Abstract
MicroRNAs (miRNAs) are potential biomarkers for cancer detection including esophageal squamous cell carcinoma (ESCC); however, little is known about their expression profile and diagnostic impact in esophageal squamous cell intraepithelial neoplasia, the pathological precancerous lesion of ESCC. In this study, we examined the expression levels of eight miRNAs that were reported to be deregulated in ESCC, including miR-25, let-7a, miR-100, miR-133a, miR-223, miR-375, miR-483-5p and miR-1322, in 30 pairs of esophageal squamous cell neoplasia lesion tissues and corresponding adjacent normal tissues using quantitative real-time PCR (qRT-PCR). Differential expression of miRNAs was further examined by in situ hybridization. Furthermore, the deregulated miRNAs were also measured in serum and serum exosome samples of these patients. miR-25, an oncomir that had been reported to be upregulated in ESCC tissues, were found to be overexpressed in esophageal squamous cell intraepithelial neoplasia lesions (66.7%, 20/30) compared to adjacent normal tissues (P < 0.05), while the other seven miRNAs did not show a significant difference between the lesions and controls. The miR-25 signal was stronger in lesion tissues than in normal tissues according to in situ hybridization. The concentrations of miR-25 in both serum and exosome samples of patients were not significantly different from those of healthy individuals. These findings suggested that the overexpression of miR-25 in esophageal squamous cell intraepithelial neoplasia lesions might be a promising early biomarker candidate for the prediction of ESCC.
Keywords: Esophageal squamous cell carcinoma, esophageal squamous cell intraepithelial neoplasia, miRNA, miR-25, qRT-PCR, in situ hybridization
Introduction
Esophageal cancer, which was the eighth most common cancer and the sixth leading cause of cancer related mortality in 2008, has become a worldwide health concern, with 455,800 new cases and 400,200 deaths estimated in 2012 [1-3]. The main histological pathological types of esophageal cancer are squamous cell carcinoma and adenocarcinoma [1,2]. For northern and central China, 90% of esophageal cancer cases are squamous cell carcinomas [1,3,4]. Esophageal squamous cell carcinoma (ESCC) is characterized by an insidious onset without major signs or symptoms, leading to late diagnoses of advanced ESCC, which partially accounts for the extremely poor prognosis, high mortality and high recurrence observed in this disease [5]. ESCC develops through progression from normal esophageal epithelium to low grade intraepithelial neoplasia (LGIN), high grade intraepithelial neoplasia (HGIN) and then ESCC with an accumulation of genetic and epigenetic abnormalities [6,7]. Esophageal squamous cell intraepithelial neoplasia, considered as precancerosis of ESCC, substantially elevates the risk of progression to ESCC and approximately 65% of HGIN cases progress to ESCC within 3.5 years [8-10]. Some studies have investigated precancerous lesions and got valuable results [11-13]; however, molecular markers for early diagnosis and prediction of prognosis or treatment responses are still limited [4]. Thus, further identify novel molecular markers in precancerous patients for the prevention and early detection of ESCC could help to limit the lethality of this disease.
MicroRNAs (miRNAs) are small noncoding RNAs that are 21-25 nucleotides in length and regulate the translation of many genes by binding to the untranslated region (3’UTR) of target mRNAs. miRNAs are involved in a variety of physiological and pathological processes, in particular, cancer development [14,15]. Accumulating evidence shows that miRNAs are aberrantly expressed in many types of cancers and these dysregulated miRNAs help in the diagnosis and prognosis of patients [16-18]. A number of miRNAs expression profiling studies have revealed that sets of miRNAs are upregulated (such as miR-25, miR-223, miR-483-5p and miR-1322) [19-27] or downregulated (such as miR-133a, miR-100, miR-375 and let-7a) [28-36] in ESCC tissues or cells. Furthermore, our group and others also identified some specific miRNAs that were increased or decreased in serum or plasma samples from patients with ESCC [37-48]. These features support the feasibility of miRNAs as biological markers for ESCC. Recently, it was found that miR-224 was significantly overexpressed not only in ESCC tissues, but also in esophageal intraepithelial neoplasia, which would predict the progression to ESCC [49]. We wondered whether other miRNAs that were dysregulated in ESCC tissues were also aberrantly expressed in precancerous lesions.
Therefore, in this study we measured the expression levels of eight miRNAs that had been previously reported as dysregulated in ESCC, namely, miR-25, miR-223, miR-483-5p, miR-1322, miR-100, miR-375, miR-133a and let-7a [16-36], in the lesions of esophageal squamous cell intraepithelial neoplasia and corresponding adjacent normal tissues using a TaqMan based qRT-PCR assay. Furthermore, in situ hybridization was performed to validate the results achieved. In addition, the levels of differentially expressed miRNAs in serum and serum exosome samples were also examined.
Materials and methods
Study design and patients
The present study enrolled 30 patients newly diagnosed with esophageal squamous cell intraepithelial neoplasia who were treated at the Department of Gastroenterology and Hepatology of Jinling Hospital (Nanjing, China) between February 2014 and October 2015. Patients age ranged from 50 to 80 years old, with a mean of 62.3 years old. No adjunctive therapy was carried out before the endoscopy and biopsy. The operations were classified as curative surgery, and pathology specimens from all patients enrolled in the study were reviewed and diagnosed according to the current WHO classification scheme. Patients with acute infections or other types of cancer were excluded from our study to eliminate inconclusive confounders. Clinical and pathological information were recorded in considerable detail in patients’ medical charts.
