Abstract
There is a general expectation that sexual selection should align with natural selection to aid the purging of deleterious mutations, yet experiments comparing purging under monogamy versus polygamy have provided mixed results. Recent studies suggest that this may be because the simplified mating environments used in these studies reduce the benefit of sexual selection through males and hamper natural selection through females by increasing costs associated with sexual conflict. To test the effect of the physical mating environment on purging, we use experimental evolution in Drosophila melanogaster to track the frequency of four separate deleterious mutations in replicate populations that experience polygamy under either a simple or structurally complex mating arena while controlling for arena size. Consistent with past results suggesting a greater net benefit of polygamy in a complex environment, two of the mutations were purged significantly faster in this environment. The other two mutations showed no significant difference between environments.
Keywords: Drosophila melanogaster, monogamy, polygamy, sexual selection
1. Introduction
Healthier and more vigorous (i.e. higher condition) individuals are likely to have greater reproductive success, generating sexual selection that should align with natural selection to favour alleles that increase condition [1]. Because most loci are likely to affect an individual's condition, much of the genome should be subject to sexual selection that promotes adaptation and the purging of deleterious mutations [2–5]. One approach that has been used to test for such a benefit of sexual selection compares the outcome of experimental evolution under polygamy (sexual selection permitted) with that under enforced monogamy (sexual selection absent). However, manipulations of mating systems may affect not only the opportunity for sexual selection, generally through males, but also natural selection through females. If male sexual attention is harmful to females (i.e. as a result of interlocus sexual conflict [6]) and males prefer high condition females, then the increased harm these females attract can reduce their fitness relative to low condition females, generating a ‘cost of sexual attractiveness’ that decreases the variance in female fitness and thus weakens natural selection through them [7]. The effects of polygamy may thus reflect opposing contributions from a strengthening of sexual selection through males and a weakening of natural selection through females. Two recent empirical studies that failed to detect a benefit of polygamy provided evidence consistent with a weakening of natural selection through females [8,9], demonstrating that such costs can be important.
The majority of monogamy versus polygamy studies have used various species of Drosophila. Relative to a more natural setting, the small and structurally simple environments common to such experiments may both reduce the benefit of sexual selection through males and cause a more pronounced weakening of natural selection through females. With respect to sexual selection, low condition males may suffer less when housed in close proximity with females that cannot escape and that are exhausted from persistent male harassment. In contrast, in a larger and more structurally complex environment it may be more difficult to locate and mate females, making male reproductive success more condition-dependent. Consistent with this, MacLellan et al. [10] showed that sexual selection against deleterious mutations in D. melanogaster males was stronger on average in a larger mating environment. With respect to natural selection through females, in small and structurally simple environments both the opportunity for male choice and the extent of male harm may be exaggerated, increasing the effects of differential male harm and thus causing a considerable weakening of natural selection. Indeed, in a recent study in D. melanogaster, Yun et al. [11] demonstrated that sexual interactions were more frequent, were biased toward high over low condition females, and overall male harm was greater in a smaller and simpler environment compared to a somewhat larger and more structurally complex one. The end result was that the opportunity for male choice (as occurs under polygamy) significantly weakened natural selection through females in the simple environment, consistent with past studies [7–9], but this effect vanished in the more complex environment [11].
If increased space and structural complexity reduce the cost of sexual attractiveness in females and/or strengthen sexual selection though males, then polygamy should be more beneficial to purging in a complex compared to a simple environment. We test this prediction in D. melanogaster using four deleterious mutations from a previous study that failed to detect a benefit of polygamy over monogamy with respect to their purging in a simple mating environment [8]. We manipulate environmental complexity only, both because we are interested in its effects alone and to better control for unintended changes in abiotic conditions that may be associated with manipulations of space (e.g. through the use of different containers). For each mutation, we maintain replicate populations in simple versus complex mating environments and track mutant allele frequency across generations.
2. Material and methods
We tested four mutations with recessive and visible phenotypic effects in adult D. melanogaster: three autosomal (brown, sepia, plexus) and one X-linked (white). Brown, sepia and white influence vision and eye colour whereas plexus affects wing veins. In a previous study, these mutations were found to be deleterious to non-sexual fitness but there was no evidence that the addition of sexual selection (in a simple environment) aided in their purging [8]. All four mutations were obtained from the Bloomington Stock Center and were separately introgressed into an outbred, laboratory-adapted stock population [8].
