Abstract
MicroRNAs (miRNAs) are widely up-regulated or down-regulated in a variety of tumors, including lung cancer, liver cancer, and colorectal cancer (CRC). Furthermore, miRNAs can function as tumor suppressors or proto-oncogenes by controlling the growth and metastasis of cancer cells. In the present study, we found a significant increase in miR19b-3p levels in CRC compared to tumor tissue and revealed the role of miR19b-3p in CRC growth and metastasis. The exogenous overexpression of miR19b-3p induced the proliferation, migration, and invasion of CRC cells in vitro. In addition, the nude mouse xenograft model showed that miR19b-3p overexpression promoted CRC growth and lung metastasis in vivo, whereas silencing miR19b-3p showed opposite results. Mechanistic studies have shown that the integrin beta-8 (ITGB8) transcript is one of the direct targets of miR19b-3p, and the expression of ITGB8 in CRC specimens was positively correlated with miR19b-3p. Finally, ectopic expression of ITGB8 rescued cell proliferation and invasion, which was inhibited by down-regulation of miR19b-3p. In addition, knockdown of ITGB8 neutralized the effects of miR19b-3p overexpression on cell growth and metastasis in CRC cells. Together, these results suggest that the miR19b-3p/ITGB8 axis plays an important role in the growth and metastasis of CRC.
Keywords: Colorectal cancer, miR19b-3p, ITGB8, metastasis
Introduction
Colorectal cancer (CRC) is considered to be one of the most important causes of cancer-associated mortalities worldwide, and the incidence of CRC is expected to further increase [1]. Despite improvements in treatment strategies for CRC, the overall survival (OS) of patients with CRC after surgical resection remains unsatisfactory owing to its recurrence and metastasis [2]. In addition, the molecular mechanism of CRC metastasis has not been well elucidated. Hence, a better understanding of the molecular and cellular mechanism responsible for CRC metastasis is required to improve the prognosis of patients. Substantive studies have demonstrated that a variety of microRNAs (miRNAs) are associated with human cancer progression, including growth and metastasis [3]. MiRNAs are small non-coding RNAs that are involved in the post-transcriptional regulation of genes. Importantly, its intricate regulatory network consists of a series of miRNAs that not only regulate the levels of multiple target genes, but also regulate genes via a combination of miRNAs [4]. For example, several miRNAs regulate the expression of tumor suppressor genes or oncogenes to play a functional role in the progression of cancer. In this study, we found that miR19b-3p was significantly up-regulated in CRC compared to that in adjacent non-tumor tissues [5].
MiR19b-3p belongs to both miR17-92 and miR106-363 clusters, which play significant roles in proliferation and cell survival [6]. A previous study revealed that miR19b-3p is upregulated in nasopharyngeal carcinoma and served as an independent predictor for reduced patient survival [7]. MiR19b-3p overexpression resulted in decreased sensitivity of nasopharyngeal carcinoma cells to irradiation, whereas miR19b-3p down-regulation resulted in increased sensitivity to irradiation in vitro [8]. Mechanistically, we found that miR19b-3p increased nasopharyngeal carcinoma cell radio-resistance by activating the TNFAIP3/NF-κB axis [9]. In addition, miR19b-3p may be a prospective biomarker for detecting gastric cancer and to assess its progression [10]. However, the roles of miR19b-3p in the progression of CRC have yet to be studied.
Integrins exist on the surface of cancer cells and promote the metastatic potential of cancer cells via mediating cell-cell adhesion and invasion [11]. The different integrin subfamilies are determined by the β subunit. Integrin β-8 (ITGB8) binds to the αv subunit [12], and has been shown to be up-regulated in many types of cancer, including lung cancer, breast cancer, laryngeal cancer, and gastric carcinoma [13]. In addition, ITGB8 showed higher expression in highly metastatic tumors and is considered a clinical metastasis-related gene and a potential target for the treatment of metastatic cancers [14]. However, the mechanism of ITGB8 in CRC metastasis and the relationship between ITGB8 and miR19b-3p is still not very clear.
In the present study, we demonstrated that miR19b-3p was up-regulated in CRC samples and cell lines. Overexpression of miR19b-3p induced the proliferation, colony formation, and tumor growth of CRC HCT116 and SW480 cells. In addition, overexpression of miR19b-3p significantly accelerated the metastasis of CRC cells in vitro and in vivo. However, silencing endogenous miR19b-3p in HCT116 and SW480 cells resulted in the opposite outcomes. In addition, we identified one of the miR19b-3p target genes, ITGB8, to play a functional role in the growth and metastasis of CRC cells regulated by miR19b-3p. Together, these results provide novel insights into the mechanism of the miR19b-3p/ITGB8 axis in CRC metastasis.
Materials and method
Colorectal cancer specimens and cell lines
The colorectal cancer samples and adjacent tissues were obtained from the Sixth Affiliated Hospital, Sun Yat-sen University. All informed consents from patients with colorectal cancer before surgery were obtained from the Regional Ethics Committee of the Sixth Affiliated Hospital, Sun Yat-sen University. Association of miR-19b-3p expression with clinic-pathological characteristics was summarized in Supplementary Table 2. Colorectal cancer cell lines, including HCT116, HT-29, SW480, LOVO and human colonic epithelial cells HCoEpiC were purchased from Guang Zhou Jennio Biotech Co.,Ltd. All cell lines were maintained at in the Dulbecco’s Modified Eagle medium (DMEM) or RPMI-1640 supplemented with 10% FBS, at 37°C in 5% CO2 atmosphere.
miRNA mimic, inhibitor and shITGB8 transfection
MiR-19b-3p mimics and negative control double helix (named miR-NC), miR-19b-3p inhibitor oligonucleotide (designated miR-19b-3p inhibitor) and the inhibitor of negative control oligonucleotide (named as inhibitor NC) were used for the transient gain of functional and loss of functional studies. Small interfering RNA duplex (shRNA) for ITGB8 (named shITGB8) was used for ITGB8 knock-down. Lipofectamine 2000 reagent (Invitrogen, USA) was used for transient transfection.
