Abstract
Osteosarcoma (OS) is the most common primary bone malignancies. Long noncoding RNAs (lncRNAs) are key regulatory RNAs which takes part in several biological processes. LncRNA neuroblastoma associated transcript 1 (NBAT1) is a newly identified functional lncRNA. NBAT1 functions as a tumor suppressor in some cancers. However, the expression pattern, the biological function and the mechanisms of NBAT1 in OS progress have not been elucidated. In this study, for the first time, we found that NBAT1 expression is downregulated in OS tissues and cell lines and is associated with clinical stage, distant metastasis and poor prognosis. Loss- and gain-of-function assays showed that NBAT1 played a negative regulatory role in OS growth and metastasis in vitro and in vivo. Further investigation demonstrated that NBAT1 physically interacted with miR-21 and then suppressed its expression. NBAT1 also regulated downstream genes targeted by miR-21, including PTEN, PDCD4, TPM1 and RECK. These findings may extend the function of NBAT1 in tumor progression and provide a novel target for OS treatment.
Keywords: NBAT1, miR-21, growth, metastasis
Introduction
Osteosarcoma (OS) is the most common primary bone malignancies, which is the leading cause of cancer-related deaths in children and young adolescents [1]. Previous studies have demonstrated that some pathogenic factors, such as genetic mutations and dysregulation of transcription regulation, were thought to be critical in OS progress [2,3]. Even through the improvements in diagnosis and therapy of OS have been made in the past decades, the mortality rate is still high due to the OS invasion and distant metastasis [4]. Therefore, revealing the underlying mechanisms involved in OS development and metastasis is urgently needed.
The genome-wide studies have demonstrated that only less than 2% of the human genome codes for protein [5,6]. The other noncoding RNAs (ncRNAs), including short and long noncoding RNAs (lncRNAs), are key regulatory RNAs which takes part in several biological processes [7]. MicroRNAs (miRNAs) are endogenous small ncRNAs (17-22 nucleotides). The functions of microRNAs (miRNAs) have gained a lot of interest. Studies have demonstrated that abnormal expression of miRNAs is involved in cancer progression and metastasis through association with 3’ untranslational region (3’UTR) of target genes. For example, miR-21 is one of the most common increased miRNAs in several cancers [8,9]. It has been reported that miR-21 positively regulated cell proliferation, migration and invasion. The target gene of miR-21 includes some well-known tumor suppressor, such as PTEN, PDCD4, TPM1 and RECK [8]. LncRNAs are ncRNAs longer than 200 nucleotides in length. Emerging evidences has reported that some lncRNAs are also aberrantly expressed in cancers and regulates malignant phenotypes of cancer cells. Mechanistically, lncRNAs are involved in all aspects of gene regulation, including gene imprinting, epigenetic modification, nuclear and cytoplasmic trafficking, transcription control, mRNA splicing and degradation [10]. However, to date, only a few functional lncRNAs have been well characterized in OS. LncRNA neuroblastoma associated transcript 1 (NBAT1) is a newly identified functional lncRNA. NBAT1 could be taken as a biomarker significantly predictor of neuroblastoma prognosis. Downregulation of NBAT1 promotes cellular proliferation and invasion [11]. However, the expression pattern, the biological function and the mechanisms of NBAT1 in OS progress have not been elucidated. In this study, we detected the expression level of NBAT1 in OS tissues, and investigated the functional role and molecular mechanisms of NBAT1 in OS malignant phenotypes.
Materials and methods
Cell culture
OS cell lines (MG63, U2OS, 143b, LM7 and KHOS) and a normal osteoblast cell line (Nhost) were purchased from China Center for Type Culture Collection (Wuhan, China). All cell types were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, America) at 37°C.
Tissue samples
The OS tissues and corresponding noncancerous tissues were were surgically retrieved from OS patients from Cangzhou Central Hospital. All participating patients signed consent forms. All experiments involving human subjects were reviewed and approved by the Clinical Research & Ethics Committee at Cangzhou Central Hospital.
Lentiviral vector construction, production and transfection
LncRNA NBAT1 full-length cDNAs were amplified by PCR from the mRNA of MG63 cells. Then, NBAT1 cDNA was inserted into a lentiviral vector pLV. Empty vector was used as a control. In addition, the shRNAs agatinst NBAT1 were designed, and scramble shRNA was used as a control. The target sequences for NBAT1 were as follows: sh1: CCCACAGAGATGAAGTAAC, sh2: GAGACAAACAGGGTCAACT. The shRNAs were inserted into pLKO.1 vector. The constructed vectors and the lentiviral packaging vectors were cotransfected into HEK293T cells for 48 hrs. The lentiviral particles were collected. Cells were transfected with above lentiviral particles and selected by puromycin.
