Abstract
Idiopathic pulmonary fibrosis (IPF) is a devastating disease and the pathogenesis of IPF remains unclear. Our previous study indicated that miR-5100 promotes the proliferation and metastasis of lung epithelial cells. In this study, we investigated the effect and mechanism of miR-5100 on bleomycin (BLM)-induced mouse lung fibrosis and transforming growth factor β (TGF-β1) or epidermal growth factor (EGF) induced EMT-model in A549 and Beas-2B cells. The elevated level of miR-5100 was observed in both the mouse lung fibrosis tissues and EMT cell model. Furthermore, the exogenous expression of miR-5100 promoted the EMT-related changes, enhanced TGF-β1 or EGF-induced EMT and activated the smad2/3 in lung epithelial cells, while silencing miR-5100 had the converse effects. In addition, transwell assay showed that miR-5100 can enhance cell migration. Using target prediction software and luciferase reporter assays, we identified TOB2 as a specific target of miR-5100 and miR-5100 can decrease the accumulation of endogenous TOB2 in A549 and Beas-2B cells. Moreover, the exogenous expression of TOB2 relieves the promotion of miR-5100 on EMT process and migration ability. Taken together, our results indicate that miR-5100 promotes the EMT process by targeting TOB2 associated with activating smad2/3 in lung epithlium cells. Our findings may provide novel insights into the pathogenesis of IPF.
Keywords: miR-5100, epithelial-mesenchymal transition (EMT), TOB2, smad2/3, lung epithelial cell, idiopathic pulmonary fibrosis (IPF)
Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive and devastating disease for which lung transplantation is considered the only viable intervention [1]. The pathogenesis of IPF remains largely unknown. Accumulated evidences demonstrated that epithelial-mesenchymal transition (EMT) plays a key role on pathogenesis of IPF [2,3].
Epithelial-mesenchymal transition (EMT), which is characterized by the loss of epithelial characteristics (E-cadherin) and the acquisition of a mesenchymal phenotype [N-cadherin, vimentin, and α-smooth muscle actin (α-SMA)], is a major contributor to the accumulation of myofibroblasts and fibroblasts on the site of tissue injury [4]. Accumulated myofibroblasts produce excessive extracellular matrix (ECM) components leading to progressive lung fibrosis [3]. In addition, as an inducer of EMT, transforming growth factor β (TGF-β1) has long been regarded as a central mediator of tissue fibrosis in many organs, including the lung [5-7]. Moreover, large studies reported that the EMT was induced by TGF-β1 in various lung epithelial cell lines [8-10], indicating that the TGF-β1-induced EMT in epithelial cells can be used as an in vitro model to study the pathogenesis of pulmonary fibrosis.
MiRNA regulation is critical in the process of EMT and IPF [11-14]. In previous study, we demonstrated that miR-5100 is overexpressed in lung cancer and it promoted cell proliferation in vitro and tumor growth in vivo by targeting Rab6 [15]. However, whether miR-5100 plays a role in the pulmonary fibrosis and the EMT process remain unknown. In this study, we first detected the expression of miR-5100 in BLM-induced mouse lung tissue and TGF-β1-induced cells, and then transfection of miR-5100 mimics or inhibitor, the expression of EMT-related markers was detected in lung epithelial (A549) cells and human bronchial epithelium (Beas-2B) cells both in basal condition and exposure to TGF-β1 by qRT-PCR and western blot. Cell immunofluorescence staining and transwell were also evaluated in miR-5100-overexpressing cells. Then, we identified TOB2 as a direct target of miR-5100, and TOB2 can reverse miR-5100 induced EMT process in A549 and Beas-2B cells.
Materials and methods
Cell culture
Human lung epithelial cell lines (A549) and human bronchial epithelial cells (Beas-2B) were obtained from the Shanghai Cell Institute Country Cell Bank (Shanghai, China). All cell lines were cultured in RPMI-1640 medium (Gibco/BRL, MD, USA) supplemented with 10% fetal bovine serum (Gibco/BRL, MD, USA), 100 U/ml penicillin G, and 100 ug/ml streptomycin (Sigma-Aldrich Corp., St.Louis, MO, USA) cultured at 37°C in a humidified atmosphere containing 5% CO2.
Bleomycin-induced lung fibrosis
To induce lung fibrosis, C57BL/6 male mice (8-12 weeks old, Guangdong Experimental Animal Center, China) were anesthetized and subjected to intratracheal administration of saline or a 5 mg/kg bleomycin sulfate saline solution. (CLEA Japan, Inc., Tokyo, Japan). After 21 days, lung tissues were quickly removed and processed for further analysis. The present study was performed in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal use protocol was reviewed and approved by the Institutional Animal Care and Use Committee of Guangdong Medical University (Zhanjiang, China).
The isolation of primary rat type II alveolar epithelial cell
Primary rat type II alveolar epithelial (ATII) cells were isolated from fetal rats lung tissues. Lung tissues of 19-day fetal rats were digested with trypsin and collagenase, then purified for AT-II cells with different centrifugal force and repeated attachment. Purified AT-II cells were cultured in DMEM-F12 medium (Gibco/BRL, MA, USA) with 10% fetal bovine serum and for subsequent experiments.
RNA extraction and quantitative real-time PCR assay
The total RNA was extracted from cultured cells or lung tissues using TRIZOL reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol. Following extraction, all RNA samples were stored at -80°C until use. The cDNA was synthesized using a miRcute miRNA First-strand cDNA Synthesis kit (TIANGEN BIOTECH, Beijing, China) and real-time PCR was performed using the miRcute miRNA qPCR Detection kit (SYBR Green) (TIANGEN BIOTECH, Beijing, China) on an ABI PRISM 7500 instrument (Applied Biosystems, Foster, CA, USA). All primers were designed by TIANGEN BIOTECH Co, Ltd. miR-5100 expression was normalized with the endogenous control snRNA U6 and the fold-change of the target genes was calculated using the 2-ΔΔCT method.
