Abstract
Evidence suggests that genetic variation might influence structural brain alterations in psychotic disorders. Longitudinal genetic neuroimaging (G-NI) studies are designed to assess the association between genetic variants, disease progression and brain changes. There is a paucity of reviews of longitudinal G-NI studies in psychotic disorders. A systematic search of PubMed from inception until November 2016 was conducted to identify longitudinal G-NI studies examining the link between Magnetic Resonance Imaging (MRI) and Diffusion Tensor Imaging (DTI)-based brain measurements and specific gene variants (SNPs, microsatellites, haplotypes) in patients with psychosis. Eleven studies examined seven genes: BDNF, COMT, NRG1, DISC1, CNR1, GAD1, and G72. Eight of these studies reported at least one association between a specific gene variant and longitudinal structural brain changes. Genetic variants associated with longitudinal brain volume or cortical thickness loss included a 4-marker haplotype in G72, a microsatellite and a SNP in NRG1, and individual SNPs in DISC1, CNR1, BDNF, COMT and GAD1. Associations between genotype and progressive brain changes were most frequently observed in frontal regions, with five studies reporting significant interactions. Effect sizes for significant associations were generally of small or intermediate magnitude (Cohen’s d < 0.8). Only two genes (BDNF and NRG1) were assessed in more than one study, with great heterogeneity of the results. Replication studies and studies exploring additional genetic variants identified by large-scale genetic analysis are warranted to further ascertain the role of genetic variants in longitudinal brain changes in psychosis.
Introduction
Psychotic disorders comprise a subset of psychiatric illnesses associated with severe consequences for affected individuals and a high global burden of disease.1 In these illnesses, genetic and environmental risk factors may interact with each other and with age-dependent neurodevelopmental processes to affect illness pathology and prognosis.2–4 Longitudinal studies are particularly important as they allow for probing the interaction between risk factors and brain changes. Meta-analyses and reviews of longitudinal studies in adult-onset schizophrenia samples have demonstrated a great variability in results, even between studies using cohorts with similar characteristics.5,6 The most consistent results across studies are a progressive decline in cortical gray matter (GM) volume and progressive increases in lateral ventricle (LV) volume in patients when compared to healthy controls.5–8 Similarly, a recent meta-analysis of early-onset psychosis (EOP, first appearance of psychotic symptoms before the age of 18 years) reported a significant progressive decrease in frontal GM volume in patients compared to controls during the first years following onset of psychotic symptoms.9 However, results between individual EOP studies are heterogeneous,10,11 and some studies of psychotic cohorts report no progressive structural changes compared to healthy controls.12,13
The striking disparities between published results of longitudinal neuroimaging (NI) studies may be partially explained by the effect of genetic variation on brain changes. Schizophrenia (SCZ) demonstrates high heritability,14,15 with a significant portion of heritability attributable to the cumulative effect of many single nucleotide polymorphisms (SNPs).16 Indeed, genetic association studies (including studies of previously-specified candidate SNPs, genome-wide association studies [GWAS], and studies using polygenic scores) have begun to identify SNPs in candidate genes that may contribute to the risk of developing a psychotic disorder.16,17
Genetic Neuroimaging (G-NI) studies analyze genetic variants and brain imaging measures in the same cohort, allowing for the assessment of associations between geno- and phenotype.18–20 Large-scale analyses by collaborative consortiums such as Enhancing Neuro Imaging Genetics through Meta-Analysis (ENIGMA) have suggested that genetic variation may impact structural measures such as intracranial volume (ICV) and various subcortical volumes across healthy and patient cohorts.20,21 In patients with psychotic disorders, genetic variants in certain genes (BDNF, COMT, RSG4, and ZNF804A, among several others) have been associated with average differences in GM and white matter (WM) brain volume, ventricular volume, and cortical thickness and folding patterns between patient genotype groups or patient vs. control genotype groups.22,23 However, the majority of published G-NI studies employed a cross-sectional design, which excludes the possibility of evaluating potential interactions between genetic measures and structural brain changes over time. This limitation precludes any conclusion about whether and which gene variants are associated with atypical brain changes in individuals with psychotic disorders.
Previous reviews of G-NI studies in psychosis have either covered SNPs and neuroimaging in cross-sectional studies with no or only a few longitudinal studies mentioned;22,24–29 have limited the scope to one or two candidate genes;23,30,31 or have geared the focus toward the description and use of specific neuroimaging methods rather than the integration and analysis of study results.32,33 Since characterizing whether and which genetic variants contribute to structural brain changes in psychosis is deemed highly relevant to the overall understanding of the disease, and many future studies will likely employ a longitudinal design, a review of longitudinal G-NI studies to date is warranted. Therefore, a systematic review was performed with the goal of synthesizing current knowledge of the association between genetic variants and progressive brain changes in psychotic disorders.
Results
Subject characteristics
Of the eleven studies selected for review, five studies were conducted specifically in patients experiencing a first episode of psychosis (FEP).34–38 Three studies evaluated patients with childhood-onset schizophrenia (COS).39–41 Two studies were conducted in patients with recent-onset schizophrenia spectrum disorders (recent-onset SSD) (duration of illness < 5 years), of which the percentage of FEP patients was not specified.42,43
Patient cohort size at baseline ranged from 58 to 119 participants. Gender distribution ranged from 58 to 79% male, and mean AAO from 10 to 30.6 years old. Eight of the eleven studies included a healthy control (HC) group for genetic,37,42,43 structural MRI, or both types of analysis,34–36,40,41,44 and one of these studies also included a sibling comparison group.41
Treatment with antipsychotic (AP) medication was described in eight studies.34–38,42–44 Ethnic distribution of the samples was either all Caucasian,35–38,44 95% Caucasian,42,43 or 50–72% Caucasian with the remaining portion accounted for by African American, Hispanic, and Asian participants, and participants of mixed ethnicities.34,39–41 Additional details regarding clinical characteristics of the study samples can be found in Supplementary Table 1.
Genetic measures
Genes examined were BDNF,34,35,42,44 COMT,41 NRG1,36,40 DISC1,38 CNR1,37 GAD1,39 and G72.43 All studies genotyped individual SNPs, three studies additionally performed haplotype analysis,39,40,43 and one study analyzed a microsatellite.40 Information about genes, specific variants, and descriptions of function can be found in Table 1.
Table 1.