CCK-8 assay
Cell proliferation was detected by a Cell Counting Kit-8 (CCK-8) assay (Dojindo) as manufacturer’s instructions. Cells were seeded into 96-well plates (2 × 103 cells/well) for 1, 2, 3, 4 and 5 days, respectively. Each day, cells were treated with 10 µl CCK-8 for 1.5 hr. The optical density (OD) at 450 nm was evaluated using a microplate reader.
Migration and invasion assay
Transwell chambers (Corning Costar, USA) were used in the migration and invasion assays. For migration assay, 1 × 105 cells incubated in 200 µl serum-free medium were seeded in the upper chamber, while 500 μl DMEM supplemented with 10% FBS was added to the lower chamber. For the invasion assay, the upper chambers were pre-coated with Matrigel (BD Biosciences, USA) and 3 × 105 cells incubated in 200 µl serum-free medium were seeded in the upper chamber. After 24 hrs, the migratory and invasive cells were fixed in 4% paraformaldehyde and stained in 0.1% crystal violet solution. The migratory and invasive cells across the membrane were photographed and counted.
RNA isolation and real-time PCR (RT-PCR)
Total RNA was extracted from tissues or cells using TRIZOL reagent (Invitrogen) as manufacturer’s instructions. 1 µg RNA was reverse transcribed to cDNA by using a RevertAid First Strand cDNA Synthesis kit (Thermo Fisher). Real-time PCR analyses were performed with SYBR-Green PCR Master Mix kit (Takara). GAPDH was used as an endogenous control to normalize the data. For miR-21 expression detection, reverse transcription was performed following Applied Biosystems TaqMan MicroRNA Assay protocol. The relative expression of miR-21 was normalized to U6. Primer sequences used in this study were the following: NBAT1-F: ACTGAAACCCACAGAGATGAAG, NBAT1-R: CCCGTCATGTAGAGCAATATCC; PTEN-F: CCCACCACAGCTAGAACTTATC, PTEN-R: TCGTCCCTTTCCAGCTTTAC; PDCD4-F: GAGTACCAGTGTTGGCAGTATC, PDCD4-R: GTCCCACAAAGGTCAGAAAGA; TPM1-F: GAGAGTGAGAGAGGCATGAAAG, TPM1-R: CGGGCCACCTCTTCATATTT; RECK-F, GCTCGGTTTGTTGCAGTTATG, RECK-R: ATCTGAGATGGACCAGGAGAA. The 2-ΔΔCt method was used to determine the relative quantification of gene expression levels.
Transfection
miR-21 mimic and corresponding negative control miRNAs (miR-NC) were purchased from RiboBio (Guangzhou, China). Transfection of miR-21 or miR-NC was performed using Lipofectamine 3000 (Invitrogen) as manufacturer’s instructions.
RNA immunoprecipitation
RIP assays were performed as previously described [12]. In brief, NBAT1 or a mutant NBAT1 with mutation in miR-21 binding site (NBAT1-mut) was inserted into pcDNA3.1-MS2 (Addgene). Cells were co-transfected with pcDNA3.1-MS2 or pcDNA3.1-MS2-NBAT1 or pcDNA3.1-MS2-NBAT1-mut and pMS2-GFP (Addgene). After 48 hrs of transfection, cells were used to perform RIP experiments by using Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Millipore) as manufacturer’s instructions. 5 μg of GFP antibody (Abcam) or negative control IgG (Abcam) and magnetic beads were added to the cell lysate and incubated for overnight at 4°C with gentle rotation. The coprecipitated RNAs were reverse-transcribed into first strand cDNA using reverse transcriptase and detected by RT-PCR. For anti-AGO2 RIP, cells were transfected with negative control miRNA (miR-NC) or miR-21. After 48 hrs of transfection, cells were used to perform RIP assay by using an anti-AGO2 antibody (Millipore) as described above.
Luciferase reporter assay
NBAT1 or a mutant NBAT1 with mutation in miR-21 binding site (NBAT1-mut) was inserted into pmirGLO reporter vector, respectively. The pmirGLO containing nothing, NBAT1 or NBAT1-mut was transfected with miR-21 mimic or miR-NC into cells by Lipofectamine® 3000 (Invitrogen). After 48 hrs, the luciferase activity was detected. The relative luciferase activity was normalized to Renilla luciferase activity.