Cell transfection
The miRNA mimics, inhibitor and negative controls were synthesized by Gene Pharma (Shanghai, China). The cells were performed in serum-free medium 24 h after plating, then transfected with miR-5100 mimics, miR-5100 inhibitor, or a negative control (NC) using Lipofectamine 2000 (Invitrogen, USA). Mimic sequences were: miR-5100 mimic sense, 5’-UUCAGAUCCCAGCGGUGCCUCU-3’ and anti-sense, 5’-AGGCACCGCUGGGAUCUGAAUU-3’; miR-5100 inhibitor sense 5’-AGAGGCACCGCUGGGAUCUGAA-3’, negative control sense, 5’-UUCUCCGAACGUGUCACGUTT-3’ and anti-sense, 5’-ACGUGACACGUUCGGAGAATT-3’. The mixture was then added to cells; after incubation at 37°C for 4 h, the medium was changed to complete medium and cultured for a further 48 h. The cells were then collected for further analysis.
EMT induction
The cells were kept for overnight serum starvation after they reached 70-80% confluence or transfected 24 h later, and then treated with TGF-β1 (PeproTech, USA) or EGF (PeproTech, USA) at final concentration of 5 ng/ml or 25 ng/ml for 24 h or 48 h. The total RNA and proteins were collected for use in subsequent experiments.
Immunofluorescence staining
Cells were washed with cold PBS and then fixed with 4% paraformaldehyde for 15 min at room temperature. After fixation, the cells blocking in TBS solution supplemented with 1% Triton X-100 (TBS-Triton) and 2.5% milk for 20 min. Then cells were incubated with primary antibodies against SMA (1:100 dilutions, Samta Cruz, USA) overnight at 4°C. Followed by five washes with PBS, cells then incubated with goat anti-mouse CY5 secondary antibody (1:400 dilutions, BOSTER Biological Technology, USA) for 1h at room temperature. The nucleus was stained with 4’, 6-diamidino-2-phenylindole (DAPI) (Beyotime Biotechnology, Shanghai, China) for 5 min. Images were recorded with a confocal microscope (Leica, German).
Transwell assay
Cell transwell was examined using 24-well transwell chambers (BD Falcon, USA), according to the manufacturer’s protocol. In total, 5×104 A549 or Beas-2B cells were seeded into the upper well of the chamber in RPMI-1640 medium without fetal bovine serum. RPMI-1640 medium containing 10% fetal bovine serum was added to the lower well to stimulate transwell. Following incubation for 18 h, the cells remaining on at the upper well were removed while the bottom cells were fixed with 4% Paraformaldehyde (Sangon Biotech, Shanghai, China) and then stained with 0.1% crystal violet. The number of transwelled cells was calculated with an inverted microscope (DMI3000B, Leica, Germany) using 5 randomly-selected fields of view at a ×10 magnification.
Western blot analysis
Cultured cells were washed twice using ice-cold PBS and lysed on ice using RIPA buffer containing protease inhibitors (Beyotime Biotechnology, Shanghai, China). And the homogenate was centrifuged at 10,000 g for 15 min at 4°C. The protein concentration of the lysates was quantified using a BCA Protein Assay kit (Beyotime Biotechnology, Shanghai, China). Equal amounts of the protein lysates (40 ug) from each sample was electrophoretically separated by 12% SDS-PAGE gels and transferred to PVDF membranes (Millipore, MA, USA). The membranes were blocked with 5% non-fat milk at room temperature for 2 h,and then incubated overnight at 4°C with the primary antibodies: E-cadherin (1:1000 dilution; Cell Signaling Technology, USA), N-cadherin (1:1000 dilution; ABGENT, USA), Vimentin (1:1000 dilution; Cell Signaling Technology, USA), α-SMA (1:500 dilution; Santa Cruz Biotechnology, USA) and GAPDH (1:1000 dilution; ABGENT, USA). After washing 3 times, horseradish peroxidase-conjugated secondary antibody was added for 1 h at room temperature, the protein bands were visualized with enhanced chemiluminescence reagents (Pierce, CA, USA). GAPDH was used as the loading control.
MiRNA target prediction
Online program miRBase (http://www.mirbase.org/) were used to predict potential targets of miR-5100.
Luciferase assays
293T cells were seeded in 96-well plates at 3000 cells per well, the day before transfection. The predicted miR-5100 binding site in the 3’UTR of the TOB2 mRNA was cloned into psiCHECK2 reporter vector and a corresponding mutant was constructed. A mixture of 100 ng psi-TOB2 3’UTR and 200 ng of NC or miR-5100 mimics was transfected into 293T cells with Lipofectamine 2000. 48 hours later, Firefly and Renilla luciferase activities were measured with a Dual-Luciferase Reporter System (Promega Corporation, USA) according to the manufacturer’s protocol.
Statistical analysis
The statistical analysis results are expressed as the means ± standard deviation (SD) at least from three independent experiments. The Student’s t test was used to determine significant differences between various experimental groups. P value <0.05 was considered statistically significant.
