Abstract
To date, several species of Asteraceae have been considered as Cd-accumulators. However, little information on the Cd tolerance and associated mechanisms of Asteraceae species Cosmos bipinnatus, is known. Presently, several physiological indexes and transcriptome profiling under Cd stress were investigated. C. bipinnatus exhibited strong Cd tolerance and recommended as a Cd-accumulator, although the biomasses were reduced by Cd. Meanwhile, Cd stresses reduced Zn and Ca uptake, but increased Fe uptake. Subcellular distribution indicated that the vacuole sequestration in root mainly detoxified Cd under lower Cd stress. Whilst, cell wall binding and vacuole sequestration in root co-detoxified Cd under high Cd exposure. Meanwhile, 66,407 unigenes were assembled and 41,674 (62.75%) unigenes were annotated in at least one database. 2,658 DEGs including 1,292 up-regulated unigenes and 1,366 down-regulated unigenes were identified under 40 μmol/L Cd stress. Among of these DEGs, ZIPs, HMAs, NRAMPs and ABC transporters might participate in Cd uptake, translocation and accumulation. Many DEGs participating in several processes such as cell wall biosynthesis, GSH metabolism, TCA cycle and antioxidant system probably play critical roles in cell wall binding, vacuole sequestration and detoxification. These results provided a novel insight into the physiological and transcriptome response to Cd in C. bipinnatus seedlings.
Introduction
Cadmium (Cd), a non-essential heavy metal, causes a distinct toxicity in both plants and humans1. In planta, Cd directly or indirectly causes several toxicities, such as inducing oxidative stress2–4, altering the chloroplast ultrastructure5, damaging chlorophyll synthesis, impairing photosynthetic efficiency6,7, and reducing mineral nutrient uptake such as Zn, Fe, and Ca8, finally inhibiting plant growth and causing death9–11. However, some Cd- tolerance plants or hyper-accumulators such as Thlaspi caerulescens 12, Sedum alfredii 13, Viola baoshanensis 14, and Solanum nigrum 15 accumulate high Cd concentrations in shoots without or having only mild toxicity symptoms16, which therefore have been/being used for phytoremediation of Cd. Meanwhile, their physiological and molecular mechanisms of Cd tolerance have been/being substantially revealed17–19. However, different species exhibit different Cd uptake, translocation, detoxification and their associated mechanisms. Thus, it is crucial to identify new Cd accumulators or hyper-accumulators, and understand their physiological and molecular mechanism.
Several species of the Asteraceae family, such as Crassocephalum crepidioides 20, Bidens pilosa, Kalimeris integrifolia 21, Chromolaena odorata 22, Elephantopus mollis 23, and Picris divaricata 24, are recommended as Cd-accumulators, which are used for phytoremediation. Cosmos (Cosmos bipinnatus Cav.), an annual species of Asteraceae, possesses ornamental value in its leaves and flowers, as well as strong adaption and plasticity traits in adverse environments. Thus, it is now widely cultivated in China. Previous study indicated that C. bipinnatus is a potential chromium (Cr) hyper-accumulator in plants25. Whether is it a Cd hyperaccumulator/accumulator, and possesses unique physiological and molecular mechanisms?
With the advent of next-generation sequencing (NGS) technology, RNA sequencing (RNA-Seq) has been/being widely used to reveal molecular mechanisms under abiotic stresses and to enrich our transcriptional evidence for plants26,27. Increasing studies using RNA-Seq have revealed Cd response in different plants and understood their associated molecular mechanism28–31. For example, compared with low-Cd-accumulation (LCA) genotypes, transcriptomic evidence indicated that high-Cd-accumulation (HCA) genotypes have more complicated mechanisms when exposed to Cd32–34. Additionally, RNA-Seq has also been used to screen candidate genes for Cd hyper-accumulator and provide a novel perspective on the molecular mechanisms, such as in Noccaea caerulescens 35 and Solanum nigrum 36. However, the transcriptome information for C. bipinnatus under Cd stress, is still unknown.
In the present study, we identified a new Cd accumulator, C. bipinnatus, from the Asteraceae family, and aimed to reveal its potential physiological and molecular mechanisms using metal subcellular distribution, several biochemical indexes, and RNA-Seq. Additionally, due to lack of genomic information of C. bipinnatus, construction of the transcriptome of the C. bipinnatus would facilitate its molecular research.
Results
Plant growth
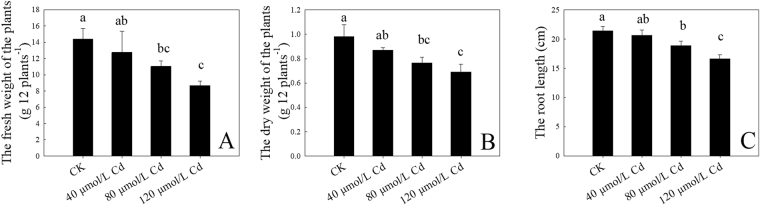
Compared with control, the seedlings treated with 40 μmol/L Cd did not show obvious toxicity symptoms after 9 days of treatment, while 80 and 120 μmol/L Cd treatment exhibited visibly toxic symptoms, such as the decreased leaf number, and the reddened stems at 120 μmol/L (Fig. 1). 40 μmol/L Cd did not significantly affect the fresh and dry weight of plant and the length of root (Fig. 2A–C). However, the biomass was significantly reduced by 80 and 120 μmol/L Cd (Fig. 2A and B). Since several significant changes were observed between 40 and 80 μmol/L (Fig. 1 and 2), 40 μmol/L Cd should be recommended as the threshold of normal growth.
Figure 1.
Growth of C. bipinnatus treated with different Cd concentrations.
Figure 2.
The growth of C. bipinnatus exposed to Cd. A: the fresh weight of the plants; B: the dry weight of the plants, and C: the root length. Values were means ± standard deviation (n = 3); values followed by different lowercase letters show significant differences at P < 0.05 .
Cd accumulation and distribution
The Cd concentration was not observed in all samples under 0 μmol/L Cd stress (Table 1). The Cd concentrations of all samples increased significantly with increasing Cd concentration. Cd accumulated highly in the roots, followed by the stems and the leaves (Table 1). Translocation factor (TF) values of the stems ranged from 0.56–0.64 was higher than those of the leaves ranged from 0.19–0.29 (Table 1), suggesting that most of Cd in aboveground was sequestrated in the stems.
Table 1.
The concentration of Cd in dry tissues and translocation factor (TF) of C. bibinnatus seedlings treated with different levels of Cd.
| Treatment | Leaf (μg/g DW) | Stem (μg/g DW) | Root (μg/g DW) | TF | |
|---|---|---|---|---|---|
| Stem | Leaf | ||||
| CK | N.D. | N.D. | N.D. | N.D. | N.D. |
| 40 μmol/L Cd | 60.36 ± 2.17a | 321.15 ± 16.04b | 576.65 ± 41.48b | 0.56 | 0.19 |
| 80 μmol/L Cd | 93.41 ± 8.29b | 414.23 ± 25.64ab | 648.98 ± 55.83b | 0.64 | 0.23 |
| 120 μmol/L Cd | 145.87 ± 6.73c | 499.05 ± 87.54a | 806.07 ± 36.26a | 0.62 | 0.29 |
Values are mean ± standard deviation (n = 3). Values within a column followed by different lowercase letters show significant differences at P < 0.05. N.D., not detected under the detection limit of Cd: 2.5 μg/g, the same as below. TF = [the mean value of concentration in stems]/[the mean value of concentration in roots] for stems and [the mean value of concentration in leaves]/[the mean value of concentration in roots] for leaves.
In order to understand whether different Cd stresses exhibited different Cd detoxifications or toxicity, we analyzed the Cd subcellular distribution mainly in the roots under these three Cd stresses. Under 40 μmol/L Cd stress, more than 80% Cd was accumulated in the soluble fraction, approximate 15% Cd was accumulated in the cell wall fraction, and only 5% Cd was transported into the organelle fraction (Fig. 3). Although Cd in soluble fraction was dramatically reduced with the increasing Cd concentrations, more than 55% Cd was still sequestrated in this fraction when treated with 120 μmol/L Cd (Fig. 3). Meanwhile, Cd in cell wall fractions were significantly increased with the increasing Cd concentrations, up to 40% Cd was binding in cell wall fraction when treated with 120 μmol/L Cd. These results indicated that the sequestration of Cd into soluble fraction is the main Cd detoxification mechanism under low Cd stress, while the sequestration of Cd into soluble fraction and the binding of Cd in the cell wall fraction represent a coaction for Cd detoxification with the increasing Cd concentrations.
Figure 3.
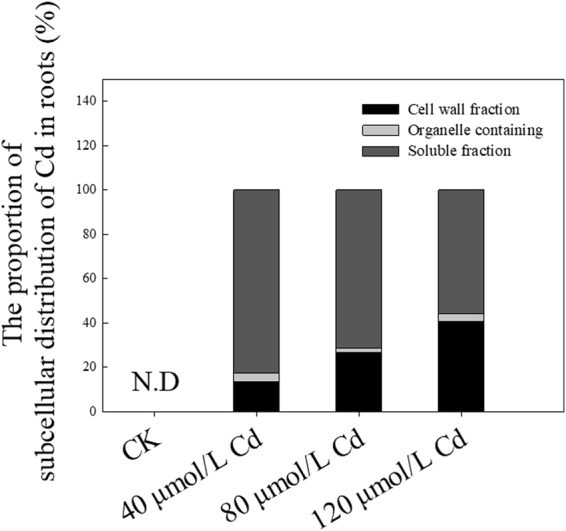
The subcellular distribution of Cd under different concentrations of Cd.