MTT proliferation assay and colony formation assay
3 x 105 HCT116 or SW480 cells were in plated into 96-well plates for 1, 2, 3 or 4 days. 100 μl of MTT resolution was added to the wells, and the cells were incubated at 37°C for 3 h. Cell supernatant was removed and 200 μl DMSO was added. Finally, the optical density (OD) value was measured at 490 nm. In colony formation assay, 1 x 102 HCT116 or SW480 cells were inoculated in complete medium and cultured for 2 weeks. The cell colonies were stained with 0.1% crystal violet and the number of colonies was analyzed.
Wound healing migration and Transwell invasion assays
1 x 105 HCT116 or SW480 cells were seeded into 6-well plates. The confluent monolayers of cells were scratched with 100 μl pipette tip, and the wound closures of cells was observed by microscopy, and by measuring the size of the initial wound and comparing the size of the wound after 24 h. 1 x 105 HCT116 or SW480 cells were seeded into upper chamber of Transwell that coated 8 μm pore size, and the Transwell chamber was placed into 24 well culture plate. After 24 h, the cells invaded into the lower chamber were stained with crystal violet and counted by microscopy [15].
Western blotting and quantitative RT-PCR
Cells were collected and split on the ice using cell lysis buffer for 15 minutes, and the supernatant was denatured in 5 x loading buffer containing SDS. PVDF membranes were blocked and antibody dilution using TBST. The following antibodies were prepared: MMP-2 (Cell Signaling Technology, 1:1000), MMP-9 (Cell Signaling Technology, 1:1000), TIMP-1 (Cell Signaling Technology, 1:1000), GAPDH (Cell Signaling Technology, 1:1000), ITGB8 (Cell Signaling Technology, 1:1000), goat anti-rabbit IgG-HRP (Santa Cruz, 1:5000). The target proteins were detected by ECL system (Millipore, Germany). Total RNA was extracted from colorectal cancer cells or clinical tissues using Trizol. RNA (1 ug) was reverse transcribed into cDNA by ReverTra Ace qPCR RT Kit. The qRT-PCR was performed to analysis mRNA of miR-19b-3p or ITGB8 using AceQ qPCR SYBR Green Master Mix. The formula for the relative expression value was calculated as 2-ΔΔCt. The primer sequences of GAPDH are 5’-CTCACCGGATGCACCAATGTT-3’ and 5’-CGCGTTGCTCACAATGTTCAT-3’; ITGB8 is 5’-CGTGACTTTCGTCTTGGATTTGG-3’ and 5’-TCCTTTCGGGGTGGATGCTAA-3’.
Luciferase reporter assay
Vector carrying the WT FOXL2 promoter sequence Luc-ITGB8 and the MUT Luc-ITGB8 were synthesized (GeneCopoeia). 1 × 105 cells/well was seeded in 24-well plate. After 24 h, the cells were co-transfected with vector plasmids and miR-19b-3p using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. 24 h later, the transfected cells were lysed and the resulting lysates were centrifuged. Luciferase activity was measured by using luciferase assay system (E2920, Promega) [16].
In vivo tumorigenesis and metastasis
All mouse experiments were performed according to the standard procedure approved by the Sixth Affiliated Hospital, Sun Yat-sen University. 1 x 106 Control cells or miR-19b-3p overexpression/down-regulated cells was subcutaneously injection of into the lateral ventral. The tumor size was measured once a week with the caliper and calculated as tumor volume = 0.5 x length × width2. The tumor tissue from mice was fixed with formalin and paraffin-embedded [17]. Sliced samples were performed immunohistochemistry staining for Ki67. In the lung metastasis assay, 1 x 106 HCT116 or SW480 cells were injected into BALB/c mice by tail vein. After 3 weeks, the nude mice were killed and the lungs were collected. H&E stains were performed on sections from embedded lung. Animal experiments were approved by the Animal Care Use Committee of the Sixth Affiliated Hospital, Sun Yat-sen University.
Statistical analysis
The results are expressed as the mean ± SD deviation of at least three repetitions in each group, and the student’s t test is performed to compare the differences between the two groups. P < 0.05 considered as significant.
Results
The expression of miR19b-3p is down-regulated in CRC
YM500v2 meta-analysis [18] was performed to identify miRNAs that were differentially expressed in colonic adenocarcinoma tissue and normal solid tissue (Figure 1A). A total of 273 miRNAs were significantly altered in solid tumors (128 were up-regulated, 145 were down-regulated) compared to that in normal tissue (Supplementary Table 1). Of the 128 up-regulated miRNAs, 26 miRNAs were identified as up-regulated in infinitely increased levels of very low expression levels in the normal group of these miRNAs. For other miRNAs, we set a threshold of 42 for the basal mean of the primary solid tumor, which was the average of the basic mean of these miRNAs. Above the threshold, we found 22 significantly up-regulated miRNAs, among which miR19b-3p was ranked first in the list. To investigate the differences in miR19b-3p expression in CRC, we identified the levels of miR19b-3p in the clinical CRC tissues (n = 40) and adjacent non-tumor tissues by qRT-PCR. As shown in Figure 1B, compared with the normal counterparts, the levels of miR19b-3p were significantly up-regulated in 35 of the 40 CRC tissues (87%). When compared with the controls, miR19b-3p expression was increased by nearly three-fold in the CRC tissues (Figure 1C). We then performed a Kaplan-Meier survival analysis to assess the prognostic value of miR19b-3p expression in the OS of patient with CRC. The results showed that shorter OS was associated with higher expression of miR19b-3p (Figure 1D). Finally, we analyzed the miR19b-3p levels in several CRC cell lines, including HT-29, HCT116, SW480, and LOVO, which were then compared to levels in human epithelial cell HCoEpiC cells by qRT-PCR. Similarly, the expression levels of miR19b-3p in the CRC cell lines were significantly higher than that in HCoEpiC cells (Figure 1E). These results suggest that miR19b-3p levels were positively correlated with CRC malignancies.