Genes and specific variants assessed in the genetic neuroimaging studies included in the review
| Gene | Chromosome | Protein | Function | Polymorphisms included in this review | SNP Location | Functional consequences of the SNP | References |
|---|---|---|---|---|---|---|---|
| BDNF | 11p13 | Brain derived neurotrophic factor | Neurotrophin that plays a role in neurodevelopmental processes, synaptic and cognitive plasticity, and neurotransmission | Val66Met (rs6265) | Exon 2 | Met allele associated with impaired secretion and lower distribution of BDNF protein in neurons | Buckley et al.;71 Chen et al.;72 Egan et al.73 |
| NRG1 | 8p12-21 | Neuregulin 1 | Mediates cell-cell signaling, plays a role in neural development, neurotransmission and synaptic plasticity | SNP8NRG221533 (rs35753505) | Intronic, 5’ flanking region | Unknown | Harrison and Weinberger;74 Stefansson et al.75 |
| Microsatellite: 420M9-1395 | 5’ region | Unknown | |||||
| DISC1 | 1q42.1 | Disrupted in schizophrenia 1 | Neurodevelopment and synaptic regulation | Leu607Phe (rs6675281) | Exon 9 | Allele Phe607 and Cys704 affect expression of alternative transcripts of DISC1 that are expressed in early human brain development | Hennah and Porteus;76 Nakata et al.77 |
| Cys704Ser (rs821616) | Exon 11 | ||||||
| CNR1 | 6q14-15 | Cannabinoid receptor 1 | GPCR for cannabinoids, mediates cannabinoid-induced CNS effects by modulating synaptic function and neurotransmission | rs1049353 | Exon 4 | Unknown | Castillo;82 Parsons and Hurd83 |
| rs2023239 | Intron 2 | Unknown | |||||
| GAD1 | 2q31.1 | GAD67 (glutamic acid decarboxylase) | Enzyme that catalyzes GABA synthesis in inhibitory neurons | rs2270335 | Intron 1 | Unknown | Blum and Mann;78 Straub et al.79 |
| rs2241165 | Intron 2 | Unknown | |||||
| G72 | 13q32-34 | D-amino acid oxidase activator | May regulate glutamate neurotransmission through interaction with DAAO. One isoform may play a role in mitochondrial function | rs3916965 | 5’ region | NA (sample divided and analyzed by haplotype, not individual SNPs) | Chumakov et al.;80 Kvajo et al.81 |
| rs3916967 | 5’ region | ||||||
| rs2391191 (Arg30Lys) | Exon 2 | ||||||
| rs778294 | Intron 3 | ||||||
| Hap 1: AGAG | |||||||
| Hap 2: GAGG | |||||||
| Hap 3: GAGA | |||||||
| COMT | 22q11 | Catechol-O-methyl transferase | Enzyme that plays a role in the breakdown of catecholamines, including dopamine | Val158Met (rs4680) | Exon 4 | Met allele encodes a lower-activity COMT variant, leading to impaired dopamine catabolism | Chen et al.;72 Harrison and Weinberger74 |
Only SNPs and haplotypes that demonstrated an association with structural brain changes in the studies included in the review are shown
Arg arginine, CNS central nervous system, Cyscysteine, DAAO D-amino acid oxidase, Hap haplotype, Leu leucine, Lys lysine, Met methionine, Phe phenylalanine, Ser serine, NA not applicable, SNP single nucleotide polymorphism, Val valine
Neuroimaging measures
No DTI studies fulfilled the complete criteria for inclusion in the review. The eleven studies identified by the search criteria used structural MRI to measure brain changes, and MRI assessments were performed on a 1.5T scanner in all studies. Further details regarding image processing software, types of sequence, slice thickness and voxel size in each study can be found in Supplementary Table 1. Eight of the studies exclusively measured volume changes, with outcomes across the studies including changes in total or regional GM or WM volume, total or regional cortical or subcortical volume (structures included the hippocampus, thalamus, caudate nucleus, and cerebellum), LV volume, and cerebrospinal fluid (CSF) volume.34–37,39,40,42–44 Two studies measured cortical thickness (CT) changes,38,41 and one study measured volume, CT, and cortical surface area (CSA) changes.37
Quality assessment
Quality analysis showed that the studies included in the review were of high (64%) or moderate-high (36%) quality. Ratings for individual checklist items according to each study can be found in Supplementary Table 2.
Genetic x structural MRI findings
Table 2 shows the main genetic x structural MRI findings from the eleven longitudinal G-NI studies retrieved. Five studies compared longitudinal brain changes between genotype or haplotype subgroups of patients with psychosis,37–40,42,43 while six studies compared longitudinal brain changes between genotype or haplotype groups in both patient and HC groups.34–36,40,41,44
Table 2.
Main genetic neuroimaging findings
| Author, Year cohort | Gene and SNP examined | Diagnosis N | Number of scans Follow-up | Morphological longitudinal measurements | Combined genetic and longitudinal NI results | EES, [95% CI] |
|---|---|---|---|---|---|---|
| Ho 200742 Iowa Longitudinal study of recent onset schizophrenia | BDNF rs6265 (Val66Met) | Recent-onset SSD N = 119 (74 Val/Val; 45 Met-carriers) | 2 scans 3 (1.6) years) | Frontal GM volume | Met-carriers ↓ Val/Val n.s. | 0.38, [0.01, 0.75]) |
| Temporal GM volume | n.s. | |||||
| Parietal GM volume | n.s. | |||||
| Occipital GM volume | n.s. | |||||
| LV volume | Met-carriers ↑ | 0.51, [0.14, 0.89] | ||||
| Val/Val n.s. | ||||||
| Sulcal CSF volume | Met-carriers ↑ | 0.48, [0.11, 0.86] | ||||
| Val/Val n.s. | ||||||
| Sulcal frontal CSF volume | Met-carriers ↑ | ≥ 0.49 | ||||
| Val/Val n.s. | ||||||
| Sulcal temporal CSF volume | Met-carriers ↑ | ≥ 0.49 | ||||
| Val/Val n.s. | ||||||
| Sulcal parietal and occipital CSF volume | n.s. | |||||
| Koolschijn 201044 | BDNF rs6265 (Val66Met) | SSD N = 87 (N = 68 with two scans) (44 Val/Val; 24 Met-carriers) HC N = 90 (N = 84 with two scans) (55 Val/Val; 29 Met-carriers) | 2 scans (mean: SSD = 4.9 (0.5) years; HC = 4.9 (0.3) years) | Hippocampal volume | n.s. | |
| Smith 201234 | BDNF rs6265 (Val66Met) | FEP N = 58 (N = 41 with two scans) (25 Val/Val, 16 Met-carriers) HC N = 39 (N = 23 with two scans) (11 Val/Val; 12 Met-carriers) | 2 scans (mean: 46 weeks) | Total brain volume Hippocampal volume | n.s.n.s. | |
| Suárez-Pinilla 201335 PAFIP | BDNF rs6265 (Val66Met) | Non-affective FEP N = 80 (60 Val/Val; 20 Met-carriers) HC N = 54 (45 Val/Val; 9 Met-carriers) | 2 scans (3 years) | Total brain volume Total GM volume Total WM volume | n.s. n.s. n.s. | |
| LV volume | n.s. | |||||
| Total CSF volume | n.s. | |||||
| Caudate nucleus volume | n.s. | |||||
| Thalamus volume | n.s. | |||||
| Addington 200740 NIMH | NRG1 56 NRG1 markers: - 54 SNPs, 2 microsatellites - 2 erbB4 SNPs (genetic analysis only) Risk allele carriers: possession of microsatellite 420M9-1395 | COS (onset before age 12) N = 59a (75% with ≥ 2 scans) (35 risk allele carriers; 24 no risk) HC N = 165a (65% with ≥ 2 scans) (131 risk allele carriers; 34 no risk allele) | Variable (approximately 2 years) | Total GM volumeFrontal GM volume | Greater ↓ in COS risk allele carriers than COS non-carriersn.s. | 0.58 [0.05, 1.11] |
| Temporal GM volume | Greater ↓ in COS risk allele carriers than COS non-carriers | 0.72 [0.18, 1.25] | ||||
| Parietal GM volume | Greater ↓ in COS risk allele carriers than COS non-carriers | 0.66 [0.13, 1.20] | ||||
| Occipital GM volume | Greater ↓ in COS risk allele carriers than COS non-carriers | 0.59 [0.06, 1.12] | ||||
| Greater ↓ in HC risk allele carriers than HC non-carriers | 0.38 [0.001, 0.76] | |||||
| Occipital WM volume | COS N.S. Greater ↓ in HC risk allele carriers than HC non-carriers | 0.48 [0.10, 0.46] | ||||
| Total WM volume, WM volumes of other lobes | n.s. | |||||
| Caudate nucleus volume | n.s. | |||||
| Cerebellum volume | n.s. | |||||
