Abstract
Chromobox homolog 8 (CBX8), also known as human polycomb 8, is a repressor that maintains the transcriptionally repressive state in various cellular genes, and has been reported to promote tumorigenesis. In the present study, we examined CBX8 expression in eight pairs of muscle invasive bladder cancer tissues and adjacent non‐tumor tissues, and found that CBX8 was frequently upregulated in muscle invasive bladder cancer tissues when compared to adjacent non‐tumor tissues. Analysis showed that high expression of CBX8 in 152 muscle invasive bladder cancer specimens was associated with progression of the T, N, and M stages (P = 0.004, 0.005, <0.001, respectively). Furthermore, Kaplan–Meier survival analysis and log–rank test showed that muscle invasive bladder cancer patients with high CBX8 expression had a poor rate of overall survival (P < 0.001) and 5‐year recurrence‐free survival (P < 0.001) compared to patients with low CBX8 expression. High CBX8 expression predicted poor overall survival and 5‐year recurrence‐free survival in T and N stages of muscle invasive bladder cancer patients. Moreover, knockdown of CBX8 inhibited cell proliferation of urothelial carcinoma of the bladder both in vitro and in vivo. In addition, CBX8 depletion resulted in cell cycle delay of urothelial carcinoma cells of the bladder at the G2/M phase by the p53 pathway. The data suggest that high expression of CBX8 plays a critical oncogenic role in aggressiveness of urothelial carcinoma cells of the bladder through promoting cancer cell proliferation by repressing the p53 pathway, and CBX8 could be used as a novel predictor for muscle invasive bladder cancer patients.
Keywords: CBX8, cell cycle, p53, survival, urothelial carcinoma of the bladder
Urothelial carcinoma of the bladder is the most common malignancy involving the genitourinary system and is responsible for the increased risk in 2015 in annual mortality in the Chinese population.1 UCB represents the fifth most common human cancer in the USA, causing 74 690 new cases and 25 580 deaths in 2014.2 Approximately 20% ~ 30% of UCB cases are MIBC at initial presentation. Despite extensive cystectomy and adjuvant therapies including chemotherapy and radiotherapy, most patients experience recurrence or die. Unlike breast, colorectal and prostate cancer, UCB lacks clinically useful biomarkers for prediction of cancer occurrence and recurrence. Thus, there is an urgent need to explore novel biomarkers that may be used for the diagnosis and prediction of UCB.
CBX8 was first described in 2000 as a member of the PcG family.3 PcG were initially identified in Drosophila as repressors that silence Hox genes during development.4, 5, 6, 7 Homologous proteins in mammals have subsequently been reported.8, 9, 10 CBX8 functions as a transcriptional repressor that interacts with the RING protein and colocalizes with BMI1 in the PcG body.10 In addition, CBX8 directly binds to the INK4A‐ARF locus to promote cell proliferation and to bypass senescence.11, 12 Recent reports have shown that CBX8 is positively associated with glioblastoma, colorectal cancer, and leukemia.13, 14, 15 For example, CBX8 upregulation is associated with TNM staging in esophageal carcinoma, and it acts as a novel DNA repair protein that promotes cell proliferation and protects cancer cells from sensitivity to ionizing radiation or hydrogen peroxide.16 Knockdown of CBX8 inhibits cell proliferation and cell cycle progression by increasing the phosphorylation of p21, Wee1, and CHK1. Tan et al. reported that CBX8 participates in leukemogenesis through MLL‐AF9‐mediated transcriptional activation by interacting with MLL‐AF9 and TIP60.15 However, Tang et al. reported that CBX8 also plays a paradoxical role in colorectal cancer wherein its upregulation is associated with low risk for distant metastasis and good prognosis.14 Knockdown of CBX8 also results in the inhibition of cell proliferation by accelerating the p53 pathway.
Although there are mounting studies suggesting the critical roles of CBX8 in human tumorigenesis and cancer progression, the roles of CBX8 in UCB remain unclear. The present study investigated the expression pattern of CBX8 in MIBC tissues and demonstrated the correlation of CBX8 expression with clinicopathological features of MIBC patients. Furthermore, knockdown of CBX8 resulted in the inhibition of UCB cell proliferation and cell cycle arrest at the G2/M phase by the p53 pathway.
Materials and Methods
Analysis of UCB data on CBX8 CNV from TCGA database
The CNV segment mean (n = 408) of CBX8 from TCGA level 3 data was downloaded from the Broad Institute TCGA Genome Data Analysis Center.17 Analysis was carried out by comparing survival distribution of two groups using the log–rank test as provided in X‐tile software.18 A cut‐off value was generated, and the patient cohort was then divided into a “CNV‐high” group and a “CNV‐low” group as previously described elsewhere.19 Then, we tested the difference in the outcomes between the two groups.
Patients and tissue specimens
Paraffin‐embedded specimens from 152 MIBC patients who underwent radical cystectomy at the Sun Yat‐Sen University Cancer Center between 2000 and 2010 were collected for IHC analysis. Two groups of eight paired fresh MIBC tissues and adjacent non‐tumor tissues from the same patient were stored in liquid nitrogen and 4% neutral formalin for quantitative RT‐PCR and IHC experiment, respectively. All samples were classified according to the 2010 American Joint Committee on Cancer TNM classification.20 In addition, all MIBC tissues were histologically identified to be urothelial carcinomas. The medical ethics committee of Sun Yat‐Sen University Cancer Center approved this study, and all patients provided consent for use of their clinical specimens.