Figure 1.
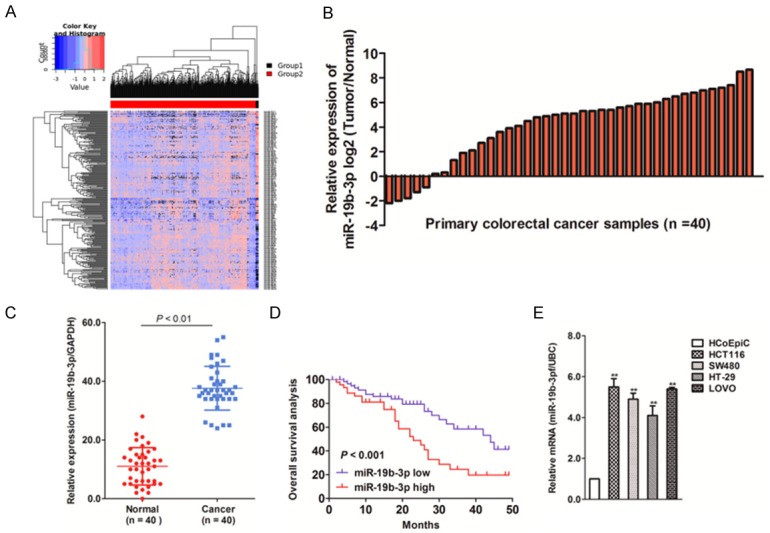
miR19b-3p expression up-regulated in colorectal cancer (CRC). A. MicroRNA (miRNA) meta-analysis in YM500v2 (http://ngs.ym.edu.tw/ym500v2/index.php) was performed to detect differentially expressed miRNAs in colon cancer compared to that in control solid tissue. Group 1: Normal tissues, Group 2: Colon cancer. B. Total RNA was extracted from CRC as well as normal tissues and then reverse transcribed into cDNA. miR19b-3p levels were detected by qRT-PCR. N represents the total number of patients with CRC. C. The levels of miR19b-3p in human colon cancer tissues and corresponding normal tissues were measured by qRT-PCR. D. A Kaplan-Meier survival analysis was performed to assess the prognostic value of miR19b-3p levels in the overall survival of patients with CRC. E. The levels of miR19b-3p in four different colorectal cancer cell lines were measured qRT-PCR. The PCR values were normalized to the levels of GAPDH. The data are expressed as the mean ± SD of three independent measurements.
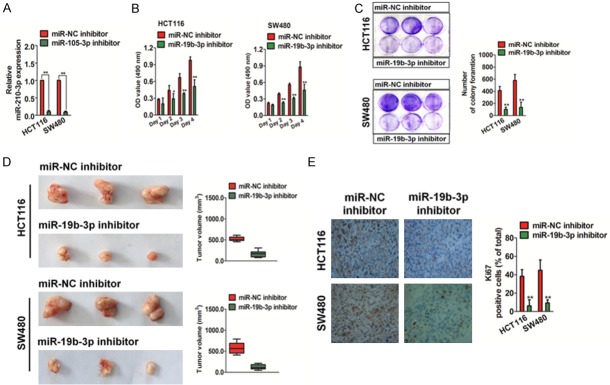
Down-regulation of miR19b-3p inhibits cell growth
In order to investigate the biological effects of miR19b-3p in CRC, we transfected HCT116 and SW480 cells with a miR19b-3p inhibitor to evaluate the effect of miR19b-3p down-regulation on CRC cell proliferation. MiR19b-3p was effectively inhibited in HCT116 and SW480 cells, as assessed by qRT-PCR (Figure 2A). Then, an MTT assay was performed and significant inhibition of CRC cell proliferation was observed in vitro (Figure 2B). miR19b-3p down-regulation also remarkably inhibited colony formation in HCT116 and SW480 cells compared to that in cells transfected with the control inhibitor (designated as miR-NC inhibitor) (Figure 2C). Next, we investigated the role of miR19b-3p in the growth of HCT116 and SW480 CRC cells in vivo. The miR19b-3p-down-regulated HCT116 or SW480 cells were subcutaneously implanted into the nude mice. As shown in Figure 2D, miR19b-3p down-regulation inhibited tumor growth significantly in vivo, and the tumor volume derived from miR19b-3p-downregulated cells was remarkably smaller than tumors from control cells. An immunohistochemical staining assay indicated that the Ki-67 index of tumors from miR19b-3p-downregulated cells was lower than that of control cells (Figure 2E). In conclusion, these results indicated that the down-regulation of miR19b-3p inhibits CRC cell growth in vitro and in vivo.
Figure 2.
Down-regulation of miR19b-3p inhibits cell growth. A. Transfection of HCT116 and SW480 cells with a miR-NC inhibitor or miR19b-3p inhibitor. The total RNA was isolated and the mRNA level of miR19b-3p was detected by qRT-PCR. Data are expressed as mean ± SD. **compared with miR-NC, P < 0.01. B. Colorectal cancer (CRC) cells transfected with the miR-NC inhibitor or miR19b-3p inhibitor were inoculated in 96-well plates and cultured for 24, 48, 72 or 96 h. MTT assay was performed and the viable cells were measured at 490 nm. C. The number of colonies in miR19b-3p inhibitor-transfected HCT116 and SW480 cells was significantly lower than that in miR-NC-inhibitor transfected cells. Data are expressed as mean ± SD. **P < 0.01, compared with miR-NC inhibitor. D. The miR-NC inhibitor- or miR19b-3p inhibitor-transfected CRC cells were implanted in the nude mice, and the tumor volume was measured once every three days. E. Xenotransplantation tumors were embedded in paraffin and preceded to IHC. Low Ki-67-positive expression was detected in tumors from CRC cells transfected with the miR19b-3p inhibitor. Data are expressed as mean ± SD. **P < 0.01, compared with miR-NC inhibitor.