Effects of Cd on Zn, Ca, and Fe concentrations in C. bipinnatus
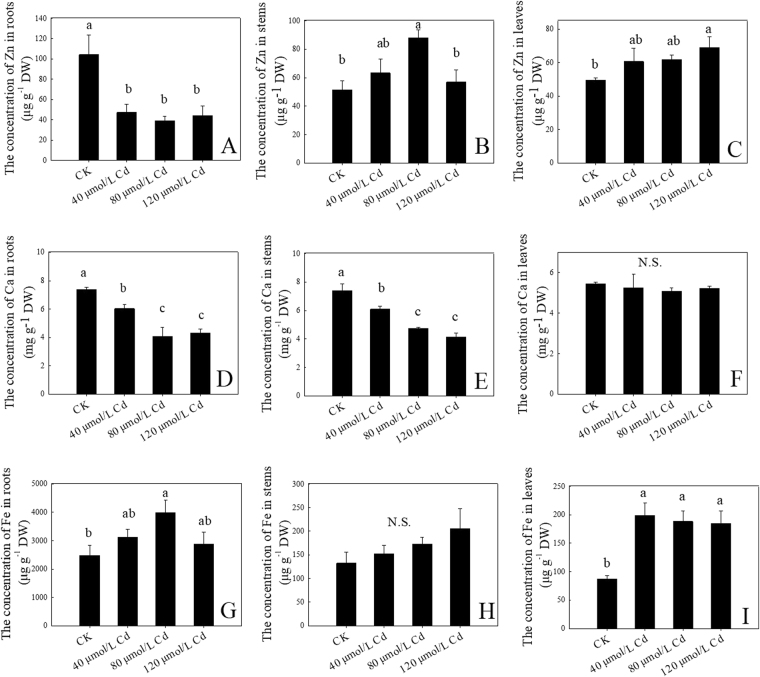
After 9 days of treatments, the Cd stresses significantly decreased the uptake of Zn in the roots when compared with CK (Fig. 4A). Interestingly, Zn concentration in the stems and leaves were mainly increased, although some decreased at 120 μmol/L Cd in stems (Fig. 4B and C). These results indicated that Cd inhibited the uptake of Zn in the roots, but promoted the translocation of Zn from root to shoot. Meanwhile, Cd stresses significantly decreased the Ca concentration in the stems and the roots (Fig. 4D and E), but did not affect the Ca concentration in the leaves (Fig. 4F). Cd increased Fe concentrations in roots and stems leaves, although the differences were not significant (Fig. 4G and H). But it significantly increased the Fe concentrations in leaves (Fig. 4I). These results indicated that Cd treatment may differentially affect the uptake of metal nutrients in C. bipinnatus.
Figure 4.
The concentration of metals under different Cd treatments of C. bipinnatus seedlings. A, B, and C: the concentration of Zn; D, E, and F: the concentration of Ca; G, H, and I: the concentration of Fe. N.S., no difference in various treatments. Values were means ± standard deviation (n = 3); values followed by different lowercase letters show significant differences at P < 0.05.
MDA concentrations and the activity of several antioxidant enzymes
In order to understand whether Cd induces biochemical damage, we investigated several biochemical indexes which are involved in oxidative stress. Compared with CK, Cd significantly increased the MDA concentrations in leaves (except of 40 μmol/L, Fig. 5A) and roots (Fig. 5B). Meanwhile, the POD activity in the leaves and roots were dramatically increased (Fig. 5C and D). Interestingly, Cd stresses did not affect the CAT activity in the leaves (Fig. 5E) and roots (except of 40 μmol/L, Fig. 5F). Except of 120 μmol/L Cd in the leaves, the SOD activity in the leaves and roots were increased by all three Cd treatments (Fig. 5G and H). Although 40 μmol/L Cd did not affect the GR activity, 80 and 120 μmol/L Cd significantly increased the GR activity in leaves and roots (Fig. 5I and J). These results indicated that lower Cd treatment (40 μmol/L) did not cause severe oxidative stresses, while higher Cd treatment (80–120 μmol/L) produced more oxidative damages.
Figure 5.
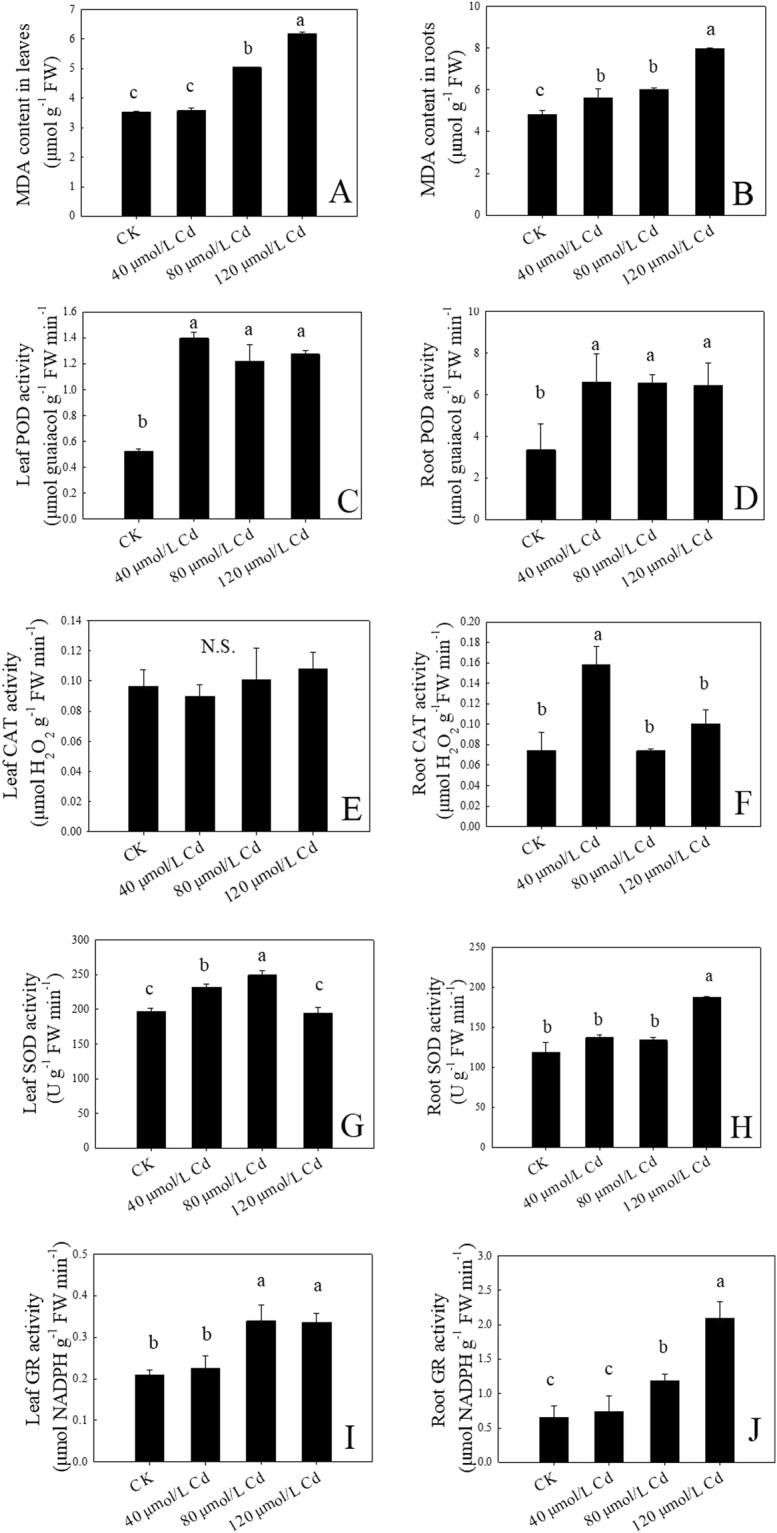
Physiological parameters of C. bipinnatus under different concentrations of Cd. A and B: the concentration of MDA; C and D: the POD activity; E and F: the CAT activity; G and H: the SOD activity; I and J: the GR activity. N.S., no difference in various treatment. Values were means ± standard deviation (n = 3); values followed by different lowercase letters show significant differences at P < 0.05.
Transcriptome sequence and de novo assembly
Cd stresses changed the accumulation of some metal nutrients, the subcellular distribution of Cd, and the concentration and activity of some biochemical indexes. Meanwhile, some significantly changes were induced by 40 μmol/L Cd and no obvious toxic symptom was observed. Root samples treated with 40 μmol/L Cd was interestingly used to transcriptome analysis to reveal molecular response. 14.9 Gb nucleotides were generated (Table 2), which was deposited in the Sequence Read Archive (SRA) database with the accession numbers SRR3546768 and SRR3546769. 66,407 unigenes with the means length of 817 bp and N50 value of 1,344 bp were assembled. Among these assembled unigenes, the length of 18,491 unigenes (27.8% of all the unigenes) was more than 1,000 bp (Table 2). These results suggested that RNA sequencing and assembled unigenes had well quality and could be used for further transcriptome analysis.
Table 2.
Overview of the reads and assembly.
| Items | Number |
|---|---|
| Total nucleotides (nt) | 14,940,933,000 |
| Unigenes | 66,407 |
| Total length of unigenes (bp) | 54,271,910 |
| Mean length of unigenes (bp) | 817 |
| N50 length of unigenes (bp) | 1,344 |
| Length range more than 1000 bp | 18,491 |
Functional annotation and classification
41,674 (62.76%) unigenes were functionally annotated in at least one of the five databases: GO, KEGG, COG, Swissprot and NR (Table 3). 24,733 (37.24%) unigenes were not annotated in any public database. Among these annotated unigenes, 15,481 unigenes were classified into 25 COG categories. In detail, the major group of COG was ‘general functions prediction only’, followed by ‘translation, ribosomal structure and biogenesis’, ‘transcription’ and ‘replication, recombination and repair’ (SFig. 1). 24,639 unigenes were classified into three major categories of GO classification. ‘Cell’, ‘organelle part’ and ‘cell part’ represented the largest proportion in the cellular component category, while ‘catalytic activity’ and ‘cell part’ represented the most abundant categories in molecular function category. Moreover, the most abundant categories were ‘metabolic process’, ‘cellular process’ and ‘single-organism process’ in biological process category (SFig. 2). A total of 18,496 unigenes were annotated in the KEGG database and were classified into 128 KEGG pathways (STable 2). Briefly, ‘ribosome’ pathway (ko03010) contains the most abundant unigenes, followed by ‘carbon metabolism’ (ko01200), ‘biosynthesis of amino acids’ (ko01230), and ‘protein processing in endoplasmic reticulum’ (ko04141). All sequences and functional information were deposited in the NCBI Transcriptome Shotgun Assembly database with accession number GEZQ00000000.
Table 3.
Result of unigne annotation.
| Annotation Database | Annotated Number | The percentage of annotated unignenes in total unigenes (%) |
|---|---|---|
| GO Annotation | 24,639 | 37.10 |
| KEGG Annotation | 18,496 | 27.85 |
| COG Annotation | 15481 | 23.31 |
| Swissprot Annotation | 27,069 | 40.76 |
| NR Annotation | 41,145 | 61.96 |
| Annotated in at least one database | 41,674 | 62.76 |
Identification and functional characterization of differentially expressed genes in response to Cd stress
In present study, a log-fold expression change (log2FC) >2 or <−2 with P values <0.0005 and FDR <0.001 was used to determine the differentially expressed genes (DEGs). Compared with CK, 2,658 unigenes including 1,292 up-regulated unigenes and 1,366 down-regulated unigenes were induced by Cd (SFig. 3). Based on GO annotation, a total of 2,460 DEGs were classified into three GO major categories (SFigure 4A–C). Moreover, a total of 1,884 DEGs were annotated in KEGG pathway (STable 3).