Immunohistochemistry analysis
Slides were heated for 3 ~ 4 h at 65°C, deparaffinized in xylene and hydrated in an alcohol gradient, and endogenous peroxidase activity blocked with 3% hydrogen peroxide for 10 min. Antigen retrieval was completed by boiling the slides in EDTA buffer (pH 8.0) for 5 min in a pressure cooker. Slides were incubated with 10% normal goat serum for 15 min at room temperature to block non‐specific binding, followed by incubation with anti‐CBX8 antibody (1:500 in PBS; Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C. The slides were washed with PBS three times, then incubated with secondary goat anti‐mouse antibody at a dilution of 1:100 at 37°C for 30 min. The slides were then immersed in a 3,3′‐diaminobenzidine (DAB) solution for 5 min and counterstained with 10% Meyer's hematoxylin for 3 min. The slides were then polarized with 70% ethyl alcohol containing 0.1% hydrochloric acid for 10 s. PBS was used as negative control, whereas IHC‐positive CBX8 staining slides of an esophageal cancer case were used as positive control.
Immunohistochemistry assessment
Degree of immunostaining of sections was reviewed and independently scored by two pathologists based on staining intensity and percentage of positively stained cells. Using the method of judging the degree of staining in MIBC IHC of Zhang et al.,21 two scoring systems were used. The scoring system for positively staining cancer cells was as follows: 0 for 0% positively staining cells, 1 for <10% positively staining cells, 2 for 10% ~ 40%, 3 for 40 ~ 70%, and 4 for >70%. Scores for staining intensity were as follows: 0 for no staining, 1 for weak staining, 2 for moderate staining, and 3 for strong staining. The staining index was calculated by multiplying the score for positively staining cells by the score for staining intensity. Using this method, we assessed CBX8 expression in MIBC and adjacent normal tissues by determining the staining index with scores of 0, 1, 2, 3, 4, 6, 8, 9, and 12. CBX8 cut‐off values were based on heterogeneity measurements with log–rank test statistical analysis with respect to OS and RFS. Optimal cut‐off used in this study was as follows: a staining index score <6 was considered low CBX8 expression, whereas ≥6 was considered high CBX8 expression.
FISH
Two‐color FISH was applied to the sections of formalin‐fixed, paraffin‐embedded MIBC tissues using spectrum red‐labeled BAC clone (RP11‐353N14) containing the CBX8 gene and a spectrum green‐labeled chromosome 17 centromere (Vysis, Downers Grove, IL, USA) was used as internal control. Details of FISH procedures were described previously.22 In brief, the sections were deparaffinized and treated with proteinase K (400 μg/mL) at 37°C for 45 min, followed by denaturation in 70% formamide and 2× SSC at 75°C for 7 min. Mixture containing 50 ng of each probe and 20 μL hybridization compound (55% formamide, 2 μg human Cot1 DNA, and 2× SSC) was prepared to be denatured at 75°C for 6 min. After this step, the mixture was hybridized to the prepared MIBC sections at 37°C for 24 h. Next, the sections were counterstained with 1 μg/mL DAPI in an anti‐fade solution. FISH signals from 300 cells in each sample were counted. Criteria for CBX8 gene amplification was defined as the presence of more than three‐fold as many gene signals as centromere signals in chromosome 17. Samples with weak target signals or with strong signal background were treated as non‐informative cases.
Cell culture
UCB cell lines T24 and BIU were cultured at 37°C with 5% CO2 in RPMI‐1640 media supplemented with 10% FBS, penicillin (100 U/mL), and streptomycin (100 μg/mL).
Western blot analysis
Frozen tissues were ground in liquid nitrogen and then lysed in RPMI‐RIPA lysis solution containing 1× protease inhibitors. The lysates were centrifuged at 12 000 g for 15 min at 4°C to obtain supernatant. The cultured cells were harvested and lysed with RPMI‐RIPA lysis solution, followed by centrifugation using the same conditions as that for the lysed frozen tissues.
After determining the protein content of the mixed lysates, protein extracts were separated by 10% SDS‐PAGE, transferred onto a PVDF membrane at 250 mA for 2 h at room temperature. The membrane was blocked with 5% BSA or defatted milk for 1 h and incubated with primary antibodies (CBX8; p53, cdc2, cyclinB1, CDK4 and cdc25A; Cell Signaling Technology) overnight at 4°C. The membranes were washed with PBS containing 0.1% Tween (PBST) three times, followed by incubation with a secondary rabbit anti‐mouse antibody for 1 h at room temperature. Hybridization signals were quantified by using an ECL detection system (Tanon, Shanghai, China).
Quantitative real‐time polymerase chain reaction (qRT‐PCR) assay
Frozen tissues were ground in liquid nitrogen, then total RNA was extracted using TRIzol reagent (Invitrogen Life Technologies, Waltham, MA, USA) according to the manufacturer's recommendations. Approximately 1 μg RNA was used in first‐strand cDNA synthesis using random primers. A 15‐μL reaction system, which included the CBX8‐specific primers, cDNA, and SYBR Green PCR mixture (Applied Biosystems, Foster City, CA, USA), was prepared for amplification of the CBX8 cDNA. The qRT‐PCR reaction conditions were as follows: initial denaturation at 95°C for 30 s, followed by annealing at 55°C for 1 min, and extension for 1 min at 72°C, for a total of 30 cycles. qRT‐PCR was carried out in triplicate on an ABI Prism 7000 Sequence Detection System (Applied Biosystems). Relative level of gene expression was expressed as ΔCt = Ctgene−Ctreference; the 2−ΔΔCt method was used to calculate the fold change of gene expression. GAPDH was used as a control and for normalization. Primer sequences are as follows: for CBX8: forward, 5′‐TCGTGAAATGGAAGGGATGG‐3′; reverse, 5′‐AGGTTTTGGGCTTGGGTC‐3′; and for GAPDH: forward, 5′‐CGGAGTCAACGGATTTGGTCGTAT‐3′ and reverse, 5′AGCCTTCTCCATGGTGGTGAAGAC‐3′.
