Abstract
MicroRNAs (miRNAs) have been identified as regulators of tumor proliferation, invasion, and metastasis in hepatocellular carcinoma (HCC). In the current study, we determined the clinical significance and biological role of miR-488 in HCC. Our results demonstrated that the expression of miR-488 was notably downregulated in HCC tissues compared with paired adjacent normal tissues. Lower miR-488 expression was positively associated with tumor size, vascular invasion, and shorter overall survival (OS) in HCC patients. Furthermore, gain-and-lost function assays showed that upregulation of miR-488 significantly inhibited cell proliferation, colony formation, cell invasion, and the epithelial-to-mesenchymal transition (EMT) process. We showed that ADAM9 served as a direct target for miR-488 and mediated lower miR-488 expression, thus inducing cell proliferation and invasion in HCC. Moreover, we found that lncRNA HULC is a target of miR-488 in HCC cells and miR-488 inhibited the expression of HULC by sponging to HULC in HCC. Thus, our results suggest that miR-488 functions as a tumor suppressor in HCC and may be a potential target for HCC treatment.
Keywords: Hepatocellular carcinoma, miR-488, ADAM9, HULC, cell proliferation
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-associated deaths and > 700,000 patients die from HCC worldwide every year [1]. Tumor invasion, metastasis, and recurrence are the main causes of death in HCC patients [2]. Despite advances in understanding of the molecular mechanisms underlying HCC and improved therapeutic methods for HCC, the 5-year overall survival (OS) is still unsatisfactory [3]. Thus, identification of novel therapeutic strategies for HCC is urgently needed.
MicroRNAs (miRNAs), a variety of small single-stranded non-coding RNA molecules, affect multiple biological processes, including cell development, differentiation, and carcinogenesis [4]. With respect to tumor progression, miRNAs regulate the expression of tumor suppressors or oncogenes by binding to the 3’-untranslated region (3’-UTR) of targeted mRNA [5]. A number of microRNAs are involved in HCC progression. Specifically, microRNA-147 suppresses cell proliferation, migration, and chemosensitivity by inhibiting HOXC6 in human HCC [6]. MiR-221 promotes tumor growth and invasion of HCC cells by constitutive activation of NFκB [7]. MiR-1299 suppresses cell proliferation of HCC by targeting CDK6 [8]. MiR-296 inhibits proliferation and induces apoptosis by targeting FGFR1 in human HCC [9]. Additionally, an increasing number of miRNAs have been shown to play a crucial role in HCC progression [10].
MiR-488 has been reported to act as a tumor suppressor involved in some tumors types; miR-488 inhibits cell proliferation and cisplatin sensitivity by activating the eIF3a-mediated NER signaling pathway in non-small-cell lung cancer (NSCLC) [11]. MiR-488 acts as a tumor suppressor gene to suppress cell proliferation, the cell cycle, colony information, and migration in gastric cancer [12]. MicroRNA-488-3p sensitizes malignant melanoma cells to cisplatin by targeting PRKDC [13]; however, the clinical significance and functional effects of miR-488 in HCC is unknown.
In the current study we showed that miR-488 was significantly down-regulated in HCC tissues. Up-regulation of miR-488 inhibited cell proliferation, colony formation, cell invasion, and EMT in vitro. We showed that ADAM9 serves as a direct target for miR-488 and mediates lower miR-488 expression, thus inducing cell proliferation and invasion in HCC. Moreover, we found that HULC is a target of miR-488 in HCC cells and miR-488 inhibits the expression of HULC by sponging HULC in HCC. Thus, our results suggest that miR-488 functions as a tumor suppressor in HCC and may be a potential target for treatment in HCC.
Methods
Clinical tissue samples
Sixty HCC and adjacent normal tissue samples were acquired from patients who underwent surgical resection from August 2009 to September 2014 in the Department of Hepatobiliary Surgery of The Cixi People’s Hospital of Zhejiang Province. None of the patients received radiotherapy or chemotherapy before surgery. The tissues were frozen immediately after resection and stored at -80°C until further analysis. Written consent was collected from all patients and the study was approved by the Ethics Committee of The Cixi People’s Hospital of Zhejiang Province. All experiments were performed in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki).
Cell lines culture, cell transfection, and plasmid construction
The human HCC cell lines, HepG2, Huh-7, MHCC97H, and MHCC97L, and a normal cell line, LO2, were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin (Gibco BRL, NY, USA) at 37°C in humidified atmosphere with 5% CO2. A miR-488 mimic, miR-488 inhibitor, and miR-negative control (NC) were purchased from GenePharma (Shanghai, China). The ADAM9 overexpression plasmid was constructed and purchased from GenePharma and were used following the manufacturer’s protocol. Small interfering RNAs (siRNAs) targeting HULC cells (si-HULC-1, 5’-AAC CTC CAG AAC TGT GAT CCA-3’; si-HULC-2, 5’-GCCTTTACAAGGGAATGAAGA-3’) were transfected using Lipofectamine 2000 (Invitrogen). Total RNA and protein were isolated 48 h after transfection.
Cell proliferation assay
A total of 2000 cells/well were seeded in 96-well plates and cell proliferation was evaluated by the MTT assay, according to the manufacturer’s instructions. MTT reagent solution was added to each well and incubated at 37°C in 5% CO2 for 4 h. Cell proliferation was assessed at 0, 24 h, 48 h, and 72 h. Then, the absorbance was read at 490 nm in a microplate absorbance reader.