Noteworthy DEGs and metabolic pathways related to Cd uptake, transportation and detoxification
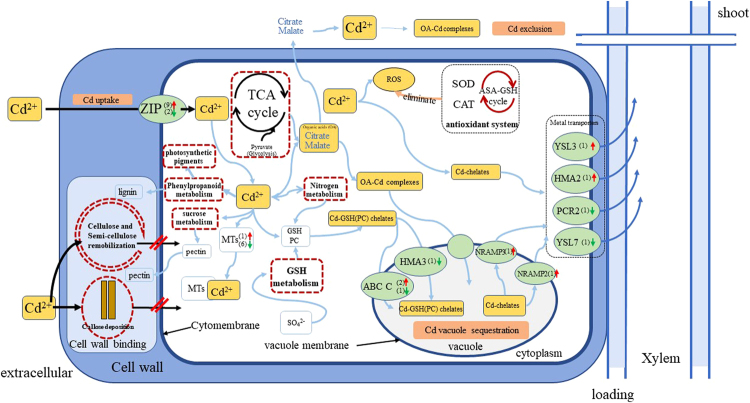
As shown in Fig. 6, a network associated to Cd uptake, transport, translocation, and detoxification were aggregated. Cd in extracellular could be obstructed by cell wall structures for toxicity reduction, while part of Cd would be transported by some metal transporters. When Cd entered into the C. bipinnatus cells, various metabolic processes could be induced for Cd detoxification. GSH, MTs and organic acid would be bound with Cd and then sequestrated into vacuoles. Cd also induced unigenes of antioxidant enzymes for oxidative defense. Finally, some Cd-chelates would be uploaded into xylem for transportation by some metal transporters.
Figure 6.
The presumed transcriptional network related to Cd uptake, translocation, and detoxification in C. bipinnatus root. The red arrows represent up-regulated genes, while the green arrows represent down-regulated genes. The dotted red boxes represent noteworthy mechanism pathways.
Briefly, this network was mainly composited by the several following processes. Firstly, total 49 metal transporters (34 up-regulated and 15 down-regulated) including zinc transporters (ZIPs), ATP-binding cassette (ABC) family members, heavy-metal ATPases (HMAs) like HMA3, HMA2, the family of natural resistance-associated macrophage protein (NRAMP) members such as NRAMP2, NRAMP3, yellow stripe-like (YSL) family members, and plant cadmium resistance protein PCR2 were observed (Table 4), suggesting that these metal transporters might participate in Cd uptake, transport and translocation. 29 DEGs (14 up-regulated and 15 down-regulated) involved in sulfate and GSH metabolism such as gene encoding adenylyl-sulfate kinase, sulfate adenylyl-transferase, serine acetyltransferase, adenylyl-sulfate reductase, sulfite reductase, serine acetyl-transferase, cysteine synthase, ornithine decarboxylase, spermidine synthase, glutathione S-transferase, and γ-glutamyl-transpeptidase 3 were regulated by Cd, suggesting the potential biosynthesis of GSH and PCs and chelation of Cd (Table 4, SFig. 5). 7 DEGs (1 up-regulated and 6 down-regulated) were metal chelates such as metallothioneins (MTs), suggesting MTs also play role in Cd chelation (Table 4). 30 DEGs (13 up-regulated and 17 down-regulated) were involved in cell wall metabolism, such as UDP-glucose 6-dehydrogenase1 (UGDH1), UDP-glucose pyrophosphorylase2 (UGP2), glucose-6-phosphate isomerase (GPI), sucrose synthase, pectinesterase, UDP-glucuronate-4-epimerase, fructokinase, beta-fructofuranosidase, xyloglucan endotransglucosylase/hydrolase protein, expansin, glucan 1,3-alpha-glucosidase, cellulose synthase, callose synthase, and laccase, suggesting the compositions of cell wall were changed, mainly focused on pectin, callose and cellulose (Table 4). In addition, 12 DEGs (1 up-regulated and 11 down-regulated) participated in phenylpropanoid metabolism were regulated by Cd (Table 4). 17 DEGs (9 up-regulated and 8 down-regulated) were participated in tricarboxylic acid cycle (TCA) pathway, which potentially enhanced glucose metabolism and produced more organic acids such as citrate and malate for Cd binding (Table 4, SFig. 6). 11 DEGs (4 up-regulated and 7 down-regulated) were related to nitrogen metabolism, including genes encoding high-affinity nitrate transporter, nitrate reductase, ferrddoxin-nitrite reductase, glutamine synthetase, and glutamate synthase [NADPH/NADH] (Table 4). Moreover, 14 DEGs (4 up-regulated and 10 down-regulated) involved in antioxidant system, like superoxide dismutase, catalase isozyme, phospholipid hydroperoxide glutathione peroxidase, monodehydroascorbate reductase, and ascorbate peroxidase were differentially regulated by Cd (Table 4 ).
Table 4.
Noteworthy DEGs and metabolic pathways related to Cd uptake, transportation and detoxification.
| Unigenes ID | Log2FC | Description |
|---|---|---|
| Metal transporter | ||
| c69105_c0 | 9.44 | ABC transporter A family member 7 |
| c84507_c0 | 8.64 | Zinc transporter 5 |
| c65267_c0 | 8.04 | ABC transporter B family member 4 |
| c39965_c0 | 7.91 | ABC transporter G family member 17 |
| c83650_c0 | 7.18 | ABC transporter G family member 43 |
| c70109_c0 | 6.61 | Zinc transporter 7 |
| c54092_c1 | 6.42 | Zinc transporter ZTP29 |
| c10112_c0 | 6.20 | ABC transporter F family member 4 |
| c77338_c0 | 6.15 | ABC transporter C family member 2 |
| c59065_c0 | 5.97 | Zinc transporter ZTP29 |
| c65853_c2 | 5.84 | ABC transporter F family member 1 |
| c80839_c1 | 5.82 | ABC transporter B family member 21 |
| c32003_c0 | 5.77 | ABC transporter F family member 4 |
| c82788_c0 | 5.36 | ABC transporter B family member 11 |
| c37195_c0 | 5.16 | Metal transporter Nramp3 |
| c87790_c0 | 5.09 | Metal transporter Nramp2 |
| c35172_c0 | 5.04 | ABC transporter G family member 22 |
| c37809_c0 | 5.01 | ABC transporter A family member 1 |
| c56776_c0 | 4.72 | Cadmium/zinc-transporting ATPase HMA2 |
| c90431_c0 | 4.69 | Zinc transporter 3 |
| c66824_c0 | 4.69 | Zinc transporter 1 |
| c77338_c1 | 4.26 | ABC transporter C family member 14 |
| c59283_c0 | 3.60 | Zinc transporter 5 |
| c60866_c0 | 3.30 | ABC transporter G family member 14 |
| c76227_c0 | 3.16 | ABC transporter A family member 2 |
| c81234_c0 | 3.13 | Zinc transporter 4 |
| c73636_c0 | 2.89 | Zinc transporter 4 |
| c78055_c0 | 2.74 | ABC transporter G family member 1 |
| c60915_c0 | 2.59 | ABC transporter B family member 11 |
| c60185_c0 | 2.59 | ABC transporter F family member 4 |
| c68969_c0 | 2.52 | Metal-nicotianamine transporter YSL3 |
| c62568_c0 | 2.51 | ABC transporter G family member 14 |
| c82493_c0 | 2.29 | ABC transporter G family member 16 |
| c81263_c0 | 2.19 | ABC transporter G family member 22 |
| c23361_c0 | −2.43 | ABC transporter F family member 4 |
| c74639_c0 | −2.50 | ABC transporter F family member 4 |
| c68390_c0 | −2.55 | Cadmium/zinc-transporting ATPase HMA3 |
| c28965_c0 | −2.67 | Metal-nicotianamine transporter YSL7 |
| c66361_c0 | −2.76 | Metal-nicotianamine transporter YSL14 |
| c57525_c0 | −2.79 | ABC transporter F family member 1 |
| c27812_c0 | −3.28 | ABC transporter F family member 4 |
| c67525_c0 | −3.75 | ABC transporter F family member 3 |
| c97488_c0 | −4.38 | ABC transporter B family member 4 |
| c3952_c0 | −4.63 | ABC transporter B family member 1 |
| c98738_c0 | −4.78 | ABC transporter C family member 2 |
| c85857_c0 | −5.30 | Zinc transporter 8 |
| c53195_c0 | −5.34 | Plant cadmium resistance protein 2 PCR2 |
| c27275_c0 | −5.68 | Zinc transporter 5 |
| c89759_c0 | −6.01 | ABC transporter G family member 40 |
| Sulfate, GSH metabolism | ||
| c85588_c0 | −6.46 | glutathione S-transferase |
| c57929_c0 | −6.28 | Monodehydroascorbate reductase |
| c85420_c0 | −6.23 | Glutathione S-transferase F9 |
| c85507_c0 | −6.18 | Glutathione S-transferase U8 |
| c66368_c0 | −6.09 | L-ascorbate peroxidase 1 |
| c85932_c0 | −6.07 | Glutathione S-transferase F6 |
| c88021_c0 | −5.71 | glutathione S-transferase |
| c92596_c0 | −5.71 | Cysteine synthase |
| c87506_c0 | −5.71 | Glutathione S-transferase U17 |
| c32193_c0 | −5.39 | glutathione S-transferase parC |
| c77199_c0 | −5.06 | L-ascorbate peroxidase 2 |
| c94817_c0 | −4.84 | Glutathione S-transferase U17 |
| c69512_c0 | −2.84 | Ornithine decarboxylase |
| c73219_c0 | −2.53 | Ornithine decarboxylase |
| c66300_c0 | −2.43 | Ornithine decarboxylase |
