Abstract
The differentiated cell types of the mature ureter arise from the distal ureteric bud epithelium and its surrounding mesenchyme. Uncommitted epithelial cells first become intermediate cells from which both basal and superficial cells develop. Mesenchymal progenitors give rise to separated layers of adventitial fibrocytes, smooth muscle cells and lamina propria fibrocytes. How progenitor expansion and differentiation are balanced is poorly understood. Here, we addressed the role of retinoic acid (RA) signaling in these programs. Using expression analysis of components and target genes, we show that pathway activity is restricted to the mesenchymal and epithelial progenitor pools. Inhibition of RA signaling in ureter explant cultures resulted in tissue hypoplasia with a relative expansion of smooth muscle cells at the expense of lamina propria fibroblasts in the mesenchyme, and of superficial cells at the expense of intermediate cells in the ureteric epithelium. Administration of RA led to a slight reduction of smooth muscle cells, and almost completely prevented differentiation of intermediate cells into basal and superficial cells. We identified cellular programs and transcriptional targets of RA signaling that may account for this activity. We conclude that RA signaling is required and sufficient to maintain mesenchymal and epithelial progenitors in early ureter development.
Introduction
The mammalian ureter is a tubular organ dedicated to the efficient removal of the urine from the renal pelvis to the bladder. Its outer mesenchymal coat is highly flexible and peristaltically active due to a three-layered organization of an outer fibroblastic tunica adventitia, a medial accumulation of smooth muscle cells (SMCs) and an inner lamina propria with fibroblasts. The inner epithelial compartment, the urothelium, is a three-tiered dilatable yet sealing tissue with a single layer of cuboidal basal cells (B-cells), one or two layers of similarly shaped intermediate cells (I-cells), and a luminal layer of large squamous superficial cells (S-cells)1.
The ordered array of differentiated cell types of the mature ureter arises by a complex program of patterning, proliferation and differentiation from epithelial and mesenchymal precursor cells in early embryogenesis. In the mouse, a diverticulum of the nephric duct emerges around E10.5 at the level of the future hindlimbs and invades the adjacent mesenchymal tissue. While the proximal aspect of the bud and the surrounding mesenchyme will form the different tissues of the kidney, the distal cell populations are specified towards a ureteric fate. From E12.5 to E14.5, the ureteric mesenchyme is subdivided into an inner and outer region that will differentiate from E15.5 onwards in a proximal to distal wave into SMCs and adventitial fibroblasts, respectively. The lamina propria can be recognized from E16.5 onwards as a cell-sparse layer in between the ureteric epithelium and the SMCs. The ureteric epithelium remains single-layered and undifferentiated until E14.5. Around this stage, I-cell markers are switched on and stratification occurs. At E16.5, the luminal layer of epithelial cells starts S-cell differentiation while individual cells of the basal layer initiate B-cell differentiation. Around birth, three distinct layers with differentiated B-, I- and S-cells can be distinguished2.
While the signals and factors that maintain the precursor character of the ureteric tissues have remained enigmatic, embryological and genetic experiments have shown that mesenchymal and epithelial differentiation is coupled by trans-acting signaling activities. Epithelial WNT signals are required for SMC differentiation while epithelial SHH induces a mesenchymal FOXF1-BMP4 module to direct SMC and urothelial differentiation3–6.
Retinoic acid (RA) signaling is a conserved paracrine pathway with diverse functions in cellular proliferation and differentiation programs both in embryogenesis and in adult tissue homeostasis7,8. RA is locally produced from dietary retinol (vitamin A) by two consecutive oxidation steps catalyzed by retinol and retinaldehyde dehydrogenases8. RA binds to nuclear RA receptors (RARs) that form heterodimers with retinoid X receptors (RXRs) which triggers the exchange of co-repressor against coactivator complexes to activate the expression of target genes9,10. Previous studies indicated diverse roles of RA signaling in the excretory system. RA from the pericloacal mesenchyme regulates ureter-bladder connectivity by inducing apoptosis of the most distal aspect of the nephric duct11. RA signaling is active in the renal stroma to regulate branching morphogenesis12,13. Studies in the bladder have recently uncovered a role for the signal in urothelial specification14.
Here, we analyze the functional involvement of RA signaling in the development of the murine ureter. We uncover a critical role for this pathway in the maintenance of progenitor cells in the early ureter and identify RA-responsive genes that may account for this function.
Results
RA signaling is active in early ureter development
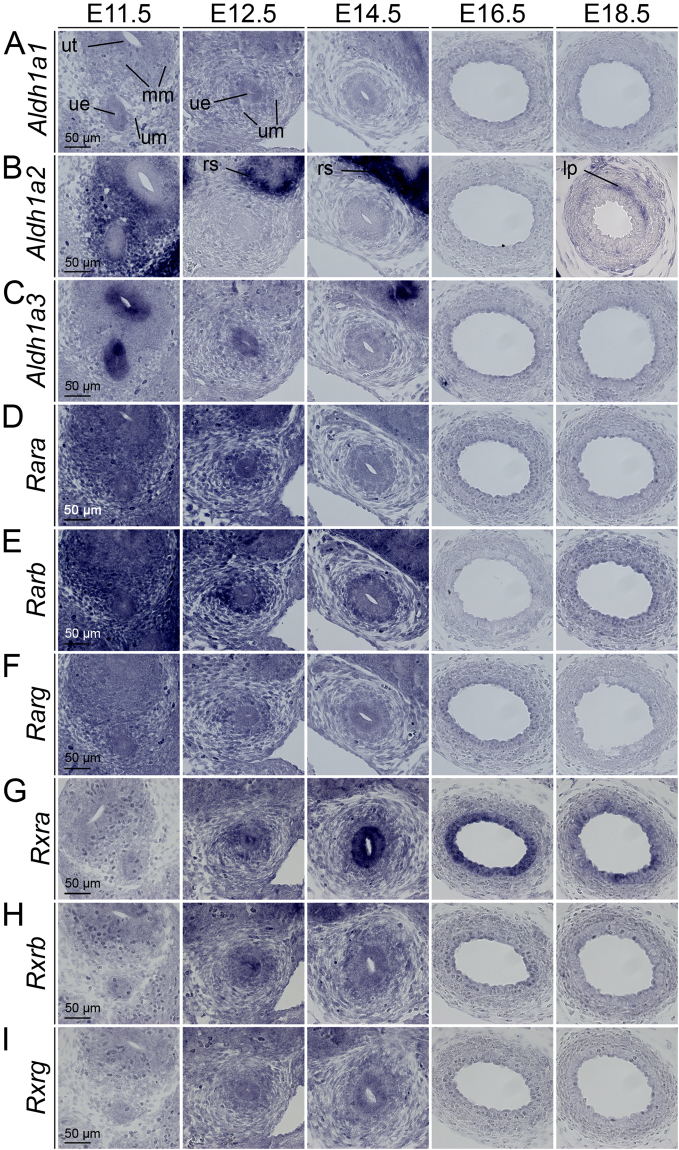
To determine the spatiotemporal profile of RA signaling in ureter development, we first analyzed by in situ hybridization on proximal ureter sections the expression of genes responsible for the local production of RA from retinaldehyde (Aldh1a1, Aldh1a2, Aldh1a3). Aldh1a2 was found throughout the ureteric mesenchyme at E11.5. Expression was absent in the proximal ureter from E12.5 to E16.5 but was reactivated in the lamina propria at E18.5 (Fig. 1A). Analysis of whole urogenital systems showed that Aldh1a2 expression persisted in the mesenchyme of the distal most aspect of the ureter close to the forming bladder (Supplementary Fig. S1). Additional expression of Aldh1a2 was found in the renal stroma (Fig. 1A, Supplementary Fig. S1 and data not shown). Aldh1a3 expression was restricted to the epithelium of the nephric duct and the ureteric bud at E11.5. Expression was maintained in the renal collecting duct system but was lost in the ureter except its distal most aspect at subsequent stages (Fig. 1A–C, Supplementary Fig. S1). We next screened for expression of genes encoding RARs (Rara, Rarb, Rarg) and RXRs (Rxra, Rxrb, Rxrg) that account for transcriptional activation of target genes upon RA binding. Expression of all three Rar genes was found in the ureteric epithelium and mesenchyme at E11.5 and E12.5. Expression of Rarb, a direct transcriptional target of this pathway15, was maintained at E14.5, particularly in the inner mesenchymal domain whereas expression of Rara and Rarg was down-regulated at this stage (Fig. 1D–F). Expression of all three Rxr genes was detected at E11.5 to E14.5 both in the ureteric epithelium and mesenchyme. Rxra was increased in the ureteric epithelium at E14.5 and was maintained in this domain until E18.5 (Fig. 1G–I). Together, this analysis shows that RA is synthesized in both tissue compartments of the ureter at E11.5, and that RA signaling occurs therein until E14.5, i.e. prior to the onset of ureteric cell differentiation.