Construction of stable cell lines
Lenti‐Pac HIV expression packaging kit (GeneCopoeia, Rockville, MD, USA) was used to construct stable low CBX8‐expression level cell lines according to the manufacturer's recommendations. Commercialized lentiviral vectors expressing CBX8 shRNAs were purchased and transfected into 293T cells to obtain CBX8 cDNAs. Then the virus solution including CBX8 cDNAs was isolated to transfect T24 or BIU cells. Stable cells were screened by using 0.1% puromycin (Gibco, Invitrogen, Darmstadt, Germany).
CCK‐8 assay
Cell viability was measured using the CCK‐8 reagent ((Dojindo, Kumamoto, Japan) according to the manufacturer's recommendations. Briefly, a total of 1 × 103 cells were seeded into 96‐well plates, and their viability was determined at different time points starting from 12 to 60 h. Absorbance of each well was measured at a wavelength of 450 nm using a microplate spectrophotometer (SpectraMax M5; Molecular Devices, Sunnyvale, CA, USA). Cell viability was expressed as the fold change normalized to that of the control (0 h).
Colony formation assay
Approximately 500 cells were seeded in each well in a six‐well plate and incubated for 7 days. Colonies were fixed with methanol for 30 min and stained with 0.1% crystal violet for 1 h.
Cell cycle analysis
A cell cycle analysis kit (Beyotime, Shanghai, China) was used to determine cell cycle distribution. Cells were harvested and washed with PBS and fixed in chilled 70% ethanol overnight at 4°C. The cells were then incubated with RNase A (50 μg/mL) for 30 min, followed by incubation with propidium iodide (50 μg/mL) for 30 min. Cell cycle determination was carried out using a flow cytometry system (MoFlo XDP; Beckman Coulter, CA, USA).
Xenograft assay
Four‐week‐old BALB/c nude mice were purchased from Charles River Laboratories (Beijing, China). The animal ethics committee of Sun Yat‐sen University, Guangzhou, China approved all animal experiments carried out in this study. The mice were randomly divided into two groups of six mice, respectively. A total of 5 × 106 T24 cells (control and CBX8 shRNA) were s.c. inoculated into the right flank of each mouse. Weight and volume of tumors were determined at the end of the study. The following formula was used to measure tumor volume: Tumor volume = 1/2 L × W 2, where L represents the length and W represents the width.
Statistical analysis
Correlation between CBX8 expression and clinicopathological features was analyzed using the chi‐squared test. Kaplan–Meier survival analysis and log–rank test were carried out to analyze associations of CBX8 expression with the subgroups of T and N stages. Significance of the variables for survival was determined by using multivariate Cox proportional hazards regression analysis. All statistical conclusions were determined using SPSS v.19.0 statistical software (SPSS, Chicago, IL, USA). Data derived from the experiments were expressed as mean ± SE and compared by using Student's t‐test. P‐value < 0.05 was considered to be statistically significant.
Results
Amplification of the CBX8 gene and upregulation of CBX8 in MIBC tissues
Bioinformatics analysis was first carried out to evaluate CBX8 gene alterations in UCB using TCGA database. We analyzed CNV of the CBX8 gene included in TCGA database. In UCB, a lower OS rate is associated with a higher number of CNV involving the CBX8 gene (HR = 2.091, log–rank P = 0.0056; Fig. 1a), whereas a lower DFS is associated with a higher number CNV in the CBX8 gene (HR = 2.764, log–rank P = 0.0031; Fig. 1b). TCGA database shows that more frequent CBX8 amplification predicts poor outcome in UCB patients, indicating that CBX8 may act as an oncogene in UCB.
Figure 1.
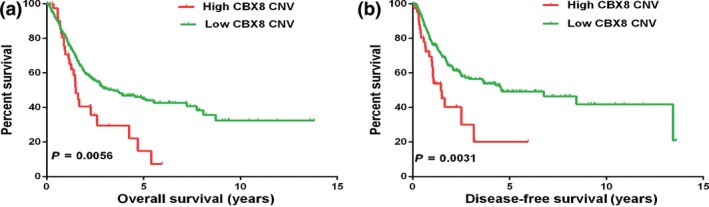
Kaplan–Meier survival plot of urothelial carcinoma of the bladder (UCB) patients using copy number variation (CNV) segment mean (n = 408) from The Cancer Genome Atlas (TCGA) database. (a) UCB patients with high chromobox homolog 8 (CBX8) CNV showed poor overall survival compared to patients with low CBX8 CNV. *P = 0.0056. (b) Percent survival of disease‐free survival in UCB patients with low CBX8 CNV was significantly higher than that in patients with high CBX8 CNV (*P = 0.0031).
Because of its high risk for morbidity and mortality, the present study focused on MIBC. To verify the role of CBX8 expression in the clinical features of MIBC, we first measured expression of CBX8 mRNA in eight pairs of MIBC samples collected from our center. Figure 2a shows that all tested MIBC tissues exhibited significant upregulation of CBX8 mRNA expression compared to the adjacent non‐tumor tissues (P < 0.05). Western blot analysis revealed that the protein levels of CBX8 in the MIBC tissues were elevated compared to those in non‐tumor tissues (Fig. 2b). These findings were confirmed by IHC analysis of MIBC tissues and non‐tumor tissues (Fig. 2c).
Figure 2.