| Suárez-Pinilla 201536 PAFIP | NRG1 a) SNP8NRG221132 (rs73235619) b) SNP8NRG221533 (rs35753505) c) SNP8NRG243177 (rs6994992) | Non-affective FEP N = 59 (31 SNP8NRG221533 C allele-carriers; 28 T/T) HC N = 14 (9 SNP8NRG221533 C allele-carriers; 5 T/T) | 3 scans (1 year; 2 years) | Total brain volume | n.s. | |
| Total GM volume | n.s. | |||||
| Total WM volume | SNP8NRG6221533 C allele carriers: ↓ WM volume compared to T homozygotes at 3-year scan, but no significant genotype by time interaction. | 0.55 [0.03, 1.07] | ||||
| LV volume | SNP8NRG6221533 C allele carriers: 3-year ↑ compared to T homozygotes. | 0.59 [0.07, 1.11] | ||||
| Total CSF volume | n.s. | |||||
| Caudate nucleus volume | n.s. | |||||
| Thalamus volume | n.s. | |||||
| Vázquez-Bourgon 201538 PAFIP | DISC1 (a) rs6675281 (Leu607Phe) (b) rs821616 (Ser704Cys) | Non-affective FEP N = 79 [N = 63 genotyped for rs6675281 (46 Leu/Leu; 17 Phe-carriers) and N = 60 genotyped for rs821616 (4 Ser/Ser; 56 Phe-carriers)] | 2 scans (3 years) | Total CT Frontal CT | rs6675281: Phe-carriers ↑ Leu/Leu N.S. | 0.84 [0.27, 1.42] |
| rs6675281 Phe + rs821616 Cys-carriers ↑ | ||||||
| rs6675281 Phe-carriers ↑ Leu/Leu N.S. | 0.79 [0.21, 1.36] | |||||
| rs6675281 Phe + rs821616 Cys-carriers ↑ | ||||||
| Temporal CT | rs6675281 Leu/Leu ↓ rs6675281 Phe-carriers ↑ | 0.96 [0.38, 1.54] | ||||
| Parietal CT | n.s. | |||||
| Occipital CT | rs6675281 Phe + rs821616 Cys-carriers ↑ | |||||
| Suárez-Pinilla 201537 PAFIP | CNR1 (a) rs1049353 (b) rs1535255 (c) rs2023239 | Non-affective FEP N = 65 (rs1049353: 36 G/G; 29 A-carriers rs2023239: 49 T/T; 16 T/C) HC N = 12 (genetic analysis only) | 2 scans (3 years) | Total GM volume Total WM volume | n.s.n.s. | |
| CSF volume | n.s. | |||||
| LV volume | n.s. | |||||
| Thalamus volume | rs2023239: T/C patients ↓ | 0.59 [0.02, 1.16] | ||||
| T/T patients ↓ | ||||||
| T/C patients ↓ >T/T patients | ||||||
| Caudate nucleus volume | rs1049353: Both A-carrier and G/G reduction was N.S. A-carriers 3.27 times greater ↓ than G/G patients. | 0.76 [0.26, 1.27] | ||||
| CT | n.s. | |||||
| Surface area | n.s. | |||||
| Addington 200539 NIMH | GAD1 (a) 14 SNPs (b) one 4-marker combination | COS N = 72 (N = 39 with ≥ 2 scans) | 2 scans (between 2 and 8 years) | Total GM volume | rs2270335 and rs2241165 were most strongly associated with progressive ↓ | |
| Frontal GM volume | rs2270335 and rs2241165 were most strongly associated with progressive ↓ | |||||
| Hartz 201043 Iowa longitudinal study of recent onset psychoses | G72 4-marker combination: rs3916965, rs3916967, rs2391191, rs778294. Hap 1: AGAG Hap 2: GAGG Hap 3: GAGA | Recent-onset SSD N = 110 [(37/57/16) had (0/1/2) Hap 1 alleles; (60/41/9) had (0/1/2) Hap 3 alleles] HC N = 120 (genetic analysis only) | At least 2 scans (mean: 3.0 (1.6) years) | Total cerebral cortex volume | Hap 1 had a significant effect on volume change | |
| Total GM volume | n.s. | |||||
| Total WM volume | Increased in Hap 3 homozygotes compared to other groups | 1.31 [0.61, 2.01] | ||||
| Total frontal lobe volume | Greater ↓ in Hap 1 homozygotes than other Hap 1 groups. N.S. changes in other Hap 1 groups. | 0.84 [0.30, 1.39] | ||||
| Frontal GM volume | Greater ↓ in Hap 1 homozygotes than other Hap 1 groups. | 0.59 [0.06, 1.13] | ||||
| Frontal WM volume | Greater ↓ in Hap 1 homozygotes than other Hap 1 groups. | 0.54 [0.01, 1.08] | ||||
| Hap 3 had a significant effect on volume change. | ||||||
| Total parietal, temporal, occipital volume | n.s. | |||||
| Parietal, temporal, and occipital WM volume | Hap 1 had a significant effect on occipital WM change. Hap 3 had an effect on change in parietal WM, temporal WM, and occipital WM. Significant ↑ in Hap 3 homozygotes compared to other Hap 3 groups in all three regions. | |||||
| Parietal, temporal, and occipital GM volume | Hap 1 had an effect on parietal GM change. | |||||
| COS (onset before age 13) N = 83 (N = 61 with ≥ 2 scans) (29 Val/Val, 42 Val/Met, 12 Met/Met) | COS and SIB: Increased Val dose associated with accelerated CT thinning, particularly in fronto-temporal regions. HC: Increased Val dose associated with attenuation of CT loss in similar areas: bilateral PFC, temporal and superior parietal regions. | |||||
| Raznahan 201141NIMH | COMT Rs4680 (Val158Met) | SIB N = 62 (N = 33 with ≥ 2 scans) (19 Val/Val, 30 Val/Met, 13 Met/Met) HC N = 208 (N = 141 with ≥ 2 scans) (57 Val/Val, 91 Val/Met, 60 Met/Met) | Variable, (approximately 2 years) | Cortical thickness | COS vs. HC: Significant differences in the relationship between Val dose and CT changes (i.e., accelerated thinning in COS and attenuated thinning in HC) were most prominent in left dlPFC and bilateral cingulate. Val dose-related CT decreases in dlPFC in COS compared to HC continued into adulthood. | |
| SIB vs. HC: Significant differences in the relationship between Val dose and CT changes (i.e., accelerated thinning in SIB and attenuated thinning in HC) were most prominent in left IPS, bilateral IFG, AC and lateral temporal regions. Val-dose related CT decreases in SIB compared to HC resolved by age 20. |
AAO age at onset; AC anterior cingulate; COS childhood-onset schizophrenia; CSF cerebrospinal fluid; CT cortical thickness; dlPFC dorsolateral prefrontal cortex; EES estimated effect size, Cohen’s d with 95% confidence intervals calculated for significant findings based on data provided by the papers; FEP first-episode psychosis; GM gray matter; Hap haplotype, HC healthy controls; IFG inferior frontal gyrus; IPS intraparietal sulcus; LV lateral ventricles; NIMH National Institute of Mental Health; NR not reported; n.s.not significant; PAFIP Prospective Longitudinal Study on First-Episode Psychosis; PFC prefrontal cortex; SCZ schizophrenia; SIB siblings; SSD schizophrenia spectrum disorders (schizophrenia, schizophreniform disorder); TBV total brain volume; WM white matter.
a For Addington et al.40 sample size for longitudinal neuroimaging data was variable at each time point. EES were thus based on the numbers of patients in each genotype group with baseline neuroimaging data available
BDNF
Out of four longitudinal G-NI studies evaluating BDNF, only one reported a significant association between BDNF variants and progressive brain changes. In an adult FEP sample, Met-carriers demonstrated significant progressive loss in frontal GM volume and progressive increase in LV and CSF volume over the three-year follow-up, while Val homozygotes did not show any significant changes during this period.42 No significant associations were observed between BDNF genotype and hippocampal volume change in either patients or HC in a FEP and HC study34 and a SCZ and HC study,44 or between BDNF genotype and changes in total brain volume (TBV), GM, WM, CSF, LV, thalamus and caudate nucleus volume in a FEP and HC study.35
NRG1
In an adult FEP sample, patient C allele carriers for SNP8NRG6221533 showed a significant increase in LV volume over 3 years compared to patient T homozygotes.36 Patient C allele carriers also showed decreased total WM volume compared to T homozygote patients at the 3-year time point, but the time x genotype interaction was not significant.36
In a second NRG1 study, family-based transmission disequilibrium tests (TDT) showed that a microsatellite (420M9-1395) was strongly associated with COS.40 COS risk allele carriers experienced a steeper rate of total GM volume loss into adolescence compared to COS non-carriers, a pattern observed throughout the temporal, parietal and occipital lobes. In the HC group, 420M9-1395 risk allele status affected GM and WM volume change in the occipital lobe only.