| c67901_c1 | 2.37 | γ-glutamyl-transpeptidase 3 |
| c80321_c1 | 2.64 | γ-glutamyl-transpeptidase 3 |
| c60771_c0 | 2.68 | Adenylyl-sulfate kinase 3 |
| c54934_c0 | 3.59 | Sulfate adenylyl-transferase |
| c56904_c0 | 4.02 | Monodehydroascorbate reductase |
| c40445_c0 | 4.69 | Spermidine synthase 1 |
| c88608_c0 | 4.79 | L-ascorbate peroxidase 7 |
| c67547_c0 | 4.86 | adenylyl-sulfate reductase 3 |
| c65681_c0 | 4.97 | Cysteine synthase |
| c31632_c0 | 5.15 | Serine acetyltransferase 5 |
| c51737_c1 | 5.88 | Glutathione S-transferase L2 |
| c69922_c0 | 5.99 | Glutathione S-transferase F9 |
| c52639_c0 | 6.36 | Glutathione S-transferase F13 |
| c27387_c0 | 6.85 | Sulfite reductase |
| Metallothioneins(MTs) | ||
| c83366_c0 | −9.45 | Metallothionein-like protein type 2 |
| c83540_c0 | −8.79 | Metallothionein-like protein type 3 |
| c39380_c0 | −8.50 | Metallothionein-like protein type 3 |
| c84110_c0 | −7.91 | Metallothionein-like protein type 2 |
| c10641_c0 | −7.39 | Metallothionein-like protein 1 |
| c85025_c0 | −5.39 | Metallothionein-like protein type 2 |
| c25697_c0 | 5.27 | Metallothionein-like protein type 2 |
| Phenylpropanoid metabolism | ||
| c84209_c0 | −7.69 | Peroxidase 42 |
| c62400_c0 | −7.33 | Peroxidase 4 |
| c52362_c0 | −6.53 | Caffeic acid 3-O-methyltransferase COMT |
| c88098_c0 | −6.44 | Trans-cinnamate 4-monooxygenase |
| c84029_c0 | −6.41 | Peroxidase 42 |
| c87932_c0 | −6.09 | Peroxidase 15 |
| c86814_c0 | −5.39 | Phenylalanine ammonia-lyase PAL |
| c90791_c0 | −5.14 | Cinnamyl alcohol dehydrogenase 1 CAD1 |
| c90727_c0 | −4.84 | Trans-cinnamate 4-monooxygenase |
| c86953_c0 | −4.71 | Caffeic acid 3-O-methyltransferase COMT |
| c98254_c0 | −4.47 | Caffeic acid 3-O-methyltransferase COMT |
| c79992_c0 | 2.10 | 4-coumarate–CoA ligase-like 4CL |
| Cell wall metabolism | ||
| c52549_c0 | 6.75 | Beta-fructofuranosidase |
| c55905_c0 | −5.85 | Fructokinase-4 |
| c22672_c0 | 4.75 | Glucose-6-phosphate isomerase GPI |
| c63434_c0 | 3.15 | Pectinesterase 2 |
| c88310_c0 | −6.09 | Pectinesterase 3 |
| c92007_c0 | −5.03 | Pectinesterase U1 |
| c63222_c0 | −7.80 | Sucrose synthase 1 |
| c25867_c0 | −5.98 | Sucrose synthase 2 |
| c88935_c0 | −5.25 | Sucrose synthase 2 |
| c87544_c0 | −5.56 | Sucrose synthase 3 |
| c86190_c0 | −4.97 | Sucrose synthase 3 |
| c53972_c0 | −6.18 | UDP-glucose 6-dehydrogenase 1 UGDH1 |
| c43360_c0 | 6.03 | UDP-glucose pyrophosphorylase UGP2 |
| c86968_c0 | −5.92 | UDP-glucuronate 4-epimerase 1 |
| c36225_c0 | −6.37 | UDP-glucuronate 4-epimerase 4 |
| c32187_c0 | −5.60 | UDP-glucuronate 4-epimerase 6 |
| c86761_c0 | −5.64 | UDP-glucuronate 4-epimerase 6 |
| c84524_c0 | 2.21 | Xyloglucan endotransglucosylase/hydrolase protein 23 |
| c84832_c0 | 2.87 | Xyloglucan endotransglucosylase/hydrolase protein 32 |
| c87800_c0 | −5.09 | Xyloglucan endotransglucosylase/hydrolase protein 6 |
| c87242_c0 | −5.09 | Xyloglucan endotransglucosylase/hydrolase protein B |
| c64115_c0 | 2.67 | Xyloglucan endotransglucosylase/hydrolase protein 32 |
| c27718_c0 | −5.75 | Xyloglucan endotransglucosylase/hydrolase protein 9 (Precursor) |
| c75386_c0 | 5.02 | Callose synthase 7 |
| c80747_c0 | 2.55 | Cellulose synthase-like protein G3 |
| c49538_c0 | 2.97 | Expansin-A1 |
| c50143_c0 | 2.46 | Expansin-A10 |
| c63814_c0 | 5.05 | Glucan 1,3-alpha-glucosidase |
| c66127_c0 | 3.21 | Laccase-12 |
| TCA cycle | ||
| c87855_c0 | −6.01 | Aconitate hydratase ACO |
| c88735_c0 | 5.48 | ATP-citrate synthase alpha chain protein 1 ACLA |
| c89044_c0 | −6.62 | Dihydrolipoyl dehydrogenase 1 LPD1 |
| c91756_c0 | 3.72 | Dihydrolipoyl dehydrogenase 1 LPD1 |
| c33952_c0 | 6.55 | Dihydrolipoyl dehydrogenase LPD |
| c71205_c0 | 2.53 | Dihydrolipoyllysine-residue acetyltransferase DLAT |
| c87987_c0 | 3.98 | Isocitrate dehydrogenase IDH3 |
| c56559_c0 | −5.92 | Malate dehydrogenase MDH |
| c59906_c0 | −5.52 | Malate dehydrogenase MDH |
| c99710_c0 | −4.84 | Malate dehydrogenase MDH |
| c61978_c0 | −4.71 | Malate dehydrogenase MDH |
| c91986_c0 | −5.09 | Succinate dehydrogenase |
| c23134_c0 | −5.82 | Malate dehydrogenase1 MDH1 |
| c52564_c0 | 3.59 | Pyruvate dehydrogenase PDHA |
| c85739_c0 | 4.49 | Pyruvate dehydrogenase PDHA |
| c87218_c0 | 4.93 | Pyruvate dehydrogenase PDHA |
| c57876_c0 | 5.79 | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit 1 SDHB |
| nitrogen metabolism | ||
| c86348_c0 | −6.61 | Glutamate synthase [NADH] |
| c85579_c0 | −6.18 | Glutamine synthetase |
| c86171_c0 | −5.26 | Glutamine synthetase |
| c39565_c0 | −5.17 | Glutamine synthetase |
| c88704_c0 | −5.09 | Nitrate reductase [NADH] |
| c91005_c0 | −4.97 | Glutamate synthase 1 [NADH] |
| c24081_c0 | −4.47 | Glutamine synthetase |
| c72948_c0 | 3.64 | Ferredoxin–nitrite reductase |
| c66268_c0 | 5.30 | High-affinity nitrate transporter 2.2 |
| c53175_c0 | 5.54 | Glutamine synthetase |
| c87775_c0 | 5.70 | Nitrate reductase [NADH] |
| antioxidant system | ||
| c85366.c0 | −7.03 | Superoxide dismutase [Cu-Zn] |
| c88413.c0 | −4.55 | Superoxide dismutase [Cu-Zn] |
| c58063.c0 | −5.60 | Superoxide dismutase [Mn] |
| c86851.c0 | −4.93 | Catalase isozyme 1 |
| c85554.c0 | −6.15 | Catalase isozyme 2 |
| c88142.c0 | −4.63 | Catalase isozyme 2 |
| c86608.c0 | −4.38 | Catalase-2 |
| c31799_c0 | 5.07 | Phospholipid hydroperoxide glutathione peroxidase 1 |
| c33214_c0 | 4.83 | Probable phospholipid hydroperoxide glutathione peroxidase 6 |
| c56904_c0 | 4.02 | Monodehydroascorbate reductase |
| c57929_c0 | −6.28 | Monodehydroascorbate reductase |
| c66368_c0 | −6.09 | L-ascorbate peroxidase 1 |
| c77199_c0 | −5.06 | L-ascorbate peroxidase 2 |
| c88608_c0 | 4.79 | L-ascorbate peroxidase 7 |
RT-qPCR validation
To confirm the differential expression profiles of DEGs identified from RNA-Seq analysis, a total of 14 candidate DEGs were randomly selected from RNA-Seq and their expression levels in CK and Cd were examined by quantitative RT-PCR. As expected, the expression pattern of those unigenes obtained from qRT-PCR was similar with the differential expressions from RNA-Seq (Fig. 7).
Figure 7.
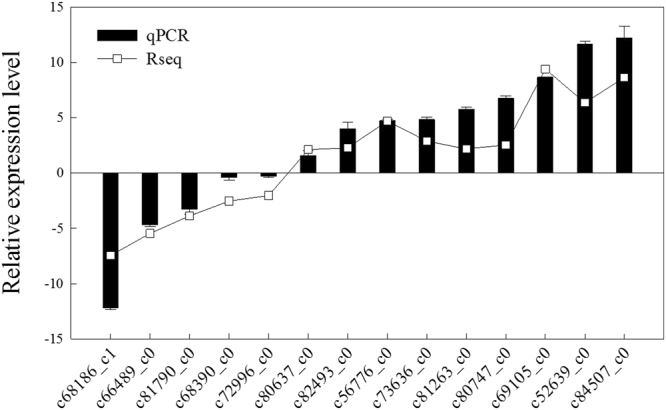
Quantitative RT-PCR of selected DEGs under control and Cd treatment in C. bipinnatus. The black bar represents the result of Rseq calculated by FPKM. The black bar with standard deviation represents the relative expression level determined by qPCR analysis.
Discussion
Under Cd stress, if the plants accumulate more than 100 μg/g Cd in dry aerial tissue, the value of TF is more than 1, and with normal growth, these plants are recommended as Cd hyper-accumulators37–39. However, most plants exhibit Cd toxicity when the leaves accumulate more than 5–10 μg/g Cd40. In the present study, C. bipinnatus accumulated 60.36 ± 2.17 μg/g Cd in the leaves, 321.15 ± 16.04 μg/g Cd in the stems, and 576.65 ± 41.48 μg/g Cd in the roots under 40 μmol/L Cd treatment without showing obvious toxic symptoms (Table 1 and Fig. 1), indicated that C. bipinnatus has strong tolerance to Cd. Although the TF values of C. bipinnatus were less than 1 (ranged from 0.66–0.79), the Cd concentrations of leaves and stems individually reached to 60.36 and 321.15 μg/g. Thus, C. bipinnatus should be a Cd accumulator, which would be potentially used for phytoremediation under mild Cd stress. However, the mechanism of strong tolerance and high Cd accumulation of C. bipinnatus was unknown. The physiological parameters and transcriptome analysis would help us revealing the mechanism.