Figure 1.
RA signaling is active in early ureter development. In situ hybridization analysis on transverse sections of the proximal ureter of wild-type embryos for expression of genes encoding RA synthesizing genes (A–C), RA receptors (D–F) and retinoid X receptors (G–I). Stages and probes are as indicated. For explanations see main text. lp, lamina propria; mm, metanephrogenic mesenchyme; rs, renal stroma; ue, ureteric epithelium; um, ureteric mesenchyme; ut, ureter tip.
RA signaling controls differentiation of the ureteric mesenchyme
To interrogate the functional involvement of RA signaling in ureter development, we used pharmacological pathway inhibition and activation approaches in explant cultures of kidney and ureter rudiments. Such an approach does not only allow for strong reduction and activation of the pathway, it also provides the possibility to analyze the temporal window of pathway activity with greater ease and less animal consumption compared to a genetic approach that has to consider redundancy of Rar and Rxr gene function. For inhibition of RA signaling, we used the pan RAR antagonist BMS49316 in a concentration of 1 µM after showing that this concentration strongly reduced expression of the RA target gene Rarb in E11.5 explants after 18 h of treatment. (Over-)activation of the pathway was reached by 1 µM RA in the same interval (Supplementary Fig. S2). Notably, similar or identical concentrations of these compounds were previously successfully used in kidney explants and other organ contexts to manipulate RA signaling13,16–18.
We explanted ureter rudiments at E11.5, E12.5, E14.5, E16.5 and E18.5 and cultured them for 14, 12, 10, 8 and 6 days to obtain similar end-points for histological and molecular analyses. We first investigated the effect of RA manipulation on mesenchymal cell types. We have recently shown that the T-box transcription factor gene Tbx18 is expressed in the undifferentiated ureteric mesenchyme, and that the descendants of this expression domain constitute the ureteric mesenchymal wall throughout development and in adulthood19,20. We therefore used mice double heterozygous for a cre knock-in the Tbx18 locus and the Rosa26 mTmG reporter line (Tbx18 cre/+; Rosa26 mTmG/+)21,22 for visualization of the ureteric mesenchyme by GFP expression.
Brightfield microscopical analysis throughout the culture period showed that the ureter elongated and became peristaltically active under all conditions. The layer of strong GFP epifluorescence surrounding the ureteric epithelium appeared decreased under BMS treatment and increased under RA treatment at the end of the culture period of ureter explants at E11.5, E12.5 and E14.5 while no changes were recognized for explants from E16.5 and E18.5 embryos (Supplementary Figs S3–7). Quantification of the thickness of the mesenchymal wall and of its cellularity confirmed a trend for opposing effects of BMS and RA treatment on these parameters which, however, only gained significance for both parameters in BMS treated explants of E12.5 embryos cultured for 12 days (Supplementary Figs S8,9).
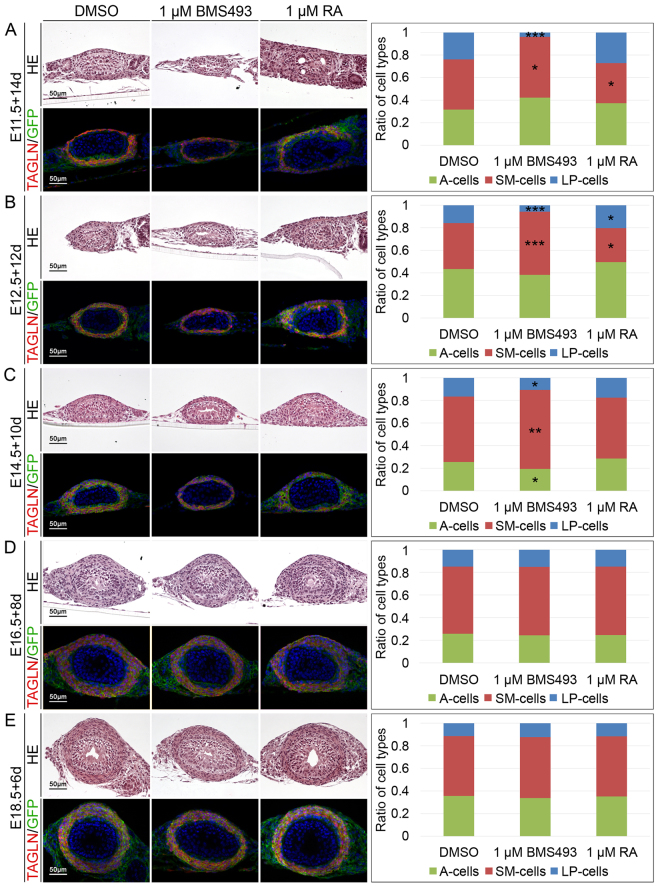
To detect and quantify cell differentiation in the ureteric mesenchyme, we analyzed co-expression of the SMC marker Transgelin (TAGLN) with the GFP reporter by immunofluorescence on sections of ureter explants of Tbx18 cre/+ ; Rosa26 mTmG/+ mice. Adventitial fibroblasts were defined as GFP+TAGLN− outer ring cells, SMCs as GFP+TAGLN+ intermediary cells and lamina propria cells as GFP+TAGLN− inner ring cells of the mesenchymal wall. Inhibition of RA signaling in cultures of E11.5 to E14.5 ureter explants resulted in a relative increase of SMCs largely at the expense of lamina propria cells. At the same stages RA treatment led to a reduction of SMCs and increased lamina formation (most obviously at E12.5). In E16.5 and E18.5 explants, neither BMS nor RA treatment affected the composition of differentiated cell types in the mesenchymal wall of the ureter (Fig. 2, Supplementary Table S1). Hence, RA signaling is required from E11.5 to E14.5 for expansion of the ureteric mesenchyme, and for promotion of lamina fibrocytes at the expense of SMC fates.