Results
Expressions of miR-5100 is increased in BLM-induced mouse pulmonary fibrosis tissues and TGF-β1 or EGF-induced cells
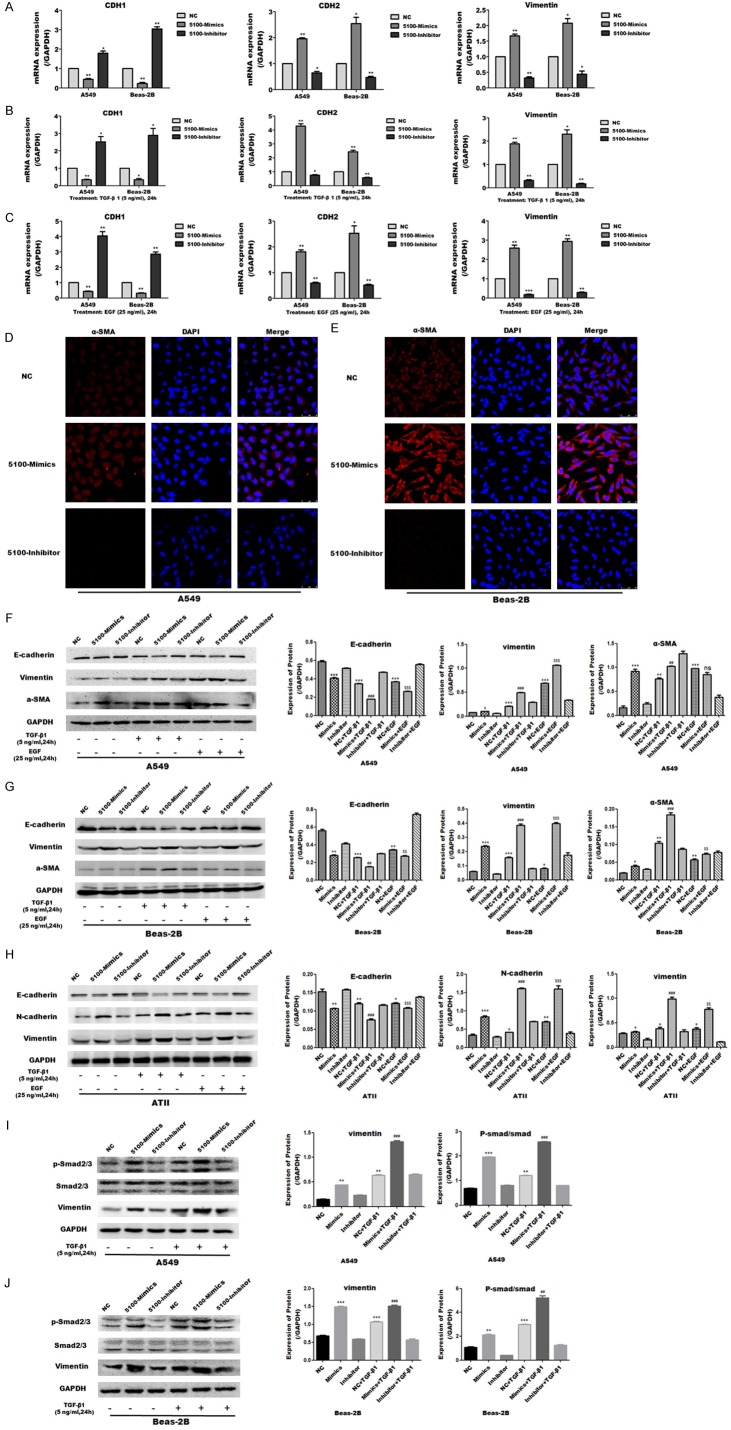
To study the mechanism of miR-5100 in the context of pulmonary fibrosis, we first detected the expression of miR-5100 in BLM-induced mouse pulmonary fibrosis tissues by qRT-PCR. The elevated level of miR-5100 was observed in the BLM-induced mouse pulmonary fibrosis tissues compared with the control tissues (Figure 1A). Then we used the TGF-β1 and EGF-induced cell EMT models, as EMT can be induced by a variety of growth factors, including TGF-β1 and EGF [16,17]. A549 and Beas-2B cells were treated with TGF-β1 or/and EGF, western blotting analysis showed that the expression of E-carderin was downregulated in TGF-β1 or/and EGF treated group compared with the control group, and the expression of N-carderin and vimentin was upregulated (Figure 1B, 1C), indicating that TGF-β1 or EGF-induced EMT in lung epithelial cells were consistent with previous study [8-10,18]. Additionally, qRT-PCR analysis showed that the expression of miR-5100 is upregulated in A549 and Beas-2B cells following TGF-β1 and EGF treatment for 24 h and 48 h (Figure 1D, 1E). Collectively, these results indicate that miR-5100 was increased in BLM-induced mouse pulmonary fibrosis tissues and TGF-β1 or EGF-induced cells.
Figure 1.
Expression of miR-5100 in BLM-induced mouse lung tissue and TGF-β1 or EGF- induced lung cells [Human lung epithelial cell lines A549 and human bronchial epithelial cells Beas-2B]. (A) qRT-PCR analysis of miR-5100 expression in saline or belomycin induced mouse lung tissues. Data are presented as the means ± SD, n=6. ***P<0.001 versus control. (B) A549 and Beas-2B cells were first stimulated with TGF-β1 (5 ng/ml) or EGF (25 ng/ml) for 24 h. Representative western blot show the protein expression of N-cadherin, E-cadherin and vimentin. (C) The results in panel B were quantified using Image J software. (D, E) qRT-PCR analysis of miR-5100 expression in A549 and Beas-2B cells treated with TGF-β1 or/and EGF for 24 h or 48 h. U6 was used as an internal control. Data are presented as the means ± SD n=3. (*P<0.05, **P<0.01, ***P<0.001 versus CK).