Cell colony assay
A cell colony formation assay was used to evaluate cell proliferation. A total of 400 cells/well were seeded in 6-well plates and cultured with DMEM supplemented with 10% fetal bovine serum at 37°C in humidified atmosphere with 5% CO2 for 2 weeks. Cell colonies were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. The number of cell colonies was evaluated by photomicroscopy (Olympus, Japan), then counted.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using Trizol reagent (Invitrogen). First strand cDNA was reversed using Reverse EasyScript One Step gDNA Removal and cDNA Synthesis SuperMix (TAKALA, Dalian, China). The qRT-PCR assays were carried out using SYBR Green Master Mixture (Roche) reagent on an ABI7500 system (Applied Biosystems, USA). GAPDH or U6 was used as an internal control. The relative mRNA levels of gene expression were assessed using the 2-ΔΔCt method. The primer sequences were as follows: ADAM9, forward (5’-GCTAGTTGGACTGGAGATTTGG-3’) and reverse (5’-TTATTACCACAGGAGGGAGCAC-3’); highly up-regulated in liver cancer (HULC), forward (5’-TCATGATGGAATTGGAGCCTT-3’) and reverse (5’-CTCTTCCTGGCTTGCAGATTG-3’); and GAPDH, forward (5’-GCACCGTCAAGGCTGAGAAC-3’) and reverse (5’-TGGTGAAGACGCCAGTGGA-3’).
Western blot analysis
Total cell protein was extracted using RIPA lysis buffer (Beyotime, Shanghai, China). The protein samples were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto PVDF membranes (Bio-Rad Laboratories, USA). The membranes were blocked in 5% skim milk, then incubated with specific primary antibodies with PCNA (1:1000; CST, USA), E-cadherin (1:1500; CST), N-cadherin (1:1000; CST), Snail (1:1500; Abcam, USA), GAPDH (1:1500; Abcam), and ADAM9 (1:2000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) at 4°C overnight. The membranes were incubated for 1 h with horseradish peroxidase-conjugated secondary antibody. Protein bands were detected using a Molecular Imager ChemiDoc XRS System (Bio-Rad Laboratories).
Luciferase reporter assay
The wild and mutant type 3’UTRs of ADAM9 or wild type HULC (HULC-WT) and mutant type HULC (HULC-MUT) containing the binding site sequences of miR-488 were cloned into the pMIRGLO Reporter Vector (Ambion). MHCC97H cells were seeded in 96-well plates and co-transfected with empty vector or pmirGLO-luciferase reporter comprising type and mutant type 3’UTRs of ADAM9 or HULC-WT and HULC-MUT and miR-NC or miR-488 mimic using Lipofectamine 2000 (Invitrogen). Transfected cells were harvested 48 h after transfection. The luciferase activity was evaluated using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol.
Statistical analysis
Statistical analysis was performed using SPSS 18.0 (SPSS, Inc., Chicago, IL, USA) and are shown as the mean ± standard deviation. A t-test was used to analyze the difference between two groups. A P-value < 0.05 was considered to indicate a statistically significant difference.
Results
Relative expression of miR-488 is lower in HCC tissues and cells
To determine the expression of miR-488 in frozen HCC and adjacent normal tissues, we performed qRT-PCR assays. The expression of miR-488 was aberrantly down-regulated in HCC tissues compared to adjacent normal tissues (Figure 1A). To validate whether or not aberrant expression of miR-488 was associated with clinicopathologic factors, we divided 60 HCC patients into 2 subgroups based on the relative mean value (0.425) of miR-488 in HCC tissue samples. As shown in Table 1, the level of miR-488 expression was significantly associated with tumor size (P = 0.031, Figure 1B) and vascular invasion (P = 0.004, Figure 1C), but there was no significant association with age, gender, differentiation, and tumor number (P > 0.05, Table 1). We also demonstrated that lower miR-488 expression was positively associated with OS in HCC patients (log rank = 10.026, P < 0.05, Figure 1D). Furthermore, we determined miR-488 expression in four human HCC cell lines (HepG2, Huh-7, MHCC97H, and MHCC97L) and a normal cell line (LO2). As shown in Figure 1E, we demonstrated that the relative expression of miR-488 in HCC cell lines was aberrantly lower compared to LO2 cells.
Figure 1.
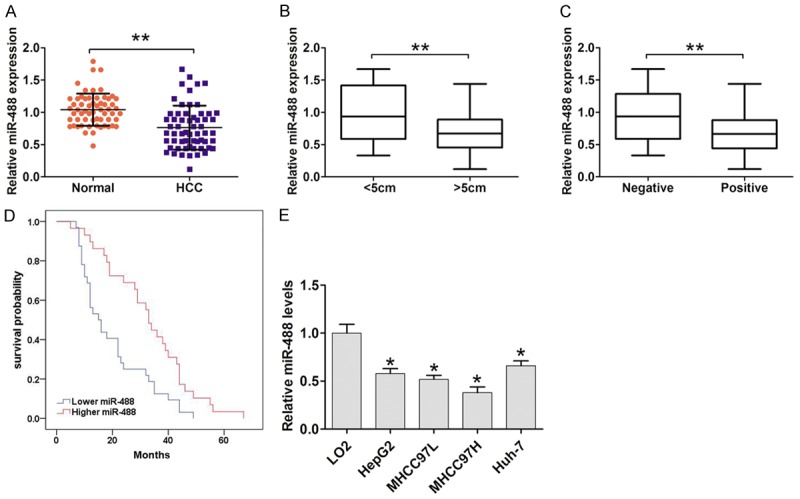
MiR-488 was lower in HCC tissues and cells. A. QRT-PCR results showed that miR-488 expression in HCC tissues was significantly reduced compared to adjacent normal tissues (n = 60). U6 served as an internal control. B. The association between miR-488 expression and tumor size in HCC patients. C. The association between miR-488 expression and vascular invasion in HCC patients. D. Patients with lower miR-488 expression had a poor survival time compared with higher miR-488 expression by Kaplan-Meier analysis and log rank test. E. QRT-PCR results showed that miR-488 expression in HCC cell lines, including HepG2, Huh-7, MHCC97H, and MHCC97L, was higher compared with the normal cell line, LO2. U6 served as an internal control. The results are the average value of three independent tests. *P < 0.05, **P < 0.01.
