Abstract
In biological systems, evolutionary innovations can spread not only from parent to offspring (i.e. vertical transmission), but also ‘horizontally’ between individuals, who may or may not be related. Nowhere is this more apparent than in bacteria, where novel ecological traits can spread rapidly within and between species through horizontal gene transfer (HGT). This important evolutionary process is predominantly a by-product of the infectious spread of mobile genetic elements (MGEs). We will discuss the ecological conditions that favour the spread of traits by HGT, the evolutionary and social consequences of sharing traits, and how HGT is shaped by inherent conflicts between bacteria and MGEs.
This article is part of the themed issue ‘Process and pattern in innovations from cells to societies’.
Keywords: lateral gene transfer, horizontal gene transfer, accessory genome, pan-genome
1. Introduction
The process of evolution by natural selection has generated diverse, elegant and efficacious innovations. Where such innovations have a genetic basis, they can be transmitted ‘vertically’ from parent to offspring, giving rise to a model of evolution represented as a bifurcating tree. However, it has long been recognized that genetic information can also be transferred between individuals not connected by inheritance, a process referred to as ‘horizontal gene transfer’ (HGT) [1,2]. Here, we define HGT as the movement of heritable genetic information that cannot be explained by vertical parent–offspring transmission [3] (thus explicitly excluding processes such as introgression, which is covered elsewhere [4]). Like single nucleotide mutations and gene duplications, HGT is essentially a process that introduces variation, which is then subject to natural selection. Such variation can be dramatic, such as the single-step acquisition of complex metabolic pathways [5,6], and can stand in contrast with historical formulations of evolution that imply gradual change [7]. Innovation, in the context of HGT, primarily implies the acquisition of novelties by a recipient lineage, though these traits may have existed for a long time in the donor lineage. Nevertheless, this can have profound consequences for how organisms evolve, making possible substantial shifts in their biology. Furthermore, HGT provides potent opportunities for the rapid spread of genetic innovations into new backgrounds: rare mutational events need occur only once before spreading broadly into various new lineages. However, HGT is not just a process for organisms to acquire novelties. As with mutations, transferred genes can be neutral or deleterious, and many of the agents that facilitate HGT are entities evolving in their own right. Their interactions with the donors and recipients of HGT can set the scene for evolutionary conflicts.
HGT can occur across vast phylogenetic distances, and these events may have important ecological consequences. For example, Hypothenemus hampei, a species of beetle, appears to have acquired a gene for mannanase degradation from Bacillus, enabling it to become an economically important pest of coffee plantations [8]. In other cases, HGT appears more opportunistic, and the adaptive consequences are less clear. For example, some strains of the human pathogen Neisseria gonorrhoeae have acquired a 685 bp region of the Long Interspersed Nuclear Element (LINE1), a fragment of a retroelement gene found in the human genome that has no clear function in the recipient bacterium [9]. Inter-kingdom gene transfer can occur surprisingly often given the right ecological conditions, as is shown by the independent acquisition of the bacterial gene acdS by 15 different lineages of fungi and other eukaryotes [10]. The amount of DNA transferred can be considerable—in one case, almost an entire bacterial genome was found to have transferred into the nuclear genome of a Drosophila [11]. Events like these demonstrate that species boundaries can be more permeable than is often assumed, and that genetic information can in principle move between even highly divergent lineages. It has even been proposed that no insurmountable barrier to HGT exists [12]. HGT into metazoan genomes is striking, the more so because it has disrupted long-held assumptions about the nature of inheritance and evolution in complex organisms. However, while metazoan HGT clearly occurs, the complexity of eukaryotic genomes, and the ease by which samples can become contaminated with bacterial DNA creating false positives, have thrown some of the more extreme claims into doubt [13]. The consensus, therefore, is that successful HGT into metazoan taxa is relatively rare, albeit with potentially huge impact for phenotype and fitness where it does occur [14].
By contrast, in the microbial world, HGT is a fact of life. For bacteria in particular, HGT is a major mode of adaptation, making a significant contribution to genome evolution and structure [1,15,16]. Bacterial HGT has a central role in adaptation to environmental challenges, like colonization of new environments, exploitation of novel carbon sources and resistance to toxins [17]. The increasing evidence placing humans in the midst of an essentially microbial world [18–20], where bacteria have fundamental roles in biogeochemical cycles, health and disease, and food security, makes it all the more important to understand their evolution and ecology. Furthermore, bacteria are increasingly used as model systems for understanding general evolutionary processes [21]. In this review, we will focus on HGT between bacteria, although many of the themes we discuss are likely to apply when considering the horizontal transfer of traits more generally.
2. Horizontal gene transfer is central to bacterial evolution
Bacteria have several features that may make them especially well suited for HGT-mediated evolution. All cells are generally reproductively proficient, i.e. germ line, meaning that mutations and acquired genes can be easily passed down to subsequent generations. Unlike eukaryotes, bacteria lack membrane-bound nuclei, meaning that their genomes are more accessible to incoming DNA. This can enhance the acquisition and integration of new genes [12]. Bacteria can evolve rapidly, owing to their potential for huge population sizes and short generation times, which means that infrequent gene transfer events are more likely to occur and selectively advantageous events less likely to be lost due to drift. Bacteria have a truly cosmopolitan distribution, inhabiting and adapting to a vast range of environments and performing reactions the benefits of which may be limited spatially, such as degradation of exotic carbon sources. Migration is thought to occur readily [22,23], and consequently myriad bacterial species can coexist in a community [24]. This represents diverse genetic material that can potentially transfer between, and be of value to, the members of the community.
Comparative analyses have revealed the pervasiveness of HGT in bacterial evolution and genome dynamics. Even closely related bacterial strains can vary greatly in genome content [25], and increased sequencing shows that only a minority of genes carried by a species might be shared across all members [25,26]. This set of genes represents a ‘core genome’, and can be contrasted with the ‘variable’ or ‘accessory’ genome which represents genes present only in a subset of strains [27]. The total of all the unique genes in a species, termed the ‘pan-genome’, generally increases in size with each new strain sequenced (though this can vary between species [28]), a pattern that emerges because different lineages within a species acquire and lose variable genes from other species in their local communities. The size of the pan-genome and its distribution among strains hint therefore at a large ‘pool’ or ‘library’ of genetic material that is available for acquisition—a genetic resource on which evolving bacteria can draw for adaptation. This library is apparently well-used. Comparative studies show that, in several bacterial lineages, genes are acquired and lost at rates comparable to or even greater than nucleotide substitutions [29,30].
3. Machines for spreading genes
In principle, HGT requires two physical processes. First, genetic information must cross biological membranes into the recipient species. Second, the genes must be linked to a functioning origin of replication in a germ line cell to ensure subsequent vertical transmission in the recipient. There are several well-defined mechanisms that facilitate gene transfer between bacteria by enhancing one or both of these processes (figure 1), with novel mechanisms still being characterized and, in all likelihood, more yet to be discovered.