Figure 2.
RA signaling controls ureteric mesenchymal differentiation. Hematoxylin and eosin (upper panel) and co-immunofluorescence analysis of GFP and TAGLN (lower panel) on transverse proximal sections of E11.5 + 14d (A), E12.5 + 12d (B), E14.5 + 10d (C), E16.5 + 8d (D) and E18.5 + 6d (E) Tbx18 cre/+; R26 mTmG/+ ureter explants that were treated with DMSO, 1 µM BMS493 or 1 µM RA. The bar graph displays ratios of differentiated cell types that were quantified based on the immunofluorescence analysis. RA signaling acts from E11.5 to E14.5 to maintain lamina propria cells at the expense of SMCs. For numbers and statistics see Supplementary Table S1. *p ≤ 0.01; **p ≤ 0.001; ***p ≤ 0.0001.
RA signaling controls urothelial differentiation
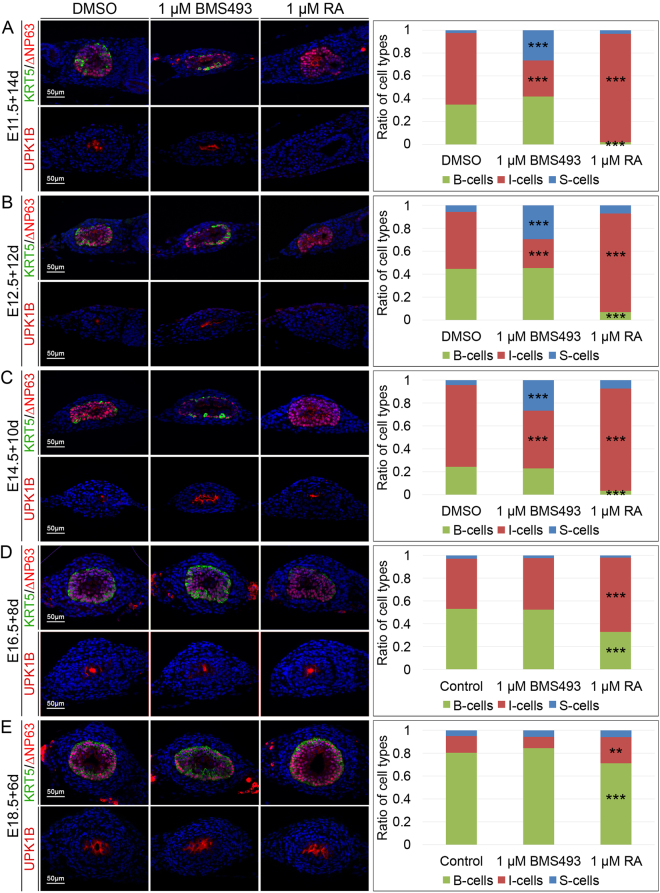
The three cell types of the mature ureteric epithelium can be distinguished by combinatorial expression of the intracytoplasmic protein KRT5, the nuclear protein ∆NP63 and the cell surface protein UPK1B. B-cells are KRT5+∆NP63+UPK1B−, I-cells are KRT5−∆NP63+UPK1B+low and S-cells are KRT5−∆NP63−UPK1B+high 2. Inhibition of RA signaling by BMS493 treatment resulted in E11.5, E12.5 and in E14.5 but not in E16.5 and E18.5 explants in a large expansion of S-cells at the expense of I-cells. Enhanced and prolonged RA signaling by RA administration prevented B-cell differentiation, reduced S-cell differentiation and expanded the pool of I-cells. In E16.5 and E18.5 explants, RA treatment expanded I-cells at the expense of B-cells (Fig. 3, Supplementary Table S2). BMS treatment did not affect tissue thickness and cellularity in any of these explants whereas RA treatment resulted in a small but significant increase in these parameters at E14.5 only (Supplementary Figs S8,9). Given our recent finding that in the ureter I-cells are precursors for both B- and S-cells2, this analysis suggests that RA signaling is dispensable for epithelial expansion but is required from E11.5 to at least E14.5 to prevent premature differentiation of I- into S- and B-cells.
Figure 3.
RA signaling controls urothelial differentiation. Co-immunofluorescence analysis of KRT5 and ∆NP63 (upper panel) and UPK1B (lower panel) on transverse proximal sections of E11.5 + 14d (A), E12.5 + 12d (B), E14.5 + 10d (C), E16.5 + 8d (D) and E18.5 + 6d (E) wildtype ureter explants that were treated with DMSO, 1 µM BMS493 or 1 µM RA. The bar diagram displays ratios of differentiated cell types that were quantified based on the immunofluorescence analysis. RA signaling acts from E11.5 to E14.5 to maintain I-cells at the expense of S-cells. Throughout all stages analyzed ectopic RA prevents the differentiation of B-cells. For numbers and statistics see Supplementary Table S2. **p ≤ 0.001; ***p ≤ 0.0001.
RA signaling has a minor function in proliferation control
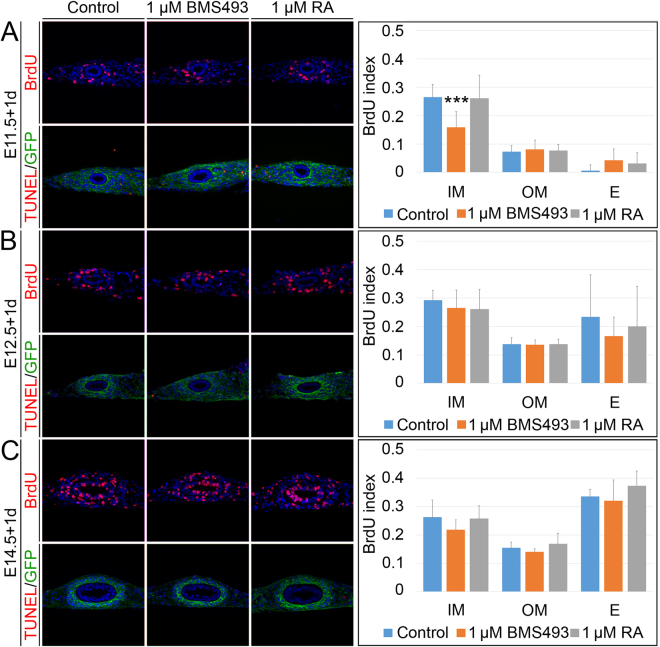
We next addressed a potential function of RA signaling in the proliferation and survival of the epithelial and mesenchymal tissue compartments of the ureter by performing bromodeoxyuridine (BrdU) incorporation assays (Fig. 4A–C, upper row, Supplementary Table S3) and terminal dUTP nick end-labeling (TUNEL) assays (Fig. 4A–C, lower row) on E11.5, E12.5 and E14.5 Tbx18 cre/+ ; R26 mTmG/+ explants that were cultured in the presence of 1 µM BMS493 or 1 µM RA for 1 day. At E11.5, inhibition of RA signaling resulted in a significant decrease of proliferation of the inner mesenchymal compartment of the ureter but left the outer mesenchymal and epithelial cells unaltered. Activation of RA signaling showed no effect on proliferation rates, as did inhibition or activation experiments at E12.5 and E14.5. Moreover, apoptosis was not altered under RA loss- and gain-of-function conditions. We conclude that RA signaling has a minor function in proliferation control at E11.5.