Experimental populations were founded by crossing individuals both within and between the mutant and non-mutant stock populations to create individuals carrying two, one or zero copies of the mutant allele (representing mutant homozygotes, heterozygotes, and non-mutant homozygotes respectively, recognizing that the non-mutant stock was outbred and may thus have been segregating more than one non-mutant allele at this locus). From these individuals, six replicate populations were founded for each mutation, at approximate Hardy–Weinberg proportions, by selecting 210 individuals of each sex and of the appropriate genotypes to achieve an initial mutant allele frequency of 0.66. However, due to limited availability of some genotypes, starting allele/genotype frequencies varied slightly among some populations (allele frequency range: 0.65–0.68; electronic supplementary material, table S1). Three of these populations were assigned to each of two treatments that manipulated the structural complexity of the arena in which mating occurred. In both cases an arena contained 30 males and 30 females and consisted of a 1650 ml round plastic container (electronic supplementary material, figure S1). The ‘simple’ arena held a single Petri dish of standard yeast-agar medium. The ‘complex’ arena followed that used by Yun et al. [11] and had pipe cleaners protruding into the interior space from the lid and the medium was distributed among five small cups.
Populations were maintained on a 12 L : 12 D cycle at 25°C and 60% humidity. Each generation, flies spent 6 days in their respective mating arena after which males were discarded and females from a given population were distributed evenly among 10 vials (15 from generation six onward to reduce larval densities) for egg laying. After 18–24 h, females were discarded. In generations subsequent to their founding, 11 days following the start of egg laying the resulting adult offspring were mixed among vials for a given population and 150 randomly chosen individuals of each sex were collected using light CO2 and distributed equally among each of five mating arenas for the 6-day mating phase. From the remaining offspring, 200 females were randomly scored for phenotype, allowing the homozygote mutant frequency to be tracked across generations.
During experimental evolution, mutant allele frequency was estimated 1–3 times in each population by placing 100 virgin females of wild-type phenotype in individual vials together with a homozygous mutant male. The pair was allowed to mate and lay eggs for 48 h and then discarded. Maternal genotype was inferred by the presence/absence of mutant offspring, denoting a mother that carried one or no mutant alleles respectively. The ratio of these two types of mothers was used to infer the genotypes of females with wild-type phenotype in the sample of 200 from that generation, allowing mutant allele frequency to be estimated.
Populations were maintained for 8–14 generations. To test whether final mutant allele frequency differed significantly between mating treatments, we fit a generalized linear model with binomial distribution and logistic link function, treating populations as replicates and including starting allele frequency as a covariate. Models were fit via maximum likelihood using the GLIMMIX procedure in SAS, version 9.4 (SAS Institute Inc., Cary, NC, USA). Results were qualitatively unchanged if instead, for each population, we calculated the change in allele frequency (initial − final) and then used a two-sample t-test to determine whether the magnitude of this change differed between treatments.
3. Results
For all four mutations, average allele frequency declined during experimental evolution (figure 1), implying selection against them. This is consistent with past results demonstrating deleterious effects of these mutations on non-sexual fitness [8]. Final mutant allele frequency was also significantly lower in the populations evolving in the complex compared to simple mating arenas for two mutations, plexus (F1,3 = 13.24, p = 0.036) and white (F1,3 = 37.47, p = 0.009). For the other two, final mutant frequency did not differ significantly between treatments: brown (F1,3 = 0.89, p = 0.416) and sepia (F1,3 = 0.97, p = 0.398). Changes in the frequency of mutant homozygotes across generations were concordant with these results for all mutations (figure 1).
Figure 1.