Table 1.
The Correlation between miR-488 expression and clinicopathological factors in 60 cases HCC patients
| Clinicopathological factors | Number of Patients (n = 60) | MiR-488 levels | P-value | |
|---|---|---|---|---|
|
| ||||
| Low (n = 32) | High (n = 28) | |||
| Age | 0.464 | |||
| ≤ 55 | 40 | 20 | 20 | |
| > 55 | 20 | 12 | 8 | |
| Gender | 0.370 | |||
| Male | 46 | 26 | 20 | |
| Female | 14 | 6 | 8 | |
| Tumor size | 0.031a | |||
| < 5 cm | 34 | 14 | 20 | |
| > 5 cm | 26 | 18 | 8 | |
| Differentiation | 0.491 | |||
| Well | 22 | 10 | 12 | |
| Morderate | 24 | 15 | 9 | |
| Low | 14 | 7 | 7 | |
| Tumor number | 0.714 | |||
| 1 | 20 | 10 | 10 | |
| > 1 | 40 | 22 | 18 | |
| Vascular invasion | 0.004a | |||
| Negative | 31 | 11 | 20 | |
| Positive | 29 | 21 | 8 | |
| TNM stage | 0.329 | |||
| I/II | 39 | 19 | 20 | |
| III/IV | 21 | 13 | 8 | |
P < 0.05.
MiR-488 inhibits cell proliferation and invasion and epithelial-mesenchymal transition (EMT) in HCC
To investigate the biological function of miR-488 expression in HCC, cells were transfected with miR-488 mimic, miR-488 inhibitor, or miR-NC in MHCC97H and Huh-7 cells. MTT assay results demonstrated that cell proliferation was suppressed after up-regulation of miR-488 by transfection with miR-488 mimic in MHCC97H cells (Figure 2A). Conversely, cell proliferation was enhanced after down-regulation of miR-488 by transfection with miR-488 inhibitor in Huh-7 cells (Figure 2B). Furthermore, cell colony assays demonstrated that the relative number of cell colonies was fewer and smaller after up-regulation of miR-488 in MHCC97H cells, but was greater and larger after down-regulation of miR-488 in Huh-7 cells (Figure 2C-F). Proliferating cell nuclear antigen (PCNA) was down-regulated by up-regulation of miR-488 in MHCC97H cells, but was increased after down-regulation of miR-488 in Huh-7 cells (Figure 2G and 2H).
Figure 2.
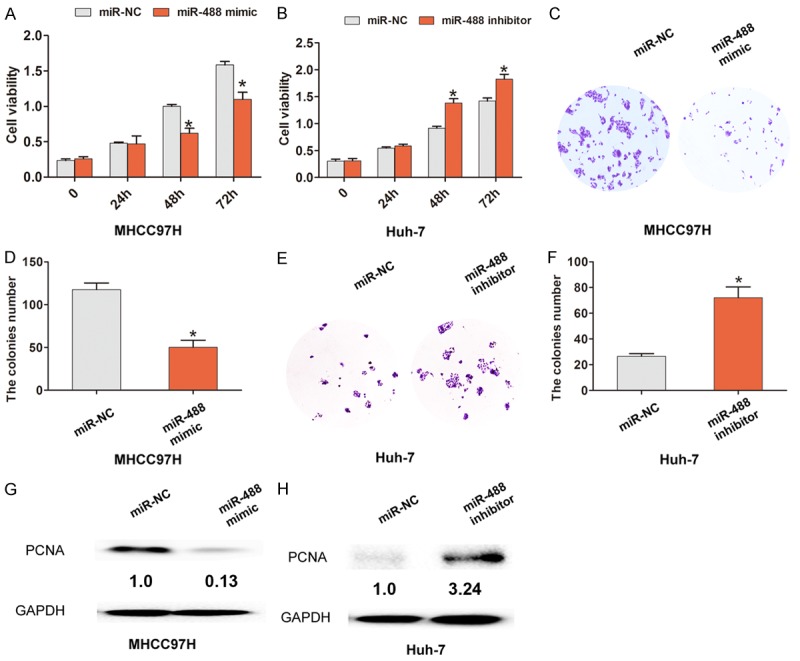
Increased miRNA-488 inhibits cell proliferation in HCC cells. A. The MTT assay showed that an increased level of miR-488 expression inhibited cell proliferation compared with the miR-NC group in MHCC97H cells. B. The MTT assay showed that a decreased level of miR-488 expression enhanced cell proliferation compared with the miR-NC group in Huh-7 cells. C, D. A cell colony assay and cell colony number analysis showed that the up-regulated level of miR-488 expression reduced the number of cell colonies compared with the miR-NC group in MHCC97H cells. E, F. A cell colony assay and cell colony number analysis showed that the down-regulated level of miR-488 expression increased the number of cell colonies compared with the miR-NC group in Huh-7 cells. G, H. Western blot analysis showed that PCNA expression was down-regulated after cells were transfected with miR-488 mimic in MHCC97H cells, but up-regulated after cells were transfected with miR-488 inhibitor in Huh-7 cells. The results are the average value of three independent tests. *P < 0.05.
