Abstract
Dilated cardiomyopathy (DCM) is an important cause of heart failure. Single gene mutations in at least 50 genes have been proposed to account for 25–50% of DCM cases and up to 25% of inherited DCM has been attributed to truncating mutations in the sarcomeric structural protein titin (TTNtv). Whilst the primary molecular mechanism of some DCM-associated mutations in the contractile apparatus has been studied in vitro and in transgenic mice, the contractile defect in human heart muscle has not been studied. In this study we isolated cardiac myofibrils from 3 TTNtv mutants, and 3 with contractile protein mutations (TNNI3 K36Q, TNNC1 G159D and MYH7 E1426K) and measured their contractility and passive stiffness in comparison with donor heart muscle as a control. We found that the three contractile protein mutations but not the TTNtv mutations had faster relaxation kinetics. Passive stiffness was reduced about 38% in all the DCM mutant samples. However, there was no change in maximum force or the titin N2BA/N2B isoform ratio and there was no titin haploinsufficiency. The decrease in myofibril passive stiffness was a common feature in all hearts with DCM-associated mutations and may be causative of DCM.
Introduction
Historically, the prevalence of DCM was considered to be in the range of 1 in 2500–3000, however, newer modalities and heightened diagnostic awareness now estimate the prevalence of DCM of up to 1 in 5001. Dilated cardiomyopathy is inherited in about 25–50% of cases and more than 50 single genes are linked to inherited DCM, including genes encoding cytoskeletal, Z-disc, nucleo-cytoskeletal, mitochondrial, and calcium handling proteins making DCM a genetically heterogeneous disease. However “pure” inherited DCM, not associated with other symptoms, such as conduction disease, is most often caused by mutations in sarcomeric proteins. Recently titin truncating (TTNtv) mutations have been found in 25% of putative familial DCM (fDCM) cases2 and a disease causing role of these mutations in A band and Z-disk titin has been asserted3. Despite the emerging importance of TTNtv mutations as a cause of familial DCM (fDCM) very little is known about the functional effects of the mutations in human heart muscle.
In a previous study of human heart tissue samples we identified 26 potentially disease-causing mutations in 14 genes, 5 of which were in contractile proteins, 4 were in OBSCN and 6 were truncating mutations in the titin gene (TTNtv)4. The high frequency of potentially disease-causing mutations in end-stage DCM patients confirms the finding of Roberts et al., who studied a large population and demonstrated a >96% probability of pathogenicity in end-stage DCM patients with TTN truncation mutations3. In this study we have directly investigated TTNtv heterozygous mutations in human heart tissue samples in comparison with donor heart and DCM-causing mutations in other sarcomeric proteins. Several human samples were analysed on total titin expression level, N2BA/N2B isoform expression ratio, as well as titin phosphorylation level. To determine the mechanism of diastolic dysfunction, the maximum tension amplitude, length dependence of maximum force, and the rates of tension development and relaxation were measured in DCM samples carrying mutations in TTN (p. R23464T fs*41 and p. Y18923*), TNNI3 (K36Q), TNNC1 (G159D) and MYH7 (E1426K).
Results
Samples and sequences
A series of 30 explanted failing heart samples from selected patients with end-stage heart failure, most of which were diagnosed with familial DCM, was analysed by whole exome sequencing. We found six unique variants in the TTN gene predicted to cause chain termination that are potentially causative of DCM4. The presence of the variants was confirmed by direct sequencing (Fig. 1 and Supplementary Table S1).
Figure 1.
Identification of TTNtv mutations in human heart muscle. Clinical and genetic data on the heart muscle samples and location of mutations in the titin molecule, based on the model of Linke and Hamdani29. D6 and D7 have the same mutation but there is no evidence of their relationship except for common ethnicity. The calculated MW of TTNtv are 2611 kDa for D6/D7, 2102 kDa for D9, 125 kDa for D13, 2353 kDa for D23 and 2539 kDa for D28. Abbreviations: NA, not applicable; LVEF, left ventricular ejection fraction; NYHA, New York Heart Association; CO, cardiac output; CI, cardiac index; LVEDD, left ventricular end-diastolic diameter; LVESD, left ventricular end-systolic diameter; FS, fractional shortening; CD, cardiomyopathy; CAD, coronary artery disease; IHD, Ischaemic heart disease; LV, left ventricle; RV, right ventricle; IDCM, idiopathic cardiomyopathy.
TTNtv mutations and contractile function
To investigate whether there was any mechanical dysfunction caused by putative DCM mutations we isolated cardiac myofibrils and measured their passive stiffness and contractility using an apparatus for single myofibril force studies. The three left ventricular TTNtv samples (D6, D7 and D9) were studied in comparison with myofibrils from donor heart and left ventricular muscle samples with mutations in contractile proteins of the sarcomere: troponin C (TnC) G159D, troponin I (TnI) K36Q and myosin heavy chain E1426K (D1, D2 and D15 respectively). These are termed familial DCM (fDCM) since linkage has been proven with these mutations5–7. Results are summarised in Table 1.
Table 1.
Myofibril active force sarcomere length dependence, kinetics of force development and relaxation, and the parameters describing the passive mechanical myofibril properties. Data are shown as mean ± SEM.
