Abstract
Peptides are ubiquitous in nature and useful in many fields, from agriculture as pesticides, in medicine as antibacterial and antifungal drugs founded in the innate immune systems, to medicinal chemistry as hormones. However, the concept of peptides as materials was not recognized until 1990 when a self-assembling peptide as a repeating segment in a yeast protein was serendipitously discovered. Peptide materials are so called because they have bona fide materials property and are made from simple amino acids with well-ordered nanostructures under physiological conditions. These structures include well-ordered nanofibres, nanotubes and nanovesicles. These peptide materials have been used for: (i) three-dimensional tissue cell cultures of primary cells and stem cells, (ii) three-dimensional tissue printing, (iii) sustained releases of small molecules, growth factors, monoclonal antibody and siRNA, (iv) accelerated wound healing in reparative and regenerative medicine as well as tissue engineering, (v) used to stabilize membrane proteins including difficult G-protein coupled receptors and photosystem I for designing nanobiodevices, (vi) a few self-assembling peptides have been used in human clinical trials for accelerated wound healings in surgical uses and (vii) in human clinical trials for siRNA delivery for treatment of cancers. It is likely that these self-assembling peptides will open doors for more and more diverse uses. The field of self-assembling peptides is growing in a number of directions in areas of materials, synthetic biology, and clinical medicine and beyond.
Keywords: materials, self-assembling, peptides, three-dimensional tissue cell culture, sustained molecular releases
1. Introduction
The field of self-assembling peptides has undergone a substantial growth since early 1990s. There are numerous groups that are actively pursuing various self-assembling peptides for a wide range of applications including new biological materials, surface coating materials and semi-conducting devices, as well as a new class of antibiotics to combat drug resistance problems. The field is still expanding at an accelerating pace. It is impossible for me to capture the entire field in a few pages. The interested reader should read other original reports, more comprehensive reviews in the literature, and search online using ‘Self-assembling peptides’. Here I will only focus this article from my own personal experience and from my own laboratory research activities over the last 25 years (boxes 1 and 2).
Box 1. In order to initiate and promote a new research field, it is very important to bring people from diverse fields to be under one roof in order to openly discuss new ideas, to cross-fertilize a new field in a relaxed and somewhat isolated setting, such as the Capsis Hotel in Crete, Greece [73]. I did precisely that. Through organizing five small workshops, every other year, by inviting many leading scientists in their own fields to mix them in an intimate environment. There, these leading scientists in the world spent lot of time with young and energetic students, postdocs and researchers. These intimate workshops had a big impact and accelerated the development of a new field. All participants enjoyed and had fond memories of such small meetings even many years after.
Figure B1.
One of the Crete workshops on self-assembling peptides and proteins. I initiated and co-organized five workshops of ‘Self-assembly peptides and proteins’ in Capsis Hotel, Crete, Greece in summers of 1999, 2001, 2003, 2005 and passed the torch to Bill DeGrado and Joel Schneider in 2007. We invited people from diverse fields including peptide chemistry, protein science, materials science, mathematics, engineering, computer science, clinical medicine and more. The invited speakers include Alexander Rich, Nobel laureate Carleton Gajdusek, Mouad Lamrani, Benoit Mandelbrot, future Nobel laureate Martin Karplus, Alan Fersht, Chris Dobson, Susan Lindquist, Carol Robinson, Meir Wilchek, Carl Brädén, David Eisenberg, Joel Sussman, George Klein, Eva Klein, Peter Klein, Andrew Szent-Györgyi, Ingemar Ernberg, Xiaojun Zhao, Horst Vogel, Uwe Sleytr, Jeff Kelly, Bill DeGrado, Jonathan Weissman, Michael Hecht, Joseph Jacobson, Matthew Tirrell, Reza Ghadiri, Miheal Polymeropoulos, Alan Windle, Amalia Aggeli, Neville Boden, Anton Middleberg, Sheena Radford, Tim Deming, Vince Conticello, Stefan Wölfl, Anna Mitraki, Aphrodite Kapurniotu, Dek Woolfson, Tom McLeish, Ted Atkins, Alan Cooper, Nick Gay, Jianren Lu, Ehud Gazit, Sam Stupp, Joanna Aizenberg, Bruce Tidor, Phil Messersmith, Rutledge Ellis-Behnke, Amy Keating, Roger Kamm, Angie Belcher, Per Westmark, Gunilla Westermark, Joel Schnur, Yechiel Shai, Hilal Lashuel, Sylvie Blondelle, Donald Hilvert, Thomas Scheibel, David Kaplan, David Lynn, Dan Urry, Charlotte Hauser, Matt Tirrell, Dan Kirschner, Joel Schneider, Michal Dadlez, Lars Baltzer, Sandra Burkett, Kenji Yamamoto, Hisakazu Mihara, Shiroh Futaki, Tomi Sasaki, Itaru Hamachi, Susumu Yoshikawa, Margherita Morpurgo, Diego Mantovani, Georgios Archontis, Peter Butko, Yuan Luo, Eser Elcin, Murat Elcin, Thomas Zemb and Jack Aviv as well as many young and energetic students and postdocs including Rein Ulijn, Louise Serpell, Cait McPhee, Mark Krebs, Jesús Zurdo, Dominik Rünzler, Meital Reches, Brian Chow, Sawyer Fuller, Wonmuk Hwang, Steve Yang, Sloan Kulper, Davide Marini, Patrick Keilly, Andrea Lomander, Hidenori Yokoi, Beau Peelle, Andreas Mershin, Brian Cook, Liselotte Kasier and many more. These students and postdocs presented their research in front of the distinguished scientists. They exchanged ideas and received sound advice. These intimate workshops (approx. 70–80 people each time) mixed people from different disciplines and stimulated further interests in pursuing the new field. This self-assembling peptide material field is not only thriving and advancing at rapid pace but it also generates many novel products that have been commercialized in diverse fields.
Box 2. Since my unexpected discovery of the self-assembling peptide EAK16-II in yeast, zuotin, I made a detour to pursue something unusual. This has truly been a wonderful journey of curiosity-driven research that eventually led to some real-world applications. This journey itself was full of risk, met many downturns, setbacks, and ridicules, numerous grant applications were rejected and papers were turned down. But I persevered and persisted to pursue it. As Max Perutz remarked ‘In Science truth always wins’. We have come a long way, from initial surprises, puzzlement, no understanding at all, to in outline, not only gradually understand the design principles at the molecular level, the molecular and fine material structures, interactions of the peptides, the dynamic self-assembly behaviours, but also how we can further improve their designs [34–39]. From there, we not only subsequently expanded designer materials using 20 natural l-amino acids or some non-natural d-amino acids, but also we proceeded to optimize their sequence for three-dimensional tissue cell cultures [37–43] delivering bioactive therapeutics such as drugs and growth factors and antibodies [54–59] to make the scaffold for surgical uses [44–46] to research into repair the spinal cord [52,53]. Recent advances in fictionalization have also led to the development of better synthetic tissue culture bioactive scaffolds that promote cell proliferation, migration and differentiation for regenerative medicine and for three-dimensional tissue printing. Furthermore, these self-assembling peptides have become an enabling medical technology that will find a wide range of uses in surgery, emergency care, accelerated wound healing and more [28–36].
Figure B2.
The timeline of developing process and diverse applications of designer chiral self-assembling peptides. It spans over 20 years from the curiosity-driven research and serendipitous discovery of the first self-assembling peptide EAK16-II in 1990 to successful human clinical trials in 2011. In 1993, shortly after I discovered the self-assembling peptides, I met Marvin Caruthers at a conference and mentioned to him about my discovery. I was concerned about the expensive peptide materials for applications. He gave me a sound advice: ‘Find as many applications as possible, the economics will take care of itself’. Marvin Caruthers invented the phosphate amide nucleic acid synthesis chemistry, and now all nucleic acid synthesis uses his invention. In the 1980s, each nucleotide base cost $25, and to make 10 base oligonucleotides cost $250 in an academic laboratory! Now each base costs $0.05 and 10 base oligonucleotides cost $0.50. It is a reduction by 500 fold!.
Figure B3.
The timeline of self-assembling peptides discovery and development: from a serendipitous discovery to benefit of society. It took over 20 years from the initial discovery in 1990 to successful human clinical trials in 2011. There have been a lot of ups and downs. After gaining full understanding and knowledge of various detailed aspects of amino acid chemistry, peptide structure properties, dynamic molecular self-assembly behaviours of the self-assembling peptides, we started to design new peptides with active biological functions that further enhance their usefulness for a wide range of applications. Seizing the opportunity, I took the license and co-founded a startup biotech company 3DMatrix to translate the technology into new economy because a multi-billion-dollar US polymer company failed to move it forward, 6 years after it took the license and paid over a million dollars licensing fee. Currently, the self-assembling peptide nanofibre scaffold hydrogel has been used for surgical applications, slow releases, three-dimensional tissue printing and accelerated wound healing. To sum up the path from the discovery to a new medical technology: (i) to stimulate, encourage and support curiosity-driven scientific research in order to gain scientific knowledge, (ii) to take risk and not be afraid of failure to translate scientific knowledge and research into enabling technologies, and (iii) be undeterred and be persistent to develop the knowledge-based economy for the benefit of mankind.
2. Discovery of the first self-assembling peptide
While I was working on yeast genetics and protein chemistry and trying to understand a left-handed Z-DNA structure in the laboratory of Alexander Rich at Massachusetts Institute of Technology in 1989, I identified a protein zuotin (Zuo in Chinese means left and there is a Z in it) for its ability to bind to left-handed Z-DNA in the presence of 400-fold molar excess of sheared salmon DNA that contains ubiquitous right-handed B-DNA and other form DNA structures. In zuotin, there is a curious repeat segment with the sequence n-AEAEAKAKAEAEAKAK-c, thus I named EAK16 for its amino acid composition and peptide length (figure 1) [1,2].
Figure 1.