RNA pull-down
RNA pull-down was performed as previously described [13]. In vitro biotin-labeled RNAs (NBAT1 and NBAT1-mut) were transcribed with the biotin RNA labeling mix (Roche) and T7 RNA polymerase (Roche) treated with RNase-free DNase I (Promega) and purified with RNeasy Mini Kit (QIAGEN). 1 mg of whole-cell lysates from cells were incubated with 5 μg of purified biotinylated transcripts for 1 hr at 25°C; complexes were isolated with streptavidin agarose beads (Invitrogen). The RNA present in the pull-down material was detected by RT-PCR analysis.
Statistical analysis
Statistical analysis was performed with SPSS 17 software (SPSS, Chicago). All experiments were repeated three times. Differences were assessed using a two-tailed student’s t-test or one-way ANOVA and the correlation between NBAT1 and miR-21 expression was analyzed with Pearson’s correlation.
Results
NBAT1 is decreased in both OS cell lines and OS tissues
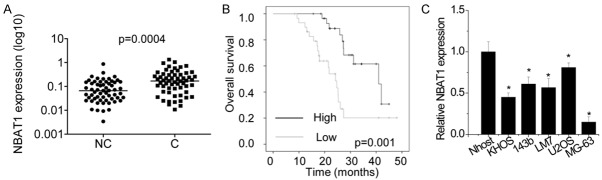
To reveal the expression pattern of NBAT1 in OS, RT-PCR was performed to examine the expression levels of NBAT1 in 60 OS tissues and adjacent normal tissues. RT-PCR analysis showed that NBAT1 xpression was significantly decreased in OS tissues compared to that in adjacent normal tissues (P<0.05, Figure 1A). We next determined the clinical significance of NBAT1 expression in OS patients. As shown in Table 1, no significant differences were observed between NBAT1 expression and gender, age, or tumor size of OS patients (P > 0.05; Table 1). Interestingly, NBAT1 expression in OS tissues was significantly associated with clinical stage and distant metastasis (P < 0.05; Table 1). Furthermore, Kaplan-Meier analysis showed that OS patients with low-level NBAT1 expression had a shorter median survival time than those with high-level NBAT1 (Figure 1B). NBAT1 expression was examined in five OS cell lines (KHOS, 143b, LM7, U2OS, and MG-63) and a normal osteoblast cell line Nhost by RT-PCR. Among these OS cell lines, U2OS (highest endogenous NBAT1 expression) and MG63 (lowest endogenous NBAT1 expression) cells were used for subsequent experiments (Figure 1C).
Figure 1.
NBAT1 is decreased in both OS cell lines and OS tissues. A. The NBAT1 expression in 60 pairs of OS tissues (C) and adjacent noncancerous bone tissues (NC) was measured using RT-PCR. B. Kaplan-Meier survival curve and log-rank test were used to evaluate the association of NBAT1 expression with overall survival rate. Patients were segregated into NBAT1-low and NBAT1-high group as the median of NBAT1 expression in OS tissues. C. The NBAT1 expression in five OS cell lines (KHOS, 143b, LM7, U2OS and MG-63) and normal osteoblast cells (Nhost) was detected by RT-PCR. The expression of NBAT1 in Nhost. *p<0.05.
Table 1.
The relationship between NBAT1 expression and clinicopathological variables in OS patients
| Variables | NBAT1 expression levels | P | |
|---|---|---|---|
|
| |||
| Low | High | ||
| Gender | |||
| Male | 14 | 13 | 0.795 |
| Female | 16 | 17 | |
| Age | |||
| >20 | 15 | 13 | 0.605 |
| ≤20 | 15 | 17 | |
| Location | |||
| Femur/Tibia | 18 | 17 | 0.793 |
| Elsewhere | 12 | 13 | |
| Tumor size (cm) | |||
| ≤5 | 17 | 20 | 0.426 |
| >5 | 13 | 10 | |
| Clinical stage | |||
| I+IIA | 7 | 21 | <0.001 |
| IIB/III | 23 | 9 | |
| Distant Metastasis | |||
| Yes | 24 | 8 | <0.001 |
| No | 6 | 22 | |
P value was acquired by Pearson chi-square test. The median expression level was used as the cutoff.
NBAT1 suppresses OS cell proliferation, migration and invasion in vitro
To investigate the functional role of NBAT1 expression in the development and progression of OS, we next performed gain- and loss-of-function studies in OS cells. We developed MG63 cells with stably overexpressed NBAT1 (Figure 1A), and U2OS with stably silenced NBAT1 expression (Figure 2B). To determine the effect of NBAT1 on OS proliferation, we performed the CCK-8 assays in vitro, we found that NBAT1 overexpression decreased the proliferative capacity of MG63 cells, compared to that of parallel stable cell lines containing the empty vector (Figure 2C). In contrast, knockdown endogenous NBAT1 expression dramatically increased the proliferaction of U2OS cells (Figure 2D). These results suggested that NBAT1 could inhibit cell proliferation in OS in vitro.