| Sample ID | Fmax at the SL shown below | Time-course parameters of myofibril activation and relaxation at SL 2.0 µm | Mechanical Parameters | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.8 µm (kPa) | 2.0 µm (kPa) | 2.2 µm (kPa) | kACT (s−1) | kTR (s−1) | tLIN (ms) | kLIN (s−1) | kREL (s−1) | Slack SL (µm) | A | ||
| Donor | KN1 | 47 ± 4 (10) | 101 ± 7 (20) | 109 ± 8 (13) | 0.73 ± 0.06 (19) | 0.63 ± 0.03 (19) | 105 ± 7 (17) | 0.44 ± 0.16 (17) | 16.8 ± 1.7 (15) | 1.89 ± 0.04 (9) | 544 ± 66 (9) |
| KN2 | 55 ± 8 (6) | 102 ± 9 (7) | 120 ± 16 (5) | 0.91 ± 0.13 (7) | 0.69 ± 0.07 (6) | 86 ± 11 (6) | 0.63 ± 0.18 (6) | 18.08 ± 1.2 (6) | 1.93 ± 0.03 (13) | 529 ± 66 (13) | |
| NL | NA | NA | NA | NA | NA | NA | NA | NA | 1.88 ± 0.04 (5) | 561 ± 46 (5) | |
| NK | NA | NA | NA | NA | NA | NA | NA | NA | 1.90 ± 0.04 (5) | 663 ± 123 (5) | |
| Combined donors | 50 ± 4 (16) | 101 ± 5 (27) | 112 ± 7 (18) | 0.78 ± 0.06 (26) | 0.64 ± 0.03 (25) | 100 ± 6 (23) | 0.49 ± 0.13 (23) | 17.1 ± 1.2 (21) | 1.91 ± 0.02 (32) | 559 ± 38 (32) | |
| fDCM | D1 | 58 ± 5 (4) | 107 ± 10 (4) | 134 ± 8 (4) | 0.72 ± 0.06 (4) | 0.62 ± 0.08 (4) | 71 ± 6 (4)* | 0.17 ± 0.07 (4) | 20.0 ± 1.5 (3) | 1.90 ± 0.03 (14) | 329 ± 33 (14)*** |
| D2 | 40 ± 5 (9) | 104 ± 11 (13) | 123 ± 13 (11) | 0.91 ± 0.10 (13) | 0.67 ± 0.04 (13) | 70 ± 5 (12)** | 0.68 ± 0.28 (11) | 22.32 ± 1.6 (12)* | 1.94 ± 0.02 (21) | 324 ± 36 (21)*** | |
| D15 | 49 ± 6 (8) | 88 ± 10 (10) | 109 ± 7 (7) | 0.88 ± 0.06 (11) | 0.76 ± 0.06 (10)* | 79 ± 7 (9)* | 0.37 ± 0.20 (9) | 23.1 ± 1.7 (9)* | 1.90 ± 0.06 (6) | 368 ± 41 (6)* | |
| TTNtv | D6 | 52 ± 4 (6) | 96 ± 6 (7) | 114 ± 7 (7) | 0.82 ± 0.05 (7) | 0.68 ± 0.07 (7) | 97 ± 7 (7) | 0.52 ± 0.10 (7) | 15.5 ± 0.8 (7) | 1.86 ± 0.03 (18) | 354 ± 37 (18)*** |
| D7 | 38 ± 4 (8) | 98 ± 12 (8) | 108 ± 7 (8) | 0.87 ± 0.12 (8) | 0.61 ± 0.05 (8) | 98 ± 4 (8) | 0.09 ± 0.19 (8) | 19.3 ± 1.6 (8) | 1.90 ± 0.02 (27) | 367 ± 46 (27)*** | |
| D9 | 52 ± 4 (17) | 94 ± 4 (18) | 116 ± 6 (18) | 0.90 ± 0.07 (18) | 0.74 ± 0.03 (18)* | 98 ± 7 (17) | 0.52 ± 0.6 (18) | 20.0 ± 0.9 (17) | 1.90 ± 0.03 (24) | 348 ± 40 (24)*** | |
The number of measurements obtained on different myofibrils is indicated in parentheses. *P < 0.05, **P < 0.01, ***P < 0.001 (one-way ANOVA analysis) vs combined donors. The fDCM samples have contractile protein mutations, D1 (TNNC3 G159D), D2 (TNNI1 K36Q) and D15 (MYH7 E1426K). NA, data were not acquired.
Maximum isometric force depended on sarcomere length, being about maximal at 2.2 µm. The mean maximum force in the donor heart muscle myofibrils was 112 ± 7 kPa (n = 18), reduced to 50 ± 4 kPa (n = 16) at 1.8 µm sarcomere length (Fig. 2B and Supplementary Fig. S1). These values are comparable with previous measurements from ourselves and others8,9. In comparison, all the familial DCM muscle samples with contractile protein or TTNtv mutations produced the same maximum force at all three sarcomere lengths tested10,11, although there are some cases where force increases whilst Ca2+-sensitivity does not12.
Figure 2.
Myofibril contractility measurements. (A) The time course of contraction and relaxation in single myofibrils, showing the measured parameters. (B) Dependence of Fmax on sarcomere length. Myofibril force was measured at sarcomere lengths of 1.8, 2.0 and 2.2 μm. (C) Effects of DCM-associated mutations on the kinetics of myofibril relaxation; duration of slow relaxation phase. (D) Effects of DCM-associated mutations on the kinetics of myofibril relaxation; rate of fast relaxation phase. (E) Myofibril passive elastic response. Tension trace (top) in response to incremental step changes in sarcomere length (bottom). The parabolic equation was fitted (dashed line) to the stress-strain data. (F) Experimental data sets for samples KN2 and D6 plotted as stress versus sarcomere length. (G) Effects of DCM-associated mutations on the parameter A. (H) Effects of DCM-associated mutations on elastic modulus. The horizontal dotted lines in C, D and G indicate the means of the combined controls (2–4 donor hearts, n=21–32 myofibril samples). Data are shown as means ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001 (one-way ANOVA analysis) versus the combined controls.
Neither the activation (kACT and kTR) nor relaxation kinetics (tLIN, kLIN, kREL) were dependent on sarcomere length at maximally activating Ca2+ concentrations (Supplementary Fig. S1). This data suggests that sarcomere-length itself does not determine myosin crossbridge kinetics and number of force-generating myosin heads.
The rate of force increase following Ca2+ jump, kACT, was 0.78 ± 0.06 s−1 (n = 26, SL = 2.0 µm) with the donor myofibrils, a value compatible with previous measurements, and was not significantly different in any of the DCM mutant heart myofibrils (TTNtv kACT = 0.86 ± 0.02 s−1, 3 heart samples; other fDCM kACT = 0.84 ± 0.06 s−1, 3 heart samples). kTR, which measures the rate of active crossbridge attachment, was 0.64 ± 0.03 s−1 (n = 25) for the donor heart myofibrils and very similar for the DCM mutant myofibrils (mean kTR = 0.68 ± 0.04); there was a trend to a slightly higher kTR in the mutant myofibrils that was statistically significant for D9 (1.16-fold higher) and D15 (1.18-fold higher). kTR was consistently lower than kACT, especially during the first contraction-relaxation cycle, however the difference was diminished with every new cycle. Fmax, after quick release-restretch, also did not fully recover. This shows that the state was progressively altered. Stretching may depress myofibril activation kinetics and force development in human cardiac myofibrils, but it is also possible that this effect could be related to a deterioration in the myofibril samples (Table 1).