Tissue and serum samples
Lesion tissues and the corresponding adjacent normal tissues were taken from 30 patients with esophageal squamous cell intraepithelial neoplasia after endoscopic biopsy resection. The tissue samples examined in this study were the tissue remaining after diagnostic biopsies and were frozen in liquid nitrogen immediately after collection, then stored at -80°C until RNA extraction. Serum samples were collected from 21 patients with esophageal squamous cell intraepithelial neoplasia. The 21 control serum samples were from individuals seeking a routine health check up in the Healthy Physical Examination Center of Jinling Hospital. Controls were selected from people who presented no evidence of cancers or others diseases. No significant difference in the distribution of age, gender, nation, or territory presented between patients and controls. Serum samples from patients were collected before endoscopic biopsy resection. All samples were stored at -80°C until analysis. The ethics committee of Jinling Hospital approved this study protocol in accordance with the Helsinki Declaration (as revised in Fortaleza, Brazil, October 2013), and written informed consent was obtained from all participants.
H&E staining
The excision biopsy samples of esophageal squamous cell intraepithelial neoplasia plaque lesions were fixed with formalin solution and paraffin embedded. Then, the samples were cut into 6 μm sections and stained with H&E.
miRNA in situ hybridization (MISH)
Tissues for MISH analysis were frozen in liquid nitrogen immediately after being obtained. An oligonucleotide probe complementary to human mature hsa-miR-25-3p was used for MISH. The probe sequence was 5’-CATTGCACTTG TCTCGGTCTGA-3’, and the 3’terminal was labeled with biotin (VETEC, Sigma, Germany). Specimens were incubated with the miR-25 probe (3 ng/μL) at 20°C overnight. All procedures were performed under RNase free conditions according to the standard procedure for ISH. Counterstaining was performed with DAPI. Further analysis was completed using an inverted fluorescence microscope (NIKON ECLIPSE TI-SR, NIKON, Japan) and NIKON DS-U3 imaging system ((NIKON, Japan). A scrambled probe, instead of the miR-25 specific probe, served as a negative control.
RNA extraction
Total RNA was dissolved and extracted from each tissue sample with TRI Reagent® (Sigma, Life Science, USA) in accordance with the manufacturer’s protocols. The aqueous phase was subjected to 3 TRIzol/chloroform purification steps to eliminate protein residues from the RNA before isopropyl alcohol precipitation. Then, the RNA dreg was washed with 75% ethyl alcohol. The resulting total RNA was dissolved in 20 μL of DEPC water and stored at -80°C until further analysis. RNA isolated from the serum samples was used for acid phenol extraction, and the extracted total RNA was dissolved in 22 μL of DEPC water. We confirmed the quantity and purity of the RNA by measuring optical density values at 260 and 280 nm with a spectrometer.
Exosome isolation
Serum samples were transferred to sterile tubes and the appropriate RiboTM Exosome Isolation Reagent for plasma or serum (RiboBio, Guangzhou, China) was added to isolate exosomes according to the manufacturer’s protocols. Approximately, 100 μL of serum and an equal volume of reagent were perfectly mixed and left standing overnight at 4°C. The mixture was then separated by sedimentation at 1,500×g for 30 min at 4°C, and the supernatant was aspirated. Finally, exosome pellets were collected, and RNA was immediately extracted according to the protocol above.
Individual qRT-PCR assays
Expression levels of miRNAs in examined samples were determined using TaqMan® probe based qRT-PCR according to the manufacturer’s instructions (7300 Sequence Detection System; Applied Biosystems). Briefly, the reverse transcription reaction was carried out in 10 μL volumes, and real-time PCR was performed with an Applied Biosystems 7300 Sequence Detection System with a final volume of 20 μL. The specific steps of the protocol were strictly completed as described in our previous study [38]. All reactions were performed in triplicate. The tissue miRNAs were normalized to U6 snRNA, an endogenous control. An exogenous reference gene, plant miRNA MIR2911, was spiked into each serum sample during RNA isolation at a final concentration of 106 fmol/L and served to normalize miRNA expression. Relative miRNA expression was calculated using the comparative Cq method (2-ΔCq).
Prediction of miR-25 target genes
In this study, we employed three independent databases, miRanda (www.microrna.org/microrna/home.do), Target Scan (www.targetscan.org) and PicTar (pictar.Mdc-berlin.de/), to generate a list of the most reliable miR-25 targets. We selected the most common genes, cancer and ESCC associated genes as our predicted targets. The Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.7 (http://david.Abcc.ncifcrf.gov/) was queried for the functional assignment of the target genes.
Statistical analysis
Statistical testing was conducted with SPSS 22.0 software. Levels of miRNA expression are presented as the means ± SEM and are representative of at least three independent experiments. Statistical significance was defined as P value < 0.05. Significant differences between lesion tissues and adjacent normal tissues were evaluated using the paired t-test. Comparisons between two groups were made using Mann-Whitney test.
Results
Demographic and clinical data of patients
Table 1 summarizes the demographic and clinical data of 30 patients with esophageal squamous cell intraepithelial neoplasia. Tissue biopsy confirmed LGIN in 12 patients and HGIN in 18 patients.
Table 1.
Clinicopathological features of esophageal squamous epithelium neoplasia patients enrolled in the study
| Characteristics | Patients No. | % |
|---|---|---|
| Age (mean) | 50-80 (62.3) | |
| Gender | ||
| Male | 25 | 83.3 |
| Female | 5 | 16.7 |
| Stage of intraepithelial neoplasia | ||
| High | 18 | 60.0 |
| Low | 12 | 40.0 |
| Tumor location | ||
| Upper | 4 | 13.3 |
| Middle | 11 | 36.7 |
| Lower | 15 | 50.0 |
| Smoking status | ||
| Ever or current | 9 | 30.0 |
| Never | 17 | 56.7 |
| Unknown | 4 | 13.3 |
| Alcohol consumption | ||
| Ever or current | 9 | 30.0 |
| Never | 17 | 56.7 |
| Unknown | 4 | 13.3 |
| Family history of cancer | ||
| Esophageal cancer | 2 | 6.7 |
| Gastric cancer or cardiac cancer | 3 | 10.0 |
| No | 21 | 70.0 |
| Unknown | 4 | 13.3 |
| Complication | ||
| Gastritis | 16 | 53.3 |
| Oesophagitis | 2 | 6.7 |
| Ulceration | 4 | 13.3 |
Representative cases of HGIN and LGIN
Endoscopic and histopathological findings in representative cases of HGIN and LGIN diagnosed by narrow band imaging (NBI) are shown in Figure 1. Lesions appeared as a pale area in NBI. After endoscopic mucosal resection (EMR), histopathological examination of the resected lesion led to a diagnosis of HGIN or LGIN according to the proportion of neoplastic cells with atypical nuclei and prominent nucleoli. Analysis was limited to the basilemma. Neoplastic cells were found > 50% in the thickness of the epithelium in HGIN and < 50% in LGIN.