Figure 2.
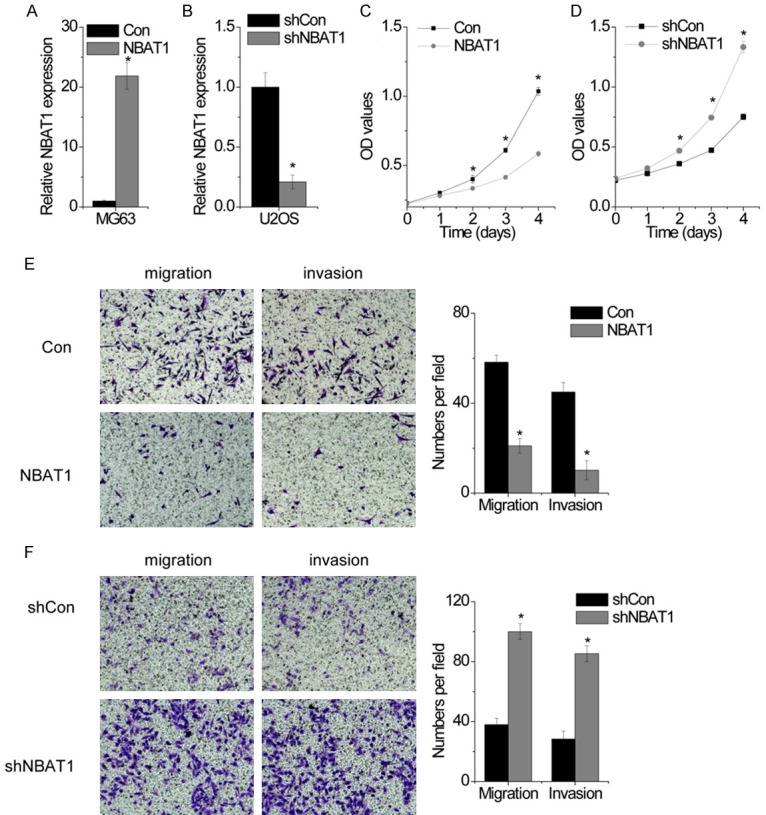
NBAT1 suppresses OS cell proliferation, migration and invasion in vitro. A. MG63 cells were transfected with lentiviral particles expressing empty vector (con) and NBAT1, and the relative expression of NBAT1 was detected by RT-PCR. B. U2OS cells were transfected with lentiviral particles expressing scamble shRNA (shcon) and NBAT1 shRNA, and the relative expression of NBAT1 was detected by RT-PCR. C. The cell proliferation of control and NBAT1 overexpressed MG63 cells was determined by CCK-8 assay. D. The cell proliferation of control and NBAT1 silenced U2OS cells was determined by CCK-8 assay. E. The representitave images (left) and number (right) of the migrated and invaded MG63 cells in the control and NBAT1-overexpressed group. F. The representitave images (left) and number (right) of the migrated and invaded U2OS cells in the control and NBAT1-silenced group. All experiments were repeated three times. *p<0.05.
Next, we detected the influence of NBAT1 on OS migration and invasion. We performed transwell and Matrigel transwell assay to measure the migratory and invasive capacities of OS cells. The results showed that ectopic expression of NBAT1 led to significantly decreased migration and invasion of the MG63 cells (Figure 2E). In contrast, silence of NBAT1 dramatically enhanced the migratory and invasive ability of U2OS cells compared with the control cells (Figure 2F). These results indicate a functional role of NBAT1 in suppressing OS metastasis.
NBAT1 inhibits OS growth and metastasis in vivo
Our above findings demonstrated that NBAT1 suppressed cell proliferation, migration and invasion, we next investigated the effects of ZFAS1 on OS growth and metastatic ability in vivo. Xenograft tumors grown from cells overexpressing NBAT1 had smaller mean volumes, and formed more slowly, than tumors grown from control cells (Figure 3A). In contrast, silence of NBAT1 significantly promoted tumor growth (Figure 3B). To establish a metastatic cancer model in vivo, above OS cells were labled with firefly luciferase and injected into tail vein of nude mice. A bioluminescent signal detection was performed at 10 weeks after tail vein injection. We found that the incidence of lung metastasis in the NBAT1-upregulated group is significantly decreased compared with the control group (Figure 3C), whereas NBAT1 knockdown showed the opposite effect (Figure 3D). Taken together, these data demonstrated that ZFAS1 promotes both growth and metastasis of OS tumors in vivo.
Figure 3.