Figure 4.
RA signaling affects proliferation in a minor way. Determination of cellular proliferation by BrdU incorporation assay (upper row) and apoptosis by TUNEL/GFP assay (lower row) on transverse sections of the proximal ureter of E11.5 (A), E12.5 (B) and E14.5 (C) Tbx18 cre/+ ; R26 mTmG/+ explants that were cultured in the presence of DMSO, 1 µM BMS493 or 1 µM RA for 1 day. RA signaling is required to maintain proliferation of inner mesenchymal cells at E11.5. The bar diagrams display the BrdU incorporation indices in arbitrarily defined compartments of the ureter under given conditions. For numbers and statistics see Supplementary Table S3. ***p ≤ 0.0001. E, epithelium; IM, inner mesenchyme; OM, outer mesenchyme.
RA signaling prevents precocious differentiation of ureteric progenitors
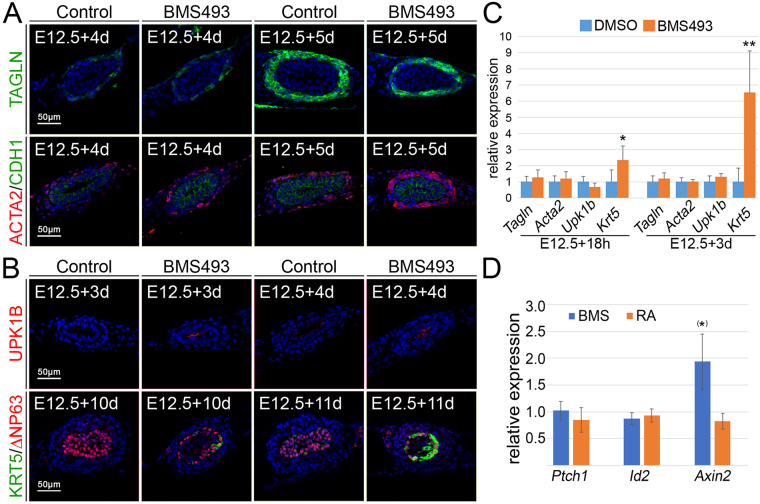
To further test whether RA signaling contributes to the maintenance of precursor cells, we explanted E12.5 ureters and cultured them for different intervals to score the onset of cell differentiation in the mesenchymal and epithelial compartments. SMC differentiation, as analyzed by TAGLN (Fig. 5A, upper row) and ACTA2 (Fig. 5A, lower row) immunofluorescence, was initiated after 4 days in control cultures. TAGLN showed a comparable expression in BMS493 treated cultures after 4 and 5 days, but ACTA2 expression was considerably stronger under these conditions at both time-points. Expression of UPK1B and KRT5 indicated onset of S-cell and B-cell differentiation after 4 and 11 days, respectively, in control cultures. Both markers were expressed 1 day earlier in BMS493 treated cultures (Fig. 5B, upper and lower row). To confirm these findings in an independent assay, we performed RT-PCR analysis on RNA isolated from E12.5 ureters treated for 18 h and 3 days, respectively, with 1 µM BMS. After 18 h, Upk1b was slightly decreased, expression of Tagln and Acta2 was weakly but not significantly increased, levels of Krt5 mRNA were strongly and significantly increased. After three days, there was a weak trend for increased Upk1b, Tagln and Acta2 expression. Krt5 expression was even more enhanced under BMS treatment (Fig. 5C, Supplementary Table S4).
Figure 5.
RA signaling prevents precocious differentiation in the ureter. (A) Immunofluorescence analysis for TAGLN (upper row) and co-immunofluorescence analysis of ACTA/CDH1 (lower row) on E12.5 ureter explants that were cultured for 3 and 4 days in presence of DMSO or 1 µM BMS493 shows that SMCs markers are slightly enhanced in their expression when RA signaling is abrogated. (B) Immunofluorescence analysis for UPK1B (upper row) and co-immunofluorescence analysis of KRT5/∆NP63 (lower row) on E12.5 ureter explants that were cultured for 3 and 4 days (UPK1B) or 10 and 11 days (KRT5/∆NP63) in the presence of DMSO or 1 µM BMS493. Markers for S-cells (UPK1B) and B-cells (KRT5) are prematurely expressed when RA signaling is inhibited. (C) RT-PCR analysis of expression of the SMC marker Tagln and Acta2, the S-cell marker Upk1b and the B-cell marker Krt5 in ureters treated for 18 h and 3 days with DMSO or the RA signaling inhibitor BMS493. Note the strong increase of expression of Krt5 at both stages. (D) RT-PCR analysis of expression of the SHH target gene Ptch1, of the WNT target gene Axin2 and the BMP target gene Id2 in ureters treated for 18 h with DMSO, 1 µM BMS493 or 1 µM RA. Note increase of expression of Axin2 almost reaching significance as indicated by (*). For numbers and statistics (for C and D) see Supplementary Table S4 and 5. *p ≤ 0.01; **p ≤ 0.001.
Previous work implicated SHH and BMP4 in both epithelial and mesenchymal differentiation, and WNT signaling in mesenchymal cell differentiation in ureter development5,6,23. To analyze whether dysregulation of any of these pathways underlies the observed premature cytodifferentiation when RA signaling is inhibited, we determined expression of the bona fide target genes of these pathways, Ptch1, Axin2 and Id2 24–26, in E12.5 ureters cultured for 18 h with BMS493. While Ptch1 and Id2 expression was unchanged, Axin2 expression was two-fold upregulated. Addition of 1 µM RA led to a slight but not significant reduction of expression of all of these genes (Fig. 5D, Supplementary Table S5). Together, these findings suggest a role for RA signaling in suppressing differentiation of epithelial progenitors into S- and most strongly into B-cells, and to attenuate SMC differentiation of mesenchymal progenitors. Since canonical WNT signaling acts in the mesenchyme to induce SMCs from precursors23, its increased activity (as indicated by increased Axin2 expression) may at least partly contribute to premature SMC differentiation when RA signaling is inhibited.
Characterization of the RA-responsive transcriptome in early ureter development
To get more insights into the phenotypic changes and the underlying molecular programs that are controlled by RA signaling in the early ureter, we treated E12.5 ureters with 1 µM BMS493 or 1 µM RA for 18 h and performed microarray analysis of global differential gene expression compared to untreated controls.