DISC1
Of two DISC1 SNPs tested in a 3-year follow-up study of adult FEP patients, rs6675281 (Leu607Phe) was associated with longitudinal changes in total, frontal and temporal CT.38 While patients homozygous for the Leu allele showed a significant progressive CT decrease in temporal cortex, patient Phe allele carriers showed a progressive increase in total, frontal, and temporal CT. A significant association between rs6675281(Leu607Phe) and rs821616 (Ser704Cys) combined genotypes was also observed, with increases in total, frontal and occipital CT found only in Phe + Cys-carrier patients.38
CNR1
Two of three CNR1 SNPs tested in a study in a FEP sample were associated with greater longitudinal decrease in volumes of subcortical structures.37 Caudate nucleus volume reductions over time were non-significant, but patient rs1049353 A-carriers demonstrated a 3.27 times greater decrease in caudate nucleus volume than G/G patients. For rs2023239, T/C patients showed a significantly greater longitudinal decrease in thalamic volume than T/T patients.
GAD1
Family-based TDT in COS patients and their parents indicated a significant association between 3 GAD1 SNPs (rs3749034, rs2270335, rs2241165) and COS phenotype. Quantitative TDT (QTDT) showed that two of these SNPs (rs2270335, rs2241165) demonstrated the strongest associations with longitudinal total and frontal GM volume loss of the SNPs tested. For both SNPs, the common alleles over-represented in COS were also the ones associated with greater GM volume loss.39
G72
Of three four-marker haplotypes representing G72 SNPs rs3916965, rs3916967, rs2391191, and rs778294, homozygosity for haplotype 1 (AGAG) was associated with greater total frontal volume loss over 3 years when compared to other haplotype 1 groups in a sample of recent-onset SSD patients. Homozygosity for haplotype 3 (GAGA) was associated with progressive increases in subcortical white matter volume across all lobes compared to other haplotype 3 groups.43
COMT
The COMT rs4680 (Val158Met) Val allele was over-represented in COS patients, and was associated with different patterns of CT development among COS, sibling (SIB), and HC comparison groups.41 Increased Val dose (having 2 vs. 1 vs. 0 Val alleles) was associated with an acceleration of cortical thinning in the COS and SIB group and an attenuation of CT loss in the HC group, particularly in fronto-temporal cortical regions. Differences were observed in the timing and regions most affected by Val dose in the COS and SIB groups (see Table 2).
Association of G-NI results with measures of clinical severity and cognitive functioning
Three studies found an association between genetic variants linked with progressive brain changes and clinical or cognitive measures.37,40,43 In an adult recent-onset SSD sample, a four-marker G72 haplotype associated with increased frontal volume loss was also associated with greater severity of psychotic symptoms at follow-up (measured by the Scale for the Assessment of Positive Symptoms (SAPS) hallucinations and delusions domains).43 In a CNR1 FEP sample, rs2023239 was associated both with the rate of thalamic volume reduction and with the amount of improvement observed in positive and negative symptomatology during follow-up.37 Specifically, rs2023239 T/C carriers showed greater progressive decreases in thalamic volume as well as less improvement in positive (SAPS) and negative (SANS) symptoms than their T/T counterparts. Finally, in a COS cohort, possession of an NRG1 microsatellite (420M9-1395) risk allele was associated with steeper rates of GM volume loss, and this same risk allele was also associated with poorer premorbid functioning (as measured by the Premorbid Adjustment Scale).40
Discussion
This is, to our knowledge, the first systematic review to explore the association between genetic variants and longitudinal structural brain changes in psychosis. Most of the genes yielding significant associations were evaluated in only one study (G72, DISC1, CNR1, GAD1, and COMT). Genes examined in more than one study were NRG1 and BDNF, and results from those studies were difficult to compare due to different genetic variants being tested (NRG1 studies) or considerable differences in demographic, clinical, and methodological variables (BDNF studies). A significant association between longitudinal volume or CT changes and specific genetic variants was most consistently reported in the frontal lobe, with five studies, out of seven specifically assessing this region, detecting a significant association. Results were mixed for the temporal, parietal and occipital lobes. This is consistent with meta-analyses usually reporting frontal regions as those showing the greatest longitudinal changes in studies measuring progressive brain changes in psychosis.5,6,9 Thus, the observation that genetic variants were primarily associated with changes in frontal regions may be linked with the increased vulnerability of this area to disease-associated longitudinal changes, particularly during the early phases of the illness and sensitive periods of neurodevelopment. Except for one study finding a significant association between polymorphisms in CNR1 and longitudinal changes in the caudate nucleus and thalamus, most of the studies assessing subcortical structures did not detect significant associations between genetic variants and changes in these regions. This is also consistent with meta-analyses of longitudinal brain changes in psychosis, which generally have not reported significant changes in subcortical structures over time,45,46 although one meta-analysis did report greater progressive decline in left caudate nucleus volume in SCZ patients compared to controls.5 In the studies yielding significant associations between genetic variants and longitudinal changes, effect sizes were usually of small or intermediate magnitude,36–38,40,42,43 with large effects (Cohen’s d ≥ 0.8) only found for change in total WM and frontal lobe volume in the study assessing G7243 and for change in total and temporal CT for the rs6675281 SNP in the study assessing DISC1.38 Since many genetic and environmental variables might impact deviant structural changes related to psychosis, it is to be expected that specific candidate genes may only account for very small amounts of variance in longitudinal brain changes seen in psychosis.
The most frequently studied gene among studies identified by our search criteria was BDNF. Structural alterations associated with BDNF Val66Met genotype were frontal lobe GM, LV and sulcal CSF volume in one recent-onset SSD study,42 but a separate study in non-affective FEP reported no interactions with these same regions despite similar cohort characteristics, length of follow-up, and image processing techniques.35 A possible explanation for the differing results may be the younger age at onset and shorter duration of illness in the recent-onset SSD study. Another explanation could be potential differences in AP treatment status at baseline and during follow-up between the samples, as antipsychotics may have an effect on brain changes and different genetic backgrounds may interact with that effect.42
The other gene examined in multiple longitudinal G-NI studies was NRG1, with results suggesting an effect for a 5’ SNP on LV changes in FEP patients and a 5’ microsatellite on the trajectory of brain changes in COS patients and controls.36,40 These two NRG1 studies and a COMT study were the only studies in this review to report that certain genetic variants were associated with greater progressive brain changes in patients but not in controls.36,40,41 These results suggest that certain genetic variants may have specific effects on progressive brain changes in psychosis. Another possibility is that certain variants have more marked effects on patients with psychosis, which make it easier to detect changes in these groups.
In the COMT study, findings in COS and SIB groups were similar during adolescence, but by early adulthood Val dose-related CT differences between SIB and HC groups had disappeared while Val dose-related CT differences between COS and HC groups remained. This suggests that CT alterations associated with COMT genotype may exist on a continuum between psychotic patients, their siblings, and healthy controls, with brain changes at certain developmental stages only detectable in the psychotic sample. These results underscore the importance of including comparison groups in G-NI studies, as effects of genetic variants on structural patterns may differ between patients, relatives, and controls. Longitudinal comparison of these groups could help identify at-risk phenotypes in addition to characterizing actual disease progression.
Along the lines of the COMT study, results from several other studies in this review suggest that the effect of certain genetic variants on brain changes might vary according to developmental stage or illness progression. NRG1 and DISC1 are highly relevant genes for neurodevelopmental and brain maturation processes. In the three studies examining these genes, cross-sectional results were either non-significant at some or all time points (as in the NRG1 FEP and DISC1 FEP studies),36,38 or only represented part of the developmental trajectory of volume changes (as in the NRG1 COS study).40 The effect of a CNR1 SNP on caudate nucleus reduction rate was also undetectable at cross-sectional analysis. These findings suggest that certain genes may affect the progression or trajectory of structural brain changes during the early years of illness and support the use of a longitudinal design to appropriately assess their association.