The simple molecular models of the designer amphiphilic self-assembling peptides. These peptides have two distinctive sides, one hydrophobic and the other hydrophilic. The hydrophobic side forms a double sheet inside of the fibre and the hydrophilic side forms the outside of the nanofibres that interact with water molecules, forming an extremely high water content hydrogel that contains as high as 99.9% water. At least three types of molecules can be made, with −, +, −/+ on the hydrophilic side. The individual self-assembling peptide molecules are approximately 6 nm long. The first such peptide, EAK16-II, was discovered from a yeast protein, zuotin [1–2]. This peptide inspired us to design a large class of self-assembling peptide designer motifs. When they are dissolved in water in the presence of salt, they spontaneously assemble into well-ordered nanofibres and then further into nanofibre scaffold hydrogels. The right panel shows the EM image of yeast cells. Yeast cells are approximately 5–10 µm in size depending their cell phases, resting, growing and dividing phases; resting phase, cells are smaller and in the growing and dividing phases, cells are larger.
I initially used computer modelling and primitive simulation to examine this EAK16 sequence. These results showed the EAK16 structure to be a typical α-helix: its lysines and glutamic acids on the side-chains with i, i + 3 and i, i + 4 arrangements that could form potential ionic bonds. This prediction is in agreement with two previously published papers that similar peptides with A, E, K compositions form stable α-helices under a variety of conditions [3,4].
I wondered if this peptide could be synthesized and studied to satisfy my scientific curiosity. I asked my mentor Alexander Rich if I could order the EAK16 peptide for further study that would cost over $1000. Alex Rich asked me ‘Are you certain you want to pursue these new experiments?’ I immediately replied, ‘Yes’ without hesitation. He agreed to order the actual EAK16 peptide. I quickly ordered a custom synthesis through the Biopolymers Laboratory at Massachusetts Institute of Technology.
I believe if my discovery had been made in another laboratory other than Alex Rich's, the observation perhaps would not have been pursued further because there was no grant funding to support it. Thus, the entire self-assembling peptide materials field would have had to wait for a few more years. Alex said to me ‘You have made an unauthorized discovery’. Alex Rich always had an open mind and allowed people in his laboratory to pursue unexpected discoveries. Furthermore, his laboratory was well funded from a variety of sources. For Alex Rich's open-mindedness, his laboratory made numerous discoveries and contributed a lot of fundamental knowledge to mankind [5].
At the time in 1990, I never thought and predicted that my curiosity would lead me into an entirely unexplored field of peptide materials and nanobiotechnology for over a decade. I decidedly made a detour of my research on Z-DNA biology and went into an uncharted territory to pursue the self-assembling peptides. The late Ephraim Katzir of Weizmann Institute of Science invited me to visit him several times and remarked to me one time during my visit at his home: ‘You have opened a new direction for peptide materials research, no one had thought about’. Katzir was a great pioneer peptide biochemist in the 1940s in John Edsall's laboratory at Harvard University. They together contributed enormously to our understanding of peptide science in 1940s–1950s.
When the EAK16 peptide was first studied following the reported method using Aviv circular dichroism spectroscopy, an unexpected result occurred. Instead of showing a spectrum of the computed modelled α-helix, the peptide showed an exceedingly stable β-sheet structure. The EAK16 not only was stable in pH 1–11 with little circular dichroism spectrum changes, but also was resistant to heat treatment, 1% sodium dodecylsulfate, 8 M urea and 6 M guanidine HCl [6]. Upon adding salt, a thin layer membrane-like substance and transparent materials occurred in the Petri dish visible to the naked eye [2,7].
Alex Rich was a very close friend of the late legendary Francis Crick since early 1950s. They published several collagen structure papers together in the 1950s and early 1960s. Alex Rich introduced me to Francis Crick during his visit to Alex Rich's laboratory in September 1988, approximately three months after I joined Alex Rich's laboratory.
In August 1991, I visited Francis Crick in his office at the Salk Institute and I told him about my discovery. Crick first suggested to do X-ray diffraction. I told him this EAK16 peptide does not form crystals easily as I had already tried several times. He then suggested me to look at it under a scanning electron microscope (SEM). I did in late 1991 and early 1992. It took me more than 1 year to understand how the seemingly soluble short peptides underwent self-assembly to form well-ordered nanofibre scaffold that was visible to the naked eye. My colleagues and I published the paper where we reported the first self-assembling peptide that formed visible nanofibre material [2,6,7]; since then self-assembling peptide field has been expanded in a number of directions over past two decades [8–72].
3. Self-assembling peptide materials
Designer materials that are self-assembled molecule-by-molecule or atom-by-atom to produce novel supramolecular architectures belong to ‘bottom-up’ instead of ‘top-down’ approach. This approach requires a deep understanding of individual molecular building blocks, their structures and dynamically assembly properties [10,13,24–27]. These self-assembled materials have become an integral part of broad spectra of designer and fine materials.
These self-assembling peptides have alternating hydrophobic, namely, alanine, valine, leucine, isoleucine and phenylalanine, and hydrophilic sides, which include positively charged lysine, arginine, histidine and negatively charged aspartic acids and glutamic acids [10,13,24–27].
The complementary ionic sides have been classified into modulus I, II, III, IV and mixed moduli. This classification is based on the hydrophilic surface of the molecules that have alternating positively and negatively charged amino acid residues, either alternating by 1, 2, 3, 4 and so on. For example, charge arrangements for the different moduli are as follows: modulus I, −+−+−+− +; modulus II, −−++−−++; modulus III, −−−+++; and modulus IV, −−−−++++ (figures 1 and 2) [28]. The charge orientation can also be designed in reverse orientations that yield entirely different molecules with distinct molecular behaviours. These well-defined sequences allow them to undergo ordered self-assembly, resembling some situations found in well-studied polymer assemblies. This simple idea is the basis of the self-assembling peptide building blocks.
Figure 2.

Self-assembling peptide RADA16-I nanofibre scaffold hydrogel. (Upper panel) Amino acid sequence of RADA16-I, molecular model of a single RADA16-I peptide, the calculated peptide dimensions are approximately 6 nm long depending on end capping, 1.3 nm wide and 0.8 nm thick; hundreds of thousands or millions of individual peptides self-assemble into a nanofibre depending on the fibre length as revealed by the SEM image of RADA16-I nanofibre scaffold. Note the scale bar, 0.5 μm or 500 nm (SEM image courtesy of Fabrizio Gelain). RADA16-I peptide forms nanofibres in aqueous solution that further form hydrogel with extremely high water content (99.5–99.9% w/v water).
4. Basic chemical properties of the self-assembling peptide systems
The peptide synthesis using conventional mature solid phase or solution peptide synthesis chemistry has become more and more affordable. The peptide production cost directly correlates with the motif length, purity of peptides, skill of the manufactures and the chirality of amino acids. Most self-assembling peptides are readily soluble in water because their amino acid molecules consist of alternating hydrophilic and hydrophobic regions that contain 50% charged residues with distinct polar and non-polar surfaces and periodic repeats of two to four times. The self-assembly is accelerated by millimolar salt concentration under physiological pH conditions to form ordered nanostructure such as nanofibre, nanotube and nanovesicle [2,13,21–27].
For example, RADA16-I and RADA16-II with arginine and aspartate residues replacing lysine and glutamate were designed. The alanines form overlapping hydrophobic interactions in water; both positive Arg and negative Asp charges are packed together through intermolecular ionic interactions in a checkerboard-like manner. They self-assemble to form nanofibres approximately 10 nm in diameter, and theses nanofibres interweave into scaffolds that retain extremely high hydration, greater than 99% in water (1–10 mg ml−1, w/v; figure 3) [13].
Figure 3.

Peptide RADA16-I. (a) Amino acid sequence and molecular model of RADA16-I, the dimensions are approximately 5 nm long, 1.3 nm wide and 0.8 nm thick. Atomic force microscopy images of RADA16-I nanofibre scaffold are shown here (b–d). The sizes of the entire images are: (b) 8 µm2, (c) 2 µm2 and (d) 0.5 µm2. Note the different height of the nanofibre, approximately 1.3 nm in (d) thus suggesting a double layer structure. Photographs of RADA16-I hydrogel at various conditions: (e) 0.5 wt% (pH 7.5), (f) 0.1 wt% (pH 7.5, Tris–HCl), (g) 0.1 wt% (pH 7.5, PBS) before sonication and (h) re-assembled RADA16-I hydrogel after four times of sonication (image courtesy of Hidenori Yokoi) [13].
The formation of the scaffold and its mechanical properties are influenced by several factors: (i) amino acid sequence, (ii) the level of hydrophobicity, (iii) length of the peptides, and (iv) self-assembling time. For example, the extent of the hydrophobic residues Ala, Val, Ile, Leu, Tyr, Phe, Trp (or single letter code, A, V, I, L, Y, P, W) can significantly influence the mechanical properties of the scaffolds and the speed of their self-assembly. The higher the content of hydrophobicity, the easier it is for scaffold formation and the better for their mechanical properties [11].
5. The proposed general assembly of nanofibre formations
The detailed mechanism of how self-assembling peptides self-organize themselves in water at low concentration still remains inadequately understood at present; nevertheless, various self-assembling peptides as biological materials have been used for diverse applications.
Several plausible ideas for different nanostructures have been proposed. First, for nanofibres, a molecular model to interpret the formation of EAK16 and RADA16 was proposed. These two peptides are representative of a class of peptides that undergo self-assembly into ordered nanofibres: (i) numerous intermolecular hydrogen bonds between the peptides –C=O----H–N– in the conventional β-sheets on the peptide backbones, (ii) the side chains of positively and negatively charged residues form intermolecular ionic bonds in a checkerboard manner, (iii) hydrophobic interactions between the peptides from the amino acid residues are pushed by water molecules, and (iv) alternating polar and non-polar surface interaction. It is known that salt ions facilitate the self-assembly; however, it is not yet clear where the monovalent ions may coordinate the charged residues in a higher order of geometry [2,13].