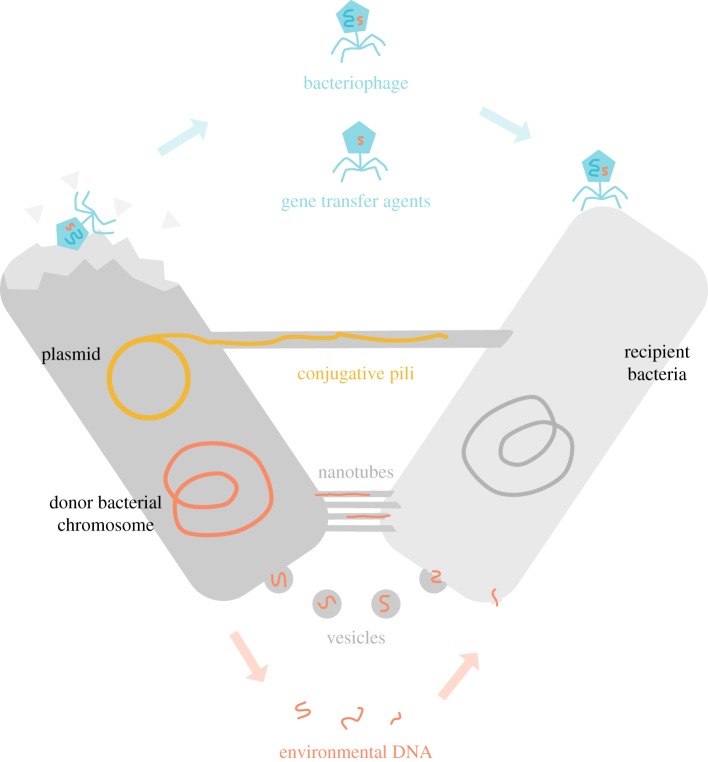
Figure 1.
The many routes for horizontal gene transfer. DNA can be transferred between individuals by multiple mechanisms falling broadly into three categories. In transduction (blue text), DNA is transferred either as part of the phage genome itself or as additional DNA packaged into phage particles or gene transfer agents. In conjugation (yellow text), donor cells form conjugative pili, typically encoded on plasmids or integrative conjugative elements, through which DNA is transferred. Finally, transformation (red text) is the process by which DNA in the environment is actively taken up by the donor cell. In addition, recent studies have shown that bacteria can also transfer DNA fragments in membrane-bound vesicles and via nanotubes.
Traditionally, there are three canonical mechanisms of DNA transfer in bacteria: conjugation, transduction and transformation. Conjugation occurs when a donor bacterium expresses a multicomponent macromolecular membrane-traversing structure—a ‘conjugative pilus’—which provides a physical link for DNA to move between donor and recipient [31,32]. Transferred DNA contains an ‘origin of transfer’ (oriT), a sequence of bases that is recognized by the conjugative machinery [33]. Transduction occurs when bacteriophage from a donor bacterium package non-phage DNA in viral particles which then infect other bacteria [34,35]. Both conjugation and transduction provide protection for DNA from environmental damage after it leaves the donor cell. Natural transformation is the process by which bacteria take up DNA from their environment. This occurs by the retraction of cell surface fibres (pili) which pull double-stranded DNA close to the cell membrane, allowing uptake via a conserved membrane pore. The DNA substrates for transformation may be actively secreted into the environment by donors [36], or released by dying bacteria (e.g. following lysis by phage, [37]). It has been recorded that bacteria can take up genes released by neighbours they themselves have killed [38]. More recent research has identified other mechanisms by which DNA can transfer between hosts. Gene transfer agents (GTAs) are DNA-containing particles that resemble phage, but are incapable of carrying the genes for particle production [39]. Genes can also be transferred between bacteria that form intercellular connections via nanotubes [40] or membrane fusion [41]. Some bacteria release DNA-containing membrane-bound vesicles that can carry genetic information to new hosts [42]. Mycobacteria undergo an unusual form of conjugation that appears to be regulated in part by the recipient, does not require oriT and results in transconjugants whose genomes are a patchwork of their parents' [43,44].
Once DNA has entered a recipient, it must replicate to avoid loss by segregation during cell division. Incoming DNA can carry its own origin of replication thus replicating separately from the chromosome of its new host. Such is the case for plasmids: pieces of DNA, usually circular, that remain physically distinct from the chromosome [45]. Alternatively, the incoming DNA must recombine with a resident element to gain access to an origin of replication, either on the chromosome or on an extrachromosomal replicon like a plasmid. This can happen via general mechanisms of recombination, but is enhanced by an assortment of enzymes, which catalyse the integration, excision and recombination of DNA [46].
Although the machineries discussed in this section enable the horizontal transfer of genetic innovations, it should be noted that they have not necessarily evolved ‘for’ that purpose. For example, transformation tends to cause the replacement of longer stretches of DNA with shorter ones, and hence is more prone to reducing, rather than expanding, genome content [47]. Various restrictions on assimilating DNA from unrelated strains suggest that transformation may tend towards being a more conservative than innovative process [48,49]. But perhaps more importantly, many of these mechanisms are not in fact under the control of the bacteria but are instead controlled by semi-autonomous segments of DNA which, far from being functional tools for bacterial gene exchange, have their own self-interest at heart.
4. Mobile genetic elements: perpetrators of horizontal gene transfer
From the perspective of a gene, HGT represents another opportunity for reproduction, and thus is subject to natural selection. The microbial world is teeming with mobile genetic elements (MGEs), genetic entities that are adapted to transferring between strands of DNA and between different bacterial hosts [50]. The bestiary of MGEs is rich, and the elements involved are dynamic, modular and nested. For example, transposable elements (TEs), DNA sequences that carry genes enabling them to hop between DNA strands, can be found on larger elements such as plasmids which carry an origin of replication [51]. Plasmids may carry their own set of genes for conjugative transfer (i.e. conjugative plasmids), or, if they have a compatible oriT, may use the conjugative machinery encoded by a different replicon (i.e. are mobilizable) [33,52]. Plasmid gene content is dynamic, and plasmids that acquire new genes from their hosts, perhaps through the activity of TEs, carry these genes onwards when they conjugate [53,54]. Integrative and conjugative elements (ICEs) resemble plasmids in many ways, except that they carry enzymes that catalyse insertion into the host chromosome and thus do not need to carry their own origin of replication [55]. Bacteriophage can be either purely virulent, killing their hosts quickly in order to reproduce, or ‘temperate’ phage that, similarly to ICEs, can insert their genomes into the bacterial chromosome. Both types of phage can mediate HGT. For temperate phage, there is an opportunity for bacterial genes or transposons to become integrated into the phage genome and thus be co-inserted into the bacterial genome when they integrate, called lysogenic conversion [56]. However, for all phage, bacterial DNA or other MGEs can be packaged in phage particles and become transferred by transduction [57]. Indeed, some integrative elements specialize in repurposing phage capsids for their own transfer [58]. Collaborations and conflicts between MGEs can therefore enhance their ability to spread within and between hosts.
MGEs can carry genes other than those necessary for transfer and replication. These genes—or where they can be functionally grouped, ‘modules’ [59]—may help the selfish vertical transmission of the element. For example, partitioning systems segregate plasmids between daughter cells, reducing the frequency of plasmid-free offspring, and toxin–antitoxin systems impose a large (usually lethal) cost on daughter cells that have lost the MGE, favouring MGE carriers by removing such competitors from the population [60]. The evolutionary benefits of such modules are easy to appreciate. Many MGEs also carry ‘accessory genes’, which do not play a direct role in their vertical or horizontal transmission. Instead, accessory genes may have effects at higher organizational levels, boosting the success of the element indirectly. For example, the spread of antimicrobial resistance (AMR) genes is facilitated by the carriage of these genes on elements such as transposons, ICEs and plasmids [61]. Acquiring these accessory genes can allow a bacterial host to flourish in otherwise deadly environments, with concomitant positive effects on the elements that it carries. The benefits of accessory gene carriage can be seen in the success of integrons, elements first identified on MGEs that appear adapted for the acquisition, assembly, and expression of accessory genes [62,63]. Yet, the function of many accessory genes is not known [64], and the selective factors that favour their mobility and co-occurrence are similarly unclear. Nevertheless, MGEs probably constitute the means by which most genes travel through bacterial communities, and are therefore potent agents of HGT [65,66].