Three studies assessing genes involved in neurotransmission (GAD1, COMT, and G72) reported that genetic variants associated with progressive brain changes were also over-transmitted to patients or associated with the disease phenotype.39,41,43 Two 5’ GAD1 SNPs were associated with both COS and exaggerated cortical frontal GM loss.39 The Val allele at COMT Val158Met was overrepresented in COS patients (but not in their siblings) compared to controls, and was associated with accelerated CT thinning in patients. A G72 haplotype associated with increased frontal volume loss in a recent- onset SSD cohort 43 was consistent with alleles reported to be over-represented in SCZ cohorts in previous G72 genetic studies and meta-analyses.47–49 This G72 haplotype was also associated with more severe psychotic symptoms at follow-up.43 Aside from G72, genetic variants in two other genes (NRG1 and CNR1) were also associated with both progressive brain changes and clinical measures. The association of the same genetic variants with progressive brain changes, increased disease risk, and/or clinical variables point to their potential role in the pathophysiology of schizophrenia and other psychoses, and to alterations in brain structure as a core component of the illness. Future studies should further explore the links between genetic variants, brain changes, and clinical progression. A better understanding of these relationships may eventually contribute to increased accuracy in predicting disease course and greater individualization of clinical care in patients with psychosis.
Several limitations must be considered in interpreting the results of this review. First, the majority of the results have not been replicated, and since few studies evaluated a common genetic variant it was not possible to perform a quantitative synthesis and meta-analysis. Second, the overall number of studies included was low, all studies had relatively small sample sizes, and several of them were conducted by the same research groups. This means certain samples overlapped significantly and were comprised of participants from similar backgrounds that may not be representative of other populations. Considering studies carried out by different teams, several important factors such as cohort type, specific diagnosis, length of follow-up, ethnic composition, and genetic variant and NI measurement evaluated complicated direct comparison of results. Furthermore, besides those examining BDNF, almost every study reported a significant association between a genetic variant and progressive brain changes. This raises the question of whether associations with other brain regions may have been tested as well, but only positive associations were reported. Regarding the significance of findings, some of the studies did not provide sufficient information to calculate effect sizes and thereby quantify changes. Several of the studies did not include a healthy comparison group for NI changes. Another limitation was that cortical regions were assessed more often than subcortical regions during NI analysis, raising the possibility of region-reporting bias. Finally, most of the studies did not report on laterality when describing brain changes, making it difficult to determine whether changes were bilateral or whether any hemispheric differences were present.
In the studies that yielded significant results, potential confounding factors must be considered. AP treatment may have an effect on certain brain volumes,19 and these effects may differ depending on type (i.e., typical vs. atypical) of AP received.50 Research has suggested AP treatment may not only influence brain structure independently but also interact with genes evaluated in these studies, with animal studies showing different effects for atypical and typical APs on BDNF levels and cell proliferation in the hippocampus.51,52 Participants received different AP medications in several studies included in this review, but not all controlled for AP treatment in statistical analysis. Several other environmental factors may have affected results through interactions with genes, brain structure, or both, such as childhood trauma, stress, and cannabis use.53,54 For example, studies in cohorts of patients with psychosis have indicated interactions between childhood trauma, BDNF expression or genotype, and hippocampal volumes.55–57 The CNR1 study in this review reported interactions between CNR1 SNPs, and WM and LV volume changes in cannabis non-consumers, but not in cannabis consumers.37 Finally, given the limited variety of genes studied thus far, the lack of cohesive results may be partially due to the possibility that certain genetic variants relevant to progressive brain changes have not been studied yet. For example, genetic variants in genes related to inflammation and oxidative stress processes (such as genes involved in the synthesis and metabolism of glutathione, manganese superoxide dismutase, interleukins, major histocompatibility complex [MHC], or complement components) have increasingly been implicated in the pathophysiology of schizophrenia and other psychotic disorders,58,59 but are yet to be explored in a longitudinal G-NI study. Copy number variation (CNV) is another area that is yet to be extensively researched in relation to psychotic disorders. Certain copy number variants (such as 22q11.2 deletion, 16p11.2 duplication, and 3q29 deletion) have been implicated in the genetic architecture of psychotic disorders 60–62 and may be associated with structural brain differences in high-risk populations or patients with SCZ,63,64 suggesting these variants may be promising avenues of investigation in longitudinal G-NI research.
Candidate gene association studies in general are limited when used to address complex disorders and their associated outcomes, as significant associations are common but often fail to be replicated.65 While the candidate gene approach has had some successes in identifying genetic variants associated with complex brain diseases—one example being the association between APOE variation and Alzheimer’s disease risk and brain structure66–68—similar results have not been achieved in schizophrenia, and recent research using GWAS and meta-analysis suggests that many historical candidate genes for schizophrenia (including BDNF, COMT, DISC1, and NRG1) fail to reach genome-wide significance.16,66 One promising approach to address the issue of low reproducibility of candidate gene association studies is conducting large scale meta-analysis via consortia such as ENIGMA, which will likely play an increasing role in the identification and replication of genetic variants relevant to brain changes and psychiatric illnesses.19,69 Thus far, large-scale genetic research in schizophrenia has pointed to the extended MHC and several genes involved in glutamatergic synaptic function and calcium signaling are important loci for future research, and has also highlighted the potential value of polygenic risk score analysis.16,17,66 Even with these advances, individual studies remain important since large-scale analysis is impossible without the contribution of individual datasets, and certain hypotheses and causal questions may be better addressed, at least initially, in smaller studies.69
Despite the mentioned limitations, this paper represents the first systematic review and qualitative analysis of studies of genetic variants and progressive structural brain changes in psychotic cohorts. Results from this review suggest that several genes related to neurodevelopment, brain maturation processes, neurotransmission and plasticity might impact progressive structural alterations in psychosis. However, sparse studies and discrepancies in cohort characteristics, genes and specific variants examined and study methodology make forming substantial conclusions difficult. In future studies, research groups should attempt to reproduce results in larger samples and different populations so that replicability is established. Researchers should pay particular attention to consistency of design, genes evaluated, and imaging parameters used, so that a more cohesive literature base addressing the role of these factors in psychosis can be developed. Furthermore, an effort should be made to include longitudinal clinical and cognitive measures in order to increase the applicability of genetic neuroimaging findings to other facets of psychotic illness.
Methods
Search strategy
A systematic two-step literature search was performed according to the guidelines described in the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) statement.70 First, a PubMed database search from inception until November 2016 was performed with the following search terms: (gene OR SNP OR nucleotide OR polymorphism) AND (MRI OR “magnetic resonance imaging” OR neuroimaging OR cortical OR imaging OR DTI OR volume) AND (psychosis OR schizophrenia OR psychotic) AND (Humans[Mesh] AND English[lang]). Second, the reference lists of selected studies were manually reviewed to identify relevant studies missed by the initial computerized search. The original search yielded 1960 results, and five additional studies were identified during manual review.
Selection criteria
1965 abstracts were assessed and considered for review if the study described met the following hierarchical eligibility criteria:
The study was published as an original peer-reviewed article written in English.
The study was performed in humans.
The study included a patient group with a diagnosis of schizophrenia or other psychotic disorders according to the Diagnostic and Statistical Manual of Mental Disorders (DSM) criteria (DSM-III, DSM-III-R, DSM-IV, DSM-IV-TR, DSM-5).
The study assessed both a genetic variant (SNP, microsatellite, or haplotype) and longitudinal changes in at least one structural and/or DTI-based MRI measure, including volume (total or regional cortical or subcortical brain volume, total or regional GM or WM volume, LV or cerebrospinal fluid [CSF] volume), cortical thickness (CT), cortical surface area (CSA), fractional anisotropy (FA), mean diffusivity (MD), radial diffusivity (RD), and axial diffusivity (AD).