We first addressed our hypothesis that RA signaling maintains the progenitor status in the mesenchymal and epithelial compartments of the ureter by examining transcripts that were negatively regulated by RA signaling. Using an intensity threshold of 100, we identified in three independent pools of treated and untreated ureters a set of 554 genes that were consistently more than 1.2 fold increased in their expression upon BMS treatment and 612 genes that were consistently more than 1.2 fold decreased in RA treated ureters yielding a set of 261 genes that were negatively regulated by RA signaling (Supplementary Fig. S10A, Supplementary Tables S6-8). GO enrichment analysis did not provide conclusive insight into causative changes of cellular or molecular pathways (Supplementary Table S9). Among the most affected transcripts were two cytokeratins which are known to be expressed in urothelial B-cells, namely Krt6a (4.2x) and Krt5 (2.1x)2,27 as well as transcripts encoding for structural components of SMCs, including Actg2 (1.6x), Acta1 (1.5x), Actc1 (1.4x) and Tagln (1.3x) (Supplementary Fig. S10B), providing further evidence for a functional requirement of RA signaling in preventing precocious cell differentiation in the ureter. To validate the RA-responsive expression of top-deregulated genes and to characterize their spatial confinement we performed RNA in situ hybridization analysis on explants of E12.5 kidney/ureter rudiments cultured for 1 day in the presence of 1 µM BMS493. We found weak upregulation of Rgcc, Serpine1 and Dkk1 in the ureteric epithelium (Supplementary Fig. S10C).
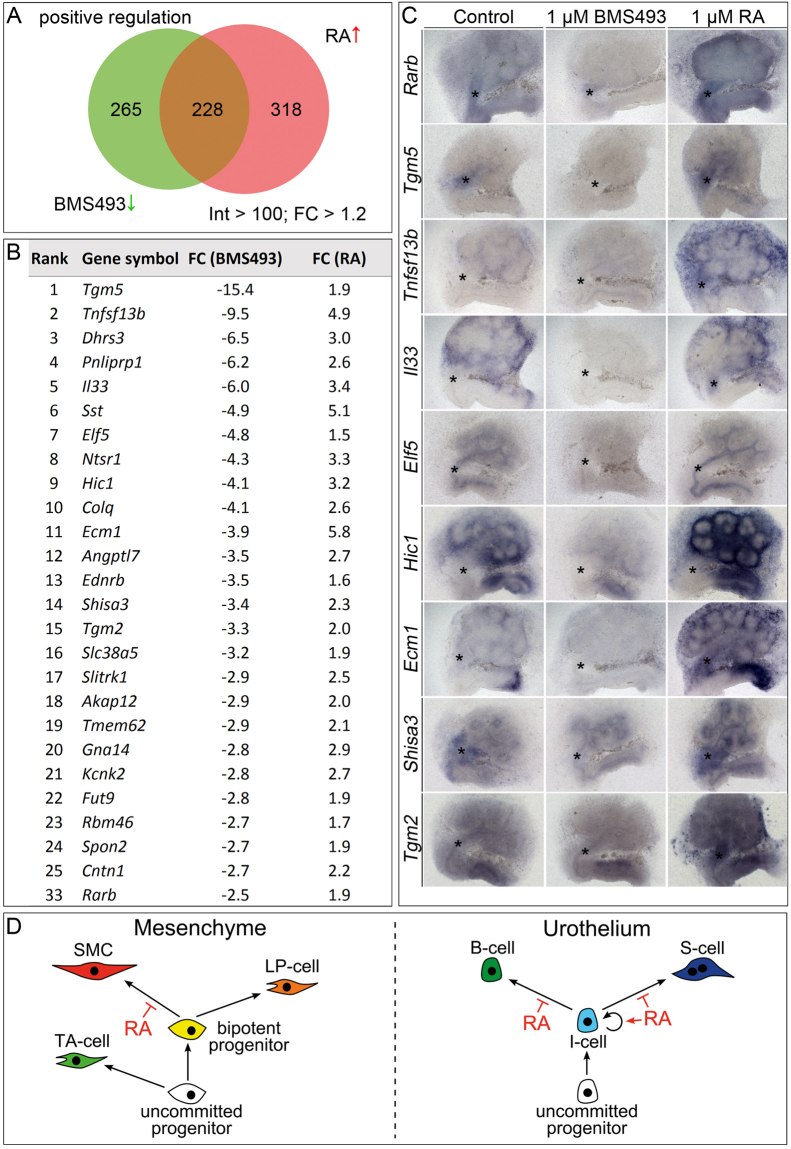
To identify potential target genes of the RAR/RXR transcriptional activator that may account for this function, we next focused on positively regulated transcripts. Using the same parameters as above (intensity > 100, fold change > 1.2 in each experiment), we identified 493 transcripts that decreased upon BMS treatment and 546 genes that were increased in their expression in RA treated ureters yielding a set of 228 genes that are positively regulated by RA signaling (Fig. 6A, Supplementary Tables S10–12). GO enrichment analysis reflected the diversity of roles in which RA signaling plays a role in the organisms but did not provide conclusive evidence about major or causative changes in pathways or factors (Supplementary Table S13).
Figure 6.
Identification of genes positively regulated by RA signaling in microarray experiments. Summary of the results from the microarray analysis of E12.5 ureters explanted and treated with DMSO or 1 µM BMS493 or 1 µM RA for 18 h filtered with an intensity (Int) threshold of 100 and a fold change (FC) cut-off of 1.2. (A) The Venn diagram displays transcripts which were positively regulated by RA signaling, i.e. down-regulated upon BMS493 treatment (493, green) or up-regulated upon RA treatment (546, red). The intersection shows the common group of positively regulated transcripts (228). (B) Top 25 of the common positively regulated genes were ranked according to their FC down-regulation upon BMS493 treatment. (C) In situ hybridization analysis of expression of selected genes which were positively regulated by RA signaling on E11.5 kidney explant cultures, treated with 1 µM BMS493 or 1 µM RA for 1 day. (D) Schematic illustration of RA signaling function in the maintenance of ureteric progenitors. In the ureteric mesenchyme (left) uncommitted progenitors diversify into adventitial fibrocytes (AF-cells) and bipotent progenitors of SMCs and lamina propria fibrocytes (LP-cells). RA specifically acts in the bipotent progenitors to prevent precocious differentiation towards the SMC lineage. In the ureteric epithelium (right) uncommitted progenitors give rise to I-cells which are maintained by RA signaling by preventing precocious differentiation towards the B- and S-cell lineage.
Among the most deregulated transcripts were the well-established RA target genes Dhrs3 and Rarb (Fig. 6B)15,28. To validate the RA-responsive expression of candidate genes and to characterize their spatial confinement we performed RNA in situ hybridization analysis on explants of E11.5 kidney and ureter rudiments cultured for 1 day in the presence of 1 µM BMS493 or 1 µM RA (Fig. 6C). As previously shown, Rarb expression in the ureteric mesenchyme and renal stroma was strongly RA dependent. To our surprise, some genes including Tnfsf13b, Il33, Ecm1 and Hic1 were expressed in the renal stroma but not in the ureter of control explants. BMS493 treatment abolished expression of these genes, while RA treatment enhanced stromal expression and induced expression in the ureteric mesenchyme. Expression of Tgm5 and Shisa3 was strong in the ureteric mesenchyme of control explants. BMS493 abolished, RA treatment slightly enhanced this expression. The weak expression of Shisa3 in the ureteric epithelium and renal collecting ducts was not RA-responsive. Tgm2 showed a weak and ubiquitous expression in control explants which was slightly reduced upon BMS493 treatment. Administration of RA augmented the overall expression level of Tgm2 and induced a strong expression in the ureteric mesenchyme. Expression of Elf5 was restricted to the epithelium of the nephric duct, the ureter and the collecting ducts and was strongly reduced and enhanced upon BMS493 and RA treatment, respectively.