Myofibrillar relaxation has a lag phase of almost constant force followed by an exponential decrease in force (Fig. 2A). In donor heart samples the duration of the slow phase was 100 ± 6 ms and the rate of the fast relaxation phase was 17.1 ± 1.2 s−1. The TTNtv mutation myofibrils had rates of relaxation indistinguishable from donor but the three contractile protein fDCM mutations relaxed about 27% faster (Fig. 2C and D).
We then measured passive stiffness. Myofibrils in relaxing solution containing 30 mM BDM were stretched in several cumulative steps. We found that the slack sarcomere length was 1.91 ± 0.02 µm (n = 32) in the donor heart samples and was not significantly different in any of the mutant samples. The increase in passive tension with length was less than donor for all of the mutant samples. For the donor heart samples, the parameter A was 559 ± 38 kPa (n = 32), corresponding to a Young’s modulus of 170 kPa at 2.2 µm sarcomere length (Fig. 2G and H). We found substantial reductions in passive stiffness in all of the fDCM samples including the TTNtv mutants. On average the fDCM myofibrils had a 38% lower passive stiffness (Young’s modulus) (Fig. 2H).
Troponin function in TTNtv mutant hearts
Previous studies of heart samples with fDCM-related mutations in thin filament proteins have indicated that the troponin is abnormal, with a consistent loss of modulation of myofilament Ca2+-sensitivity by PKA-mediated phosphorylation of TnI at Ser 22 and 23 and it has been proposed that this uncoupling is sufficient to induce the DCM phenotype13,14. We therefore investigated the Ca2+-regulatory properties of phosphorylated and unphosphorylated troponin isolated from ventricular or atrial muscle using the in vitro motility assay. Donor heart troponin showed the normal modulation of Ca2+-sensitivity. Troponin from the heart samples with mutations in contractile proteins was uncoupled; this has been previously shown for TNNI3 K36Q and TNNC1 G159D in vitro14–16, but uncoupling due to a myosin heavy chain mutation (MYH7 E1426K) is novel. Three TTNtv heart samples showed normal modulation of Ca2+-sensitivity by TnI phosphorylation, while D6 and D7 showed a blunted relationship of TnI phosphorylation to Ca2+-sensitivity (Supplementary Fig. S2 and Table S2). This suggests that it is unlikely that un-coupling could be a primary cause of the DCM phenotype associated with titin mutations.
Titin expression and modification in human heart
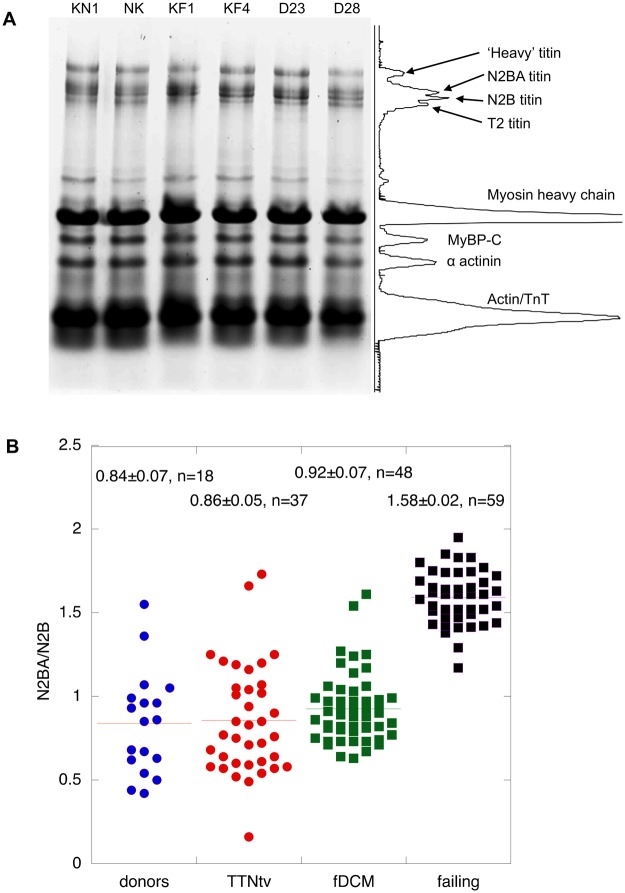
In idiopathic heart failure it has been noted that the proportion of the less stiff N2BA isoform relative to N2B is increased and may therefore account for the reduced passive stiffness observed17,18. We found that there were no significant differences between the N2BA/N2B ratio of donor heart or any of the mutant DCM heart muscle samples, although we did observe a higher ratio in myofibrils from idiopathic DCM as expected (Figs 3 and 4).
Figure 3.
Titin isoform expression. (A) 2% agarose/2% polyacrylamide-SDS gel electrophoresis of human heart myofibrils showing separation of titin isoforms, identified in the scanned profile: Two donors, KN1, NK, two failing heart, KF1, KF4, and two TTNtv mutant samples are shown. (B) Comparison of N2BA/N2B ratio measured in donors, TTNtv, other fDCM and idiopathic DCM. Pooled data from multiple gels similar to A. Data shown for donor heart represents KN1 (17 replicates), NK (13 replicates) and NH (8 replicates). Data shown for TTNtv represents D6 (7 replicates), D7 (8 replicates), D9 (6 replicates), D23 (15 replicates), D28 (15 replicates) and D29 (4 replicates). Data for other fDCM represents D1 (5 replicates), D2 (8 replicates), D4 (4 replicates), D11 (9 replicates), D12 (9 replicates) and D15 (9 replicates). Data shown for idiopathic failing heart represents FG (13 replicates), FH (11 replicates), FI (7 replicates), KF1 (8 replicates), KF2 (3 replicates), KF3 (9 replicates) and KF4 (12 replicates). Only idiopathic DCM samples have a significantly higher N2BA/N2B ratio than donor (1.9x).