The study had a longitudinal design, with participants undergoing at least two structural MRI assessments over a period of time.
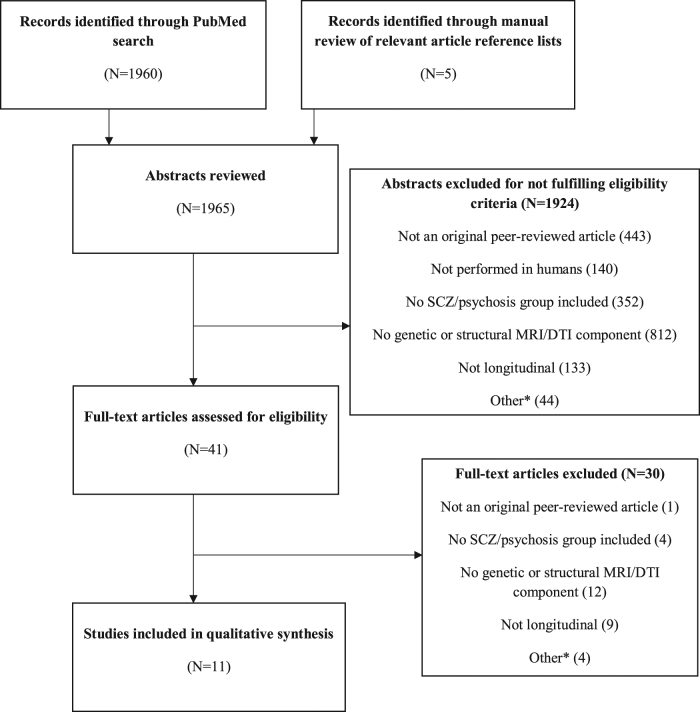
The full text was consulted in cases where information provided by the abstract was not sufficient to evaluate eligibility or where no abstract was available. After full text review, a final group of eleven studies using T1 weighted images were included in the review. Figure 1 provides a flowchart summarizing the selection process.
Fig. 1.
Flowchart of study selection. *Articles excluded for reasons not encompassed by other categories (N < 5; results had not yet been reported; psychosis was induced by medication or another biological syndrome; the topic of the article was not relevant. SCZ schizophrenia; MRI magnetic resonance imaging; DTI diffusion tensor imaging
Data extraction, synthesis, and analysis
Data for the following variables were retrieved for each study (see Supplementary Table 1): author names, year of publication, number of participants, diagnosis and diagnostic criteria, demographic variables (age at onset [AAO], age at baseline, duration of untreated illness [DUI], proportion of male subjects, medication, genetic data (gene and location; SNP, haplotype or microsatellite and location), structural neuroimaging data (number of scans, between-scan interval, structural brain measurements, software, scanner strength, type of T1-weighted sequence, slice thickness), genetic x neuroimaging outcome, and additional clinical or cognitive outcomes. Further information regarding genes and SNPs examined (i.e., function of gene, functional consequence of specific SNP) was collected from additional sources.71–84 For the purpose of synthesis and analysis, studies were grouped by gene examined and thereafter by year of publication. For individual studies providing sample size and relevant statistics for significant genetic x neuroimaging findings, estimated effect sizes (Cohen’s d) with 95% confidence intervals were calculated using Comprehensive Meta-Analysis Software version 3 (Biostat, Inc., Englewood, NJ).
Quality assessment
The quality of the studies was assessed using an item-checklist designed specifically for this review based on a previously published quality assessment of longitudinal neuroimaging studies in psychosis.45 For each item, quality was assessed with a range of 0 (minimum) to 2 (maximum) points. The eleven studies were rated according to the sum of the points for each item, and then categorized as high (>80% of the maximum possible points), moderate-high (60–79%), moderate (40–59%), moderate-low (20–39%), or low (<19%) quality (see Supplementary Tables 2 and 3 for further details).
Data availability
The authors declare that all data supporting the findings of this review are available within the paper and supplementary files.
Electronic supplementary material
Acknowledgements
This work was supported by the Spanish Ministry of Economy and Competitiveness. Instituto de Salud Carlos III, co-financed by ERDF Funds from the European Commission, “A way of making Europe” (PI12/01303, PI13/02112, PI4/00815), CIBERSAM. Madrid Regional Government (S2010/BMD-2422 AGES), European Union Structural Funds, European Union Seventh Framework Program under grant agreements FP7-HEALTH-2009-2.2.1-2-241909 (Project EU-GEI), FP7-HEALTH-2009-2.2.1-3-242114 (Project OPTiMiSE), FP7- HEALTH-2013-2.2.1-2-603196 (Project PSYSCAN) and FP7- HEALTH-2013-2.2.1-2-602478 (Project METSY), and European Union H2020 Framework Program under the Innovative Medicines Initiative 2 Joint Undertaking (grant agreement No 115916, project PRISM); Fundación Alicia Koplowitz and Fundación Mutua Madrileña. CD-C has previously held a grant from Instituto de Salud Carlos III, Spanish Ministry of Economy and Competitiveness, and from Fundación Alicia Koplowitz. Thanks to FIS PI16/00998 and the Comissionat per a Universitat i Recerca del DIUE (2014SGR1636).
Author contributions
J.H. performed the literature review and extraction of data under the supervision of CD-C., J.H., CD-C., B.A., J.J., K.M., and C.A. contributed to the selection of variables of interest, the interpretation of findings, and the writing of the manuscript. All authors had full access to the data, take responsibility for their integrity and accuracy and approved the final version of the manuscript.
Competing interests
The authors declare that they have Competing financial interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies the paper on the npj Schizophrenia website (10.1038/s41537-017-0036-2).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bárbara Arias, Email: barbara.arias@ub.edu.
Celso Arango, Email: carango@hggm.es.
References
- 1.Mathers, C., Fat, D. M., Boerma, J. T., & World Health Organization. The global burden of disease: 2004 update (World Health Organization, 2008).