Thus, there exist many potential routes by which genes can transfer between bacteria, and although rates for processes have been measured experimentally (e.g. [67–69]), understanding the relative importance of these mechanisms in situ is complicated by the fact that efficacies are likely to vary between species and environments [45,70]. This can stem from physical limitations, for example, mechanical agitation can inhibit conjugative transfer [71] and different environments are more or less harsh to extracellular DNA, affecting opportunities for transformation [72]. It is also likely to be driven by the ecology of both bacteria and MGEs, for example, lysogenic conversion is more common among bacteria with fast growth rates [73], which probably reflects the conditions that favour the life-history strategy of temperate phage. Genome analyses suggest that plasmids are better-connected ‘hubs’ in networks of gene exchange than phage, for example [66], but it can be difficult to identify how genes have moved by analysing their sequences [74]. A clear priority for future research is to quantify the relative importance of HGT mechanisms and how such rates vary with taxonomy and ecology.
5. Costs, benefits and conflicts in horizontal gene transfer
Horizontally transferred genes have the potential to provide their recipients with striking benefits. However, HGT is not a benign process, and gene exchange can impose significant fitness costs on both donor and recipient. Indeed, the distribution of fitness effects of HGT has been proposed to be more dispersed than that of nucleotide mutations [29], with potential costs, as well as potential benefits, likely to be more extreme. Costs emerge in the short term from a variety of mechanisms [75]. Incoming genes represent additional DNA that draws on cellular resources for replication, transcription, and translation. The sequence composition of acquired DNA may be poorly optimized for the host's expression machinery, resulting in stalled ribosomes, misfolded proteins, and triggering of stress responses, while expressed genes may interfere with cellular homeostasis by disrupting metabolic or signalling processes. Acquired TEs can proliferate in the chromosome, damaging the genes into which they insert and causing gene loss through recombination. Donors are also affected. The production of conjugative machinery is metabolically expensive, and exposes its bearers to ‘male-specific’ bacteriophage which recognize and use the conjugative pilus as a receptor [76]. The release of capsids to secrete genetic material, through generalized transduction or GTAs, can require lysis [39]. Unlike the metaphors that refer to transferred genes as swappable ‘smartphone apps’ [77], HGT may in reality be a more traumatic experience.
Several mechanisms have evolved that can inhibit HGT, allowing cells to escape this disruption [45]. For example, DNA restriction–modification systems and CRISPR-Cas (CRISPR, clustered regularly interspaced short palindromic repeats; Cas, CRISPR-associated) loci represent ‘immunity’ systems which recognize and selectively degrade foreign DNA. Though they probably evolved as a means of resisting highly antagonistic agents, such as bacteriophage, these systems may also impact potentially beneficial MGEs [78,79]. They, therefore, have the potential to cut lineages off from the flow of adaptive innovations. Experimental studies exploring this tension show that where pressure to acquire plasmid-borne genes is strong enough, bacteria tend to jettison their CRISPR-Cas immunity loci completely [80], enabling gene acquisition. However, immunity loci are also horizontally transferred, so it is possible to reacquire them. Indeed, comparative studies show no correlation between the degree of CRISPR-Cas immunity and recent HGT acquisitions [81], suggesting that immunity to HGT is likely to be dynamic, with transient periods of susceptibility and resistance. Besides physically degrading foreign DNA, bacteria can also exert some control over the expression of recent acquisitions. Newly arrived DNA tends to be relatively AT-rich in comparison with the resident chromosome, possibly reflective of an itinerant lifestyle [82]. The histone-like nucleoid-structuring protein (H-NS), a regulator encoded by bacteria, binds to and silences AT-rich DNA, preventing costly and maladapted expression of foreign genes [83,84], and representing a form of ‘censorship’ by the established genome.
An important aspect of the MGEs that facilitate gene exchange is that they themselves reproduce and mutate, and are subject to natural selection. They therefore have their own fitness ‘interests’, which may not necessarily be aligned with those of their hosts. For example, plasmids are under selection to increase copy number within a cell, but high copy number imposes a high cost on that cell [85]. This can generate significant evolutionary conflict between hosts and MGEs. Plasmid carriage, for example, can exert a considerable toll on host fitness, and selection might favour hosts that have managed to shed their plasmid burden [86]. Meanwhile, to prevent their loss, MGEs acquire modules to ensure their maintenance, such as those involved in plasmid partitioning, or genes that disable the host's CRISPR-Cas immunity loci [87]. Vertical and horizontal modes of MGE transmission are likely to trade-off against one another: adaptations that improve the ability of an MGE to move across lineages are likely to make that MGE costlier to the host it is in, while decreases in cost are likely to come from repressing horizontal transfer [88]. Hosts are under pressure to ‘domesticate’ or shed fractious MGEs, while MGEs are under pressure to maintain autonomy. In this context, it is interesting that of the three canonical mechanisms of DNA transfer, only one (natural transformation) is under the direct control of bacteria; the others are encoded by the semi-autonomous MGEs that inhabit them. The mobile gene pool may be akin to a library, but the books are alive.
Considering the potential for conflict between MGEs and their hosts, carriage by MGEs of potentially useful accessory genes, such as those involved in AMR or virulence, is difficult to explain. The benefits of accessory genes are likely to be highly context-dependent, varying with the chemical, physical and social environment [89–91]. Plasmids, for example, carry genes for resistance to environmental pollutants even in pristine habitats [92]. Under such conditions, where accessory genes are not beneficial, plasmids persist as parasitic entities, and would be expected to become more efficient parasites, streamlining their genomes through accessory gene excision and an increased transfer rate. Positive selection for accessory genes could offset the costs of plasmid carriage, but under such conditions selection would favour integration of the beneficial traits into the host chromosome and loss of the plasmid backbone. Regardless of selective conditions, accessory gene carriage by MGEs, though widespread, appears problematic. This puzzle has been termed the ‘plasmid paradox’ [93], but it can be generalized to include other MGEs such as transposons and integrative elements which maintain accessory gene mobility.
6. Keeping genes moving: resolving the plasmid paradox
Experimental evolution studies are providing some answers to this problem, at least for plasmids. Coevolution between plasmid and host can rapidly ameliorate the major costs of plasmid carriage, reducing the effects of purifying selection and maintaining gene mobility. Compensatory evolution can occur in the chromosome [94–96] or in the plasmid [97,98], and may be specific to that host–plasmid pairing or represent a more general adaptation. Interestingly, in some cases plasmid cost emerges from conflicts with other horizontally transferred elements. In Pseudomonas aeruginosa PAO1, cytotoxic gene expression from the small plasmid pNUK73 is induced by two recently acquired chromosomal genes. Disruption of one or both of these genes alleviates plasmid cost, resulting in maintenance of the plasmid and the antibiotic resistance gene it carries [99]. Some plasmids deploy their own H-NS-like genes, which reduces the burden they place on their hosts by repressing plasmid gene expression [100]. Evolution of gene regulators may prove to be a general theme in the accommodation of acquired genes, as comparative analyses show that gene regulatory regions tend to correlate with the accessory compartment rather than the core genome [26].
Alternatively, where rates of conjugation outweigh the costs of carriage and imperfect transmission to daughter cells, plasmids can be maintained in a population through infectious transfer [101]. Though there has been considerable debate over whether they are achieved in nature, high infection rates have been shown to sustain carriage in several laboratory experiments, at least over short periods [102–104]. Persistence through infection leads to a more antagonistic relationship between plasmids and their hosts: hosts are predicted to develop adaptations for actively resisting (re)-infection, whereas plasmids are likely to lose accessory genes to become better parasites.