Figure 4.
Analysis of titin expression and phosphorylation. (A) Representative 4–20% SDS-PAGE gels of myofibrillar fraction or whole tissue extracts of human heart muscle showing titin. Left: The myofibrillar fraction stained with SYPRO Ruby protein stain. Middle: Western blots of whole tissue extracts were probed with 9D10 anti-titin antibody. Right: The myofibrillar fraction counterstained with Pro-Q Diamond phosphoprotein stain. Plots B, C and D are based on densitometry of a large number of replicate gels. Yellow arrows indicate positions of MW markers, red arrows indicate myofibrillar proteins. (B) N2BA/N2B titin isoform expression ratio was estimated from 4–20% SDS-PAGE. (C) Total titin protein level normalised to MyBP-C was assessed by SYPRO Ruby staining of 4–20% SDS-PAGE. The titin bands were identified by Western blotting. (D) The total titin phosphorylation level was assessed from Pro-Q Diamond staining relative to Sypro Ruby stain. Gel to gel variations were normalised to KN1 sample, loaded in all gels. The bars show the means and SEM for replicate measurements on the same sample (n = 3–10). No significant differences were between the means of the donor group and the means of the patient groups (one-way ANOVA, P > 0.05).
The study of Roberts et al. likewise found the same N2BA/N2B ratio in donor and TTNtv mutant muscle3. We also investigated the possibility of reduced titin expression. Using methodology similar to that which previously identified obscurin haploinsufficiency, we compared donor heart myofibrils with the TTNtv samples and also with the other mutant DCM samples and found no significant haploinsufficiency either in myofibrils or whole tissue extracts19 (Fig. 4). Finally, using Pro-Q Diamond staining for phosphorylation that successfully identified hypophosphorylation of titin in HFpEF20 we found no systematic differences in the phosphorylation level of titin from donor, TTNtv or other mutant DCM myofibrils (Fig. 4). This finding does not exclude the possibility of equal and opposite changes of phosphorylation at the individual phosphorylation sites of titin.
Morphological findings
In ultrastructure studies, interstitial fibrosis was significantly increased in all the DCM samples compared to donor hearts, and TUNEL analysis showed strongly increased apoptosis of cardiomyocytes in all the fDCM samples (Fig. 5 and Supplementary Table S3). Comparison of the sarcomere length in donor hearts with that in DCM heart samples did not show any consistent differences.
Figure 5.
Immunofluorescence microscopy of TTNtv and fDCM heart muscle samples. (A) Examples of immunofluorescent microscopy using antibodies against α-actinin to show sarcomere structure, collagen to show fibrosis, and TUNEL to show apoptosis. The scale bars are 30 µm. (B) Quantification of sarcomere length, fibrosis and apoptosis (see Supplementary Table S3). Data are shown as means ± SEM, One-way ANOVA analysis, ***P < 0.001.
Discussion
We have performed a functional analysis of sarcomeric protein mutations using explanted tissue from patients with a diagnosis of fDCM. The mutations in TnC, TnI and myosin heavy chain have been studied in vitro and the familial dilated cardiomyopathy phenotype is well established5–7,15. In this study we have measured contractility and passive stiffness in human cardiac myofibrils with the sarcomeric fDCM mutations and also putative fDCM due to TTN truncating mutations3.
Our measurements of the rate of donor myofibril activation are similar to those previously published; we observed a fast relaxation rate similar to9 but considerably faster than reported by Poggesi’s group8; the differences probably relate to different experimental conditions such as phosphate concentration, Ca2+ concentration and temperature.
The direct measurements of contractile function show that the DCM-related mutations studied here can be distinguished from human heart tissue with hypertrophic cardiomyopathy (HCM). In contrast to the studies on human heart tissue from HCM patients21, we found the maximum isometric force was not diminished, and length dependent activation of maximal force was not altered in any of the DCM mutants. This is consistent with our detailed measurements using myofibrils from the ACTC E361G DCM transgenic mouse11,21–23.
It is also apparent that the contractile protein fDCM and TTNtv mutant phenotypes are distinct from idiopathic DCM or HFpEF since neither N2BA/N2B ratios or contractile protein and titin phosphorylation levels are different from donor heart3,17–19,24.
It is important to note that the changes in contractile properties of adult human tissue samples with TTNtv mutations are very different from TTNtv mutations incorporated into iPSC-derived cardiomyocytes which exhibit sharply reduced contractility, haploinsufficiency, expression of truncated peptides and decreased sarcomere length25, none of which were observed in our adult myofibril samples. The different abnormalities in TTNtv hiPSC-CM and adult myofibrils is most likely due to the immaturity of iPSC-derived cardiomyocytes which functionally resemble foetal muscles26. In contrast, the properties of muscle from transgenic mice with a heterozygous TTNtv mutation19,27 and our human heart samples are comparable, suggesting that human iPSC-derived cardiomyocytes may indicate congenital effects of TTNtv mutations but do not reproduce the disease-causing process of DCM due to TTNtv observed in adult patients.
It is noteworthy that the DCM-associated mutations in contractile proteins are functionally distinct from DCM-related TTNtv mutant tissue. The faster relaxation rates of the contractile protein mutations D1, D2 and D15 suggest a reduced Ca2+-sensitivity, since kREL increases with increasing EC5011. This was not observed for the TTNtv mutations. In addition, uncoupling provides another potential causative mechanism for contractile protein mutations14,28. Several recent studies have demonstrated that mutations in thin filament proteins lead to uncoupling of TnI phosphorylation by protein kinase A from the change in Ca2+-sensitivity measured in isolated thin filaments and myofibrils. This leads to a blunting of the adrenergic response that was demonstrated to be sufficient to cause DCM under stress13,14,28. Whilst the contractile protein mutations in samples D1, D2 and D15 showed uncoupling, uncoupling was not a consistent feature of the TTNtv mutations suggesting that it is unlikely that uncoupling contributes to the DCM phenotype associated with titin truncating mutations.