Tbx18, Tshz3, Six1 and Sox9 that were previously characterized as transcriptional regulators of SMC differentiation19,29–31 were not represented in the set of genes positively or negatively regulated by RA signaling. Bmp4, the transcriptional target of its signaling activity Id2, Shh and its transcriptional target Ptch1 were also unchanged arguing against the fact that RA signaling function is mediated by altered activity of these factors or pathways.
Discussion
Ureteric SMCs arise in a temporally and spatially tightly controlled manner from the adluminal mesenchymal cell layer. Differentiation starts around E15.5 in the proximal region of the ureter and progresses distally within the following day2. The program depends on epithelial signals, SHH and members of WNT family, that activate autocrine BMP4 signaling in the mesenchymal compartment3,5,6,23,32. Intriguingly all of these signaling pathways are active from E11.5 onwards but culminate at E14.5 only in transcriptional activation of Myocd, which encodes the master regulator of the SMC differentiation program33. While it is conceivable that the delayed expression of Myocd results from a gradual build-up of positive signaling inputs, an alternative scenario features the presence of inhibitory activities in the undifferentiated ureteric mesenchyme.
Our findings suggest that RA constitutes such an inhibitory signal that acts in the undifferentiated ureteric mesenchyme to temporally and spatially restrict differentiation, possibly through modulation of the WNT signaling pathway. First, a comprehensive expression analysis of components and target genes of the pathway revealed that RA signaling is active in the ureteric mesenchyme from E11.5 to E14.5, i.e. until the onset of SMC differentiation. Second, RA synthesizing enzyme genes Aldh1a2 and Aldh1a3 were transiently expressed in the proximal ureter but maintained in its distal aspect in line with the delayed onset of SMC differentiation at this site2. An independent function of this distal expression domain for bladder insertion of the distal ureter can, however, not be excluded11. Third, our pharmacological loss- and gain-of-function experiments delimited the functional requirement of RA to the interval E11.5 to E14.5. Furthermore, a relative expansion of SMCs at the expense of lamina propria fibrocytes under RA loss-of-function conditions suggests that the mesenchymal progenitors of the ureter prematurely and preferentially differentiated towards the SMC lineage. This hypothesis was further supported by the relative expansion of lamina propria fibrocytes under RA gain-of-function conditions. Importantly, the analysis of differentiation markers revealed a precocious differentiation of SMCs when RA signaling was inhibited.
Microarray analysis characterized the RA-responsive transcriptome in the early ureter. Interestingly, a considerable number of identified genes including Tgm5, Tnfsf13b, Il33, Elf5 and Ecm1 were previously reported to be RA-responsive in a similar assay performed for the renal stroma of the embryonic kidney13. Tnfsf13b and Il33 activate NFκB signaling and have been reported to control renal branching morphogenesis, while Ecm1 exerts a similar function via control of GNDF/RET signaling12,13. The present study newly identified the RA-responsive expression of two genes encoding antagonists of the WNT signaling pathway in the ureter. Hic1 encodes for a transcriptional repressor that attenuates canonical WNT signaling by sequestration of TCF-4 and CTNNB1 in specific nuclear compartments34. Shisa3 encodes a member of the SHISA family of proteins that inhibit WNT signaling by retention of WNT receptors in the endoplasmic reticulum and/or by promoting CTNNB1 degradation35,36. Importantly, we observed an upregulation of Axin2 expression, i.e. of canonical WNT signaling in ureter explants treated with BMS493. Thus, a negative modulation of mesenchymal WNT signaling by RA represents a potential molecular mechanism to control or at least partly contribute to the timing of ureteric SMC differentiation.
Since uncommitted mesenchymal progenitors first diversify into adventitial fibrocytes and bipotent progenitors of SMCs and lamina propria fibrocytes, we conclude that RA is specifically required in the bipotent progenitor to prevent precocious differentiation towards the SMC lineage. The late re-expression of Aldh1a2 in the lamina propria suggests that this cell population constitutes a SMC progenitor in homeostasis and regeneration (Fig. 6C).
Our previous work has shown that I-cells arise in the ureteric epithelium at E13.5 and differentiate into S-cells and B-cells at E15.5 and E16.5, respectively2. How this strict temporal sequence of differentiation is molecularly regulated has remained elusive. Studies in the developing and regenerating bladder urothelium reported a function of RA signaling in urothelial specification of endodermal precursors14. Given the distinct developmental origin of the ureter urothelium from the intermediate mesoderm, it has remained unclear whether this finding also accounts for the ureter. The present study suggests that RA signaling in the ureter urothelium is involved in the temporal control of cellular differentiation rather than in the specification of progenitor cells. First, pharmacological inhibition of RA signaling in ureter explant cultures between E11.5 and E14.5 resulted in a relative expansion of S-cells at the expense of I-cells. Moreover, compared to control specimens S-cells and B-cells differentiated one day earlier under RA loss-of-function conditions. Second, pharmacological overactivation of RA signaling between E11.5 and E14.5 reduced S-cell differentiation, completely blocked B-cell differentiation and resulted in an expansion of I-cells. Even at later time points was excessive RA sufficient to decrease the abundance of B-cells and to expand I-cells.
A negative effect of RA signaling on epithelial differentiation and especially on the expression of cytokeratins has been reported in the context of keratinocytes, tracheal and bronchial epithelial cells and the salivary gland epithelium37–39. In vitro studies of the Krt5 and Krt14 promotor identified an unconventional mechanism of RA-mediated direct transcriptional repression that involves ligand bound homodimers of RARs40. The precocious upregulation of several cytokeratin genes under RA loss-of-function conditions indicates that such a mechanism may be utilized to control the timing of B-cell differentiation in the urothelium. Alternatively, RA may maintain urothelial progenitors indirectly via intermediate factors. Our transcriptome analysis revealed RA-dependent expression of Elf5 in the undifferentiated ureteric epithelium. The paralogous gene Elf3 has been reported to activate urothelial differentiation downstream of PPARG signaling41. Recent in vitro studies identified an enrichment of ELF5 binding sites in regulatory elements of urothelial differentiation genes42. Interestingly, misexpression of Elf5 in the lung epithelium interferes with terminal differentiation43. Moreover, Elf5 regulates stem cell self-renewal and counteracts precocious differentiation in the trophoblast44,45. It is conceivable that, in contrast to Elf3, Elf5 negatively regulates the urothelial differentiation program. Our molecular analysis uncovered that RA signaling suppresses the expression of Dkk1 which encodes a WNT signaling antagonists in the ureteric epithelium46. Since WNT signaling is thought to be absent from E12.5 in the ureteric epithelium23, the significance of this molecular regulation remains unclear. We conclude that RA signaling acts specifically in urothelial progenitors to prevent precocious differentiation of B- and S-cells and to control progenitor self-renewal and maintenance in development and possibly in homeostasis (Fig. 6D).