Cell invasion ability was markedly inhibited after up-regulation of miR-488 in MHCC97H; however, down-regulation of miR-488 promoted cell invasion in Huh-7 cells (Figure 3A-D). Moreover, the EMT relative marker, E-cadherin, was up-regulated, but the transcription factor (Snail) and mesenchymal marker (N-cadherin) were down-regulated after up-regulation of miR-488 in MHCC97H; however, the opposite results were demonstrated after down-regulation of miR-488 in Huh-7 cells (Figure 3E and 3F). Taken together, our results indicate that miR-488 functions as a tumor suppressor to inhibit cell proliferation, invasion, and EMT in HCC.
Figure 3.
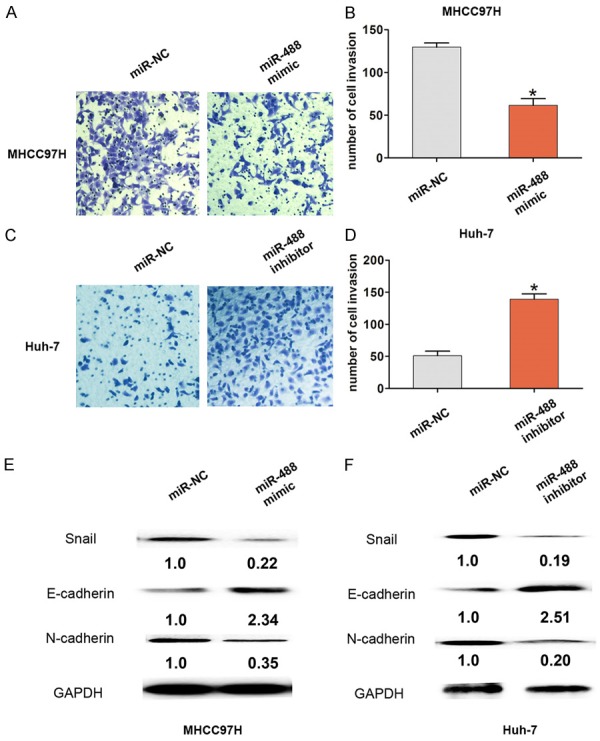
Increased miRNA-488 suppressed cell invasion in HCC cells. A, B. A cell invasion assay and cell invasion number analysis showed that the level of up-regulated miR-488 expression reduced the cell invasion number compared with the miR-NC group in MHCC97H cells. C, D. A cell invasion assay and cell invasion number analysis showed that the down-regulated level of miR-488 expression increased the cell invasion number compared with the miR-NC group in Huh-7 cells. E, F. Western blot analysis showed the expression of Snail, E-cadherin, and N-cadherin protein after cells were transfected with miR-488 mimic in MHCC97H cells or after cells were transfected with miR-488 inhibitor in Huh-7 cells compared with the miR-NC group. The results are the average value of three independent tests. *P < 0.05.
ADAM9 is a direct target of miR-488 in HCC
We further showed that ADAM9 expression was up-regulated in HCC tissues and cells compared with adjacent normal tissues and LO2 cells, respectively (Figure 4A and 4B). Moreover, we demonstrated that lower miR-488 expression had a significantly negative association with higher ADAM9 in HCC tissues by Pearson correlation analysis (r = -0.380, P < 0.05, Figure 4C). We speculate that ADMA9 may be a potential regulated target of miR-488. By using three bioinformatics databases (TargetScan, miRanda, and PicTar) to predict potential targets of miR-488, we reasoned that there is a putative miR-488 binding site located in the 3’-UTR of the ADAM9 transcript. To confirm whether or not ADMA9 is a direct target of miR-488 in HCC, we constructed wild type ADMA9 3’UTR (ADAM9-WT) or mutant type ADMA9 3’UTR (ADAM9-MUT) vectors containing the binding sites of miR-488 (Figure 4D). The dual-luciferase reporter assay showed that miR-488 mimic significantly inhibited the luciferase activity of the ADAM9-WT 3’UTR-binding site, whereas luciferase activity was not significantly changed in the ADAM9-MUT 3’UTR-binding site (Figure 4E). Additionally, we demonstrated that the mRNA and protein levels of ADMA9 were dramatically decreased when miR-488 was overexpressed in MHCC97H cells, but significantly increased when miR-488 was down-regulated in Huh-7 cells (Figure 4F-H). These results suggest that miR-488 directly targeted ADAM9 and regulated the expression in HCC.
Figure 4.