Figure 1.
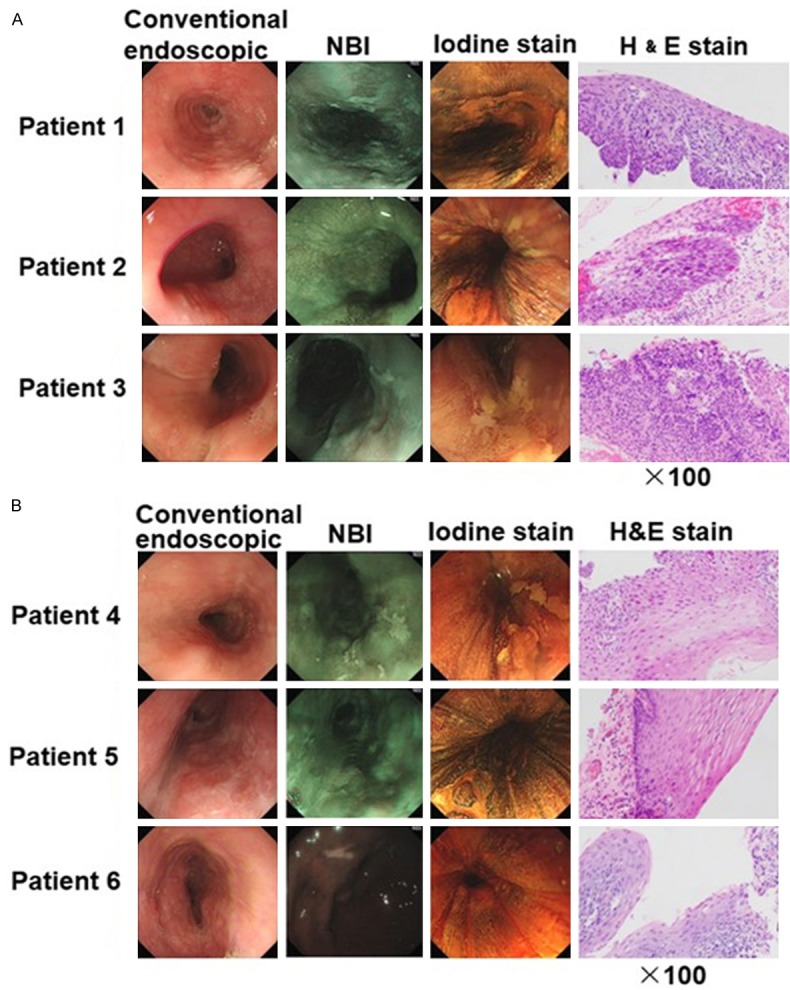
Routine endoscopy and narrow band imaging (NBI) images of representative cases of HGIN patients and LGIN patients. Conventional endoscopic view: a slightly depressed lesion with iodine unstained areas. Resected specimens with H&E (×100) stain were shown. A. Specimens were diagnosed as HGIN if stained with neoplastic cells > 50% in the thickness of the epithelium. B. Specimens were diagnosed as LGIN if stained with neoplastic cells < 50% in the thickness of the epithelium.
Expression of miRNAs in esophageal squamous cell intraepithelial neoplasia lesion tissues
We measured the expression levels of eight miRNAs in 30 paired samples of esophageal squamous cell intraepithelial neoplasia tissues and corresponding adjacent normal tissue. According to qRT-PCR results, statistically significantly amplified expression of miR-25 was found in lesion tissues compared with control tissues (P = 0.031), identifying miR-25 as a candidate for further analysis but not the other seven miRNAs which presented no significant difference (Figure 2A-H). The expression of miR-25 was up regulated in 66.7% (20/30) of patients’ lesions (Figure 2I). Furthermore, we compared the expression levels of miR-25 between HGIN and LGIN cases but found no significant difference between the two groups (Figure 3). These results suggested that the expression level of miR-25 might be a potential biomarker for esophageal squamous cell intraepithelial neoplasia of various grades.
Figure 2.