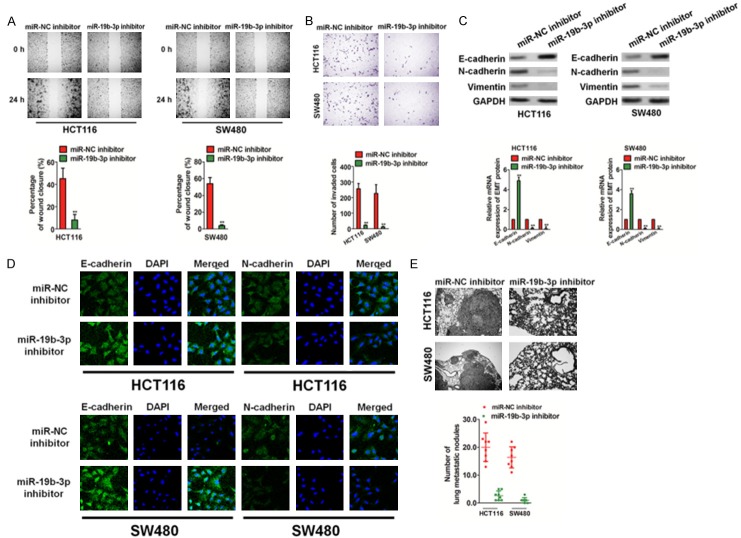
Down-regulation of miR19b-3p inhibits CRC cell metastasis
We examined the functional role of miR19b-3p in the migration and invasion of CRC cells, since metastasis has been associated with the poor prognosis of CRC. Down-regulation of miR19b-3p significantly inhibited the migration of HCT116 and SW480 cells in a wound healing analysis (Figure 3A). Similarly, the down-regulation of miR19b-3p dramatically suppressed the invasion of CRC cells in vitro (Figure 3B). Epithelial-mesenchymal transition (EMT) of cancer cells has been recognized to be associated with the initiation of metastasis. Immunoblotting analyses, qRT-PCRs, and immunofluorescence assays were performed to investigate whether miR-1296 down-regulation resulted in the inhibition of EMT in CRC cells. As shown in Figure 3C and 3D, the miR19b-3p inhibitor resulted in increased expression of epithelial markers (E-cadherin) and decreased expressions of intermediate markers (N-cadherin and vimentin). Finally, we tested the effect of miR19b-3p on cancer cell metastasis in vivo. The miR19b-3p inhibitor or miR-NC inhibitor transfected cells were injected into nude mice via the tail vein. Hematoxylin and eosin (H&E) staining of pulmonary slices confirmed that the lung tissue of mice injected with the miR-NC inhibitor-transfected cells had a higher number of tumor lesions than those with the miR19b-3p inhibitor-transfected cells (Figure 3E). Based on the above evidence, we demonstrated that down-regulation of miR19b-3p leads to the inhibition of CRC cell metastasis in vitro and in vivo.
Figure 3.
Down-regulation of miR19b-3p inhibits colorectal cancer (CRC) cell metastasis. A. HCT116 and SW480 cell transplantation with miR19b-3p inhibitor or miR-NC inhibitor was performed to evaluate the cell migration ability. Data are expressed as mean ± SD. **P < 0.01 compared with the miR-NC inhibitor. B. The invasive abilities of HCT116 and SW480 cells were assessed by the Transwell invasion assay. Quantitative statistics for invasive cells are shown on the right panel. Data are expressed as mean ± SD. **P < 0.01 compared with miR-NC inhibitor. C. HCT116 and SW480 cells were transfected with the miR19b-3p inhibitor or miR-NC inhibitor, and western blotting (upper panel) and qRT-PCR (lower panel) assays were used to detect the expression of E-cadherin, N-cadherin, and vimentin. D. HCT116 and SW480 cells were incubated with E-cadherin and N-cadherin. Cells were immunized with anti-rabbit FITC-conjugated secondary antibody and then stained with DAPI. The core is shown in blue, and the target protein is shown in green. The scale bar represents 50 μm. E. Representative hematoxylin and eosin staining of lung tissue from mice injected with miR-NC inhibitor cells and miR19b-3p inhibitor cells.
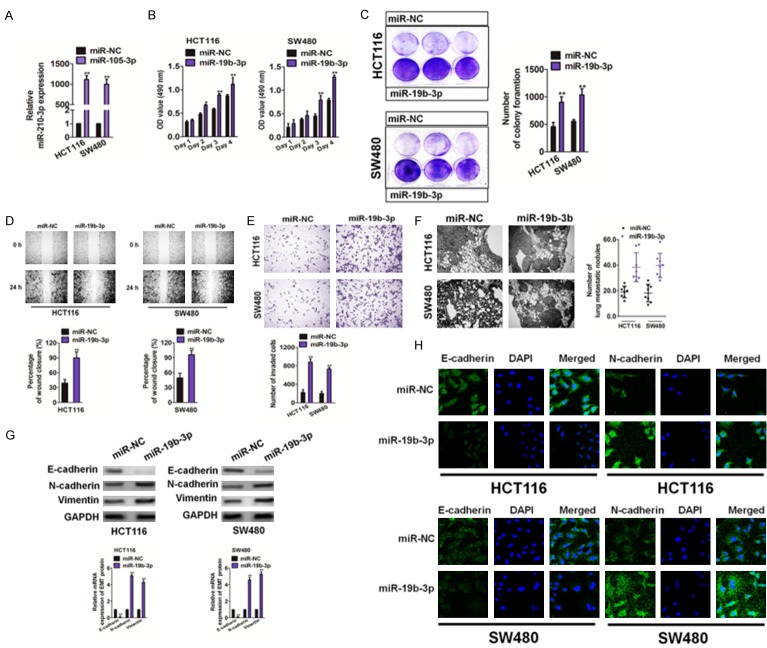
Overexpression of miR19b-3p promotes the growth and metastasis of CRC cells
We used precursor miR19b-3p clones to mimic overexpression of miR19b-3p in HCT116 and SW480 cells. After miR19b-3p was transfected into the cells, miR19b-3p mRNA levels were measured by qRT-PCR (Figure 4A). Both MTT and colony formation assay results (Figure 4B and 4C, respectively) demonstrated that overexpression of miR19b-3p significantly increased CRC cell growth in vitro. Furthermore, overexpression of miR19b-3p effectively induced the migration (Figure 4D) and invasion (Figure 4E) of HCT116 and SW480 cells. MiR19b-3p-overexpressing HCT116 or SW480 cells were injected into mice via the tail vein. Histological analyses of the lung tissue showed that lung tissues of nude mice injected with miR19b-3p-overexpressing cells had a higher number of tumor lesions (Figure 4F). Finally, an immunoblotting analysis, qRT-PCR, and immunofluorescence assay were performed to reveal whether MIR1296 overexpression accelerated EMT in CRC cells. As shown in Figure 4G and 4H, miR19b-3p resulted in decreased expression of epithelial markers (E-cadherin) and increased expressions of intermediate markers (N-cadherin and vimentin). These results showed that overexpression of miR19b-3p accelerated colon cancer growth and metastases.