6. Dynamic reassembly of self-assembling peptides
The self-assembling peptides form stable β-sheet structure in water. The interactions between the peptides in β-sheets are: (i) non-covalent hydrogen bonds along the backbones, (ii) arrays of ionic + and – charge interactions, (iii) alanine hydrophobic interactions and van der Waals interactions, and (iv) water-mediated hydrogen bond formations. Thus, the nanofibres can be disrupted mechanically using sonication [13]. The self-assembling process is reversible and dynamic because these peptides are short and simple. Numerous individual peptides can be readily self-organized through these weak interactions. However, they can undergo dynamic reassembly repeatedly (figure 4), similar to the material self-healing process. The driving energy of the assembly in water is not only through hydrophobic van der Waals interactions, but also the arrays of ionic interactions as well as the peptide backbone hydrogen bonds. This phenomenon can be further exploited for production and fabrication of many self-assembling peptide materials [13].
Figure 4.

AFM images of RADA16-I nanofibre at various time points after sonication. The observations were made using AFM immediately after sample preparation. (a) 1 min after sonication, (b) 2 min, (c) 4 min, (d) 8 min, (e) 16 min, (f) 32 min, (g) 64 min, (h) 2 h, (i) 4 h and (j) 24 h. Note the elongation and reassembly of the peptide nanofibres over time. By approximately 1–2 h, these self-assembling peptide nanofibres have nearly fully reassembled (image courtesy of Hidenori Yokoi, PNAS) [13]. The process (sonication and re-assembly) can be repeated many times, thus truly demonstrating the power of self-assembly.
Unlike processed polymer microfibres in which the fragments of polymers cannot undergo reassembly without addition of catalysts or through material processing, the supramolecular self-assembly and re-assembly event is likely to be widespread in many unrelated fibrous biological materials where numerous weak interactions are involved. Self-assembly and re-assembly are a very important property for fabricating novel materials, and it is necessary to fully understand its detailed process in order to design and to improve biological materials.
Atomic force microscopy (AFM) images revealed that the nanofibres range from several hundred nanometres to a few micrometres in length before sonication. After sonication, the fragments were broken into approximately 20–100 nm. The kinetics of the nanofibre reassembly is closely followed at 1, 2, 4, 8, 16, 32 and 64 min as well as 2, 4 and 24 h (figure 4). The nanofibre length reassembly is a function of time: by 2 h, the peptide nanofibres have essentially reassembled to their original length. The β-sheet structure had little change because the β-sheets at the molecular level remain unchanged despite the nanofibre length change [13].
7. Molecular modelling of the self-assembly process
We carried out molecular modelling of the self-assembly process. For molecular modelling clarity, the RADA16-I β-sheet is presented as a non-twisted strand. It is known that these peptides form stable β-sheet structure in water. They form intermolecular hydrogen bonding on the peptide backbones, similar to the conventional β-sheet. They also have two distinctive sides, one hydrophobic with array of overlapping alanines (figure 5, green colour sandwiched inside), similar to what is found in silk fibroin or spider silk assemblies; the other side of the backbones has negatively charged (−) amino acids, represented as red, and positively charged (+) amino acids, represented as blue.
Figure 5.

Based on the experimental observations (figures 3 and 4), we proposed molecular sliding diffusion model for dynamic reassembly of a single-peptide nanofibre consisting of thousands of individual peptides. When the peptides self-assemble into stable β-sheets in water, they form intermolecular hydrogen bonds along the peptide backbones. The β-sheet structure has two distinctive sides, one hydrophobic with an array of alanines and the other with negatively charged and positively charged amino acids. These peptides form anti-parallel β-sheet structures. The alanines form overlap packed hydrophobic interactions in water, a structure that is found in silk fibroin from silkworm and spiders. On the charged sides, both positive and negative charges are packed together through intermolecular ionic interactions in a checkerboard-like manner. When the fragments of nanofibre first meet, the hydrophobic sides may not fit perfectly but with gaps. However, the non-specific hydrophobic interactions permit the nanofibre to diffuse in a sliding manner along the fibre in either direction that minimizes the exposure of hydrophobic alanines and eventually fill the gaps. The sliding diffusion phenomenon was also proposed for nucleic acids of polyA and polyU in 1956 [14,15]. For clarity, these β-sheets are not presented as twisted strands. Colour code: green, alanines; red, negatively charged amino acids; blue, positively charged amino acids (image courtesy of Hidenori Yokoi) [13].
The alanines form packed hydrophobic interactions in water, which can be disrupted mechanically during sonication. However, these hydrophobic cohesive ends could find each other quickly in water because the exposure of hydrophobic alanine arrays to water is energetically unfavourable. Since the hydrophobic alanines' interaction is non-specific, they can slide diffuse along the nanofibre, like trains sliding along train tracks. The same sliding diffusion phenomenon was also observed in early studies of nucleic acids where polyA and polyU form complementary base pairings that can slide diffuse along the double helical chains [14,15]. If however, the bases are heterogonous, containing G, A, T, C, then the bases cannot undergo sliding diffusion. Likewise, if the hydrophobic side of the peptides does not always contain alanine, containing residues such as valine and isoleucine, it would become more difficult for sliding diffusion to occur due to the structural constraint.
On the charged side, both positive and negative charges are packed together through intermolecular ionic interactions in a checkerboard-like manner. Likewise, the collectively complementary positive and negative ionic interactions may also facilitate reassembly. Similar to restriction-digested DNA fragments, these nanofibre fragments could form various assemblies, like blunt and protruding ends. The fragments with various protruding ends as well as blunt ends can reassemble readily through hydrophobic and ionic interactions (figure 5).
8. General self-assembling peptide materials
There are general peptide materials that are generic with no specific biological active motifs, in other words, not tailor-made for a specific purpose. In an analogy, the general peptide materials are like pieces of bread without butter, cream spread or jam; or a bowl of pure rice without any added flavours.
There are also several classes of peptide materials: (i) Lego-like peptides: they self-assemble to form nanofibres; (ii) lipids-like or surfactants-like peptides: they self-assemble into nanotubes and nanovesicles; (iii) biological paint-like peptides: they self-assemble on surfaces to modify the surfaces on a nanometre scale.
9. Self-assembling peptide nanofibres
The first self-assembling peptide EAK16 and subsequently designed RADA16 are general peptides without any active motifs. Here, RADA16 made the first milestone for new economic development. RADA16 has become commercial products for various three-dimensional tissue cell cultures. It has also been successfully developed for various applications as medical devices including surgical applications, for instance, to stop bleeding in a few seconds.
These peptides form stable second structures including α-helix and β-sheet. But some peptide secondary structures are more dynamic under various environmental factors: (i) the amino acid sequence arrangements (even with the identical composition), (ii) the molecular size of the peptide, (iii) peptide concentration, (iv) pH of the solution, (v) temperature, (vi) the medium composition, such as solvent or substrate, (vii) ionic strength, and (viii) the presence of de-naturation agents, such as sodium dodecylsulfate, urea and guanidium.HCl. These factors can significantly influence the dynamic behaviours of peptide secondary structures and also affect the process of self-assembly [2,10,11,13,28].
10. Specific self-assembling peptide materials
Although self-assembling peptides are promising materials, they show no specific cell interaction because their sequences are not naturally found in living systems. In order to introduce the specific needs for individuals, the next logical step is to tailor-make the materials by directly coupling biologically active and functional peptide motifs onto the generic peptide (figure 6). Accordingly, the second generation of designer scaffolds will have significantly enhanced interactions with cells and tissues as well as other molecules.
Figure 6.

Molecular and schematic models of the designer peptides and of the scaffolds. Direct extension of the self-assembling peptide sequence by adding different functional motifs. Light turquoise cylinders represent the self-assembling backbone and the yellow, pink and tan lines represent various functional peptide motifs. Molecular model of a self-assembling peptide nanofibre with functional motifs flagging from both sides of the double β-sheet nanofibres. Either few or more functionalized and active peptide can be mixed at the same time. The density of these functionalized peptides can be easily adjusted by simply mixing them in various ratios, 1 : 1–1 000 000 or more before the assembling step. They then will be part of the self-assembled scaffold [37–43]. The left panel is the enlargements of a small part of the peptide nanofibres in the right panel. The enlarged parts show more details of co-assembly of peptides with biologically active motifs, either different peptides (upper left panel), or different proteins (lower left panel: A–C).
We have made a wide range of designer self-assembling peptide nanofibre scaffolds for both three-dimensional tissue cell cultures and sustained molecular releases [7,8,12,16–20,37–43,50–59].
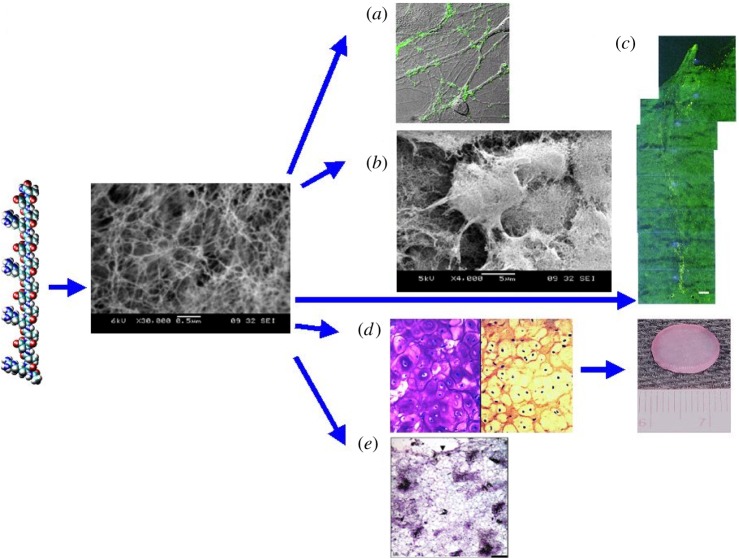
The designer self-assembling peptides with additional active motifs not only play a much more important biological role, but they also become more attractive biological materials with added functionalities and values. Although additional active motifs were appended to the generic peptide backbone, the nanofibre structures remain quite similar, as shown by SEM (figure 7) [37,38].
Figure 7.