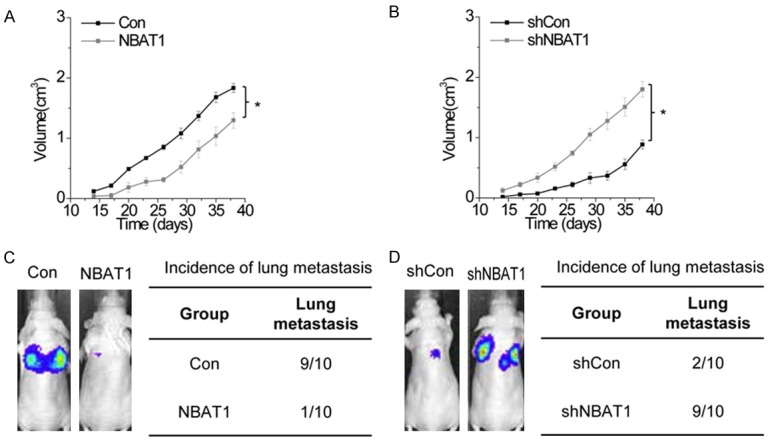
NBAT1 suppresses OS growth and metastasis in vivo. A. Effects of NBAT1 overexpression on tumor growth in vivo. Tumor growth curves measured after injection of MG63 cells expressing control and NBAT1. *p<0.05. B. Effects of NBAT1 overexpression on tumor growth in vivo. Tumor growth curves measured after injection of U2OS cells expressing scamble and NBAT1 shRNA. *p<0.05. C. Left: Luciferase signal intensities of the mice in each group over time after tail vein injection with 106 indicated MG63 cells expressing control and NBAT1. Right: Incidence of lung metastasis in each group of nude mice. D. Left: Luciferase signal intensities of the mice in each group over time after tail vein injection with 106 indicated U2OS cells expressing scramble and NBAT1 shRNA. Right: Incidence of lung metastasis in each group of nude mice.
NBAT1 is physically associated with miR-21
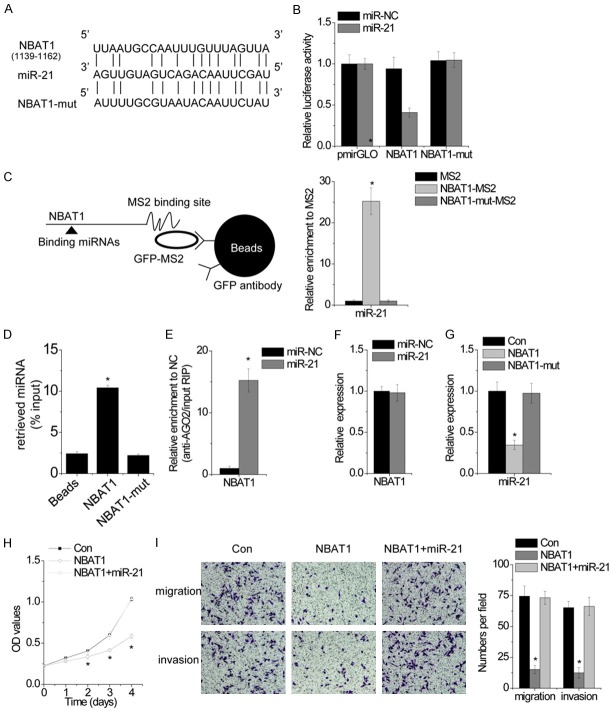
Emerging evidences have demonstrated that the interaction between lncRNAs and microRNAs is a novel important manner for gene regulation. Using Microinspector software, we found a set of miRNAs that putatively bind to NBAT1. Among these miRNA candidates, we found that miR-21 may directly bind to NBAT1 (Figure 4A). Dual-luciferase assays showed a significant decrease in luciferase activities following cotransfection of miR-21 and the wild-type NBAT1 expression, but not a mutant NBAT1 (NBAT1-mut, mutant in miR-21 binding site) (Figure 4B). To further confirm the direct association between NBAT1 and miR-21 at endogenous levels, we performed an RIP assay to pull down endogenous microRNAs associated with NBAT1 and demonstrated via RT-PCR analysis that the NBAT1 RIP in U2OS cells was significantly enriched for miR-21 compared to the empty vector (MS2), IgG, nontargeting microRNA (miR-200) and NBAT1-mut (Figure 4C). The specific association between miR-200s and lncRNA-ATB was further validated by affinity pull-down of endogenous miR-21 using in vitro transcribed biotin-labeled NBAT1 (Figure 4D). The microRNAs are known to bind their targets and cause translational repression and/or RNA degradation in an AGO2-dependent manner. To determine whether NBAT1 was regulated by miR-21 in such a manner, we performed anti-AGO2 RIP in U2OS cells transiently transfected with miR-21. Endogenous NBAT1 was significantly pulled down by AGO2 in miR-21-transfected cells (Figure 4E), supporting that miR-21 are NBAT1-targeting microRNAs.
