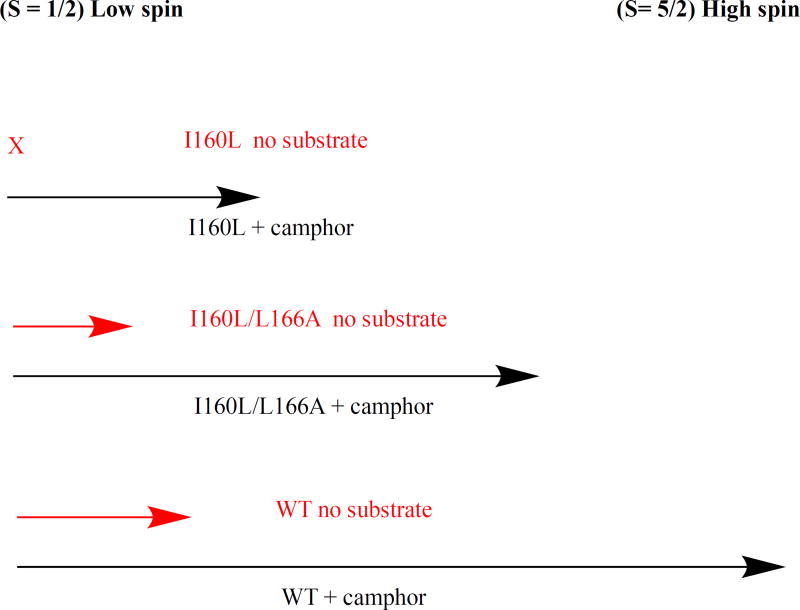
Diagram of effect of substrate d-camphor 1 binding on spin state equilibrium in CYP101A1. Black arrows represent degree of high-spin (S= 5/2) ferric form (to the right on the diagram) upon binding of 1, while red arrows represent expected degree of high-spin form in the absence of substrate. Binding of 1 to WT CYP101A1 shifts conformational ensemble equilibrium in favor of > 95% high-spin. The I160L/L166A mutant binding of 1 shifts equilibrium to ~80% high-spin. Binding of 1 to I160L mutant shifts equilibrium less than 50% towards high-spin. Only WT and I160L/L166A have sufficient residual high-spin in the absence of 1 (ns = no substrate, red arrows) to support reduction of 6.
Scheme II.
Compounds discussed in this work.
Highlights.
Some variants of CYP101A1 catalyze the reduction of nitroacetophenones to the corresponding anilines (NRAse activity).
Mutants that do not catalyze the reduction exhibit less than 50% high spin form upon binding of native substrate.
NRAse activity is proposed as a probe for residual high spin in cytochromes P450.
Acknowledgments
This work was supported in part by NIH grant R01-GM44191 (T.C.P.). EPR spectrometry was performed by M.-E.P. using the facilities at the Pennsylvania State University Dept. of Chemistry (State College, PA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sligar SG. Coupling of spin, substrate and redox equilibria in cytochrome P450. Biochemistry. 1976;15:5399–5406. doi: 10.1021/bi00669a029. [DOI] [PubMed] [Google Scholar]
- 2.Gunsalus IC, Sligar SG. Redox regulation of cytochrome P450cam mixed-function oxidation by putidaredoxin and camphor ligation. Biochimie. 1976;58:143–147. doi: 10.1016/s0300-9084(76)80364-1. [DOI] [PubMed] [Google Scholar]
- 3.Simgen B, Contzen J, Schwarzer R, Bernhardt R, Jung C. Substrate Binding to 15 Beta-Hydroxylase (Cyp106A2) Probed by FT Infrared Spectroscopic Studies of the Iron Ligand CO Stretch Vibration. Biochemical and Biophysical Research Communications. 2000;269:737–742. doi: 10.1006/bbrc.2000.2348. [DOI] [PubMed] [Google Scholar]
- 4.Wang A, Stout CD, Zhang QH, Johnson EF. Contributions of ionic interactions and protein dynamics to cytochrome P450 2D6 (CYP2D6) substrate and inhibitor binding. Journal of Biological Chemistry. 2015;290:5092–5104. doi: 10.1074/jbc.M114.627661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell SG, Chen XH, Sowden RJ, Xu F, Williams JN, Wong LL, Rao ZH. Molecular recognition in (+)-alpha-pinene oxidation by cytochrome P450(cam) Journal of the American Chemical Society. 2003;125:705–714. doi: 10.1021/ja028460a. [DOI] [PubMed] [Google Scholar]
- 6.Colthart AM, Tietz DR, Ni Y, Friedman JL, Dang M, Pochapsky TC. Detection of substrate-dependent conformational changes in the P450 fold by nuclear magnetic resonance. Scientific Reports. 2016;6:22035. doi: 10.1038/srep22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guengerich FP. Common and Uncommon Cytochrome P450 Reactions Related to Metabolism and Chemical Toxicity. Chemical Research in Toxicology. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 8.Harada N, Omura T. Participation of cytochrome P450 in the reduction of nitro compounds by rat liver microsomes. Journal of Biochemistry. 1980;87:1539–1554. doi: 10.1093/oxfordjournals.jbchem.a132895. [DOI] [PubMed] [Google Scholar]
- 9.Wang B, Yang LP, Zhang XZ, Huang SQ, Bartlam M, Zhou SF. New insights into the structural characteristics and functional relevance of the human cytochrome P450 2D6 enzyme. Drug Metabolism Reviews. 2009;41:573–643. doi: 10.1080/03602530903118729. [DOI] [PubMed] [Google Scholar]