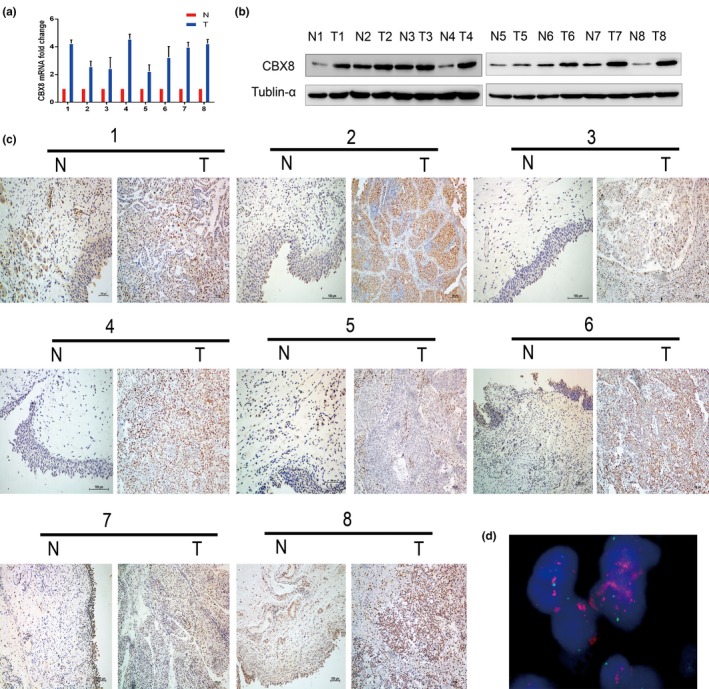
Expression of chromobox homolog 8 (CBX8) in muscle invasive bladder cancer (MIBC) tissues and adjacent non‐tumor tissues. (a) Upregulated expression of CBX8 mRNA was detected by qRT‐PCR in eight pairs of MIBC tissues compared to that of adjacent non‐tumor tissues. (b) Upregulated expression of CBX8 expression was measured with western blot in eight paired MIBC tissues and adjacent non‐tumor tissues. (c) Immunohistochemistry was examined in eight pairs of MIBC tissues and adjacent non‐tumor tissues. Each pair of samples was derived from the same patient. N, adjacent non‐tumor tissues; T, MIBC tissues. (d) Amplification of the CBX8 gene was observed by FISH in MIBC sections. Detection of CBX8 gene signals (pink) was at least three‐fold greater than detection of signals on chromosome 17 centromere (blue). Original magnification, ×1000.
Amplification of CBX8 examined by FISH in MIBC tissues
To verify amplification of CBX8 in MIBC tissues, FISH analysis was carried out in all 152 specimens, of which 38.2% (58/152) of samples were informative whereas the other 61.8% (94/152) of samples were non‐informative. 10.3% (6/58) of samples were determined to have CBX8 amplification (Fig. 2d). In the remaining 52 informative samples without CBX8 amplification, 38 samples displayed normal CBX8 expression whereas the other 14 samples showed CBX8 overexpression. A significant correlation between overexpression and amplification of CBX8 was evaluated (Table1).
Table 1.
Association of CBX8 expression and amplification in MIBC tissues
| CBX8 gene | Informative samples | CBX8 expression | P‐valuea | |
|---|---|---|---|---|
| Normal | Overexpression | |||
| No amplification | 52 | 38 (73.1) | 14 (26.9) | 0.0119 |
| Amplification | 6 | 1 (16.7) | 5 (83.3) | |
Fisher's exact test. MIBC, muscle invasive bladder cancer.
CBX8 expression is associated with clinicopathological features of MIBC patients
IHC was carried out in 152 specimens to examine the expression of CBX8 in MIBC patients who underwent cystectomy. Table 2 shows that 77 (50.7%) of the samples showed low expression, whereas the other 75 (49.3%) samples displayed high expression. Chi‐squared testing suggested that high CBX8 expression was significantly associated with pT stage, pN stage, M stage, and recurrence status of MIBC patients. However, no significant association between CBX8 expression and other clinical features such as age, gender, smoking history, and gross hematuria was observed.
Table 2.
Correlation between CBX8 expression and clinicopathological features in 152 samples of MIBC
| Variable | All samples (n = 152) | CBX8 expression (%) | P‐valuea | |
|---|---|---|---|---|
| Low expression (n = 77) | High expression (n = 75) | |||
| Age (years) | ||||
| <60 | 64 | 31 (48.4%) | 33 (51.6%) | 0.762 |
| ≥60 | 88 | 46 (52.3%) | 42 (47.7%) | |
| Sex | ||||
| Male | 137 | 68 (49.6%) | 69 (50.4%) | 0.624 |
| Female | 15 | 9 (60.0%) | 6 (40.0%) | |
| pT stage | ||||
| pT2 | 86 | 63 (73.3%) | 23 (26.7%) | 0.0035 |
| pT3 | 36 | 9 (25.0%) | 27 (75.0%) | |
| pT4 | 30 | 5 (16.7%) | 25 (83.3%) | |
| pN stage | ||||
| pN– | 119 | 68 (57.1%) | 51 (42.9%) | 0.0045 |
| pN+ | 33 | 9 (27.3%) | 24 (72.7%) | |
| Smoking history | ||||
| Yes | 97 | 46 (47.4%) | 51 (52.6%) | 0.373 |
| No | 55 | 31 (56.4%) | 24 (43.6%) | |
| Gross hematuria | ||||
| Yes | 97 | 42 (43.3%) | 55 (56.7%) | 0.106 |
| No | 55 | 35 (63.6%) | 20 (36.4%) | |
| Metastasis | ||||
| Yes | 28 | 4 (14.3%) | 24 (85.7%) | 0.0001 |
| No | 124 | 73 (58.9%) | 51 (41.1%) | |
| Recurrence | ||||
| Yes | 72 | 21 (29.2%) | 51 (70.8%) | 0.0000 |
| No | 80 | 56 (70.0%) | 24 (30.0%) | |
chi‐squared test. MIBC, muscle invasive bladder cancer.
CBX8 serves as an independent predictor for MIBC patient outcomes
Combined with Kaplan–Meier analysis and log–rank test, the analysis showed a significant association of CBX8 expression with OS (P < 0.001), 5‐year RFS (P < 0.001), pT2 (P = 0.002), pT3‐4 (P < 0.001), pN− (P < 0.001), and pN+ (P = 0.024), suggesting that MIBC patients with high CBX8 expression had lower survival rates than patients with low CBX8 expression (Fig. 3a–f, respectively). Moreover, univariate and multivariate analyses both showed that CBX8 expression, T stage, N stage, and M stage were independent prognostic factors for OS rates of MIBC patients (Table3). These findings suggest that CBX8 may potentially be used as an independent prognosticator for survival of MIBC patients.