These studies confirm the expectation that familial DCM caused by mutations in different genes can work through a number of different mechanisms that cause loss of function, in contrast to the apparent single mechanism for the gain of function in HCM.
The most striking abnormality found in our samples was a 38% reduced passive stiffness for all the fDCM samples. In the case of the TTNtv mutations this was the only functional abnormality we detected and is thus likely to be indicative of the disease-causing abnormality. Our data showing a decrease in passive stiffness are in accord with other studies, notably of samples from idiopathic DCM hearts, and it has been proposed that this phenomenon could be explained by an increased expression of the N2BA TTN isoform or altered titin phosphorylation levels17,18,20. In our fDCM and TTNtv samples we observed reduced passive stiffness with no difference in isoform expression (N2BA/N2B or presence of foetal isoforms) and no haploinsufficiency; nor was there incorporation of predicted truncated peptides into the myofibrils in common with other researchers3,19. There was no significant difference in overall titin phosphorylation level in the fDCM and TTNtv samples but we cannot exclude a change in the relative phosphorylation at sites in titin that enhance stiffness (PKC) and sites that reduce stiffness (PKA, PKG)29. At present we have no explanation for why the fDCM mutations cause reduced passive stiffness other than the possibility of reduced Ca2+-sensitivity may promote relaxation. For the TTNtv mutations it is possible that the mutations cause changes in exon usage that modulate elasticity as was observed with RBM20 ablation30 whilst causing no detectable change in molecular size. It is likely that the decreased passive stiffness would have an effect when the heart muscle is shortening under load that would not be apparent in the isometric contraction31. Such changes could be related to disruption of the dynamics of myosin head activation by stretch leading to reduced contractility32 or alteration of the “slow force response” by modulating of mechanosensitive signalling pathways that affect [Ca2+]i homeostasis and promote apoptosis and fibrosis as observed in our samples33. In concert such processes could impair contractility and potentially trigger a DCM phenotype.
Materials and Methods
Human heart samples
We used anonymised tissue samples from explanted hearts in the Sydney Heart Bank34. Twenty-eight patients with a diagnosis of familial or idiopathic DCM requiring a heart transplant at a young age were selected for sequencing and further studies of which 13 are listed in Supplementary Table S1. As controls we studied muscle from eight donor hearts and two myectomy samples from patients with HCM. Donor hearts were obtained when no suitable transplant recipient was found. The donors had no history of cardiac disease, the hearts showed no structural cardiac pathology and had normal electrocardiogram and the heart ventricular function. Donor heart samples KN1 and KN2 were from University of Kentucky35. Patients were consented under ethical approvals obtained from the Australian Red Cross Blood Service (donor hearts) (HREC #2012/2814) and St Vincent’s Hospital, Darlinghurst, the University of Sydney (failing hearts: HREC #H91/048/1a; donor hearts: HREC #2016/923), and the University of Kentucky, USA (08–03338-F2L). The collection and use of control samples used for immunofluorescence microscopy was approved by the ethics committee of the Medical Council of the State of Hessen. The investigations conform to the principles of the Declaration of Helsinki. The functional characteristics of some of the donor heart and myectomy samples have been previously reported24,36 (see Supplementary Table S1).
Sequencing and gel electrophoresis
DNA extraction, whole exome sequencing, was performed as previously reported4,24,37–39.
Pyrosequencing
TTNtv mutations were confirmed by standard pyrosequencing according to manufacturer’s instructions on a PSQ™ 96 HS instrument (Qiagen, Hilden, Germany) and analyzed by pyrosequencing software.
Gel electrophoresis
Whole tissue extracts were obtained for gel electrophoresis using T-Per Protein extraction reagent (Thermo Scientific 78510) including 1 µg/ml E-64, chymostatin and leupeptin protease inhibitors according to the manufacturer’s protocol. The myofibrillar fraction of heart muscle was prepared by our standard protocol24. For the best resolution myofibril samples were dissolved in gel buffer without heating. High molecular weight proteins were separated on 4–15% SDS-PAGE gradient gels (Criterion, Bio-Rad) with a running time of 2.5 hours at 100 volts or on 2% agarose/2% polyacrylamide “titin gels”.
To assess protein phosphorylation 4–15% SDS-PAGE gradient gels were stained with Pro-Q Diamond phosphoprotein specific stain followed by SYPRO Ruby total protein stain. Western blots were made in a wet blotting apparatus for 5 hours at 250 milliamps at 4 °C. Blotted proteins were reversibly stained with MemCode Reversible stain kit (Thermo Scientific) and then probed with 9D10 monoclonal antibody to titin, specific to the I band region38 (Developmental Studies Hybridoma Bank, Iowa) and visualised with ECL plus (GE Healthcare).
Titin gel preparation and electrophoresis
The method was slightly adapted from that of Tatsumi & Hattori39. Agarose (0.3 g) was stirred in 20 mls of deionized water and dissolved by heating in a microwave for 1 min. The molten agarose was poured into a pre-warmed falcon tube (water bath set at 50 °C) and left to cool. The acrylamide solution was prepared as follows: 17.1 ml (deionized water), 3.6 ml 22.2% acrylamide (acrylamide: N,N’-methylenebiscarylamide = 20:1), 4 ml gel buffer (0.4 M tris, 0.2 M sodium acetate, 20 mM EDTA, 1% SDS, pH 7.4) and 15% ammonium persulphate, 200 µl (mixed in this order). The mixture was warmed in the water bath for 5 mins. 13.3 ml of the cooled agarose was added to the acrylamide solution and gently mixed, 30 µl of TEMED was added to the acrylamide/agarose mix and gently inverted. The gel solution was rapidly poured into a glass gel cassette (approx. 80 × 75 × 1.5 mm) and the combs were added instantly. The gels were then cooled at 4 °C for 7–10 min, then returned to room temperature for 2 hours. The samples were loaded and the gel was run at 4 °C at 7.5 mA for 30 mins, then raised to 15 mA (total time about 4 hours).