- 2.Woods BT. Is schizophrenia a progressive neurodevelopmental disorder? Toward a unitary pathogenetic mechanism. Am. J. Psychiatr. 1998;155:1661–1670. doi: 10.1176/ajp.155.12.1661. [DOI] [PubMed] [Google Scholar]
- 3.McGrath JJ, Feron FP, Burne TH, Mackay-Sim A, Eyles DW. The neurodevelopmental hypothesis of schizophrenia: a review of recent developments. Annu. Med. 2003;35:86–93. doi: 10.1080/07853890310010005. [DOI] [PubMed] [Google Scholar]
- 4.Rapoport JL, Giedd JN, Gogtay N. Neurodevelopmental model of schizophrenia: update 2012. Mol. Psychiatr. 2012;17:1228–1238. doi: 10.1038/mp.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olabi B, et al. Are there progressive brain changes in schizophrenia? A meta-analysis of structural magnetic resonance imaging studies. Biol. Psychiatr. 2011;70:88–96. doi: 10.1016/j.biopsych.2011.01.032. [DOI] [PubMed] [Google Scholar]
- 6.Vita A, De Peri L, Deste G, Sacchetti E. Progressive loss of cortical gray matter in schizophrenia: a meta-analysis and meta-regression of longitudinal MRI studies. Transl. psychiatr. 2012;2:e190. doi: 10.1038/tp.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kempton MJ, Stahl D, Williams SC, DeLisi LE. Progressive lateral ventricular enlargement in schizophrenia: a meta-analysis of longitudinal MRI studies. Schizophr. Res. 2010;120:54–62. doi: 10.1016/j.schres.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 8.Hulshoff Pol HE, Kahn RS. What happens after the first episode? A review of progressive brain changes in chronically ill patients with schizophrenia. Schizophr. Bull. 2008;34:354–366. doi: 10.1093/schbul/sbm168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraguas D, Diaz-Caneja CM, Pina-Camacho L, Janssen J, Arango C. Progressive brain changes in children and adolescents with early-onset psychosis: A meta-analysis of longitudinal MRI studies. Schizophr. Res. 2016;173:132–139. doi: 10.1016/j.schres.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 10.Keller A, et al. Corpus callosum development in childhood-onset schizophrenia. Schizophr. Res. 2003;62:105–114. doi: 10.1016/S0920-9964(02)00354-7. [DOI] [PubMed] [Google Scholar]
- 11.Johnson SL, et al. Absence of anatomic corpus callosal abnormalities in childhood-onset schizophrenia patients and healthy siblings. Psychiatry. Res. 2013;211:11–16. doi: 10.1016/j.pscychresns.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James AC, Javaloyes A, James S, Smith DM. Evidence for non-progressive changes in adolescent-onset schizophrenia: follow-up magnetic resonance imaging study. Bri. J. Psychiatr. 2002;180:339–344. doi: 10.1192/bjp.180.4.339. [DOI] [PubMed] [Google Scholar]
- 13.Haukvik UK, et al. No progressive brain changes during a 1-year follow-up of patients with first-episode psychosis. Psychol. Med. 2016;46:589–598. doi: 10.1017/S003329171500210X. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatr. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 15.Cardno AG, et al. Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Arch. Gen. Psychiatry. 1999;56:162–168. doi: 10.1001/archpsyc.56.2.162. [DOI] [PubMed] [Google Scholar]
- 16.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci Nature511 421–427 (2014). [DOI] [PMC free article] [PubMed]
- 17.Ripke S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer-Lindenberg A. Imaging genetics of schizophrenia. Dialogues. Clin. Neurosci. 2010;12:449–456. doi: 10.31887/DCNS.2010.12.4/amlindenberg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto R, et al. Imaging genetics and psychiatric disorders. Curr. Mol. Med. 2015;15:168–175. doi: 10.2174/1566524015666150303104159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hibar DP, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams HH, et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat. Neurosci. 2016;19:1569–1582. doi: 10.1038/nn.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurung R, Prata DP. What is the impact of genome-wide supported risk variants for schizophrenia and bipolar disorder on brain structure and function? A systematic review. Psychol. Med. 2015;45:2461–2480. doi: 10.1017/S0033291715000537. [DOI] [PubMed] [Google Scholar]
- 23.van Haren NE, Bakker SC, Kahn RS. Genes and structural brain imaging in schizophrenia. Curr. Opin. Psychiatry. 2008;21:161–167. doi: 10.1097/YCO.0b013e3282f4f25b. [DOI] [PubMed] [Google Scholar]
- 24.Kurnianingsih YA, et al. Neurocognitive-genetic and neuroimaging-genetic research paradigms in schizophrenia and bipolar disorder. J. Neur. Trans. 2011;118:1621–1639. doi: 10.1007/s00702-011-0672-z. [DOI] [PubMed] [Google Scholar]
- 25.Najjar S, Pearlman DM. Neuroinflammation and white matter pathology in schizophrenia: systematic review. Schizophr. Res. 2015;161:102–112. doi: 10.1016/j.schres.2014.04.041. [DOI] [PubMed] [Google Scholar]
- 26.Voineskos AN. Genetic underpinnings of white matter ‘connectivity’: heritability, risk, and heterogeneity in schizophrenia. Schizophr. Res. 2015;161:50–60. doi: 10.1016/j.schres.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 27.Fineberg AM, Ellman LM. Inflammatory cytokines and neurological and neurocognitive alterations in the course of schizophrenia. Biol. Psychiatry. 2013;73:951–966. doi: 10.1016/j.biopsych.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisenberg DP, Berman KF. Executive function, neural circuitry, and genetic mechanisms in schizophrenia. Neuropsychopharmacology. 2010;35:258–277. doi: 10.1038/npp.2009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birur B, Kraguljac NV, Shelton RC, Lahti AC. Brain structure, function, and neurochemistry in schizophrenia and bipolar disorder—a systematic review of the magnetic resonance neuroimaging literature. npj Schizophrenia. 2017;3:15. doi: 10.1038/s41537-017-0013-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duff BJ, Macritchie KA, Moorhead TW, Lawrie SM, Blackwood DH. Human brain imaging studies of DISC1 in schizophrenia, bipolar disorder and depression: a systematic review. Schizophr. Res. 2013;147:1–13. doi: 10.1016/j.schres.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 31.Notaras M, Hill R, van den Buuse M. A role for the BDNF gene Val66Met polymorphism in schizophrenia? A comprehensive review. Neurosci. Biobehav. Rev. 2015;51:15–30. doi: 10.1016/j.neubiorev.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 32.Roffman JL, Weiss AP, Goff DC, Rauch SL, Weinberger DR. Neuroimaging-genetic paradigms: a new approach to investigate the pathophysiology and treatment of cognitive deficits in schizophrenia. Harv. Rev. Psychiatr. 2006;14:78–91. doi: 10.1080/10673220600642945. [DOI] [PubMed] [Google Scholar]
- 33.Bearden CE, van Erp TG, Thompson PM, Toga AW, Cannon TD. Cortical mapping of genotype-phenotype relationships in schizophrenia. Hum. Brain. Mapp. 2007;28:519–532. doi: 10.1002/hbm.20404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith GN, et al. Hippocampal volume and the brain-derived neurotrophic factor Val66Met polymorphism in first episode psychosis. Schizophr. Res. 2012;134:253–259. doi: 10.1016/j.schres.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 35.Suarez-Pinilla P, et al. BDNF Val66Met variants and brain volume changes in non-affective psychosis patients and healthy controls: a 3 year follow-up study. Prog. Neuropsychopharmacol. Biol. Psychiatr. 2013;45:201–206. doi: 10.1016/j.pnpbp.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 36.Suarez-Pinilla P, et al. Progressive structural brain changes and NRG1 gene variants in first-episode nonaffective psychosis. Neuropsychobiology. 2015;71:103–111. doi: 10.1159/000370075. [DOI] [PubMed] [Google Scholar]
- 37.Suarez-Pinilla P, et al. Brain structural and clinical changes after first episode psychosis: Focus on cannabinoid receptor 1 polymorphisms. Psychiatry. Res. 2015;233:112–119. doi: 10.1016/j.pscychresns.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Vazquez-Bourgon, J. et al. Variations in disrupted-in-Schizophrenia 1 gene modulate long-term longitudinal differences in cortical thickness in patients with a first-episode of psychosis. Brain. Imaging. Behav.10.1007/s11682-015-9433-1 (2015). [DOI] [PubMed]
- 39.Addington AM, et al. GAD1 (2q31.1), which encodes glutamic acid decarboxylase (GAD67), is associated with childhood-onset schizophrenia and cortical gray matter volume loss. Mol. Psychiatr. 2005;10:581–588. doi: 10.1038/sj.mp.4001599. [DOI] [PubMed] [Google Scholar]
- 40.Addington AM, et al. Neuregulin 1 (8p12) and childhood-onset schizophrenia: susceptibility haplotypes for diagnosis and brain developmental trajectories. Mol. Psychiatr. 2007;12:195–205. doi: 10.1038/sj.mp.4001906. [DOI] [PubMed] [Google Scholar]