Figure 3.
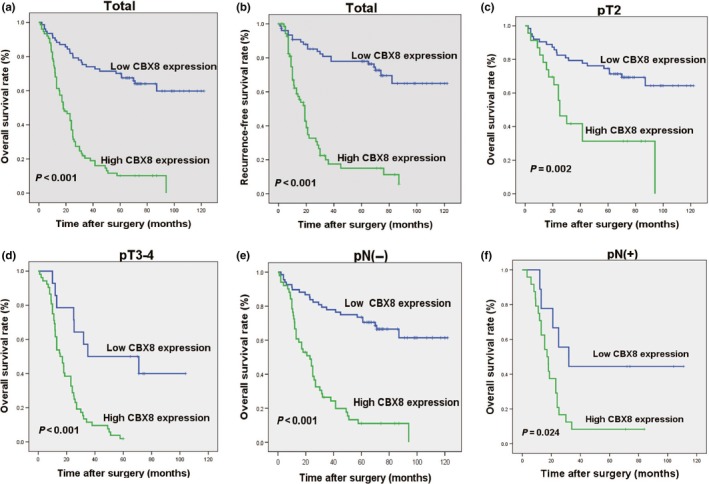
Kaplan–Meier survival analysis of chromobox homolog 8 (CBX8) expression in the entire cohort and different subgroups of muscle invasive bladder cancer (MIBC) patients. (a) Low CBX8‐expressing patients showed a higher overall survival (OS) rate than high CBX8‐expressing patients (P < 0.001). (b) Recurrence‐free survival (RFS) rate in patients with low CBXB expression was significantly higher than that in patients with high CBX8 expression (P < 0.001). (c–f) MIBC patients with low CBX8 expression had a higher OS rate than patients with high CBX8 expression in different T stages (P = 0.002, P < 0.001, respectively) and N stages (P < 0.001, P = 0.024, respectively).
Table 3.
Univariate and multivariate analyses of various prognostic parameters in MIBC patients
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P‐value | Hazard ratio | 95% CI | P‐value | |
| Age, years (≥60 vs <60) | 1.481 | 0.970–2.262 | 0.069 | 1.577 | 1.007–2.470 | 0.046 |
| Gender (Male vs Female) | 1.496 | 0.692–3.232 | 0.306 | 1.030 | 0.467–2.270 | 0.942 |
| No. tumors (≥2 vs 1) | 0.890 | 0.786–1.008 | 0.066 | 0.978 | 0.862–1.109 | 0.731 |
| T stage (T2 vs T3‐4) | 3.913 | 2.547–6.012 | 0.000 | 1.846 | 1.100–3.096 | 0.020 |
| N stage (N+ vs N–) | 1.640 | 1.256–2.140 | 0.000 | 1.054 | 0.952–1.670 | 0.106 |
| M stage (M– vs M+) | 2.055 | 1.425–2.965 | 0.000 | 1.380 | 0.874–2.179 | 0.016 |
| CBX8 score (High vs Low) | 5.190 | 3.273–8.231 | 0.000 | 3.849 | 2.314–6.404 | 0.000 |
CI, confidence interval; MIBC, muscle invasive bladder cancer.
Knockdown of CBX8 inhibits UCB cell proliferation
Further in vitro investigations involving knocking down CBX8 expression were carried out in the present study to explore the role of CBX8 in promoting UCB cell proliferation. We also constructed stable T24 and BIU cells that were permanently repressed from expressing CBX8 (Fig. 4a). Then, CCK‐8 and colony‐forming assays were carried out to detect whether the ability of proliferation was affected after knocking down CBX8. Figure 4b shows that the CBX8‐silenced cells displayed a significant decrease in viability compared to that in the control cells. The CBX8‐knockdown cells generated significantly fewer colonies compared to that in the control cells (Fig. 4c). Taken together, these findings indicate that knockdown of CBX8 inhibited UCB cell proliferation.
Figure 4.
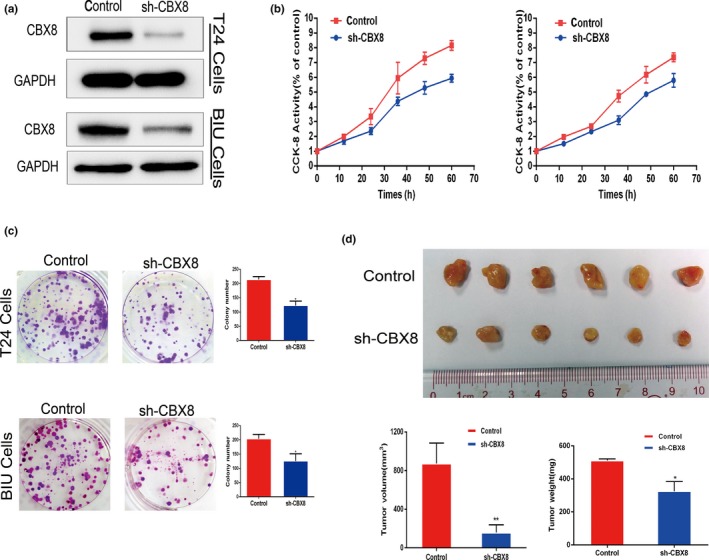
Knockdown of chromobox homolog 8 (CBX8) inhibited the proliferation of urothelial carcinoma of the bladder (UCB) cell lines both in vitro and in vivo. (a) Western blot was carried out in T24 and BIU cell lines to examine the efficacy of knocking down CBX8 when transfected with CBX8‐shRNAs. (b) Cell counting kit‐8 (CCK‐8) assay was carried out to measure the viability of control cells or CBX8‐shRNA cells in T24 and BIU cells at different time points. (c) Colony forming assay was carried out in T24 and BIU cells transfected with shRNA to measure the colony forming ability after knocking down CBX8. Error bars represent the standard deviation of three independent experiments. *P < 0.05. (d) T24 cells transfected with control or CBX8‐shRNA were inoculated into nude mice to construct xenograft models in vivo to assess proliferation rates after knocking down CBX8. Tumors from mice inoculated with control cells exhibited higher volumes and weights than tumors from mice inoculated with CBX8‐shRNAs (*P < 0.05, **P < 0.01, respectively).