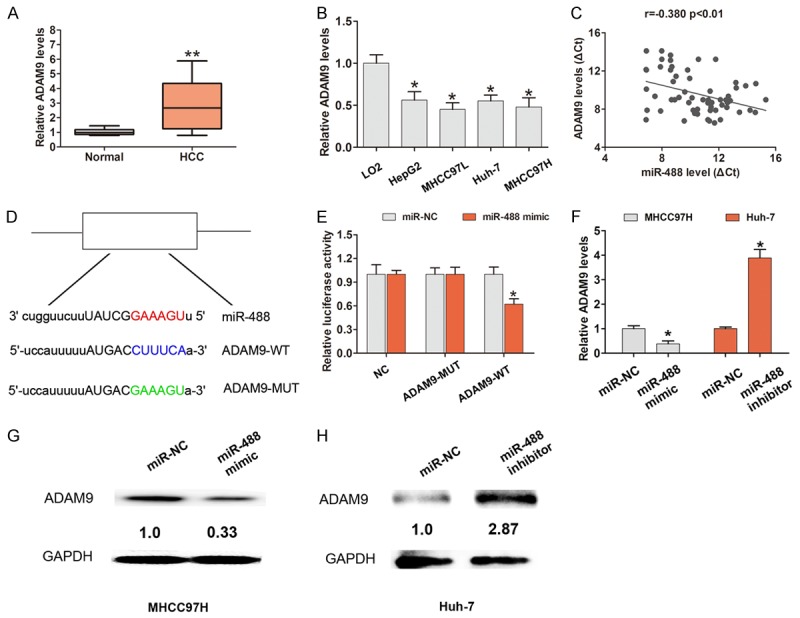
ADAM9 is a direct target of miR-488 in HCC cells. A. QRT-PCR showed that ADAM9 expression in HCC tissues was significantly higher compared to adjacent normal tissues (n = 60). GAPDH served as an internal control. B. QRT-PCR showed that ADAM9 expression in HCC cell lines, including HepG2, Huh-7, MHCC97H, and MHCC97L, was higher compared with the normal cell line, LO2. GAPDH served as an internal control. C. The association between miR-488 and ADAM9 expression was analyzed by Pearson correlation analysis. D. The predicted binding sites between miR-488 and ADAM9 mRNA through complementary base-pairs. The wild-type ADMA9 3’UTR (ADAM9-WT) or mutant-type ADMA9 3’UTR (ADAM9-MUT) vectors containing the binding sites of miR-488 were constructed. E. The dual-luciferase reporter assay showed that miR-488 mimic significantly inhibited the luciferase activity of the ADAM9-WT 3’UTR-binding site, but was not significantly changed in the ADAM9-MUT 3’UTR-binding site. F-H. QRT-PCR and Western blot analysis showed the expression of mRNA and protein of ADAM9 after cells were transfected with miR-488 mimic in MHCC97H cells or after cells were transfected with miR-488 inhibitor in Huh-7 cells compared with the miR-NC group. The results are the average value of three independent tests. *P < 0.05.
Upregulated ADAM9 reverses the tumor suppressive effect of miR-488 in HCC
To detect whether or not ADAM9 mediates decreased miR-488-induced cell proliferation and invasion in HCC, we overexpressed ADAM9 by transfection with pcDNA3.0-ADAM9 into MHCC97H cells, which resulted in increased ADAM9 expression in MHCC97H cells (Figure 5A). Furthermore, we demonstrated that cell proliferation and cell colony formation abilities were significantly inhibited by transfection with miR-488 mimic into MHCC97H cells, but ameliorated by co-transfection with miR-488 mimic and pcDNA3.0-ADAM9 (Figure 5B-D). Moreover, cell invasion ability was significantly inhibited by transfection with miR-488 mimic into MHCC97H cells, but negated by co-transfection with miR-488 mimic and pcDNA3.0-ADAM9 (Figure 5E and 5F). These results indicated that ADAM9 mediated lower miR-488-induced cell proliferation and invasion in HCC.
Figure 5.
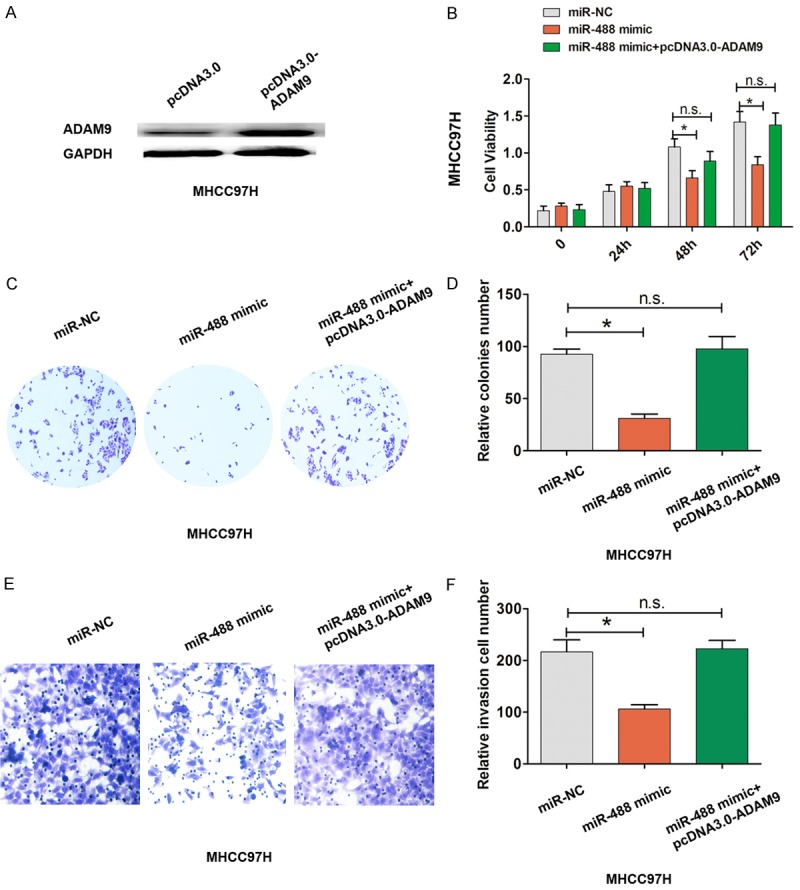
Up-regulated ADAM9 reversed the tumor suppressive effect of miR-488 in HCC. A. ADAM9 was over-expressed by transfection with pcDNA3.0-ADAM9 into MHCC97H cells. B. Cell proliferation was evaluated using the MTT assay after transfection with miR-NC, miR-488 mimic, miR-488 mimic, and pcDNA3.0-ADAM9 into MHCC97H cells. C, D. The cell colony number was evaluated after transfection with miR-NC, miR-488 mimic, miR-488 mimic, and pcDNA3.0-ADAM9 into MHCC97H cells. E, F. A cell invasion assay and the cell invasion number were evaluated after transfection with miR-NC, miR-488 mimic, miR-488 mimic, and pcDNA3.0-ADAM9 into MHCC97H cells. The results are the average value of three independent tests. *P < 0.05; n.s., not significant.