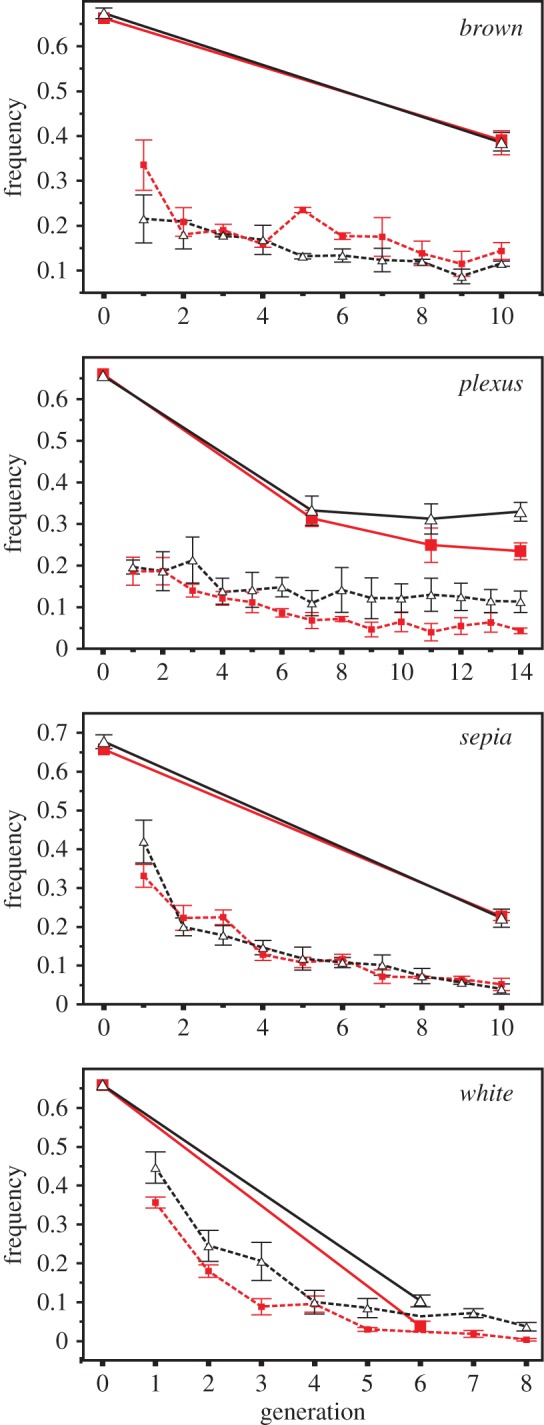
Average ± s.e. frequency of homozygote mutants (dashed lines) and mutant alleles (solid lines) from three replicate populations evolving under simple (black, open triangles) and complex (red, closed squares) mating arenas for four separate mutations. Points/s.e. for homozygote white in generation 6 are omitted for clarity. Statistical analysis for each mutation tested for a difference between treatments in the mutant allele frequency in the final generation (see Methods). (Online version in colour.)
4. Discussion
Faster purging is expected in the complex arenas if increased structural complexity reduces differential male harm of females (i.e. the ‘cost of sexual attractiveness’) and/or strengthens sexual selection though males. Yun et al. [11] showed that, in D. melanogaster, a joint increase in the volume and complexity of the mating environment eliminated the hampering effect of differential male harm on natural selection through females. Their complex arena was the same as we used here, strongly suggesting that the more rapid purging we observed was due, at least in part, to a reduction in the cost of attractiveness to females in the complex environment. Stronger sexual selection through males may have also contributed as successful reproduction in a more complex environment should depend on the condition-dependent expression of additional traits involved in locating and mating females. Increased space has been shown to have such an effect in D. melanogaster [10] although structural complexity has not been directly tested. We cannot exclude other differences in abiotic factors between our simple and complex arenas that may have altered the strength of selection on these mutations, although our use of the same containers should minimize such unintended differences.
Using a small and structurally simple mating environment, Arbuthnott & Rundle [8] found no evidence of faster purging of these four mutations under polygamy compared to monogamy, with evidence that polygamy actually hampered purging for sepia and some indication for brown and plexus as well. Taken together, the faster purging of white in the current study suggests that the net effect of polygamy in a complex environment is beneficial to the purging of this mutation, although direct comparison with a monogamy treatment would be required to confirm this. Whether the more rapid purging of sepia found here would produce a net positive effect of polygamy would also require further study.
Why an increase in complexity of the mating environment did not affect the purging of brown or sepia is not known. When variation in starting mutant frequency is accounted for, declines in both were slightly greater in the complex compared to simple environments, suggesting power might be an issue. The importance of different aspects of the mating environment (e.g. size versus complexity) may also vary among mutations. A parallel study tested the effect of simple versus complex mating environments on the purging of 22 gene disruption mutations [12] in D. melanogaster. In this experiment the mating arenas differed in both size and structural complexity; although the complex arena was the same as that used here, the simple arena was a standard fly vial. Results were similar in that more rapid purging was observed in the complex environment for 18 of the 22 mutations tested. The potentially stronger signal (18/22 compared to 2/4) could reflect an additional contribution of arena size and could also result from the different nature of the mutations tested (i.e. those exhibiting versus lacking highly visible phenotype effects). Additional mutations would need to be tested to explore these possibilities further. Nevertheless, the more rapid purging of two mutations in the current study implies that the structural complexity of the mating environment can matter for a class of mutations that have featured prominently in past work on this topic. It also suggests that care must therefore be taken in extrapolating results of laboratory studies to the effects of sexual selection in more natural environments.