Knockdown of CBX8 inhibits xenograft tumor growth in vivo
Xenograft BALB/c nude mice were used to investigate whether knockdown of CBX8 inhibits UCB cells proliferation in vivo. Figure 4d shows that mice infected with T24‐CBX8 shRNA generated tumors that were smaller in size and lower in weight than those observed in T24‐control mice, indicating that knockdown of CBX8 significantly inhibited tumor growth in vivo.
Knockdown of CBX8 arrests cell cycle transition
A recent study involving human esophageal carcinoma EC109 cells showed that depletion of CBX8 results in cell cycle delay at the G2/M phase.16 Therefore, we measured the cell cycle transition to detect whether knockdown of CBX8 affects cell cycle distribution in the UCB cell lines T24 and BIU. The results showed that knockdown of CBX8 resulted in an accumulation of cells in the G2 phase from 6.45% and 4.81% in the control cells to 20.67% and 19.24% in the T24 and BIU cells transfected with CBX8 shRNA, respectively (Fig. 5a). Adversely, the percentage of cells in the G1 phase decreased from 57.87% to 68.06% in the control cells to 40.75% and 58.79% in the T24 and BIU cells transfected with CBX8 shRNA, respectively. These findings indicate that knockdown of CBX8 leads to inhibition of G2‐M phase transition, which, in turn, contributes to the inhibition of UCB cell proliferation.
Figure 5.
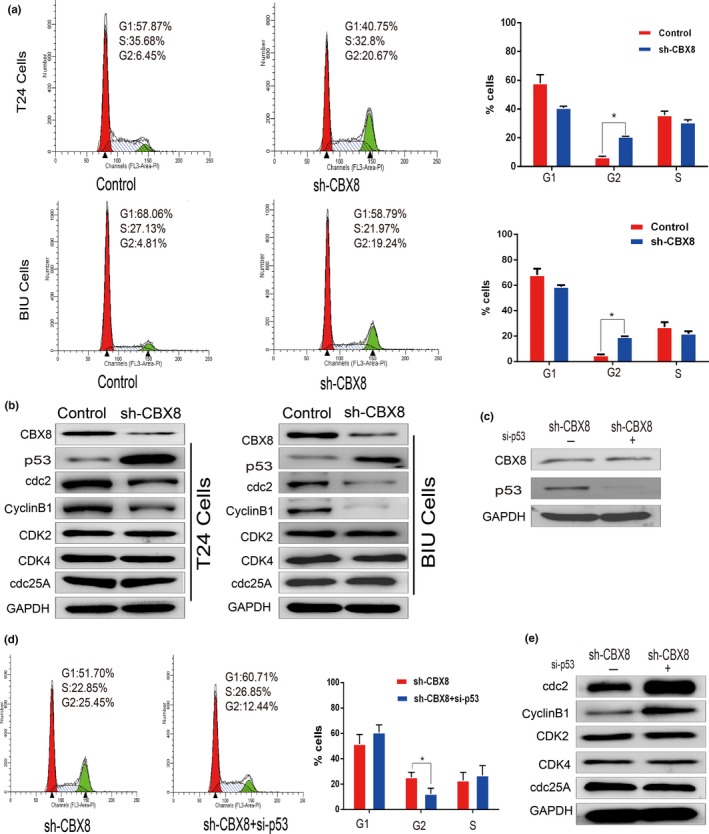
Knockdown of chromobox homolog 8 (CBX8) results in cell cycle arrest at the G2/M phase by the p53 pathway. (a) Flow cytometry was carried out in T24 and BIU cell lines transfected with control or CBX8 shRNA to assess cell cycle distribution. Error bars represent the mean ± SD of three independent experiments. *P < 0.05. (b) Western blot was used to detect protein levels of CBX8 and indicated proteins (left panel). (c) Western blot was carried out in T24‐CBX8 shRNA cells to determine the effectiveness of p53‐siRNAs. (d) Measurement of cell cycle distribution was repeated in T24‐CBX8 shRNA cell lines transfected with or without p53‐siRNAs. (e) Western blot assay showed an increase in cdc2/cyclinB1 levels when p53 expression was repressed in CBX8‐silenced T24 cells.
Knockdown of CBX8 arrests cell cycle transition by the p53‐dependent pathway
A recent study showed that depletion of CBX8 delays cell cycle progression and results in the inhibition of proliferation of CRC by the p53 pathway.14 We wondered whether a similar pathway is involved in UCB when CBX8 is depleted. Thus, we detected the protein expression level of p53 and its downstream regulators, cdc2 and cyclinB1, which are implicated in the transition from G2 to M phase,23, 24 in the two UCB cell lines, T24 and BIU. Figure 5b shows that the protein expression level of p53 was elevated, whereas that of cdc2/cyclinB1 decreased after knocking down CBX8 expression. In contrast, protein levels of CDK2, CDK4, and cdc25A, which are involved in the transition from the G1 to S phases,23 were not affected.
To validate the effect of p53 on cell cycle arrest, we carried out a rescue experiment by repressing p53 expression in T24‐CBX8 shRNA cells with siRNA (Fig. 5c) to determine whether their cell cycle distribution was affected. Figure 5d shows that the percentage of CBX8‐silenced cells in G2 decreased from 25.45% to 12.44% after transfection with p53‐siRNA, indicating that knocking down CBX8 inhibits cell cycle progression and is p53‐dependent. Meanwhile, protein levels of cdc2/cyclinB1 were elevated in CBX8‐silenced cells transfected with p53‐RNA, whereas the protein levels of CDK2, CDK4 and cdc25A were not affected (Fig. 5d).