Whether and how conflict with MGEs is resolved varies between hosts, as differing gene content between strains offers different opportunities and constraints to conflict resolution. In some species, plasmids are highly unstable owing to poor vertical transmission, in others they are unstable owing to a high cost [86], with differing evolutionary outcomes. For example, the IncP-1 plasmid derivative pMS0506 evolved increased stability in Shewanella odeidensis through mutations in trfA, which reduced plasmid cost [105], but in Pseudomonas moraviensis stability was increased by the acquisition, from another plasmid, of a transposon carrying a toxin–antitoxin system, which effectively reduced plasmid loss [106] (an example of mobile elements interacting to enhance plasmid maintenance). Moreover, although beneficial accessory genes can become ‘captured’ by the chromosome under positive selection, with consequent loss of the plasmid, this phenomenon varies between species [107].
MGEs are not just the traits they carry, and the relationship between these elements and their hosts may be multifaceted and more subtle than the effects of their accessory genes. Temperate phage, for instance, can be effective ‘weapons’ in bacterial warfare [108], while plasmid encoded conjugative pili can help bacterial hosts to form biofilms [109]. Besides these ecological effects, MGEs can alter bacterial evolution and gene regulation in ways beyond gene acquisition. Integrative elements like temperate phage, transposons and other IS elements can jump into genes and regulatory regions. This sledge-hammer approach to gene disruption can lead to rapid adaptation of the host to new environments [110,111]. Recent advances may also suggest that integrative elements may actively integrate into and drop out of bacterial genes to act as functional ‘switches’ in turn disrupting and restoring gene function [112]. Furthermore, the multicopy nature of many of these elements increases the opportunity for mutations in the genes they carry [113], and, in the case of plasmids, constitutes a responsive platform for altering gene dosage by varying copy number. During infection, Yersinia requires an increased gene dose of its type 3 secretion system (T3SS) for efficient colonization. This is achieved by a transient increase in copy number of the virulence plasmid pIBX, which carries the T3SS, from 1 to 3 per cell [114]. Aureimonas species carry plasmid-borne ribosomal RNA genes, potentially enabling rapid change in copy number which might provide selective benefit under changing environmental conditions [115].
Plasmid maintenance in a species, through amelioration or infectious transfer, may resonate through a community. In species-rich microbial communities, a subset of members able to maintain plasmids may act as a ‘source’ species for otherwise unfavourable hosts [107], a pattern that is reflected in the fact that the ability of a plasmid to invade a microbial community is correlated with existing plasmid maintenance [116]. Plasmids can still conjugate from hosts that have completely ameliorated their cost [96], and the ability of a plasmid to invade a diverse fraction of a community [117] means that a few source species could maintain community-wide gene mobility.
7. The impacts of horizontal gene transfer on genome evolution
Genes can be rapidly lost as well as gained, and gene loss frequently acts to pare down bacterial genomes [118]. This balance between acquisition and loss gives rise to the patterns of HGT that become apparent in large-scale genome analyses. These infer HGT by identifying genes shared between lineages and thus detect both recent gene transfer events and those that occurred long ago [30]. Shared genes represent not only successful transfer, but also maintenance in the donor and recipient lineages [119]. There is a bias in the types of gene detected by these studies, leading to theories about why certain genes are overrepresented among shared genes (i.e. are more ‘transferable’ than others). The ‘Complexity Hypothesis’ suggests that a gene's associated biological process is the main determinant of its transferability, with ‘informational’ genes (involved in transcription, translation and replication) less likely to be successfully transferred than ‘operational’ genes (involved in functions such as metabolism and regulation) [15]. This hypothesis has been refined to show that the primary factor impacting transferability is the number of protein–protein interactions the gene product is involved in [120]: genes highly integrated with many partners in one cell will be unlikely to provide benefit in a different cellular environment. Innovations that perform distinct, specific tasks are thus more likely to be maintained in a new lineage.
The apparently high rates of gene gain and loss detected when comparing recently diverged lineages with more ancient branches [29], suggests that transferred genes ‘live fast, die young’, undergoing constant turnover [30]. Those that are beneficial are retained by selection, while others, excised by the pervasive razor of gene deletion, are lost [118]. A study of HGT among human-associated microbes showed that although genes with plasmid-, phage- or transposon-related functions were identified, they comprised only a small fraction of the transfers detected [121]. This suggests that while MGEs can enhance HGT, they are not required for long-term maintenance of transferred genes in the recipient. Longer timescales are likely to see retention of beneficial genes and loss of their means of entry [122]. Indeed, the genetic context of transferred genes varies considerably between individuals and between populations [74] owing to recombination breaking linkage. This is consistent with a model of HGT whereby horizontal gene spread is lubricated by the activity of MGEs, but over longer periods the signatures of these elements is gradually erased as the functional genes become integrated into the physiology of their new hosts.
Where there is sufficient HGT, selection appears to act on genes, and sweeps can carry a particular allele to fixation in a population without purging other loci of their diversity, as was observed in a marine Vibrio population [123]. However, if advantageous alleles arise where there is a relatively low rate of HGT, genetic diversity is lost in a ‘genome-wide’ selective sweep, resulting in a much more clonal population. A nine-year study of 30 bacterial populations in a freshwater lake found gradual purging of genome-wide diversity in one species of green sulfur bacterium owing to a selective sweep [124], though these dynamics were not shared by other species at the same site. Interestingly, propensity to undergo gene-specific or genome-wide selective sweeps may be a stable trait, with low-diversity populations with evidence of prior genome-wide sweeps more likely to undergo future diversity-purging sweeps [125]. This suggests that the flow of genes varies within and between species, and is structured by consistent barriers. These barriers are probably determined in large part by the peculiarities of the elements involved, for example, carriage and compatibility of MGEs, or the presence of cognate restriction–modification systems [45,126].
8. Horizontal gene transfer shapes bacterial populations
The opportunity for gene transfer can have significant effects on the genetic structure of bacterial populations. Related bacteria end up with fewer genes in common, as lineages acquire different sets of genes from their local neighbours [127]. Meanwhile, horizontally transferred genes are associated more with ecological conditions or geographical locations rather than the phylogenetic lineages in which they are found [25,74,121]. In some cases, these genes have a clear relationship to the local environment, encoding, for example, degradation of locally occurring carbohydrate sources [74,128], or biofilm formation and host colonization [123]. Where bacteria can migrate, MGEs may be under selection to maintain mobility of these ‘niche-specific gene pools’ [129], perhaps by transfer to and assembly on a readily exchangeable plasmid or ICE, because transfer of these locally beneficial traits to potentially competitive newcomers benefits the success of the MGE [101,130].
Within communities HGT promotes diversity, rescuing unrelated species from purifying selection by the spread of ecologically relevant traits [101]. At the same time, HGT increases relatedness at specific loci, creating conditions conducive to the evolution and success of ‘cooperative’ traits (which may likewise be niche-specific). Cooperative traits, in this context, are costly actions that provide a benefit not only to the individual performing them, but also to their neighbours. Where neighbours do not reciprocate, cooperative traits are difficult to explain, because cooperators, burdened by the cost of their actions and sharing the benefits, are out-competed. Cooperative traits are thus expected to succeed where the recipients of the cooperative action are likely to be cooperators too. By spreading the genes involved in cooperation between otherwise unrelated individuals, plasmids and other MGEs can favour cooperation. In other words: from a plasmid's perspective, inducing cooperative behaviour in their hosts is beneficial, because it enhances the success of neighbours that are likely to become plasmid hosts too, through HGT [131]. Consistent with this, plasmids and other MGEs are overrepresented in accessory genes that encode traits regarded as cooperative: secreted functions the benefits of which are shared as ‘public goods’ among neighbours [132]. Experimental and modelling studies on a synthetic plasmid system also suggest that HGT can favour cooperative traits [133].