Materials and Methods
Animals
Gt(ROSA)26So tm4(ACTB-tdTomato-EGFP)Luo (synonym: R26 mTmG)21 and Tbx18 tm4(cre)Akis (synonym: Tbx18 cre)22 mouse lines were maintained on an NMRI outbred background. Embryos for gene expression and microarray analysis were derived from matings of NMRI wildtype mice. Tbx18 cre/+ ; R26 mTmG/+ embryos were obtained from mating Tbx18 cre/+ males with R26 mTmG/mTmG females. For timed pregnancies, vaginal plugs were checked in the morning after mating, and noon was defined as embryonic day (E) 0.5. Embryos and urogenital systems were dissected in PBS. Specimens were fixed in 4% PFA/PBS, transferred to methanol and stored at −20 °C prior to immunofluorescence or in situ hybridization analyses. PCR genotyping was performed on genomic DNA prepared from yolk sac or tail biopsies. All animal work conducted for this study was approved by A. Bleich, state head of the animal facility at Medizinische Hochschule Hannover and performed according to European and German legislation.
Organ cultures
Cultures of embryonic ureter and kidney explants were performed as described2. BMS493 (#3509, Tocris) or RA (#0695, Tocris) were dissolved in DMSO and added to the medium at a final concentration of 1 µM. Culture medium was replaced every day.
Histological analysis
Embryos, urogenital systems or explant cultures were paraffin-embedded and the proximal ureter was sectioned to 5 µm. Hematoxylin and eosin staining was performed according to standard procedures.
In situ hybridization analysis
Whole-mount in situ hybridization was performed following a standard procedure with digoxigenin-labeled antisense riboprobes47. Stained specimens were transferred in 80% glycerol prior to documentation. In situ hybridization on 10-µm paraffin sections was performed essentially as described48. For each marker, at least three independent specimens were analyzed.
Immunofluorescent detection of antigens
Immunofluorescent analysis on 5-µm paraffin sections was done as described2. At least three embryos of each genotype were used for each analysis.
Cell proliferation and apoptosis assays
Cell proliferation rates in explant cultures (n = 3 per condition) were investigated by the detection of incorporated BrdU on 5 µm paraffin sections similar to published protocols49. Explant cultures where incubated for 2 h with 3.3 µg/ml BrdU in the culture medium. For each specimen 12 sections of the proximal ureter were assessed. The BrdU-labeling index was defined as the number of BrdU-positive nuclei relative to the total number of nuclei as detected by 4′,6-diamidino-2-phenylindole (DAPI) counterstaining in arbitrarily defined compartments of the ureter. Data were expressed as mean ± standard deviation. Apoptosis was analyzed on 5 µm paraffin sections using the ApopTag Plus Fluorescein In Situ Apoptosis Detection Kit (Chemicon).
Quantitative reverse transcription PCR
Three independent pools of 10 E12.5 ureters were cultured with DMSO or 1 μM BMS493 for 18 h or 3 days. Total RNA was extracted with peqGOLD RNApure (PeqLab) and first-strand cDNA synthesis was performed with RevertAid reverse transcriptase (Fermentas). For quantitative PCR, the number of cycles was adjusted to the mid-logarithmic phase. The experiments were performed with three biological and two technical replicates. Quantification was performed with ImageJ50. Oligos and PCR conditions are provided upon request.
Statistical analysis
Statistical analysis was performed using the two-tailed Student’s t-test. Data were expressed as mean ± standard deviation. For relative analyses wildtype values were set to 1. Differences were considered significant with a P-value below 0.05 (p < 0.05, *), highly significant (p ≤ 0.005, **) and extremely significant (p ≤ 0.0005, ***).
Microarray
Three independent pools of 50 E12.5 ureters were cultured with DMSO or 1 μM BMS493 or 1 μM RA for 18 h. Total RNA was extracted with peqGOLD RNApure (PeqLab) and was sent to the Research Core Unit Transcriptomics of Hannover Medical School where RNA was Cy3-labeled and hybridized to Agilent Whole Mouse Genome Oligo v2 (4 × 44 K) Microarrays. To identify differentially expressed genes, normalized expression data was filtered using Excel based on an intensity threshold of 100 and a more than 1.2 fold change in all pools.
Image documentation
Sections and organ cultures were photographed using a Leica DM5000 microscope with Leica DFC300FX digital camera or a Leica DM6000 microscope with Leica DFC350FX digital camera. Figures were prepared with Adobe Photoshop CS4.
Data Availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
This work was supported by a grant from the German Research Foundation (DFG KI728/9-1) to A.K.
Author Contributions
Study concept and design, T.B. and A.K. Acquisitions of data, T.B., M.L., A.-C.W., T.H.L., M.-O.T. Drafting of the manuscript T.B. and A.K. Critical revision of manuscript, all authors. Statistical analysis, T.B., T.H.L., M.-O.T.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14790-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Velardo, J. T. In The ureter (ed. H. Bergman. (Springer-Verlag, 1981).
- 2.Bohnenpoll T, et al. Diversification of Cell Lineages in Ureter Development. J Am Soc Nephrol. 2017;28:1792–1801. doi: 10.1681/ASN.2016080849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J, Carroll TJ, McMahon AP. Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development. 2002;129:5301–5312. doi: 10.1242/dev.129.22.5301. [DOI] [PubMed] [Google Scholar]
- 4.Miyazaki Y, Oshima K, Fogo A, Ichikawa I. Evidence that bone morphogenetic protein 4 has multiple biological functions during kidney and urinary tract development. Kidney Int. 2003;63:835–844. doi: 10.1046/j.1523-1755.2003.00834.x. [DOI] [PubMed] [Google Scholar]
- 5.Mamo TM, et al. BMP4 uses several different effector pathways to regulate proliferation and differentiation in the epithelial and mesenchymal tissue compartments of the developing mouse ureter. Hum Mol Genet. 2017;26:3553–3563. doi: 10.1093/hmg/ddx242. [DOI] [PubMed] [Google Scholar]
- 6.Bohnenpoll T, et al. A SHH-FOXF1-BMP4 signaling axis regulating growth and differentiation of epithelial and mesenchymal tissues in ureter development. PLoS Genet. 2017;13:e1006951. doi: 10.1371/journal.pgen.1006951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duester G. Retinoid signaling in control of progenitor cell differentiation during mouse development. Semin Cell Dev Biol. 2013;24:694–700. doi: 10.1016/j.semcdb.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhinn M, Dolle P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 9.Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res. 2002;43:1773–1808. doi: 10.1194/jlr.R100015-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Rochette-Egly C, Germain P. Dynamic and combinatorial control of gene expression by nuclear retinoic acid receptors (RARs) Nucl Recept Signal. 2009;7:e005. doi: 10.1621/nrs.07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batourina E, et al. Distal ureter morphogenesis depends on epithelial cell remodeling mediated by vitamin A and Ret. Nat Genet. 2002;32:109–115. doi: 10.1038/ng952. [DOI] [PubMed] [Google Scholar]