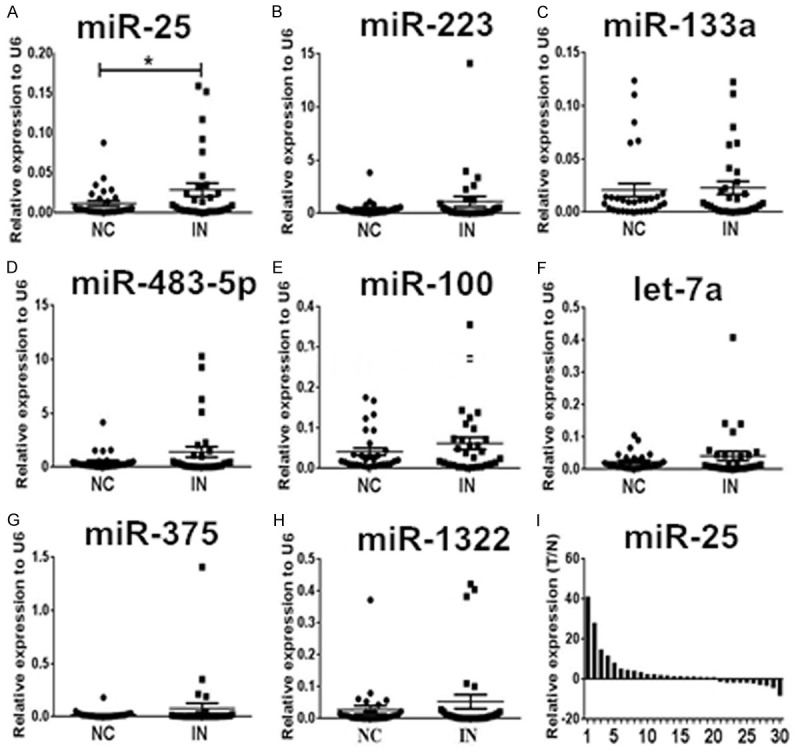
Expression levels of the eight miRNAs examined in esophageal squamous cell intraepithelial neoplasia (IN) plaque tissues (n = 30) and the matched adjacent normal control (NC). A-H. Only miR-25 was up regulated with a significant difference. The relative expression levels of miRNAs were normalized to U6 snRNA. I. Expression levels of miR-25 were ranged from the largest to the smallest. Relative expression was calculated using the 2-Δcq method, and N refers to the normal control. *P < 0.05.
Figure 3.
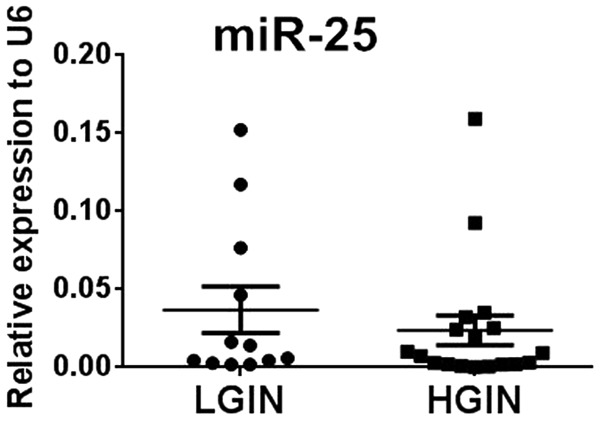
Expression levels of miR-25 in LGIN and HGIN tissues analyzed using qRT-PCR.
MISH detection of the miR-25 expression pattern
We further examined miR-25 expression in esophageal squamous cell intraepithelial neoplasia lesion tissues using MISH. The result demonstrated that miR-25 expression levels (labeled with BIO) were higher in the plaques of esophageal squamous cell intraepithelial neoplasia tissues than that in normal control tissues (Figure 4A). The green fluorescence signals of the miR-25 probe were dispersed uniformly throughout the cytoplasm of the intraepithelial neoplasia cells. While no fluorescence signals could be detected in cells hybridized with the scramble probe control (Figure 4B).
Figure 4.
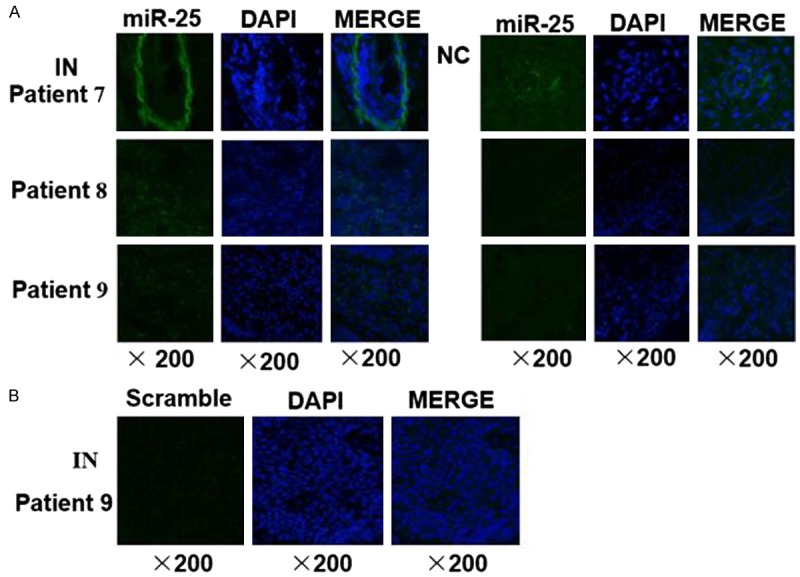
Representative photographs of miR-25 ISH results in esophageal squamous cell intraepithelial neoplasia. A. The levels of miR-25 (green signal) were higher in the plaques of intraepithelial neoplasia (IN) tissues than that in matched adjacent normal controls (NC). B. As negative control, a scrambled probe was used instead of the miR-25 specific probe. Original magnification: 20×objective.
Expression levels of circulating miR-25 in patients
We also examined miR-25 levels in serum and serum exosomes from 21 patients with esophageal squamous cell intraepithelial neoplasia (Table 2). The expression levels of both serum and serum exosomal miR-25 from patients were not different from those in the 21 age and sex matched healthy controls (P = 0.183 and P = 0.340, respectively) (Figure 5).
Table 2.
Demographic and clinical features of the esophageal squamous cell intraepithelial neoplasia patients and normal individuals in serum and serum exosomal miRNA study
| Variables | Patients (n = 21) | Normal controls (n = 21) | P-value | ||
|---|---|---|---|---|---|
|
|
|||||
| No. | % | No. | % | ||
| Average age (years) | 63.2 | 59.3 | |||
| Age (years) | |||||
| ≤60 | 7 | 33.3 | 10 | 47.6 | 0.53 |
| >60 | 14 | 66.7 | 11 | 52.4 | |
| Sex | |||||
| Male | 14 | 66.7 | 14 | 66.7 | 1.00 |
| Female | 7 | 33.3 | 7 | 33.3 | |
| Stage of intraepithelial neoplasia | |||||
| High | 15 | 71.4 | |||
| Low | 6 | 28.6 | |||
Figure 5.