Normally, Cd was intake, translocated and accumulated using other metal transporters, such as Zn, Fe and Cu41. Although ZIP genes mainly transport Zn, some ZIPs participate in Cd transport in Arabidopsis and Thlaspi caerulescens 42. We found that Cd up-regulated ZIP1, ZIP3, ZIP5, ZIP7, and ZTP29 (Table 4), suggesting that these genes were involved in Cd and Zn transport, thus the Zn concentrations in roots were reduced (Fig. 4A). HMA2, a Cd/Zn transporter, loads Cd/Zn into xylem43,44. The increased expression level of HMA2 in Arabidopsis 45, rice44, and barley43 induced Cd or Zn xylem uploading for translocation to the shoot. HMA3, another P-type ATPase gene, segregates Cd or Zn into the root vacuolar to limit the Cd xylem loading46,47. Down-regulation of HMA3 resulted in a decreased concentration of Cd in the root48. Certainly, when HMA3 is localized at the leaf vacuolar, it also transports Cd into the leaf vacuolar, finally producing Cd hyper-accumulator of Thlaspi caerulescens 16. Therefore, in the present study, up-regulation of HMA2 and down-regulation of HMA3 not only implied most of Cd was uploaded to aerial tissues, which resulted in high root-shoot translocation (Table 1), but also suggested a certain amount of Zn was uploaded to shoots, so that higher Zn concentrations accumulated in aerial tissues under Cd treatment (Fig. 4B and C). NRAMP (nature resistance associated with microphage) family members display poor selectivity towards divalent mental cations, which are responsible for heavy metal ions uptake and transport49. Previous studies have found that NRAMP1, NRAMP3, NRAMP4 and NRAMP6 transport Fe and Cd49–52. NRAMP2 and NRAMP3 involved in metal efflux from the vacuole49,53,54. In Arabidopsis, increased expression levels of NRAMP3 result in an increased metal output from the vacuole55. Two NRAMP family genes (NRAMP2 and NRAMP3) were both up-regulated by Cd in our research, suggesting that they involved in Fe or Cd efflux from vacuole, thereby leading to high accumulation of Fe in aerial tissues (Fig. 4H and I). Higher Fe concentration in shoots alleviated the Cd toxicity in Arabidopsis 56. Therefore, the increased concentration of Fe in leaves of C. bipinnatus may also be associated with the detoxification of the plant. Additionally, NRAMP2 and NRAMP3 may also have a role in Cd transport processes. YSL genes participate in Fe-nicotinamide (Fe-NA) complex root-to-shoot transport57. Cd mediated the expression of YSL3 and YSL7, suggesting Cd affected Fe transport in the plant. Previous study also found that YSL3 in Solanum nigrum and YSL7 in Brassica juncea were induced by Cd as well58,59. However, it remains unknown whether YSL family genes are involved in transport of Cd-NA complexes transport, and further study must be conducted to analyze the function of YSL genes under Cd stress.
The subcellular distribution of Cd in the root is associated with the accumulation, translocation, and detoxification of Cd60. Cd bound in the cell wall fraction is an important mechanism for Cd tolerance61–63. A large part of Cd was bunded in root cell wall of C. bipinnatus, especially at high Cd concentration treatment (Fig. 3). Cell wall is comprised of polysaccharide (including cellulose, semi-cellulose and pectate) and protein64,65. Containing abundant hydroxyl (OH-) for metal binding66, cellulose and semi-cellulose are both essential components of primary and secondary cell walls of higher plants31,67. In addition, cellulose synthase plays important role in cellulose formation, while xyloglucan endotransglucosylase (XTH) are involved in cell wall extension by cutting loosened xyloglucan strands and integrating new xyloglucans into the cell walls68. Present study found that the genes encoding cellulose synthase and XTH are up-regulated by Cd, suggesting the synthase and remobilization of cellulose and semi-cellulose play critical role in Cd binding. Moreover, the existence of pectin enhances the binding capacity of cell wall69,70. The synthase of pectin is associated with glucose metabolism. Several DEGs involved in UGP2, GPI, beta-fructofuranosidase and pectinesterase were up-regulated by Cd treatment (Table 4, SFig. 7), suggesting that Cd induce formation of pectin, thereby enhancing the capacity of Cd accumulation in cell walls, resulting high Cd tolerance of C. bipinnatus. Moreover, callose functions as a mechanical barrier to prevent ions penetration52,71–73, and the multi-copper-containing glycoprotein laccases involved in cell wall lignification74. The unigenes encoding callose synthase and laccase were up-regulated by Cd treatments in our study, implied that C. bipinnatus accumulated callose deposition and enhance cell wall lignification in root to prevent Cd entering the protoplasts under Cd stress. These results indicated that cell wall obstruction is one of important detoxification mechanism in C. bipinnatus.
After Cd entered into cytoplasm, it would be bound with metal chelates and then sequestrated into vacuoles to reduce their toxicity. The result of Cd subcellular distribution demonstrated that a large proportion of Cd was found in soluble fractions, suggesting the vacuole was the predominant detoxification sink for Cd in C. bipinnatus root. Vacuole possesses abundant sulphur-rich peptides such as GSH and PCs75–77, and organic acids78, which binds heavy metals and decreases their migration to reduce toxicity. Interestingly, unigenes involved in glutathione (GSH) metabolism, including serine acetyltransferase, cysteine synthase, ornithine decarboxylase, spermidine synthase, glutathione S transferase, γ-glutamyltranspeptidase, and unigenes from ABCC family were up-regulated under Cd treatment (Table 4). Meanwhile, ABCC1, ABCC2 and ABCC3 are major vacuolar PC-Cd transporters in other plants75,79, which were also up-regulated. Thus, Cd in cytoplasm turned into GSH (PC)-toxic compounds, finally transported by ABCC transporters without displaying cytotoxic to plant cell80–82. Vacuole sequestration of Cd also plays main role in Phytolacca Americana 83 and Arachis hypogaea 84. Moreover, GSH is also one of important antioxidants in plants. AsA -GSH cycle system can be able to eliminate ROS in many plants. Genes encoding enzymes involved in AsA-GSH cycle like glutathione peroxidase, monodehydroascorbate reductase and L-ascorbate peroxidase were up-regulated by Cd. Similarly, the activity of GR under Cd stress significantly increased compared with CK. These results indicated that enhancement of AsA-GSH cycle improved Cd tolerance of C. bipinnatus.
MDA is the product of lipid peroxidation, and its concentration reflects the degree of oxidative damage. In our study, 40 μmol/L Cd did not increase the MDA concentrations in leaves, while MDA concentration increased in roots of C. bipinnatus with higher accumulation of Cd (Fig. 5A and B), indicating that C. bipinnatus had strong tolerance under lower Cd but suffered cellular oxidative stress at higher Cd. Activities of antioxidant enzymes are induced by oxidative stress, and increased antioxidant level prevent oxidative damages85. Previous studies illustrated their functions in scavenging ROS in plants86,87. The enzyme SOD alters O2 − to H2O2 and oxygen88. CATs convert H2O2 to water and molecular oxygen, while PODs have a more elevated affinity to H2O2 than CATs89. The coordination between different enzymes can alleviate oxidative stress in the plant. In our study, compared with CK, the activity of POD and SOD in leaves were significantly increased under 40 μmol/L Cd treatment (Fig. 5C and G), while other two enzymes did not show significant changes (Fig. 5E and I), suggested that the activities of SOD and POD possessed sufficient capacity to scavenging ROS under lower Cd treatment. Therefore, the MDA concentration did not increase under 40 μmol/L Cd. However, generation of ROS were increased with the increased of Cd concentrations, exceeding the limits of POD and SOD scavenging ability (Fig. 5I). Meanwhile, the GR activity increased, complementing the ability of ROS scavenging. In addition, two unignenes encoding peroxidase were up-regulated under Cd treatment. These results demonstrated that antioxidative enzymes indeed play an important role in Cd detoxification and enhance tolerance of C. bipinnatus under adverse environment.
Summary, C. bipinnatus was recommended as a “Cd-accumulator” that would be potentially used for phytoremediation under mild Cd stress. Subcellular distribution of Cd displayed different detoxification mechanisms under different levels of Cd stress. C. bipinnatus initiated diverse defense and detoxify response to keep strong tolerance when treated with Cd stress. RNA-Seq analysis revealed that ZIPs, NRAMPs, HMAs, and ABC transporters were involved in Cd uptake, translocation and accumulation. Meanwhile, several processes such as cell wall biosynthesis, glutathione (GSH) metabolism, TCA cycle and the antioxidant system probably played critical roles in cell wall binding, vacuole sequestration and detoxification.
Materials and Methods
Plant culture and Cd treatment
Cosmos seeds (Cosmos bipinnatus Cav.) were sterilized with 2% NaClO for 20 min then rinsed with deionized water. The sterilized seeds were germinated on clean sand at 25 °C. After 2 weeks, the uniform seedlings were transplanted into plastic pots with 2.5 L half-strength Hoagland nutrient (60 plants per pot, pH 6.5) for 7 days and this was then replaced with full Hoagland nutrient solution. The plastic pots were randomly divided into 4 groups, each in triplicate. The four groups were treated with control, 40, 80, and 120 μmol/L CdCl2, respectively. All plants grew in a growth chamber with a daily temperature of 25 °C, a relative humidity of 70% and a photon flux density of 500 μmol/m2·S. Leaf, stem and root samples of all the treatments were collected on the 9th day. The root samples from 10 plants (10 plants per replicate, three biologic repeats) were collected and rapidly frozen in liquid nitrogen and then stored at −80 °C for RNA extraction.
Phenotype characterization
On the 9th day after treatment, the plant height and root length were measured (12 plants per biological replicate, three biologic repeats). The fresh weight of root, stem and leaf were also weighted. After weighing, all tissues were then dried at 80 °C for two days for dry weight calculation and metal concentration measurement.
Measurement of metal concentrations and calculation of translocation factor (TF)
The concentration of Cd, Zn, Fe, and Ca was measured as described by Wang et al.90 with some modifications90. Briefly, approximately 0.2 g dried plant samples were ground into powder which was digested at 320 °C with mixed acid [HNO3 + HClO4 (4:1, v/v)]. The concentration of Cd, Zn, Fe, and Ca in the digestions was detected by FAAS (flame atomic absorbance spectrometry, Shimadzu AA-6300, Kyoto, Japan). The limit for Cd, Zn, Fe, and Ca detection was 0.02 mg/L and a reference standard solution was purchased from Fisher Scientific Ltd. (China). The translocation factor was calculated as described by Li et al.91.
Malondialdehyde (MDA) determination
MDA in the roots and leaves was determined according to the method of Wang and Jin (2005) with some modifications92. Briefly, 0.2 g of fresh sample was homogenized in 6 mL 20% trichloroacetic acid (TCA) and centrifuged at 4000 r/min for 10 min at 4 °C. The mixture containing 2 mL of the supernatant and 2 mL of 0.6% thiobarbituric acid (TBA) in 10% TCA was incubated at 95 °C for 30 min and cooled immediately, then centrifuged at 4000 r/min for 5 min. The absorbance of the supernatant was recorded at 450, 532, and 600 nm. The concentration of MDA was calculated according to the following equation:
Determination of four enzymatic activities
Approximately 0.5 g of fresh leaf or root sample was homogenized in 5 mL of pre-cooled 50 mmol/L Tris-HCl buffer (pH 7.0) containing 1 mmol/L EDTA, 1 mmol/L DTT, 5 mmol/L MgCl2, 1 mmol/L AsA, and 1 mmol/L GSH93. Then the homogenate was centrifuged at 12000 r/min for 20 min at 4 °C and the extract was used for the enzyme assay.