Manipulation and measurement of TnI phosphorylation level
Troponin isolated from human heart samples was treated to change the level of phosphorylation. They were dephosphorylated by treatment with shrimp alkaline phosphatase (Sigma, P9088) or phosphorylated by treatment with protein kinase A (PKA) catalytic subunit (Sigma, P2645–400) as previously described14,40. TnI phosphorylation level in isolated troponin was measured by phosphate affinity SDS-PAGE as described by Messer et al.41.
Quantitative in vitro motility assay
Troponin isolated from human heart samples was treated to change the level of phosphorylation (see above). Thin filaments were reconstituted with 10 nM rabbit skeletal muscle α-actin (labelled with TRITC phalloidin), tropomyosin (40–60 nM) and troponin (60 nM) to study Ca2+-regulation of filament motility14,42. Thin filament movement over a bed of immobilised rabbit fast skeletal muscle heavy meromyosin (100 μg/ml) was compared in dual channel motility cells in which troponin varied by a single factor (mutation or phosphorylation state). The temperature was set to 29 °C. Filament movement was recorded and analysed as previously described43, yielding two parameters, the fraction of filaments moving and the speed of moving filaments. The fraction motile and the sliding speeds were measured over a range of Ca2+ concentrations to generate Ca2+-activation curves as shown previously14,44. The data were fitted to the 4-variable Hill equation to yield a value for EC50 and nH.
Mechanical measurements on myofibrils
The apparatus and methodology for the measurement of force and passive stiffness in single myofibrils has been described45. Single myofibrils were isolated from samples of human left ventricular tissue45 and kept on ice for use in experiments for up to 3 days. Myofibrils were isolated from the human heart samples and suspended horizontally using specially prepared microneedles. Contraction and relaxation were initiated by a rapid Ca2+ concentration jump achieved by a fast-solution switch system implementing a double-barrelled micropipette moving across the mounted myofibril. Relaxing (0.01 µM Ca2+ or no added Ca2+) and activating (10 µM Ca2+) solutions contained (mM): MOPS 10, MgATP 5, free Mg2+ 1, DTT 5, phosphocreatine 10, creatine kinase (200 Units/mL), bacterial purine nucleoside phosphorylase (0.1 Units/mL), 7-methylguanosine 0.3, pH 7.0. The Ca-EGTA: EGTA ratio was set to obtain 10 mM total EGTA and the desired free [Ca2+]. Potassium propionate and sodium sulphate were added to adjust the ionic strength of the solution to 200 mM. The relaxing solution in the bath chamber was supplemented with (in μM): chymostatin 10, pepstatin 5, leupeptin 40, E-64 10, PMSF 200. The rate of force increase at 10 µM Ca2+, the maximum achieved isometric force, the rate of force regain after a quick release and re-stretch (kTR) and the slow and fast phase component rates of relaxation (tLIN, kLIN and kREL) at 0.01 µM Ca2+ were measured. Additionally, 30 mM of 2,3-butanedione monoxime (BDM) was added to the relaxing solution with no added Ca2+ during myofibril passive stiffness measurements. The experiments were performed at 17 °C.
Data were fitted using Levenberg-Marquardt nonlinear least square algorithm in LabVIEW. The rate constants for the exponential force development (kACT) and redevelopment (kTR) and for the fast phase of relaxation (kREL) were evaluated by curve-fitting using the equation . The slow phase was fit with a linear fit and characterized by the rate constant kLIN calculated from the slope value of the linear fit by normalizing it to the maximum tension (). A parabola was fit to the stress-strain data, where is stress and ε is strain (, where is slack sarcomere length). Young’s modulus was determined as the derivative of the function and is defined by the equation . Statistics analysis (one-way ANOVA) and graphs were prepared using GraphPad Prism 6 (GraphPad Software, San Diego, CA).
Immunofluorescence microscopy
For these studies control tissues consisted of: LV samples from four patients subjected to subvalvular myectomy during surgical aortic valve replacement, and LV samples from two donor hearts with normal LV function which could not be used for transplantation. Patients with aortic valve stenosis showed preserved cardiac function (ejection fraction ≥ 60%), and the resected myocardial tissue did not present signs of ischemic damage or myocardial hypertrophy.
5 μm cryosections were air dried and fixed with 4% paraformaldehyde. A rabbit polyclonal primary antibody against collagen I (Rockland) and a mouse monoclonal antibody against sarcomeric α-actinin (clone EA-53, Sigma) were used. Secondary detection system consisted of anti-rabbit or anti-mouse IgG and streptavidin linked Cy 3 (Amersham). TRITC Phalloidin was purchased from Sigma and used to stain F-actin; DAPI (nuclear staining) was purchased from Vector Laboratories. Apoptosis was analyzed using the in situ Cell Death Detection Kit (Roche, #116847959110) according to the manufacturer’s instructions. Tissue sections were examined by laser scanning confocal microscopy (Leica TCS SP2 and Leica TCS SP8). A series of confocal optical sections were taken using a Leica Plan Apo × 63/1.32 objective lens. After data acquisition, the images were transferred to a Windows 7 workstation for restoration and three-dimensional reconstruction using Imaris 7.7 multichannel image processing software (Bitplane, Zürich, Switzerland) described below.
Analysis of confocal microscopy
Quantification of collagen I was performed as described previously46,47. In brief, cryosections from at least two different levels in each tissue sample were used. All samples were immunolabeled simultaneously with identical conditions of fixation and dilutions of primary and secondary antibodies. Sections exposed to PBS instead of primary antibodies served as negative controls. For each heart at least 10 random fields of vision were analyzed using image analysis (Leica) and Image J software (NIH). The quantification process was performed blinded to the type of section, having on the screen only one channel showing F-actin labelling. For each quantification, a specific confocal setting was established and kept constant in all measurements. Quantification was performed by measurements of fluorescence intensity by using a range of 0 to 255 grey values. The area of collagen I was calculated as percent of positive labelling per tissue area. Quantification of apoptotic cells was performed as described48. Sarcomere length was measured by intensity profiles of α-actinin in XYZ axes using the three-dimensional extended “Section” mode of the Imaris 7.7 program.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the British Heart Foundation (RG/11/20/29266 and PG/17/5/32705). R.K. was supported by the Fondation LeDucq. We thank the patients and staff of St. Vincent’s Hospital Sydney, Kerckhoff Heart Center, Bad Neuhein, Germany, and the University of Kentucky Hospital, Lexington, KY.