SEM images of Matrigel and various designer peptide nanofibre scaffolds. (a) Matrigel, (b) RADA16, (c) RADA16-BMHP1, (d) RADA16-BMHP2 nanofibre scaffolds assembled in PBS solutions. Matrigel nanostructures are comparable in size to nanofibres found after self-assembly of the designer peptides. Clusters and aggregates of the unidentified naturally derived proteins in Matrigel (a) are absent in the pure peptide scaffolds shown in (b–d). The interwoven nanofibres are approximately 10 nm in diameter in each of the peptide scaffolds with approximately 5–200 nm pores. The appended functional motifs did not prevent peptide self-assembly [37,38].
In some cases, when the active sequences become longer, the self-assembling speed and mechanical property are affected [39–42]. The three-dimensional nanofibre materials are superb scaffolds for diverse tissue cells, not only for differentiation, proliferation, migration, but also for long-term maintenance [39–42].
Interestingly, the nanofibre structure is similar to the widely used Matrigel nanostructure except that Matrigel is not pure, but with many other substances and growth factors in it (figure 7a).
In order to fully understand how cells behave in the three-dimensional microenvironment, three-dimensional gradient diffusion, three-dimensional cell migration and three-dimensional cell–cell contact interactions and in tissue engineering and regenerative medicine, it is important to develop a well-controlled three-dimensional tissue culture system where every single ingredient is known.
For over 100 years since the Petri dish was invented and used for tissue culture studies, almost all tissue cells have been studied on the two-dimensional Petri dish and various formats of coated two-dimensional surfaces. However, this two-dimensional surface is rather unlike three-dimensional tissue and the body's microenvironment [30]. Thus, it is important to develop a true three-dimensional microenvironment to mimic the real tissue and body situation. The commonly used biomaterials are inadequate due to their microfibre and micropore size. Animal derived collagen gel and Matrigel contain other residue materials that are not always adequate for finely controlled studies. Thus, designer scaffolds become more and more desirable [30] (figure 8).
Figure 8.
SEM image of adult mouse NSCs embedded in designer peptide nanofibre scaffold RADA16-BMHP1 (1% v/w) after 14 day in vitro cultures. Cluster of three visible mouse NSCs embedded in three-dimensional self-assembling RADA16-BMHP1. It is important to point out that the nanoscales of peptide scaffold and the extracellular matrix made by cells are indistinguishable. This is totally unlike the many processed biopolymer microfibres that are often 10–50 µm in diameter, which is 1000–5000 larger (many more orders of magnitude larger in three dimensions) than the self-assembling peptide nanofibres [37,38].
In order to achieve fine-tuning and control, we have designed tailor-made peptides to suit specific individual needs of studies or applications through appending specific active motifs onto the basic peptide, such as RADA16-I or EAK16-II.
From a synthetic organic chemistry aspect, both peptide C- or -N termini could be attached to the modified motifs. However, the functional motifs should be always located on the C-termini, because solid-phase peptide synthesis initiates synthesis from the C-termini and proceeds towards N-terminus. The longer the peptide sequence is made, the more probable the coupling error would occur. Thus, in order to avoid peptide synthesis errors, the active sequence motifs should always be at the C-terminus without exception (figure 6) [37–42].
Usually a spacer comprising 2-glycine residues is added to guarantee flexible and correct exposure of the motifs to cell surface receptors. If one combines a few designer peptides with different active motifs, these different functional motifs in various ratios can be incorporated in the same scaffold (figure 6). Upon exposure to solution at neutral pH, the functionalized sequences self-assemble, leaving the added motifs on both sides of each nanofibre (figure 6). Nanofibres take part in the overall scaffold, thus providing functionalized microenvironments with specific biological stimuli (figure 6).
Self-assembling peptide scaffolds with functional motifs can be commercially produced at a reasonable cost. Thus, this method can be readily adopted for widespread uses including the study of cell interactions with their local- and micro-environments, cell migrations in three dimensions, tumour and cancer cell interactions with normal cells, cell processes and neurite extensions, cell-based drug screen assays and other diverse applications.
We have produced different designer peptides from a variety of functional motifs with different lengths. We showed that the addition of motifs in some cases to the self-assembling peptide RADA16-I did not significantly inhibit self-assembling properties. Furthermore, one can combine the RADA16-I nanofibre with the active designer self-assembling peptides by mixing the modified peptides. Although their nanofibre structures are indistinguishable from the RADA16-I scaffold, the appended functional motifs significantly influence cell behaviours.
Using the designer self-assembling peptide nanofibre system, every ingredient of the scaffold can be defined. Furthermore, it can be combined with multiple functionalities including soluble factors. Cells reside in a three-dimensional environment where the extracellular matrix receptors on cell membranes can bind to the functional ligands appended to the peptide scaffolds. It is likely that higher tissue architectures with multiple cell types, rather than monolayers, could be constructed using these designer three-dimensional self-assembling peptide nanofibre scaffolds.
Even if only a fraction of functionalized motifs on the three-dimensional scaffold are available for cell receptor binding, cells may likely receive more external stimuli than when in contact with coated two-dimensional Petri dishes or RGD-coated (or other motifs) polymer microfibres, which is substantially larger than the cell surface receptors and in most cases, larger than the cells themselves. There, cells are not in a real three-dimensional environment, but rather, they are on a two-dimensional surface wrapping around the microfibre polymers with a curvature that depends on the diameter of the polymers. It is plausible in a two-dimensional environment, where only one side of the cell body is in direct contact with the surface, that receptor clustering at the attachment site may be induced; on the other hand, the receptors for growth factors, cytokines, nutrients and signals may be on the other sides that are directly exposed to the culture media. Perhaps cells may become partially polarized. In the three-dimensional environment, the functional motifs on the nanofibre scaffold surround the whole cell body in all dimensions. Thus, growth factors may form a gradient in three-dimensional nanoporous microenvironment.
In our search for additional functional motifs, we found that a class of bone marrow homing peptides BMHP is one of the most promising active motifs for stimulating adult mouse neural stem cell (NSC) adhesion and differentiation. This observation suggests a new class of designer self-assembling peptides for three-dimensional cell biology studies [38].
We attached several functional motifs including cell adhesion, differentiation and bone marrow homing motifs on the C-termini as well as matrix metalloproteinase-sensitive cleavage motif in the middle of the peptide [43] to modulate peptide mechanical property. We used them to study NSC, and these functionalized peptides underwent self-assembly into nanofibre structures as well. More functionalized self-assembling peptides have been shown to promote specific cellular responses and long-term cell survival, proliferation, migration and morphological differentiation, and thus these peptide scaffolds are excellent for many applications [37–43].
Designer self-assembling peptide scaffolds also show interesting interactions with functional proteins for study of molecular releases since the release kinetics results suggested that protein diffusion through nanofibre scaffolds primarily depended on the size of proteins [53–58].
11. Diverse uses of self-assembling peptide nanofibres
A wide range of diverse uses in various areas has been developed from these self-assembling peptide materials. They include: (i) reparative and regenerative medicine, (ii) accelerated wound healing in human clinical and surgical applications, (iii) sustained molecular releases (small molecules, protein growth factors and monoclonal antibodies), (iv) stabilization of diverse membrane proteins in solution and dry surface for nanobiotechnological device fabrications, and (v) production of G protein-coupled receptors. Some of they are highlighted below.
12. Reparative and regenerative medicine
The self-assembling peptides are easy to use in tissue engineering, wound healing, addressing chronic wound problems and regenerative medicine (figure 9). Holmes et al. showed the primary neurites extend to great distance from the cell body and form active synapses with very high density in RADA16-I nanofibre scaffold (figure 9a) [8]. Inspired by this finding, Ellis-Behnke and colleagues used RADA16-I to repair injured rat and mouse brain structures, and their result showed the peptide scaffold hydrogel was excellent, not only for axon regeneration through the site of an acute brain injury, but also to knit the injured brain tissue together seamlessly (figure 9c) [44]. This work represents an enabling nanobiomedical technology for tissue brain repair and restoration (figure 9) [44].
Figure 9.
From designer peptide to scaffold to tissues. (a) Active synapses on the peptide surface. Primary rat hippocampal neurons form active synapses on peptide scaffolds. The confocal images show bright discrete green dot labelling indicative of synaptically active membranes after incubation of neurons with the fluorescent lipophilic probe FM-143. FM-143 can selectively trace synaptic vesicle turnover during the process of synaptic transmission. The active synapses on the peptide scaffold are fully functional, indicating that the peptide scaffold is a permissible material for neurite outgrowth and active synapse formation. (b) Adult mouse NSCs embedded in three-dimensional scaffold (image courtesy of Fabrizio Gelain). (c) Brain damage repair in hamster. The peptide scaffold was injected into the optical nerve area of brain that was first severed with a knife (image courtesy of Rutledge Ellis-Behnke). The gap was sealed by the migrating cells after a few days. A great number of neurons form synapses. (d) Peptide KLD12 (KLDLKLDLKLDL), chondrocytes in the peptide scaffold and cartilage. The chondrocytes are stained with TB showing abundant GAG production (left panel) and antibody to type II collagen demonstrating abundant type II collagen production (right panel). A piece of pre-moulded cartilage with encapsulated chondrocytes in the peptide nanofibre scaffold. The cartilage formed over a three to four week period after the initial seeding of the chondrocytes (image courtesy of John Kisiday). (e) Von Kossa staining showing transverse sections of primary osteoblast cells on HA-PHP-RADA16-I self-assembling peptide nanofibre scaffold. Scale bar, 0.1 mm. The intensely stained black areas represent bone nodules forming.
Ellis-Behnke and colleagues found that the self-assembling peptide scaffolds are useful in repairing the damaged brain [44]. They also found that the peptide scaffold hydrogel instantly stopped bleeding in a few seconds during the procedure of repairing injured brain. They then went on to study homeostasis of brain, spinal cord, femoral artery and liver of rat [45,46].
The early self-assembling peptide scaffold work has inspired others to expand it to different kinds of tissues, organs and animals [48–53]. RADA16-I functionalized with biologically active motifs also induced favourable repair in injured spinal cords [52,53].