Figure 4.
NBAT1 is physically associated with miR-21. A. Schematic outlining the predicted binding sites of miR-21 on NBAT1 or NBAT1-mut. B. Luciferase activity in U2OS cells cotransfected with miR-NC or miR-21 and luciferase reporters containing nothing, NBAT1 or NBAT1 mutant (NBAT1-mut) transcript. Data are presented as the relative ratio of firefly luciferase activity to renilla luciferase activity. C. The RIP assay was performed to pull down the miRNAs associated with NBAT1. D. MG63 cell lysates were incubated with biotin-labeled NBAT1; after pull-down, miR-21 was assessed by RT-PCR. E. Anti-AGO2 RIP was performed in U2OS cells transiently overexpressing miR-NC or miR-21, followed by RT-PCR to detect NBAT1 associated with AGO2. F. The relative expression of NBAT1 in U2OS cells transfected with miR-NC or miR-21 was detected by RT-PCR. G. The relative expression of miR-21 in control or NBAT1 or NBAT1-mut overexpressed MG63 cells. H. The cellular proliferation of MG63 cells expressing NBAT1 with and without miR-21. I. The migration and invasion of MG63 cells expressing NBAT1 with and without miR-21. All experiments were repeated three times. *p<0.05.
We further clarified the regulatory relationship between NBAT1 and miR-21. Transfection of miR-21 mimic did not affect NBAT1 expression, indicating that miR-21 bound to NBAT1 but did not induce the degradation of NBAT1 (Figure 4F). Upregulation of NBAT1, but not the mutant NBAT1, significantly suppressed miR-21 expression (Figure 4G), suggesting that NBAT1 functions upstream of miR-21. In addition, miR-21 overexpression abolished the suppression of cellular proliferation, migration and invasion induced by NBAT1 overexrpression (Figure 4H and 4I). Collectively, these results demonstrated that NBAT1 promotes cell proliferation, migration and invasion through directly interaction with miR-21.
NBAT1 is inversely correlated with miR-21 levels in OS tissues
We next examined miR-21 levels by RT-PCR in the same set of 60 OS tissues and adjacent normal tissues. As shown in Figure 5A, miR-21 was significantly upregulated in OS tissues compared to that in adjacent normal tissues. In addition, an inverse correlation between miR-21 and NBAT1 levels was further confirmed by Pearson correlation analysis in 60 OS tissues (p<0.0001, r=-0.5049; Figure 5B). This result further supported that miR-21 was regulated by NBAT1.
Figure 5.
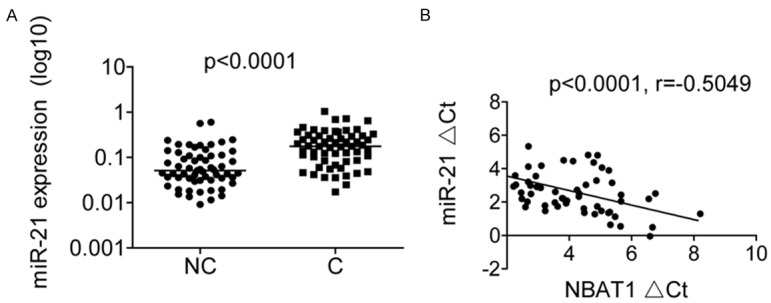
NBAT1 is inversely correlated with miR-21 levels in OS tissues. A. The miR-21 expression in 60 pairs of OS tissues (C) and adjacent noncancerous bone tissues (NC) was measured using RT-PCR. B. The correlation between NBAT1 and miR-21 expression in 60 OS tissue samples.
NBAT1 reverses the miR-21-mediated suppression of target genes
The target gene of miR-21 includes PTEN, PDCD4, TPM1 and RECK [8]. We suspected that NBAT1 may regulate these genes expression through miR-21. Overexpression of NBAT1 increased PTEN, PDCD4, TPM1 and RECK transcript. Ectopic expression of miR-21 abrogated this upregulation (Figure 6A). In addition, we inhibited miR-21 in NBAT1-silenced U2OS. The depletion of NBAT1 decreased PTEN, PDCD4, TPM1 and RECK mRNA levels, while inhibition of miR-21 overcame the suppression of PTEN, PDCD4, TPM1 and RECK (Figure 6B).
Figure 6.