Exogenous miR-5100 promotes cell migration ability
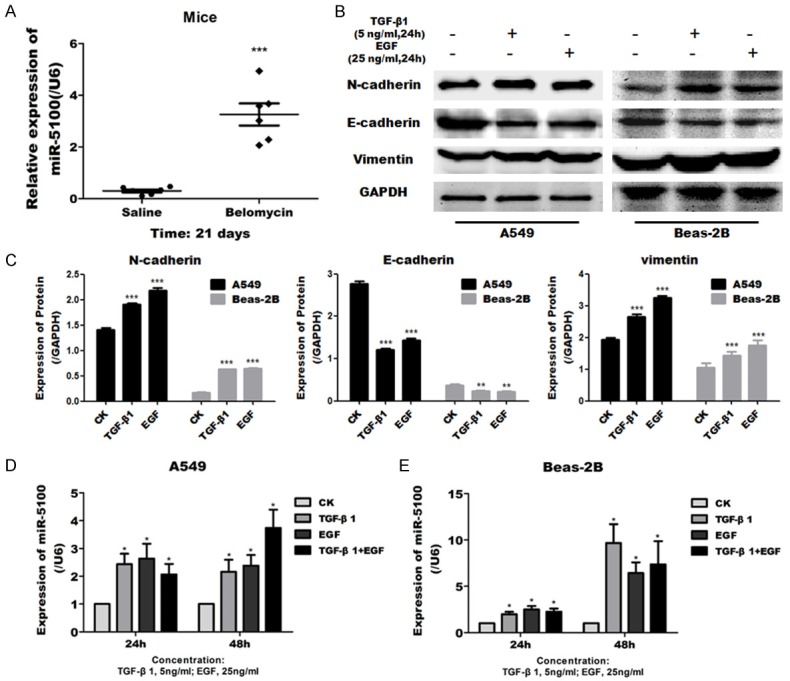
To evaluate the potential function of miR-5100 on cell motility and migration, we assessed the effect of miR-5100 in lung epithelial cells by transwell assay. Control non-targeting miRNA (NC), miR-5100 mimics, or miR-5100 inhibitor were used to transfected in cells and the efficiency of mimics and inhibitor in modulating miR-5100 expression were determined by qRT-PCR (Figure 2A). In both A549 and Beas-2B cells, exogenous expression of miR-5100 promoted the cell migration as assessed by traswell assay, while miR-5100 inhibitor caused a slight inhibition of cell migration (Figure 2B, 2C). These data indicate that miR-5100 plays a role on promoting the migration of cells.
Figure 2.
Effects of miR-5100 on cell migration of lung cells. A549 and Beas-2B cells were transfected with negative control (NC), miR-5100 mimics (5100-Mimics), or miR -5100 inhibitor (5100-Inhibitor). (A) qRT-PCR analysis of miR-5100 expression in A549 and Beas-2B cells. U6 was used as an internal control and data were standardized to an average of 1.0 in NC-transfected cells. (B, C) Cell migration ability of A549 and Beas-2B was assessed by transwell assay at the indicated times. The data are expressed as the means ± SD (n=3). (**P<0.01, **P<0.01 versus NC).
The mimics of miR-5100 promoted the EMT in A549 and Beas-2B cells and rats’ primary ATII cells
To investigate the effects of miR-5100 on EMT process in vitro model, A549 and Beas-2B cells were transfected with miR-5100 mimics or inhibitor and the expression of EMT-related markers were measured by qRT-PCR and western blot analysis. The qRT-PCR data showed that E-cadherin (CDH1) was significantly decreased whereas N-cadherin (CDH2) and vimentin were significantly increased after overexpression of miR-5100 in the cells. And the preceding markers showed the opposite trend after transfection of miR-5100 inhibitor (Figure 3A). When the cells were treated with TGF-β1 or EGF, similar results were observed, that is miR-5100 mimics promoted the EMT-related changes and the miR-5100 inhibitor suppressed the EMT-related changes (Figure 3B, 3C). Furthermore, immunofluorescence staining results showed the protein expression of α-SMA was increased by miR-5100 mimics and decreased by miR-5100 inhibitor in A549 and Beas-2B cells (Figure 3D, 3E).
Figure 3.
MiR-5100 promotes EMT process in lung epithelial cells. Lung epithelial cells were transfected with miR-5100 mimics or miR-5100 inhibitor before treated with TGF-β1 (5 ng/ml) or EGF (25 ng/ml) for 24 h. (A-C) The transcription level of epithelial phenotypic marker E-cadherin (CDH1) and mesenchymal phenotypic markers N-cadherin (CDH2) and vimentin were measured by qRT-PCR in A549 and Beas-2B cells. (D, E) Immunofluorescence staining of A549 and Beas-2B cells showing positive staining for α-SMA. Scale bars =50 um. Red indicates the protein of SMA, and blue indicates the cell nucleus. (F-H) The protein expression of E-cadherin, vimentin, N-cadherin or α-SMA was measured by western blot in A549, Beas-2B, and primary ATII cells. GAPDH was tested as a loading control. (I, J) Expression of miR-5100 significantly affects protein expression of P-smad2/3 as assesed by western blot in A549 and Beas-2B cells. Quantification of E-cadherin, vimentin, N-cadherin, α-SMA and P-smad2/3 in lung cells were carried out using Image J software. Mean optical densities were measured. All data are shown as the mean ± SD (n=3). (*P<0.05, **P<0.01, ***P<0.001 versus NC; ##P<0.01, ###P<0.001 versus NC+TGF-β1; $$P<0.01, $$$P<0.001 versus NC+EGF).