Figure 4.
Overexpression of miR19b-3p promotes the growth and metastasis of colorectal cancer (CRC) cells. A. HCT116 and SW480 cells were transfected with miR19b-3p or miR-NC. The levels of miR19b-3p were detected by qRT-PCR. Data are expressed as mean ± SD. **P < 0.01, compared with miR-NC. B. The relative cell viability of HCT116 and SW480 cells transfected with the miR19b-3p or miR-NC was evaluated by the MTT assays. C. A colony formation analysis was performed to evaluate the rates of cells transfected with miR19b-3p. Data are expressed as mean ± SD. **P < 0.01, compared with miR-NC. D. Overexpression of miR19b-3p promoted the migration of CRC cells. Data are expressed as mean ± SD. **P < 0.01, compared with miR-NC. E. Overexpression of miR19b-3p promoted the invasion of CRC cells in the Transwell invasion assays (left panel). Quantitative analysis of invasive cells was shown (right panel). Data are expressed as mean ± SD. **P < 0.01, compared with miR-NC. F. Representative hematoxylin and eosin stains of lung tissue from mice injected with miR-NC and miR19b-3p overexpressing HCT116 and SW480 cells. Scale bar: 100 μm. G. Cells were transfected with miR19b-3p or miR-NC, and western blotting (upper panel) and qRT-PCR (lower panel) assays were used to detect the expression of E-cadherin, N-cadherin and vimentin. H. Cells were incubated with E-cadherin and N-cadherin. Cells were immunized with anti-rabbit FITC-conjugated secondary antibody and then stained with DAPI. Blue depicted the core and green depicted the target protein.
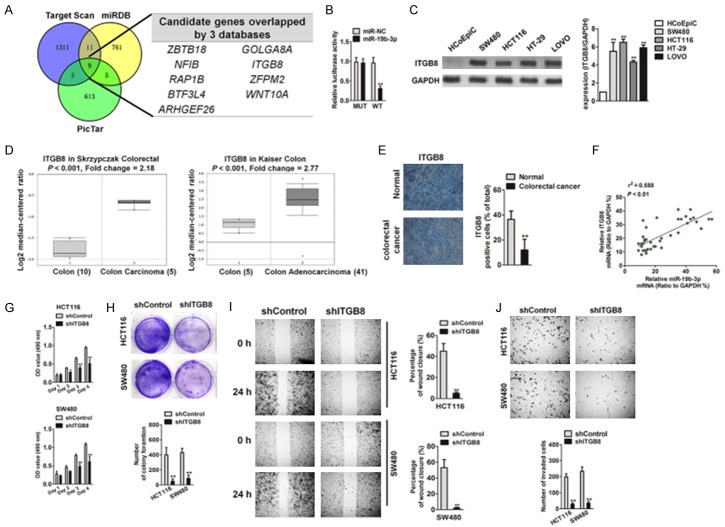
miR19b-3p binds to the 3’UTR of ITGB8
To investigate the target genes of miR19b-3p, three prediction websites (Targetscan, MIRDB, and PicTar) were used to predict its target genes. Notably, 9 genes (ZBTB18, NFIB, RAP1B, BTF3L4, ARHGEF26, ITGB8, GOLGA8A, ZFPM2, and WNT10A) were present in the databases (Figure 5A). Among these candidates, we focused on ITGB8 because its role in CRC growth and metastasis was unknown. In order to validate the 3’-UTR of ITGB8 as a direct target of miR19b-3p, ITGB8 3’-UTR and wild-type miR19b-3p binding sites (WT) were inserted into the p-MIR report vector, and a vector with a corresponding mutation binding site (MUT) was also constructed. The relative luciferase activities in 293T cells co-transfected with miR-19b-3p mimics and WT vectors were significantly inhibited, whereas the luciferase activity in cells transfected with the miR19b-3p mimics and MUT vector were not affected (Figure 5B). In addition, ITGB8 expressions in several CRC cell lines were higher than that in HCoEpiC cells (Figure 5C). Meanwhile, the Oncomine microarray databases [19,20] indicated that ITGB8 was substantially up-regulated in CRC compared to that in normal tissue (Figure 5D). We then attempted to analyze the correlation between ITGB8 and miR19b-3p expression in CRC and found that ITGB8 was overexpressed in CRC (Figure 5E) and was positively associated with miR19b-3p levels (Figure 5F). To determine whether ITGB8 was involved in the growth and metastasis of CRC, short hairpin RNA (shRNA) was used to knockdown ITGB8 expression in HCT116 and SW480 cells (Supplementary Figure 1). Then, we checked the effect of ITGB8 knockdown on tumor cell proliferation (Figure 5G), colony formation (Figure 5H), migration (Figure 5I), and invasion (Figure 5J). As expected, the stable knockdown of ITGB8 inhibited the proliferation, migration, and invasion of CRC cells in vitro, which suggests that ITGB8 functions as an oncogenic gene in CRC growth and metastasis.
Figure 5.