MiR-488 inhibits HULC expression by sponging to HULC in HCC cells
HULC has recently been reported to be involved in HCC development and progression [14]. Our results demonstrated that up-regulation of miR-488 inhibits HULC expression; however, down-regulation of miR-488 enhanced the expression of HULC in MHCC97H cells (Figure 6A). Knockdown of HULC in MHCC97H significantly increased miR-488 expression (Figure 6B). By bioinformatics database (starbase v2.0 and miRanda) analysis, we demonstrated that there is a putative miR-488 binding site with HULC. We constructed the wild-type HULC (HULC-WT) or mutant-type HULC (HULC-MUT) vectors containing the binding sites of miR-488 (Figure 6C). The luciferase reporter assay showed that miR-488 mimic significantly inhibited the luciferase activity of the HULC-WT binding site, but was not significantly changed in the HULC-MUT binding site (Figure 6D). We chose si-HULC-2 for the following experiments due to its higher efficiency of HULC knockdown in MHCC97H cells. We demonstrated that cell colony and cell invasion ability were significantly enhanced by transfection with miR-488 inhibitor into MHCC97H cells, but was ameliorated by co-transfection with miR-488 inhibitor and si-HULC-2 (Figure 6E-H). The results indicated that miR-488 suppresses cell proliferation by sponging to HULC and regulates HULC expression in HCC.
Figure 6.
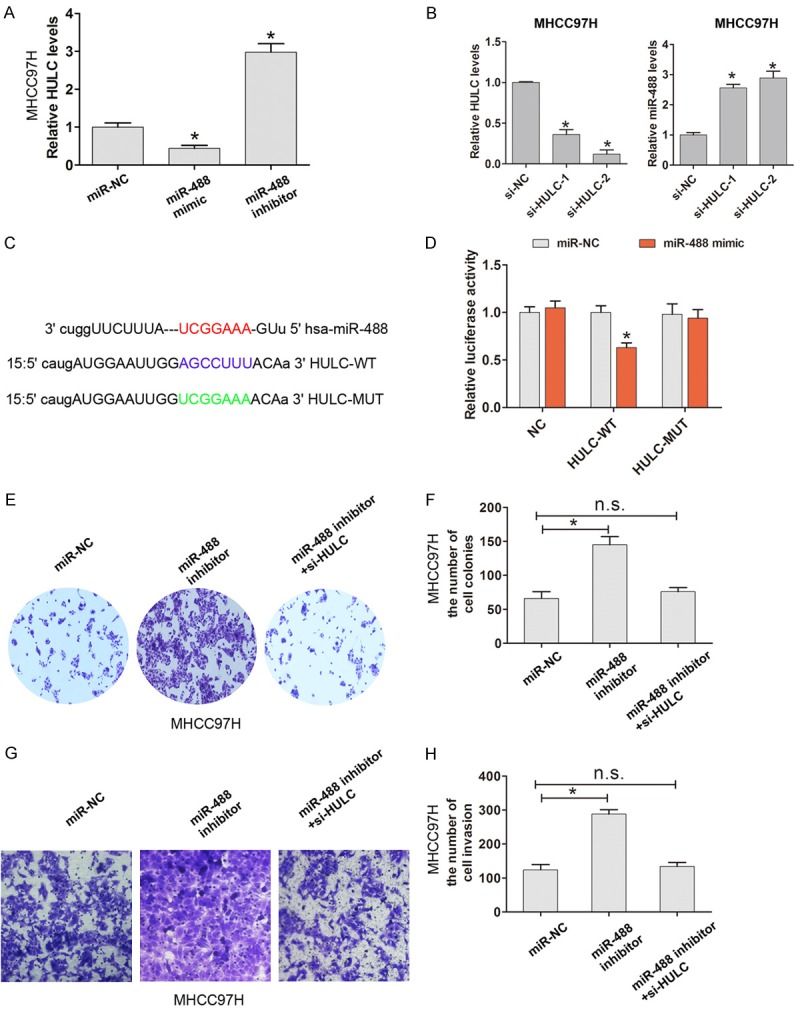
MiR-488 inhibits HULC expression by sponging to HULC in HCC cells. A. The relative HULC expression after transfection with miR-NC, miR-488 mimic, and miR-488 inhibitor into MHCC97H cells was analyzed by qRT-PCR. B. The relative HULC and miR-488 expression after transfection with si-NC, si-HULC-1, and si-HULC-2 into MHCC97H cells was analyzed by qRT-PCR. C. The predicted binding bites between miR-488 and HULC mRNA through complementary base-pairs. The wild-type HULC (HULC-WT) or mutant-type HULC (HULC-MUT) vectors containing the binding sites of miR-488 were constructed. D. The dual-luciferase reporter assay showed that miR-488 mimic significantly inhibited the luciferase activity of the HULC-WT binding site, whereas luciferase activity was not significantly changed in the HULC-MUT binding site. E, F. The cell colony number was evaluated after transfection with miR-NC, miR-488 inhibitor, miR-488 inhibitor, and si-HULC into MHCC97H cells. G, H. The cell invasion assay and invasion number were evaluated after transfection with miR-NC, miR-488 inhibitor, miR-488 inhibitor, and si-HULC into MHCC97H cells. The results are the average value of three independent tests. *P < 0.05; n.s., not significant.