Taken together, these findings indicate that knockdown of CBX8 results in cell cycle arrest at the G2/M phase by the p53 pathway.
Discussion
Despite extensive treatment involving radical cystectomy and adjuvant chemotherapy or radiotherapy, MIBC patients continue to show poor 5‐year survival (<35%) and a high risk for regional or distal metastasis.25 Complications following extended cystectomy such as urinary diversion and urinary tract infection may also result in a decrease in patient quality of life. Unlike other cancers such as liver cancer and prostate cancer, UCB lacks available biomarkers for diagnosis and monitoring. Although numerous research studies have been conducted to identify biomarkers for UCB, none have been widely used in the clinic because the biological behavior of bladder cancer is relatively complicated. Hence, the identification of novel biomarkers for UCB diagnosis and prediction is warranted.
CBX8, a member of the CBX family that consists of other members such as CBX2, CBX4, CBX6, and CBX7, is capable of repressing gene activity by interacting with RING and colocalizing with BMI1 in the PcG body.3, 26, 27 PcG is a protein complex which includes two core components of PC1 and PC2. Creppe et al. reported that CBX8 was part of the PC1 complex and coexisted with H3K27me3 on CBX8 target genes, which is apparently required for gene repression.28 Chung et al. reported that knockdown of CBX8 significantly decreases tumor sphere formation in mouse and human breast cancer cells, and the number of invasive colonies is significantly reduced when CBX8 is repressed.29 Recent studies have revealed the roles of CBX8 in cancer; however, how CBX8 functions in UCB remains unclear.
We initially assessed variations in CBX8 copy number in UCB using the online TCGA database. Our findings indicated that patients with a higher number of CBX8 CNV had a poor OS and DFS compared to patients with lower CBX8 CNV. This observation prompted us to investigate CBX8 expression patterns in UCB tissues and cell lines. qRT‐PCR and western blot analyses demonstrated that CBX8 mRNA and protein expression levels increased in eight paired MIBC tissues. This observation was further validated by IHC in another eight paired MIBC tissues and adjacent non‐tumor tissues. To correlate CBX8 expression with the clinicopathological features of MIBC patients, IHC was carried out in 152 MIBC specimens. Chi‐squared testing showed that CBX8 expression was correlated with TNM stage, and high CBX8 expression was associated with cancer progression as indicated by the T, N, and M stages. Kaplan–Meier survival analysis and log–rank test showed that patients with high CBX8 expression levels had poorer OS and RFS than patients with low CBX8 expression levels, and a T or N stage with a high CBX8 expression level was associated with poor OS compared to low CBX8 expression. Multivariate analysis revealed that CBX8 expression and TNM stage were independent prognostic indicators of MIBC.
Together, these results have motivated us to further investigate the critical role of CBX8 in UCB cell proliferation. We observed that knockdown of CBX8 decreases cell viability and clonogenicity of the UCB cell lines in vitro. Furthermore, xenografting nude mice proved that knockdown of CBX8 inhibits cell growth in vivo. A previous study has shown that knockdown of CBX8 represses cancer cell proliferation in vitro and in vivo and results in cell cycle arrest.4, 16 Thus, a flow cytometry assay was carried out to determine cycle distribution, which showed that CBX8 silencing accelerates the G1 to G2 phase transition, which possibly contributes to the inhibition of UCB cell proliferation. Western blotting indicated upregulation of p53 during CBX8 silencing, indicating that CBX8 that arrests the G2/M transition is p53‐dependent. To identify this molecular event involved in the G2/M transition, we measured the cell cycle distribution when p53 was repressed in T24‐CBX8 shRNA cells and found that the rate of transition from G1 to G2 decreased. This result thus indicates that CBX8 arrests the G2/M transition and is p53‐dependent.
In the current study, we mainly investigated the expression of CBX8 in MIBC tissues and identified that CBX8 expression is involved in UCB growth and is associated with UCB patient outcomes. Our findings showed that high expression of CBX8 plays a critical oncogenic role in UCB aggressiveness by promoting cancer cell proliferation by repressing the p53 pathway, and CBX8 could be used as a novel predictor for human UCB patients. Nevertheless, there are limitations that have to be addressed. For example, the detailed mechanisms of CBX8 involved in the carcinogenesis of UCB were not explored in this study. Second, we did not measure the discrepant CBX8 expression between MIBC and non‐MIBC tissues. Third, more clinical and laboratory efforts are required to determine the potential value of CBX8 as a biomarker for UCB.
Disclosure Statement
Authors declare no conflicts of interest for this article. All data in this study have been recorded at Sun Yat‐sen University Cancer Center (Number RDDB2017000109).
Abbreviations
- BAC
bacterial artificial chromosome
- CBX8
chromobox homolog 8
- CCK‐8
cell counting kit‐8
- CNV
copy number variation
- CRC
colorectal carcinoma
- DFS
disease‐free survival
- HR
hazard ratio
- IHC
immunohistochemistry
- MIBC
muscle invasive bladder cancer
- OS
overall survival
- PcG
polycomb group
- RFS
recurrence‐free survival
- TCGA
The Cancer Genome Atlas
- UCB
urothelial carcinoma of the bladder
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (No. 2017YFC1309001, 2016YFC1302305), the National Natural Science Foundation of China (No. 81572359, 81602233, 81672530), the Natural Science Foundation of Guangdong Province (2016A030310219), the Chinese Postdoctoral Science Foundation, the Foundation of Shenzhen science and Technology Innovation Committee (JCYJ20160429104111283).
Cancer Sci 108 (2017) 2166–2175
Funding Information
National Key Research and Development Program of China (No. 2017YFC1309001, 2016YFC1302305), National Natural Science Foundation of China (Nos. 81572359, 81602233, 81672530), Natural Science Foundation of Guangdong Province (2016A030310219), Chinese Postdoctoral Science Foundation, and Foundation of Shenzhen Science and Technology Innovation Committee (JCYJ20160429104111283).