The acquisition of new traits by HGT can have decisive effects on the evolutionary trajectory of the recipient lineage with consequences that extend into human society. Pathogenic lineages often owe their devastating behaviour to genes harboured on MGEs. The ymt gene, which provided the agent of plague, Yersinia pestis, with an arthropod vector by allowing it to colonize the guts of fleas, was acquired when a TE transposed into a plasmid sometime in the Late Bronze Age [134]. The lysogenic phage CTX-phi converts Vibrio cholerae hosts from non-pathogenic to pathogenic by the expression of the cholera toxin, which was acquired horizontally by CTX-phi, exemplifying the nested levels of gene mobility in microbes [135]. Meanwhile the efficiency and flexibility of HGT, amplified by this nested structure, can be observed in the spread of AMR genes in hospital-acquired infections, where resistance genes carried on transposons are able to hop between different resident plasmids each able to infect multiple host lineages [54]. On a larger scale, HGT between more divergent participants lies at the root of key evolutionary transitions—although a matter of current debate in the literature, it has been suggested that major phylogenetic transitions in Archaea are associated with HGT from Eubacteria [136,137]. New acquisitions causing dramatic shifts in phenotype space offer opportunities to take up a very different lifestyle.
9. Concluding remarks
HGT among microbes may prove to be a good model for understanding the spread of innovations at other scales. Certainly, microbes have various idiosyncrasies that make them exceptionally susceptible to HGT as an engine of evolutionary change, and harbour well-described elements that are adapted for the job. But though the mechanisms facilitating it remain unclear (though some candidates have been described, [4,138]), the occurrence of HGT within and between multicellular eukaryotes is becoming increasingly apparent [14]. It will be interesting to see how far the ecological drivers of HGT in microbes similarly promote gene exchange among other life forms. The interaction between selection and drift in shaping patterns of gene exchange [127] and the ability for HGT to facilitate rapid adaptation of migrants [130] may have particular relevance.
Generalizing further, bacterial HGT demonstrates that, for the spread of innovations, ‘the medium is the message’ [139]—at least in the short term. High impact innovations are usually carried into cells by selfish MGEs, and though their signal may eventually be masked by gene loss, recombination, and domestication, the peculiarities of these strange semi-autonomous biological entities affect both the ways that genes flow through communities, and the consequences of this flux. Understanding the negotiations between MGEs and their hosts may therefore be as important as understanding the ecological or clinical significance of the genes that they transfer.
Acknowledgements
We thank Cagla Stevenson and two anonymous reviewers for comments and suggestions on the manuscript.
Data accessibility
This article has no additional data.
Competing interests
We declare that we have no competing interests.
Funding
J.P.J.H., M.A.B. and E.H. are supported by ERC Consolidator Grant Agreement no. 311490-COEVOCON to M.A.B.
References
- 1.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304. ( 10.1038/35012500) [DOI] [PubMed] [Google Scholar]
- 2.Syvanen M. 1994. Horizontal gene transfer: evidence and possible consequences. Annu. Rev. Genet. 28, 237–261. ( 10.1146/annurev.ge.28.120194.001321) [DOI] [PubMed] [Google Scholar]
- 3.Goldenfeld N, Woese C. 2007. Biology's next revolution. Nature 445, 369–369 ( 10.1038/445369a) [DOI] [PubMed] [Google Scholar]
- 4.Soucy SM, Huang J, Gogarten JP. 2015. Horizontal gene transfer: building the web of life. Nature 16, 472–482. ( 10.1038/nrg3962) [DOI] [PubMed] [Google Scholar]
- 5.Dennis JJ. 2005. The evolution of IncP catabolic plasmids. Curr. Opin. Biotechnol. 16, 291–298. ( 10.1016/j.copbio.2005.04.002) [DOI] [PubMed] [Google Scholar]
- 6.Monier A, Pagarete A, de Vargas C, Allen MJ, Read B, Claverie J-M, Ogata H. 2009. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Res. 19, 1441–1449. ( 10.1101/gr.091686.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koonin EV, Wolf YI. 2009. Is evolution Darwinian or/and Lamarckian? Biol. Direct. 4, 42 ( 10.1186/1745-6150-4-42) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acuña R, et al. 2012. Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc. Natl Acad. Sci. USA 109, 4197–4202. ( 10.1073/pnas.1121190109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson MT, Seifert HS. 2011. Opportunity and means: horizontal gene transfer from the human host to a bacterial pathogen. mBio 2, e00005-11 ( 10.1128/mBio.00005-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruto M, Prigent-Combaret C, Luis P, Moënne-Loccoz Y, Muller D. 2014. Frequent, independent transfers of a catabolic gene from bacteria to contrasted filamentous eukaryotes. Proc. R. Soc. B 281, 20140848 ( 10.1098/rspb.2014.0848) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hotopp JCD, et al. 2007. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756. ( 10.1126/science.1142490) [DOI] [PubMed] [Google Scholar]
- 12.Huang J. 2013. Horizontal gene transfer in eukaryotes: the weak-link model. Bioessays 35, 868–875. ( 10.1002/bies.201300007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koutsovoulos G, et al. 2016. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proc. Natl Acad. Sci. USA 113, 5053–5058. ( 10.1073/pnas.1600338113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boto L. 2014. Horizontal gene transfer in the acquisition of novel traits by metazoans. Proc. R. Soc. B 281, 20132450 ( 10.1098/rspb.2013.2450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain R, Rivera MC, Lake JA. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA 96, 3801–3806. ( 10.1073/pnas.96.7.3801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koonin EV, Makarova KS, Aravind L. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55, 709–742. ( 10.1146/annurev.micro.55.1.709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiedenbeck J, Cohan FM. 2011. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35, 957–976. ( 10.1111/j.1574-6976.2011.00292.x) [DOI] [PubMed] [Google Scholar]
- 18.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583. ( 10.1073/pnas.95.12.6578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sunagawa S, et al. 2015. Ocean plankton. Structure and function of the global ocean microbiome. Science 348, 1261359 ( 10.1126/science.1261359) [DOI] [PubMed] [Google Scholar]
- 20.Funkhouser LJ, Bordenstein SR. 2013. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 11, e1001631 ( 10.1371/journal.pbio.1001631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buckling A, Craig Maclean R, Brockhurst MA, Colegrave N. 2009. The Beagle in a bottle. Nature 457, 824–829. ( 10.1038/nature07892) [DOI] [PubMed] [Google Scholar]
- 22.Kellogg CA, Griffin DW. 2006. Aerobiology and the global transport of desert dust. Trends Ecol. Evol. 21, 638–644. ( 10.1016/j.tree.2006.07.004) [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson DM. 2010. Have we underestimated the importance of humans in the biogeography of free-living terrestrial microorganisms? J. Biogeogr. 37, 393–397. ( 10.1111/j.1365-2699.2009.02236.x) [DOI] [Google Scholar]