Effect of ITGB8 knockdown on CRC cells growth and metastasis. A. miR19b-3p target genes were predicted by in silico analyses. The Venn graph represented the number of candidate genes determined by three prediction algorithms. B. The relative luciferase activity in 293T cells co-transfected with the miR19b-3p mimic and WT vector or MUT vector was analyzed by luciferase reporter assay. C. RNA was extracted from CRC cells and the levels of ITGB8 were determined by qRT-PCR (upper panel). Data are expressed as mean ± SD. *P < 0.05, **P < 0.01, compared with HCoEpiC cells. The expression of ITGB8 was detected by western blotting of CRC cell lines (lower panel). D. ITGB8 was overexpressed in CRC tissues compared to that in normal tissues, based on the datasets from Oncomine. E. ITGB8 levels in CRC tissues were measured by IHC. F. The positive correlation analysis of miR19b-3p and ITGB8 in CRC tissues. G. Cell proliferation activity was measured by MTT assay. H. The growth capacity was determined in the colony formation assay. I. HCT116 and SW480 cell migration was analyzed by the wound healing assay as described above. J. HCT116 and SW480 cell invasion capacity was analyzed by Transwell Matrigel invasion assay as described above. Data are expressed as mean ± SD. *P < 0.05, **P < 0.01, compared with control cells.
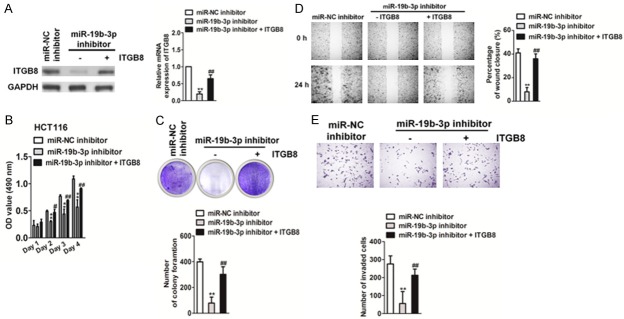
Confirmation of the role of ITGB8 in miR19b-3p-induced colorectal cancer cell growth and metastasis
To investigate whether miR19b-3p regulates the proliferation and metastasis of CRC by targeting ITGB8, we constructed miR19b-3p-downregulated CRC cell lines that overexpressed ITGB8. Both qRT-PCR and western blot analysis showed that ectopic expression of ITGB8 rescued its levels in HCT116 and SW480 cells that were inhibited by the miR19b-3p inhibitor (Figure 6A). Both the MTT and soft agar growth assays (Figure 6B, 6C) showed that overexpression of ITGB8 promoted CRC cell proliferation when compared to that in cells transfected with the miR19b-3p inhibitor alone. Wound healing and Transwell invasion (Figure 6D, 6E) showed that overexpression of ITGB8 restored CRC cell migration and invasion compared with that in cells transfected with miR19b-3p inhibitor alone. To further confirm the function of ITGB8 in miR19b-3p-induced growth and metastasis, we co-transfected cells with shITGB8 and miR19b-3p (Figure 7A). As expected, ITGB8 knockdown counteracted the accelerated effects of miR19b-3p in CRC cell proliferation and growth (Figure 7B, 7C). Wound healing and Transwell invasion assays also showed that knockdown of ITGB8 reversed the promotion of cell migration and invasion caused by miR19b-3p overexpression (Figure 7D, 7E). In summary, these results demonstrated the important role of ITGB8 in miR19b-3p-induced CRC cell growth and metastasis.
Figure 6.
Effect of miR19b-3p inhibitor on colorectal cancer growth and metastasis is rescued by ITGB8 overexpression. A. The mRNA levels of ITGB8 were detected by qRT-PCR (right panel). Western blotting was performed to determine the expression levels of ITGB8 (left panel). B. HCT116 and SW480 cell proliferation was measured by MTT assay as described. C. Growth capacities of the indicated cells were detected by the soft agar colony formation assay as described above. Data are expressed as mean ± SD. **P < 0.05 compared with miR-NC inhibitor, ##P < 0.01 compared with miR19b-3p inhibitor. D. Wound healing assay as described was used to evaluate the cell migration ability in vitro. E. Cell invasion was assessed by Transwell Matrigel invasion assay. Data are expressed as mean ± SD. **P < 0.05 compared with miR-NC inhibitor, ##P < 0.01 compared with the miR19b-3p inhibitor.
Figure 7.
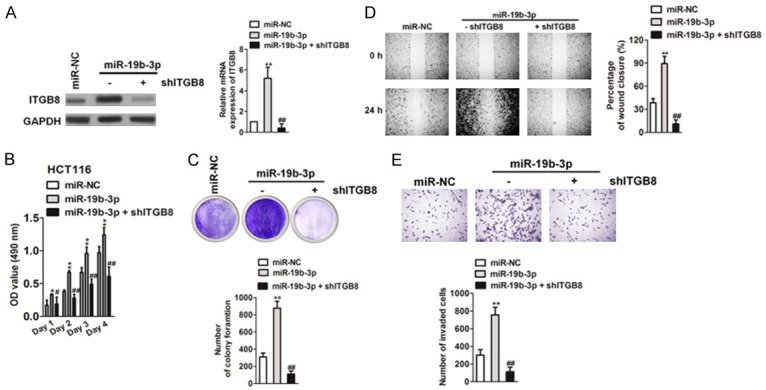
Effect of miR19b-3p on colorectal cancer (CRC) cell growth and metastasis is blocked by ITGB8 knockdown. A. The levels of ITGB8 in were detected by qRT-PCR (right panel) and western blotting analysis (left panel). B. Cell proliferation activity was measured by MTT assay as described. C. CRC growth was measured by the colony formation assay as described in vitro. The image (upper panel) and quantitative analysis of tumor cells colonies (lower panel) are shown. Data are expressed as mean ± SD. **P < 0.05 compared to miR-NC. ##P < 0.01 compared to miR19b-3p. D. Cell migration ability was measured by the wound healing assay. The image (left panel) and quantitative analysis of the percentage of closure (right panel) are shown. E. Cell invasion was analyzed by the Transwell Matrigel invasion assay. The image (upper panel) and quantitative analysis of the total invasive cells (lower panel) are shown. Data are expressed as mean ± SD. **P < 0.05 compared to miR-NC. ##P < 0.01 compared to miR19b-3p.