Author Contributions
P.G.V. designed and performed the single myofibril studies and did the data analysis. M.R.T. and R.K. did the sequencing. C.M., A.B.M., N.S. and O.C. performed the Western blots and gel electrophoresis. N.S. and A.E.M. performed the in vitro motility assay experiments. S.K. did the confocal microscopy. C.G.dR., A.L., M.R., C.B., M.G. and K.S.C. provided the material. S.B.M. and R.K. designed the study. All authors wrote and reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
9/24/2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
Contributor Information
Petr G. Vikhorev, Email: p.vikhorev@imperial.ac.uk
Steven B. Marston, Email: s.marston@imperial.ac.uk
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13675-8.
References
- 1.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 2.Herman DS, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts AM, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med. 2015;7:270ra276. doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marston S, et al. OBSCN mutations associated with dilated cardiomyopathy and haploinsufficiency. PLoS One. 2015;10:e0138568. doi: 10.1371/journal.pone.0138568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaski JP, Burch M, Elliott PM. Mutations in the cardiac troponin C gene are a cause of idiopathic dilated cardiomyopathy in childhood. Cardiol Young. 2007;17:675–677. doi: 10.1017/S1047951107001291. [DOI] [PubMed] [Google Scholar]
- 6.Carballo S, et al. Identification and functional characterization of cardiac troponin I as a novel disease gene in autosomal dominant dilated cardiomyopathy. Circ Res. 2009;105:375–382. doi: 10.1161/CIRCRESAHA.109.196055. [DOI] [PubMed] [Google Scholar]
- 7.Villard E, et al. Mutation screening in dilated cardiomyopathy: prominent role of the beta myosin heavy chain gene. Eur Heart J. 2005;26:794–803. doi: 10.1093/eurheartj/ehi193. [DOI] [PubMed] [Google Scholar]
- 8.Piroddi N, et al. Tension generation and relaxation in single myofibrils from human atrial and ventricular myocardium. Pflugers Arch. 2007;454:63–73. doi: 10.1007/s00424-006-0181-3. [DOI] [PubMed] [Google Scholar]
- 9.Walker JS, Walker LA, Margulies K, Buttrick P, de Tombe P. Protein kinase A changes calcium sensitivity but not crossbridge kinetics in human cardiac myofibrils. Am J Physiol Heart Circ Physiol. 2011;301:H138–146. doi: 10.1152/ajpheart.00838.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar M, et al. Cardiac myosin-binding protein C and troponin-I phosphorylation independently modulate myofilament length-dependent activation. J Biol Chem. 2015;290:29241–29249. doi: 10.1074/jbc.M115.686790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vikhorev PG, et al. The dilated cardiomyopathy-causing mutation ACTC E361G in cardiac muscle myofibrils specifically abolishes modulation of Ca(2+) regulation by phosphorylation of troponin I. Biophys J. 2014;107:2369–2380. doi: 10.1016/j.bpj.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dvornikov AV, et al. Restrictive cardiomyopathy troponin I R145W mutation does not perturb myofilament length-dependent activation in human cardiac sarcomeres. J Biol Chem. 2016;291:21817–21828. doi: 10.1074/jbc.M116.746172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Messer A, Marston S. Investigating the role of uncoupling of troponin I phosphorylation from changes in myofibrillar Ca2+-sensitivity in the pathogenesis of cardiomyopathy. Front Physiol. 2014;5:315. doi: 10.3389/fphys.2014.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Memo M, et al. Familial dilated cardiomyopathy mutations uncouple troponin I phosphorylation from changes in myofibrillar Ca(2)(+) sensitivity. Cardiovasc Res. 2013;99:65–73. doi: 10.1093/cvr/cvt071. [DOI] [PubMed] [Google Scholar]
- 15.Dyer EC, et al. Functional analysis of a unique troponin c mutation, GLY159ASP, that causes familial dilated cardiomyopathy, studied in explanted heart muscle. Circ Heart Fail. 2009;2:456–464. doi: 10.1161/CIRCHEARTFAILURE.108.818237. [DOI] [PubMed] [Google Scholar]
- 16.Biesiadecki BJ, Kobayashi T, Walker JS, Solaro RJ, de Tombe PP. The troponin C G159D mutation blunts myofilament desensitization induced by troponin I Ser23/24 phosphorylation. Circ Res. 2007;100:1486–1493. doi: 10.1161/01.RES.0000267744.92677.7f. [DOI] [PubMed] [Google Scholar]
- 17.Nagueh SF, et al. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004;110:155–162. doi: 10.1161/01.CIR.0000135591.37759.AF. [DOI] [PubMed] [Google Scholar]
- 18.Makarenko I, et al. Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ Res. 2004;95:708–716. doi: 10.1161/01.RES.0000143901.37063.2f. [DOI] [PubMed] [Google Scholar]
- 19.Schafer S, et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat Genet. 2017;49:46–53. doi: 10.1038/ng.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamdani N, Bishu KG, von Frieling-Salewsky M, Redfield MM, Linke WA. Deranged myofilament phosphorylation and function in experimental heart failure with preserved ejection fraction. Cardiovasc Res. 2013;97:464–471. doi: 10.1093/cvr/cvs353. [DOI] [PubMed] [Google Scholar]
- 21.Sequeira V, et al. Perturbed length-dependent activation in human hypertrophic cardiomyopathy with missense sarcomeric gene mutations. Circ Res. 2013;112:1491–1505. doi: 10.1161/CIRCRESAHA.111.300436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belus A, et al. The familial hypertrophic cardiomyopathy-associated myosin mutation R403Q accelerates tension generation and relaxation of human cardiac myofibrils. J Physiol. 2008;586:3639–3644. doi: 10.1113/jphysiol.2008.155952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrara C, et al. The HCM-associated cardiac troponin T mutation K280N increases the energetic cost of tension generation in human cardiac myofibrils. Biophysical Journal. 2013;104:187a. doi: 10.1016/j.bpj.2012.11.1056. [DOI] [Google Scholar]