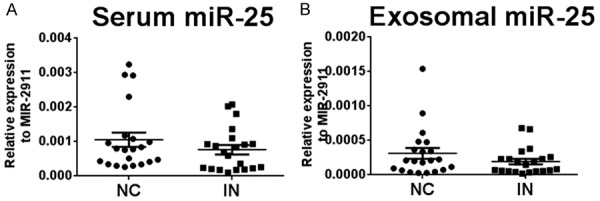
Expression levels of miR-25 in serum and isolated exosomes in esophageal squamous cell intraepithelial neoplasia (IN) and healthy normal controls (NC). A. Expression levels of miR-25 in serum samples. B. Expression levels of miR-25 in exosome samples. The relative levels of miR-25 were normalized to miR-2911 and calculated using the 2-Δcq method.
Target prediction and functional analysis of miR-25
To explore the possible role and molecular basis of altered miR-25 expression in the occurrence of esophageal squamous cell intraepithelial neoplasia, we predicted the potential target genes of miR-25 through a bioinformatics analysis using the Target Scan, miRanda and PicTar databases. We also searched the DAVID functional annotation bioinformatics microarray analysis database to explore the functional significance of up regulated miR-25 expression in tumorigenesis and progression. Computational analysis revealed that several of the predicted target genes participated in the regulation of the cell signal pathway, gene transcription, cell cycle arrest and apoptosis, tumorigenicity, cell metabolism, and more (Table 3).
Table 3.
Target genes of miR-25 and their function in relation to tumorigenesis
| miRNA | Target gene | Description | Validated or not |
|---|---|---|---|
| miR-25 | DSC2 | Intercellular desmosome junction | YES |
| CDH1 | Tumor metastasis suppressor | YES | |
| RECK | Reversed oncogenic effect | YES | |
| E2F1 | A transcription factors | YES | |
| MOAP1 | Involved in regulation of cell apoptosis | YES | |
| TOB1 | An anti-proliferative protein | YES | |
| BTG2 | YES | ||
| CDC42 | A member of Rho GTPase family | YES | |
| β-TRCP2 | A ubiquitin ligase | YES | |
| RGS3 | Regulator of G protein signaling 3 | YES | |
| EP300 | A transcriptional activator of E-cadherin | YES | |
| PTEN | A tumor suppressor | YES | |
| LATS2 | YES | ||
| FBXW7 | YES | ||
| TSC1 | NO | ||
| Smad7 | Antagonist of signaling by TGF-beta | YES | |
| Bim | A pro-apoptotic protein | YES | |
| SYT1 | Tumor suppressor gene involved in the synaptic activity regulation | NO | |
| MAP2K4 | Dual specificity kinasethat activates the JUN kinases MAPK8 (JNK1) and MAPK9 (JNK2) as well as MAPK14 (p38) | NO | |
| CUX1 | As a repressor of developmentally regulated gene expression | NO | |
| GPRC5A | Involved in modulating differentiation and maintaining homeostasis of epithelial cells | NO | |
| ZEB2 | A transcriptional inhibitor of E-cadherin | NO | |
| MBD2 | A transcriptional repressor | NO | |
| MDM4 | Inhibit p53- and p73-mediated cell cycle arrest and apoptosis | NO | |
| NEO1 | Involved as a regulatory protein in the cell differentiation | NO | |
| TP63 | Acted as transcriptional activator or repressor | NO |
Discussion
A specific set of miRNAs have been demonstrated to be dysregulated in ESCC. For example, miR-25 [20-22], miR-223 [23-25], miR-483-5p [26], miR-100 [31,32], and miR-1322 [27] are up regulated; let-7a [36], miR-133a [28-30], and miR-375 [33-35] are down regulated. However, whether these miRNAs are also aberrantly expressed in precancerous lesion, remain unknown. In this study, we measured the expression levels of above eight miRNAs in esophageal squamous cell intraepithelial neoplasia tissues using the qRT-PCR method and found that miR-25 was up regulated in plaque tissues relative to matched normal tissues. MISH further confirmed that miR-25 was overexpressed in lesion tissues. Our results demonstrate for the first time that miR-25 is upregulated before the occurrence of ESCC.
As tumor inhibiting or carcinogenic factors, miRNAs have extensive value in tumor diagnostic applications and prognosis predicting. Specifically, miR-25 is a highly conserved miRNA belonging to the miR-106b~25 cluster together with the miR-106b and miR-93, and its up regulation has been reported in many tumors, including digestive system tumors such as ESCC [20-22], esophageal adenocarcinoma [50], gastric cancer [51,52], and other tumors like non-small cell lung cancer [53] and ovarian cancer [54]. In most of these cancers, miR-25 functions as an oncogene and promotes the proliferation and migratory ability of cells. The upregulation of miR-25 was significantly correlated with the status of lymph node metastasis and TNM (Tumor, Node and Metastasis) stage of ESCC [21]. In this study, we found that miR-25 was also overexpressed in precancerous lesions of ESCC, which furthers our knowledge of miRNAs involving in the procession to ESCC. We suspect that the combination of H&E staining results and miR-25 expression level may have greater potential to detect esophageal squamous cell intraepithelial neoplasia. In this study, we also compared miR-25 levels in high grade lesion tissues with those of low grade lesion tissues but no significant difference was found. The upregulation of miR-25 in precancerous lesion of various grades suggest that miR-25 might be a promising and beneficial biomarker candidate for early detection of ESCC. However, because the number of patients in each subgroup was small, further studies of larger sample size are necessary to validate this.