In addition, western blot analysis revealed similar results to those observed in the qRT-PCR results. The expression of E-caderin was decreased, whereas the vimentin, N-cadherin or α-SMA were increased in A549, Beas-2B and primary ATII cells following transfection with miR-5100 mimics, compared with the control group, and miR-5100 mimics can aggravate the EMT processes when the cells were treated with TGF-β1 or EGF (Figure 3F-H). Taken together, these findings demonstrated that miR-5100 promoted the EMT at both the transcriptional and translational level in A549, Beas-2B and primary rat ATII cells.
The effect of miR-5100 on the activation of TGF-β1/Smad2/3 signaling pathway was also evaluated by western blot analysis. The expression of the mesenchymal protein vimentin and the p-Smad2/3 was significantly upregulated in A549 (Figure 3I) and Beas-2B (Figure 3J) cells following miR-5100 mimics transfection, and the above proteins were increased more obviously when the cells treated with TGF-β1, whereas no changes were observed in Smad3 expression. These results suggested that miR-5100 promoted the EMT process probably by activating the TGF-β1/Smad pathway.
MiR-5100 translationally represses TOB2 by targeting specific sequences in the 3’UTR of TOB2
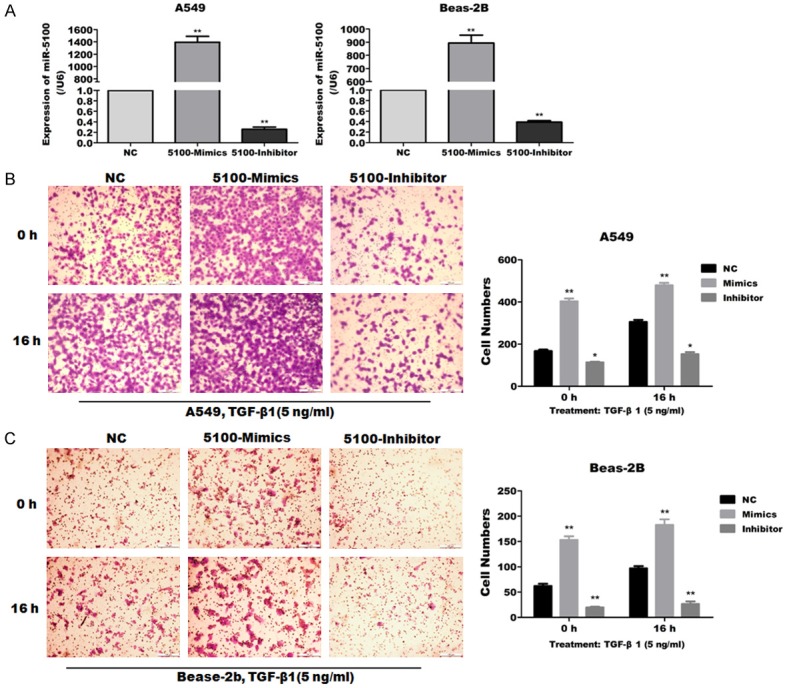
To investigate the underlying molecular mechanisms by which miR-5100 mediate the regulation of EMT, the potential targets of miR-5100 were identified by the miRNA target prediction tool miRBase. TOB2 was predicted to be a potential target of miR-5100 (Figure 4A). To validate TOB2 is the target of miR-5100; we first performed a luciferase activity assay. We constructed and verified the wild-type Tob2 3’UTR luciferase reporter plasmid and the mutant, which were then used for cotransfection of 293T cells with miR-5100 mimics or the negative control. Compared with the negative control cells, the cells cotransfected with the wild type and the miR-5100 mimics exhibited significantly inhibited luciferase activity after 48 h (Figure 4B). Otherwise, we used western blot to further evaluate the target, and we found that the overexpression of miR-5100 significantly suppressed the TOB2 protein level, whereas the inhibition of miR-5100 had no effect on the expression of TOB2 in A549 and Beas-2B cells (Figure 4C, 4D). These results indicate that TOB2 was the target of negative regulation by miR-5100, and affecting the translational level.
Figure 4.
MiR-5100 inhibits the expression of TOB2 by binding its 3’UTR. (A) A putative miR-5100 binding site in the TOB2 3’UTR is indicated with overstriking characters. This site was identified using miRBase. (B) Luciferase reporter assay of 293T cells transfected with a psiCheck2 vector with an inserted TOB2 3’UTR sequence (TOB2 3’UTR) or a vector with an inserted mutated 3’UTR sequence (3’UTR mutant), together with either negative control miRNA (NC) or miR-5100 (mimics). The luciferase activity was normalized to firefly luciferase activity. (C) The protein levels of TOB2 was examined by Western blotting for two cell lines infected with NC, 5100-Mimic or 5100-Inhibitor. (D) Quantification of the results in panel C were carried out using Image J software. The data are expressed as the means ± SEM (n=3), (**P<0.01 versus NC).
Exogenous expression of TOB2 compromises the effects of miR-5100 on cell migration and EMT process
In order to verify that miR-5100 regulates EMT through its target TOB2, we constructed the overexpression plasmid of pCMV-TOB2-HA and then cotransfected A549 and Beas-2B cells with miR-5100 mimics. The results showed that the migration ability of miR-5100 mimics transfected cells was significantly increased in A549 and Beas-2B cells (Figure 5A, 5B), compared with the control group. The migration ability of the cotransfected group was significantly decreased compared with the miR-5100 mimics alone group. And this process was more obvious in the cells treated with TGF-β1 for 16 h.
Figure 5.