- 24.Messer AE, Jacques AM, Marston SB. Troponin phosphorylation and regulatory function in human heart muscle: dephosphorylation of Ser23/24 on troponin I could account for the contractile defect in end-stage heart failure. J Mol Cell Cardiol. 2007;42:247–259. doi: 10.1016/j.yjmcc.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Hinson JT, et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pioner JM, et al. Isolation and mechanical measurements of myofibrils from human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Reports. 2016;6:885–896. doi: 10.1016/j.stemcr.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gramlich M, et al. Stress-induced dilated cardiomyopathy in a knock-in mouse model mimicking human titin-based disease. J Mol Cell Cardiol. 2009;47:352–358. doi: 10.1016/j.yjmcc.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilkinson R, Song W, Smoktunowicz N, Marston S. A dilated cardiomyopathy mutation blunts adrenergic response and induces contractile dysfunction under chronic angiotensin IIstress. American Journal of Physiology - Heart and Circulatory Physiology. 2015;309:H1936–H1946. doi: 10.1152/ajpheart.00327.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linke WA, Hamdani N. Gigantic business: titin properties and function through thick and thin. Circ Res. 2014;114:1052–1068. doi: 10.1161/CIRCRESAHA.114.301286. [DOI] [PubMed] [Google Scholar]
- 30.Methawasin M, et al. Experimentally increasing titin compliance in a novel mouse model attenuates the Frank-Starling mechanism but has a beneficial effect on diastole. Circulation. 2014;129:1924–1936. doi: 10.1161/CIRCULATIONAHA.113.005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beqqali A, et al. A mutation in the glutamate-rich region of RNA-binding motif protein 20 causes dilated cardiomyopathy through missplicing of titin and impaired Frank-Starling mechanism. Cardiovasc Res. 2016;112:452–463. doi: 10.1093/cvr/cvw192. [DOI] [PubMed] [Google Scholar]
- 32.Fusi L, Brunello E, Yan Z, Irving M. Thick filament mechano-sensing is a calcium-independent regulatory mechanism in skeletal muscle. Nat Commun. 2016;7:13281. doi: 10.1038/ncomms13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ait-Mou Y, Zhang M, Martin JL, Greaser ML, de Tombe PP. Impact of titin strain on the cardiac slow force response. Prog Biophys Mol Biol. 2017 doi: 10.1016/j.pbiomolbio.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lal S, et al. Best practice biobanking of human heart tissue. Biophys Rev. 2015;7:399–406. doi: 10.1007/s12551-015-0182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haynes P, et al. Transmural heterogeneity of cellular level power output is reduced in human heart failure. J Mol Cell Cardiol. 2014;72:1–8. doi: 10.1016/j.yjmcc.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mollova M, et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci USA. 2013;110:1446–1451. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warren CM, Krzesinski PR, Greaser ML. Vertical agarose gel electrophoresis and electroblotting of high-molecular-weight proteins. Electrophoresis. 2003;24:1695–1702. doi: 10.1002/elps.200305392. [DOI] [PubMed] [Google Scholar]
- 38.Wang SM, et al. Studies on cardiac myofibrillogenesis with antibodies to titin, actin, tropomyosin, and myosin. J Cell Biol. 1988;107:1075–1083. doi: 10.1083/jcb.107.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatsumi R, Hattori A. Detection of giant myofibrillar proteins connectin and nebulin by electrophoresis in 2% polyacrylamide slab gels strengthened with agarose. Anal Biochem. 1995;224:28–31. doi: 10.1006/abio.1995.1004. [DOI] [PubMed] [Google Scholar]
- 40.Bayliss CR, et al. Myofibrillar Ca(2+) sensitivity is uncoupled from troponin I phosphorylation in hypertrophic obstructive cardiomyopathy due to abnormal troponin T. Cardiovasc Res. 2013;97:500–508. doi: 10.1093/cvr/cvs322. [DOI] [PubMed] [Google Scholar]
- 41.Messer A, et al. The use of phosphate-affinity SDS-PAGE to measure the troponin I phosphorylation site distribution in human heart muscle. Proteomics Clin Appl. 2009;3:1371–1382. doi: 10.1002/prca.200900071. [DOI] [PubMed] [Google Scholar]
- 42.Fraser ID, Marston SB. In vitro motility analysis of actin-tropomyosin regulation by troponin and calcium. The thin filament is switched as a single cooperative unit. J Biol Chem. 1995;270:7836–7841. doi: 10.1074/jbc.270.14.7836. [DOI] [PubMed] [Google Scholar]
- 43.Marston SB, Fraser ID, Bing W, Roper G. A simple method for automatic tracking of actin filaments in the motility assay. J Muscle Res Cell Motil. 1996;17:497–506. doi: 10.1007/BF00123365. [DOI] [PubMed] [Google Scholar]
- 44.Papadaki M, Vikhorev PG, Marston SB, Messer AE. Uncoupling of myofilament Ca2+ sensitivity from troponin I phosphorylation by mutations can be reversed by epigallocatechin-3-gallate. Cardiovasc Res. 2015;108:99–110. doi: 10.1093/cvr/cvv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vikhorev PG, Ferenczi MA, Marston SB. Instrumentation to study myofibril mechanics from static to artificial simulations of cardiac cycle. MethodsX. 2016;3:156–170. doi: 10.1016/j.mex.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polyakova V, et al. Fibrosis in endstage human heart failure: severe changes in collagen metabolism and MMP/TIMP profiles. Int J Cardiol. 2011;151:18–33. doi: 10.1016/j.ijcard.2010.04.053. [DOI] [PubMed] [Google Scholar]
- 47.Knoll R, et al. A common MLP (muscle LIM protein) variant is associated with cardiomyopathy. Circ Res. 2010;106:695–704. doi: 10.1161/CIRCRESAHA.109.206243. [DOI] [PubMed] [Google Scholar]
- 48.Buyandelger B, et al. ZBTB17 (MIZ1) Is important for the cardiac stress response and a novel candidate gene for cardiomyopathy and heart failure. Circ Cardiovasc Genet. 2015;8:643–652. doi: 10.1161/CIRCGENETICS.113.000690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.