Our study indicates that miR-25 may play a role in the development of esophageal squamous cell intraepithelial neoplasia. A full understanding of miR-25 target genes will help clarify the pathogenesis of esophageal squamous cell intraepithelial neoplasia and broaden miR-25 clinical applications. Analysis of the resulting data sets based on multiple target prediction algorithms highlighted that several of the predicted target genes of miR-25 are involved in the regulation of the cell signal pathway, cell cycle arrest and apoptosis. For example, it has been demonstrated that miR-25 significantly promotes the proliferation, invasion, and migration of gastric cancer cells in vitro [51,52]. Furthermore, miR-25 repressed F-box and WD-40 domain protein 7 (FBXW7) and TOB1 expression, and inverse correlations were observed between expression of miR-25 and these genes in primary gastric cancer tissues [51,52]. In ESCC, miR-25 was also reported to promote cell migration and invasion by suppressing E-cadherin (CDH1), a very important tumor metastasis suppressor, and desmocollin-2 (DSC2), a desmosomal cadherin protein that was associated with enhanced tumor metastasis and poor prognosis [21,55]. Due to the small size of the lesion tissues and the inability to acquire additional tissue (the samples used in this study were the remaining tissues from diagnostic biopsies), we did not have enough tissues to examine the candidate target genes further. Additional studies are therefore needed to clarify the effects of miR-25 expression and its underlying molecular mechanisms in esophageal squamous cell intraepithelial neoplasia.
To evaluate whether the deregulated expression of miR-25 in lesion tissues is reflected in the circulation, we also examined the levels of miR-25 in serum and serum exosome samples from patients with esophageal squamous cell intraepithelial neoplasia. No significant difference was found in miR-25 levels in serum or serum exosome samples between patients and normal controls. Based on the characteristic that the small size of neoplasia lesions examined in our study limited to the basilemma of epithelium without vascular contact [6,7], detectable amounts of miR-25 in circulation might not be passively released or actively secreted from lesion tissues into the circulation. The exact reason remains to be clarified.
In conclusion, we showed that miR-25 was up regulated in esophageal squamous cell intraepithelial neoplasia tissue. The aberrant expression of miR-25 in lesion tissues may contribute to the pathogenesis of esophageal squamous cell intraepithelial neoplasia and could be used as an auxiliary biomarker candidate for the prediction of ESCC.
Acknowledgements
This work was supported by the National Basic Research Program of China [2014CB542300]; the Public Welfare Industry of Health of China [201302018]; the National Natural Science Foundation of China [81472021, 81672102 and 81401257], the Natural Science Foundation of Jiangsu Province [BK20140730] and State Key Laboratory of Analytical Chemistry for Life Science [5431ZZXM1601].
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 4.Tran GD, Sun XD, Abnet CC, Fan JH, Dawsey SM, Dong ZW, Mark SD, Qiao YL, Taylor PR. Prospective study of risk factors for esophageal and gastric cancers in the Linxian general population trial cohort in China. Int J Cancer. 2005;113:456–463. doi: 10.1002/ijc.20616. [DOI] [PubMed] [Google Scholar]
- 5.Ruol A, Castoro C, Portale G, Cavallin F, Sileni VC, Cagol M, Alfieri R, Corti L, Boso C, Zaninotto G, Peracchia A, Ancona E. Trends in management and prognosis for esophageal cancer surgery: twenty-five years of experience at a single institution. Arch Surg. 2009;144:247–254. doi: 10.1001/archsurg.2008.574. [DOI] [PubMed] [Google Scholar]
- 6.Guindi M, Riddell RH. The pathology of epithelial pre-malignancy of the gastrointestinal tract. Best Pract Res Clin Gastroenterol. 2001;15:191–210. doi: 10.1053/bega.2001.0169. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu M, Nagata K, Yamaguchi H, Kita H. Squamous intraepithelial neoplasia of the esophageal: past, present, and future. J Gastroenterol. 2009;44:103–112. doi: 10.1007/s00535-008-2298-y. [DOI] [PubMed] [Google Scholar]
- 8.Dawsey SM, Lewin KJ, Wang GQ, Liu FS, Nieberg RK, Yu Y, Li JY, Blot WJ, Li B, Taylor PR. Squamous esophageal histology and subsequent risk of squamous cell carcinoma of the esophagus. A prospective follow-up study from Linxian, China. Cancer. 1994;74:1686–1692. doi: 10.1002/1097-0142(19940915)74:6<1686::aid-cncr2820740608>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 9.Wang GQ, Abnet CC, Shen Q, Lewin KJ, Sun XD, Roth MJ, Qiao YL, Mark SD, Dong ZW, Taylor PR, Dawsey SM. Histological precursors of oesophageal squamous cell carcinoma: results from a 13 year prospective follow up study in a high risk population. Gut. 2005;54:187–192. doi: 10.1136/gut.2004.046631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin XF, Sun QY, Chai TH, Li SH, Guo YL. Clinical value of multiband mucosectomy for the treatment of squamous intraepithelial neoplasia of the esophagus. J Gastroenterol Hepatol. 2013;28:650–655. doi: 10.1111/jgh.12111. [DOI] [PubMed] [Google Scholar]