The superoxide dismutase (SOD) activity was determined according the method described earlier94. The reaction mixture consisted of 50 mmol/L Tris-HCl buffer (pH 7.8), 0.1 mmol/L EDTA, 0.1 mmol/L nitroblue tetrazolium (NBT), 13.37 mmol/L methionine, and 0.1 mmol/L riboflavin and enzyme extract. The reaction was initiated by adding the riboflavin. The mixture was first placed under light then transferred into darkness immediately and the absorbance recorded at 560 nm. One unit of SOD activity was defined as the amount of enzyme that inhibited 50% of NBT photoreduction.
The catalase (CAT) activity was assayed in a reaction mixture containing 2.9 mL 50 mmol/L Tris-HCl buffer (pH 7.0), 50 μL 750 mmol/L H2O2, and 50 μL enzyme extract as per the method of Aebi (1984)95. Activity was measured by following the decomposition of H2O2 at 240 nm.
The peroxidase (POD) activity was determined according to the guaiacol method96 with some modifications. The reaction mixture was 50 mmol/L Tris-HCl buffer (pH 7.0) containing 0.1 mmol/L EDTA, 10 mmol/L guaiacol, 5 mmol/L H2O2 and 100 μL enzyme extract. The reaction was initiated by adding the extract. Guaiacol oxidation was determined based on an increase in the absorbance at 470 nm. One unit of POD activity was expressed as units (μmol guaiacol decomposed per minute) per mg of fresh weight (FW).
The glutathione reductase (GR) activity was assayed as described by Foyer and Halliwell (1976) with some modifications97. The reaction mixture consisted of 450 μL of the enzyme extract, 2.34 mL 50 mmol/L Tris-HCl buffer (containing 0.1 mmol/L EDTA, 5 mmol/L MgCl2 pH 7.5), 60 μL 10 mmol/L NADPH and 150 μL 10 mmol/L oxidized glutathione (GSSG). The reaction was initiated by adding the extract, NADPH, and GSSG. The NADPH oxidation rate was determined by recording the decrease in absorbance at 340 nm. The GR activity was expressed as the amount of enzyme needed to oxidize 1 μmol of NADPH /min· mg FW.
Subcellular distribution of Cd in the C. bipinnatus root
Cd subcellular distribution was determined according to Su et al.84 with some modifications84. The frozen root samples (1 g) were ground into powder with a pre-cold extraction buffer [50 mmol/L Tris-HCl buffer solution (pH 7.5), 250 mmol/L sucrose, 1.0 mmol/L DTE (C4H10O2S2) and 5.0 mmol/L ascorbic acid]. The homogenate was centrifuged at 4000 r/min for 15 min and the precipitate was designated as a cell wall fraction consisting mainly of cell walls and cell wall debris. The supernatant solution was further centrifuged at 16000 r/min for 45 min. The resultant deposit and supernatant solution were designated as the organelle-containing fraction and the soluble fraction, respectively. All fractions were dried and then digested in 5 mL HNO3. The Cd concentrations in the different fractions were analyzed by FAAS.
RNA extraction
The total RNA of each root sample (CK, 40 μmol/L Cd treatment) was extracted by using the Quick RNA isolation Kit (Huayueyang Biotech Co., Ltd., Bejing, China) according to the instruction manual. RNase-free DNasel (TaKaRa Biotech Co., Ltd., Dalian, China) was used for removing residual DNA in the extracted RNA. The quality of the total RNA sample was measured by 1% agarose gels, and the concentrations of the total RNA samples were assayed with an Agilent 2011 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA).
Library construction and illumina sequencing
High-quality RNA samples from C. bipinnatus root were prepared for cDNA library construction and sequencing. The mRNA was purified from total RNA using oligo (dT) magnetic beads and poly (A) tails. The RNA sequencing libraries were generated using the TruSeq RNA sample Prep Kit (Illumina, San Diego, CA) with multiplexing primers, according to the manual. The cDNA library was constructed with average inserts of 250 bp, with non-stranded library preparation. The QIAquick PCR extraction kit (Qiagen, Inc., Hilden, Germany) was used for cDNA purifying. The short cDNA fragments were subjected to end repair, adapter ligation, and agarose gel electrophoresis filtration. Subsequently, the appropriate fragments were selected as templates for PCR amplification. Sequencing was performed via a paired-end 125 cycle rapid run on 2 lanes of the Illumina HiSeq. 2500 system, generating pairs of reads of great quality as intended.
Transcriptome assembly
Adapter-related and low-quality reads including ambiguous reads (‘N’), duplicated sequences were removed from the raw reads to obtain the clean reads. Trinity software (http://trinityrnaseq.sourceforge.net/) was used for the de novo assembled transcriptomes. In brief, the contigs were formed by combining the certain overlap length into long fragments without N (contigs) and then they were clustered using the TGICL software to produce unigines (without N) and finally the redundancies were removed to obtain non-redundant unigenes98.
Unigene functional annotation
A series of databases and software were used for putative unigenes annotations. BLAST software99 was used to align the unigene with the NR100, Swiss-Prot101, GO102, COG103, and KEGG databases104 (E-value ≤ 1E−5) to retrieve protein functional annotations based on sequence similarity. The ESTScan software was used to decide the sequence direction of the unigenes that could not be aligned to any of the above databases105. Functional categories of putative unigenes were grouped using the GO and KEGG databases.
Differential expression analysis
FPKM values were used to compare gene expression differences between the two samples. The DESeq package was used to obtain the base mean value for identifying DEGs. FDR ≤0.01 and the absolute values of log2 ratio ≥1 were set as the thresholds for the significance of the gene expression difference between the two samples.
Real-time quantitative (qRT-PCR) validation of partial DEGs
qRT-PCR was performed in a 96-well plate with the CFX-96 real-time system (Bio-Rad, CA, USA). Each reaction of 15 μL contained 6.3 μL (30 ng/μL) cDNA, 0.6 μL (4 pmol/μL) for each forward and reverse primer, and 7.5 μL iQ SYBR Green Supermix (Bio-Rad, CA, USA). Each cDNA sample was amplified in triplicates. The PCR reaction conditions were 95 °C for 5 min, 39 cycles of 95 °C for 15 s, 56 °C for 30 s, and 72 °C for 10 s, followed by the generation of a dissociation curve by increasing the temperature starting from 65 °C to 95 °C to check for the specificity of amplification. Actin was used to standardize the transcript levels in each sample. The relative expression level was calculated with the 2−△△CT formula106. The primers that were designed and used in the RT-qPCR analyses are shown in STable 1.
Data analysis
The SPSS version 22.0 software was used for statistical analyses. The mean and standard deviation (SD) of three replicates were calculated. Duncan’s test was used to determine the significant differences between means (p < 0.05)107. Besides, the figures were drawn with Sigmaplot 12.5.
Electronic supplementary material
Acknowledgements
The authors would like to thank the National Natural Science Foundation of China (No. 31470305 and 31570700).
Author Contributions
Yujing Liu, Yi Wang and Xiaofang Yu conceived and designed the research, and wrote the article. Yujing Liu, Yimei Feng, Chao Wang, Xiaolu Wang, Yulin Jiang, Mali Tong, Shuxiang Zhang, Chenghuan Cai and Yuxuan Mo conducted experiments. Yi Wang, Chao Zhang, Jian Zeng, Zhuo Huang, Houyang Kang, Xing Fan, Lina Sha, Haiqin Zhang, Yonghong Zhou, Suping Gao, and Qibing Chen had provided valuable suggestions and comments to the manuscript. Yujing Liu and Xiaofang Yu analyzed data. All authors read and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Yujing Liu and Xiaofang Yu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14407-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wan L, Zhang H. Cadmium toxicity: Effects on cytoskeleton, vesicular trafficking and cell wall construction. Plant Signal Behav. 2012;7:345–348. doi: 10.4161/psb.18992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuypers A, et al. Cadmium stress: An oxidative challenge. Biometals. 2010;23:927–940. doi: 10.1007/s10534-010-9329-x. [DOI] [PubMed] [Google Scholar]
- 3.Ogawa I, Nakanishi H, Mori S, Nishizawa NK. Time course analysis of gene regulation under cadmium stress in rice. Plant Soil. 2009;325:97–108. doi: 10.1007/s11104-009-0116-9. [DOI] [Google Scholar]
- 4.Ranieri A, et al. Oxidative stress and phytochelatin characterization in bread wheat exposed to Cadmium excess. Plant Physiol. Biochem. 2005;43:45–54. doi: 10.1016/j.plaphy.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Castro AV, et al. Morphological, biochemical, molecular and ultrastructural changes induced by Cd toxicity in seedlings of Theobroma cacao L. Ecotoxicol. Environ. Saf. 2015;115:174–186. doi: 10.1016/j.ecoenv.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Liu C, et al. Effects of cadmium and salicylic acid on growth, spectral reflectance and photosynthesis of castor bean seedlings. Plant Soil. 2011;344:131–141. doi: 10.1007/s11104-011-0733-y. [DOI] [Google Scholar]
- 7.Tian S, et al. Cellular sequestration of cadmium in the hyperaccumulator plant species sedum alfredii. Plant Physiol. 2011;157:1914–1925. doi: 10.1104/pp.111.183947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rizwan M, et al. Cadmium minimization in wheat: A critical review. Ecotoxicol. Environ. Saf. 2016;130:43–53. doi: 10.1016/j.ecoenv.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Anjum SA, et al. Cadmium toxicity in maize (Zea mays L.): Consequences on antioxidative systems, reactive oxygen species and cadmium accumulation. Environ. Sci. Pollut. Res. 2015;22:17022–17030. doi: 10.1007/s11356-015-4882-z. [DOI] [PubMed] [Google Scholar]