- 24.Lozupone CA, Knight R. 2007. Global patterns in bacterial diversity. Proc. Natl Acad. Sci. USA 104, 11 436–11 440. ( 10.1073/pnas.0611525104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowell RW, Green S, Laue BE, Sharp PM. 2014. The extent of genome flux and its role in the differentiation of bacterial lineages. Genome Biol. Evol. 6, 1514–1529. ( 10.1093/gbe/evu123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNally A, et al. 2016. Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS Genet. 12, e1006280 ( 10.1371/journal.pgen.1006280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young JPW, et al. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7, R34 ( 10.1186/gb-2006-7-4-r34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tettelin H, et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ‘pan-genome’. Proc. Natl Acad. Sci. USA 102, 13 950–13 955. ( 10.1073/pnas.0506758102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vos M, Hesselman MC, Beek TA, van Passel MWJ, Eyre-Walker A. 2015. Rates of lateral gene transfer in prokaryotes: high but why? Trends Microbiol. 23, 598–605. ( 10.1016/j.tim.2015.07.006) [DOI] [PubMed] [Google Scholar]
- 30.Hao W, Golding GB. 2006. The fate of laterally transferred genes: life in the fast lane to adaptation or death. Genome Res. 16, 636–643. ( 10.1101/gr.4746406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guglielmini J, de la Cruz F, Rocha EPC. 2013. Evolution of conjugation and type IV secretion systems. Mol. Biol. Evol. 30, 315–331. ( 10.1093/molbev/mss221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7, 703–714. ( 10.1038/nrmicro2218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EPC, de la Cruz F. 2010. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. ( 10.1128/MMBR.00020-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Novick RP. 2009. Phage-mediated intergeneric transfer of toxin genes. Science 323, 139–141. ( 10.1126/science.1164783) [DOI] [PubMed] [Google Scholar]
- 35.Modi SR, Lee HH, Spina CS, Collins JJ. 2013. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222. ( 10.1038/nature12212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Draghi JA. 2006. DNA secretion and gene-level selection in bacteria. Microbiology 152, 2683–2688. ( 10.1099/mic.0.29013-0) [DOI] [PubMed] [Google Scholar]
- 37.Keen EC, Bliskovsky VV, Malagon F, Baker JD, Prince JS, Klaus JS, Adhya SL. 2017. Novel ‘superspreader’ bacteriophages promote horizontal gene transfer by transformation. mBio 8, e02115-16 ( 10.1128/mBio.02115-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. 2015. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347, 63–67. ( 10.1126/science.1260064) [DOI] [PubMed] [Google Scholar]
- 39.Lang AS, Zhaxybayeva O, Beatty JT. 2012. Gene transfer agents: phage-like elements of genetic exchange. Nat. Rev. Microbiol. 10, 472–482. ( 10.1038/nrmicro2802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dubey GP, Ben-Yehuda S. 2011. Intercellular nanotubes mediate bacterial communication. Cell 144, 590–600. ( 10.1016/j.cell.2011.01.015) [DOI] [PubMed] [Google Scholar]
- 41.Naor A, Lapierre P, Mevarech M, Papke RT, Gophna U. 2012. Low species barriers in halophilic Archaea and the formation of recombinant hybrids. Curr. Biol. 22, 1444–1448. ( 10.1016/j.cub.2012.05.056) [DOI] [PubMed] [Google Scholar]
- 42.Fulsundar S, Harms K, Flaten GE, Johnsen PJ, Chopade BA, Nielsen KM. 2014. Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl. Environ. Microbiol. 80, 3469–3483. ( 10.1128/AEM.04248-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mortimer TD, Pepperell CS. 2014. Genomic signatures of distributive conjugal transfer among mycobacteria. Genome Biol. Evol. 6, 2489–2500. ( 10.1093/gbe/evu175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray TA, Clark RR, Boucher N, Lapierre P, Smith C, Derbyshire KM. 2016. Intercellular communication and conjugation are mediated by ESX secretion systems in mycobacteria. Science 354, 347–350. ( 10.1126/science.aag0828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. ( 10.1038/nrmicro1234) [DOI] [PubMed] [Google Scholar]
- 46.Craig NL, Chandler M, Gellert M, Lambowitz AM, Rice PA, Sandmeyer SB. 2015. Mobile DNA III. Washington, DC: American Society for Microbiology. [Google Scholar]
- 47.Croucher NJ, Mostowy R, Wymant C, Turner P, Bentley SD, Fraser C. 2016. Horizontal DNA transfer mechanisms of bacteria as weapons of intragenomic conflict. PLoS Biol. 14, e1002394 ( 10.1371/journal.pbio.1002394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ambur OH, Engelstädter J, Johnsen PJ, Miller EL, Rozen DE. 2016. Steady at the wheel: conservative sex and the benefits of bacterial transformation. Phil. Trans. R. Soc. B 371, 20150528 ( 10.1098/rstb.2015.0528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engelmoer DJP, Donaldson I, Rozen DE. 2013. Conservative sex and the benefits of transformation in Streptococcus pneumoniae. PLoS Pathog. 9, e1003758 ( 10.1371/journal.ppat.1003758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jalasvuori M, Koonin EV. 2015. Classification of prokaryotic genetic replicators: between selfishness and altruism. Ann. NY Acad. Sci. 1341, 96–105. ( 10.1111/nyas.12696) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leclercq S, Cordaux R. 2011. Do phages efficiently shuttle transposable elements among prokaryotes? Evolution 65, 3327–3331. ( 10.1111/j.1558-5646.2011.01395.x) [DOI] [PubMed] [Google Scholar]
- 52.Ramsay JP, Firth N. 2017. Diverse mobilization strategies facilitate transfer of non-conjugative mobile genetic elements. Curr. Opin. Microbiol. 38, 1–9. ( 10.1016/j.mib.2017.03.003) [DOI] [PubMed] [Google Scholar]
- 53.He S, Chandler M, Varani AM, Hickman AB, Dekker JP, Dyda F. 2016. Mechanisms of evolution in high-consequence drug resistance plasmids. mBio 7, e01987-16 ( 10.1128/mBio.01987-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheppard AE, et al. 2016. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob. Agents Chemother. 60, 3767–3778. ( 10.1128/AAC.00464-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wozniak RAF, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8, 552–563. ( 10.1038/nrmicro2382) [DOI] [PubMed] [Google Scholar]
- 56.Obeng N, Pratama AA, van Elsas JD. 2016. The significance of mutualistic phages for bacterial ecology and evolution. Trends Microbiol. 24, 440–449. ( 10.1016/j.tim.2015.12.009) [DOI] [PubMed] [Google Scholar]
- 57.McCarthy AJ, Loeffler A, Witney AA, Gould KA, Lloyd DH, Lindsay JA. 2014. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol. Evol. 6, 2697–2708. ( 10.1093/gbe/evu214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quiles-Puchalt N, Carpena N, Alonso JC, Novick RP, Marina A, Penadés JR. 2014. Staphylococcal pathogenicity island DNA packaging system involving cos-site packaging and phage-encoded HNH endonucleases. Proc. Natl Acad. Sci. USA 111, 6016–6021. ( 10.1073/pnas.1320538111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toussaint A, Merlin C. 2002. Mobile elements as a combination of functional modules. Plasmid 47, 26–35. ( 10.1006/plas.2001.1552) [DOI] [PubMed] [Google Scholar]
- 60.Sengupta M, Austin S. 2011. Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect. Immun. 79, 2502–2509. ( 10.1128/IAI.00127-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wintersdorff CJH, Penders J, van Niekerk JM, Mills ND, Majumder S, van Alphen LB, Savelkoul PHM, Wolffs PFG. 2016. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7, 173 ( 10.3389/fmicb.2016.00173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Partridge SR, Tsafnat G, Coiera E, Iredell JR. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33, 757–784. ( 10.1111/j.1574-6976.2009.00175.x) [DOI] [PubMed] [Google Scholar]
- 63.Lacotte Y, Ploy M-C, Raherison S. 2017. Class 1 integrons are low-cost structures in Escherichia coli. ISME J. 11, 1535–1544. ( 10.1038/ismej.2017.38) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. ( 10.1038/nrmicro1235) [DOI] [PubMed] [Google Scholar]