In order to better understand the individual molecular and material building blocks, their structures, assembly properties, dynamic behaviours and application for rapid homeostasis, we also used d-amino acids; the chiral self-assembling peptide d-EAK16 also forms three-dimensional nanofibre scaffold. This study not only provided insights into understanding the chiral assembly properties for rapid homeostasis, but also to aid in further design of self-assembling d-form peptide scaffolds for clinical trauma emergency [47–48].
13. Sustained molecular releases
Since the self-assembling peptides form nanofibre scaffolds with nanopores and the process is dynamic over the time, it is possible to use it load small molecules and large proteins for control drug delivery in molecular medicine [54–60].
Using EAK16 II, RAD16-II and RAD16-I as a model to slow release hydrophobic molecules showed that these types of self-assembling peptide scaffolds encapsulated hydrophobic drug. Various dye molecules including phenol red, bromophenol blue, 8-hydroxypyrene-1,3,6-trisulfonic acid tri-sodium salt, 1,3,6,8-pyrenetetrasulfonic acid tetra-sodium salt, and Coomassie Brilliant Blue G-250 through RADA16 hydrogels, providing an alternative route of controlled release of small molecules [54]. Furthermore, nanofibre encapsulated camptothecin or ellipticine have been confirmed to inhibit tumour growth.
The scaffolds have also been used for sustained release of proteins including lysozyme, trypsin inhibitor, BSA, MMP-13 and monoclonal antibody IgG (figure 10) [56] active cytokines β-FGF, TGF, VEGF and BDNF [57]. Furthermore, they can be used for sustained release from a few days to over 100 days (greater than three months) when the experiments were terminated [58]. It is likely that the sustained release was much longer because the content was not decreased significantly when the experiment results were collected (figure 11) [58].
Figure 10.
Molecular representation of lysozyme, trypsin inhibitor, BSA and IgG as well as of the Ac-N-(RADA)4-CONH2 peptide monomer and of the peptide nanofibre. Colour scheme for proteins and peptides: positively charged (blue), negatively charged (red) and hydrophobic (light blue). Protein models were based on known crystal structures. Image courtesy of Sotirios Koutsopolous [56].
Figure 11.

The release profiles during the entire three-month period for IgG through hydrogels of different peptides and different peptide nanofibre densities. Hydrogels consisted of the self-assembling peptides (i) Ac-N(RADA)4-CONH2 with concentration 0.5% w/v (light blue), 1.0% w/v (blue) and 1.5% w/v (dark blue) and of (ii) ac-(KLDL)3-CONH2 with concentration 0.3% w/v (red) and 0.6% w/v (magenta). Release experiments were performed in PBS, pH 7.4 at room temperature. Data points represent the average of five samples. Image courtesy of Sotirios Koutsopolous [58].
Our results not only provide evidence for long-term sustained molecular release from self-assembling peptide scaffolds, but also inspire others to design more self-assembling peptides to control molecular release for clinical applications.
14. Self-assembling peptide hydrogel for human surgical uses of accelerated wound healing
Human wound healing clinical processes still seem to be similar to procedures used many years ago, use of sutures during operations, trying to prevent infection and other treatment measures. Accelerated wound healing will benefit both patients and clinical workers. However, accelerated surgical and trauma wound healing, especially chronically diabetic ulcer wound healing are still problematic, and the self-assembling peptide medical technology may alleviate the problem and put it to good practice.
The self-assembling peptide scaffolds have been found to be permissible for all tissue cell cultures including human cells tested, and the degradation products of the scaffolds are natural amino acids that pose no harm to the human body. Furthermore, a variety of stringent animal tests with rigorous controls showed that the peptides did not elicit noticeable immune responses, nor caused measurable inflammatory reactions through injections and surgical procedures at various tissue sites. It thus encouraged people to carry out human clinical trials for accelerated wound healing indication.
Subjects have been treated for various surgical wounds including cardiovascular surgeries for coronary artery bypass and synthetic blood vessel replacement, gastrointestinal surgery for partial hepatectomy and gastrointestinal treatment for endoscopic excision of mucosa. Those clinical results are very encouraging and beneficial for patients. There were no observed adverse and undesirable side effects so far. This is not surprising because the designer self-assembling peptide scaffolds are totally pure synthetic amino acid-based materials; there are no animal-derived impurities, no chemical and biological contaminations, no organic solvents and no toxic compounds.
Since the successful human clinical trials, additional trials for several indications have been planned or launched for human tooth wound healing, skin wound healing from other diseases and injuries in various parts of the world. Based on the previous successful clinical trials, it is anticipated that these clinical trials will likely be successful. It is hoped that the self-assembling peptide scaffolds will become an enabling medical technology that will truly benefit society.
15. Surface modification self-assembling peptides
In the late 1990s, I also designed a second class of self-assembling peptides that specifically self-assemble on various surfaces to instantly change the physical, chemical and biological characteristics of those surfaces [9]. In an analogy, it is like a grass lawn to cover the soil, or a layer of paint to change the colour and texture of solid surfaces. Except in this case, the coating is a nanometre layer of biological active peptides that can specifically interact with other molecules, or cells. Not only can we pattern the cell for arbitrary shapes, but cells can also be confined in the region where they are seeded. An example of cells on the patterned surface is shown in figure 12.
Figure 12.
Cells are patterned on a peptide-coated surface. (a) Molecular models of the surface self-assembling peptides. This type of peptide has three distinct segments: a biologically active segment where it interacts with other proteins and cells; a linker segment that can not only be flexible or stiff, but also sets the distance from the surface; and an anchor for covalent attachment to the surface [9]. These peptides can be used as ink for an inkjet printer to directly print on a surface, instantly creating any arbitrary pattern, as shown here. (b) Mouse neural cells are seeded on the coated surface. The cells only attach where the adhesion surfaces and not the areas with non-adhesive substrate, such as oligoPEG. The entire frame of the cell pattern is 4 × 1 mm = 4 mm2.
16. Lipid-like self-assembling peptide surfactant materials
The third class of peptides are lipid-like peptide surfactants. The chemical properties of these peptides are similar to natural phospholipids, except they have tunable hydrophobic tails to various degrees of hydrophobic amino acids such as alanine, valine, isoleucine or leucine; and a hydrophilic head with either negatively charged aspartic and glutamic acids, or positively charged histidine, lysine or arginine (figures 13 and 14) [21–27]. These lipid-like peptides undergo self-assembly to form well-ordered nanostructures including nanotubes, nanovesicles and micelles (figures 15 and 16).
Figure 13.
The designer lipid-like peptides. These lipid-like peptides all have a hydrophilic head and a hydrophobic tail, much like lipids or detergents. They sequester their hydrophobic tail inside of micelles, vesicles or nanotube structures and their hydrophilic heads are exposed to water. At least three kinds of molecules can be made, with −, +, −/+ heads and in two orientations.
Figure 14.

Molecular models of peptide detergents at neutral pH. (a) Ac-AAAAAAD-COOH. (b) Ac-AAAAAAK-CONH2. (c) DAAAAAA-CONH2. (d) KAAAAAA-CONH2. (e) Ac-VVVD-COOH. (f) Ac-VVVK-CONH2. (g) Ac-IIID-COOH. (h) Ac-IIIK-CONH2. (i) Ac-LLLD-COOH. (j) Ac-LLLK-CONH2. (k) Ac-GAVILEE. (l) Ac-GAVILRR. Aspartic acid (D) is negatively charged and lysine (K) is positively charged. The hydrophobic tails of the peptide detergents consist of alanine (A), valine (V), isoleucine (I) and leucine (L). Each peptide is approximately 2–2.5 nm long, similar size to biological phospholipids. Colour code: teal, carbon; red, oxygen; blue, nitrogen; white, hydrogen.
Figure 15.
High-resolution transmission electron microscopy images of Ac-G6D2 showing different structures and dynamic behaviours of these structures. (a) A pair of finger-like structures branching off from the stem. (b) Enlargement of the box in (a), the detail opening structures are clearly visible. (c) The openings (arrows) from the nanotube which may have resulted in the growth of the finger-like structures. Some nanovesicles are also visible. (d) The nanovesicles may undergo fission (arrows). Image courtesy of Dr Steve Yang [22].
Figure 16.
(a) Transmission electron microscopy image of nanotubes and nanovesicles of lipid-like peptide ac-VVVVVVD-OH in water. Micelles are also present (image courtesy of Dr Steve Yang) [21,22]. (b) AFM image of nanotubes of A6K lipid-like self-assembling peptides [25]. When the solution pH is less than the lysine pKa of 10, the peptide bears a positive charge. The openings of peptide nanotubes are clearly visible [21,22]. These nanotube structures can also undergo structural changes depending on various conditions, particularly pH changes, ionic strength of salts, temperature and incubation time. The other sheet like materials are likely the un-assembled peptides at the time of the image being collected.
17. A class of lipid-like self-assembling peptides
The lipid-like peptide is a new class of short peptides. Cationic, anionic and zwitterionic peptide detergents were designed [21–27]. These lipid-like peptides are a class of molecules with properties similar to surfactants. They have hydrophilic heads comprising 1–2 residues, and hydrophobic tails 3–6 residues long. They are about 2–3 nm in length, and their ionic character and strength can be controlled by selecting appropriate amino acids or by capping the termini. Lysine or aspartic acid was used for the hydrophilic head. To control the detergent ionic nature, each peptide was capped by acetylation at the N-terminus, or selective amidation at the C-terminus when required. Alanine, valine, leucine and isoleucine were used for the hydrophobic tails (figures 13 and 14).
These lipid-like peptides behave comparably to traditional detergents, but offer several advantages over other detergents. Their chemical properties are similar to commonly used detergents, they can be systematically designed and produced at high purity, and they remain stable for long periods of time.