- 10.Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 11.Li XM, Ma LJ, Bu N, Li YY, Zhang LH. Effects of salicylic acid pre-treatment on cadmium and/or UV-B stress in soybean seedlings. Biol. Plant. 2014;58:195–199. doi: 10.1007/s10535-013-0375-4. [DOI] [Google Scholar]
- 12.Brown SL, Angle JS, Chaney RL, Baker AJM. Zinc and cadmium uptake by hyperaccumulator Thlaspi caerulescens grown in nutrient solution. Soil Sci. Soc. Am. J. 1995;59:125–133. doi: 10.2136/sssaj1995.03615995005900010020x. [DOI] [PubMed] [Google Scholar]
- 13.Yang XE, Long X, Ni W, Fu C. Sedum alfredii H: A new Zn hyperaccumulating plant first found in China. Chinese Sci. Bull. 2002;47:1634–1637. doi: 10.1007/BF03184113. [DOI] [Google Scholar]
- 14.Liu W, Shu W, Lan C. viola baoshansis, a plant that hyperaccumulates cadmium. Chinese Sci. bull. 2004;1:29–32. doi: 10.1007/BF02901739. [DOI] [Google Scholar]
- 15.Sun Y, Zhou Q, Diao C. Effects of cadmium and arsenic on growth and metal accumulation of Cd-hyperaccumulator Solanum nigrum L. Bioresour. Technol. 2008;99:1103–1110. doi: 10.1016/j.biortech.2007.02.035. [DOI] [PubMed] [Google Scholar]
- 16.Ueno D, et al. Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Plant J. 2011;66:852–862. doi: 10.1111/j.1365-313X.2011.04548.x. [DOI] [PubMed] [Google Scholar]
- 17.Pence NS, et al. The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc. Natl. Acad. Sci. USA. 2000;97:4956–4960. doi: 10.1073/pnas.97.9.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun R, Zhou Q, Sun F, Jin C. Antioxidative defense and proline/phytochelatin accumulation in a newly discovered Cd-hyperaccumulator, Solanum nigrum L. Environ. Exp. Bot. 2007;60:468–476. doi: 10.1016/j.envexpbot.2007.01.004. [DOI] [Google Scholar]
- 19.Zhou W, Qiu B. Effects of cadmium hyperaccumulation on physiological characteristics of Sedum alfredii Hance (Crassulaceae) Plant Science. 2005;169:737–745. doi: 10.1016/j.plantsci.2005.05.030. [DOI] [Google Scholar]
- 20.Yamato M, Yoshida S, Iwase K. Cadmium accumulation in Crassocephalum crepidioides (Benth.) S. Moore (Compositae) in heavy-metal polluted soils and Cd-added conditions in hydroponic and pot cultures. Soil Sci. Plant Nutr. 2008;54:738–743. doi: 10.1111/j.1747-0765.2008.00295.x. [DOI] [Google Scholar]
- 21.Wei S, Zhou Q. Screen of Chinese weed species for cadmium tolerance and accumulation characteristics. Int. J. Phytorem. 2008;10:584–597. doi: 10.1080/15226510802115174. [DOI] [PubMed] [Google Scholar]
- 22.Tanhan P, Kruatrachue M, Pokethitiyook P, Chaiyarat R. Uptake and accumulation of cadmium, lead and zinc by Siam weed [Chromolaena odorata (L.) King & Robinson] Chemosphere. 2007;68:323–329. doi: 10.1016/j.chemosphere.2006.12.064. [DOI] [PubMed] [Google Scholar]
- 23.Silveira FS, Azzolini M, Divan AM. Scanning cadmium photosynthetic responses of Elephantopus mollis for potential phytoremediation practices. Water, Air, & Soil Pollut. 2015;226:359. doi: 10.1007/s11270-015-2625-x. [DOI] [Google Scholar]
- 24.Broadhurst CL, et al. Accumulation of zinc and cadmium and localization of zinc in Picris divaricata Vant. Environ. Exp. Bot. 2013;87:1–9. doi: 10.1016/j.envexpbot.2012.08.010. [DOI] [Google Scholar]
- 25.Santiago-Cruz MA, et al. Exploring the Cr(VI) phytoremediation potential of Cosmos bipinnatus. Water, Air, & Soil Pollut. 2014;225:1–8. doi: 10.1007/s11270-014-2166-8. [DOI] [Google Scholar]
- 26.Peng H, et al. Transcriptomic changes during maize roots development responsive to cadmium (Cd) pollution using comparative RNAseq-based approach. Biochem. Biophys. Res. Commun. 2015;464:1040–1047. doi: 10.1016/j.bbrc.2015.07.064. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, et al. Transcriptome sequencing and differential gene expression analysis in Viola yedoensis Makino (Fam. Violaceae) responsive to cadmium (Cd) pollution. Biochem. Biophys. Res. Commun. 2015;459:60–65. doi: 10.1016/j.bbrc.2015.02.066. [DOI] [PubMed] [Google Scholar]
- 29.He J, et al. A transcriptomic network underlies microstructural and physiological responses to Cadmium in Populus x canescens. Plant Physiol. 2013;162:424–439. doi: 10.1104/pp.113.215681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu L, et al. De novo sequencing of root transcriptome reveals complex cadmium-responsive regulatory networks in radish (Raphanus sativus L.) Plant Sci. 2015;236:313–323. doi: 10.1016/j.plantsci.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Yang J, et al. Characterization of early transcriptional responses to cadmium in the root and leaf of Cd-resistant Salix matsudana Koidz. BMC Genomics. 2015;16:705. doi: 10.1186/s12864-015-1923-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H, Hong C, Xiao M, Chen X, Chen H. Real-time kinetics of cadmium transport and transcriptomic analysis in low cadmium accumulator Miscanthus sacchariflorus. Planta. 2016;244:1289–1302. doi: 10.1007/s00425-016-2578-3. [DOI] [PubMed] [Google Scholar]
- 33.Huang Y, et al. Comparative transcriptome analysis of two Ipomoea aquatica Forsk. cultivars targeted to explore possible mechanism of genotype-dependent accumulation of cadmium. J. Agric. Food Chem. 2016;64:5241–5250. doi: 10.1021/acs.jafc.6b01267. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Q, et al. Comparative transcriptome analysis between low- and high-cadmium-accumulating genotypes of pakchoi (Brassica chinensis L.) in response to cadmium stress. Environ. Sci. Technol. 2016;50:6485–6494. doi: 10.1021/acs.est.5b06326. [DOI] [PubMed] [Google Scholar]
- 35.Halimaa P, Lin YF, Ahonen VH, Blande D, Clemens S. Gene expression differences between Noccaea caerulescens ecotypes help to identify candidate genes for metal phytoremediation. Environ. Sci. Technol. 2014;48:3344–3353. doi: 10.1021/es4042995. [DOI] [PubMed] [Google Scholar]
- 36.Xu J, Sun J, Du L, Liu X. Comparative transcriptome analysis of cadmium responses in Solanum nigrum and Solanum torvum. New Phytol. 2012;196:110–124. doi: 10.1111/j.1469-8137.2012.04235.x. [DOI] [PubMed] [Google Scholar]
- 37.Chaney RL, et al. Phytoremediation of soil metals. Curr. Opin. Chem. Biol. 1997;8:279–284. doi: 10.1016/s0958-1669(97)80004-3. [DOI] [PubMed] [Google Scholar]
- 38.Pollard AJ, Reeves RD, Baker AJM. Facultative hyperaccumulation of heavy metals and metalloids. Plant Sci. 2014;1:8–17. doi: 10.1016/j.plantsci.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 39.van der Ent A, Baker AJM, Reeves RD, Pollard AJ, Schat H. Hyperaccumulators of metal and metalloid trace elements: Facts and fiction. Plant soil. 2013;362:319–334. doi: 10.1007/s11104-012-1287-3. [DOI] [Google Scholar]
- 40.White PJ, Brown PH. Plant nutrition for sustainable development and global health. Ann. Bot. 2010;105:1073–1080. doi: 10.1093/aob/mcq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clemens S. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie. 2006;88:1707–1719. doi: 10.1016/j.biochi.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Grotz N, Fox T, Connolly E, Park W, Guerinot ML. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc. Natl. Acad. Sci. USA. 1998;98:7220–7224. doi: 10.1073/pnas.95.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mills RF, Peaston KA, Runions J, Williams LE. HvHMA2, a P(1B)-ATPase from barley, is highly conserved among cereals and functions in Zn and Cd transport. PLoS One. 2012;7:e42640. doi: 10.1371/journal.pone.0042640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satoh-Nagasawa N, et al. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant and Cell Physiol. 2012;53:213–224. doi: 10.1093/pcp/pcr166. [DOI] [PubMed] [Google Scholar]
- 45.Chong KEW, Jarvis RS, Sherson SM, Cobbett CS. Functional analysis of the heavy metal binding domains of the Zn/Cd-transporting ATPase, HMA2, in Arabidopsis thaliana. New Phytol. 2009;181:79–88. doi: 10.1111/j.1469-8137.2008.02637.x. [DOI] [PubMed] [Google Scholar]
- 46.Morel M, et al. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2008;149:894–904. doi: 10.1104/pp.108.130294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Yu K, Poysa V, Shi C, Zhou Y. A single point mutation in GmHMA3 affects cadimum (Cd) translocation and accumulation in soybean seeds. Mol. Plant. 2012;5:1154–1156. doi: 10.1093/mp/sss069. [DOI] [PubMed] [Google Scholar]
- 48.Miyadate H, et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011;189:190–199. doi: 10.1111/j.1469-8137.2010.03459.x. [DOI] [PubMed] [Google Scholar]
- 49.Pottier M, et al. Identification of mutations allowing natural resistance associated macrophage proteins (Nramp) to discriminate against cadmium. Plant J. 2015;83:625–637. doi: 10.1111/tpj.12914. [DOI] [PubMed] [Google Scholar]
- 50.Cailliatte R, Lapeyre B, Briat JF, Mari S, Curie C. The Nramp6 metal transporter contributes to cadmium toxicity. Biochem. J. 2009;422:217–228. doi: 10.1042/BJ20090655. [DOI] [PubMed] [Google Scholar]
- 51.Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Natl. Acad. Sci. USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei W, Chai TY, Zhang YX, Han L, Xu J. The Thlaspi caerulescens Nramp homologue TcNramp3 is capable of divalent cation transport. Mol. Biotechnol. 2009;41:15–21. doi: 10.1007/s12033-008-9088-x. [DOI] [PubMed] [Google Scholar]
- 53.Nathalie V, Christian H, Henk S. Molecular mechanisms of metal hyperaccumulation in plants. New Phytol. 2008;180:759–776. doi: 10.1111/j.1469-8137.2008.02748.x. [DOI] [PubMed] [Google Scholar]
- 54.Oomen, R. et al. Functional characterization of Nramp3 and Nramp4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytol. 637–650 (2009). [DOI] [PubMed]
- 55.Lanquar V, Bolte SSCECH, Alcon C, Neumann D. Mobilization of vacuolar iron by AtNramp3 and AtNramp4 is essential for seed germination on low iron. Embo. J. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu H, et al. Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 2012;158:790–800. doi: 10.1104/pp.111.190983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishimaru Y, et al. Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 2010;62:379–390. doi: 10.1111/j.1365-313X.2010.04158.x. [DOI] [PubMed] [Google Scholar]