- 65.Norman A, Hansen LH, Sorensen SJ. 2009. Conjugative plasmids: vessels of the communal gene pool. Phil. Trans. R. Soc. B 364, 2275–2289. ( 10.1126/science.1093857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. 2010. Network analyses structure genetic diversity in independent genetic worlds. Proc. Natl Acad. Sci. USA 107, 127–132. ( 10.1073/pnas.0908978107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dahlberg C, Bergström M, Hermansson M. 1998. In situ detection of high levels of horizontal plasmid transfer in marine bacterial communities. Appl. Environ. Microbiol. 64, 2670–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kenzaka T, Tani K, Nasu M. 2010. High-frequency phage-mediated gene transfer in freshwater environments determined at single-cell level. ISME J. 4, 648–659. ( 10.1038/ismej.2009.145) [DOI] [PubMed] [Google Scholar]
- 69.McDaniel LD, Young E, Delaney J, Ruhnau F, Ritchie KB, Paul JH. 2010. High frequency of horizontal gene transfer in the oceans. Science 330, 50–50 ( 10.1126/science.1192243) [DOI] [PubMed] [Google Scholar]
- 70.Sørensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3, 700–710. ( 10.1038/nrmicro1232) [DOI] [PubMed] [Google Scholar]
- 71.Zhong X, Krol JE, Top EM, Krone SM. 2010. Accounting for mating pair formation in plasmid population dynamics. J. Theor. Biol. 262, 711–719. ( 10.1016/j.jtbi.2009.10.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mao D, Luo Y, Mathieu J, Wang Q, Feng L, Mu Q, Feng C, Alvarez PJJ. 2014. Persistence of extracellular DNA in river sediment facilitates antibiotic resistance gene propagation. Environ. Sci. Technol. 48, 71–78. ( 10.1021/es404280v) [DOI] [PubMed] [Google Scholar]
- 73.Touchon M, Bernheim A, Rocha EP. 2016. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 10, 2744–2754. ( 10.1038/ismej.2016.47) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brito IL, et al. 2016. Mobile genes in the human microbiome are structured from global to individual scales. Nature 535, 435–439. ( 10.1038/nature18927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baltrus DA. 2013. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489–495. ( 10.1016/j.tree.2013.04.002) [DOI] [PubMed] [Google Scholar]
- 76.May T, Tsuruta K, Okabe S. 2010. Exposure of conjugative plasmid carrying Escherichia coli biofilms to male-specific bacteriophages. ISME J. 5, 771–775. ( 10.1038/ismej.2010.158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Young JPW. 2016. Bacteria are smartphones and mobile genes are apps. Trends Microbiol. 24, 931–932. ( 10.1016/j.tim.2016.09.002) [DOI] [PubMed] [Google Scholar]
- 78.Vasu K, Nagamalleswari E, Nagaraja V. 2012. Promiscuous restriction is a cellular defense strategy that confers fitness advantage to bacteria. Proc. Natl Acad. Sci. USA 109, E1287–E1293. ( 10.1073/pnas.1119226109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garneau JE, et al. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. ( 10.1038/nature09523) [DOI] [PubMed] [Google Scholar]
- 80.Jiang W, Maniv I, Arain F, Wang Y, Levin BR, Marraffini LA. 2013. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet. 9, e1003844 ( 10.1371/journal.pgen.1003844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gophna U, Kristensen DM, Wolf YI, Popa O, Drevet C, Koonin EV. 2015. No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales. ISME J. 9, 2021–2027. ( 10.1038/ismej.2015.20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Daubin V, Lerat E, Perrière G. 2003. The source of laterally transferred genes in bacterial genomes. Genome Biol. 4, R57 ( 10.1186/gb-2003-4-9-r57) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JCD. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2, e81 ( 10.1371/journal.ppat.0020081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238. ( 10.1126/science.1128794) [DOI] [PubMed] [Google Scholar]
- 85.Paulsson J. 2002. Multileveled selection on plasmid replication. Genetics 161, 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.De Gelder L, Ponciano JM, Joyce P, Top EM. 2007. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 153, 452–463. ( 10.1099/mic.0.2006/001784-0) [DOI] [PubMed] [Google Scholar]
- 87.Maxwell KL. 2016. Phages fight back: inactivation of the CRISPR-Cas bacterial immune system by anti-CRISPR proteins. PLoS Pathog. 12, e1005282 ( 10.1371/journal.ppat.1005282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Turner PE, Cooper VS, Lenski RE. 1998. Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution 52, 315–329. ( 10.7717/peerj.2060) [DOI] [PubMed] [Google Scholar]
- 89.Hall JPJ, Harrison E, Lilley AK, Paterson S, Spiers AJ, Brockhurst MA. 2015. Environmentally co-occurring mercury resistance plasmids are genetically and phenotypically diverse and confer variable context-dependent fitness effects. Environ. Microbiol. 17, 5008–5022. ( 10.1111/1462-2920.12901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI. 2014. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio 5, e01918-14 ( 10.1128/mBio.01918-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ellis RJ, Lilley AK, Lacey SJ, Murrell D, Godfray HCJ. 2007. Frequency-dependent advantages of plasmid carriage by Pseudomonas in homogeneous and spatially structured environments. ISME J. 1, 92–95. ( 10.1038/ismej.2007.11) [DOI] [PubMed] [Google Scholar]
- 92.Mindlin S, Minakhin L, Petrova M, Kholodii G, Minakhina S, Gorlenko Z, Nikiforov V. 2005. Present-day mercury resistance transposons are common in bacteria preserved in permafrost grounds since the Upper Pleistocene. Res. Microbiol. 156, 994–1004. ( 10.1016/j.resmic.2005.05.011) [DOI] [PubMed] [Google Scholar]
- 93.Harrison E, Brockhurst MA. 2012. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 20, 262–267. ( 10.1016/j.tim.2012.04.003) [DOI] [PubMed] [Google Scholar]
- 94.Modi RI, Adams J. 1991. Coevolution in bacterial-plasmid populations. Evolution 45, 656 ( 10.2307/2409918) [DOI] [PubMed] [Google Scholar]
- 95.Dahlberg C, Chao L. 2003. Amelioration of the cost of conjugative plasmid carriage in Escherichia coli K12. Genetics 165, 1641–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harrison E, Guymer D, Spiers AJ, Paterson S, Brockhurst MA. 2015. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 25, 2034–2039. ( 10.1016/j.cub.2015.06.024) [DOI] [PubMed] [Google Scholar]
- 97.Porse A, Schønning K, Munck C, Sommer MOA. 2016. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol. Biol. Evol. 33, msw163 ( 10.1093/molbev/msw163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bedhomme S, Perez Pantoja D, Bravo IG. 2017. Plasmid and clonal interference during post horizontal gene transfer evolution. Mol. Ecol. 26, 1832–1847. ( 10.1111/mec.14056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.San Millan A, Toll-Riera M, Qi Q, MacLean RC. 2015. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat. Commun. 6, 6845 ( 10.1038/ncomms7845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ. 2007. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science 315, 251–252. ( 10.1126/science.1137550) [DOI] [PubMed] [Google Scholar]
- 101.Bergstrom CT, Lipsitch M, Levin BR. 2000. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155, 1505–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stevenson C, Hall JP, Harrison E, Wood AJ, Brockhurst MA. 2017. Gene mobility promotes the spread of resistance in bacterial populations. ISME J. 63, 1577 ( 10.1038/ismej.2017.42) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bahl MI, Hansen LH, Sørensen SJ. 2007. Impact of conjugal transfer on the stability of IncP-1 plasmid pKJK5 in bacterial populations. FEMS Microbiol. Lett. 266, 250–256. ( 10.1111/j.1574-6968.2006.00536.x) [DOI] [PubMed] [Google Scholar]