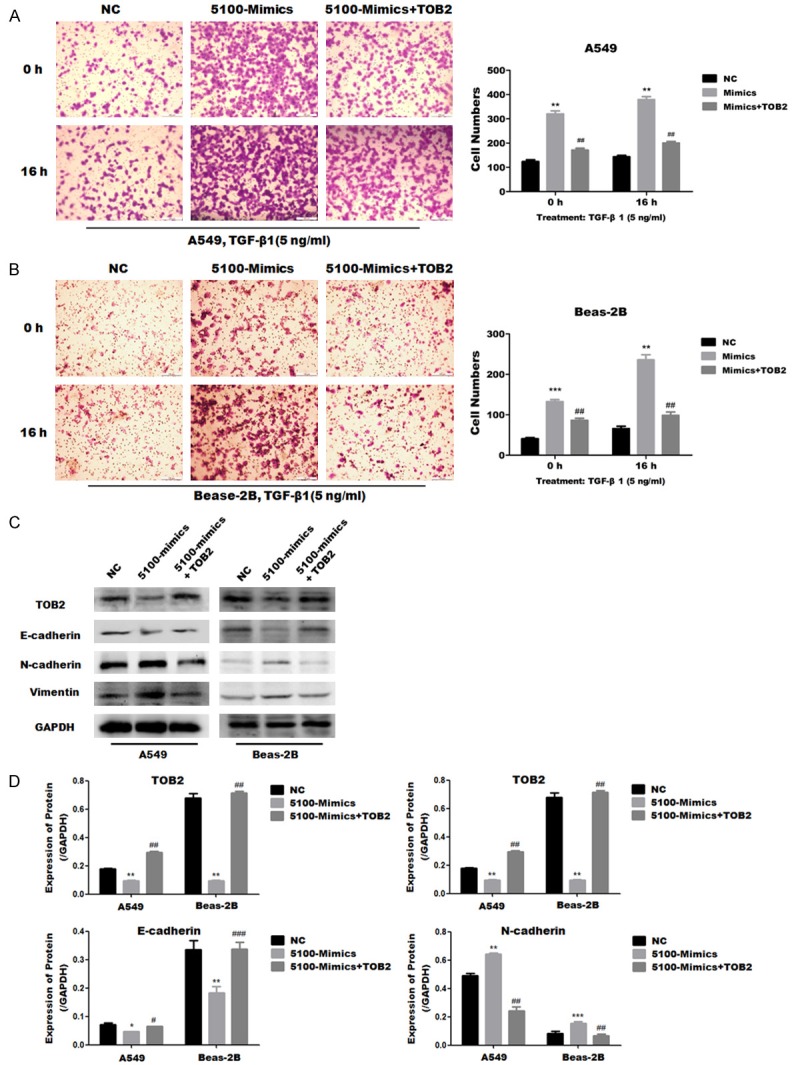
TOB2 attenuated miR-5100 induced EMT-relatd changes in A549 and Beas-2B cells. A549 and Beas-2B cells were transfected with a negative control (NC), miR-5100 mimics (5100-mimics) or mi5100-mimics and TOB2 expression plasmid (5100-mimics+TOB2). (A, B) Cell migration ability of A549 and Beas-2B cells was assessed by an transwell assay at the indicated times. (C) Expression of E-cadherin, N-cadherin and vimentin was assesed by western blot in A549 and Beas-2B cells. (D) Quantification of the results in C was carried out using Image J software. The data are expressed as the mean ± SD (n=3). (**P<0.01, ***P<0.001 versus NC; ##P<0.01 versus 5100-mimics).
In addition, western blot was used to examine the effects of upregulating TOB2 on the EMT-related proteins. The expression of E-cadherin was downregulated, and the interstitial marker proteins N-cadherin and vimentin were upregulated in A549 and Beas-2B cells transfected with miR-5100 mimics compared with the control group (Figure 5C, 5D); but miR-5100 mimics and TOB2 cotransfected cells reverse the EMT process which caused by miR-5100 mimics. These results suggest that exogenous expression of TOB2 compromises the ability of miR-5100 to promote cell migration and EMT process.
Discussion
Recently, numerous miRNAs have been reported to involve in the pathogenesis of IPF in various pathways. EMT reportedly plays a critical role in the pathogenesis of pulmonary fibrosis, and miRNAs regulate lung fibrosis via EMT [19,20]. BLM-induced mouse model of pulmonary fibrosis is the most commonly used model of IPF for studying disease pathogenesis and for testing novel pharmaceutical compounds [21]. In the current study, we observed that miR-5100 was upregulated in BLM-induced mouse lung tissue and TGF-β1 or EGF-induced cells (Figure 1), indicating that miR-5100 may plays an important role in pulmonary fibrosis. Previously we have reported that miR-5100 promoted the tumor growth and metastasis in lung cancer [15]. The cancer metastasis was also involved the EMT process and EMT is an accepted biological process in cancer progression and is an appealing concept in IPF because of the unique features of fibroblastic foci [22]. Thus, we hypothesis that miR-5100 play a regulatory role in the pathogenesis of pulmonary fibrosis by influencing the EMT process.
To confirm the hypothesis, we examined the role of miR-5100 on EMT in lung epithelial cells including human lung cancer cell lines A549 and human bronchial epithelium cells Beas-2B. We found that miR-5100 promoted cell migration (Figure 2) and the EMT-related proteins were changed (Figure 3A-G) in A549 and Beas-2B cells both in basal condition and exposure to TGF-β1 or EGF. In this study, we also observed that miR-5100 had the similar effects on the EMT process in primary rat ATII cells (Figure 3H).