18. Structure of nanotube and nanovesicle formations
The lipid-like self-assembling peptides can form ordered-nanotubes and nanovesicles (figures 15 and 16) [21–22], but the mechanism is not yet clear. As a plausible path from the monomer state to the assemblies, two peptides form dimer tail-to-tail packing to form a bilayer, monomeric peptides form small segments of the bilayer ring, with hydrophobic tails packing together to avoid water and hydrophilic heads exposed to water on the inner and outer portion of the tube subsequently stack through non-covalent interactions to form longer nanotubes (figure 17).
Figure 17.
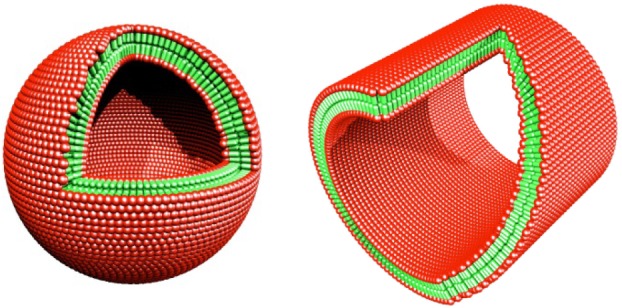
Molecular modelling of cut-away structures formed from the peptides with negatively charged heads and non-polar tail. Peptide nanotube with an area sliced away. Peptide nanovesicle. Colour code: red, negatively charged aspartic acid heads; green, non-polar tail. The non-polar tails are packed inside of the bilayer away from water and the aspartic acids are exposed to water, much like other surfactants. The modelled dimension is 50–100 nm in diameter.
I also designed the cone-shaped lipid-like peptide Ac-GAVILRR-CONH2 with large size amino acid arginine at the C-terminus and progressively reduced the size to glycine at the N-terminus [27]. It has a hydrophilic head with two positive charges and a relatively large head size and a hydrophobic tail with decreasing hydrophobicity and side-chain size with a cone shape, and can self-assemble into an interesting nano-donut structure [27].
Further study of the mechanism is of great importance because these lipid-like peptides have successfully stabilized diverse membrane proteins including G-protein coupled receptors (GPCRs) and used for molecular deliveries [66–72].
19. Stabilize membrane proteins
Membrane proteins play vital roles in all living systems. They involve in energy conversions, cell–cell and cell–environmental communications and sensing, specific ion channels and pumps, transporters and all sorts of transports. Membrane proteins are also essential for our senses: sight, hearing, smell, taste, touch and temperature sensing; and GPCRs are crucial in learning, memory, stem cell renewal and differentiation, body-plan development, the immune system, ageing and more. However, our understanding of their structures and function falls far behind that of soluble proteins.
Over the past few years, our teams have gradually overcome some of the obstacles of membrane protein production and purification (figure 18). We solubilized and stabilized several classes of membrane proteins using the lipid-like peptides, including E. coli glycerol-3-phosphate dehydrogenase [66], the multi-domain protein complex Photosystem-I (PSI) on surface in dry form [67,68] and in aqueous solution [69], GPCR bovine rhodopsin (figure 19) [70] and many olfactory receptors [71,72].
Figure 18.
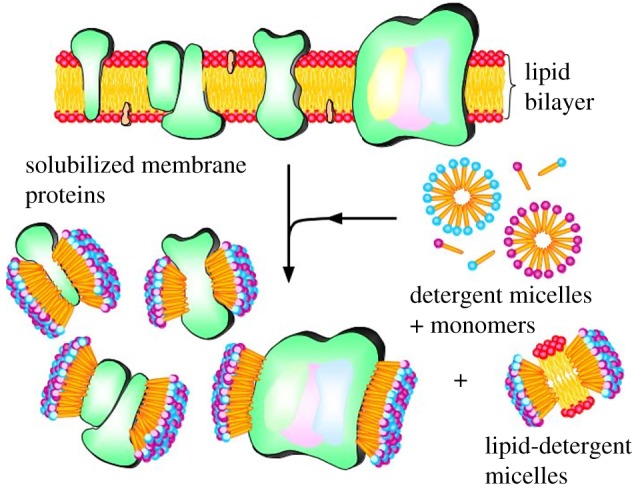
A proposed scheme for how the designer lipid-like peptides stabilize membrane proteins. These simple designer self-assembling lipid-like peptides have been used to solubilize, stabilize and crystallize membrane proteins. These peptides have a hydrophilic head and a hydrophobic tail, much like other biological lipids. They use their tail to sequester the hydrophobic part of membrane proteins, and the hydrophilic heads are exposed to water. Thus, they make membrane proteins soluble and stable outside of their native cellular lipid milieu. These lipid-like peptides are very important for overcoming the barrier of high resolutions of molecular structure for challenging membrane proteins.
Figure 19.
Designer self-assembling peptides stabilize membrane proteins. (a) A proposed model of peptide surfactants. (b) Quick-freeze/deep-etch transmission electron microscopy image of peptide surfactant (V6D) dissolved in water. (c) Stability kinetics of rhodopsin (Rho) in different surfactants; kinetics of bovine rhodopsin under different conditions. Stability of rhodopsin in the absence of OG at different temperatures. Half-life of rhodopsin was as follows: not available in 2.5 mM A6D at 40°C, 50°C and 55°C (left); decay of A500 in de-lipid rhodopsin in the absence of OG. Half-life of rhodopsin was as follows: 122 min in 1.25 mM A6D, 47 min in PBS and 27 min 1% OG (middle); stability of de-lipid rhodopsin at 40°C (right). (d) Lipid-like peptides stabilize functional photosystem I [67,69].
20. Lipid-like peptides used in cell-free production of high-yield membrane proteins
Cell-free production of proteins is a robust method and capable of molecular fabrications quickly, often in a few hours. It can be industrially standardized with rigorous quality controls. The technology is simple and versatile, and an unskilled person can be trained in a few hours to perform the task. Various functional proteins including membrane proteins, particularly difficult GPCRs and other 7-transmembrane proteins have been made for a variety of studies and for a wide range of applications.
Recently, cell-free protein productions have become cost effective and competitive. Gram amounts of functional proteins and materials can be made in a few hours instead of weeks. Cell-free system can be integrated into other devices, because of its simplicity and robustness.
Selecting the right surfactant is thus crucial because bottlenecks in elucidating the structure and function of membrane proteins are the difficulty of producing large quantities of functional receptors. Various self-assembling peptide surfactants in commercial E. coli cell-free systems can rapidly produce milligram quantities of soluble GPCRs that include the human formyl peptide receptor, human trace amine-associated receptor, vomeronasal type 1 receptor 1 and other olfactory receptors [71,72].
Furthermore, using short, designer lipid-like peptides as surfactants, we not only produced 12 unique mammalian olfactory receptors, but also solubilized and stabilized to maintain their structure and function (figure 20) [71,72]. These simple and inexpensive lipid-like peptide surfactants will likely make significant contributions to facilitate the production of GPCRs, and perhaps other membranes.
Figure 20.
Designer lipid-like peptides are used in cell-free systems. Tens of functional olfactory receptor purification and secondary structure studies have been achieved. Olfactory receptors are soluble in Brij-35 and peptide detergents. Each receptor was expressed in the presence of Brij-35 or a peptide detergent using a commercial E. coli cell-free expression system (Qiagen, RiNA and Invitrogen). (a) The presence of a detergent was necessary to solubilize the olfactory receptors, and all of the peptide detergents were able to solubilize four unique receptors. (b) The detergent peptides and Brij-35 were able to solubilize similar fractions of protein. Peptides that were positively charged or had longer tails tended to solubilize higher fractions of receptors. (c) Detergent peptides can yield milligram quantities of solubilized olfactory receptors, and the maximum yield of the monomeric form of all tested olfactory receptors expected in a 10 ml reaction. Only results from the most effective detergent peptide are shown. (d) Circular dichroism spectra of Brij-35 and peptide detergent-produced olfactory receptors to Olfr226, mOR174-4, mOR174-9 and mOR103-15 (courtesy of Karolina Corin) [71,72].
21. Perspective
In science, there are numerous examples of curiosity-driven research and unintentional (unauthorized) discoveries that eventually lead to technological breakthroughs and new economic development. It is extremely difficult to imagine not pursuing curiosity-driven research and explorations. Curiosity-driven research not only provides new insights into mysteries of nature and generates new knowledge, but it also can translate the knowledge into new enabling technology that will eventually be developed as knowledge-based economy. It is clear that new knowledge also leads to new economic development. The lack of knowledge inevitably hinders the new economic development.
There are a few examples: the discovery of X-rays, the structure of DNA double helix, DNA–RNA and RNA–RNA hybridizations, reverse transcription, RNA splicing, RNA as enzymes, telomeres, natural killer cells, programmed cell death, microRNA, RNA interference, S-layer proteins, Kuru disease, carbon 60, carbon nanotubes, carbon graphene and the indispensible World Wide Web, and the latest CRISPR for simple and elegant gene and genome editing. The discovery of the self-assembling peptide is another good example where an unexpected curiosity-driven discovery led to the development of an enabling medical technology that benefits society. The recent examples of successful clinical trials of the self-assembling peptide scaffolds for accelerating wound healing and regenerative medicine and surgical uses provide a glimpse of what is coming for widespread uses of self-assembling peptides. Therefore, curiosity-driven research must be strongly encouraged and fully supported, despite the current emphasis of application-driven research.
Acknowledgements
I gratefully acknowledge supports from ARO, ONR, AFRO, DARPA, NSF, NIH, Whitaker Foundation, John Simon Guggenheim Foundation, Menicon Co. Ltd, Mitsubishi Chemicals, Mitsui Chemical, ROHM and 3DMatrix. I also sincerely thank former students, postodcs and visiting scientists in my laboratory who made invaluable contributions to the successful studies of self-assembling peptides and translation into new knowledge-based economy.
Data accessibility
This article has no additional data.
Competing interests
I declare I have no competing interests.
Funding
I received no funding for this study.