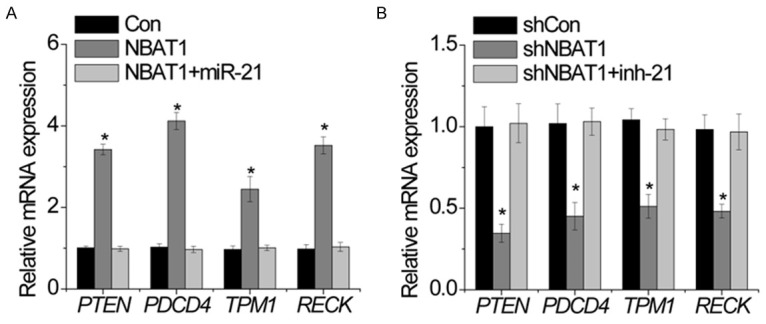
NBAT1 reverses the miR-21-mediated suppression of target genes. A. The relative expression of indicated genes in MG63 cells expressing NBAT1 with and without miR-21 upregulation was determined by RT-PCR. B. The relative expression of indicated genes in U2OS cells expressing shNBAT1 with and without miR-21 inhibitor (inh-21) was determined by RT-PCR. All experiments were repeated three times. *p<0.05.
Discussion
Over the past decade, emerging evidences have highlighted how regulatory RNAs such miRNAs and lncRNAs play important roles in the development of diseases. Dysregulation of lncRNAs has been demonstrated to play crucial roles in OS progression [14-16]. Understanding of biological functions of different lncRNAs may contribute to the developing more effective therapeutic targets for OS patients. In this study, for the first time, we found that NBAT1 expression is downregulated in OS tissues and cell lines and is associated with clinical stage, distant metastasis and poor prognosis. Further investigation revealed that NBAT1 plays a negative regulatory role in OS growth and metastasis via suppression of miR-21.
Recently, lncRNA NBAT1 was found to be downregulated and act as a tumor suppressor in some cancers, such as neuroblastoma, renal cancer, ovarian cancer and breast cancer [11,17-19]. NBAT1 regulates growth and metastasis through different mechanisms. For example, NBAT1 was significantly decreased in neuroblastoma and suppressed cellular proliferation and invasion through epigenetic silencing of target genes and affected neuronal differentiation through activation of the neuronal-specific transcription factor NRSF/REST [11]. Yan et al found that NBAT-1 was obviously downregulated in ovarian cancer compared to normal ovarian tissue. Overexpression of NBAT1 significantly inhibited cell proliferation, invasion, and migration by targeting the ERK1/2 and AKT signaling pathways [18]. Hu et al also reported that reduced NBAT1 in breast cancer was associated with tumor metastasis and poor patient prognosis. Upregulation of NBAT1 inhibited migration and invasion of breast cancer cells through association with PRC2 member EZH2 and regulation of global gene expression, such as DKK1 [19]. These studies indicated that NBAT1 was an bona fide tumor suppressor. However, the functional role of NBAT1 in OS and whether NBAT1 functions as a sponge of microRNAs remains unclear. In the present study, the expression of NBAT1 in OS tissues and cell lines was observed to be significantly decreased in OS tissues and cell lines compared with corresponding normal tissues and the normal cell line. The biological functions of NBAT1 in OS were then detected. We found that NBAT1 inhibited OS growth and metastasis in vitro and in vivo. These results suggest that NBAT1 may act as a tumor suppressor in OS. Furthermore, luciferase reporter, RIP and RNA pull-down assays demonstrated a direct interaction between NBAT1 and miR-21. NBAT1 suppressed malignant phnotypes in a miR-21-dependent manner.
miRNAs are key players in gene regulation, which bind their targets and cause translational repression and/or RNA degradation. miR-21 is one of the most commonly observed aberrant miRNAs in human cancers and is one of the first miRNAs to be described as an oncomir. A large-scale miRNA analysis on 540 samples in six different types of solid tumors has shown that miR-21 was the only miRNA upregulated in all cancer types [20]. It is well known that miRNA function relies primarily on miRNA target genes. For example, inhibition of miR-21 in liver cancer cells increased the expression of PTEN tumor suppressor, and decreased cell proliferation, migration, and invasion [21]. Inhibition of miR-21 leads to an increase in TPM1 expression levels to suppress migration and invasion in breast cancer [22]. miR-21 promotes proliferation, migration, and invasion of lung cancer cells by suppression of PDCD4 [23]. miR-21 also promotes metastasis through inhibition of RECK [24]. In this study, for the first time, we demonstrated that NBAT1 increased the target genes of miR-21, including PTEN, PDCD4, TPM1 and RECK, while overexpression of miR-21 abolished this effect. These results suggest that NBAT1 functions as a competitive endogenous (ceRNA) against miR-21.