- 10.Oshima R, Fushinobu S, Su F, Zhang L, Takaya N, Shoun H. Structural evidence for direct hydride transfer from NADH to cytochrome P450nor. J Mol Biol. 2004;342:207–217. doi: 10.1016/j.jmb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Advanced Chemistry Development Software. ACD/Labs, ACS SciFinder; 2016. [Google Scholar]
- 12.Pochapsky TC, Kazanis S, Dang M. Conformational plasticity and structure/function relationships in cytochromes P450. Antioxidants & Redox Signaling. 2010;13:1273–1296. doi: 10.1089/ars.2010.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asciutto EK, Dang M, Pochapsky SS, Madura JD, Pochapsky TC. Experimentally Restrained Molecular Dynamics Simulations for Characterizing the Open States of Cytochrome P450(cam) Biochemistry. 2011;50:1664–1671. doi: 10.1021/bi101820d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asciutto EK, Young MJ, Madura JD, Pochapsky SS, Pochapsky TC. Solution structural ensembles of substrate-free cytochrome P450cam. Biochemistry. 2012;51:3383–3393. doi: 10.1021/bi300007r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nihei M, Shiga T, Maeda Y, Oshio H. Spin crossover iron(III) complexes. Coordination Chemistry Reviews. 2007;251:2606–2621. [Google Scholar]
- 16.Raag R, Poulos TL. The structural basis for substrate-induced changes in redox potential and spin equilibrium in cytochrome P450CAM. Biochemistry. 1989;28:917–922. doi: 10.1021/bi00428a077. [DOI] [PubMed] [Google Scholar]
- 17.Rajarathnam K, Lamar GN, Chiu ML, Sligar SG, Singh JP, Smith KM. H-1 NMR hyperfine shift pattern as a probe for ligation state in high-spin ferric hemoproteins- water binding to metmyoglobin mutants. Journal of the American Chemical Society. 1991;113:7886–7892. [Google Scholar]
- 18.Liptak MD, Wen X, Bren KL. NMR and DFT Investigation of Heme Ruffling: Functional Implications for Cytochrome c. Journal of the American Chemical Society. 2010;132:9753–9763. doi: 10.1021/ja102098p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karunakaran V, Denisov I, Sligar SG, Champion PM. Investigation of the Low Frequency Dynamics of Heme Proteins: Native and Mutant Cytochrome P450(cam) and Redox Partner Complexes. J. Phys. Chem. B. 2011;115:5665–5677. doi: 10.1021/jp112298y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petzold DR, Rein H, Schwarz D, Sommer M, Ruckpaul K. Relation between the structure of benzphetamine analogues and their binding properties to cytochrome P-450 LM2. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1985;829:253–261. doi: 10.1016/0167-4838(85)90195-5. [DOI] [PubMed] [Google Scholar]
- 21.Nickerson DP, Wong LL. The dimerization of Pseudomonas putida cytochrome P450(cam): Practical consequences and engineering of a monomeric enzyme. Protein Engineering. 1997;10:1357–1361. doi: 10.1093/protein/10.12.1357. [DOI] [PubMed] [Google Scholar]
- 22.Gunsalus IC, Wagner GC, Sidney Fleischer P, Lester . Methods in Enzymology. Academic Press; 1978. [17] Bacterial P-450cam methylene monooxygenase components: Cytochrome m, putidaredoxin, and putidaredoxin reductase; pp. 166–188. Place Published. [DOI] [PubMed] [Google Scholar]
- 23.OuYang B, Pochapsky SS, Dang M, Pochapsky TC. A functional proline switch in cytochrome P450(cam) Structure. 2008;16:916–923. doi: 10.1016/j.str.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raag R, Poulos TL. Crystal structure of the carbon monoxide-substrate-cytochrome P450CAM ternary complex. Biochemistry. 1989;28:7586–7592. doi: 10.1021/bi00445a013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.