- 12.Paroly SS, et al. Stromal protein Ecm1 regulates ureteric bud patterning and branching. PLoS One. 2013;8:e84155. doi: 10.1371/journal.pone.0084155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takayama M, Miyatake K, Nishida E. Identification and characterization of retinoic acid-responsive genes in mouse kidney development. Genes Cells. 2014;19:637–649. doi: 10.1111/gtc.12163. [DOI] [PubMed] [Google Scholar]
- 14.Gandhi D, et al. Retinoid signaling in progenitors controls specification and regeneration of the urothelium. Dev Cell. 2013;26:469–482. doi: 10.1016/j.devcel.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendelsohn C, Ruberte E, LeMeur M, Morriss-Kay G, Chambon P. Developmental analysis of the retinoic acid-inducible RAR-beta 2 promoter in transgenic animals. Development. 1991;113:723–734. doi: 10.1242/dev.113.3.723. [DOI] [PubMed] [Google Scholar]
- 16.Chazaud C, Dolle P, Rossant J, Mollard R. Retinoic acid signaling regulates murine bronchial tubule formation. Mech Dev. 2003;120:691–700. doi: 10.1016/S0925-4773(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 17.Desai TJ, Malpel S, Flentke GR, Smith SM, Cardoso WV. Retinoic acid selectively regulates Fgf10 expression and maintains cell identity in the prospective lung field of the developing foregut. Dev Biol. 2004;273:402–415. doi: 10.1016/j.ydbio.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 18.Fernandes-Silva, H. et al. Retinoic acid regulates avian lung branching through a molecular network. Cell Mol Life Sci (2017). [DOI] [PMC free article] [PubMed]
- 19.Airik R, Bussen M, Singh MK, Petry M, Kispert A. Tbx18 regulates the development of the ureteral mesenchyme. J Clin Invest. 2006;116:663–674. doi: 10.1172/JCI26027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohnenpoll T, et al. Tbx18 expression demarcates multipotent precursor populations in the developing urogenital system but is exclusively required within the ureteric mesenchymal lineage to suppress a renal stromal fate. Dev Biol. 2013;380:25–36. doi: 10.1016/j.ydbio.2013.04.036. [DOI] [PubMed] [Google Scholar]
- 21.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 22.Trowe MO, et al. Loss of Sox9 in the periotic mesenchyme affects mesenchymal expansion and differentiation, and epithelial morphogenesis during cochlea development in the mouse. Dev Biol. 2010;342:51–62. doi: 10.1016/j.ydbio.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Trowe MO, et al. Canonical Wnt signaling regulates smooth muscle precursor development in the mouse ureter. Development. 2012;139:3099–3108. doi: 10.1242/dev.077388. [DOI] [PubMed] [Google Scholar]
- 24.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 25.Jho EH, et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999;274:19838–19845. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- 27.Choi W, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng L, Hernandez RE, Waxman JS, Yelon D, Moens CB. Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Dev Biol. 2010;338:1–14. doi: 10.1016/j.ydbio.2009.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caubit X, et al. Teashirt 3 is necessary for ureteral smooth muscle differentiation downstream of SHH and BMP4. Development. 2008;135:3301–3310. doi: 10.1242/dev.022442. [DOI] [PubMed] [Google Scholar]
- 30.Nie X, Sun J, Gordon RE, Cai CL, Xu PX. SIX1 acts synergistically with TBX18 in mediating ureteral smooth muscle formation. Development. 2010;137:755–765. doi: 10.1242/dev.045757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Airik R, et al. Hydroureternephrosis due to loss of Sox9-regulated smooth muscle cell differentiation of the ureteric mesenchyme. Hum Mol Genet. 2010;19:4918–4929. doi: 10.1093/hmg/ddq426. [DOI] [PubMed] [Google Scholar]
- 32.Wang GJ, Brenner-Anantharam A, Vaughan ED, Herzlinger D. Antagonism of BMP4 signaling disrupts smooth muscle investment of the ureter and ureteropelvic junction. J Urol. 2009;181:401–407. doi: 10.1016/j.juro.2008.08.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci USA. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valenta T, Lukas J, Doubravska L, Fafilek B, Korinek V. HIC1 attenuates Wnt signaling by recruitment of TCF-4 and beta-catenin to the nuclear bodies. EMBO J. 2006;25:2326–2337. doi: 10.1038/sj.emboj.7601147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto A, Nagano T, Takehara S, Hibi M, Aizawa S. Shisa promotes head formation through the inhibition of receptor protein maturation for the caudalizing factors, Wnt and FGF. Cell. 2005;120:223–235. doi: 10.1016/j.cell.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 36.Chen CC, et al. Shisa3 is associated with prolonged survival through promoting beta-catenin degradation in lung cancer. Am J Respir Crit Care Med. 2014;190:433–444. doi: 10.1164/rccm.201312-2256OC. [DOI] [PubMed] [Google Scholar]
- 37.Gilfix BM, Eckert RL. Coordinate control by vitamin A of keratin gene expression in human keratinocytes. J Biol Chem. 1985;260:14026–14029. [PubMed] [Google Scholar]
- 38.Lee HY, et al. Retinoic acid receptor- and retinoid X receptor-selective retinoids activate signaling pathways that converge on AP-1 and inhibit squamous differentiation in human bronchial epithelial cells. Cell Growth Differ. 1996;7:997–1004. [PubMed] [Google Scholar]
- 39.Abashev TM, Metzler MA, Wright DM, Sandell LL. Retinoic acid signaling regulates Krt5 and Krt14 independently of stem cell markers in submandibular salivary gland epithelium. Dev Dyn. 2017;246:135–147. doi: 10.1002/dvdy.24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jho SH, et al. J Invest Dermatol. 2005. The book of opposites: the role of the nuclear receptor co-regulators in the suppression of epidermal genes by retinoic acid and thyroid hormone receptors; pp. 1034–1043. [DOI] [PubMed] [Google Scholar]
- 41.Bock M, et al. Identification of ELF3 as an early transcriptional regulator of human urothelium. Dev Biol. 2014;386:321–330. doi: 10.1016/j.ydbio.2013.12.028. [DOI] [PubMed] [Google Scholar]
- 42.Fishwick C, et al. Heterarchy of transcription factors driving basal and luminal cell phenotypes in human urothelium. Cell Death Differ. 2017;24:809–818. doi: 10.1038/cdd.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metzger DE, Stahlman MT, Shannon JM. Misexpression of ELF5 disrupts lung branching and inhibits epithelial differentiation. Dev Biol. 2008;320:149–160. doi: 10.1016/j.ydbio.2008.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Latos PA, et al. Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev. 2015;29:2435–2448. doi: 10.1101/gad.268821.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearton DJ, et al. Elf5 counteracts precocious trophoblast differentiation by maintaining Sox2 and 3 and inhibiting Hand1 expression. Dev Biol. 2014;392:344–357. doi: 10.1016/j.ydbio.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 46.Glinka A, et al. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 47.Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-W. [DOI] [PubMed] [Google Scholar]
- 48.Moorman AF, Houweling AC, de Boer PA, Christoffels VM. Sensitive nonradioactive detection of mRNA in tissue sections: novel application of the whole-mount in situ hybridization protocol. J Histochem Cytochem. 2001;49:1–8. doi: 10.1177/002215540104900101. [DOI] [PubMed] [Google Scholar]
- 49.Bussen M, et al. The T-box transcription factor Tbx18 maintains the separation of anterior and posterior somite compartments. Genes Dev. 2004;18:1209–1221. doi: 10.1101/gad.300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.