Discussion
MiRNAs have emerged recently as potential regulators of cell development, cell proliferation, apoptosis, and tumorigenesis. In previous studies, miRNAs have been identified as predictors of prognosis in HCC patients. High expression of miR-105-1 is positively correlated with clinical prognosis in patients with HCC by targeting the oncogene, NCOA1 [15]. Up-regulation of miR-522 is associated with poor outcome in patients with HCC [16]. Decreased expression of serum miR-424 correlates with poor prognosis in patients with HCC [17]. In the current study, we confirmed that miR-488 is down-regulated in HCC tissues compared with adjacent normal tissues. Lower miR-488 was positively associated with larger tumor size, vascular invasion, and shorter OS in HCC patients.
MiRNAs bind to the specific 3’UTR region of target mRNA and are involved in post-transcriptional regulation, which plays a crucial role in a variety of biological processes, including HCC progression. Specifically, miR-34a targets HDAC1 and regulates expression to affect cell proliferation and apoptosis in HCC [18]. MiR-199a-3p inhibits cell proliferation and induces cell apoptosis by targeting YAP1, suppressing Jagged1-Notch signaling in HCC [19]. MiR-22 targets YWHAZ to inhibit metastasis of HCC, the down-regulation of which predicts poor survival [20]. In the current study, gain-and-lost function assays showed that up-regulation of miR-488 inhibits cell proliferation, colony formation, cell invasion, and EMT. ADAM9 targets miR-488 in HCC cells. Up-regulated miR-488 suppressed the expression of ADAM9 in HCC cells; however, down-regulated miR-488 increased the expression of ADAM9.
ADAM9 has been shown to have a tumor-promoting role. ADAM9 up-regulates N-cadherin via miR-218 suppression in lung adenocarcinoma cells [21]. ADAM9 regulates lung cancer metastasis to the brain by facilitating tPA-mediated cleavage of CDCP1 [22]. ADAM9 enhances CDCP1 protein expression by suppressing miR-218 for lung tumor metastasis [23]. MiR-203 suppresses the proliferation and metastasis of HCC by targeting the oncogene, ADAM9 [24]. In the current study, up-regulated miR-488 was shown to suppress cell proliferation and invasion in HCC cells, but was reversed by up-regulated ADAM9 expression, which indicated that ADAM9 mediates decreased miR-488-induced cell proliferation and invasion in HCC.
LncRNA HULC has been reported to function as a tumor-promoting, long non-coding RNA involving in HCC progression. HULC enhances EMT to promote tumorigenesis and metastasis of HCC via the miR-200a-3p/ZEB1 signaling pathway [14]. Elevation of HULC by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulation of p18 [25]. HULC promotes tumor angiogenesis in liver cancer via miR-107/E2F1/SPHK1 signaling [26]. Our results showed that HULC is a direct target of miR-488 and up-regulated miR-488 inhibits HULC expression in HCC cells. Cell proliferation and invasion was enhanced by down-regulation of miR-488 expression, but reversed by HULC knockdown in HCC.
In conclusion, our results showed that miR-488 was lower in HCC and lower miR-488 predicted a poor prognosis in HCC patients. Moreover, miR-488 inhibited cell proliferation, cell invasion, and EMT. MiR-488 inhibited cell invasion by targeting ADAM9 and sponging to HULC in HCC. Thus, our results imply that miR-488 functions as a tumor suppressor in HCC and may be a potential target for treatment in HCC.