- 58.Feng S, et al. Isolation and characterization of a novel cadmium-regulated yellow stripe-like transporter (SnYSL3) in Solanum nigrum. Plant Cell Rep. 2017;36:281–296. doi: 10.1007/s00299-016-2079-7. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, Li Y, Zhang Y, Chai T. Molecular cloning and characterization of a Brassica juncea yellow stripe-like gene, BjYSL7, whose overexpression increases heavy metal tolerance of tobacco. Plant Cell Rep. 2013;32:651–662. doi: 10.1007/s00299-013-1398-1. [DOI] [PubMed] [Google Scholar]
- 60.Zhao Y, et al. Subcellular distribution and chemical forms of cadmium in the edible seaweed, Porphyra yezoensis. Food Chem. 2015;168:48–54. doi: 10.1016/j.foodchem.2014.07.054. [DOI] [PubMed] [Google Scholar]
- 61.Cobbett CS. Heavy metals and plants-model systems and hyperaccumulators. New Phytol. 2003;159:289–293. doi: 10.1046/j.1469-8137.2003.00832.x. [DOI] [PubMed] [Google Scholar]
- 62.Wang J, et al. Comparisons of cadmium subcellular distribution and chemical forms between low-Cd and high-Cd accumulation genotypes of watercress (Nasturtium officinale L. R. Br.) Plant Soil. 2015;396:325–337. doi: 10.1007/s11104-015-2580-8. [DOI] [Google Scholar]
- 63.Wu FB, Dong J, Qian QQ, Zhang GP. Subcellular distribution and chemical form of Cd and Cd-Zn interaction in different barley genotypes. Chemosphere. 2005;60:1437–1446. doi: 10.1016/j.chemosphere.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 64.Stolt JP, Sneller F, Bryngelsson T, Lundborg T, Schat H. Phytochelatin and cadmium accumulation in wheat. Envion. Exp. Bot. 2003;49:21–28. doi: 10.1016/S0098-8472(02)00045-X. [DOI] [Google Scholar]
- 65.Toppi L, Gabbrielli R. Response to cadmium in higher plants. Environ. Exp. Bot. 1999;41:105–130. doi: 10.1016/S0098-8472(98)00058-6. [DOI] [Google Scholar]
- 66.Li T, Tao Q, Shohag M, Yang X, Sparks DL. Root cell wall polysaccharides are involved in cadmium hyperaccumulation in Sedum alfredii. Plant Soil. 2015;389:387–399. doi: 10.1007/s11104-014-2367-3. [DOI] [Google Scholar]
- 67.Yang H, Yan R, Chen H, Lee DH, Zheng C. Characteristics of hemicellulose, cellulose and lignin pyrolysis. Fuel. 2007;86:1781–1788. doi: 10.1016/j.fuel.2006.12.013. [DOI] [Google Scholar]
- 68.Van Sandt VS, Suslov D, Verbelen JP, Vissenberg K. Xyloglucan endotransglucosylase activity loosens a plant cell wall. 2007;100:1467–1473. doi: 10.1093/aob/mcm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiong J, An LY, Lu H, Zhu C. Exogenous nitric oxide enhances cadmium tolerance of rice by increasing pectin and hemicellulose contents in root cell wall. Planta. 2009;230:755–765. doi: 10.1007/s00425-009-0984-5. [DOI] [PubMed] [Google Scholar]
- 70.Zhu XF, et al. Exogenous auxin alleviates cadmium toxicity in Arabidopsis thaliana by stimulating synthesis of hemicellulose 1 and increasing the cadmium fixation capacity of root cell walls. J. Hazard. Mater. 2013;263:398–403. doi: 10.1016/j.jhazmat.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 71.Cai X, Davis EJ, Ballif J, Liang M, Bushman E. Mutant identification and characterization of the laccase gene family in Arabidopsis. J. Exp. Bot. 2006;57:2563–2569. doi: 10.1093/jxb/erl022. [DOI] [PubMed] [Google Scholar]
- 72.Samardakiewicz S, Krzesłowska M, Bilski H, Bartosiewicz R, Woźny A. Is callose a barrier for lead ions entering Lemna minor L. root cells? Protoplasma. 2012;249:347–351. doi: 10.1007/s00709-011-0285-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zavaliev R, Ueki S, Epel BL, Citovsky V. Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma. 2011;248:117–130. doi: 10.1007/s00709-010-0247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaneda M, et al. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J. Exp. Bot. 2011;62:2063–2077. doi: 10.1093/jxb/erq416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brunetti P, et al. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 2015;66:3815–3829. doi: 10.1093/jxb/erv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gallego SM, et al. Unravelling cadmium toxicity and tolerance in plants: Insight into regulatory mechanisms. Environ. Exp. Bot. 2012;83:33–46. doi: 10.1016/j.envexpbot.2012.04.006. [DOI] [Google Scholar]
- 77.Gupta OP, Sharma P, Gupta RK, Sharma I. MicroRNA mediated regulation of metal toxicity in plants: Present status and future perspectives. Plant Mol. Biol. 2014;84:1–18. doi: 10.1007/s11103-013-0120-6. [DOI] [PubMed] [Google Scholar]
- 78.Bhatia NP. Detection and quantification of ligands involved in nickel detoxification in a herbaceous Ni hyperaccumulator Stackhousia tryonii Bailey. J. Exp. Bot. 2005;56:1343–1349. doi: 10.1093/jxb/eri135. [DOI] [PubMed] [Google Scholar]
- 79.Song WY, Park J, Mendozacózatl DG, Sutergrotemeyer M, Shim D. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. USA. 2010;107:21187–21192. doi: 10.1073/pnas.1013964107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin CY, Trinh NN, Fu SF, Hsiung YC, Chia LC. Comparison of early transcriptome responses to copper and cadmium in rice roots. Plant Mol. Biol. 2013;81:507–522. doi: 10.1007/s11103-013-0020-9. [DOI] [PubMed] [Google Scholar]
- 81.Moons A. Osgstu3 andosgtu4, encoding tau class glutathione S -transferases, are heavy metal- and hypoxic stress-induced and differentially salt stress-responsive in rice roots. FEBS Letters. 2003;553:427–432. doi: 10.1016/S0014-5793(03)01077-9. [DOI] [PubMed] [Google Scholar]
- 82.Smiri M, Chaoui A, Rouhier N, Gelhaye E, Jacquot JP. Cadmium affects the glutathione/glutaredoxin system in germinating pea seeds. Biol. Trace Elem. Res. 2011;142:93–105. doi: 10.1007/s12011-010-8749-3. [DOI] [PubMed] [Google Scholar]
- 83.Fu X, Dou C, Chen Y, Chen X, Shi J. Subcellular distribution and chemical forms of cadmium in Phytolacca americana L. J. Hazard. Mater. 2011;186:103–107. doi: 10.1016/j.jhazmat.2010.10.122. [DOI] [PubMed] [Google Scholar]
- 84.Su Y, et al. Effects of iron deficiency on subcellular distribution and chemical forms of cadmium in peanut roots in relation to its translocation. Environ. Exp. Bot. 2014;97:40–48. doi: 10.1016/j.envexpbot.2013.10.001. [DOI] [Google Scholar]
- 85.Yasemin E, Deniz T, Beycan A. Effects of cadmium on antioxidant enzyme and photosynthetic activities in leaves of two maize cultivars. J. Plant Physiol. 2008;165:600–611. doi: 10.1016/j.jplph.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 86.He J, et al. Net Cadmium flux and accumulation reveal tissue-specific oxidative stress and detoxification in Populus × canescens. Physiol. Plant. 2011;143:50–63. doi: 10.1111/j.1399-3054.2011.01487.x. [DOI] [PubMed] [Google Scholar]
- 87.Jiang Q, et al. Effect of Funneliformis mosseae on the growth, cadmium accumulation and antioxidant activities of Solanum nigrum. Appl. Soil Ecol. 2016;98:112–120. doi: 10.1016/j.apsoil.2015.10.003. [DOI] [Google Scholar]
- 88.Shah K, Kumar RG, Verma S, Dubey RS. Effect of cadmium on lipid peroxidation, superoxide anion generation and activities of antioxidant enzymes in growing rice seedlings. Plant Soil. 2001;161:1135–1144. [Google Scholar]
- 89.Noctor G, Foyer CH. Ascobate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 90.Wang Y, et al. Cadmium treatment alters the expression of five genes at the Cda1 locus in two soybean cultivars [Glycine Max (L.) Merr]. Sci. World J. 2014;2014:1–8. doi: 10.1155/2014/979750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li S, et al. Cadmium-induced oxidative stress, response of antioxidants and detection of intracellular cadmium in organs of moso bamboo (Phyllostachys pubescens) seedlings. Chemosphere. 2016;153:107–114. doi: 10.1016/j.chemosphere.2016.02.062. [DOI] [PubMed] [Google Scholar]
- 92.Wang H, Jin JY. Photosynthetic rate, chlorophyll fluorescence parameters, and lipid peroxidation of maize leaves as affected by Zinc deficiency. Photosynthetica. 2005;43:591–596. doi: 10.1007/s11099-005-0092-0. [DOI] [Google Scholar]
- 93.Knorzer OC, Durner J, Boger P. Alterations in the antioxidative system of suspension-cultured soybean cells (Glycine max) induced by oxidative stress. Physiol. Plant. 1996;97:388–396. doi: 10.1034/j.1399-3054.1996.970225.x. [DOI] [Google Scholar]
- 94.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 95.Aebi H. Catalase in Vitro. Methods in Enzymol. 1984;105:121–126. doi: 10.1016/S0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 96.Chance B, Maehly AC. Assay of catalase and peroxidases. Enzymol. 1955;2:764–775. doi: 10.1016/S0076-6879(55)02300-8. [DOI] [Google Scholar]
- 97.Halliwell B, Foyer CH. Ascorbic acid, metal ions and the superoxide radical. Biochem. J. 1976;133:697–700. doi: 10.1042/bj1550697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pertea G, et al. TIGR gene indices clustering tools (TGICL): A software system for fast clustering of large est datasets. βBioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 99.Altschul SF, et al. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deng Y, et al. Integrated nr database in protein annotation system and its localization. Computer Engineering. 2006;32:71–72. [Google Scholar]
- 101.Apweiler R, et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004;32:115–119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ashburner M, et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori AM. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Iseli C, Jongeneel CV, Bucher P. ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;99:138–148. [PubMed] [Google Scholar]
- 106.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 107.Duncan DB. Multiple range and multiple F-Test. Biometrics. 1955;11:1–42. doi: 10.2307/3001478. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.