Overall, our present study that lncRNA NBAT1 expression is downregulated in OS tissues and cell lines and that decreased NBAT1 expression in OS is closely associated with clinical stage, lymph node metastasis and poor prognosis. Moreover, we also showed that NBAT1 suppresses OS growth and metastasis in vitro and in vivo via association with miR-21. Our experimental data indicated that NBAT1 may be a promising therapeutic target for OS treatment.
Disclosure of conflict of interest
None.
References
- 1.Leichter AL, Sullivan MJ, Eccles MR, Chatterjee A. MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours. Mol Cancer. 2017;16:15. doi: 10.1186/s12943-017-0584-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13:480–491. doi: 10.1038/nrendo.2017.16. [DOI] [PubMed] [Google Scholar]
- 3.Yang G, Yuan J, Li K. EMT transcription factors: implication in osteosarcoma. Med Oncol. 2013;30:697. doi: 10.1007/s12032-013-0697-2. [DOI] [PubMed] [Google Scholar]
- 4.Reed DR, Hayashi M, Wagner L, Binitie O, Steppan DA, Brohl AS, Shinohara ET, Bridge JA, Loeb DM, Borinstein SC, Isakoff MS. Treatment pathway of bone sarcoma in children, adolescents, and young adults. Cancer. 2017;123:2206–2218. doi: 10.1002/cncr.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moraes F, Goes A. A decade of human genome project conclusion: scientific diffusion about our genome knowledge. Biochem Mol Biol Educ. 2016;44:215–223. doi: 10.1002/bmb.20952. [DOI] [PubMed] [Google Scholar]
- 6.Malladi VS, Erickson DT, Podduturi NR, Rowe LD, Chan ET, Davidson JM, Hitz BC, Ho M, Lee BT, Miyasato S, Roe GR, Simison M, Sloan CA, Strattan JS, Tanaka F, Kent WJ, Cherry JM, Hong EL. Ontology application and use at the ENCODE DCC. Database (Oxford) 2015;2015 doi: 10.1093/database/bav010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127:761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markou A, Zavridou M, Lianidou ES. miRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer (Auckl) 2016;7:19–27. doi: 10.2147/LCTT.S60341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lv C, Hao Y, Tu G. MicroRNA-21 promotes proliferation, invasion and suppresses apoptosis in human osteosarcoma line MG63 through PTEN/Akt pathway. Tumour Biol. 2016;37:9333–9342. doi: 10.1007/s13277-016-4807-6. [DOI] [PubMed] [Google Scholar]
- 10.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 11.Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S, Ostensson M, Akyurek LM, Abrahamsson J, Pfeifer S, Larsson E, Shi L, Peng Z, Fischer M, Martinsson T, Hedborg F, Kogner P, Kanduri C. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, Shang JL, Gao CF, Zhang FR, Wang F, Sun SH. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 13.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Liu Z, Wu S. Long non-coding RNA CTA sensitizes osteosarcoma cells to doxorubicin through inhibition of autophagy. Oncotarget. 2017;8:31465–31477. doi: 10.18632/oncotarget.16356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv GY, Miao J, Zhang XL. Long non-coding RNA XIST promotes osteosarcoma progression by targeting Ras-related protein RAP2B via miR-320b. Oncol Res. 2017 doi: 10.3727/096504017X14920318811721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang CL, Zhu KP, Ma XL. Antisense lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. Cancer Lett. 2017;396:66–75. doi: 10.1016/j.canlet.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Xue S, Li QW, Che JP, Guo Y, Yang FQ, Zheng JH. Decreased expression of long non-coding RNA NBAT-1 is associated with poor prognosis in patients with clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2015;8:3765–3774. [PMC free article] [PubMed] [Google Scholar]
- 18.Yan C, Jiang Y, Wan Y, Zhang L, Liu J, Zhou S, Cheng W. Long noncoding RNA NBAT-1 suppresses tumorigenesis and predicts favorable prognosis in ovarian cancer. Onco Targets Ther. 2017;10:1993–2002. doi: 10.2147/OTT.S124645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu P, Chu J, Wu Y, Sun L, Lv X, Zhu Y, Li J, Guo Q, Gong C, Liu B, Su S. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget. 2015;6:32410–32425. doi: 10.18632/oncotarget.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Meng H, Peng Q, Yang X, Gan R, Zhao L, Chen Z, Lu J, Meng QH. Downregulation of microRNA-21 expression restrains non-small cell lung cancer cell proliferation and migration through upregulation of programmed cell death 4. Cancer Gene Ther. 2015;22:23–29. doi: 10.1038/cgt.2014.66. [DOI] [PubMed] [Google Scholar]
- 24.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]