Supplementary Material
Supplementary Material
Acknowledgements
We thank A.F. Agrawal for insightful discussions on the subject.
Data accessibility
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.42g20 [13].
Authors' contributions
H.D.R. conceived the project with input from J.C., and J.C., D.W. and H.S.S. conducted the experiments. H.D.R. analysed the data and wrote the paper, with input from the other authors. All authors agree to be held accountable for the content of this work and approve the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Funding
This project was supported by a grant from the Natural Sciences and Engineering Research Council of Canada.
References
- 1.Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London, UK: John Murray; [PMC free article] [PubMed] [Google Scholar]
- 2.Houle D. 1991. Genetic covariance of fitness correlates: what genetic correlations are made of and why it matters. Evolution 45, 630–648 ( 10.1111/j.1558-5646.1991.tb04334.x) [DOI] [PubMed] [Google Scholar]
- 3.Rowe L, Houle D. 1996. The lek paradox and the capture of genetic variance by condition dependent traits. Proc. R. Soc. Lond. B 263, 1415–1421. ( 10.1098/rspb.1996.0207) [DOI] [Google Scholar]
- 4.Whitlock MC. 2000. Fixation of new alleles and the extinction of small populations: drift load, beneficial alleles, and sexual selection. Evolution 54, 1855–1861 ( 10.1111/j.0014-3820.2000.tb01232.x) [DOI] [PubMed] [Google Scholar]
- 5.Whitlock MC, Agrawal AF. 2009. Purging the genome with sexual selection: reducing mutation load through selection on males. Evolution 63, 569–582. ( 10.1111/j.1558-5646.2008.00558.x) [DOI] [PubMed] [Google Scholar]
- 6.Arnqvist G, Rowe L. 2005. Sexual conflict. Princeton, NJ: Princeton University Press. [Google Scholar]
- 7.Long TA, Pischedda A, Stewart AD, Rice WR. 2009. A cost of sexual attractiveness to high-fitness females. PLoS Biol. 7, e1000254 ( 10.1371/journal.pbio.1000254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arbuthnott D, Rundle HD. 2012. Sexual selection is ineffectual or inhibits the purging of deleterious mutations in Drosophila melanogaster. Evolution 66, 2127–2137. ( 10.1111/j.1558-5646.2012.01584.x) [DOI] [PubMed] [Google Scholar]
- 9.Chenoweth SF, Appleton NC, Allen SL, Rundle HD. 2015. Genomic evidence that sexual selection impedes adaptation to a novel environment. Curr. Biol. 25, 1860–1866. ( 10.1016/j.cub.2015.05.034) [DOI] [PubMed] [Google Scholar]
- 10.MacLellan K, Whitlock MC, Rundle HD. 2009. Sexual selection against deleterious mutations via variable male search success. Biol. Lett. 5, 795–797. ( 10.1098/rsbl.2009.0475) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yun L, Chen PJ, Singh A, Agrawal AF, Rundle HD. 2017. The physical environment mediates male harm and its effect on selection in females. Proc. R. Soc. B 284, 20170424 ( 10.1098/rspb.2017.0424) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh A, Agrawal AF, Rundle HD. In press. Environmental complexity and the purging of deleterious alleles. Evolution. ( 10.1111/evo.13334) [DOI] [PubMed] [Google Scholar]
- 13.Colpitts J, Williscroft D, Sekhon HS, Rundle HD. 2017. Data from: The purging of deleterious mutations in simple and complex mating environments Dryad Digital Repository. ( 10.5061/dryad.42g20) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Colpitts J, Williscroft D, Sekhon HS, Rundle HD. 2017. Data from: The purging of deleterious mutations in simple and complex mating environments Dryad Digital Repository. ( 10.5061/dryad.42g20) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.42g20 [13].