Discussion
CRC is the one of the most serious forms of human malignancies and is the main cause of global cancer-related deaths [21]. Although the treatments of CRCs have improved, the overall prognosis of patients with CRC remains poor. Consequently, in order to identify biomarkers and therapeutic targets for CRC, uncovering the molecular mechanism of CRC progression is crucial [22]. Recent studies have suggested that miRNAs regulate tumor cell processes, including cell proliferation, survival, invasion, and metastasis [23]. Thus, miRNAs are considered potential regulators in cancer progression. MIR19b has been implicated in a number of human cancers. MiR19a and miR19b (miR19a/b) promote the proliferation and migration of lung cancer cells by targeting microtubule-associated tumor suppressor 1 (MTUS1) [24]. As members of the miR17-92 cluster, miR19b also functions as oncogenes in many types of cancer, including gastric cancer, pancreatic cancer, and breast cancer. Protein tyrosine phosphatase receptor type G (PTPRG) is an important tumor suppressor gene in multiple human cancers [25]. MiR19b could inhibit PTPRG expression to promote tumorigenesis in human breast cancer [25]. Collectively, these data indicate an oncogenic role of miR19b in cancer, though its clinical significance and potential roles in CRC remain uncovered. In the present study, overexpression of miR19b-3p was confirmed in CRC tissues. Our study demonstrated that miR19b-3p may serve an oncogenic role in CRC metastasis and as a potential biomarker for the prognostic prediction of CRC.
In order to test the hypothesis that miR19b-3p plays an important role in the growth and metastasis of CRC cells, we analyzed the expression levels of miR19b-3p in CRC and several cell lines. Compared with the normal tissue, we found that miR19b-3p was significantly increased in CRC. The gain-of-function and loss-of-function studies for miR19b-3p were performed in two CRC cell lines. The results showed that miR19b-3p down-regulation inhibited the proliferation and metastasis of CRC cells in vitro and in vivo. Nevertheless, overexpression of miR19b-3p resulted in opposite outcomes between the two lines, indicating that miR19b-3p was a tumor promoter in CRC. The molecular action of miR19b-3p in inducing cell growth and metastasis was partly due to the regulation of ITGB8. Furthermore, ITGB8 has been identified as a tumor promoter in several cancers. Herein, we identified ITGB8, a direct target gene for miR19b-3p, to play an important role in CRC cell metastasis. In this study, the complementary sequence of miR-19b-3p was identified to be in the 3’-UTR of ITGB8. In the luciferase reporter assay, miR-19b-3p resulted in a significant change in luciferase activity of WT ITGB8-3’-UTR without affecting the luciferase activity of MUT ITGB8-3’-UTR. Our current study showed that ITGB8 itself plays an important role in regulating tumor cell proliferation and metastasis. Our results also suggest that ITGB8 is up-regulated in all CRC cell lines, confirming that ITGB8 plays a broader role in regulating the development of CRC. In CRC cells, it was observed that ectopic expression of ITGB8 significantly accelerated proliferation, migration, and invasion, while knockdown of ITGB8 inhibited CRC cell growth and metastasis.
In conclusion, we demonstrated that miR19b-3p is overexpressed in CRC tissues and verified that overexpression of miR19b-3p accelerated CRC cell growth, invasion, and metastasis both in vitro and in vivo. Functional studies also demonstrated that ITGB8 was a direct downstream target of miR19b-3p. Collectively, these results provide novel insights for better understanding the mechanisms of miR19b-3p/ITGB8 in the regulation of CRC metastasis and for identifying potential therapeutic targets for the clinical treatment of CRC metastasis.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Qin Y, Tang B, Hu CJ, Xiao YF, Xie R, Yong X, Wu YY, Dong H, Yang SM. An hTERT/ZEB1 complex directly regulates E-cadherin to promote epithelial-to-mesenchymal transition (EMT) in colorectal cancer. Oncotarget. 2016;7:351–361. doi: 10.18632/oncotarget.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiang Y, Yao X, Chen K, Wang X, Zhou J, Gong W, Yoshimura T, Huang J, Wang R, Wu Y, Shi G, Bian X, Wang J. The G-protein coupled chemoattractant receptor FPR2 promotes malignant phenotype of human colon cancer cells. Am J Cancer Res. 2016;6:2599–2610. [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Zhu L, Fang J, Ge Z, Li X. LRG1 modulates epithelial-mesenchymal transition and angiogenesis in colorectal cancer via HIF-1alpha activation. J Exp Clin Cancer Res. 2016;35:29. doi: 10.1186/s13046-016-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li P, Xue WJ, Feng Y, Mao QS. MicroRNA-205 functions as a tumor suppressor in colorectal cancer by targeting cAMP responsive element binding protein 1 (CREB1) Am J Transl Res. 2015;7:2053–2059. [PMC free article] [PubMed] [Google Scholar]
- 5.Kurokawa K, Tanahashi T, Iima T, Yamamoto Y, Akaike Y, Nishida K, Masuda K, Kuwano Y, Murakami Y, Fukushima M, Rokutan K. Role of miR-19b and its target mRNAs in 5-fluorouracil resistance in colon cancer cells. J Gastroenterol. 2012;47:883–895. doi: 10.1007/s00535-012-0547-6. [DOI] [PubMed] [Google Scholar]