References
- 1.Zhang S, Lockshin C, Herbert A, Winter E, Rich A. 1992. Zuotin, a putative Z-DNA binding protein in Saccharomyces cerevisiae. EMBO. J. 11, 3787–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang S, Holmes T, Lockshin C, Rich A. 1993. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc. Natl Acad. Sci. USA 90, 3334–3338. ( 10.1073/pnas.90.8.3334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marqusee S, Baldwin RL. 1987. Helix stabilization by Glu-.Lys+ salt bridges in short peptides of de novo design. Proc. Natl Acad. Sci. USA 84, 8898–8902. ( 10.1073/pnas.84.24.8898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marqusee S, Robinson VH, Baldwin RL. 1989. Unusually stable helix formation in short alanine-based peptides. Proc. Natl Acad. Sci. USA 86, 5286–5290. ( 10.1073/pnas.86.14.5286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rich A. 2004. The excitement of discovery. Ann. Rev. Biochem. 37, 1–37. ( 10.1146/annurev.biochem.73.011303.073945) [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Lockshin C, Cook R, Rich A. 1994. Unusually stable beta-sheet formation of an ionic self-complementary oligopeptide. Biopolymers 34, 663–672. ( 10.1002/bip.360340508) [DOI] [PubMed] [Google Scholar]
- 7.Zhang S, Holmes T, DiPersio M, Hynes R.O, Su X, Rich A. 1995. Self-complementary oligopeptide matrices support mammalian cell attachment. Biomaterials 16, 1385–1393. ( 10.1016/0142-9612(95)96874-Y) [DOI] [PubMed] [Google Scholar]
- 8.Holmes T, Delacalle S, Su X, Rich A, Zhang S. 2000. Extensive neurite outgrowth and active neuronal synapses on peptide scaffolds. Proc. Natl Acad. Sci. USA 97, 6728–6733. ( 10.1073/pnas.97.12.6728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang S, Yan L, Altman M, Lässle M, Nugent H, Frankel F, Lauffenburger D, Whitesides G, Rich A. 1999. Biological surface engineering: a simple system for cell pattern formation. Biomaterials 20, 1213–1220. ( 10.1016/S0142-9612(99)00014-9) [DOI] [PubMed] [Google Scholar]
- 10.Marini D, Hwang W, Lauffenburger DA, Zhang S, Kamm R. 2002. Left-handed helical ribbon intermediates in the self-assembly of a β-sheet peptide. Nano Lett. 2, 295–299. ( 10.1021/nl015697g) [DOI] [Google Scholar]
- 11.Caplan MR, Schwartzfarb EM, Zhang S, Kamm RD, Lauffenburger DA. 2002. Control of self-assembling oligopeptide matrix formation through systematic variation of amino acid sequence. Biomaterials 23, 219–227. ( 10.1016/S0142-9612(01)00099-0) [DOI] [PubMed] [Google Scholar]
- 12.Kisiday J, Jin M, Kurz B, Hung H, Semino C, Zhang S, Grodzinsky AJ. 2002. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: implications for cartilage tissue repair. Proc. Natl Acad. Sci. USA 99, 9996–10001. ( 10.1073/pnas.142309999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoi H, Kinoshita T, Zhang S. 2005. Dynamic reassembly of peptide RADA16 nanofiber scaffold. Proc. Natl Acad. Sci. USA 102, 8414–8419. ( 10.1073/pnas.0407843102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rich A, Davies DR. 1956. A new two-stranded helical structure: polyadenylic acid and polyuridylic acid. J. Am. Chem. Soc. 78, 3548 ( 10.1021/ja01595a086) [DOI] [Google Scholar]
- 15.Felsenfeld G, Davies DR, Rich A. 1957. Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 79, 2023–2024. ( 10.1021/ja01565a074) [DOI] [Google Scholar]
- 16.Narmoneva DA, Oni O, Sieminski AL, Zhang S, Gertler JP, Kamm RD, Lee RT. 2005. Self-assembling short oligopeptides and the promotion of angiogenesis. Biomaterials 26, 4837–4846. ( 10.1016/j.biomaterials.2005.01.005) [DOI] [PubMed] [Google Scholar]
- 17.Davis ME, Motion JPM, Narmoneva DA, Takahashi T, Hakuno D, Kamm RD, Zhang S, Lee RT. 2005. Injectable self-assembling peptide nanofibers create intramyocardial microenvironments for endothelial cells. Circulation 111, 442–450. ( 10.1161/01.CIR.0000153847.47301.80) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis ME, Hsieh PCH, Takahashi T, Song Q, Zhang S, Kamm RD, Grodzinsky AJ, Anversa P, Lee RT. 2006. Local myocardial IGF-1 delivery with biotinylated peptide nanofibers improves cell therapy for myocardial infarction. Proc. Natl Acad. Sci. USA 103, 8155–8160. ( 10.1073/pnas.0602877103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bokhari MA, Akay G, Zhang S, Birch MA. 2005. The enhancement of osteoblast growth and differentiation in vitro on a peptide hydrogel–polyHIPE polymer hybrid material. Biomaterials 26, 5198–5208. ( 10.1016/j.biomaterials.2005.01.040) [DOI] [PubMed] [Google Scholar]
- 20.Schneider A, Garlick JA, Egles C. 2008. Self-assembling peptide nanofiber scaffolds accelerate wound healing. PLoS ONE 3, e1410 ( 10.1371/journal.pone.0001410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vauthey S, Santoso S, Gong H, Watson N, Zhang S. 2002. Molecular self-assembly of surfactant-like peptides to form nanotubes and nanovesicles. Proc. Natl Acad. Sci. USA 99, 5355–5360. ( 10.1073/pnas.072089599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santoso S, Hwang W, Hartman H, Zhang S. 2002. Self-assembly of surfactant-like peptides with variable glycine tails to form nanotubes and nanovesicles. Nano Lett. 2, 687–691. ( 10.1021/nl025563i) [DOI] [Google Scholar]
- 23.von Maltzahn G, Vauthey S, Santoso S, Zhang S. 2003. Positively charged surfactant-like peptides self-assemble into nanostructures. Langmuir 19, 4332–4337. ( 10.1021/la026526%2B) [DOI] [Google Scholar]
- 24.Yang S, Zhang S. 2006. Self-assembling behavior of designer lipid-like peptides. Supramol. Chem. 18, 389–396. ( 10.1080/10615800600658586) [DOI] [Google Scholar]
- 25.Nagai A, Nagai Y, Qu H, Zhang S. 2007. Self-assembling behaviors of lipid-like peptides A6D and A6 K. J. Nanosci. Nanotechnol. 7, 2246–2252. ( 10.1166/jnn.2007.647) [DOI] [PubMed] [Google Scholar]
- 26.Khoe U, Yang Y, Zhang S. 2008. Synergistic effect and hierarchical nanostructure formation in mixing two designer lipid-like peptide surfactants Ac-A6D-OH and Ac-A6 K-NH2. Macromol. Biosci. 8, 1060–1067. ( 10.1002/mabi.200800182) [DOI] [PubMed] [Google Scholar]
- 27.Khoe U, Yanlian Y, Zhang S. 2009. Self-assembly of nano-donut structure from cone-shaped designer lipid-like peptide surfactant. Langmuir 25, 4111–4114. ( 10.1021/la8025232) [DOI] [PubMed] [Google Scholar]
- 28.Zhang S. 2002. Emerging biological materials through molecular self-assembly. Biotechnol. Adv. 20, 321 ( 10.1016/S0734-9750(02)00026-5) [DOI] [PubMed] [Google Scholar]
- 29.Zhang S. 2003. Fabrication of novel materials through molecular self-assembly. Nat. Biotechnol. 21, 1171–1178. ( 10.1038/nbt874) [DOI] [PubMed] [Google Scholar]
- 30.Zhang S. 2004. Beyond the Petri dish. Nat. Biotechnol. 22, 151 ( 10.1038/nbt0204-151) [DOI] [PubMed] [Google Scholar]
- 31.Zhao X, Zhang S. 2006. Molecular designer self-assembling peptides. Chem. Soc. Rev. 35, 1110–1110. ( 10.1039/b511336a) [DOI] [PubMed] [Google Scholar]
- 32.Hauser CAE, Zhang S. 2010. Designer self-assembling peptide biological materials. Chem. Soc. Rev. 39, 2780–2790. ( 10.1039/b921448h) [DOI] [PubMed] [Google Scholar]
- 33.Hauser CAE, Zhang S. 2010. Peptides as biological semiconductors. Nature 468, 516–517. ( 10.1038/468516a) [DOI] [PubMed] [Google Scholar]
- 34.Koutsopoulos S, Kaiser L, Eriksson HM, Zhang S. 2012. Designer peptide surfactants stabilize diverse functional membrane proteins. Chem. Soc. Rev. 41, 1721–1728. ( 10.1039/C1CS15180K) [DOI] [PubMed] [Google Scholar]
- 35.Zhang S. 2012. Lipid-like self-assembling peptides. Acc. Chem. Res. 45, 2142–2150. ( 10.1021/ar300034v) [DOI] [PubMed] [Google Scholar]
- 36.Luo Z, Zhang S. 2012. Designer nanomaterials using chiral self-assembling peptide systems and their emerging benefit for society. Chem. Soc. Rev. 41, 4736–4754. ( 10.1039/c2cs15360b) [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, Gelain F, Zhao X. 2005. Designer self-assembling peptide nanofiber scaffolds for 3-D tissue cell cultures. Semin. Cancer Biol. 15, 413–420. ( 10.1016/j.semcancer.2005.05.007) [DOI] [PubMed] [Google Scholar]
- 38.Gelain F, Bottai D, Vescovi A, Zhang S. 2006. Designer self-assembling peptide nanofiber scaffolds for adult mouse neural stem cell 3-dimensional cultures. PLoS ONE 1, e119 ( 10.1371/journal.pone.0000119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horii A, Wang X, Gelain F, Zhang S. 2007. Biological designer self-assembling peptide scaffolds significantly enhance osteoblast proliferation, differentiation and 3-D migration. PLoS ONE 2, e190 ( 10.1371/journal.pone.0000190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Horii A, Zhang S. 2008. Designer functionalized self-assembling peptide nanofiber scaffolds for growth, migration, tubulogenesis of human umbilical vein endothelial cell. Soft Matter 4, 2388–2395. ( 10.1039/b807155a) [DOI] [Google Scholar]