- 11.Kaneko K, Katagiri A, Konishi K, Kurahashi T, Ito H, Kumekawa Y, Yamamoto T, Muramoto T, Kubota Y, Nozawa H, Makino R, Kushima M, Imawari M. Study of p53 gene alteration as a biomarker to evaluate the malignant risk of Lugol-unstained lesion with non-dysplasia in the oesophagus. Br J Cancer. 2007;96:492–498. doi: 10.1038/sj.bjc.6603582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimizu Y, Yoshida T, Kato M, Hirota J, Ono S, Nakagawa M, Kobayashi T, Kubota K, Asaka M. Low-grade dysplasia component in early invasive squamous cell carcinoma of the esophagus. J Gastroenterol Hepatol. 2010;25:314–318. doi: 10.1111/j.1440-1746.2009.06032.x. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi M, Kawachi H, Takizawa T, Uchida K, Sekine M, Kumagai J, Momma K, Nemoto T, Akashi T, Funata N, Eishi Y, Koike M. p53 mutation analysis of low-grade dysplasia and highgrade dysplasia/carcinoma in situ of the esophagus using laser capture microdissection. Oncology. 2006;71:237–245. doi: 10.1159/000106448. [DOI] [PubMed] [Google Scholar]
- 14.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 15.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Paranjape T, Slack FJ, Weidhaas JB. MicroRNAs: tools for cancer diagnostics. Gut. 2009;58:1546–1554. doi: 10.1136/gut.2009.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harada K, Baba Y, Ishimoto T, Shigaki H, Kosumi K, Yoshida N, Watanabe M, Baba H. The role of microRNA in esophageal squamous cell carcinoma. J Gastroenterol. 2016;51:520–530. doi: 10.1007/s00535-016-1161-9. [DOI] [PubMed] [Google Scholar]
- 20.Gu J, Wang Y, Wu X. MicroRNA in the pathogenesis and prognosis of esophageal cancer. Curr Pharm Des. 2013;19:1292–1300. doi: 10.2174/138161213804805775. [DOI] [PubMed] [Google Scholar]
- 21.Xu X, Chen Z, Zhao X, Wang J, Ding D, Wang Z, Tan F, Tan X, Zhou F, Sun J, Sun N, Gao Y, Shao K, Li N, Qiu B, He J. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2012;421:640–645. doi: 10.1016/j.bbrc.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 22.Zhu L, Yan W, Rodriguez-Canales J, Rosenberg AM, Hu N, Goldstein AM, Taylor PR, Erickson HS, Emmert-Buck MR, Tangrea MA. MicroRNA analysis of microdissected normal squamous esophageal epithelium and tumor cells. Am J Cancer Res. 2011;1:574–584. [PMC free article] [PubMed] [Google Scholar]
- 23.Kurashige J, Watanabe M, Iwatsuki M, Kinoshita K, Saito S, Hiyoshi Y, Kamohara H, Baba Y, Mimori K, Baba H. Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer. 2012;106:182–188. doi: 10.1038/bjc.2011.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu HL, Wu DP, Wang XF, Wang JG, Jiao F, Song LL, Xie H, Wen XY, Shan HS, Du YX, Zhao YP. Altered miRNA expression is associated with differentiation, invasion, and metastasis of esophageal squamous cell carcinoma (ESCC) in patients from Huaian, China. Cell Biochem Biophys. 2013;67:657–668. doi: 10.1007/s12013-013-9554-3. [DOI] [PubMed] [Google Scholar]
- 25.Fong LY, Taccioli C, Jing R, Smalley KJ, Alder H, Jiang Y, Fadda P, Farber JL, Croce CM. MicroRNA dysregulation and esophageal cancer development depend on the extent of zinc dietary deficiency. Oncotarget. 2016;7:10723–10738. doi: 10.18632/oncotarget.7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue L, Nan J, Dong L, Zhang C, Li H, Na R, He H, Wang Y. Upregulated miR-483-5p expression as a prognostic biomarker for esophageal squamous cell carcinoma. Cancer Biomark. 2017;19:193–197. doi: 10.3233/CBM-160506. [DOI] [PubMed] [Google Scholar]
- 27.Zhang T, Zhao D, Wang Q, Yu X, Cui Y, Guo L, Lu SH. MicroRNA-1322 regulates ECRG2 allele specifically and acts as a potential biomarker in patients with esophageal squamous cell carcinoma. Mol Carcinog. 2013;52:581–590. doi: 10.1002/mc.21880. [DOI] [PubMed] [Google Scholar]
- 28.Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. miR-145, miR-133a and miR-133b: tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:2804–2814. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- 29.Akanuma N, Hoshino I, Akutsu Y, Murakami K, Isozaki Y, Maruyama T, Yusup G, Qin W, Toyozumi T, Takahashi M, Suito H, Hu X, Sekino N, Matsubara H. MicroRNA-133a regulates the mRNAs of two invadopodia-related proteins, FSCN1 and MMP14, in esophageal cancer. Br J Cancer. 2014;110:189–198. doi: 10.1038/bjc.2013.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki S, Yokobori T, Tanaka N, Sakai M, Sano A, Inose T, Sohda M, Nakajima M, Miyazaki T, Kato H, Kuwano H. CD47 expression regulated by the miR-133a tumor suppressor is a novel prognostic marker in esophageal squamous cell carcinoma. Oncol Rep. 2012;28:465–472. doi: 10.3892/or.2012.1831. [DOI] [PubMed] [Google Scholar]
- 31.Sun J, Chen Z, Tan X, Zhou F, Tan F, Gao Y, Sun N, Xu X, Shao K, He J. MicroRNA-99a/100 promotes apoptosis by targeting mTOR in human esophageal squamous cell carcinoma. Med Oncol. 2013;30:411. doi: 10.1007/s12032-012-0411-9. [DOI] [PubMed] [Google Scholar]
- 32.Zhou S, Yang B, Zhao Y, Xu S, Zhang H, Li Z. Prognostic value of microRNA-100 in esophageal squamous cell carcinoma. J Surg Res. 2014;192:515–520. doi: 10.1016/j.jss.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Kong KL, Kwong DL, Chan TH, Law SY, Chen L, Li Y, Qin YR, Guan XY. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 34.Yi J, Jin L, Chen J, Feng B, He Z, Chen L, Song H. MiR-375 suppresses invasion and metastasis by direct targeting of SHOX2 in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai) 2017;49:159–169. doi: 10.1093/abbs/gmw131. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Li X, Li Y, Yang H, Wang L, Qin Y, Liu H, Fu L, Guan XY. Cell-specific detection of miR-375 downregulation for predicting the prognosis of esophageal squamous cell carcinoma by miRNA in situ hybridization. PLoS One. 2013;8:e53582. doi: 10.1371/journal.pone.0053582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen D, Jin L, Zhu L, Mou X, Wang S, Mao C. Expressions and correlations of let-7a and IL-6 in esophageal squamous cell carcinoma. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2013;29:1181–1184. [PubMed] [Google Scholar]