EMT is a process in which epithelial cells lose their epithelial phenotype, acquire mesenchymal properties, and display reduced cell adhesion and increased motility [2,3]. Many growth factors such as TGF-β1 and EGF have been identified to be widely associated with EMT induction [16,17]. The present study, both TGF-β1 and EGF-induced EMT process was confirmed in A549 and Beas-2B cells (Figure 1B, 1C), the endogenous expression of miR-5100 was upreregulated in TGF-β1 or EGF-induced cells (Figure 1D, 1E) and exogenous miR-5100 promoted EMT and aggravated the EMT process of the cells treated with TGF-β1 or EGF (Figure 3A-H).
As TGF-β1 is a key regulator of EMT in pulmonary fibrosis [23] and the TGF-β/smad signalling pathway plays a key role in both EMT and fibrosis [5,24], we also examed the effect of miR-5100 on p-smad2/3 protein and found that exogenous miR-5100 leds to the elevated level of p-smad2/3 in A549 and Beas-2B cells both in basal condition and exposure to TGF-β1 (Figure 3G, 3H). On the other hand, these results indicated that miR-5100 promoted the EMT process and aggravated the TGF-β1-induced EMT in pulmonary fibrosis involved in activating the TGF-β/smad pathway. Many reporters have showed that EGF induce EMT via activation of JAK/STAT3 and integrin-linked kinase (ILK) and ERK1/2-phospho-smad2/3-snail signaling pathway [25-27]. While our results showed miR-5100 activated p-smad2/3, further indicated miR-5100 regulates directly or indirectly TGF-β1-induced EMT process involving TGF-β/smad signaling pathway.
TOB2 has been reported to be regulated by a variety of miRNAs that play a role in a variety of tumors and was considered to be a potential tumor suppressor [28,29]. It has also been reported that miR-5100 promoted osteogenic differentiation by targeting TOB2 [30]. However, the function of TOB2 on the EMT or pulmonary fibrosis remains unclear. In this study, for the first time to the best of our knowledge, we confirmed that miR-5100 participated in promoting EMT process by downregulating TOB2 (Figure 4) and exogenous expression of TOB2 relieved the promotion of miR-5100 on EMT process and migration ability in A549 and Beas-2B cells (Figure 5), indicating the role of miR-5100 on EMT process and migration ability are at least partly via TOB2.
There are some limitations of our study. Firstly, it is unknown whether the dysregulation of miR-5100 is a cause or consequence of pulmonary fibrosis, as we did not verify the results from the clinical samples of patients with IPF, so clinical samples are required to confirm these data in following study. In addition, as miRNAs have multiple targets and individual gene being regulated by various miRNAs [31], the miR-5100 and TOB2 themselves could cover the regulation of EMT, the miR-5100 play a regulatory role in EMT is limited.
In conclusion, miR-5100 promoted occurrence of EMT and mediated the TGF-β1-induced EMT in lung epithelial cells through downregulation of its target, TOB2, and associated with activating the TGF-β1/Smad signaling pathway. Our findings may provide a potential target for therapy of IPF in future.
Acknowledgements
This work is supported by the Natural Science Foundation of China (grant No: NSFC81172615, 81600049 and 81570062); Guangdong Natural Science Foundation (grant No: 2016A030313681); Guangdong Medical Science Foundation (grant No: A2017010, A2017027); Guangdong medical university scientific research fund (grant No: M2016001, M2016007, M2016022).
Disclosure of conflict of interest
None.
References
- 1.Nalysnyk L, Cid-Ruzafa J, Rotella P, Esser D. Incidence and prevalence of idiopathic pulmonary fibrosis: review of the literature. Eur Respir Rev. 2012;21:355–361. doi: 10.1183/09059180.00002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112:1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen KJ, Li Q, Wen CM, Duan ZX, Zhang JY, Xu C, Wang JM. Bleomycin (BLM) induces epithelial-to-mesenchymal transition in cultured A549 cells via the TGF-beta/Smad signaling pathway. J Cancer. 2016;7:1557–1564. doi: 10.7150/jca.15566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowen T, Jenkins RH, Fraser DJ. MicroRNAs, transforming growth factor beta-1, and tissue fibrosis. J Pathol. 2013;229:274–285. doi: 10.1002/path.4119. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez IE, Eickelberg O. The impact of TGF-beta on lung fibrosis: from targeting to biomarkers. Proc Am Thorac Soc. 2012;9:111–116. doi: 10.1513/pats.201203-023AW. [DOI] [PubMed] [Google Scholar]
- 8.Takano M, Yamamoto C, Yamaguchi K, Kawami M, Yumoto R. Analysis of TGF-beta1- and drug-induced epithelial-mesenchymal transition in cultured alveolar epithelial cell line RLE/Abca3. Drug Metab Pharmacokinet. 2015;30:111–118. doi: 10.1016/j.dmpk.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Kasai H, Allen JT, Mason RM, Kamimura T, Zhang Z. TGF-beta1 induces human alveolar epithelial to mesenchymal cell transition (EMT) Respir Res. 2005;6:56. doi: 10.1186/1465-9921-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willis BC, Borok Z. TGF-beta-induced EMT: mechanisms and implications for fibrotic lung disease. Am J Physiol Lung Cell Mol Physiol. 2007;293:L525–534. doi: 10.1152/ajplung.00163.2007. [DOI] [PubMed] [Google Scholar]