Disclosure of conflict of interest
None.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Lau WY, Lai EC. Hepatocellular carcinoma: current management and recent advances. Hepatobiliary Pancreat Dis Int. 2008;7:237–257. [PubMed] [Google Scholar]
- 3.Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J. Clin. Oncol. 2009;27:1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esquela-Kerscher A, Slack FJ. OncomirsmicroRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 5.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sui CJ, Xu F, Shen WF, Dai BH, Lu JJ, Zhang MF, Yang JM. MicroRNA-147 suppresses human hepatocellular carcinoma proliferation migration and chemosensitivity by inhibiting HOXC6. Am J Cancer Res. 2016;6:2787–2798. [PMC free article] [PubMed] [Google Scholar]
- 7.Tian YW, Shen Q, Jiang QF, Wang YX, Li K, Xue HZ. Decreased levels of miR-34a and miR-217 act as predictor biomarkers of aggressive progression and poor prognosis in hepatocellular carcinoma. Minerva Med. 2017;108:108–113. doi: 10.23736/S0026-4806.16.04616-4. [DOI] [PubMed] [Google Scholar]
- 8.Zhu H, Wang G, Zhou X, Song X, Gao H, Ma C, Chang H, Li H, Liu FF, Lu J, Ma J. miR-1299 suppresses cell proliferation of hepatocellular carcinoma (HCC) by targeting CDK6. Biomed Pharmacother. 2016;83:792–797. doi: 10.1016/j.biopha.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Bo X, Zheng Q, Xiao X, Wu L, Li B. miR-296 inhibits proliferation and induces apoptosis by targeting FGFR1 in human hepatocellular carcinoma. FEBS Lett. 2016;590:4252–4262. doi: 10.1002/1873-3468.12442. [DOI] [PubMed] [Google Scholar]
- 10.Hayes CN, Chayama K. MicroRNAs as biomarkers for liver disease and hepatocellular carcinoma. Int J Mol Sci. 2016;17:280. doi: 10.3390/ijms17030280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang C, Chen YX, Wu NY, Yin JY, Li XP, Huang HS, Zhang W, Zhou HH, Liu ZQ. MiR-488 inhibits proliferation and cisplatin sensibility in non-small-cell lung cancer (NSCLC) cells by activating the eIF3a-mediated NER signaling pathway. Sci Rep. 2017;7:40384. doi: 10.1038/srep40384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Lu G, Ke X, Lu X, Wang X, Li H, Ren M, He S. miR-488 acts as a tumor suppressor gene in gastric cancer. Tumour Biol. 2016;37:8691–8698. doi: 10.1007/s13277-015-4645-y. [DOI] [PubMed] [Google Scholar]
- 13.Li N, Ma Y, Ma L, Guan Y, Ma L, Yang D. MicroRNA-488-3p sensitizes malignant melanoma cells to cisplatin by targeting PRKDC. Cell Biol Int. 2017;41:622–629. doi: 10.1002/cbin.10765. [DOI] [PubMed] [Google Scholar]
- 14.Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ, Wang CY, Zhang HM, Zhang RX, Zhang JJ, Yao Z, Shen ZY. LncRNA HULC enhances epithelialmesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431–42446. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma YS, Wu TM, Lv ZW, Lu GX, Cong XL, Xie RT, Yang HQ, Chang ZY, Sun R, Chai L, Cai MX, Zhong XJ, Zhu J, Fu D. High expression of miR-105-1 positively correlates with clinical prognosis of hepatocellular carcinoma by targeting oncogene NCOA1. Oncotarget. 2017;8:11896–11905. doi: 10.18632/oncotarget.14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi YH, Qi BB, Liu XB, Ding HM. Upregulation of miR-522 is associated with poor outcome of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2016;20:3194–3198. [PubMed] [Google Scholar]
- 17.Yao H, Liu X, Chen S, Xia W, Chen X. Decreased expression of serum miR-424 correlates with poor prognosis of patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:14830–14835. [PMC free article] [PubMed] [Google Scholar]
- 18.Sun TY, Xie HJ, Li Z, Kong LF, Gou XN, Li DJ, Shi YJ, Ding YZ. miR-34a regulates HDAC1 expression to affect the proliferation and apoptosis of hepatocellular carcinoma. Am J Transl Res. 2017;9:103–114. [PMC free article] [PubMed] [Google Scholar]
- 19.Ren K, Li T, Zhang W, Ren J, Li Z, Wu G. miR-199a-3p inhibits cell proliferation and induces apoptosis by targeting YAP1, suppressing Jagged1-Notch signaling in human hepatocellular carcinoma. J Biomed Sci. 2016;23:79. doi: 10.1186/s12929-016-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M, Hu W, Xiong CL, Qu Z, Yin CQ, Wang YH, Luo CL, Guan Q, Yuan CH, Wang FB. miR-22 targets YWHAZ to inhibit metastasis of hepatocellular carcinoma and its down-regulation predicts a poor survival. Oncotarget. 2016;7:80751–80764. doi: 10.18632/oncotarget.13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sher YP, Wang LJ, Chuang LL, Tsai MH, Kuo TT, Huang CC, Chuang EY, Lai LC. ADAM9 upregulates N-cadherin via miR-218 suppression in lung adenocarcinoma cells. PLoS One. 2014;9:e94065. doi: 10.1371/journal.pone.0094065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin CY, Chen HJ, Huang CC, Lai LC, Lu TP, Tseng GC, Kuo TT, Kuok QY, Hsu JL, Sung SY, Hung MC, Sher YP. ADAM9 promotes lung cancer metastases to brain by a plasminogen activator-based pathway. Cancer Res. 2014;74:5229–5243. doi: 10.1158/0008-5472.CAN-13-2995. [DOI] [PubMed] [Google Scholar]
- 23.Chiu KL, Kuo TT, Kuok QY, Lin YS, Hua CH, Lin CY, Su PY, Lai LC, Sher YP. ADAM9 enhances CDCP1 protein expression by suppressing miR-218 for lung tumor metastasis. Sci Rep. 2015;5:16426. doi: 10.1038/srep16426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan D, Shen S, Fu S, Preston B, Brandon C, He S, Shen C, Wu J, Wang S, Xie W, Chen B, Liya A, Guo Y, Zheng D, Zhi Q, Peng B. miR-203 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting oncogene ADAM9 and oncogenic long non-coding RNA HULC. Anticancer Agents Med Chem. 2016;16:414–23. doi: 10.2174/1871520615666150716105955. [DOI] [PubMed] [Google Scholar]
- 25.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–11. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Z, Xiao Z, Liu F, Cui M, Li W, Yang Z, Li J, Ye L, Zhang X. Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1) Oncotarget. 2016;7:241–54. doi: 10.18632/oncotarget.6280. [DOI] [PMC free article] [PubMed] [Google Scholar]