- 37.Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, Dai J, Hu Z, Zhou X, Chen L, Zhang Y, Li Y, Qiu H, Xing J, Liang Z, Ren B, Yang C, Zen K, Zhang CY. Expression profile of microRNAs in serum: A fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–1879. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 38.Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X, Zhang Y, Chen X, Wang J, Zen K, Zhang CY, Zhang C. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS One. 2014;9:e92292. doi: 10.1371/journal.pone.0092292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105:104–111. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurashige J, Kamohara H, Watanabe M, Tanaka Y, Kinoshita K, Saito S, Hiyoshi Y, Iwatsuki M, Baba Y, Baba H. Serum microRNA-21 is a novel biomarker in patients with esophageal squamous cell carcinoma. J Surg Oncol. 2012;106:188–192. doi: 10.1002/jso.23064. [DOI] [PubMed] [Google Scholar]
- 41.Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, Komatsu A, Jitsukawa M, Matsubara H. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:644–652. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirajima S, Komatsu S, Ichikawa D, Takeshita H, Konishi H, Shiozaki A, Morimura R, Tsujiura M, Nagata H, Kawaguchi T, Arita T, Kubota T, Fujiwara H, Okamoto K, Otsuji E. Clinical impact of circulating miR-18a in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:1822–1829. doi: 10.1038/bjc.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komatsu S, Ichikawa D, Hirajima S, Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Arita T, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Yagi N, Otsuji E. Plasma microRNA profiles: identification of miR-25 as a novel diagnostic and monitoring biomarker in oesophageal squamous cell carcinoma. Br J Cancer. 2014;111:1614–1624. doi: 10.1038/bjc.2014.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu C, Li M, Hu C, Duan H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol Biol Rep. 2014;41:1257–1266. doi: 10.1007/s11033-013-2970-z. [DOI] [PubMed] [Google Scholar]
- 45.He FC, Meng WW, Qu YH, Zhou MX, He J, Lv P, Ming L. Expression of circulating microRNA-20a and let-7a in esophageal squamous cell carcinoma. World J Gastroenterol. 2015;21:4660–4665. doi: 10.3748/wjg.v21.i15.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong W, Li B, Wang J, Song Y, Zhang Z, Fu C, Zhang P. Diagnostic and predictive significance of serum microRNA-7 in esophageal squamous cell carcinoma. Oncol Rep. 2016;35:1449–1456. doi: 10.3892/or.2015.4499. [DOI] [PubMed] [Google Scholar]
- 47.Wang C, Guan S, Liu F, Chen X, Han L, Wang D, Nesa EU, Wang X, Bao C, Wang N, Cheng Y. Prognostic and diagnostic potential of miR-146a in oesophageal squamous cell carcinoma. Br J Cancer. 2016;114:290–297. doi: 10.1038/bjc.2015.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Li M, Gao F, Ge X. Serum microRNA-15a level acts as a potential diagnostic and prognostic biomarker for human esophageal squamous cell carcinoma. Cancer Biomark. 2017;18:11–17. doi: 10.3233/CBM-160667. [DOI] [PubMed] [Google Scholar]
- 49.He X, Zhang Z, Li M, Li S, Ren L, Zhu H, Xiao B, Shi R. Expression and role of oncogenic miRNA-224 in esophageal squamous cell carcinoma. BMC Cancer. 2015;15:575. doi: 10.1186/s12885-015-1581-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slaby O, Srovnal J, Radova L, Gregar J, Juracek J, Luzna P, Svoboda M, Hajduch M, Ehrmann J. Dynamic changes in microRNA expression profiles reflect progression of Barrett’s esophagus to esophageal adenocarcinoma. Carcinogenesis. 2015;36:521–527. doi: 10.1093/carcin/bgv023. [DOI] [PubMed] [Google Scholar]
- 51.Li BS, Zuo QF, Zhao YL, Xiao B, Zhuang Y, Mao XH, Wu C, Yang SM, Zeng H, Zou QM, Guo G. MicroRNA-25 promotes gastric cancer migration, invasion and proliferation by directly targeting transducer of ERBB2, 1 and correlates with poor survival. Oncogene. 2015;34:2556–2565. doi: 10.1038/onc.2014.214. [DOI] [PubMed] [Google Scholar]
- 52.Gong J, Cui Z, Li L, Ma Q, Wang Q, Gao Y, Sun H. MicroRNA-25 promotes gastric cancer proliferation, invasion, and migration by directly targeting F-box and WD-40 Domain Protein 7, FBXW7. Tumour Biol. 2015;36:7831–7840. doi: 10.1007/s13277-015-3510-3. [DOI] [PubMed] [Google Scholar]
- 53.Xiang J, Hang JB, Che JM, Li HC. MiR-25 is upregulated in non-small cell lung cancer and promotes cell proliferation and motility by targeting FBXW7. Int J Clin Exp Pathol. 2015;8:9147–9153. [PMC free article] [PubMed] [Google Scholar]
- 54.Feng S, Pan W, Jin Y, Zheng J. MiR-25 promotes ovarian cancer proliferation and motility by targeting LATS2. Tumour Biol. 2014;35:12339–12344. doi: 10.1007/s13277-014-2546-0. [DOI] [PubMed] [Google Scholar]
- 55.Fang WK, Liao LD, Li LY, Xie YM, Xu XE, Zhao WJ, Wu JY, Zhu MX, Wu ZY, Du ZP, Wu BL, Xie D, Guo MZ, Xu LY, Li EM. Down-regulated desmocollin-2 promotes cell aggressiveness through redistributing adherens junctions and activating beta-catenin signalling in oesophageal squamous cell carcinoma. J Pathol. 2013;231:257–270. doi: 10.1002/path.4236. [DOI] [PubMed] [Google Scholar]