- 41.Raznahan A, et al. Catechol-o-methyl transferase (COMT) val158met polymorphism and adolescent cortical development in patients with childhood-onset schizophrenia, their non-psychotic siblings, and healthy controls. Neuroimage. 2011;57:1517–1523. doi: 10.1016/j.neuroimage.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho BC, Andreasen NC, Dawson JD, Wassink TH. Association between brain-derived neurotrophic factor Val66Met gene polymorphism and progressive brain volume changes in schizophrenia. Am. J. Psychiatr. 2007;164:1890–1899. doi: 10.1176/appi.ajp.2007.05111903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartz SM, et al. G72 Influences longitudinal change in frontal lobe volume in schizophrenia. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2010;153B:640–647. doi: 10.1002/ajmg.b.31033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koolschijn PC, et al. Effects of brain-derived neurotrophic factor Val66Met polymorphism on hippocampal volume change in schizophrenia. Hippocampus. 2010;20:1010–1017. doi: 10.1002/hipo.20699. [DOI] [PubMed] [Google Scholar]
- 45.Fusar-Poli P, et al. Progressive brain changes in schizophrenia related to antipsychotic treatment? A meta-analysis of longitudinal MRI studies. Neurosci. Biobehav. Rev. 2013;37:1680–1691. doi: 10.1016/j.neubiorev.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steen RG, Mull C, McClure R, Hamer RM, Lieberman JA. Brain volume in first-episode schizophrenia: systematic review and meta-analysis of magnetic resonance imaging studies. Bri. J. Psychiatr. 2006;188:510–518. doi: 10.1192/bjp.188.6.510. [DOI] [PubMed] [Google Scholar]
- 47.Detera-Wadleigh SD, McMahon FJ. G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biol. Psychiatr. 2006;60:106–114. doi: 10.1016/j.biopsych.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 48.Yue W, et al. Association of G72/G30 polymorphisms with early-onset and male schizophrenia. Neuroreport. 2006;17:1899–1902. doi: 10.1097/WNR.0b013e3280102ed4. [DOI] [PubMed] [Google Scholar]
- 49.Li D, He L. G72/G30 genes and schizophrenia: a systematic meta-analysis of association studies. Genetics. 2007;175:917–922. doi: 10.1534/genetics.106.061796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lieberman JA, et al. Antipsychotic drug effects on brain morphology in first-episode psychosis. Arch. Gen. Psychiatr. 2005;62:361–370. doi: 10.1001/archpsyc.62.4.361. [DOI] [PubMed] [Google Scholar]
- 51.Parikh V, Khan MM, Mahadik SP. Olanzapine counteracts reduction of brain-derived neurotrophic factor and TrkB receptors in rat hippocampus produced by haloperidol. Neurosci. Lett. 2004;356:135–139. doi: 10.1016/j.neulet.2003.10.079. [DOI] [PubMed] [Google Scholar]
- 52.Park SW, Lee SK, Kim JM, Yoon JS, Kim YH. Effects of quetiapine on the brain-derived neurotrophic factor expression in the hippocampus and neocortex of rats. Neurosci. Lett. 2006;402:25–29. doi: 10.1016/j.neulet.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 53.Holtzman CW, et al. Stress and neurodevelopmental processes in the emergence of psychosis. Neuroscience. 2013;249:172–191. doi: 10.1016/j.neuroscience.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Habets P, Marcelis M, Gronenschild E, Drukker M, van Os J. Reduced cortical thickness as an outcome of differential sensitivity to environmental risks in schizophrenia. Biol. Psychiatr. 2011;69:487–494. doi: 10.1016/j.biopsych.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 55.Aas M, et al. BDNF val66met modulates the association between childhood trauma, cognitive and brain abnormalities in psychoses. Prog. Neuropsychopharmacol. Biol. Psychiatr. 2013;46:181–188. doi: 10.1016/j.pnpbp.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 56.Aas M, et al. Interplay between childhood trauma and BDNF val66met variants on blood BDNF mRNA levels and on hippocampus subfields volumes in schizophrenia spectrum and bipolar disorders. J. Psychiatr. Res. 2014;59:14–21. doi: 10.1016/j.jpsychires.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 57.Mondelli V, et al. Stress and inflammation reduce brain-derived neurotrophic factor expression in first-episode psychosis: a pathway to smaller hippocampal volume. J. Clin. Psychiatr. 2011;72:1677–1684. doi: 10.4088/JCP.10m06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bitanihirwe BK, Woo TU. Oxidative stress in schizophrenia: an integrated approach. Neurosci. Biobehav. Rev. 2011;35:878–893. doi: 10.1016/j.neubiorev.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sekar A, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rutkowski TP, et al. Unraveling the genetic architecture of copy number variants associated with schizophrenia and other neuropsychiatric disorders. J. Neurosci. Res. 2017;95:1144–1160. doi: 10.1002/jnr.23970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cnv, Schizophrenia Working Groups of the Psychiatric Genomics, C. & Psychosis Endophenotypes International, C. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects Nat. Genet.49 27–35 (2017). [DOI] [PMC free article] [PubMed]
- 62.Rees E, et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br. J. Psychiatr. 2014;204:108–114. doi: 10.1192/bjp.bp.113.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maillard AM, et al. The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Mol. Psychiatr. 2015;20:140–147. doi: 10.1038/mp.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin AK, Robinson G, Reutens D, Mowry B. Copy number deletion burden is associated with cognitive, structural, and resting-state network differences in patients with schizophrenia. Behav. Brain. Res. 2014;272:324–334. doi: 10.1016/j.bbr.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 65.Arango C. Candidate gene associations studies in psychiatry: time to move forward. Eur. Arch. Psychiatr. Clin. Neurosci. 2017;267:1–2. doi: 10.1007/s00406-016-0765-7. [DOI] [PubMed] [Google Scholar]
- 66.Farrell MS, et al. Evaluating historical candidate genes for schizophrenia. Mol. Psychiatr. 2015;20:555–562. doi: 10.1038/mp.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zou Z, Liu C, Che C, Huang H. Clinical genetics of Alzheimer’s disease. Biomed. Res. Int. 2014;2014:291862. doi: 10.1155/2014/291862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roussotte FF, et al. Combined effects of Alzheimer risk variants in the CLU and ApoE genes on ventricular expansion patterns in the elderly. J. Neurosci. 2014;34:6537–6545. doi: 10.1523/JNEUROSCI.5236-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson PM, et al. ENIGMA and the individual:predicting factors that affect the brain in 35 countries worldwide. Neuroimage. 2017;145:389–408. doi: 10.1016/j.neuroimage.2015.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyzes: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buckley PF, Pillai A, Howell KR. Brain-derived neurotrophic factor: findings in schizophrenia. Curr. Opin. Psychiatr. 2011;24:122–127. doi: 10.1097/YCO.0b013e3283436eb7. [DOI] [PubMed] [Google Scholar]
- 72.Chen ZY, et al. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J. Neurosci. 2004;24:4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Egan MF, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/S0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 74.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol. Psychiatr. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 75.Stefansson H, et al. Neuregulin 1 and susceptibility to schizophrenia. Am. J. Hum. Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hennah W, Porteous D. The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLoS. ONE. 2009;4:e4906. doi: 10.1371/journal.pone.0004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakata K, et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc. Nat. Acad. Sci. USA. 2009;106:15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blum BP, Mann JJ. The GABAergic system in schizophrenia. Int. J. Neuropsychopharmacol. 2002;5:159–179. doi: 10.1017/S1461145702002894. [DOI] [PubMed] [Google Scholar]
- 79.Straub RE, et al. Allelic variation in GAD1 (GAD67) is associated with schizophrenia and influences cortical function and gene expression. Mol. Psychiatr. 2007;12:854–869. doi: 10.1038/sj.mp.4001988. [DOI] [PubMed] [Google Scholar]
- 80.Chumakov I, et al. Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc. Nat. Acad. Sci. USA. 2002;99:13675–13680. doi: 10.1073/pnas.182412499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kvajo M, Dhilla A, Swor DE, Karayiorgou M, Gogos JA. Evidence implicating the candidate schizophrenia/bipolar disorder susceptibility gene G72 in mitochondrial function. Mol. Psychiatr. 2008;13:685–696. doi: 10.1038/sj.mp.4002052. [DOI] [PubMed] [Google Scholar]
- 82.Castillo PE, Younts TJ, Chavez AE, Hashimotodani Y. Endocannabinoid signaling and synaptic function. Neuron. 2012;76:70–81. doi: 10.1016/j.neuron.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parsons LH, Hurd YL. Endocannabinoid signalling in reward and addiction. Nat. Rev. Neurosci. 2015;16:579–594. doi: 10.1038/nrn4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen J, et al. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am. J. Hum. Genet. 2004;75:807–821. doi: 10.1086/425589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this review are available within the paper and supplementary files.