- 6.Gu Y, Liu S, Zhang X, Chen G, Liang H, Yu M, Liao Z, Zhou Y, Zhang CY, Wang T, Wang C, Zhang J, Chen X. Oncogenic miR-19a and miR-19b co-regulate tumor suppressor MTUS1 to promote cell proliferation and migration in lung cancer. Protein Cell. 2017;8:455–466. doi: 10.1007/s13238-017-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li RK, Gao J, Guo LH, Huang GQ, Luo WH. PTENP1 acts as a ceRNA to regulate PTEN by sponging miR-19b and explores the biological role of PTENP1 in breast cancer. Cancer Gene Ther. 2017;24:309–315. doi: 10.1038/cgt.2017.29. [DOI] [PubMed] [Google Scholar]
- 8.Osip’yants AI, Knyazev EN, Galatenko AV, Nyushko KM, Galatenko VV, Shkurnikov MY, Alekseev BY. Changes in the level of circulating hsa-miR-297 and hsa-miR-19b-3p miRNA are associated with generalization of prostate cancer. Bull Exp Biol Med. 2017;162:379–382. doi: 10.1007/s10517-017-3620-6. [DOI] [PubMed] [Google Scholar]
- 9.Wu C, Cao Y, He Z, He J, Hu C, Duan H, Jiang J. Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. Tohoku J Exp Med. 2014;232:85–95. doi: 10.1620/tjem.232.85. [DOI] [PubMed] [Google Scholar]
- 10.Wu Q, Yang Z, Wang F, Hu S, Yang L, Shi Y, Fan D. MiR-19b/20a/92a regulates the selfrenewal and proliferation of gastric cancer stem cells. J Cell Sci. 2013;126:4220–4229. doi: 10.1242/jcs.127944. [DOI] [PubMed] [Google Scholar]
- 11.Anderson LR, Owens TW, Naylor MJ. Integrins in development and cancer. Biophys Rev. 2014;6:191–202. doi: 10.1007/s12551-013-0123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aso K, Goi T, Nakazawa T, Kimura Y, Hirono Y, Katayama K, Yamaguchi A. The expression of integrins is decreased in colon cancer cells treated with polysaccharide K. Int J Oncol. 2013;42:1175–1180. doi: 10.3892/ijo.2013.1832. [DOI] [PubMed] [Google Scholar]
- 13.Clezardin P. Recent insights into the role of integrins in cancer metastasis. Cell Mol Life Sci. 1998;54:541–548. doi: 10.1007/s000180050182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee A. Integrins--new frontier in cancer biology. Indian J Exp Biol. 1996;34:923–926. [PubMed] [Google Scholar]
- 15.Fan Y, Yin S, Hao Y, Yang J, Zhang H, Sun C, Ma M, Chang Q, Xi JJ. miR-19b promotes tumor growth and metastasis via targeting TP53. RNA. 2014;20:765–772. doi: 10.1261/rna.043026.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin DN, Qian L, Hu DL, Yu ZB, Han SP, Zhu C, Wang X, Hu X. Effects of miR-19b overexpression on proliferation, differentiation, apoptosis and Wnt/beta-catenin signaling pathway in P19 cell model of cardiac differentiation in vitro. Cell Biochem Biophys. 2013;66:709–722. doi: 10.1007/s12013-013-9516-9. [DOI] [PubMed] [Google Scholar]
- 17.Zhong C, Wang K, Liu Y, Lv D, Zheng B, Zhou Q, Sun Q, Chen P, Ding S, Xu Y, Huang H. miR-19b controls cardiac fibroblast proliferation and migration. J Cell Mol Med. 2016;20:1191–1197. doi: 10.1111/jcmm.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung IF, Chang SJ, Chen CY, Liu SH, Li CY, Chan CH, Shih CC, Cheng WC. YM500v3: a database for small RNA sequencing in human cancer research. Nucleic Acids Res. 2017;45:D925–D931. doi: 10.1093/nar/gkw1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, Pachlewski J, Oledzki J, Ostrowski J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5:e13091. doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser S, Park YK, Franklin JL, Halberg RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, Kong S, Sakthivel B, Xu H, Reichling T, Azhar M, Boivin GP, Roberts RB, Bissahoyo AC, Gonzales F, Bloom GC, Eschrich S, Carter SL, Aronow JE, Kleimeyer J, Kleimeyer M, Ramaswamy V, Settle SH, Boone B, Levy S, Graff JM, Doetschman T, Groden J, Dove WF, Threadgill DW, Yeatman TJ, Coffey RJ Jr, Aronow BJ. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu JJ, Qu XY, Zhou DZ. miR4262 inhibits colon cancer cell proliferation via targeting of GALNT4. Mol Med Rep. 2017;16:3731–3736. doi: 10.3892/mmr.2017.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dowling CM, Hayes SL, Phelan JJ, Cathcart MC, Finn SP, Mehigan B, McCormick P, Coffey JC, O’Sullivan J, Kiely PA. Expression of protein kinase C gamma promotes cell migration in colon cancer. Oncotarget. 2017 doi: 10.18632/oncotarget.18916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kok MG, Mandolini C, Moerland PD, de Ronde MW, Sondermeijer BM, Halliani A, Nieuwland R, Cipollone F, Creemers EE, Meijers JC, Pinto-Sietsma SJ. Low miR-19b-1-5p expression in isolated platelets after aspirin use is related to aspirin insensitivity. Int J Cardiol. 2016;203:262–263. doi: 10.1016/j.ijcard.2015.10.098. [DOI] [PubMed] [Google Scholar]
- 24.Zaporozhchenko IA, Morozkin ES, Skvortsova TE, Ponomaryova AA, Rykova EY, Cherdyntseva NV, Polovnikov ES, Pashkovskaya OA, Pokushalov EA, Vlassov VV, Laktionov PP. Plasma miR-19b and miR-183 as potential biomarkers of lung cancer. PLoS One. 2016;11:e0165261. doi: 10.1371/journal.pone.0165261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Song Y, Zhang C, Zhi X, Fu H, Ma Y, Chen Y, Pan F, Wang K, Ni J, Jin W, He X, Su H, Cui D. Circulating MiR-16-5p and MiR-19b-3p as two novel potential biomarkers to indicate progression of gastric cancer. Theranostics. 2015;5:733–745. doi: 10.7150/thno.10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.