- 11.Takano M, Nekomoto C, Kawami M, Yumoto R. Role of miR-34a in TGF-beta1- and drug-Induced epithelial-mesenchymal transition in alveolar type II epithelial cells. J Pharm Sci. 2017;106:2868–2872. doi: 10.1016/j.xphs.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Jaca A, Govender P, Locketz M, Naidoo R. The role of miRNA-21 and epithelial mesenchymal transition (EMT) process in colorectal cancer. J Clin Pathol. 2017;70:331–356. doi: 10.1136/jclinpath-2016-204031. [DOI] [PubMed] [Google Scholar]
- 13.Liang H, Liu S, Chen Y, Bai X, Liu L, Dong Y, Hu M, Su X, Huangfu L, Li X, Gu Y, Shan H. miR-26a suppresses EMT by disrupting the Lin28B/let-7d axis: potential cross-talks among miRNAs in IPF. J Mol Med (Berl) 2016;94:655–665. doi: 10.1007/s00109-016-1381-8. [DOI] [PubMed] [Google Scholar]
- 14.Wang YC, Liu JS, Tang HK, Nie J, Zhu JX, Wen LL, Guo QL. miR221 targets HMGA2 to inhibit bleomycininduced pulmonary fibrosis by regulating TGFbeta1/Smad3-induced EMT. Int J Mol Med. 2016;38:1208–1216. doi: 10.3892/ijmm.2016.2705. [DOI] [PubMed] [Google Scholar]
- 15.Huang H, Jiang Y, Wang Y, Chen T, Yang L, He H, Lin Z, Liu T, Yang T, Kamp DW, Wu B, Liu G. miR-5100 promotes tumor growth in lung cancer by targeting Rab6. Cancer Lett. 2015;362:15–24. doi: 10.1016/j.canlet.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003;278:21113–21123. doi: 10.1074/jbc.M211304200. [DOI] [PubMed] [Google Scholar]
- 17.Cheng JC, Auersperg N, Leung PC. EGF-induced EMT and invasiveness in serous borderline ovarian tumor cells: a possible step in the transition to low-grade serous carcinoma cells? PLoS One. 2012;7:e34071. doi: 10.1371/journal.pone.0034071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alipio ZA, Jones N, Liao W, Yang J, Kulkarni S, Sree Kumar K, Hauer-Jensen M, Ward DC, Ma Y, Fink LM. Epithelial to mesenchymal transition (EMT) induced by bleomycin or TFG(b1)/EGF in murine induced pluripotent stem cellderived alveolar Type II-like cells. Differentiation. 2011;82:89–98. doi: 10.1016/j.diff.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Pandit KV, Milosevic J. MicroRNA regulatory networks in idiopathic pulmonary fibrosis. Biochem Cell Biol. 2015;93:129–137. doi: 10.1139/bcb-2014-0101. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Zhao X, Shan H, Liang H. MicroRNAs in idiopathic pulmonary fibrosis: involvement in pathogenesis and potential use in diagnosis and therapeutics. Acta Pharm Sin B. 2016;6:531–539. doi: 10.1016/j.apsb.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moeller A, Ask K, Warburton D, Gauldie J, Kolb M. The bleomycin animal model: a useful tool to investigate treatment options for idiopathic pulmonary fibrosis? Int J Biochem Cell Biol. 2008;40:362–382. doi: 10.1016/j.biocel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connor JW, Gomez EW. Biomechanics of TGFbeta-induced epithelial-mesenchymal transition: implications for fibrosis and cancer. Clin Transl Med. 2014;3:23. doi: 10.1186/2001-1326-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willis BC, Liebler JM, Luby-Phelps K, Nicholson AG, Crandall ED, du Bois RM, Borok Z. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am J Pathol. 2005;166:1321–1332. doi: 10.1016/s0002-9440(10)62351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue P, Zhang X, Paladino D, Sengupta B, Ahmad S, Holloway RW, Ingersoll SB, Turkson J. Hyperactive EGF receptor, Jaks and Stat3 signaling promote enhanced colony-forming ability, motility and migration of cisplatin-resistant ovarian cancer cells. Oncogene. 2012;31:2309–2322. doi: 10.1038/onc.2011.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed N, Maines-Bandiera S, Quinn MA, Unger WG, Dedhar S, Auersperg N. Molecular pathways regulating EGF-induced epitheliomesenchymal transition in human ovarian surface epithelium. Am J Physiol Cell Physiol. 2006;290:C1532–1542. doi: 10.1152/ajpcell.00478.2005. [DOI] [PubMed] [Google Scholar]
- 27.Kim J, Kong J, Chang H, Kim H, Kim A. EGF induces epithelial-mesenchymal transition through phospho-Smad2/3-Snail signaling pathway in breast cancer cells. Oncotarget. 2016;7:85021–85032. doi: 10.18632/oncotarget.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu BL, Peng XH, Zhao FP, Liu X, Lu J, Wang L, Li G, Chen HH, Li XP. MicroRNA-378 functions as an onco-miR in nasopharyngeal carcinoma by repressing TOB2 expression. Int J Oncol. 2014;44:1215–1222. doi: 10.3892/ijo.2014.2283. [DOI] [PubMed] [Google Scholar]
- 29.Shen H, Li W, Tian Y, Xu P, Wang H, Zhang J, Li Y. Upregulation of miR-362-3p modulates proliferation and anchorage-independent growth by directly targeting Tob2 in hepatocellular carcinoma. J Cell Biochem. 2015;116:1563–1573. doi: 10.1002/jcb.25110. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Cui Y, Luan J, Zhou X, Li C, Li H, Shi L, Han J. MiR-5100 promotes osteogenic differentiation by targeting Tob2. J Bone Miner Metab. 2016 doi: 10.1007/s00774-016-0799-y. [DOI] [PubMed] [Google Scholar]
- 31.Lamouille S, Subramanyam D, Blelloch R, Derynck R. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell Biol. 2013;25:200–207. doi: 10.1016/j.ceb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]