- 104.Fox RE, Zhong X, Krone SM, Top EM. 2008. Spatial structure and nutrients promote invasion of IncP-1 plasmids in bacterial populations. ISME J. 2, 1024–1039. ( 10.1038/ismej.2008.53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yano H, Wegrzyn K, Loftie-Eaton W, Johnson J, Deckert GE, Rogers LM, Konieczny I, Top EM. 2016. Evolved plasmid-host interactions reduce plasmid interference cost. Mol. Microbiol. 101, 743–756. ( 10.1111/mmi.13407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Loftie-Eaton W, et al. 2015. Evolutionary paths that expand plasmid host-range: implications for spread of antibiotic resistance. Mol. Biol. Evol. 33, 885–897. ( 10.1093/molbev/msv339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hall JPJ, Wood AJ, Harrison E, Brockhurst MA. 2016. Source–sink plasmid transfer dynamics maintain gene mobility in soil bacterial communities. Proc. Natl Acad. Sci. USA 113, 8260–8265. ( 10.1073/pnas.1600974113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brown SP, Le Chat L, De Paepe M, Taddei F. 2006. Ecology of microbial invasions: amplification allows virus carriers to invade more rapidly when rare. Curr. Biol. 16, 2048–2052. ( 10.1016/j.cub.2006.08.089) [DOI] [PubMed] [Google Scholar]
- 109.Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412, 442–445. ( 10.1038/35086581) [DOI] [PubMed] [Google Scholar]
- 110.Davies EV, James CE, Williams D, O'Brien S, Fothergill JL, Haldenby S, Paterson S, Winstanley C, Brockhurst MA. 2016. Temperate phages both mediate and drive adaptive evolution in pathogen biofilms. Proc. Natl Acad. Sci. USA 113, 8266–8271. ( 10.1073/pnas.1520056113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Biémont C. 2010. A brief history of the status of transposable elements: from junk DNA to major players in evolution. Genetics 186, 1085–1093. ( 10.1534/genetics.110.124180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. 2015. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 13, 641–650. ( 10.1038/nrmicro3527) [DOI] [PubMed] [Google Scholar]
- 113.San Millan A, Escudero J, Gifford D, Mazel D. 2016. Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nat. Ecol. Evol. 1, 10 ( 10.1038/s41559-016-0010) [DOI] [PubMed] [Google Scholar]
- 114.Wang H, Avican K, Fahlgren A, Erttmann SF, Nuss AM, Dersch P, Fallman M, Edgren T, Wolf-Watz H. 2016. Increased plasmid copy number is essential for Yersinia T3SS function and virulence. Science 353, 492–495. ( 10.1126/science.aaf7501) [DOI] [PubMed] [Google Scholar]
- 115.Anda M, Ohtsubo Y, Okubo T, Sugawara M, Nagata Y, Tsuda M, Minamisawa K, Mitsui H. 2015. Bacterial clade with the ribosomal RNA operon on a small plasmid rather than the chromosome. Proc. Natl Acad. Sci. USA 112, 14 343–14 347. ( 10.1073/pnas.1514326112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bellanger X, Guilloteau H, Breuil B. 2014. Natural microbial communities supporting the transfer of the IncP-1β plasmid pB10 exhibit a higher initial content of plasmids from the same incompatibility group. Front. Microbiol. 5, 637 ( 10.3389/fmicb.2014.00637/abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Klümper U, Riber L, Dechesne A, Sannazzarro A, Hansen LH, Sørensen SJ, Smets BF. 2015. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 9, 934–945. ( 10.1038/ismej.2014.191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kuo CH, Ochman H. 2009. Deletional bias across the three domains of life. Genome Biol. Evol. 1, 145–152. ( 10.1093/gbe/evp016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rocha EP. 2008. Evolutionary patterns in prokaryotic genomes. Curr. Opin. Microbiol. 11, 454–460. ( 10.1016/j.mib.2008.09.007) [DOI] [PubMed] [Google Scholar]
- 120.Cohen O, Gophna U, Pupko T. 2011. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol. Biol. Evol. 28, 1481–1489. ( 10.1093/molbev/msq333) [DOI] [PubMed] [Google Scholar]
- 121.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. 2011. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244. ( 10.1038/nature10571) [DOI] [PubMed] [Google Scholar]
- 122.Wagner A. 2006. Periodic extinctions of transposable elements in bacterial lineages: evidence from intragenomic variation in multiple genomes. Mol. Biol. Evol. 23, 723–733. ( 10.1093/molbev/msj085) [DOI] [PubMed] [Google Scholar]
- 123.Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabó G, Polz MF, Alm EJ. 2012. Population genomics of early events in the ecological differentiation of bacteria. Science 336, 48–51. ( 10.1126/science.1218198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bendall ML, et al. 2016. Genome-wide selective sweeps and gene-specific sweeps in natural bacterial populations. ISME J. 10, 1589–1601. ( 10.1038/ismej.2015.241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shapiro BJ. 2016. How clonal are bacteria over time? Curr. Opin. Microbiol. 31, 116–123. ( 10.1016/j.mib.2016.03.013) [DOI] [PubMed] [Google Scholar]
- 126.Oliveira PH, Touchon M, Rocha EPC. 2016. Regulation of genetic flux between bacteria by restriction–modification systems. Proc. Natl Acad. Sci. USA 113, 5658–5663. ( 10.1073/pnas.1603257113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McInerney JO, McNally A, O'Connell MJ. 2017. Why prokaryotes have pangenomes. Nat. Microbiol. 2, 17040 ( 10.1038/nmicrobiol.2017.40) [DOI] [PubMed] [Google Scholar]
- 128.Hehemann J-H, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. 2010. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912. ( 10.1038/nature08937) [DOI] [PubMed] [Google Scholar]
- 129.Polz MF, Alm EJ, Hanage WP. 2013. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. 29, 170–175. ( 10.1016/j.tig.2012.12.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Niehus R, Mitri S, Fletcher AG, Foster KR. 2015. Migration and horizontal gene transfer divide microbial genomes into multiple niches. Nat. Commun. 6, 8924 ( 10.1038/ncomms9924) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Birch J. 2014. Gene mobility and the concept of relatedness. Biol. Phil. 29, 445–476. ( 10.1007/s10539-014-9445-z) [DOI] [Google Scholar]
- 132.Rankin DJ, Rocha EPC, Brown SP. 2011. What traits are carried on mobile genetic elements, and why? Heredity 106, 1–10. ( 10.1038/hdy.2010.24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dimitriu T, Lotton C, Benard-Capelle J, Misevic D, Brown SP, Lindner AB, Taddei F. 2014. Genetic information transfer promotes cooperation in bacteria. Proc. Natl Acad. Sci. USA 111, 11 103–11 108. ( 10.1073/pnas.1406840111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rasmussen S, et al. 2015. Early divergent strains of Yersinia pestis in Eurasia 5000 years ago. Cell 163, 571–582. ( 10.1016/j.cell.2015.10.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Faruque SM, Mekalanos JJ. 2012. Phage-bacterial interactions in the evolution of toxigenic Vibrio cholerae. Virulence 3, 556–565. ( 10.4161/viru.22351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nelson-Sathi S, et al. 2015. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517, 77–80. ( 10.1038/nature13805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Groussin M, Boussau B, Szöllõsi G, Eme L, Gouy M, Brochier-Armanet C, Daubin V. 2016. Gene acquisitions from bacteria at the origins of major archaeal clades are vastly overestimated. Mol. Biol. Evol. 33, 305–310. ( 10.1093/molbev/msv249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Graham LA, Li J, Davidson WS, Davies PL. 2012. Smelt was the likely beneficiary of an antifreeze gene laterally transferred between fishes. BMC Evol. Biol. 12, 190 ( 10.1186/1471-2148-12-190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McLuhan ML. 1964. Understanding media. London, UK: Allen Lane. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.