- 41.Kumada Y, Zhang S. 2010. Significant Type I and Type III collagen production from human periodontal ligament fibroblast in 3D peptide scaffolds without extra growth factors. PLoS ONE 5, e1030 ( 10.1371/journal.pone.0010305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumada Y, Hammond NA, Zhang S. 2010. Functionalized self-assembling short peptide scaffolds significantly promote fibroblasts proliferation and 3-D migration. Soft Matter 6, 5073 ( 10.1039/c0sm00333f) [DOI] [Google Scholar]
- 43.Chau Y, Luo Y, Cheung ACY, Nagai Y, Zhang S, Kobler JB, Zeitels SM, Langer R. 2008. Incorporation of a matrix metalloproteinase-sensitive substrate into self-assembling peptides—a model for biofunctional scaffolds. Biomaterials 29, 1713–1719. ( 10.1016/j.biomaterials.2007.11.046) [DOI] [PubMed] [Google Scholar]
- 44.Ellis-Behnke R, Liang YX, You SW, Tay D, Zhang S, So KF, Schneider G. 2006. Nano neuro knitting: peptide nanofiber scaffold for brain repair and axon regeneration with functional return of vision. Proc. Natl Acad. Sci. USA 103, 5054–5059. ( 10.1073/pnas.0600559103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellis-Behnke RG, Liang YX, Tay DKC, Kau PWF, Schneider GE, Zhang S, Wu W, So K-F. 2007. Nano hemostat solution: immediate hemostasis at the nanoscale. Nanomed. Nanotechnol. Biol. Med. 2, 207–215. ( 10.1016/j.nano.2006.08.001) [DOI] [PubMed] [Google Scholar]
- 46.Ellis-Behnke RG, Schneider GE. 2011. Peptide amphiphiles and porous biodegradable scaffolds for tissue regeneration in the brain and spinal cord. Methods Mol. Biol. 726, 259–281. ( 10.1007/978-1-61779-052-2_17) [DOI] [PubMed] [Google Scholar]
- 47.Luo Z, Zhao X, Zhang S. 2008. Structural dynamic of a self-assembling peptide d-EAK16 made of only D-amino acids. PLoS ONE 3, e2364 ( 10.1371/journal.pone.0002364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo Z, Wang S, Zhang S. 2011. Fabrication of self-assembling D-form peptide nanofiber scaffold d-EAK16 for rapid hemostasis. Biomaterials 32, 2013–2020. ( 10.1016/j.biomaterials.2010.11.049) [DOI] [PubMed] [Google Scholar]
- 49.Song H, Zhang L, Zhao X. 2011. Hemostatic efficacy of biological self-assembling peptide nanofibers in a rat kidney model. Macromol. Biosci. 10, 33–39. ( 10.1002/mabi.200900129) [DOI] [PubMed] [Google Scholar]
- 50.Meng H, Chen L, Ye Z, Wang S, Zhao X. 2008. The effect of a self-assembling peptide nanofiber scaffold (peptide) when used as a wound dressing for the treatment of deep second degree burns in rats. J Biomed. Mater. Res. B 89B, 379–391. ( 10.1002/jbm.b.31226) [DOI] [PubMed] [Google Scholar]
- 51.Kopesky PW, Vanderploeg EJ, Kisiday JD, Frisbie DD, Sandy JD, Grodzinsky AJ. 2010. Controlled delivery of transforming growth factor β1 by self-assembling peptide hydrogels induces chondrogenesis of bone marrow stromal cells and modulates Smad2/3 signaling. Tissue Eng. A 17, 83–92. ( 10.1089/ten.tea.2010.0198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gelain F, et al. 2011. Transplantation of nanostructured composite scaffolds results in the regeneration of chronically injured spinal cords. ACS Nano 5, 227–236. ( 10.1021/nn102461w) [DOI] [PubMed] [Google Scholar]
- 53.Cigognini D, Satta A, Colleoni B, Silva D, Donegà M, Antonini S, Gelain F. 2011. Evaluation of early and late effects into the acute spinal cord injury of an injectable functionalized self-assembling scaffold. PLoS ONE 6, e19782 ( 10.1371/journal.pone.0019782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagai Y, Unsworth LD, Koutsopoulos S, Zhang S. 2006. Diffusion coefficients in self-assembling peptide nanofiber scaffold hydrogel. J. Controlled Release 115, 18–25. ( 10.1016/j.jconrel.2006.06.031) [DOI] [PubMed] [Google Scholar]
- 55.Fung SY, Yang H, Chen P. 2008. Sequence effect of self-assembling peptides on the complexation and in vitro delivery of the hydrophobic anticancer drug ellipticine. PLoS ONE 3, e1956 ( 10.1371/journal.pone.0001956) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koutsopolous S, Unsworth LD, Nagai Y, Zhang S. 2009. Slow release of functional proteins through designer self-assembling peptide nanofiber hydrogel scaffolds. Proc. Natl Acad. Sci. USA 106, 4623–4628. ( 10.1073/pnas.0807506106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gelain F, Unsworth L, Zhang S. 2010. Slow and sustained release of active cytokines from designer self-assembling peptide nanofiber scaffolds for adult mouse neural stem cell cultures. J. Controlled Release 145, 231–239. ( 10.1016/j.jconrel.2010.04.026) [DOI] [PubMed] [Google Scholar]
- 58.Koutsopoulos S, Zhang S. 2012. Multi-layered injectable self-assembling peptide hydrogels for long-term sustained release of human antibodies. J. Controlled Release 160, 451–458. ( 10.1016/j.jconrel.2012.03.014) [DOI] [PubMed] [Google Scholar]
- 59.Koutsopoulos S, Zhang S. 2013. Long-term three-dimensional neural tissue cultures in functionalized self-assembling peptide hydrogels, Matrigel and Collagen I. Acta Biomater. 9, 5162–5169. ( 10.1016/j.actbio.2012.09.010) [DOI] [PubMed] [Google Scholar]
- 60.Ruan L, Zhang H, Luo H, Liu J, Tang F, Shi YK, Zhao X. 2009. Designed amphiphilic peptide forms stable nanoweb, slowly releases encapsulated hydrophobic drug, accelerates animal hemostasis. Proc. Natl Acad. Sci. USA 106, 5105–5010. ( 10.1073/pnas.0900026106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen C, Zhang S, Cao M, Wang J, Xu H, Zhao X, Lu JR. 2010. Antibacterial activities of short designer peptides: a link between propensity for nanostructuring and capacity for membrane destabilization. Biomacromolecules 11, 402–411. ( 10.1021/bm901130u) [DOI] [PubMed] [Google Scholar]
- 62.Zhao X, Pan F, Lu JR. 2008. Recent development of peptide self-assembly. Prog. Nat. Sci. 18, 653–660. ( 10.1016/j.pnsc.2008.01.012) [DOI] [Google Scholar]
- 63.Luo Z, Åkerman B, Zhang S, Nordén B. 2010. Structures of self-assembled amphiphilic peptide-heterodimers: effects of concentration, pH, temperature and ionic strength. Soft Matter 6, 2260–2270. ( 10.1039/b926962b) [DOI] [Google Scholar]
- 64.Bucak S, Cenker C, Nasir I, Olsson U, Zackrisson M. 2009. Peptide nanotube nematic phase. Langmuir 25, 4262–4265. ( 10.1021/la804175h) [DOI] [PubMed] [Google Scholar]
- 65.Castelletto V, Nutt DR, Hamley IW, Bucak S, Cenker Ç, Olsson U. 2010. Structure of single-wall peptide nanotubes: in situ flow aligning x-ray diffraction. Chem. Commun. 46, 6270–6272. ( 10.1039/c0cc00212g) [DOI] [PubMed] [Google Scholar]
- 66.Yeh JI, Du S, Tordajada A, Paulo J, Zhang S. 2005. Peptergent: peptide detergents that improve stability and functionality of a membrane protein glycerol-3-phosphate dehydrogenase. Biochemistry 44, 16 912–16 919. ( 10.1021/bi051357o) [DOI] [PubMed] [Google Scholar]
- 67.Kiley P, Zhao X, Vaughn M, Baldo M, Bruce BD, Zhang S. 2005. Self-assembling peptide detergents stabilize isolated photosystem I on a dry surface for an extended time. PLoS Biol. 3, 1181–1186. ( 10.1371/journal.pbio.0030230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mershin A, Matsumoto K, Kaiser L, Yu D, Vaughn M, Nazeeruddin MDK, Bruce B, Graetzel M, Zhang S. 2012. Self-assembled photosystem-I biophotovoltaics on nanostructured TiO2 and ZnO. Sci. Rep. 2, e234 ( 10.1038/srep00234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsumoto K, Koutsopoulos S, Vaughn M, Bruce BD, Zhang S. 2009. Designer lipid-like peptide surfactants stabilize functional photosystem I membrane complex in solution. J. Phys. Chem. 115, 75–83. ( 10.1021/jp8021425) [DOI] [PubMed] [Google Scholar]
- 70.Zhao X, Nagai Y, Revees P, Kiley P, Khorana HG, Zhang S. 2006. Designer lipid-like peptides significantly stabilize G-protein coupled receptor bovine rhodopsin. Proc. Natl Acad. Sci. USA 103, 17 707–17 712. ( 10.1073/pnas.0607167103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Corin K, Wienken CJ, Jerabek-Willemsen M, Duhr S, Braun D, Zhang S. 2011. Peptide surfactants for cell-free production of functional G protein-coupled receptors. Proc. Natl Acad. Sci. USA 118, 9049–9054. ( 10.1073/pnas.1018185108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corin K, et al. 2011. Designer lipid-like peptides: a class of detergents for studying functional olfactory receptors using commercial cell-free systems. PLoS ONE 6, e25067 ( 10.1371/journal.pone.0025067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mershin A, Cook B, Kaiser L, Zhang S. 2005. A classic assembly. Nat. Biotechnol. 23, 1379–1380. ( 10.1038/nbt1105-1379) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.