Contributor Information
Fang‐jian Zhou, Email: zhoufj@sysucc.org.cn.
Dan Xie, Email: xied@mail.sysu.edu.cn.
References
- 1. Chen W, Zheng R, Baade PD et al Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66: 115–32. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin 2014; 64: 9–29. [DOI] [PubMed] [Google Scholar]
- 3. Bárdos JI, Saurin AJ, Tissot C, Duprez E, Freemont PS. HPC3 is a new human polycomb orthologue that interacts and associates with RING1 and Bmi1 and has transcriptional repression properties. J Biol Chem 2000; 275: 28785–92. [DOI] [PubMed] [Google Scholar]
- 4. Bienz M, Müller J. Transcriptional silencing of homeotic genes in Drosophila. BioEssays 1995; 17: 775–84. [DOI] [PubMed] [Google Scholar]
- 5. Kennison JA. The Polycomb and trithorax group proteins of Drosophila: trans‐regulators of homeotic gene function. Annu Rev Genet 1995; 29: 289–303. [DOI] [PubMed] [Google Scholar]
- 6. Orlando V, Paro R. Chromatin multiprotein complexes involved in the maintenance of transcription patterns. Curr Opin Genet Dev 1995; 5: 174–9. [DOI] [PubMed] [Google Scholar]
- 7. Pirrotta V. Chromatin complexes regulating gene expression in Drosophila. Curr Opin Genet Dev 1995; 5: 466–72. [DOI] [PubMed] [Google Scholar]
- 8. Gould A. Functions of mammalian Polycomb group and trithorax group related genes. Curr Opin Genet Dev 1997; 7: 488–94. [DOI] [PubMed] [Google Scholar]
- 9. Schumacher A, Magnuson T. Murine Polycomb‐ and trithorax‐group genes regulate homeotic pathways and beyond. Trends Genet 1997; 13: 167–70. [PubMed] [Google Scholar]
- 10. van Lohuizen M. Functional analysis of mouse Polycomb group genes. Cell Mol Life Sci 1998; 54: 71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dietrich N, Bracken AP, Trinh E et al Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A‐ARF locus. EMBO J 2007; 26: 1637–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bracken AP, Kleine‐Kohlbrecher D, Dietrich N et al The Polycomb group proteins bind throughout the INK4A‐ARF locus and are disassociated in senescent cells. Genes Dev 2007; 21: 525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li G, Warden C, Zou Z et al Altered expression of polycomb group genes in glioblastoma multiforme. PLoS One 2013; 8: e80970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang J, Wang G, Zhang M et al Paradoxical role of CBX8 in proliferation and metastasis of colorectal cancer. Oncotarget 2014; 5: 10778–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tan J, Jones M, Koseki H et al CBX8, a polycomb group protein, is essential for MLL‐AF9‐induced leukemogenesis. Cancer Cell 2011; 20: 563–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao W, Ou C, Qin J et al CBX8, a novel DNA repair protein, promotes tumorigenesis in human esophageal carcinoma. Int J Clin Exp Pathol 2014; 7: 4817–26. [PMC free article] [PubMed] [Google Scholar]
- 17. Broad Institute TCGA Genome Data Analysis Center: Analysis‐ready standardized TCGA data from Broad GDAC Firehose 2016_01_28 run. Broad Institute of MIT and Harvard. Dataset. [Cited 13 May 2016.] Available from URL: https://doi.org/10.7908/c11g0km9.
- 18. Camp RL, Dolled‐Filhart M, Rimm DL. X‐tile: a new bio‐informatics tool for biomarker assessment and outcome‐based cut‐point optimization. Clin Cancer Res 2004; 10: 7252–9. [DOI] [PubMed] [Google Scholar]
- 19. Lu J, Wei JH, Feng ZH et al miR‐106b‐5p promotes renal cell carcinoma aggressiveness and stem‐cell‐like phenotype by activating Wnt/β‐catenin signalling. Oncotarget 2017; 8: 21461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edge SB, Byrd DR, Carducci MA et al AJCC Cancer Staging Handbook, 7th edn. New York: Springer, 2010. [Google Scholar]
- 21. Zhang Z, Yu C, Li Y, Jiang L, Zhou F. Utility of SAM68 in the progression and prognosis for bladder cancer. BMC Cancer 2015; 15: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xie D, Sham JS, Zeng WF et al Correlation of AIB1 overexpression with advanced clinical stage of human colorectal carcinoma. Hum Pathol 2005; 36: 777–83. [DOI] [PubMed] [Google Scholar]
- 23. Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009; 9: 153–66. [DOI] [PubMed] [Google Scholar]
- 24. Zhan Q, Antinore MJ, Wang XW et al Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53‐regulated protein Gadd45. Oncogene 1999; 18: 2892–900. [DOI] [PubMed] [Google Scholar]
- 25. DeSantis CE, Lin CC, Mariotto AB et al Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin 2014; 64: 252–71. [DOI] [PubMed] [Google Scholar]
- 26. Maertens GN, El MS, Racek T et al Several distinct polycomb complexes regulate and co‐localize on the INK4a tumor suppressor locus. PLoS ONE 2009; 4: e6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Di CL, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 2013; 20: 1147–55. [DOI] [PubMed] [Google Scholar]
- 28. Creppe C, Palau A, Malinverni R, Valero V, Buschbeck M. A Cbx8‐containing polycomb complex facilitates the transition to gene activation during ES cell differentiation. PLoS Genet 2014; 10: e1004851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chung CY, Sun Z, Mullokandov G et al Cbx8 acts non‐canonically with Wdr5 to promote mammary tumorigenesis. Cell Rep 2016; 16: 472–86. [DOI] [PMC free article] [PubMed] [Google Scholar]


